Abstract
Background
More accurate risk assessments are needed to improve prostate cancer management.
Objective
To identify blood-based protein biomarkers that provided prognostic information for risk stratification.
Design, setting, and participants
Mass spectrometry was used to identify biomarker candidates from blood, and validation studies were performed in four independent cohorts retrospectively collected between 1988 and 2015.
Outcome measurements and statistical analysis
The primary outcome objectives were progression-free survival, prostate cancer–specific survival (PCSS), and overall survival. Statistical analyses to assess survival and model performance were performed.
Results and limitation
Serum leucine-rich α-2-glycoprotein 1 (LRG1) was found to be elevated in fatal prostate cancer. LRG1 provided prognostic information independent of metastasis and increased the accuracy in predicting PCSS, particularly in the first 3 yr. A high LRG1 level is associated with an average of two-fold higher risk of disease-progression and mortality in both high-risk and metastatic patients. However, our study design, with a retrospective analysis of samples spanning several decades back, limits the assessment of the clinical utility of LRG1 in today’s clinical practice. Thus, independent prospective studies are needed to establish LRG1 as a clinically useful biomarker for patient management.
Conclusions
High blood levels of LRG1 are unfavourable in newly diagnosed high-risk and metastatic prostate cancer, and LRG1 increased the accuracy of risk stratification of prostate cancer patients.
Patient summary
High blood levels of leucine-rich α-2-glycoprotein 1 are unfavourable in newly diagnosed high-risk and metastatic prostate cancer.
Keywords: Biomarker, Leucine-rich α-2-glycoprotein 1, Noninvasive, Prostate cancer
Take Home Message
High blood levels of leucine-rich α-2-glycoprotein 1 (LRG1) is unfavourable in patients with newly diagnosed high-risk or metastatic prostate cancer. Implementation of a blood test for LRG1 alongside standard risk stratification schemes could provide higher precision in treatment decisions.
1. Introduction
Nearly 1.3 million men were estimated to be diagnosed with prostate cancer (PCa) worldwide in 2018 [1]. Several risk assessment tools are in clinical use to guide treatment and communicate prognostic information [2]. However, as these tools are primarily developed for the prediction of biochemical recurrence, a large diversity has been reported in the accuracy of predicting long-term outcomes [3]. Hence, some guidelines, including those of the National Comprehensive Cancer Network (NCCN), are now endorsing biomarker guidance upon treatment decisions to enable better precision [4], [5].
The multifocal nature and the intra- and interheterogeneity of PCa [6], [7] represent a challenge for tissue-based biomarker discovery. Biological fluids, on the contrary, are easily collected, allow for repeated sampling, and can function as an unbiased source of biomarkers to mirror the complexity of PCa. Unfortunately, the collection of biological samples has been an underfunded effort in many institutional environments; thus, paraffin-embedded prostate specimens have been the primary source of cancer biomarker research so far, which is also reflected in the types of tests approved [8]. Although valuable information can be conveyed from tissue-based tests, these are vulnerable for sampling biases, and consequently utility has been most promising in a postprostatectomy setting [4].
In recent years, the focus of biomarker research in PCa has been on reducing overdiagnosis due to low specificity of the prostate-specific antigen (PSA) test. Several promising blood-based biomarkers and models have been proposed, spanning multiple kallikreins and their isoforms (4KScore [9], Prostate Health Index [10]), and panels of single nucleotide polymorphisms and proteins (STHLM3 [11]), to circulating tumour DNA [12]. However, to date, only the 4KScore has been tested for its utility in predicting long-term outcomes, such as metastasis-free and disease-specific survival [13], [14], and at present, no noninvasive biomarkers have been reported to improve on current standard risk nomograms as predictors of treatment response.
In this study, we sought to identify blood-based proteins that would enable higher precision in predicting unfavourable PCa and long-term outcomes upon diagnosis. We have identified and validated a blood-based protein, leucine-rich α-2-glycoprotein 1 (LRG1), which associates with metastatic PCa and provides increased accuracy in predicting survival outcomes.
2. Patients and methods
2.1. Study design
This study was planned in agreement with the reporting recommendations for tumour marker prognostic studies (REMARK) guidelines, as outlined in the Supplementary material (REMARK), and a summary of the study is illustrated in Supplementary Fig. 1.
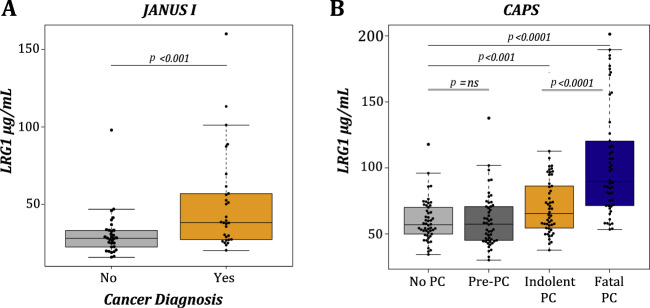
Fig. 1.
LRG1 is elevated in blood from PCa patients and associates with fatal PCa. Tukey boxplots demonstrating the concentration of LRG1 in blood from donors grouped by PCs diagnosis at the time of sampling in (A) JANUS I and (B) CAPS cohorts. The p values were established by MWU test comparing the difference between groups.
CAPS = Cancer of the Prostate Sweden; LRG1 = leucine-rich α-2-glycoprotein 1; MWU = Mann-Whitney U test; ns = not significant; PCa = prostate cancer.
2.2. Patient samples and ethical approvals
Baseline characteristics for the cohorts used are shown in Table 1, and cohorts are further described in the Supplementary material (Materials and methods). In brief, samples from four independent patient cohorts were identified. A discovery cohort (JANUS I, n = 61) was built from the prospective collected Janus Serum Bank (Norway) using a case-control design for PCa and no cancer (control). Samples were included based on cancer diagnosis at blood draw or no cancer diagnosed within 10 yr. A confirmation cohort was generated drawing samples from the prospectively collected Cancer of the Prostate Sweden (CAPS) with a case-control design to represent no PCa (n = 50), before PCa (n = 50), indolent PCa (n = 50), and fatal PCa (n = 50). The inclusion criterion was a minimum of 10 yr of follow-up (FU). Next, an explorative cohort (JANUS II, n = 82) was built to assess LRG1 levels in different NCCN risk strata and confirm association with survival. A final validation cohort (ProMPT-OUH, n = 451) was created by combining 385 samples from the ProMPT (Prostate Cancer Mechanisms of Progression and Treatment) study with 66 samples from a local study at the Oslo University Hospital (OUH).
Table 1.
Baseline characteristics of patient cohorts
| JANUS I |
CAPS |
JANUS II |
ProMPT-OUH |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pre-PC |
PC |
No PC |
Pre-PC |
Indolent PC |
Fatal PC |
PC |
PC |
|||||||||
| n | (%) | n | (%) | n | (%) | n | (%) | n | (%) | n | (%) | n | (%) | n | (%) | |
| Total | 31 | (100) | 30 | (100) | 50 | (100) | 50 | (100) | 50 | (100) | 50 | (100) | 82 | (100) | 430 | (100) |
| Year of sample collection | 1972–2003 | 1988–2009 | 2002–2003 | 2002–2003 | 2001–2004 | 2001–2004 | 1990–2011 | 1995–2015 | ||||||||
| Follow-up time (yr), median (IQR) | 17.3 | (12.5–22.8) | 1.59 | (0.9–3.6) | 9.0 | (8.6–9.6) | 9.2 | (8.7–9.7) | 8.6 | (7.4–10.0) | 0.44 | (0.25–0.63) | 6.8 | (2.7–7.7) | 6.8 | (5.4–8.8) |
| Time from sampling to diagnosis (yr), median (IQR) | 14.9 | (11.0–17.2) | –0.4 | (–1.4 to 0.0) | – | – | 4.2 | (3.0–5.4) | –0.4 | (–0.5 to –0.3) | –0.4 | (–0.6 to –0.3) | –0.3 | (–0.8 to –0.2) | 0.3 | (0.1–0.5) |
| Age at sampling (yr), median (IQR) | 43 | (41–51) | 60 | (57–64) | 65 | (61–73) | 67 | (62–72) | 76 | (74–78) | 66 | (60–73) | 61 | (59–67) | 65 | (60–69) |
| PSA (ng/mL), median (IQR) | NA | 7.0 | (4.0–8.9) | 165.5 | (45.0–511.0) | 10.1 | (6.4–37.6) | 8.6 | (5.6–15.1) | |||||||
| PSA at diagnosis (ng/mL) | ||||||||||||||||
| <10 | NA | 50 | (100) | 0 | (0.0) | 38 | (46.3) | 243 | (56.5) | |||||||
| 10–20 | NA | 0 | (0.0) | 0 | (0.0) | 13 | (15.9) | 109 | (24.2) | |||||||
| >20 | NA | 0 | (0.0) | 50 | (100) | 25 | (30.5) | 78 | (18.2) | |||||||
| Missing | 30 | (100) | 0 | (0.0) | 0 | (0.0) | 6 | (7.3) | 0 | (0.0) | ||||||
| Gleason score | ||||||||||||||||
| 6 | NA | 50 | (100) | 1 | (2.0) | 30 | (36.6) | 107 | (24.9) | |||||||
| 7 | NA | 0 | (0.00) | 11 | (22.0) | 19 | (23.3) | 204 | (47.4) | |||||||
| 8–10 | NA | 0 | (0.00) | 35 | (70.0) | 23 | (28.0) | 109 | (25.3) | |||||||
| Missing | 30 | (100) | 0 | (0.00) | 3 | (6.0) | 10 | (12.2) | 10 | (2.3) | ||||||
| T stage | ||||||||||||||||
| T1 | 4 | (13.3) | 30 | (60.0) | 2 | (4.0) | 24 | (29.3) | 47 | (10.9) | ||||||
| T2 | 3 | (10.0) | 20 | (40.0) | 6 | (12.0) | 33 | (40.2) | 145 | (33.7) | ||||||
| T3 | 12 | (40.0) | 0 | (0.00) | 26 | (52.0) | 15 | (18.3) | 202 | (47.0) | ||||||
| T4 | 1 | (3.3) | 0 | (0.00) | 13 | (26.0) | 6 | (7.3) | 9 | (2.1) | ||||||
| Missing | 10 | (33.3) | 0 | (0.00) | 3 | (6.0) | 4 | (4.9) | 27 | (6.3) | ||||||
| Metastasis | ||||||||||||||||
| Not known (N0/Nx and M0/Mx) | 3 | (10.0) | 50 | (100) | 13 | (26.0) | 55 | (67.9) | 360 | (83.7) | ||||||
| Lymph node (N1+ and M0/Mx) | 9 | (30.0) | 0 | (0.0) | 2 | (4.0) | 0 | (0.0) | 18 | (4.2) | ||||||
| Distant (N0/Nx and M1+) | 17 | (56.7) | 0 | (0.0) | 32 | (64.0) | 25 | (30.9) | 24 | (5.6) | ||||||
| Lymph node and distant (N1+ and M1+) | 1 | (3.3) | 0 | (0.0) | 3 | (6.0) | 1 | (1.2) | 28 | (6.5) | ||||||
| NCCN risk group | ||||||||||||||||
| Low | NA | 28 | (56.0) | 0 | (0.0) | 8 | (9.9) | 56 | (13.0) | |||||||
| Intermediate | NA | 22 | (44.0) | 0 | (0.0) | 30 | (37.0) | 87 | (20.2) | |||||||
| High | NA | 0 | (0.0) | 13 | (36.0) | 17 | (20.7) | 217 | (50.5) | |||||||
| Metastasis | 20 | (66.7) | 0 | (0.0) | 37 | (74.0) | 27 | (32.9) | 70 | (16.3) | ||||||
| Unknown | 10 | (33.3) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | ||||||
| Primary treatment | ||||||||||||||||
| Radical prostatectomy | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | 35 | (43.2) | 183 | (42.6) | ||||||
| Radiation with or without hormone therapy | 0 | (0.0) | 3 | (6.0) | 0 | (0.0) | 32 | (39.5) | 210 | (48.8) | ||||||
| Hormone therapy | 0 | (0.0) | 9 | (18.0) | 48 | (96.0) | 10 | (12.3) | 36 | (8.4) | ||||||
| Unknown/not recorded | 30 | (100) | 38 | (76.0) | 2 | (4.0) | 4 | (4.9) | 1 | (0.2) | ||||||
| Overall mortality | ||||||||||||||||
| Censored | 7 | (22.6) | 6 | (20.0) | 41 | (82.0) | 44 | (88.0) | 31 | (62.0) | 0 | (0.00) | 51 | (63.0) | 359 | (83.5) |
| Event | 24 | (77.4) | 24 | (80.0) | 9 | (18.0) | 6 | (12.0) | 19 | (18) | 50 | (100) | 30 | (37.0) | 71 | (16.5) |
| Cause of death | ||||||||||||||||
| Prostate cancer | 22 | (100) | 21 | (100) | 0 | (0.0) | 1 | (100) | 0 | (0.0) | 50 | (100) | 25 | (83.3) | 26 | (36.7) |
| Other cause | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | 5 | (16.7) | 7 | (9.6) |
| Unknown/Not recorded | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | 38 | (53.5) |
CAPS = Cancer of the Prostate Sweden; IQR = interquartile range; NA = not available; NCCN = National Comprehensive Cancer Network; PC = prostate cancer; PSA = prostate-specific antigen.
All patients received best of care by their treating physician, and the treatment distribution of patients in all cohorts is described in Table 1.
This study was approved by relevant ethical committees in each country, as detailed in the Supplementary material (Materials and methods). Written informed consent was obtained from all participants.
2.3. Laboratory methods
All sample handling was performed in a blinded fashion and according to institutional procedures for handling human biological material. Detailed information on sample handling and protocols performed can be reviewed in the Supplementary material (Materials and methods).
2.3.1. Mass spectrometry
Abundant proteins in full serum were depleted using ProteoSpin Abundant Serum Depletion Kit (Cat. #17300; Norgen Biotek, Thorold, Ontario, Canada) according to the protocol provided by the manufacturer.
Depleted serum proteins (2.5 µg) were used for in-solution trypsin digest, and peptides were analysed by an ESI-Orbitrap (LTQ Orbitrap XL; Thermo Scientific, Bremen, Germany) mass spectrometer coupled to a nano-LC system and label-free quantification was performed. The samples were analysed once.
2.3.2. Immunoassay
The levels of LRG1 in serum/plasma samples were measured in duplicates using a commercially available solid-phase sandwich enzyme-linked immunosorbent assay (ELISA) kit (Cat.no JP27769; IBL International GmbH, Hamburg, Germany) according to the manufacturer’s description.
2.4. Statistical analysis
A detailed description of the statistical analysis is provided in the Supplementary material (Material and methods). Two-sided p < 0.05 was considered significant. In brief, Fisher exact test and standardised mean difference (SMD) were used to assess significant differences in quantified peptide spectra from cancer samples compared with control samples. Two-sided Mann-Whitney U test (MWU) and SMD were used to measure differences in concentrations of LRG1 in grouped samples. Kaplan-Meier estimates and Cox proportional hazard model, supplemented with log-rank test and Harrell’s concordance index (c-index), respectively, was used to assess time-to-event outcomes: progression-free survival (PFS), PCa-specific survival (PCSS), and overall survival (OS). Cut-point for LRG1 dichotomisation was established through grouping LRG1 concentrations into tertiles in JANUS II, where the tertile cut-point that significantly enriched for mortality (compared with the first tertile) by log-rank test was chosen. Time-dependent receiver operator characteristic curve and decision curve analysis were performed to assess added value for predicting survival outcomes for patients.
2.5. Statistical power calculations
Sample size needed to perform a validation study was calculated assuming two-sided equality using a Cox PH model [15] and with a conservatively modified hazard ratio (HR) estimate from the third tertile in the JANUS II cohort. Assuming a 10% incidence of PCa-specific mortality at 10 yr after diagnosis and allowing a 5% type I error rate, at least 414 samples were needed to detect an HR of 3.0 with 90% power.
3. Results
3.1. LRG1 is elevated in blood and associates with aggressive PCa
A mass spectrometry (MS) analysis of 21 serum samples from the Janus Serum Bank identified peptide spectra from a total of 93 proteins (Supplementary Table 1). Of the 21 serum samples analysed, 10 donors harboured PCa at time of blood draw, whereas 11 did not get a PCa diagnosis within 10 yr of FU (median 14.9 yr [interquartile range or IQR 11.0–17.2]). Six proteins showed a significant upregulation in the cancer group (alpha-1-acid glycoprotein 2 [ORM2], complement C4-A [C4A], desmoplakin [DSP], serum amyloid A protein [SAA1], junction plakoglubulin [JUP], and LRG1). LRG1 showed that the greatest fold change (6.9; Fisher exact test with Benjamini-Hochberg multiple-testing correction: p = 0.028) in detected peptides comparing normal and PCa samples (Supplementary Table 1), ranked significant by SMDs (0.88; MWU p = 0.042), was through literature also implicated in aggressive PCa [16], [17] and was therefore selected for further exploration. The finding was confirmed by ELISA using serum from 61 samples from the JANUS I cohort (of which 21 was previously used in MS analysis). Serum LRG1 levels were significantly higher in PCa than in healthy control samples (SMD: 0.79, MWU p < 0.001; Fig. 1A). In the CAPS cohort, LRG1 concentrations were raised notably in men with PCa (MWU p < 0.001) compared with controls, and further increased in men with fatal PCa versus indolent PCa (MWU p < 0.0001; Fig. 1B and Supplementary Table 2). No difference was observed between healthy controls and men who were diagnosed with PCa within 5 yr following blood draw.
Table 2.
Univariable and multivariable Cox proportional hazard modelling for progression-free survival in the ProMPT-OUH cohort
| Univariable |
Multivariable model excluding LRG1 |
Multivariable model with LRG1 cont. |
Multivariable model with LRG1 dicho. |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | p value | HR | 95% CI | p value | HR | 95% CI | p value | HR | 95% CI | p value | |
| LRG1 (cont.) | 1.02 | (1.01–1.02) | <0.0001 | – | – | – | 1.01 | (1.00–1.01) | 0.019 | – | – | – |
| LRG1 (dicho.) | 2.08 | (1.47–2.94) | <0.0001 | – | – | – | – | – | – | 1.60 | (1.25–2.05) | <0.0001 |
| PSA (cont., ng/mL) | 1.00 | (1.00 –1.00) | <0.0001 | 1.00 | (1.00 –1.00) | 0.53 | 1.00 | (1.00–1.00) | 0.39 | 1.00 | (1.00–1.00) | 0.43 |
| GS 6/7 vs GS ≥8 | 3.09 | (2.16–4.42) | <0.0001 | 1.54 | (1.16–2.03) | 0.003 | 1.48 | (1.11–1.96) | 0.007 | 1.11 | (0.84–1.46) | 0.013 |
| T1/2 vs T ≥3 | 1.74 | (1.19–2.55) | 0.004 | 1.06 | (0.80–1.39) | 0.68 | 1.08 | (0.82–1.43) | 0.56 | 1.23 | (0.95–1.58) | 0.12 |
| Metastasis | 7.03 | (4.96–9.95) | <0.0001 | 3.39 | (2.59–4.43) | <0.0001 | 3.18 | (2.39–4.18) | <0.0001 | 3.42 | (2.559–4.50) | <0.0001 |
| C-index | 0.708 (0.657–0.759) | 0.744 (0.697–0.791) | 0.737 (0.690–0.784) | |||||||||
| p value | Ref. | 0.008 | 0.031 | |||||||||
CI = confidence interval; cont. = continuous; dicho. = dichotomised; GS = Gleason score; HR = hazard ratio; LRG1 = leucine-rich α-2-glycoprotein 1; PSA = prostate-specific antigen; Ref. = reference.
3.2. LRG1 is independent of metastasis in predicting fatal PCa
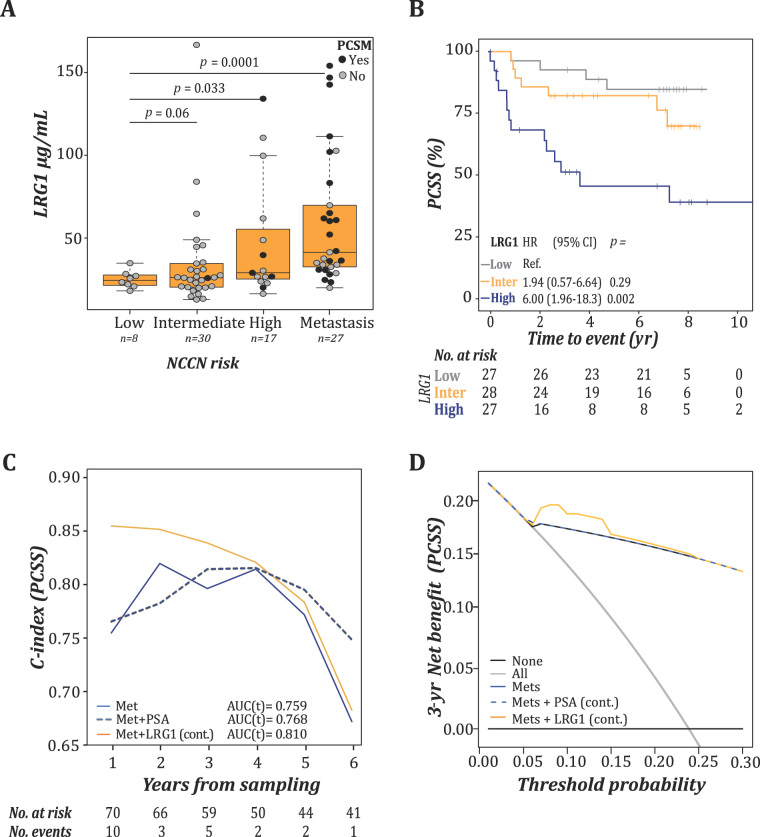
A marked elevation of LRG1 concentrations was further observed in patients with metastatic PCa and fatal PCa in a new sample set of 82 patients from the Janus Serum Bank (JANUS II; Fig. 2A and Supplementary Table 3). The new sample set included 25 (30.9%) samples from men who had died from PCa (median FU 1.3 yr [IQR 0.8–3.3]). The samples were grouped by tertiles of LRG1 concentrations (first <26 µg/mL, second 26–37 µg/ml, and third ≥38 µg/mL). Using the first tertile as a reference (median FU 7.5 yr [IQR 6.4–7.9]), a Kaplan-Meier analysis showed significantly shorter survival in patients with LRG1 levels in the third tertile (median 2.5 yr [IQR 0.7–7.3]), whereas patients with LRG1 levels in the second tertile did not demonstrate a significant difference in PCSS (median 6.4 yr [IQR 3.0–7.3]; Fig. 2B). Consequently, the third tertile was chosen for further dichotomisation of LRG1 into high (third tertile) and low levels (first and second tertiles).
Fig. 2.
LRG1 was an independent prognostic factor in the JANUS II cohort. (A) Tukey boxplot illustrating serum distribution of LRG1 grouped according to the NCCN risk criteria. Filled and open circles illustrate the PCa-specific mortality status at the last FU (median 6.8 yr, [IQR 2.5–7.7]). Significant differences were assessed by MWU testing. (B) Kaplan-Meier survival analysis and log-rank testing of prostate cancer–specific survival (PCSS) of patients grouped by LRG1 concentrations (tertiles: <26 μg/mL [first/low], 26–37 μg/mL [second/inter], and ≥38 μg/mL [third/high]). (C) Time-dependent c-index for PCSS. Metastasis (blue) was included as a covariate in the clinical model and supplemented with PSA (blue, dashed) or LRG1 (orange). (D) Time-dependent decision curve analyses of combining LRG1 and metastasis for predicting PCSS within the first 3 yr after diagnosis.
AUC = area under the curve; CI = confidence interval; HR = hazard ratio; cont. = continuous; IQR = interquartile range; LRG1 = leucine-rich α-2-glycoprotein 1; Met = metastasis; MWU = Mann-Whitney U test; NCCN = National Comprehensive Cancer Network; PCa = prostate cancer; PSA = prostate-specific antigen; Ref. = reference.
In a multivariable Cox proportional hazard model, both continuous and dichotomised LRG1 showed independence of metastasis in predicting PCa-specific mortality (HR = 1.02, [95% confidence interval {CI} 1.01–1.03], p = 0.001 and HR = 1.88 [95% CI 1.05–3.35], p = 0.033). A time-dependent ROC analysis showed that the gained accuracy of adding LRG1 to metastatic status was most apparent in the first 3 yr of FU (Fig. 2C). Decision curve analyses show that adding LRG1 to the model provides not only increased sensitivity and specificity, but also an additional clinical benefit in a 3-yr perspective (Fig. 2D).
3.3. LRG1 provides increased precision in established risk stratification
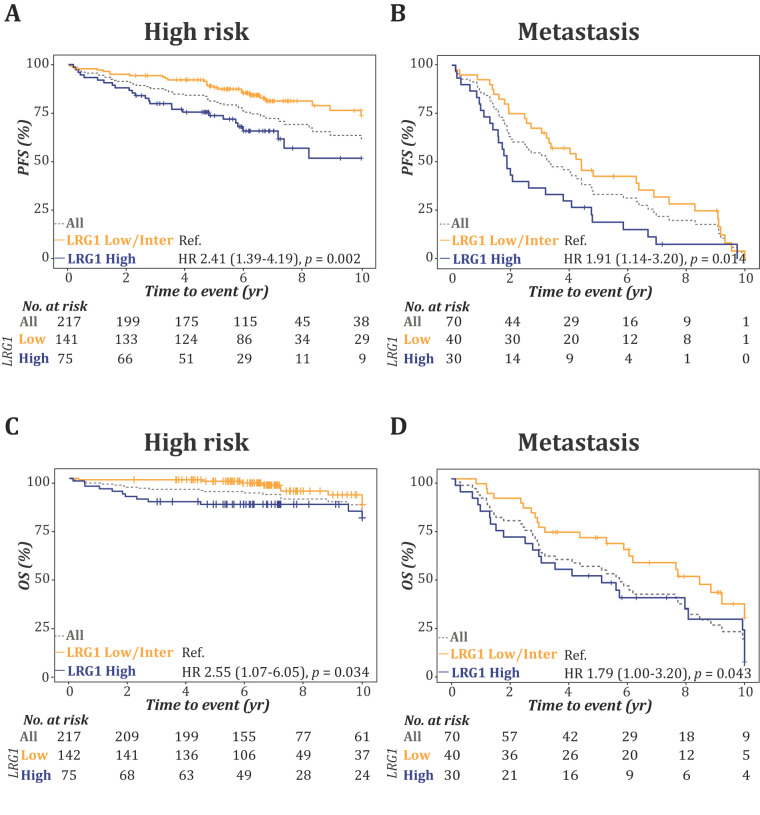
To further confirm that LRG1 is an independent prognostic biomarker, we analysed samples from the ProMPT-OUH cohort using the cut-off established in the JANUS II cohort (>38 µg/mL). The ProMPT-OUH cohort included data on progression and overall mortality, whereas confirmed cause of death was available only for 33 (46.3%). The association of LRG1 with PFS and OS in all risk strata and in metastatic patients is presented as Kaplan-Meier plots (Fig. 3A–D and Supplementary Fig. 2). Very few deaths/events occurred in the low- and intermediate-risk strata (Supplementary Fig. 2). A high LRG1 level was associated with a higher probability of both disease progression and mortality than low LRG1 levels in the NCCN high-risk strata (Fig. 3A and 3C) and among patients with metastatic disease (Fig. 3B and 3D).
Fig. 3.
LRG1 provides enhanced precision for predicting progression and mortality to established risk stratification. Kaplan-Meier plot and log-rank test for progression and overall survival in the ProMPT-OUH cohort grouped according to the NCCN risk strata, (A and C) high risk and (B and D) metastasis, which were supplemented with dichotomised LRG1 levels (high/low).
HR = hazard ratio; LRG1 = leucine-rich α-2-glycoprotein 1; NCCN = National Comprehensive Cancer Network; OS = overall survival; PFS = progression-free survival; Ref. = reference.
3.4. LRG1 enhances the subclassification of high-risk patients
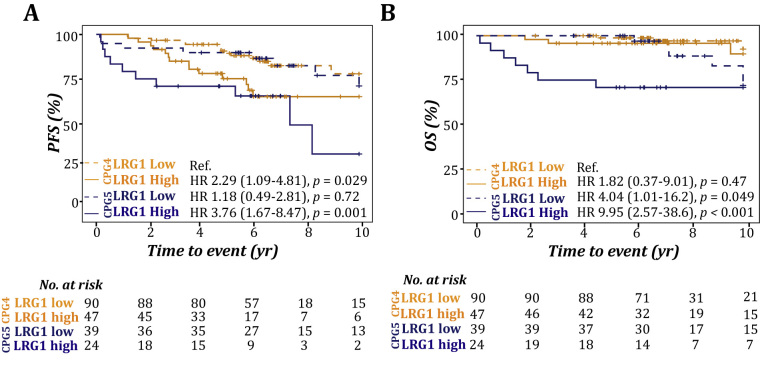
Sufficient clinical information was available for 200 NCCN high-risk patients in the ProMPT-OUH cohort to subclassify patients according to the Cambridge Prognostic Group (CPG) [18]. This model splits typical NCCN high-risk patients into two distinct groups, CPG4 and CPG5. Addition of information on LRG1 levels on top of this model demonstrated a further subclassification of high-risk patients for progression and mortality (Fig. 4).
Fig. 4.
LRG1 enhances the subclassification of high-risk patients. Kaplan-Meier plot and log-rank test for progression and overall survival in high-risk patients in the ProMPT-OUH cohort subclassified according to the Cambridge Prognostic Group (CPG4 and CPG5), which were supplemented with dichotomised LRG1 levels (high/low); (A) PFS and (B) OS.
HR = hazard ratio; LRG1 = leucine-rich α-2-glycoprotein 1; OS = overall survival; PFS = progression-free survival; Ref. = reference.
Overall, in a multivariable Cox proportional hazard model with PSA, Gleason score, T stage, and metastasis, LRG1 demonstrated an independent association with PFS and a significant improvement in accuracy both as a continuous or as a dichotomised measure (Table 2). Comparing c-indices over time, these improvements were most prominent within the first years (Supplementary Fig. 3A and 3B).
4. Discussion
In this study, we identified LRG1 as a marker of fatal PCa. This is the first study to report and validate the independent prognostic capacity and clinical significance of elevated LRG1 in PCa. We establish the clinical impact of elevated LRG1 through validation in a number of independent patient cohorts, and subsequent incorporation of LRG1 into established and novel risk stratification tools.
A particular added value to current clinical risk factors was observed in predicting early mortality (PCSS < 3 yr) in JANUS II and further confirmed in the ProMPT-OUH cohort, where similar survival proportions were observed in high-risk patients with high LRG1 levels and patients with metastasis and low LRG1 at 2-yr FU.
Recently, a head-to-head comparison of widely used risk stratification tools was reported [5]. The novel CPG risk group system outperformed other risk grouping systems, and our data demonstrate further improvement when adding LRG1 into the model. This is particularly true for predicting early progression and/or mortality.
LRG1 has previously been implicated as a promising biomarker for aggressive PCa using a PTEN knockout mouse model [16]. Later, Rehman et al [17] performed proteomic studies directly on serum samples from patients with benign hyperplasia, and localised and metastatic disease, and found LRG1 to be elevated in more aggressive PCa. LRG1 was recently incorporated into a proteomic signature for the detection of aggressive PCa [19] and has been found to be elevated in PCa patients with bone metastasis [20].
Numerous reports have associated LRG1 with unfavourable outcomes in multiple cancer types [21], [22], [23], [24], suggesting a conserved pan-cancer role for LRG1 in disease progression. Hence, a thorough understanding of the biological role of LRG1 may both improve its clinical utility and also help identify potential actionable targets. The function of LRG1 is so far largely unknown, although publications have shown that LRG1, through TGFβ signalling, regulates angiogenesis, migration, and invasion [25], [26], [27], [28], [29], [30]. Furthermore, both inhibition of apoptosis through activation of RUNX1 and interaction with cytochrome C have been reported [25], [26], [28], [29], [30], [31], [32], [33], [34], [35]. LRG1 is a key regulator of myeloid differentiation [36], [37], expressed in most myeloid linages [38], and induced in response to specific acute-phase cytokines [29], [39], with associations to autoimmune diseases such as rheumatoid arthritis, Crohn’s disease, and ulcerative cholitis [40], [41], [42], [43]. This implicates a significant biological role of LRG1 in inflammation, but also several important hallmarks of cancer development and progression, which might shed light on the highly diverse outcomes among patients diagnosed with high-risk and metastatic PCa. Potentially, this could provide guidance towards adjuvant therapies, treatment selection, and enrolment into clinical trials for a significant patient group.
However, our study has been limited in its design to assess the true relevance of LRG1 in PCa and disease progression. Several questions arise such as whether blood levels of LRG1 is primarily a contribution from the tumour, its microenvironment, or a systemic effect to a disseminated cancer, and whether LRG1 can be affected by cancer treatment and can be used to monitor treatment effect. Information on PTEN status in tumour tissue, immune cell infiltration, or activity would have brought important information regarding LRG1’s relevance in a clinical setting. Furthermore, an assessment of the association and implication of elevated LRG1 blood levels in PCa patients with autoimmune diseases will need to be further performed prior to implementing LRG1 into risk stratification tools.
The long FU times of the cohorts included in this study limit the assessment of clinical usefulness of LRG1 in today’s clinical practice. These cohorts were collected before multiparametric magnetic resonance (mpMR) imaging–guided biopsies became an option in clinical practice. mpMR imaging has high accuracy for detecting clinically significant disease, implying that fewer men are diagnosed with low-grade disease nowadays [44], [45]. However, identification of ways towards prioritising men upfront for imaging is also becoming an important task in regions where imaging modalities are scarce and noninvasive, inexpensive tests are needed [46]. Thus, our study representing preimaging cohorts shows that LRG1, similar to 4Kscore, can potentially be used to prioritise whom to offer mpMR imaging. It would be of interest to perform head-to-head comparisons with noninvasive tests aiming to reduce overdiagnosis of clinically insignificant PCa. On the contrary, the long span of the collection period is also a strength because it shows that LRG1 is a prognostic marker within multiple treatment regiments. Although we find LRG1 adding significant information on disease aggressiveness at the diagnosis of high-risk and metastatic disease, there has been vast improvements in both novel treatment strategies, optimal treatment matching and assessing treatment effects for these patients, which provide better outcomes overall. Thus, prospective studies that allow for parallel investigation of patient assessments and management with or without LRG1 values, including head-to-head comparisons with novel noninvasive tests and imaging, will be essential to gather sound evidence and facilitate clinical implementation. Moreover, although great resources have been spent to develop new tests for improving patient management in PCa, to date, very limited data are available on cost and benefits, and clinical adoption. However, so far, we have found LRG1 as a prognostic biomarker irrespective of the study design, participant selection, or study date.
5. Conclusions
We have found that elevated blood levels of LRG1 are unfavourable in patients with newly diagnosed high-risk or metastatic PCa. Thus, our study has established LRG1 as a convincing candidate for inclusion in studies aiming to improve PCa management.
Author contributions: Ingrid J. Guldvik had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Guldvik, Gnanapragasam, Wiklund, Kvåle, Taskén, Mills.
Acquisition of data: Guldvik, Lilleby, Thiede, Kvåle, Grönberg, George, Gnanapragasam.
Analysis and interpretation of data: Guldvik, Zuber, Braadland, Grytli, Ramberg, Zucknick, Lilleby, Gnanapragasam, Wiklund, Neal, Taskén, Mills.
Drafting of the manuscript: Guldvik, Zuber, Braadland, Grytli, Taskén, Mills.
Critical revision of the manuscript for important intellectual content: Guldvik, Zuber, Braadland, Grytli, Ramberg, Lilleby, Thiede, Zucknick, Saatcioglu, Gislefoss, Kvåle, George, Grönberg, Wiklund, Neal, Gnanapragasam, Taskén, Mills.
Statistical analysis: Guldvik, Zuber, Braadland, Zucknick.
Obtaining funding: Guldvik, Taskén, Mills.
Administrative, technical, or material support: Thiede, Saatcioglu, Gislefoss, George, Grönberg.
Supervision: Taskén, Mills.
Other: None.
Financial disclosures: Ingrid J. Guldvik certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: Ingrid J. Guldvik and Ian G. Mills patent EP3243077 prostate cancer biomarkers and uses thereof.
Funding/Support and role of the sponsor: This work has been supported by the Anders Jahre Foundation, University of Oslo, Movember Foundation, South-Eastern Norway Regional Health Authority, Oslo University Hospital, and the Norwegian Research Council, whereas the urology biorepository for the ProMPT (Prostate Cancer Mechanisms of Progression and Treatment) study is funded by the CRUK Cambridge Centre urology malignancy programme and the Cambridge NIHR Biomedical Campus.
Acknowledgements: We thank all the study participants in the Janus Serum Bank (Norway), CAncer of Prostate Sweden (CAPS) study, the Prostate cancer Mechanisms of Progression and Treatment (ProMPT) study, and the OUH cohort. We would further thank Frank Sætre for excellent technical support.
CRediT authorship contribution statement
Ingrid J. Guldvik: Conceptualization, Data curation, Writing - original draft, Writing - review & editing, Visualization, Project administration, Funding acquisition. Verena Zuber: Validation, Writing - original draft, Writing - review & editing. Peder R. Braadland: Formal analysis, Writing - original draft, Writing - review & editing. Helene H. Grytli: Validation, Writing - original draft, Writing - review & editing. Håkon Ramberg: Formal analysis, Writing - review & editing. Wolfgang Lilleby: Resources, Writing - review & editing. Bernd Thiede: Resources, Writing - review & editing. Manuela Zucknick: Validation, Writing - review & editing. Fahri Saatcioglu: Resources, Writing - review & editing. Randi Gislefoss: Resources, Writing - review & editing. Rune Kvåle: Resources, Data curation, Writing - review & editing. Anne George: Resources, Data curation, Writing - review & editing. Henrik Grönberg: Resources, Writing - review & editing. Fredrik Wiklund: Conceptualization, Writing - review & editing. David E. Neal: Resources, Writing - Review & Editing. Vincent J. Gnanapragasam: Conceptualization, Resources, Writing - Review & Editing. Kristin A. Taskén: Conceptualization, Funding acquisition, Project administration, Supervision, Writing - Original Draft, Writing - Review & Editing. Ian G. Mills: Conceptualization, Funding acquisition, Project administration, Supervision, Writing - Original Draft, Writing - Review & Editing.
Associate Editor: Jochen Walz
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.euros.2020.08.007.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Rodrigues G., Warde P., Pickles T. Pre-treatment risk stratification of prostate cancer patients: a critical review. Can Urol Assoc J. 2012;6:121–127. doi: 10.5489/cuaj.11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patel K.M., Gnanapragasam V.J. Novel concepts for risk stratification in prostate cancer. J Clin Urol. 2016;9:18–23. doi: 10.1177/2051415816673502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cucchiara V., Cooperberg M.R., Dall’Era M. Genomic markers in prostate cancer decision making. Eur Urol. 2018;73:572–582. doi: 10.1016/j.eururo.2017.10.036. [DOI] [PubMed] [Google Scholar]
- 5.Zelic R., Garmo H., Zugna D. Predicting prostate cancer death with different pretreatment risk stratification tools: a head-to-head comparison in a nationwide cohort study. Eur Urol. 2020;77:180–188. doi: 10.1016/j.eururo.2019.09.027. [DOI] [PubMed] [Google Scholar]
- 6.Lovf M., Zhao S., Axcrona U. Multifocal primary prostate cancer exhibits high degree of genomic heterogeneity. Eur Urol. 2019;75:498–505. doi: 10.1016/j.eururo.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 7.Yadav S.S., Stockert J.A., Hackert V., Yadav K.K., Tewari A.K. Intratumor heterogeneity in prostate cancer. Urol Oncol. 2018;36:349–360. doi: 10.1016/j.urolonc.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 8.Murphy L., Prencipe M., Gallagher W.M., Watson R.W. Commercialized biomarkers: new horizons in prostate cancer diagnostics. Expert Rev Mol Diagn. 2015;15:491–503. doi: 10.1586/14737159.2015.1011622. [DOI] [PubMed] [Google Scholar]
- 9.Parekh D.J., Punnen S., Sjoberg D.D. A multi-institutional prospective trial in the USA confirms that the 4Kscore accurately identifies men with high-grade prostate cancer. Eur Urol. 2015;68:464–470. doi: 10.1016/j.eururo.2014.10.021. [DOI] [PubMed] [Google Scholar]
- 10.Catalona W.J., Partin A.W., Sanda M.G. A multicenter study of [-2]pro-prostate specific antigen combined with prostate specific antigen and free prostate specific antigen for prostate cancer detection in the 2.0 to 10.0 ng/ml prostate specific antigen range. J Urol. 2011;185:1650–1655. doi: 10.1016/j.juro.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eklund M., Nordstrom T., Aly M. The Stockholm-3 (STHLM3) model can improve prostate cancer diagnostics in men aged 50-69 yr compared with current prostate cancer testing. Eur Urol Focus. 2018;4:707–710. doi: 10.1016/j.euf.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 12.Albitar M., Ma W., Lund L. Predicting prostate biopsy results using a panel of plasma and urine biomarkers combined in a scoring system. J Cancer. 2016;7:297–303. doi: 10.7150/jca.12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stattin P., Vickers A.J., Sjoberg D.D. Improving the specificity of screening for lethal prostate cancer using prostate-specific antigen and a panel of kallikrein markers: a nested case-control study. Eur Urol. 2015;68:207–213. doi: 10.1016/j.eururo.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sjoberg D.D., Vickers A.J., Assel M. Twenty-year risk of prostate cancer death by midlife prostate-specific antigen and a panel of four kallikrein markers in a large population-based cohort of healthy men. Eur Urol. 2018;73:941–948. doi: 10.1016/j.eururo.2018.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chow S.C., Shao J., Wang H. ed. 2. Chapman & Hall/CRC; Boca Raton, FL: 2008. Sample size calculations in clinical research. [Google Scholar]
- 16.Cima I., Schiess R., Wild P. Cancer genetics-guided discovery of serum biomarker signatures for diagnosis and prognosis of prostate cancer. Proc Natl Acad Sci U S A. 2011;108:3342–3347. doi: 10.1073/pnas.1013699108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rehman I., Evans C.A., Glen A. iTRAQ identification of candidate serum biomarkers associated with metastatic progression of human prostate cancer. PLoS One. 2012;7:e30885. doi: 10.1371/journal.pone.0030885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gnanapragasam V.J., Bratt O., Muir K. The Cambridge Prognostic Groups for improved prediction of disease mortality at diagnosis in primary non-metastatic prostate cancer: a validation study. BMC Med. 2018;16:31. doi: 10.1186/s12916-018-1019-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy K., Murphy B.T., Boyce S. Integrating biomarkers across omic platforms: an approach to improve stratification of patients with indolent and aggressive prostate cancer. Mol Oncol. 2018;12:1513–1525. doi: 10.1002/1878-0261.12348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan B., Chen B., Min S. iTRAQ-based comparative serum proteomic analysis of prostate cancer patients with or without bone metastasis. J Cancer. 2019;10:4165–4177. doi: 10.7150/jca.33497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andersen J.D., Boylan K.L., Jemmerson R. Leucine-rich alpha-2-glycoprotein-1 is upregulated in sera and tumors of ovarian cancer patients. J Ovarian Res. 2010;3:21. doi: 10.1186/1757-2215-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ladd J.J., Busald T., Johnson M.M. Increased plasma levels of the APC-interacting protein MAPRE1, LRG1, and IGFBP2 preceding a diagnosis of colorectal cancer in women. Cancer Prev Res (Phila) 2012;5:655–664. doi: 10.1158/1940-6207.CAPR-11-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandanayake N.S., Sinclair J., Andreola F. A combination of serum leucine-rich alpha-2-glycoprotein 1, CA19-9 and interleukin-6 differentiate biliary tract cancer from benign biliary strictures. Br J Cancer. 2011;105:1370–1378. doi: 10.1038/bjc.2011.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wen S.Y., Zhang L.N., Yang X.M. LRG1 is an independent prognostic factor for endometrial carcinoma. Tumour Biol. 2014;35:7125–7133. doi: 10.1007/s13277-014-1953-6. [DOI] [PubMed] [Google Scholar]
- 25.Meng H., Song Y., Zhu J. LRG1 promotes angiogenesis through upregulating the TGFbeta1 pathway in ischemic rat brain. Mol Med Rep. 2016;14:5535–5543. doi: 10.3892/mmr.2016.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song W., Wang X. The role of TGFbeta1 and LRG1 in cardiac remodelling and heart failure. Biophys Rev. 2015;7:91–104. doi: 10.1007/s12551-014-0158-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun D.C., Shi Y., Wang L.X. Leucine-rich alpha-2-glycoprotein-1, relevant with microvessel density, is an independent survival prognostic factor for stage III colorectal cancer patients: a retrospective analysis. Oncotarget. 2017;8:66550–66558. doi: 10.18632/oncotarget.16289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X., Abraham S., McKenzie J.A.G. LRG1 promotes angiogenesis by modulating endothelial TGF-beta signalling. Nature. 2013;499:306–311. doi: 10.1038/nature12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y., Xu J., Zhang X. TNF-alpha-induced LRG1 promotes angiogenesis and mesenchymal stem cell migration in the subchondral bone during osteoarthritis. Cell Death Dis. 2017;8:e2715. doi: 10.1038/cddis.2017.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J., Zhu L., Fang J., Ge Z., Li X. LRG1 modulates epithelial-mesenchymal transition and angiogenesis in colorectal cancer via HIF-1alpha activation. J Exp Clin Cancer Res. 2016;35:29. doi: 10.1186/s13046-016-0306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun D., Kar S., Carr B.I. Differentially expressed genes in TGF-beta 1 sensitive and resistant human hepatoma cells. Cancer Lett. 1995;89:73–79. doi: 10.1016/0304-3835(95)90160-4. [DOI] [PubMed] [Google Scholar]
- 32.Zhou Y., Zhang X., Zhang J., Fang J., Ge Z., Li X. LRG1 promotes proliferation and inhibits apoptosis in colorectal cancer cells via RUNX1 activation. PLoS One. 2017;12:e0175122. doi: 10.1371/journal.pone.0175122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong D., He G., Zhao S. LRG1 modulates invasion and migration of glioma cell lines through TGF-beta signaling pathway. Acta Histochem. 2015;117:551–558. doi: 10.1016/j.acthis.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 34.Codina R., Vanasse A., Kelekar A., Vezys V., Jemmerson R. Cytochrome c-induced lymphocyte death from the outside in: inhibition by serum leucine-rich alpha-2-glycoprotein-1. Apoptosis. 2010;15:139–152. doi: 10.1007/s10495-009-0412-0. [DOI] [PubMed] [Google Scholar]
- 35.Ban Z., He J., Tang Z., Zhang L., Xu Z. LRG1 enhances the migration of thyroid carcinoma cells through promotion of the epithelial mesenchymal transition by activating MAPK/p38 signaling. Oncol Rep. 2019;41:3270–3280. doi: 10.3892/or.2019.7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Donnell L.C., Druhan L.J., Avalos B.R. Molecular characterization and expression analysis of leucine-rich alpha2-glycoprotein, a novel marker of granulocytic differentiation. J Leukoc Biol. 2002;72:478–485. [PubMed] [Google Scholar]
- 37.Druhan L.J., Lance A., Li S. Leucine rich alpha-2 glycoprotein: a novel neutrophil granule protein and modulator of myelopoiesis. PLoS One. 2017;12:e0170261. doi: 10.1371/journal.pone.0170261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hao L., Xie H., Zhang B. LRG1 downregulation in allergic airway disorders and its expression in peripheral blood and tissue cells. J Transl Med. 2016;14:202. doi: 10.1186/s12967-016-0929-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shirai R., Hirano F., Ohkura N., Ikeda K., Inoue S. Up-regulation of the expression of leucine-rich alpha(2)-glycoprotein in hepatocytes by the mediators of acute-phase response. Biochem Biophys Res Commun. 2009;382:776–779. doi: 10.1016/j.bbrc.2009.03.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ha Y.J., Kang E.J., Lee S.W. Usefulness of serum leucine-rich alpha-2 glycoprotein as a disease activity biomarker in patients with rheumatoid arthritis. J Korean Med Sci. 2014;29:1199–1204. doi: 10.3346/jkms.2014.29.9.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Honda H., Fujimoto M., Miyamoto S. Sputum Leucine-Rich Alpha-2 Glycoprotein as a Marker of Airway Inflammation in Asthma. PLoS One. 2016;11:e0162672. doi: 10.1371/journal.pone.0162672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Serada S., Fujimoto M., Terabe F. Serum leucine-rich alpha-2 glycoprotein is a disease activity biomarker in ulcerative colitis. Inflamm Bowel Dis. 2012;18:2169–2179. doi: 10.1002/ibd.22936. [DOI] [PubMed] [Google Scholar]
- 43.Shinzaki S., Matsuoka K., Iijima H. Leucine-rich alpha-2 glycoprotein is a serum biomarker of mucosal healing in ulcerative colitis. J Crohns Colitis. 2017;11:84–91. doi: 10.1093/ecco-jcc/jjw132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahmed H.U., El-Shater Bosaily A., Brown L.C. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confirmatory study. Lancet. 2017;389:815–822. doi: 10.1016/S0140-6736(16)32401-1. [DOI] [PubMed] [Google Scholar]
- 45.Kasivisvanathan V., Rannikko A.S., Borghi M. MRI-targeted or standard biopsy for prostate-cancer diagnosis. N Engl J Med. 2018;378:1767–1777. doi: 10.1056/NEJMoa1801993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Auvinen A., Rannikko A., Taari K. A randomized trial of early detection of clinically significant prostate cancer (ProScreen): study design and rationale. Eur J Epidemiol. 2017;32:521–527. doi: 10.1007/s10654-017-0292-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.