Abstract
Background
Multiparametric magnetic resonance imaging (mpMRI) has shown promise to improve detection of prostate cancer over conventional methods. However, most studies do not describe whether the location of mpMRI lesions match that of cancer found at biopsy, which may lead to an overestimation of accuracy.
Objective
To quantitate the effect of mapping locations of mpMRI lesions to locations of positive biopsy cores on the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of mpMRI.
Design, setting, and participant
We retrospectively identified patients having mpMRI of the prostate preceding prostate biopsy at three centres from 2013 to 2016. Men with targetable lesions on imaging underwent directed biopsy in addition to systematic biopsy. We correlated locations of positive mpMRI lesions with those of positive biopsy cores, defining a match when both were in the same sector of the prostate. We defined positive mpMRI as Prostate Imaging Reporting and Data System (PI-RADS) score ≥4 and significant cancer at biopsy as grade group ≥2.
Outcome measurements and statistical analysis
Sensitivity, specificity, PPV, and NPV were calculated with and without location matching.
Results and limitations
Of 446 patients, 247 (55.4%) had positive mpMRI and 232 (52.0%) had significant cancer at biopsy. Sensitivity and NPV for detecting significant cancer with location matching (both 63.4%) were decreased compared with those without location matching (77.6% and 73.9%, respectively). Of the 85 significant cancers not detected by mpMRI, most were of grade group 2 (64.7%, 55/85).
Conclusions
We report a 10–15% decrease in sensitivity and NPV when location matching was used to detect significant prostate cancer by mpMRI. False negative mpMRI remains an issue, highlighting the continued need for biopsy and for improving the standards around imaging quality and reporting.
Patient summary
The true accuracy of multiparametric magnetic resonance imaging (mpMRI) must be determined to interpret results and better counsel patients. We mapped the location of positive mpMRI lesions to where cancer was found at biopsy and found, when compared with matching to cancer anywhere in the prostate, that the accuracy of mpMRI decreased by 10–15%.
Keywords: Diagnostic accuracy, Multiparametric magnetic resonance imaging, Prostate biopsy, Prostate cancer
Take Home Message
We have precisely correlated the location of positive multiparametric magnetic resonance imaging lesions with that of positive prostate biopsy cores. We report a 10–15% reduction in sensitivity and negative predictive value using this location-matching method compared with not using location matching.
1. Introduction
Multiparametric magnetic resonance imaging (mpMRI) has played an important role in the diagnosis of prostate cancer in recent years. Utilisation of mpMRI allows suspicious areas to be targeted at biopsy, with studies suggesting improved cancer detection rates in comparison with standard biopsy approaches alone [1], [2], [3], [4]. In addition, mpMRI permits the noninvasive prediction of lesion grade and aggressiveness, with higher suspicion grades on mpMRI being more likely to harbour higher-grade cancer [5]. Encouraging results have been reported in the literature, with the recent Prostate MRI Imaging Study (PROMIS) [6] reporting a sensitivity of 88% and a negative predictive value (NPV) of 76% for the prediction of Gleason score ≥7 disease. In addition, accuracies of up to 92% for the detection of index lesions at radical prostatectomy have been reported [31].
Most studies consider positive mpMRI to be correct in identifying a prostate cancer regardless of the location within the prostate where the cancer is found, reporting mpMRI as successful (true positive) even if the positive biopsy is from a different region of the prostate [6], [7], [8], [9], [10]. Location matching involves correlating the location of an mpMRI lesion with the location of a positive prostate biopsy. This is difficult to perform due to the lack of standardisation in describing the location of mpMRI lesions, which often do not correlate with operator descriptions of the location of prostate biopsy cores. However, this lack of granularity with current studies is hypothesised to overestimate the accuracy of mpMRI, with a decreased false negative rate and thus misleadingly high sensitivity reported. For mpMRI to be effective as a diagnostic tool for prostate cancer, it is important to determine the true accuracy of mpMRI to predict the presence of significant cancers in the correct location before undergoing biopsy.
We aim to assess the effect of location matching between mpMRI lesions and cancer found at prostate biopsy by examining the accuracy of mpMRI with and without “location matching”. A comparison of analyses with and without location matching will allow us to quantify the effect of location matching, allowing calibration of pre-existing studies that have not been able to implement this method. We hypothesise that location matching will result in lower sensitivity than that calculated without location matching.
2. Patients and methods
2.1. Study population
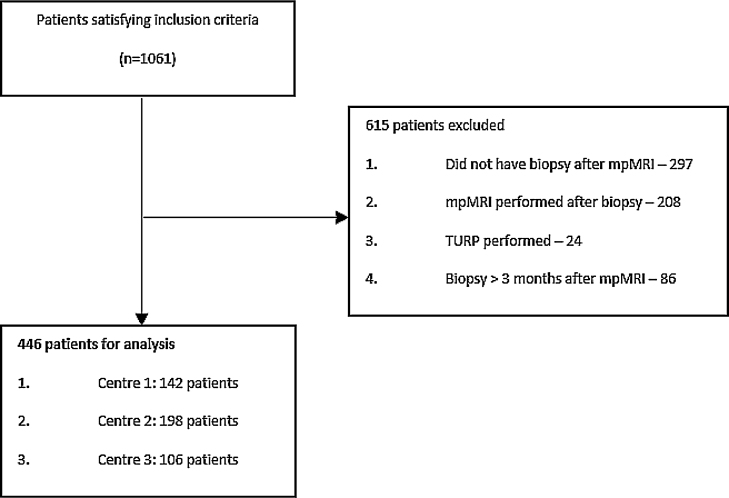
This institutional review board–approved study was designed as a nonrandomised retrospective analysis from a centralised database. Patients were recruited from three Australian centres between August 2013 and September 2016. Data collection occurred prospectively at centre 2 and retrospectively at centres 1 and 3. Patients were identified from radiology mpMRI prostate codes and included for analysis if prostate biopsy was performed for a raised prostate-specific antigen (PSA) level and/or an abnormal digital rectal examination. Patients were excluded if the mpMRI was performed after the biopsy, the biopsy was performed >3 mo after mpMRI, cancer was detected during transurethral resection of the prostate, or biopsy was not performed after mpMRI (Fig. 1). There was a heterogeneous cohort, with 346 patients (77.6%) being biopsy naïve, 37 (8.3%) having a previous negative biopsy, and 63 (14.1%) having previous prostate cancer detected on biopsy.
Fig. 1.
Flowchart of patient selection for study. mpMRI = multiparametric magnetic resonance imaging; TURP = transurethral resection of the prostate.
2.2. Imaging
Prostate mpMRI prostate was performed using a 3 T MR system (Skyra; Siemens, Forchheim, Bavaria, Germany) at centres 1 and 3, and using a 1.5 T system (Optima MR360; GE, Boston, Massachusetts, USA) at centre 2. Endorectal coil was not used. T2, diffusion-weighted imaging, and dynamic contrast enhancement were utilised. A summary of the mpMRI sequences is provided in the Supplementary material.
Results were interpreted by multiple radiologists with varying levels of experience in prostate mpMRI (all had >12 mo of experience reporting prostate MRI): centre 1—six radiologists, one of whom reported prostate mpMRI regularly; centre 2—six radiologists trained in prostate mpMRI (double reported); and centre 3—two radiologists. Lesions were graded according to the validated Prostate Imaging Reporting and Data System (PI-RADS) scoring system, either version 1 or version 2 [11], [12]; centre 1 changed to PI-RADS version 2 in January 2016 and centre 2 changed to PI-RADS version 2 in August 2015, whereas centre 3 used PI-RADS version 2 exclusively.
2.3. Biopsy protocol
Urologists at consultant and registrar levels performed all biopsies. The biopsy schema used included standard systematic prostate needle biopsies either by a 12-core standard extended peripheral zone template for the transrectal ultrasound (TRUS) approach and the modified Barzell and Melamed [13] 20-zone template, or by transperineal template mapping biopsy [14] for the transperineal ultrasound approach. If a suspicious mpMRI lesion was identified, targeted biopsy was performed either by cognitive estimation biopsy (n = 241) or by MRI/TRUS software fusion biopsy (n = 112) depending on patient and operator preference, with at least two extra cores taken. Cognitive estimation biopsy was performed using either transrectal or transperineal biopsy, which involved the operator reviewing the mpMRI images prior to correlating the suspicious areas to the real-time TRUS images. MRI/TRUS software fusion biopsy was performed using the Variseed low-dose rate treatment planning system at centre 1, the BioJet Fusion Software System (DK Technologies, Barum, Germany) combined with a transperineal grid TRUS platform (BK Medical, Herlev, Denmark) at centre 2, and the Alexus Services (Sunbury, Victoria, Australia) software program at centre 3.
2.4. Histopathology
Biopsy locations were described in a standard extended prostate biopsy template for TRUS biopsies [15] and in either the modified Barzell and Melamed [13] 20-zone template or template mapping [14] for transperineal biopsies. Histology specimens were reviewed at each centre as follows: a single pathologist at centre 1, multiple external pathology providers at centre 2, and two pathologists at centre 3. All biopsy cores were reported according to either the International Society of Urological Pathology (ISUP)-modified Gleason score [16] or the newer ISUP grade group format [17]. For the purposes of this study, cancer grading was expressed in the ISUP grade group format [17].
2.5. Location mapping between mpMRI and histopathology
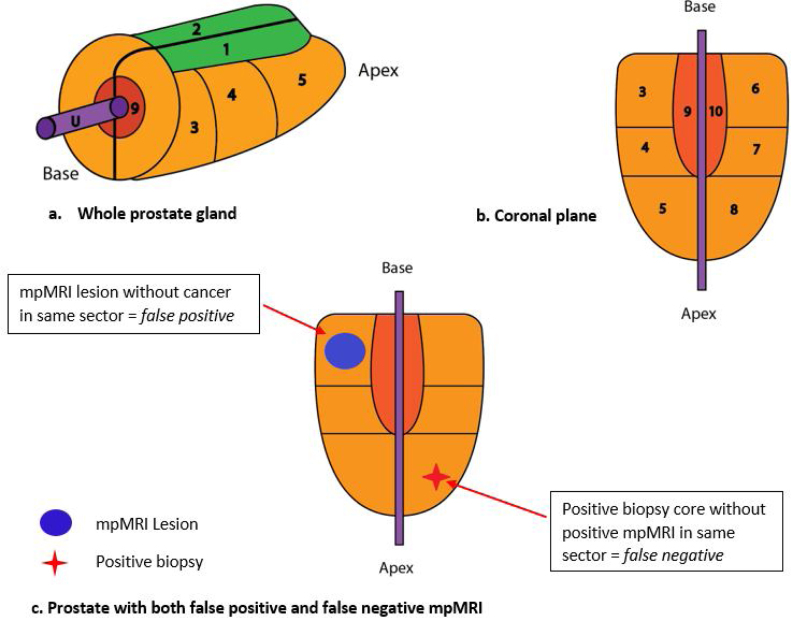
To standardise locations for analysis, the prostate was divided into 10 sectors (Fig. 2) based on the extended TRUS-biopsy template (six zones) [15] with the addition of left and right anterior and transition zones. The locations of suspicious lesions on mpMRI and positive biopsy cores were mapped according to these defined sectors. The reported biopsy locations for the modified Barzell and Melamed [13] 20-zone template approach and the transperineal template mapping approach were adapted according to our standardised 10-sector locations shown in Fig. 2.
Fig. 2.
Prostate gland divided into 10 sectors for analysis: 1—right anterior zone; 2—left anterior zone; 3—right base peripheral zone; 4—right mid peripheral zone; 5—right apex peripheral zone; 6—left base peripheral zone; 7—left mid peripheral zone; 8—left apex peripheral zone; 9—right transition zone; and 10—left transition zone. (A) Whole prostate gland. (B) Coronal view of prostate gland. (C) Coronal view of prostate gland demonstrating a false positive and a false negative result in the same patient. This patient was designated as “false negative”. mpMRI = multiparametric magnetic resonance imaging; U = urethra.
The correlation between sectors containing suspicious mpMRI lesions and positive biopsy cores was assessed. A positive match was identified when a positive mpMRI lesion and a positive biopsy core were in the exact same sector. To allow for the known geometric error associated with TRUS prostate biopsy [18], [19], we also performed a sensitivity analysis considering positive cores in a sector adjacent to a positive mpMRI lesion as a match.
2.6. Outcomes and statistical analysis
The primary outcome was to assess the ability of mpMRI to detect clinically significant prostate cancer, found at biopsy, in the correct location. A positive mpMRI lesion was defined as having a PI-RADS score of ≥4. We defined clinically significant cancer at biopsy as ISUP grade group ≥2. Sensitivity, specificity, positive predictive values (PPVs), and NPVs were calculated according to the definitions shown in Table 1.
Table 1.
Definitions of true positive (TP), false positive (FP), false negative (FN), and true negative (TN) used for location matching.
| Positive mpMRI (PI-RADS ≥4) | Negative mpMRI (PI-RADS <4) | |
|---|---|---|
| Significant cancer (Grade group ≥2) | TP (match between a positive mpMRI lesion and any significant cancer) | FN (significant cancer detected but negative mpMRI) |
| No significant cancer | FP (positive mpMRI but no significant cancer) | TN (negative mpMRI and negative for significant cancer) |
mpMRI = multiparametric magnetic resonance imaging; PI-RADS = Prostate Imaging Reporting and Data System.
The analysis was performed at the whole-prostate level and not at a per-lesion level. It is possible for patients to have both a false positive and a false negative result in a single mpMRI examination (Fig. 2). For analysis, these patients were classified as “false negatives” as this was felt to be a more significant clinical error.
Sensitivity analyses were also performed by changing the threshold for cancer (lowered to detect any-grade cancer), changing the threshold for a positive mpMRI lesion (lowered to PI-RADS score ≥3), and decreasing the stringency for location matching (lowered to accept adjacent sector as a match, and no location matching—akin to analyses in previous studies). When performing the analysis without location matching, a “true positive” was defined as having positive mpMRI and significant cancer in any sector of the prostate.
Differences in the median maximum cancer core length of significant cancer at biopsy were calculated using the equality-of-medians test.
Statistical analyses were performed using STATA 14 (StataCORP LP, TX, USA). Categorical data were presented using absolute and relative frequencies. Differences between means were determined by the two-way t test, whereas differences between proportions were calculated using the chi-square test. Differences between medians were calculated using the equality-of-medians test.
3. Results
Clinical and biopsy characteristics are reported in Table 2. The median age was 65 yr and median PSA 7.1 ng/ml. A total of 346 patients were biopsy naïve (77.6%). The median time between mpMRI and prostate biopsy was 25 d. Most patients (55.4%, 247/446) were reported to have a PI-RADS ≥4 lesion. At biopsy, 52.0% (232/446) had grade group ≥2 cancer.
Table 2.
Clinical and biopsy characteristics.
| Overall | Centre 1 | Centre 2 | Centre 3 | |
|---|---|---|---|---|
| No. of patients | 446 | 142 (31.8) | 198 (44.4) | 106 (23.8) |
| Age (yr), median (IQR) | 65 (60–70) | 64 (59–69) | 65 (60–68) | 68 (64–73) |
| PSA (ng/ml), median (IQR) | 7.1 (5.3–9.7) | 7.5 (5.4–10.5) | 6.6 (4.9–8.6) | 8.0 (6.1–10.2) |
| DRE, n (%) | ||||
| Unknown status | 101 (22.6) | 82 (57.7) | 19 (9.6) | 0 (0) |
| cT1 | 262 (58.7) | 38 (26.8) | 157 (79.3) | 67 (63.2) |
| cT2 | 74 (16.6) | 18 (12.7) | 20 (10.1) | 36 (34.0) |
| cT3 | 9 (2.1) | 4 (2.8) | 2 (1.0) | 3 (2.8) |
| Biopsy status, n (%) | ||||
| Biopsy naïve | 346 (77.6) | 95 (66.9) | 177 (89.4) | 74 (69.8) |
| Previous negative biopsy | 37 (8.3) | 18 (12.7) | 7 (3.5) | 12 (11.3) |
| Previous prostate cancer diagnosis | 63 (14.1) | 29 (20.4) | 14 (7.1) | 20 (18.9) |
| MRI prostate volume (cc), median (IQR) | 48.6 (37.0–67.8) | 50 (37–72) | 49 (35–71) | 48 (39–63) |
| Maximum PI-RADS, n (%) | ||||
| 1–2 | 102 (22.9) | 28 (19.7) | 61 (30.8) | 13 (12.3) |
| 3 | 97 (21.7) | 16 (11.3) | 61 (30.8) | 20 (18.9) |
| ≥4 | 247 (55.4) | 98 (69.0) | 76 (38.4) | 73 (68.8) |
| Days between mpMRI and biopsy, median (IQR) | 25 (10–42) | 19.5 (7–39) | 25 (11–42) | 34 (16–47) |
| Biopsy technique, n (%) | ||||
| TRUS (systematic only) | 67 (15.0) | 15 (10.6) | 45 (22.7) | 7 (6.6) |
| TRUS cognitive fusion | 187 (41.9) | 88 (62.0) | 62 (31.3) | 37 (34.9) |
| TRUS software fusion | 10 (2.2) | 4 (2.8) | 6 (3.1) | 0 (0) |
| Transperineal (systematic only) | 26 (5.8) | 3 (2.1) | 19 (9.6) | 4 (3.8) |
| Transperineal cognitive fusion | 54 (12.2) | 6 (4.2) | 1 (0.5) | 47 (44.3) |
| Transperineal software fusion | 102 (22.9) | 26 (18.3) | 65 (32.8) | 11 (10.4) |
| Highest-grade group cancer detected, n (%) | ||||
| No cancer | 130 (29.2) | 40 (28.2) | 71 (35.9) | 19 (17.9) |
| Grade group 1 | 84 (18.8) | 23 (16.2) | 39 (19.7) | 22 (20.8) |
| Grade group 2 | 89 (20.0) | 37 (26.1) | 29 (14.7) | 23 (21.7) |
| Grade group 3 | 71 (15.9) | 20 (14.1) | 29 (14.7) | 22 (20.8) |
| Grade group ≥4 | 72 (16.1) | 22 (15.4) | 16 (8.0) | 20 (18.8) |
| Cores taken at biopsy, median (IQR) | 22 (16–30) | 18 (14–27) | 22 (18–28) | 27 (20–35) |
DRE = digital rectal examination; IQR = interquartile range; mpMRI = multiparametric magnetic resonance imaging; MRI = magnetic resonance imaging; PI-RADS = Prostate Imaging Reporting and Data System; PSA = prostate-specific antigen; TRUS = transrectal ultrasound.
3.1. Ability of mpMRI to detect cancers found at biopsy (location matching)
The ability of mpMRI to detect any cancer and significant prostate cancer across all three centres is shown in Table 3. When examining the ability of mpMRI to detect significant cancer (grade group ≥2) utilising a threshold of PI-RADS 4, the sensitivity and specificity were 63.4% and 68.7%, respectively, with PPV and NPV being 68.7% and 63.4%, respectively.
Table 3.
Diagnostic accuracy of mpMRI to detect any-grade cancer and significant cancer using location matching, by centre.
| Sensitivity | Specificity | PPV | NPV | |||
|---|---|---|---|---|---|---|
| All centres | ||||||
| Any PCa | ||||||
| PI-RADS ≥3 | 67.1 | 42.3 | 73.9 | 34.6 | ||
| PI-RADS ≥4 | 58.5 | 76.9 | 86.0 | 43.3 | ||
| sPCa (grade group ≥2) | ||||||
| PI-RADS ≥3 | 71.1 | 37.4 | 55.2 | 54.4 | ||
| PI-RADS ≥4 | 63.4 | 68.7 | 68.7 | 63.4 | ||
| sPCa (grade group ≥3) | ||||||
| PI-RADS ≥3 | 76.9 | 31.0 | 34.5 | 74.0 | ||
| PI-RADS ≥4 | 70.6 | 59.1 | 44.9 | 81.0 | ||
| Centre 1 | ||||||
| Any PCa | ||||||
| PI-RADS ≥3 | 62.1 | 33.3 | 71.1 | 25.0 | ||
| PI-RADS ≥4 | 57.3 | 53.9 | 76.6 | 32.3 | ||
| sPCa (grade group ≥2) | ||||||
| PI-RADS ≥3 | 59.5 | 30.2 | 51.6 | 37.3 | ||
| PI-RADS ≥4 | 57.0 | 47.6 | 57.7 | 46.9 | ||
| sPCa (grade group ≥3) | ||||||
| PI-RADS ≥3 | 57.1 | 24.0 | 24.0 | 46.2 | ||
| PI-RADS ≥4 | 57.1 | 40.0 | 28.6 | 69.0 | ||
| Centre 2 | ||||||
| Any PCa | ||||||
| PI-RADS ≥3 | 67.5 | 50.0 | 70.2 | 46.8 | ||
| PI-RADS ≥4 | 54.8 | 90.3 | 90.8 | 53.3 | ||
| sPCa (grade group ≥2) | ||||||
| PI-RADS ≥3 | 77.3 | 45.5 | 53.1 | 71.4 | ||
| PI-RADS ≥4 | 62.5 | 82.7 | 74.3 | 73.4 | ||
| sPCa (grade group ≥3) | ||||||
| PI-RADS ≥3 | 89.5 | 41.1 | 38.1 | 90.6 | ||
| PI-RADS ≥4 | 73.7 | 76.6 | 56.0 | 87.8 | ||
| Centre 3 | ||||||
| Any PCa | ||||||
| PI-RADS ≥3 | 72.4 | 31.6 | 82.9 | 20.0 | ||
| PI-RADS ≥4 | 65.5 | 73.7 | 91.9 | 31.8 | ||
| sPCa (grade group ≥2) | ||||||
| PI-RADS ≥3 | 76.9 | 26.8 | 62.5 | 42.3 | ||
| PI-RADS ≥4 | 72.3 | 63.4 | 75.8 | 59.1 | ||
| sPCa (grade group ≥3) | ||||||
| PI-RADS ≥3 | 81.4 | 19.0 | 40.7 | 60.0 | ||
| PI-RADS ≥4 | 81.4 | 49.2 | 52.2 | 79.5 | ||
mpMRI = multiparametric magnetic resonance imaging; NPV = negative predictive value; PCa = prostate cancer; PI-RADS = Prostate Imaging Reporting and Data System; PPV = positive predictive value; sPCa = significant prostate cancer.
The effect of changing the thresholds for positive mpMRI (PI-RADS 3 vs PI-RADS 4) is shown in Table 3. Using a less stringent mpMRI threshold, the sensitivity increased (71.1%), whilst the specificity, PPV, and NPV were lower (37.4%, 55.2%, and 54.4%, respectively).
Of the 147 patients with significant cancer matched to a positive mpMRI lesion, 127 (86.4%) had the highest Gleason grade found at biopsy.
A comparison between the three centres is shown in Table 3. Sensitivity values for the detection of grade group ≥2 cancers at the PI-RADS 4 threshold for centres 1, 2, and 3 were 57.0%, 62.5%, and 72.3%, respectively. Sensitivity improved across all three centres if a PI-RADS 3 threshold was adopted.
A sensitivity analysis to compare the results of each centre was performed (Table 3), and this showed some differences between the centres (analysis of variance test and glorified t test for multiple comparisons).
To quantitate the effect of location matching, comparisons between location matching (same or adjacent sector), exact location matching, and no location matching were made for grade group ≥2 and ≥3 cancer (Table 4). When comparing location matching with that of no matching, there were marked differences in the sensitivity (63.4% vs 77.6%) and NPV (63.4% vs 73.9%) for grade group ≥2 cancer. Thus, exact location matching decreased the sensitivity and NPV by 10–15%.
Table 4.
Quantitating the effect of location matching on the diagnostic accuracy of mpMRI for grade group ≥2 and ≥3 cancers at PI-RADS ≥4 threshold.
| Sensitivity | Specificity | PPV | NPV | |
|---|---|---|---|---|
| No location matching a | ||||
| Grade group ≥2 | 77.6 | 68.7 | 72.9 | 73.9 |
| Grade group ≥3 | 86.0 | 59.1 | 49.8 | 89.9 |
| Location matching (exact location) | ||||
| Grade group ≥2 | 63.4 | 68.7 | 68.7 | 63.4 |
| Grade group ≥3 | 70.6 | 59.1 | 44.9 | 81.0 |
| Location matching (same or adjacent sector) | ||||
| Grade group ≥2 | 74.6 | 68.7 | 72.1 | 71.4 |
| Grade group ≥3 | 82.5 | 59.1 | 48.8 | 87.7 |
mpMRI = multiparametric magnetic resonance imaging; NPV = negative predictive value; PI-RADS = Prostate Imaging Reporting and Data System; PPV = positive predictive value.
No location matching = cancer present anywhere in the gland is considered a “match” for a positive mpMRI lesion.
When a more lenient approach was taken (same or adjacent sector) for grade group ≥2 cancer, a smaller effect on the sensitivity (74.6% vs 77.6%) and NPV (71.4% vs 78.4%) was seen in comparison with no matching.
3.2. Significant cancers not predicted by mpMRI (false negative mpMRI)
Of the 232 men with significant cancer, 36.6% (85/232) were unmatched to a PI-RADS ≥4 lesion in the same sector. Of these, 55 (64.7%) had grade group 2, 19 (22.4%) grade group 3, and 11 (12.9%) grade group ≥4 cancer.
There were a significantly greater proportion of grade group 2 cancers in the unmatched group (64.7%, 55/85) than in the matched group (30.6%, 45/147; p < 0.001). Conversely, there were a greater proportion of high-grade cancers (grade group ≥4) in the matched group (36.1%, 53/147) than in the unmatched group (12.9%, 11/85; p < 0.001).
The median maximum cancer core length of significant cancers matched to a PI-RADS ≥4 lesion was significantly greater than that unmatched to a PI-RADS ≥4 lesion (11 vs 5.9 mm, p < 0.001).
A re-review of 49 men with false negative mpMRI (total n = 85) from centres 1 and 2 was performed by an experienced radiologist, who was blinded to the location of positive biopsy. Even after review, 69.4% (34/49) of these patients with significant cancer remained unmatched to a PI-RADS ≥4 lesion.
4. Discussion
In this multicentre study examining 446 men who had mpMRI of the prostate followed by prostate biopsy within 3 mo, a positive mpMRI (PI-RADS ≥4) lesion predicted clinically significant cancer at biopsy (ISUP grade group ≥2) with moderate sensitivity (63.4%) and NPV (63.4%) in the exact sector when using the location-matching technique. Without matching of location, the sensitivity, PPV, and NPV for significant cancer at biopsy increased (77.6%, 72.9%, and 73.9% respectively). Location matching had no effect on specificity. Hence, we have demonstrated that results from studies that have not performed location matching are likely to overestimate mpMRI sensitivity.
Unlike previous studies, our data could match the location of mpMRI lesions to positive biopsy cores. Other studies investigating mpMRI accuracy [6], [7], [9], [20] have not analysed their data on a per-sector level. For example, a patient with a positive mpMRI lesion in the right base and significant cancer found at biopsy in the left apex would be considered a “true positive” in these studies. When examined on a per-sector basis, this “positive mpMRI” has two errors—both a false positive and a false negative (Fig. 2). Thus, their data are likely contaminated with false negative lesions, affecting the true sensitivity and NPV of mpMRI. Furthermore, the presence of false positive lesions will overestimate the specificity and PPV. In our study, location matching decreased the sensitivity (14.2%), PPV (4.2%), and NPV (10.5%) when compared with the nonmatching method. The specificity is not altered (68.7%) by location matching, as we prioritised the false negative result over the false positive result for patients with both results (Fig. 2). As our false positive rate did not change, our specificity was unaffected by location matching.
Studies have proposed that mpMRI has improved prediction of higher-grade cancers, including primary pattern Gleason 4 disease, which is known to have poorer prognosis [21], [22]. Our data (Table 4) examining grade group ≥3 cancers showed high sensitivity (86%) and NPV (89.9%) at the PI-RADS 4 threshold without location matching. However, when location matching was used, there was a large reduction in accuracy (sensitivity 70.6% and NPV 81%). Thus, the true accuracy of mpMRI for predicting high-grade cancers may be lower than that previously reported.
False negative mpMRI results remain a concern, with 36.6% (85/232) of men with significant cancer having false negative mpMRI. Most missed cancers were of grade group 2 (64.7%, 55/85), which coincides with the results reported by Borofsky et al [23]. Interestingly, of the 49 men with mpMRI available for a re-review by a single blinded experienced radiologist, 69.4% (34/49) remained false negative at the PI-RADS 4 threshold. Multiple factors contribute to false negative mpMRI. The accuracy of mpMRI is improved when reported by “expert” radiologists when compared with general radiologists [24]. This may have been a factor in our study, with up to 14 radiologists involved across multiple centres. The transition zone is known to be difficult to interpret, and lesions may be mistakenly reported as benign prostatic hyperplasia [25]. The quality of mpMRI may also affect reporting accuracy. The use of b values of <1000s/mm2 is still accepted by the European Society of Urogenital Radiology guidelines [26]; however, b values of <1000s/mm2 have poorer diagnostic performance than higher b values [27]. Small tumour foci may lead to poor visibility on mpMRI [28], [29], which may have been a factor in our study, with the median maximum cancer core length of unmatched significant cancers being lower than that of matched significant cancers (5.9 vs 11 mm, p < 0.001). Furthermore, histopathological features of some prostate cancers may affect visibility on mpMRI [30], with a lack of a discrete tumour nodule resulting in poor detection on imaging.
The limitations of our study should be considered. Owing to its retrospective nature, we cannot rule out selection and observation bias. Of note, 297 men did not undergo biopsy after mpMRI, suggesting clinician and patient selection bias to proceed with biopsy based on the mpMRI result. There was marked variability in radiologist experience, reflecting real-world experience, which resulted in interobserver variability in reporting across the three centres.
As we did not utilise radical prostatectomy as our reference standard, there was the possibility of missing tumour foci not detected by biopsy. Location matching is difficult to perform due to the lack of standardisation of both radiology reporting and biopsy needle location. To minimise error, this was performed manually by a single person (D.G.) with guidance from our radiologists. Finally, a small proportion (n = 23, 5.2%) of mpMRI scans utilised older technology (b800 values), which may have led to poorer scan quality.
5. Conclusions
We have quantified the effect of precise location mapping of positive mpMRI lesions to significant prostate cancer found at biopsy. Our results show a 10–15% reduction in sensitivity and NPV with location matching. Results published in the literature that have not been able to match the location are likely to be artificially high, and our adjustment should be taken into consideration. False negative mpMRI remains an issue, highlighting the need for improvement of standards around imaging quality and reporting.
Author contributions: Lih-Ming Wong had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Gavin, Wong, Louie-Johnsun, Krelle.
Acquisition of data: Gavin, Krelle, Wong, Kam, Koschel, Yuminaga, Kim, Aluwihare, Skinner.
Analysis and interpretation of data: Gavin, Krelle, Kam, Sutherland.
Drafting of the manuscript: Gavin, Wong, Sutherland.
Critical revision of the manuscript for important intellectual content: Sutherland, Wong.
Statistical analysis: Gavin, Jenkins, Wong.
Obtaining funding: None.
Administrative, technical, or material support: None.
Supervision: Brennan, Louie-Johnsun, Wong, Sutherland.
Other: None.
Financial disclosures: Lih-Ming Wong certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: None.
Funding/Support and role of the sponsor: None.
CRediT authorship contribution statement
Dominic James Gavin: Investigation, Resources, Writing - original draft, Writing - review & editing, Visualization. Jonathan Kam: Methodology, Investigation, Resources, Writing - original draft. Matthew Krelle: Methodology, Investigation, Resources. Mark Louie-Johnsun: Methodology, Conceptualization, Supervision. Tom Sutherland: Methodology, Writing - review & editing, Supervision. Samantha Koschel: Methodology, Investigation, Resources. Mark Jenkins: Formal analysis, Supervision. Yuigi Yuminaga: Conceptualization, Methodology, Writing - review & editing. Raymond Kim: Investigation, Supervision. Kushlan Aluwihare: Investigation, Supervision. Sarah Skinner: Investigation, Supervision. Janelle Brennan: Conceptualization, Methodology, Supervision. Lih-Ming Wong: Conceptualization, Methodology, Writing - review & editing, Supervision.
Associate Editor: Guillaume Ploussard
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.euros.2020.07.001.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Borkowetz A., Platzek I., Toma M. Comparison of systematic transrectal biopsy to transperineal magnetic resonance imaging/ultrasound-fusion biopsy for the diagnosis of prostate cancer. BJU Int. 2015;116:873–879. doi: 10.1111/bju.13023. [DOI] [PubMed] [Google Scholar]
- 2.Delongchamps N.B., Peyromaure M., Schull A. Prebiopsy magnetic resonance imaging and prostate cancer detection: comparison of random and targeted biopsies. J Urol. 2013;189:493–499. doi: 10.1016/j.juro.2012.08.195. [DOI] [PubMed] [Google Scholar]
- 3.Sonn G.A., Chang E., Natarajan S. Value of targeted prostate biopsy using magnetic resonance-ultrasound fusion in men with prior negative biopsy and elevated prostate-specific antigen. Eur Urol. 2014;65:809–815. doi: 10.1016/j.eururo.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siddiqui M.M., Rais-Bahrami S., Truong H. Magnetic resonance imaging/ultrasound-fusion biopsy significantly upgrades prostate cancer versus systematic 12-core transrectal ultrasound biopsy. Eur Urol. 2013;64:713–719. doi: 10.1016/j.eururo.2013.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Habchi H., Bratan F., Paye A. Value of prostate multiparametric magnetic resonance imaging for predicting biopsy results in first or repeat biopsy. Clin Radiol. 2014;69:e120–e128. doi: 10.1016/j.crad.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 6.Ahmed H.U., El-Shater Bosaily A., Brown L.C. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confirmatory study. Lancet. 2017;389:815–822. doi: 10.1016/S0140-6736(16)32401-1. [DOI] [PubMed] [Google Scholar]
- 7.De Rooij M., Hamoen E.H.J., Fütterer J.J., Barentsz J.O., Rovers M.M. Accuracy of multiparametric MRI for prostate cancer detection: a meta-analysis. Am J Roentgenol. 2014;202:343–351. doi: 10.2214/AJR.13.11046. [DOI] [PubMed] [Google Scholar]
- 8.Abd-Alazeez M., Kirkham A., Ahmed H.U. Performance of multiparametric MRI in men at risk of prostate cancer before the first biopsy: a paired validating cohort study using template prostate mapping biopsies as the reference standard. Prostate Cancer Prostatic Dis. 2014;17:40–46. doi: 10.1038/pcan.2013.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simmons L.A.M., Kanthabalan A., Arya M. The PICTURE study: diagnostic accuracy of multiparametric MRI in men requiring a repeat prostate biopsy. Br J Cancer. 2017;116:1159–1165. doi: 10.1038/bjc.2017.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Margel D., Yap S.A., Lawrentschuk N. Impact of multiparametric endorectal coil prostate magnetic resonance imaging on disease reclassification among active surveillance candidates: a prospective cohort study. J Urol. 2018;187:1247–1252. doi: 10.1016/j.juro.2011.11.112. [DOI] [PubMed] [Google Scholar]
- 11.Barentsz J.O., Richenberg J., Clements R. ESUR prostate MR guidelines 2012. Eur Radiol. 2012;22:746–757. doi: 10.1007/s00330-011-2377-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weinreb J.C., Barentsz J.O., Choyke P.L. PI-RADS Prostate Imaging - Reporting and Data System: 2015, version 2. Eur Urol. 2016;69:16–40. doi: 10.1016/j.eururo.2015.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barzell W.E., Melamed M.R. Appropriate patient selection in the focal treatment of prostate cancer: the role of transperineal 3-dimensional pathologic mapping of the prostate—a 4-year experience. Urology. 2007;70(6 Suppl 1):27–35. doi: 10.1016/j.urology.2007.06.1126. [DOI] [PubMed] [Google Scholar]
- 14.Kuru T.H., Wadhwa K., Chang R.T.M. Definitions of terms, Processes and a minimum dataset for transperineal prostate biopsies: a standardization approach of the Ginsburg Study Group for enhanced prostate diagnostics. BJU Int. 2013;112:568–577. doi: 10.1111/bju.12132. [DOI] [PubMed] [Google Scholar]
- 15.Presti J.C.J., O’Dowd G.J., Miller M.C., Mattu R., Veltri R.W. Extended peripheral zone biopsy schemes increase cancer detection rates and minimize variance in prostate specific antigen and age related cancer rates: results of a community multi-practice study. J Urol. 2003;169:125–129. doi: 10.1016/S0022-5347(05)64051-7. [DOI] [PubMed] [Google Scholar]
- 16.Epstein J.I., Allsbrook W.C.J., Amin M.B., Egevad L.L. The 2005 International Society of Urological Pathology (ISUP) consensus conference on Gleason grading of prostatic carcinoma. Am J Surg Pathol. 2005;29:1228–1242. doi: 10.1097/01.pas.0000173646.99337.b1. [DOI] [PubMed] [Google Scholar]
- 17.Gordetsky J., Epstein J. Grading of prostatic adenocarcinoma: current state and prognostic implications. Diagn Pathol. 2016;11:25. doi: 10.1186/s13000-016-0478-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gayet M., Van Der Aa A., Beerlage H.P., Schrier B.P., Mulders P.F.A., Wijkstra H. The value of magnetic resonance imaging and ultrasonography (MRI/US)-fusion biopsy platforms in prostate cancer detection: a systematic review. BJU Int. 2016;117:392–400. doi: 10.1111/bju.13247. [DOI] [PubMed] [Google Scholar]
- 19.Cool D.W., Zhang X., Romagnoli C., Izawa J.I., Romano W.M., Fenster A. Evaluation of MRI-TRUS fusion versus cognitive registration accuracy for MRI-targeted, TRUS-guided prostate biopsy. Am J Roentgenol. 2015;204:83–91. doi: 10.2214/AJR.14.12681. [DOI] [PubMed] [Google Scholar]
- 20.Futterer J.J., Briganti A., De Visschere P. Can clinically significant prostate cancer be detected with multiparametric magnetic resonance imaging? A systematic review of the literature. Eur Urol. 2015;68:1045–1053. doi: 10.1016/j.eururo.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 21.Lau W.K., Blute M.L., Bostwick D.G., Weaver A.L., Sebo T.J., Zincke H. Prognostic factors for survival of patients with pathological Gleason score 7 prostate cancer: differences in outcome between primary Gleason grades 3 and 4. J Urol. 2001;166:1692–1697. [PubMed] [Google Scholar]
- 22.Rasiah K.K., Stricker P.D., Haynes A.M. Prognostic significance of Gleason pattern in patients with Gleason score 7 prostate carcinoma. Cancer. 2003;98:2560–2565. doi: 10.1002/cncr.11850. [DOI] [PubMed] [Google Scholar]
- 23.Borofsky S., George A.K., Gaur S. What are we missing? False-negative cancers at multiparametric MR imaging of the prostate. Radiology. 2018;286:186–195. doi: 10.1148/radiol.2017152877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Branger N., Maubon T., Traumann M. Is negative multiparametric magnetic resonance imaging really able to exclude significant prostate cancer? The real-life experience. BJU Int. 2017;119:449–455. doi: 10.1111/bju.13657. [DOI] [PubMed] [Google Scholar]
- 25.Oto A., Kayhan A., Jiang Y. Prostate cancer : differentiation of central gland cancer from benign prostatic hyperplasia by using diffusion-weighted and dynamic contrast-enhanced MR imaging. Radiology. 2010;257:715–723. doi: 10.1148/radiol.10100021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steiger P., Thoeny H.C. Prostate MRI based on PI-RADS version 2: how we review and report. Cancer Imaging. 2016;16:9. doi: 10.1186/s40644-016-0068-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang K., Shen Y., Zhang X. Predicting prostate biopsy outcomes: a preliminary investigation on screening with ultrahigh B-value diffusion-weighted imaging as an innovative diagnostic biomarker. PLoS One. 2016;11:1–14. doi: 10.1371/journal.pone.0151176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cornud F., Khoury G., Bouazza N. Tumor target volume for focal therapy of prostate cancer—does multiparametric magnetic resonance imaging allow for a reliable estimation? J Urol. 2014;191:1272–1279. doi: 10.1016/j.juro.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 29.Turkbey B., Pinto P.A., Mani H. Prostate cancer: value of multiparametric MR imaging at 3 T for detection—histopathologic correlation. Radiology. 2010;255:89–99. doi: 10.1148/radiol.09090475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenkrantz A.B., Mendrinos S., Babb J.S., Taneja S.S. Prostate cancer foci detected on multiparametric magnetic resonance imaging are histologically distinct from those not detected. J Urol. 2012;187:2032–2038. doi: 10.1016/j.juro.2012.01.074. [DOI] [PubMed] [Google Scholar]
- 31.Radtke J., Schwab C., Wolf M., Freitag M., Alt C., Kesch C. Multiparametric Magnetic Resonance Imaging (MRI) and MRI-Transrectal Ultrasound Fusion Biopsy for Index Tumor Detection: Correlation with Radical Prostatectomy Specimen. Eur Urol. 2016;70(5):846–853. doi: 10.1016/j.eururo.2015.12.052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.