Abstract
Background
Serine-arginine protein kinase 1 (SRPK1) has been implicated in prostate cancer (PCa) progression. However, its prognostic value and association with ERG and PTEN expression, two of the most common genetic alterations, have not been explored fully.
Objective
We assessed the prognostic value of SRPK1 in association with ERG and PTEN in a cohort of patients managed nonsurgically by androgen deprivation therapy (ADT) for advanced disease.
Design, setting, and participants
The study cohort consisted of men diagnosed with PCa by transurethral resection of the prostate (TURP; n = 480). The patients were divided into three main groups: incidental (patients with Gleason score [GS] ≤7 with no prior ADT), advanced (patients with GS ≥8 with no prior ADT), and castrate-resistant PCa (patients with prior ADT).
Outcome measurements and statistical analysis
A total of 480 TURP samples were assessed by immunohistochemistry for SRPK1, ERG, and PTEN, and results were correlated with Gleason grade group (GG), overall survival (OS), and PCa-specific mortality (PCSM).
Results and limitations
High SRPK1 expression was noted in 105/455 (23%) available patient cores. Expression of SRPK1 was associated with Gleason grade grouping (p < 0.0001) with high expression detected in 22/74 (33%) with GG 5. High SRPK1 was not associated with ERG positivity (p = 0.18) but was significantly associated with PTEN intensity (p = 0.001). High SRPK1 was associated with OS (hazard ratio [HR] 1.99; confidence interval [CI]: 1.57–2.54, p < 0.0001) and PCSM (HR 1.64; CI: 1.19–2.26, p < 0.002). Adjusting for Gleason score, patients with high SRPK1 and negative PTEN had the worst clinical outcome for both OS and PCSM compared with other patients (p < 0.0001, HR: 3.02; CI: 1.87–4.88 and HR: 6.40, CI: 3.19–12.85, respectively).
Conclusions
High SRPK1 is associated with worse OS and PCSM. Moreover, patients with high SRPK1 expression and loss of PTEN had the worst clinical outcome for OS and cancer-specific mortality. Combined status of SRPK1 and PTEN may provide added value in stratifying patients into various prognostic groups.
Patient summary
The expression of serine-arginine protein kinase 1 (SRPK1) combined with PTEN has a significant prognostic role in prostate cancer patients. Patients with high SRPK1 expression and negative PTEN had the worst clinical outcome for overall survival and cancer-specific mortality.
Keywords: SRPK1, PTEN, ERG, Protein expression, Immunohistochemistry, Gleason score, Androgen deprivation therapy, Cancer-specific mortality, Overall survival
Take Home Message
The expression of serine-arginine protein kinase 1 (SRPK1) is associated with prostate cancer progression, overall survival, and cause-specific mortality. Assessing combined status of SRPK1 and PTEN is of added prognostic significance compared with either marker alone. Patients with high SRPK expression/PTEN loss show the highest prognostic value among all combinations.
1. Introduction
Prostate cancer (PCa) remains one of the most common cancers worldwide. The prevalence of PCa is highest in North America, and prediction of PCa progression remains a major clinical challenge for physicians. Characterization of clinical biomarkers is considered one of the most critical approaches to the diagnosis and prediction of PCa progression. However, even with today's advancement in molecular pathology, none of the biomarkers has been implemented into routine clinical practice for men with PCa. Several studies have been investigating and characterizing molecular signatures in search of biomarkers or signatures that could easily be implemented in routine clinical practice to stratify patients into different risk groups [1]. The expression of ERG and PTEN is among the most genomic alterations to occur in PCa and may be of potential significance in molecularly subtyping PCa.
Serine-arginine protein kinase 1 (SRPK1) is a protein kinase encoded by a gene located on chromosome 6 (6p21.31) in humans [2]. It was found that SRPK1 specifically phosphorylates proteins containing serine/arginine-rich (SR) domains. SR proteins are involved in regulating several RNA-processing pathways, including RNA stability, alternative splicing, and translation [3]. High SRPK1 expression is associated with numerous malignancies including breast, colonic, and pancreatic carcinomas [4]. Recent intravital screen identified SRPK1 as a driver of human cancer cell metastatic behavior. An immunohistochemistry (IHC) analysis showed SRPK1 overexpression in PCa-invasive zones, and SRPK1 expression knockdown decreased PCa cell migration in vitro [5]. Previous studies show an oncogenic role and overexpression of SRPK1 in breast, gastric, prostate, and lung cancers, in addition to gliomas and hepatocellular carcinoma [6], [7], [8]. Recently, a study, using Oncomine analysis and The Cancer Genome Atlas (TCGA) databases, identified that SRPK1 was among four highly expressed genes in lung adenocarcinoma and was associated with cancer progression [9]. Furthermore, antiangiogenic therapy, via SRPK1 inhibition, was proved to be a successful potential therapeutic strategy in PCa [10]. In the present study, we evaluated the prognostic significance of SRPK1 and its association with other molecular events (ERG and PTEN) in a large nonsurgically treated cohort with incidental, advanced, and castrate-resistant PCa (CRPC).
2. Patients and methods
2.1. Study population and tissue microarray construction
The study cohort consisted of a group of men diagnosed with PCa by transurethral resection of the prostate (TURP; n = 480). Patients within this cohort were either not treated actively or treated by a luteinizing hormone-releasing hormone (LHRH) agonist. Treatments were implemented after (advanced group) or prior to TURP sample assessment in patients who had a previous diagnosis of PCa to relieve symptomatic obstruction of locally advanced disease while on LHRH (CRPC group). The cohort samples were collected between 1999 and 2014, with a median follow-up of 38.14 (range: 0.30–147.55) mo. The study cohort consisted of three main groups: incidental (those with Gleason score [GS] ≤7 with no prior androgen deprivation therapy [ADT]), advanced (patients with GS ≥8 with no prior ADT), and CRPC (patients with prior ADT). Table 1 demonstrates patient demographics of the study cohort.
Table 1.
Study population demographics
| Group | N (%) | OS total, N (events) | Median follow-up | Median overall survival | p value | CSS total N (events) | Median cancer–specific survival | p value |
|---|---|---|---|---|---|---|---|---|
| Incidental | 158 (33) | 146 (64) | 59.01 (4.70–137.03) | 92.78 (67.06–118.50) | <0.0001 | 146 (4) | Not reached | <0.0001 |
| Advanced | 180 (37) | 172 (132) | 35.93 (0.30–147.55) | 38.60 (30.86–46.35) | 172 (78) | 63.97 (50.65–77.29) | ||
| Castrate resistant | 142 (30) | 134 (113) | 20.85 (2.76–109.24) | 22.24 (13.73–30.75) | 134 (78) | 33.87 (21.84–45.91) | ||
| Overall | 38.77 (0.30–154.97) | 42.12 (37.29–46.95) | Not reached |
CSS = cancer-specific survival; OS = overall survival.
Clinical follow-up information was recorded and approved by the University of Calgary, Cumming School of Medicine Ethics Review Board from the Alberta Tumour Registry for dates of therapy, overall survival (OS), and cause-specific survival. The cohort's samples were assembled on to two tissue microarrays (TMAs) with an average two cores per patient using a manual tissue arrayer (Beecher Instruments, Silver Spring, MD, USA).
2.2. SRPK1, ERG, and PTEN expression by IHC
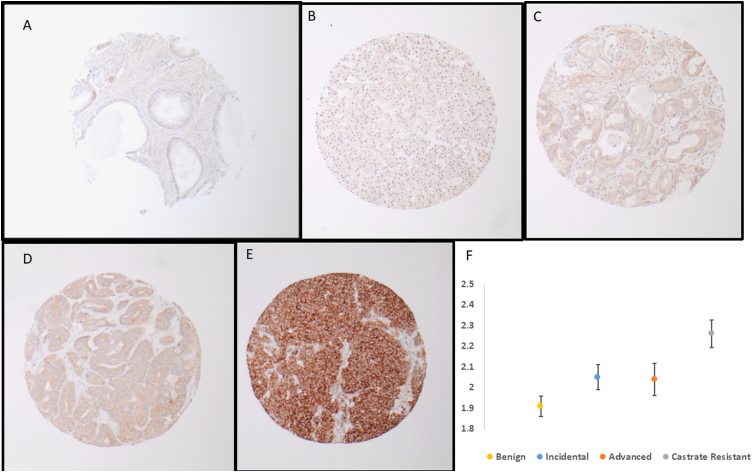
SRPK1 IHC was performed on a Dako Omnis autostainer. Briefly, 4 μ formalin-fixed paraffin-embedded sections were pretreated with citrate pH 6.0 epitope retrieval buffer. Rabbit polyclonal SRPK1 antibody was diluted to 1/10k using Dako antibody diluent and incubated for 20 min followed by 20 min secondary incubation. The FLEX DAB+ Substrate Chromogen system was used as the postincubation detection reagent. SRPK1 and PTEN IHC expression was assessed using a four-tiered system (0, negative; 1, weak; 2, moderate; and 3, high expression). ERG and PTEN IHC was assessed and stained as described previously by Huang et al [11] and Bismar et al [12]. Figure 1A–D show examples of SRPK1 intensity in various Gleason grade groups (GGs).
Fig. 1.
Examples of SRPK1 intensity in benign prostate tissue and various prostate cancer grade groups: (A) benign prostate tissue showing negative SRPK intensity, (B) Gleason grade group 2 showing weak intensity, (C) Gleason grade group 2 showing moderate intensity, (D) Gleason grade group 3 showing moderate intensity, (E) Gleason grade group 5 showing high intensity, and (F) error bars for mean intensity values of SRPK across various stages of prostate cancer progression. Incidental group: PCa detected incidentally on TURP with low volume and Gleason group 1–2, advanced group: PCa detected on TURP with GG >2 and no prior therapy, and CRPC group: PCa detected on TURP in patients with previous diagnosis of PCa and ADT therapy. ADT = androgen deprivation therapy; CRPC = castrate-resistant prostate cancer; GG = grade group; PCa = prostate cancer; SRPK1 = serine-arginine protein kinase 1; TURP = transurethral resection of the prostate.
2.3. Pathological analysis
Histological diagnoses of individual TMA cores was confirmed by the study pathologists (H.A. and T.A.B.) on the initial slides. Gleason scoring was assessed according to the 2014 World Health Organization/International Society of Urological Pathology GGs. In each patient, the two predominant patterns of PCa were sampled and included on the TMAs for analysis. Figure 1 shows several examples of variable intensity (protein expression) for SRPK1.
2.4. Statistical analysis
SPSS version 25 was used to conduct all statistical analysis (IBM SPSS Statistics for Windows, version 25.0, released 2017; IBM Corp., Armonk, NY, USA). Frequency and proportions were reported for categorical data. Mean and standard deviations were reported for normally distributed continuous data, and median and range were reported for non-normally continuous data. Chi-square test was used to compare two categorical variables and Fisher's exact test was used where the cell frequencies were <5. OS was defined as the time from diagnosis to death; patients who were alive at the end of the study period were censored. Prostate cancer–specific mortality (PCSM) was defined as death due to PCa; patients who died due to any other reason or who were alive at the end of the study period were censored. Relapse-free survival (RFS) was calculated from the date of treatment to the date of relapse; patients who did not relapse were considered censored. OS, PCSM, and RFS were analyzed using the Kaplan-Meier method. Median time and the 95% confidence interval (CI) were reported. Log-rank tests were used to compare two or more survival curves. Cox's proportional hazard models were used to determine the factors associated with OS, PCSM, and RFS; hazard ratios (HR) and the corresponding 95% CIs were reported. Adjusted Cox's models were fitted as well. A p value of <0.05 was used for statistical significance, and two-sided tests were utilized.
3. Results
3.1. SRPK1 expression and association with ERG and PTEN
SRPK1 intensity was significantly higher in PCa diagnosis than in benign prostate tissue (1.24 ± 0.446 vs 2.07 ± 0.621, p < 0.0001). SRPK1 intensity was also significantly higher in CRPC and advanced PCa than in incidental disease (2.26 ± 0.611 and 2.04 ± 0.682 vs 1.91 ± 0.558, respectively, p < 0.0001). High SRPK1 was not associated with ERG positivity (p = 0.18), but was associated with intensity and detected in 51/156 (46%) of PTEN loss cases (p < 0.001). Furthermore, SRPK1 was associated with Gleason grade grouping (p < 0.0001), and high SRPK1 intensity was noted in 74/200 (37%) of patients in GG 5 compared with 29/231 (12.5%) of patients in GG < 4 (p < 0.0001; Table 2).
Table 2.
Gleason score, and PTEN and ERG status stratified by SRPK1 in the TURP cohort (n = 480)a
| Variables | SRPK1 (0, 1, 2) | SRPK1 (3) | p value |
|---|---|---|---|
| Gleason score | |||
| ≤6 | 134 (38) | 15 (14) | <0.0001 |
| 3 + 4 | 25 (7) | 7 (7) | |
| 4 + 3 | 43 (12) | 7 (7) | |
| 8 | 23 (7) | 5 (5) | |
| 9–10 | 126 (36) | 74 (69) | |
| PTEN intensity | |||
| Negative | 105 (29) | 51 (46) | 0.001 |
| Positive | 260 (71) | 61(55) | |
| ERG dual | |||
| Negative | 271 (74) | 76 (68) | 0.184 |
| Positive | 94 (26) | 36 (112) | |
SRPK1 = serine-arginine protein kinase 1; TURP = transurethral resection of the prostate.
SRPK1 scores: 0, negative; 1, weak; 2, moderate; and 3, high.
3.2. Association of SRPK1 with patients’ clinical outcome
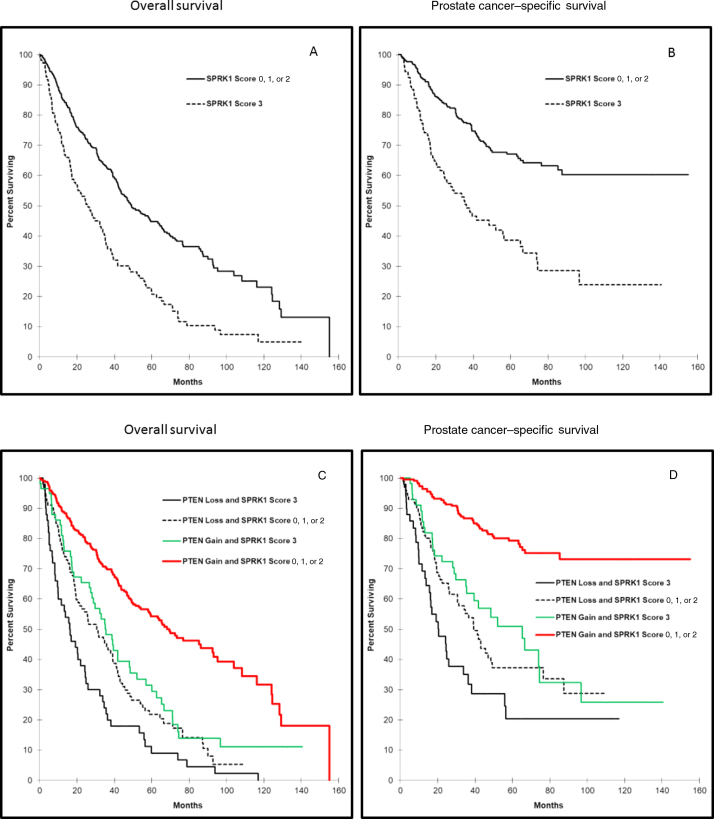
High SRPK1 expression was associated with OS and cancer-specific mortality (HR: 1.99; CI: 1.57–2.54, p < 0.0001 and HR: 1.64; CI: 1.19–2.26, p = 0.002, respectively; Table 3, and Fig. 2A and B). Since SRPK1 and PTEN were significantly associated with each other, and as both are known to be significantly associated with disease progression and patient's prognosis, we opted for investigating whether the combination of SRPK1 and PTEN status will provide an added prognostic value. In this cohort, patient's tumors with high SRPK1 and PTEN negative staining were associated with significantly lower OS and higher cancer-specific mortality, compared with patients with other combinations of SRPK1 and PTEN (HR: 4.07, CI: 2.91–5.68 p < 0.0001 and HR: 3.02, CI: 1.91–4.75, p < 0.0001, respectively). The HR for the prognostic value of the combined SRPK1/PTEN status was higher than that for PTEN or SRPK alone (Table 4). Furthermore, the combined SRPK1/PTEN signature remained significant for both OS and PCSM after adjusting for Gleason score (HR: 2.23, CI: 1.55–3.20, p < 0.0001 and HR = 2.77, CI: 0.85–2.24, p = 0.190, respectively). Patients whose tumors showed high PTEN expression and SRPK1 intensity of <high showed the best prognosis for OS and PCSM. Patients with other PTEN with SRPK1 combinations showed intermediate prognosis (Table 4 and Fig. 2C and D).
Table 3.
Hazard ratio of Gleason score, PTEN and SRPK1 intensity for overall survival and cause-specific mortality in the TURP cohorta
| Variables | Hazard ratio | 95% Confidence interval | p value |
|---|---|---|---|
| Overall survival | |||
| PTEN Positive (scores 1, 2, 3) | |||
| Negative (score 0) | 2.47 | 1.97–3.10 | <0.0001 |
| Gleason score (score ≤6) | |||
| 4 + 3 | 3.21 | 2.21–4.66 | <0.0001 |
| 3 + 4 | 1.81 | 1.24–2.64 | 0.002 |
| 8 | 5.07 | 3.44–7.47 | <0.0001 |
| 9–10 | 5.62 | 4.39–7.47 | <0.0001 |
| SRPK1 (score 0, 1, 2) | |||
| Score 3 | 1.99 | 1.57–2.54 | <0.0001 |
| Prostate cancer–specific survival | |||
| PTEN positive (scores 1, 2, 3) | |||
| Negative (score 0) | 2.01 | 1.47–2.74 | <0.0001 |
| Gleason score (score ≤6) | |||
| 4 + 3 | 14.36 | 4.75–43.40 | <0.0001 |
| 3 + 4 | 8.04 | 2.59–24.96 | <0.0001 |
| 8 | 28.67 | 9.77–84.11 | <0.0001 |
| 9–10 | 39.79 | 14.65–108.09 | <0.0001 |
| SRPK1 (scores 0, 1, 2) | |||
| Score 3 | 1.64 | 1.19–2.26 | 0.002 |
SRPK1 = serine-arginine protein kinase 1; TURP = transurethral resection of the prostate.
SRPK1 scores: 0, negative; 1, weak; 2, moderate; and 3, high.
Fig. 2.
Kaplan-Meir curves for SRPK1 (A) overall survival and (B) prostate cancer–specific mortality (SRPK1 intensity: 0, negative; 1, weak; 2, moderate; and 3, high). Kaplan-Meir curve for combined SRPK1 and PTEN (C) overall survival (D) prostate cancer–specific mortality (SRPK1 and PTEN intensity: 0, negative; 1, weak; 2, moderate; and 3, high). SRPK1 = serine-arginine protein kinase 1.
Table 4.
Multivariate analysis and HR of the different biomarker combinations for overall survival and cancer-specific mortality in the TURP cohorta
| Variables | Overall survival |
Prostate cancer–specific survival |
||
|---|---|---|---|---|
| HR (95% CI) | p value | HR (95% CI) | p value | |
| PTEN positive (scores 1, 2, 3) | ||||
| Negative (score 0) | 2.47 (1.97–3.10) | <0.0001 | 2.01 (1.47–2.74) | <0.0001 |
| SRPK1 (scores of 1, 2) | ||||
| Score 3 | 1.99 (1.57–2.54) | <0.0001 | 1.64 (1.19–2.26) | 0.002 |
| Combination of PTEN and SRPK1 (PTEN positive and SRPK1 scores 0, 1, 2) | ||||
| PTEN negative and SRPK1 score 3 | 4.07 (2.91–5.68) | <0.0001 | 3.02 (1.91–4.75) | <0.0001 |
| PTEN negative and SRPK1 scores 0, 1, 2 | 2.49 (1.89–3.30) | <0.0001 | 2.32 (1.56–3.46) | <0.0001 |
| PTEN positive and SRPK1 score 3 | 2.01 (1.44–2.80) | <0.0001 | 1.94 (1.22–3.10) | 0.005 |
| Combination of PTEN and SRPK1 (PTEN positive and SRPK1 score 0, 1, 2)b | ||||
| PTEN negative and SRPK1 score 3 | 2.23 (1.55–3.20) | <0.0001 | 1.38 (0.85–2.24) | 0.190 |
| PTEN negative and SRPK1 scores 0, 1, 2 | 1.53 (1.13–2.09) | 0.007 | 1.22 (0.80–1.87) | 0.355 |
| PTEN positive and SRPK1 score 3 | 1.31 (0.92–1.87) | 0.136 | 0.96 (0.58–1.57) | 0.859 |
CI = confidence interval; HR = hazard ratio; SRPK1 = serine-arginine protein kinase 1; TURP = transurethral resection of the prostate.
SRPK1 and PTEN scores: 0, negative; 1, weak; 2, moderate; and 3 high intensity.
Adjusted for Gleason score.
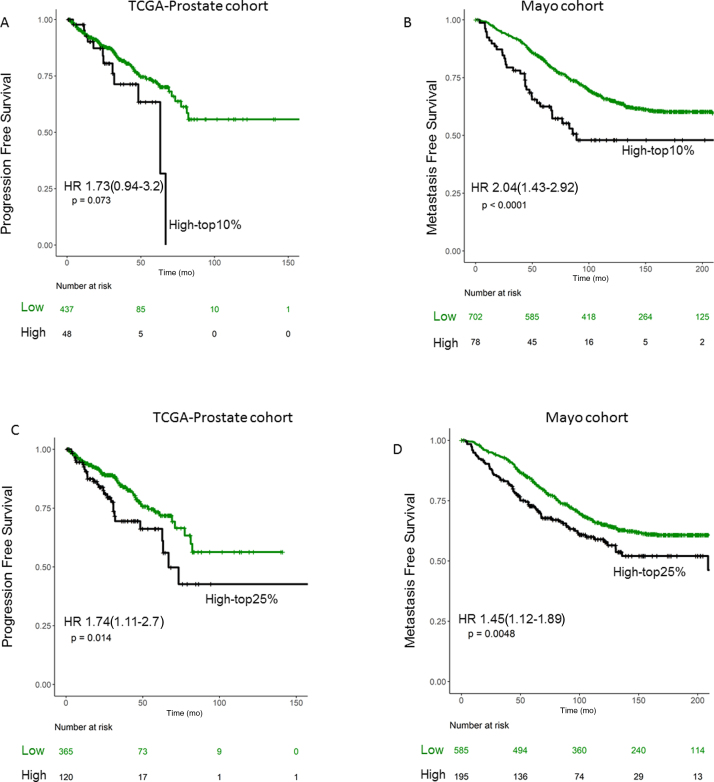
To externally validate the association of SRPK1 expression with progression- and metastasis-free survival, we used public cohorts of primary tumors from TCGA-prostate [13] and Mayo cohorts [14], [15]. TCGA data were obtained from cbioportal.org, and Mayo data were obtained from GEO (GSE62116 GSE46691). Mayo cohort is a high-risk cohort of radical prostatectomy (RP) samples (SM+ or pT3b+) from patients with no treatment prior to RP. To stratify patients based on their SRPK1 expression, we defined high SRPK1 patients as those with top 10% and 25% of SRPK1 expression, as the percentage of high SRPK1 in our TURP cohort was 22%. Using both definitions, patients with high SRPK1 were associated with poor progression- and metastasis-free survival (p < 0.05; Fig. 3A–D).
Fig. 3.
Kaplan-Meir curves for SRPK1 expression in two independent cohorts from TCGA and Mayo Clinic for progression- and metastasis-free survival for top (A and B) 10% and (C and D) 25% tumor expression. HR = hazard ratio; SRPK1 = serine-arginine protein kinase 1; TCGA = The Cancer Genome Atlas.
4. Discussion
Evaluation of molecular profiles allows for a more precise risk classification of PCa [16]. Therefore, considerable research was conducted to improve clinical insights and assist in the development of precision therapies for advanced PCa [16], [17]. Although it is well known that PCa has significant clinical heterogeneity, recent studies showed a significant correlation between ERG rearrangements and PTEN deletions in localized PCa, as reviewed by Bismar et al [12] Arora and Barbieri [17], and Cooperberg et al [18]. ERG and PTEN genomic aberrations have been shown to be prevalent (up to 40% and 50%, respectively) in PCa, and both aberrations represent a distinct molecular subtype of PCa [19], [20]. For example, disrupting PI3K/AKT signaling pathway, downstream of PTEN deletion, has been shown in almost all advanced metastatic PCa cases.
Currently, preclinical studies, using either SRPK1 inhibitors or gene silencing, have investigated the effect of SRPK1 downregulation on PCa growth and metastasis. Targeting SRPK1 disrupted diverse oncogenic pathways such as proliferation [10], metastasis [21], angiogenesis [22], and apoptosis [23]. These accumulating data strongly support the notion that SRPK1 could be a potential therapeutic target for PCa. To this end, it is becoming increasingly evident that a more refined molecular subtyping of PCa is required.
SRPK1 has been reported to be upregulated in various epithelial cancers, including lung [24], [25], breast [25], colorectal [26], stomach [27], liver [28], and esophageal [27] cancers. Along this line, recent studies have demonstrated that PCa has significantly higher SRPK1 expression and it is correlated with cancer stage [10], [29]. In this immunochemistry study with a larger patient cohort, we document that SRPK1 intensity is significantly higher in localized PCa than in benign tissue, and furthermore, its expression increases with advanced PCa Gleason GGs and in castrate-resistant disease. All these data support the crucial role of SRPK1 in the pathogenesis of PCa disease.
It has been reported that ERG expression and PTEN loss are major players in PCa, and are associated with disease progression and patient's prognosis. A major benefit of our study is that we further carried out a detailed investigation about the correlation of SRPK1 overexpression with ERG expression and PTEN loss. While we did not confirm any significant association between expression of SRPK1 and ERG, we found a significant added prognostic value combining SRPK1 and PTEN status.
In the current study, assessment of the combination of SRPK1/PTEN status showed a higher prognostic value than either one alone. Patients whose tumor showed both high SRPK1 expression and loss of PTEN expression were at the highest risk for disease progression associated with both lower OS and higher cancer-specific mortality versus patients with low SRPK1 expression and intact PTEN (as documented with high PTEN intensity). The later data support potentially incorporating the combined status of SRPK1 and PTEN to assist in better stratifying PCa patients and identify those at higher risk for lethal outcome.
Some of the study limitations stem from the population investigated as a nonsurgical cohort and also the heterogeneity spanning incidental to advanced and CRPC patients in addition to potential biomarker heterogeneity. However, our cohort represents real samples collected during practice in Canada, with PCa being detected in such samples incidentally and in advanced cases as well as CRPC patients. Additionally, we confirmed the prognostic value of SRPK1 expression in two surgical cohorts (TCGA and Mayo cohorts) of RP, which validated our finding in surgical populations.
5. Conclusions
In conclusion, our study confirms a significant prognostic role of SRPK1 in PCa patients, and this association is further substantiated by combining PTEN expression status. This adds to the previous reported data on the significance of SRPK1 in cancer in general and PCa in particular. Further studies confirming the prognostic value of SRPK1 in surgical cohorts by IHC and investigating novel methods to block SRPK1 expression may prove fruitful in combating PCa progression. The data may also provide a basis for using SRPK1 and PTEN to improve PCa prognostication and its personalized therapeutic modalities.
Author contributions: Tarek A. Bismar had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Bismar.
Acquisition of data: Assem.
Analysis and interpretation of data: Assem, Bismar.
Drafting of the manuscript: Abou-Ouf, Bismar.
Critical revision of the manuscript for important intellectual content: Lewis, Stoletov, Palanisamy, Karnes.
Statistical analysis: Ghosh.
Obtaining funding: Bismar.
Administrative, technical, or material support: Bismar.
Supervision: Bismar.
Other: None.
Financial disclosures: Tarek A. Bismar certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: None.
Funding/Support and role of the sponsor: This study was supported in part by the Prostate Cancer Foundation Young Investigator Award (Tarek A. Bismar). This work is also supported by Prostate Cancer Canada Movember TAG 2018-2060 and Funds from Ride for Dads (Tarek A. Bismar).
Acknowledgments: The authors would like to thank Mrs. Ruby Reyes for technical support in this manuscript and GenomeDX Bioscience for data access.
Associate Editor: Guillaume Ploussard
References
- 1.Matsui S., Simon R., Qu P., Shaughnessy J.D., Jr., Barlogie B., Crowley J. Developing and validating continuous genomic signatures in randomized clinical trials for predictive medicine. Clin Cancer Res. 2012;18:6065–6073. doi: 10.1158/1078-0432.CCR-12-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin J.C., Lin C.Y., Tarn W.Y., Li F.Y. Elevated SRPK1 lessens apoptosis in breast cancer cells through RBM4-regulated splicing events. RNA. 2014;20:1621–1631. doi: 10.1261/rna.045583.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Long J.C., Caceres J.F. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 4.Hayes G.M., Carrigan P.E., Miller L.J. Serine-arginine protein kinase 1 overexpression is associated with tumorigenic imbalance in mitogen-activated protein kinase pathways in breast, colonic, and pancreatic carcinomas. Cancer Res. 2007;67:2072–2080. doi: 10.1158/0008-5472.CAN-06-2969. [DOI] [PubMed] [Google Scholar]
- 5.Stoletov K., Willetts L., Paproski R.J. Quantitative in vivo whole genome motility screen reveals novel therapeutic targets to block cancer metastasis. Nat Commun. 2018;9:2343. doi: 10.1038/s41467-018-04743-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu X., Wei Y., Wang S., Luo M., Zeng H. Serine-arginine protein kinase 1 (SRPK1) is elevated in gastric cancer and plays oncogenic functions. Oncotarget. 2017;8:61944–61957. doi: 10.18632/oncotarget.18734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bullock N., Oltean S. The many faces of SRPK1. J Pathol. 2017;241:437–440. doi: 10.1002/path.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou B., Li Y., Deng Q. SRPK1 contributes to malignancy of hepatocellular carcinoma through a possible mechanism involving PI3K/Akt. Mol Cell Biochem. 2013;379:191–199. doi: 10.1007/s11010-013-1641-7. [DOI] [PubMed] [Google Scholar]
- 9.Liu W., Ouyang S., Zhou Z. Identification of genes associated with cancer progression and prognosis in lung adenocarcinoma: analyses based on microarray from Oncomine and The Cancer Genome Atlas databases. Mol Genet Genomic Med. 2019;7:e00528. doi: 10.1002/mgg3.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mavrou A., Brakspear K., Hamdollah-Zadeh M. Serine-arginine protein kinase 1 (SRPK1) inhibition as a potential novel targeted therapeutic strategy in prostate cancer. Oncogene. 2015;34:4311–4319. doi: 10.1038/onc.2014.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang K.C., Begin L.R., Palanisamy N., Donnelly B., Bismar T.A. SPINK1 expression in relation to PTEN and ERG in matched primary and lymph node metastatic prostate cancer: implications for biomarker development. Urol Oncol. 2016;34 doi: 10.1016/j.urolonc.2015.11.015. 235 e1-10. [DOI] [PubMed] [Google Scholar]
- 12.Bismar T.A., Yoshimoto M., Duan Q., Liu S., Sircar K., Squire J.A. Interactions and relationships of PTEN, ERG, SPINK1 and AR in castration-resistant prostate cancer. Histopathology. 2012;60:645–652. doi: 10.1111/j.1365-2559.2011.04116.x. [DOI] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Research N The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erho N., Crisan A., Vergara I.A. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One. 2013;8:e66855. doi: 10.1371/journal.pone.0066855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karnes R.J., Bergstralh E.J., Davicioni E. Validation of a genomic classifier that predicts metastasis following radical prostatectomy in an at risk patient population. J Urol. 2013;190:2047–2053. doi: 10.1016/j.juro.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaffenberger S.D., Barbieri C.E. Molecular subtyping of prostate cancer. Curr Opin Urol. 2016;26:213–218. doi: 10.1097/MOU.0000000000000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arora K., Barbieri C.E. Molecular subtypes of prostate cancer. Curr Oncol Rep. 2018;20:58. doi: 10.1007/s11912-018-0707-9. [DOI] [PubMed] [Google Scholar]
- 18.Cooperberg M.R., Broering J.M., Carroll P.R. Risk assessment for prostate cancer metastasis and mortality at the time of diagnosis. J Natl Cancer Inst. 2009;101:878–887. doi: 10.1093/jnci/djp122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reid A.H., Attard G., Ambroisine L. Molecular characterisation of ERG, ETV1 and PTEN gene loci identifies patients at low and high risk of death from prostate cancer. Br J Cancer. 2010;102:678–684. doi: 10.1038/sj.bjc.6605554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al Bashir S., Alshalalfa M., Hegazy S.A., Dolph M., Donnelly B., Bismar T.A. Cysteine-rich secretory protein 3 (CRISP3), ERG and PTEN define a molecular subtype of prostate cancer with implication to patients’ prognosis. J Hematol Oncol. 2014;7:21. doi: 10.1186/1756-8722-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Roosmalen W., Le Devedec S.E., Golani O. Tumor cell migration screen identifies SRPK1 as breast cancer metastasis determinant. J Clin Invest. 2015;125:1648–1664. doi: 10.1172/JCI74440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amin E.M., Oltean S., Hua J. WT1 mutants reveal SRPK1 to be a downstream angiogenesis target by altering VEGF splicing. Cancer Cell. 2011;20:768–780. doi: 10.1016/j.ccr.2011.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H., Ge W., Jiang W., Li D., Ju X. SRPK1siRNA suppresses K562 cell growth and induces apoptosis via the PARPcaspase3 pathway. Mol Med Rep. 2018;17:2070–2076. doi: 10.3892/mmr.2017.8032. [DOI] [PubMed] [Google Scholar]
- 24.Liu H., Hu X., Zhu Y., Jiang G., Chen S. Up-regulation of SRPK1 in non-small cell lung cancer promotes the growth and migration of cancer cells. Tumour Biol. 2016;37:7287–7293. doi: 10.1007/s13277-015-4510-z. [DOI] [PubMed] [Google Scholar]
- 25.Li X.H., Song J.W., Liu J.L. Serine-arginine protein kinase 1 is associated with breast cancer progression and poor patient survival. Med Oncol. 2014;31:83. doi: 10.1007/s12032-014-0083-8. [DOI] [PubMed] [Google Scholar]
- 26.Wang P., Zhou Z., Hu A. Both decreased and increased SRPK1 levels promote cancer by interfering with PHLPP-mediated dephosphorylation of Akt. Mol Cell. 2014;54:378–391. doi: 10.1016/j.molcel.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H., Wang C., Tian W., Yao Y. The crucial role of SRPK1 in IGF-1-induced EMT of human gastric cancer. Oncotarget. 2017;8:72157–72166. doi: 10.18632/oncotarget.20048. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Xu Q., Liu X., Liu Z. MicroRNA-1296 inhibits metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by targeting SRPK1-mediated PI3K/AKT pathway. Mol Cancer. 2017;16:103. doi: 10.1186/s12943-017-0675-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bullock N., Potts J., Simpkin A.J. Serine-arginine protein kinase 1 (SRPK1), a determinant of angiogenesis, is upregulated in prostate cancer and correlates with disease stage and invasion. J Clin Pathol. 2016;69:171–175. doi: 10.1136/jclinpath-2015-203125. [DOI] [PMC free article] [PubMed] [Google Scholar]