Abstract
The development of an iterative one-pot peptide ligation strategy is described that capitalises on the rapid and efficient nature of the diselenide–selenoester ligation reaction, together with photodeselenisation chemistry. This ligation strategy hinged on the development of a novel photolabile protecting group for the side chain of selenocysteine, namely the 7-diethylamino-3-methyl coumarin (DEAMC) moiety. Deprotection of this DEAMC group can be effected in a mild, reagent-free manner using visible light (λ = 450 nm) without deleterious deselenisation of selenocysteine residues, thus enabling a subsequent ligation reaction without purification. The use of this DEAMC-protected selenocysteine in iterative DSL chemistry is highlighted through the efficient one-pot syntheses of 60- and 80-residue fragments of mucin-1 as well as apolipoprotein CIII in just 2–4 hours.
A method for the rapid one-pot iterative assembly of proteins via diselenide–selenoester ligation (DSL) chemistry is described that capitalises on a novel coumarin-based photolabile protecting group for selenocysteine.
Introduction
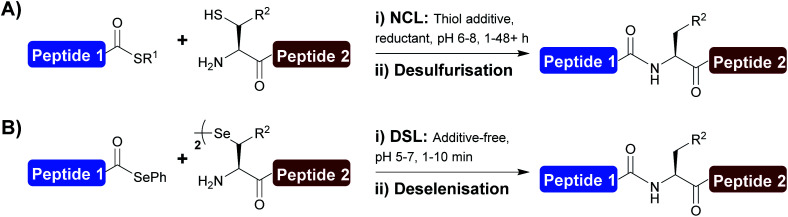
The development and application of chemoselective peptide ligation chemistry has transformed the field of chemical protein synthesis, providing rapid access to complex polypeptides and proteins.1–4 Among the several ligation technologies now available, native chemical ligation (NCL) is recognised as the ‘gold standard’ (Scheme 1A).1,5 The NCL transformation involves the reaction between a peptide bearing a C-terminal thioester and another peptide fragment with an N-terminal cysteine (Cys) leading to the generation of a native amide bond.6 The introduction of desulfurisation chemistry [that converts Cys to alanine (Ala) at the ligation junction]7–9 and the development of various thiolated amino acids4,10–16 has greatly expanded the scope of NCL and, as a result, there is no longer a prerequisite for a Cys residue at the ligation site.
Scheme 1. (A) Native chemical ligation (NCL) and desulfurisation chemistry; (B) diselenide–selenoester ligation (DSL) and deselenisation chemistry. R1 = alkyl or aryl substituent, R2 = amino acid side chain.
Despite the power of NCL, limitations remain with the methodology, namely the sluggish ligation rates at sterically hindered junctions and the need to perform the reactions at mM substrate concentrations that can be difficult to achieve with poorly soluble peptide fragments. To overcome these limitations, we recently developed the diselenide–selenoester ligation (DSL) reaction (Scheme 1B). The DSL reaction involves the chemoselective fusion of peptide selenoesters with peptides bearing an N-terminal selenocystine [the oxidised form of the 21st amino acid selenocysteine (Sec/U)].17 The superior reactivity of both reaction partners compared to the sulfur counterparts used in NCL leads to vastly improved reaction rates, with most ligations (including at sterically hindered junctions) complete within 10 min. DSL reactions are typically performed in aqueous buffer without the inclusion of additives that are required to facilitate NCL reactions, and can also be performed over a broad pH range (pH 3–7). DSL reactions also proceed efficiently at high dilution (at μM to nM concentrations) through the use of the reductant tris(2-carboxyethyl) phosphine (TCEP) and diphenyldiselenide (DPDS), a variation of the method termed reductive diselenide–selenoester ligation (rDSL).18 In recent years the scope of DSL has been expanded through the development of selenylated amino acids which can be deselenised post-ligation to afford native amino acids at the ligation site.19–24 One key benefit of the DSL methodology is that the deselenisation step used to convert Sec to Ala (or remove the selenium auxiliary from other amino acids) is chemoselective in the presence of unprotected Cys residues.25 Despite this improvement in ligation methodology, a key benefit of NCL involves the ability to perform iterative ligations of multiple fragments without intermediate purification steps. For such applications of NCL, researchers have turned to the use of various protecting groups for the side chain of Cys residues. Examples of such protection strategies include the use of thiazolidine (Thz),26 acetamidomethyl (Acm),27 and trifluoroacetamidomethyl (TfAcm)28 groups that mask the reactivity of the N-terminal Cys residue within a bifunctional fragment that also possess a reactive C-terminal thioester to prevent unwanted self-ligation of the fragment. Importantly, the omission of intermediate purification steps when performing iterative ligations in one-pot drastically reduces the time for protein synthesis, and usually often also improves the overall yield. While a small number of protecting groups have been developed and used in NCL-type reactions at Sec, e.g. selenazolidine (Sez)29,30 and p-methoxybenzyl (PMB),31,32 conditions used to remove them are harsh and often incompatible with a subsequent in situ ligation or deselenisation step and therefore intermediary purification (or desalting) and lyophilisation steps are often required.
Specifically, deprotection conditions range from treatment with strong acids and thiol additives for the PMB group31,32 to nucleophilic reagents29,30 and stoichiometric heavy metal ions, e.g. Cu ions, for Sez unmasking.33
Results and discussion
Design and synthesis of 7-diethylamino-3-methyl coumarin (DEAMC)-protected Sec
In this work we sought to develop an efficient one-pot method for the ligation of multiple fragments using DSL-deselenisation chemistry. To develop such a methodology, we required an orthogonal protecting group for the side chain of Sec that could be installed at the N-terminus of peptide selenoester fragments, and which could be removed under mild conditions that would not interfere with subsequent ligation steps. In order to meet these requirements, we envisioned the use of a photolabile group that would be stable to ligation conditions but could be rapidly and chemoselectively removed with light (and without the addition of exogenous reagents) to enable a subsequent ligation reaction without the need for work-up or intermediary purification steps. We first considered the o-nitroveratryl group that has been used to mask Cys34 and Sec35 residues in peptides. However, the UV irradiation conditions (365 nm) required for deprotecting this group has been shown to generate by-products, including dehydroalanine via an elimination pathway.35 We verified the formation of by-products when deprotecting a model peptide bearing an o-nitroveratryl-protected Sec in standard aqueous ligation buffer (see ESI† for details). We therefore turned our attention to the development of an alternate coumarin-based protecting group. Several coumarin groups exist for the protection of carboxylic acids, amines, alcohols, diols, and thiols.36–38 However, in the case of alcohols and thiols, irradiation leads to irreversible photoisomerisation of the heteroatom onto the 3-position of the coumarin.39 The installation of a 3-methyl group was therefore deemed necessary to prevent formation of this by-product (i.e. the generation of an arylselenyl species when attached to the side chain of Sec).39 It was envisioned that a 7-diethylamino-3-methyl coumarin (DEAMC) group, bearing a 7-diethylamino moiety to red-shift the absorbance spectra,40 and a 3-methyl group to prevent photoisomerisation, would meet the requirements as a protecting group for the side chain of Sec for the development of the proposed iterative one-pot DSL-deselenisation methodology.
The synthesis of the target DEAMC-protected Sec building block 1 began from commercially available ethyl 4-chloroacetoacetate 2 (Scheme 2A). Nucleophilic substitution with tert-butoxide, followed by enolisation with potassium tert-butoxide and methylation with methyl iodide generated intermediate 3 in 60% yield over two steps (Scheme 2A). Intermediate 3 was next subjected to a Pechmann condensation with 3-diethylaminophenol using 40 mol% Y(NO3)3·6H2O as a Lewis acid41 to afford the tBuO-derived DEAMC moiety which was triturated, before treating with trifluoroacetic acid (TFA) in DCM to generate the corresponding alcohol. From here, the alcohol was converted to the corresponding bromide via an Appel reaction; purification via flash chromatography generated bromide intermediate 4 in 11% yield over 3 steps. Finally, reduction of (Boc-Sec-OH)2 using sodium borohydride followed by addition of bromide 4 afforded the target Boc-Sec(DEAMC)-OH building block 1 in 83% yield.
Scheme 2. (A) Synthesis of Boc-Sec(DEAMC)-OH building block 1: (a) (i) NaH, KOtBu, THF, rt, 16 h; (ii) MeI, KOtBu, THF, rt, 16 h, 63% yield over 2 steps; (b) (i) 3-diethylaminophenol, 40 mol% Y(NO3)3·6H2O, 115 °C, 24 h; (ii) TFA, DCM, rt, 2 h; (iii) CBr4, PPh3, THF, rt, 3 h, 11% yield over 3 steps; (c) (i) NaBH4, EtOH : THF (2 : 7 v/v), (Boc-Sec-OH)2, rt, 30 min; (ii) 4, rt, 5 min, 83%. (B) Synthesis and deprotection of model DEAMC-protected selenopeptide 7. (a) Fmoc-SPPS (see ESI†) (b) Boc-Sec(DEAMC)-OH 1 (1.1 eq.), Oxyma (1.2 eq.), DIC (1.1 eq.), DMF, rt, 3 h. (c) TFA, iPr3SiH, H2O (18 : 1 : 1 v/v/v), rt, 2 h. (d) 6 M Gdn·HCl, 0.1 M Na2HPO4, pH 6.0, λ = 450 nm, 36 W LED irradiation, 10 min.
Before applying the novel photocaged Sec residue in ligation chemistry, we first prepared model peptide H-U(DEAMC)SPGYS-NH27 by Fmoc-SPPS to assess conditions for photodeprotection of the DEAMC group within a peptide (Scheme 2B). To this end, H-U(DEAMC)SPGYS-NH27 was dissolved in aqueous denaturing buffer (5 mM) commonly used in DSL ligation reactions (6 M Gdn·HCl, 0.1 M Na2HPO4, pH 6.0) and irradiated with blue light (λ = 450 nm, LED, 36 W). Pleasingly, this led to clean deprotection to peptide diselenide dimer 8 bearing an N-terminal selenocystine residue in 10 min (see ESI†).
One-pot assembly of three peptide fragments via iterative DSL chemistry
Having optimised conditions to introduce and deprotect the DEAMC-protected Sec, we shifted attention to the use of the amino acid for polypeptide and protein assembly via the proposed iterative DSL-deselenisation chemistry. Towards this end, a fragment of the variable number tandem repeat (VNTR) domain of the transmembrane glycoprotein mucin 1 (MUC1) was selected as an initial target to validate the methodology. An unglycosylated 60-residue fragment of MUC1 9 bearing three copies of the 20 amino acid VNTR was first targeted through a three-fragment assembly strategy (Scheme 3A). Disconnection at the Ser–Ala junction between each of the 20 residue VNTRs led to three differentially functionalised peptide fragments that required synthesis: (1) diselenide dimer fragment 13 bearing an N-terminal selenocystine, (2) bifunctional fragment 12 containing an N-terminal DEAMC-protected Sec and a C-terminal phenylselenoester, and (3) fragment 14 functionalised as a C-terminal phenylselenoester. We envisaged assembling these fragments by two consecutive additive-free DSL reactions, with an intervening photodeprotection of the DEAMC moiety, followed by global photodeselenisation of the two internal Sec residues (Scheme 3). Fragments 13 and 14 were prepared by standard Fmoc-SPPS conditions (see ESI† for details). For bifunctional fragment 12 a side-chain anchoring strategy was employed,42 wherein Fmoc-Ser-OAllyl was immobilised onto 2-chlorotrityl chloride resin via the unprotected alcohol side chain (Scheme 3B). Following elongation of the target peptide sequence 10 by Fmoc-SPPS, on-resin allyl deprotection followed by selenoesterification afforded the resin-bound peptide selenoester 11. Finally, Boc-Sec(DEAMC)-OH 1 was coupled at the N-terminus, followed by acidolytic cleavage and HPLC purification, furnishing the bifunctional fragment 12. With the functionalised fragments 12–14 in hand, ligation-based assembly of 9 was initiated. First, the diselenide dimer 13 (1.0 eq. of the monomer) was treated with bifunctional peptide 12 (1.1 eq.) in ligation buffer (6 M Gdn·HCl, 0.1 M Na2HPO4, pH 6.0, rt; 5 mM with respect to the monomeric selenopeptide of 13, Scheme 3C) reaching completion within 15 minutes (as judged by UPLC-MS). Without purification, the ligation product was directly subjected to photodeprotection by irradiation at 450 nm (with LEDs). Gratifyingly, clean and quantitative deprotection of the DEAMC group was achieved in 10 min, affording intramolecular diselenide 15 as the major product. Following this step, and again without purification, the N-terminal Sec residue in peptide 15 (revealed following the photodeprotection) was ligated with fragment 14 (2.0 eq. with respect to monomeric 13). This ligation reached completion after 15 min (determined by UPLC-MS analysis) to afford intramolecular diselenide 16 as the major product (Scheme 3). At this point the diphenyldiselenide (DPDS) generated during the two DSL reactions was extracted with Et2O to prevent radical quenching from this species that would inhibit the ensuing deselenisation reaction. The reaction mixture was then treated with TCEP (200 mM) and GSH (40 mM) at pH 7.0 and irradiated with a UV light source (λ = 254 nm, 35 W, 5 min) to facilitate photodeselenisation of the two internal Sec residues to afford Ala residues at the two ligation junctions. Pleasingly, purification by reverse-phase HPLC followed by lyophilisation afforded the target 60-residue MUC1 polypeptide 9 in 60% isolated yield over four steps (Scheme 3). It should be noted that the four-step process that included two DSL reactions, DEAMC deprotection and photodeselenisation was completed within 2 hours and required only a single purification step.
Scheme 3. (A) Sequence of target MUC1 VNTR trimer 9 (ligation sites indicated by dotted lines); (B) synthesis of MUC1 VNTR bifunctional fragment 12via a side chain anchoring strategy; (C) synthesis of MUC1 VNTR trimer 9via one-pot iterative DSL reactions, PG = amino acids were side chain protected in resin-bound peptide; (D) HPLC trace and MALDI-TOF MS (inset) of purified MUC1 VNTR trimer 9; (E) ESI-MS of purified MUC1 VNTR trimer 9.
One-pot assembly of four peptide fragments via iterative DSL chemistry
We next sought to extend the use of the iterative DSL approach, leveraging the DEAMC-protected Sec residue, beyond 3 fragments. Towards this end, we devised a ligation strategy to generate an 80-residue MUC1 polypeptide through three DSL reactions, two DEAMC deprotections and a global photodeselenisation in one-pot (Scheme 4). Briefly, fragments 12 and 13 were subjected to one-pot DSL-photodeprotection as before to generate the intramolecular diselenide intermediate 15. This intermediate was subjected to a further one-pot DSL-photodeprotection with 12 to afford 18.
Scheme 4. (A) Sequence of MUC1 VNTR tetramer 17 (ligation sites shown with dotted lines); (B) synthesis of MUC1 VNTR tetramer 17via one-pot iterative DSL reactions; (C) HPLC trace and MALDI-TOF MS of purified MUC1 VNTR tetramer 17; (D) ESI-MS of purified MUC1 VNTR tetramer 17.
From here, a final DSL ligation with peptide selenoester 14 and global photodeselenisation of the three Sec residues at the ligation junctions afforded the 80-residue MUC1 17 in 35% yield over 6 steps following HPLC purification (see ESI† for details). Notably, the entire multi-component assembly of 17 was accomplished in one-pot within 3 hours, highlighting the value of using DEAMC-protected Sec for iterative ligation chemistry without the need to purify intermediates.
One-pot chemical synthesis of apolipoprotein CIII
Having validated the iterative DSL methodology on the MUC1 VNTR polypeptides 9 and 17, we shifted attention towards implementing the methodology for the synthesis of a more challenging synthetic target, namely apolipoprotein C-III (ApoCIII). ApoCIII is an abundant lipoprotein that is a major surface component of chylomicrons, high density lipoproteins, and very low-density lipoproteins, and has affinity for cell surface proteoglycans and acts as an intracellular promotor for hepatic VLDL assembly within the secretory pathway. Within plasma, ApoCIII inhibits both lipolysis of very low density lipoproteins (VLDLs) and the uptake of these lipoprotein remnants in the liver.43,44 Since the protein possesses a lipophilic N-terminal sequence (residues 6–20),45 we proposed that the DSL methodology, which can be performed at higher dilution than other ligation methods (vide supra), would be uniquely suited to assemble ApoCIII.
We envisaged that the 79-residue protein could be disconnected at Thr22–Ala23 and Ser49–Leu50, thus requiring the synthesis of three suitably protected target peptide fragments (20–22) for assembly by a one-pot approach (Scheme 5). Peptide fragment 20 was synthesised by standard Fmoc-SPPS with the incorporation of a β-selenoleucine residue46 at the N-terminus (see ESI† for synthetic details). Bifunctional peptide selenoester 21 was prepared via a side chain anchoring strategy, whereas peptide selenoester 22 was prepared by standard Fmoc-SPPS, as described above (see ESI† for synthetic details).
Scheme 5. (A) Sequence of ApoCIII 19 with ligation sites indicated by dotted lines; (B) chemical synthesis of ApoCIII 19via one-pot iterative DSL reactions; (C) HPLC trace and MALDI-TOF MS of purified ApoCIII 19; (D) ESI-MS of purified ApoCIII 19; (E) CD spectrum of 19.
Synthesis of ApoCIII began with reaction of diselenide dimer fragment 20 (5 mM, 1.0 eq. with respect to monomeric selenopeptide) with the bifunctional fragment 21 (1.6 eq.) in ligation buffer (pH 6.0–6.2) under additive-free DSL conditions.
Given the sterically hindered nature of the N-terminal β-selenoleucine residue, the ligation was performed at 37 °C and reached completion after 30 min (as judged by UPLC-MS analysis). Without intermediate purification, the ligation mixture was subjected to photoirradiation (λ = 450 nm, 36 W) to unmask the N-terminal selenocysteine. Clean deprotection of the DEAMC group was observed in 10 min, providing the ligation product exclusively as the intramolecular diselenide 23. The next ligation could not be initiated without additives due to the limited solubility of the N-terminal fragment 22 in the ligation buffer. We therefore turned to the use of the reductive variant of the DSL reaction, rDSL, to facilitate the final ligation at high dilution. Accordingly, selenoester fragment 22 (2.4 eq.) was reacted with the intramolecular diselenide intermediate 23 (200 μM final concentration) in the presence of 20 mM TCEP and 30 mM DPDS at 37 °C. The ligation proceeded smoothly under these conditions, reaching completion in 2 h. Residual DPDS was extracted with Et2O and the reaction mixture was subjected to photodeselenisation (200 mM TCEP, 40 mM GSH, pH 7.0, λ = 254 nm, 35 W) which proceeded cleanly within 5 min. Gratifyingly, following reverse-phase HPLC purification, ApoCIII 19 was isolated in 60% yield over 4 steps, with the 4-step reaction sequence complete within 4 h. The synthetic ApoCIII was characterised by HPLC, ESI-MS and MALDI-TOF-MS (Scheme 5C and D). As ApoCIII is known to be predominantly α-helical in membranes, we also assessed the CD spectrum of 19 in an SDS-based buffer (10 mM Tris, 100 mM NaCl, 1.8 mM SDS, pH 7.4) at 25 °C. This CD spectrum showed strong α-helical signatures, fully consistent with recombinant ApoCIII (Scheme 5E).47
Conclusions
In summary, we have developed a novel DEAMC photolabile protecting group for Sec that can be cleanly deprotected with LED irradiation at 450 nm. The mild and reagent-free nature of the photodeprotection reaction inspired the development of an iterative DSL methodology for the one-pot assembly of larger polypeptides and proteins. The power of this new synthetic strategy was showcased through the synthesis of 60- and 80-residue MUC1 polypeptides on unprecedented time scales. Finally, we employed the iterative DSL approach to access ApoCIII, a poorly aqueous soluble and aggregation-prone protein in just 4 h over four in situ steps. Future work in our laboratory will involve the use of the DEAMC-protected Sec building block in the ligation-based assembly of post-translationally modified membrane proteins and will be reported in due course.
Data availability
All experimental procedures, analytical and spectroscopic data are provided as the ESI.
Author contributions
TSC and RJP designed the DEAMC protecting group, LK and TSC synthesised the DEAMC-protected Sec amino acid, LK synthesised the MUC1 fragments, LK performed the ligation-based assembly of the MUC1 polypeptides, LK and SSK synthesised the ApoCIII fragments and performed the one-pot protein assembly. LK, SSK and RJP wrote the paper with assistance from TSC. RJP obtained funding for the research.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
The authors acknowledge grant support from the Australian Research Council Centre of Excellence for Innovations in Peptide and Protein Science (CE200100012). The authors acknowledge technical support from Dr Charlotte Franck, The University of Sydney for circular dichroism, Dr Nicholas Proschogo, The University of Sydney for mass spectrometry and Dr Wendy Tran, Sydney Analytical, The University of Sydney for nuclear magnetic resonance.
Electronic supplementary information (ESI) available: Full experimental details, HPLC chromatograms and ESI mass spectra of crude reactions and purified peptide and protein products, NMR data and spectra. See DOI: 10.1039/d1sc02781f
References
- Kent S. B. H. Total chemical synthesis of proteins. Chem. Soc. Rev. 2009;38(2):338–351. doi: 10.1039/B700141J. [DOI] [PubMed] [Google Scholar]
- Bondalapati S. Jbara M. Brik A. Expanding the chemical toolbox for the synthesis of large and uniquely modified proteins. Nat. Chem. 2016;8(5):407–418. doi: 10.1038/nchem.2476. [DOI] [PubMed] [Google Scholar]
- Kulkarni S. S. Sayers J. Premdjee B. Payne R. J. Rapid and efficient protein synthesis through expansion of the native chemical ligation concept. Nat. Rev. Chem. 2018;2:0122. doi: 10.1038/s41570-018-0122. [DOI] [Google Scholar]
- Conibear A. C. Watson E. E. Payne R. J. Becker C. F. Native chemical ligation in protein synthesis and semi-synthesis. Chem. Soc. Rev. 2018;47(24):9046–9068. doi: 10.1039/C8CS00573G. [DOI] [PubMed] [Google Scholar]
- Dawson P. Muir T. Clark-Lewis I. Kent S. Synthesis of proteins by native chemical ligation. Science. 1994;266(5186):776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- Johnson E. C. B. Kent S. B. H. Insights into the mechanism and catalysis of the native chemical ligation reaction. J. Am. Chem. Soc. 2006;128(20):6640–6646. doi: 10.1021/ja058344i. [DOI] [PubMed] [Google Scholar]
- Wan Q. Danishefsky S. J. Free-radical-based, specific desulfurization of cysteine: a powerful advance in the synthesis of polypeptides and glycopolypeptides. Angew. Chem., Int. Ed. 2007;46(48):9248–9252. doi: 10.1002/anie.200704195. [DOI] [PubMed] [Google Scholar]
- Haase C. Seitz O. Extending the scope of native chemical peptide coupling. Angew. Chem., Int. Ed. 2008;47(9):1553–1556. doi: 10.1002/anie.200704886. [DOI] [PubMed] [Google Scholar]
- Chisholm T. S. Clayton D. Dowman L. J. Sayers J. Payne R. J. Native chemical ligation–photodesulfurization in flow. J. Am. Chem. Soc. 2018;140(29):9020–9024. doi: 10.1021/jacs.8b03115. [DOI] [PubMed] [Google Scholar]
- Malins L. R. Payne R. J. Synthetic amino acids for applications in peptide ligation–desulfurization chemistry. Aust. J. Chem. 2015;68(4):521–537. doi: 10.1071/CH14568. [DOI] [Google Scholar]
- Sayers J. Thompson R. E. Perry K. J. Malins L. R. Payne R. J. Thiazolidine-protected β-thiol asparagine: Applications in one-pot ligation–desulfurization chemistry. Org. Lett. 2015;17(19):4902–4905. doi: 10.1021/acs.orglett.5b02468. [DOI] [PubMed] [Google Scholar]
- Haase C. Rohde H. Seitz O. Native chemical ligation at valine. Angew. Chem., Int. Ed. 2008;47(36):6807–6810. doi: 10.1002/anie.200801590. [DOI] [PubMed] [Google Scholar]
- Chen J. Wan Q. Yuan Y. Zhu J. Danishefsky S. J. Native chemical ligation at valine: a contribution to peptide and glycopeptide synthesis. Angew. Chem., Int. Ed. 2008;47(44):8521–8524. doi: 10.1002/anie.200803523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson P. E. Native chemical ligation combined with desulfurization and deselenization: A general strategy for chemical protein synthesis. Isr. J. Chem. 2011;51(8-9):862–867. doi: 10.1002/ijch.201100128. [DOI] [Google Scholar]
- Thompson R. E. Chan B. Radom L. Jolliffe K. A. Payne R. J. Chemoselective peptide ligation–desulfurization at aspartate. Angew. Chem., Int. Ed. 2013;52(37):9723–9727. doi: 10.1002/anie.201304793. [DOI] [PubMed] [Google Scholar]
- Cergol K. M. Thompson R. E. Malins L. R. Turner P. Payne R. J. One-pot peptide ligation–desulfurization at glutamate. Org. Lett. 2014;16(1):290–293. doi: 10.1021/ol403288n. [DOI] [PubMed] [Google Scholar]
- Mitchell N. J. Malins L. R. Liu X. Thompson R. E. Chan B. Radom L. Payne R. J. Rapid Additive-Free Selenocystine–Selenoester Peptide Ligation. J. Am. Chem. Soc. 2015;137(44):14011–14014. doi: 10.1021/jacs.5b07237. [DOI] [PubMed] [Google Scholar]
- Chisholm T. S. Kulkarni S. S. Hossain K. R. Cornelius F. Clarke R. J. Payne R. J. Peptide Ligation at High Dilution via Reductive Diselenide-Selenoester Ligation. J. Am. Chem. Soc. 2020;142(2):1090–1100. doi: 10.1021/jacs.9b12558. [DOI] [PubMed] [Google Scholar]
- Wang X. Sanchez J. Stone M. J. Payne R. J. Sulfation of the human cytomegalovirus protein UL22A enhances binding to the chemokine RANTES. Angew. Chem., Int. Ed. 2017;56(29):8490–8494. doi: 10.1002/anie.201703059. [DOI] [PubMed] [Google Scholar]
- Malins L. R. Payne R. J. Synthesis and utility of β-selenol-phenylalanine for native chemical ligation–deselenization chemistry. Org. Lett. 2012;14(12):3142–3145. doi: 10.1021/ol3012265. [DOI] [PubMed] [Google Scholar]
- Mitchell N. J. Sayers J. Kulkarni S. S. Clayton D. Goldys A. M. Ripoll-Rozada J. Barbosa Pereira P. J. Chan B. Radom L. Payne R. J. Accelerated protein synthesis via one-pot ligation-deselenization chemistry. Chem. 2017;2(5):703–715. [Google Scholar]
- Sayers J. Karpati P. M. T. Mitchell N. J. Goldys A. M. Kwong S. M. Firth N. Chan B. Payne R. J. Construction of challenging proline-proline junctions via diselenide-selenoester ligation chemistry. J. Am. Chem. Soc. 2018;140(41):13327–13334. doi: 10.1021/jacs.8b07877. [DOI] [PubMed] [Google Scholar]
- Wang X. Corcilius L. Premdjee B. Payne R. J. Synthesis and utility of β-selenophenylalanine and β-selenoleucine in diselenide–selenoester ligation. J. Org. Chem. 2019;85(3):1567–1578. doi: 10.1021/acs.joc.9b02665. [DOI] [PubMed] [Google Scholar]
- Kulkarni S. S. Watson E. E. Premdjee B. Conde-Frieboes K. W. Payne R. J. Diselenide–selenoester ligation for chemical protein synthesis. Nat. Protoc. 2019;14(7):2229–2257. doi: 10.1038/s41596-019-0180-4. [DOI] [PubMed] [Google Scholar]
- Metanis N. Keinan E. Dawson P. E. Traceless ligation of cysteine peptides using selective deselenization. Angew. Chem., Int. Ed. 2010;49(39):7049–7053. doi: 10.1002/anie.201001900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang D. Kent S. B. H. A One-Pot Total synthesis of crambin. Angew. Chem., Int. Ed. 2004;43(19):2534–2538. doi: 10.1002/anie.200353540. [DOI] [PubMed] [Google Scholar]
- Veber D. Milkowski J. Varga S. Denkewalter R. Hirschmann R. Acetamidomethyl. A novel thiol protecting group for cysteine. J. Am. Chem. Soc. 1972;94(15):5456–5461. doi: 10.1021/ja00770a600. [DOI] [PubMed] [Google Scholar]
- Tang S. Si Y. Y. Wang Z. P. Mei K. R. Chen X. Cheng J. Y. Zheng J. S. Liu L. An efficient one-pot four-segment condensation method for protein chemical synthesis. Angew. Chem., Int. Ed. 2015;54(19):5713–5717. doi: 10.1002/anie.201500051. [DOI] [PubMed] [Google Scholar]
- Reddy P. S. Dery S. Metanis N. Chemical synthesis of proteins with non-strategically placed cysteines using selenazolidine and selective deselenization. Angew. Chem., Int. Ed. 2016;55(3):992–995. doi: 10.1002/anie.201509378. [DOI] [PubMed] [Google Scholar]
- Dery L. Reddy P. S. Dery S. Mousa R. Ktorza O. Talhami A. Metanis N. Accessing human selenoproteins through chemical protein synthesis. Chem. Sci. 2017;8(3):1922–1926. doi: 10.1039/C6SC04123J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Ashhurst A. S. Dowman L. J. Watson E. E. Li H. Y. Fairbanks A. J. Larance M. Kwan A. Payne R. J. Total synthesis of glycosylated human interferon-γ. Org. Lett. 2020;22(17):6863–6867. doi: 10.1021/acs.orglett.0c02401. [DOI] [PubMed] [Google Scholar]
- Premdjee B. Andersen A. S. Larance M. Conde-Frieboes K. W. Payne R. J. Chemical synthesis of phosphorylated insulin-like growth factor binding protein 2. J. Am. Chem. Soc. 2021;143(14):5336–5342. doi: 10.1021/jacs.1c02280. [DOI] [PubMed] [Google Scholar]
- Zhao Z. Metanis N. Copper-mediated selenazolidine deprotection enables one-pot chemical synthesis of challenging proteins. Angew. Chem., Int. Ed. 2019;58(41):14610–14614. doi: 10.1002/anie.201909484. [DOI] [PubMed] [Google Scholar]
- Karas J. A. Scanlon D. B. Forbes B. E. Vetter I. Lewis R. J. Gardiner J. Separovic F. Wade J. D. Hossain M. A. 2-Nitroveratryl as a photocleavable thiol-protecting group for directed disulfide bond formation in the chemical synthesis of insulin. Chem.–Eur. J. 2014;20(31):9549–9552. doi: 10.1002/chem.201403574. [DOI] [PubMed] [Google Scholar]
- Rakauskaitė R. Urbanavičiūtė G. Rukšėnaitė A. Liutkevičiūtė Z. Juškėnas R. Masevičius V. Klimašauskas S. Biosynthetic selenoproteins with genetically-encoded photocaged selenocysteines. Chem. Commun. 2015;51(39):8245–8248. doi: 10.1039/C4CC07910H. [DOI] [PubMed] [Google Scholar]
- Khodabakhshi S. Barium dichloride as a powerful and inexpensive catalyst for the Pechmann condensation without using solvent. Org. Chem. Int. 2012:306162. [Google Scholar]
- Lin W. Lawrence D. S. A strategy for the construction of caged diols using a photolabile protecting group. J. Org. Chem. 2002;67(8):2723–2726. doi: 10.1021/jo0163851. [DOI] [PubMed] [Google Scholar]
- So W. H. Wong C. T. Xia J. Peptide photocaging: A brief account of the chemistry and biological applications. Chin. Chem. Lett. 2018;29(7):1058–1062. doi: 10.1016/j.cclet.2018.05.015. [DOI] [Google Scholar]
- Mahmoodi M. M. Fisher S. A. Tam R. Y. Goff P. C. Anderson R. B. Wissinger J. E. Blank D. A. Shoichet M. S. Distefano M. D. 6-Bromo-7-hydroxy-3-methylcoumarin (mBhc) is an efficient multi-photon labile protecting group for thiol caging and three-dimensional chemical patterning. Org. Biomol. Chem. 2016;14(35):8289–8300. doi: 10.1039/C6OB01045H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R. Geissler D. Hagen V. Bendig J. Mechanism of photocleavage of (coumarin-4-yl) methyl esters. J. Phys. Chem. A. 2007;111(26):5768–5774. doi: 10.1021/jp071521c. [DOI] [PubMed] [Google Scholar]
- Karami B. Kiani M. Synthesis of the coumarins via Pechmann method in the presence of environmentally friendly Y(NO3)3·6H2O. J. Chin. Chem. Soc. 2014;61(2):213–216. doi: 10.1002/jccs.201200610. [DOI] [Google Scholar]
- Hanna C. C. Kulkarni S. S. Watson E. E. Premdjee B. Payne R. J. Solid-phase synthesis of peptide selenoesters via a side-chain anchoring strategy. Chem. Commun. 2017;53(39):5424–5427. doi: 10.1039/C7CC00823F. [DOI] [PubMed] [Google Scholar]
- Taskinen M.-R. Borén J. Why is apolipoprotein CIII emerging as a novel therapeutic target to reduce the burden of cardiovascular disease? Curr. Atheroscler. Rep. 2016;18(10):1–8. doi: 10.1007/s11883-016-0614-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg H. N. Brown W. V. Apolipoprotein CIII: 42 years old and even more interesting. Arterioscler., Thromb., Vasc. Biol. 2011;31(3):471–473. doi: 10.1161/ATVBAHA.110.221846. [DOI] [PubMed] [Google Scholar]
- Lins L. Flore C. Chapelle L. Talmud P. Thomas A. Brasseur R. Lipid-interacting properties of the N-terminal domain of human apolipoprotein C-III. Protein Eng. 2002;15(6):513–520. doi: 10.1093/protein/15.6.513. [DOI] [PubMed] [Google Scholar]
- Yin H. Zheng M. Chen H. Wang S. Zhou Q. Zhang Q. Wang P. Stereoselective and divergent construction of β-thiolated/selenolated amino acids via photoredox-catalyzed asymmetric Giese reaction. J. Am. Chem. Soc. 2020;142(33):14201–14209. doi: 10.1021/jacs.0c04994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferkorn C. M. Walker III R. L. He Y. Gruschus J. M. Lee J. C. Tryptophan probes reveal residue-specific phospholipid interactions of apolipoprotein C-III. Biochim. Biophys. Acta, Biomembr. 2015;1848(11):2821–2828. doi: 10.1016/j.bbamem.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All experimental procedures, analytical and spectroscopic data are provided as the ESI.