Abstract
Ortho-quinone methides (o-QMs) are reactive intermediates in biosynthesis that give rise to a variety of intra- and intermolecular cyclization/addition products in bacteria, fungi, and plants. Herein, we report a new metabolic deviation of an o-QM intermediate in a benzylic dehydrogenation reaction that links the newly described marine bacterial natural products dihydrotetrachlorizine and tetrachlorizine. We discovered these novel dichloropyrrole-containing compounds from actinomycete strain AJS-327 that unexpectedly harbors in its genome a biosynthetic gene cluster (BGC) of striking similarity to that of chlorizidine, another marine alkaloid bearing a different carbon skeleton. Heterologous expression of the homologous flavin-dependent oxidoreductase enzymes Tcz9 and Clz9 revealed their native functions in tetrachlorizine and chlorizidine biosynthesis, respectively, supporting divergent oxidative dehydrogenation and pyrrolizine-forming reactions. Swapping these berberine bridge enzyme-like oxidoreductases, we produced cyclized and dehydrogenated analogs of tetrachlorizine and chlorizidine, including a dearomatized chlorizidine analog that stabilizes an o-QM via conjugation with a 3H-pyrrolizine ring.
In the postgenomics era, the discovery of natural product molecules not only provides opportunities to explore their native and applied biological functions, but also the occasion to relate their chemistry back to their encoding genes for further biotechnological innovations.1−3 The ability to connect genes to molecules and vice versa through modern genome mining experimentation has transcended the field of microbial natural products chemistry and paved the way to new frontiers in synthetic biology,4 microbiome science,5 and biocatalysis,6 to name a few. Natural product biosynthetic enzymes are remarkable for their reaction diversity and plasticity, and as such, for their promise as biocatalysts to construct designer molecules.
Flavin adenine dinucleotide (FAD)-dependent oxidoreductases are one such family of enzymes that catalyze diverse chemical transformations with tremendous fidelity and biocatalytic utility.7,8 The berberine bridge enzyme (BBE)-like subfamily of flavoproteins are noteworthy for their ability to perform a variety of distinctive tailoring reactions in plants, fungi, and bacteria.9−12 Notably, BBE-like enzymes are involved in nicotine, cannabinoid, and berberine alkaloid biosynthesis. 13−15 Because so few BBE-like enzymes have been characterized, it is difficult for bioinformatic tools to predict the function of putative BBE-like enzymes without relying on their genomic context. The discovery of new BBE-like enzymes will yield new enzymatic reactions and improve bioinformatic predictions of BGCs containing these fascinating enzymes (Figure S1).
Recently, we isolated a taxonomically distinct marine actinomycete bacterium (strain AJS-327) that harbors in excess of 27 putative biosynthetic gene clusters (BGCs), compromising approximately 17% of its genome.16 One of these BGCs exhibited high sequence and architectural homology to the previously characterized clz BGC from Streptomyces sp. CNH-287, which is distinguished by the BBE-like enzyme Clz9 that catalyzes the formation of its unusual dihydropyrrolizine ring (Figure 1a).17 While strain AJS-327 is a prolific producer of cytotoxic secondary metabolites, including the recently discovered photopiperazines16 and ionostatin,18 we did not observe the production of chlorizidine A under several different media conditions. Instead, we identified two related tetrachlorinated metabolites in the organic extract by LC-MS analysis that were missing chlorizidine’s distinctive heteroaromatic rings.
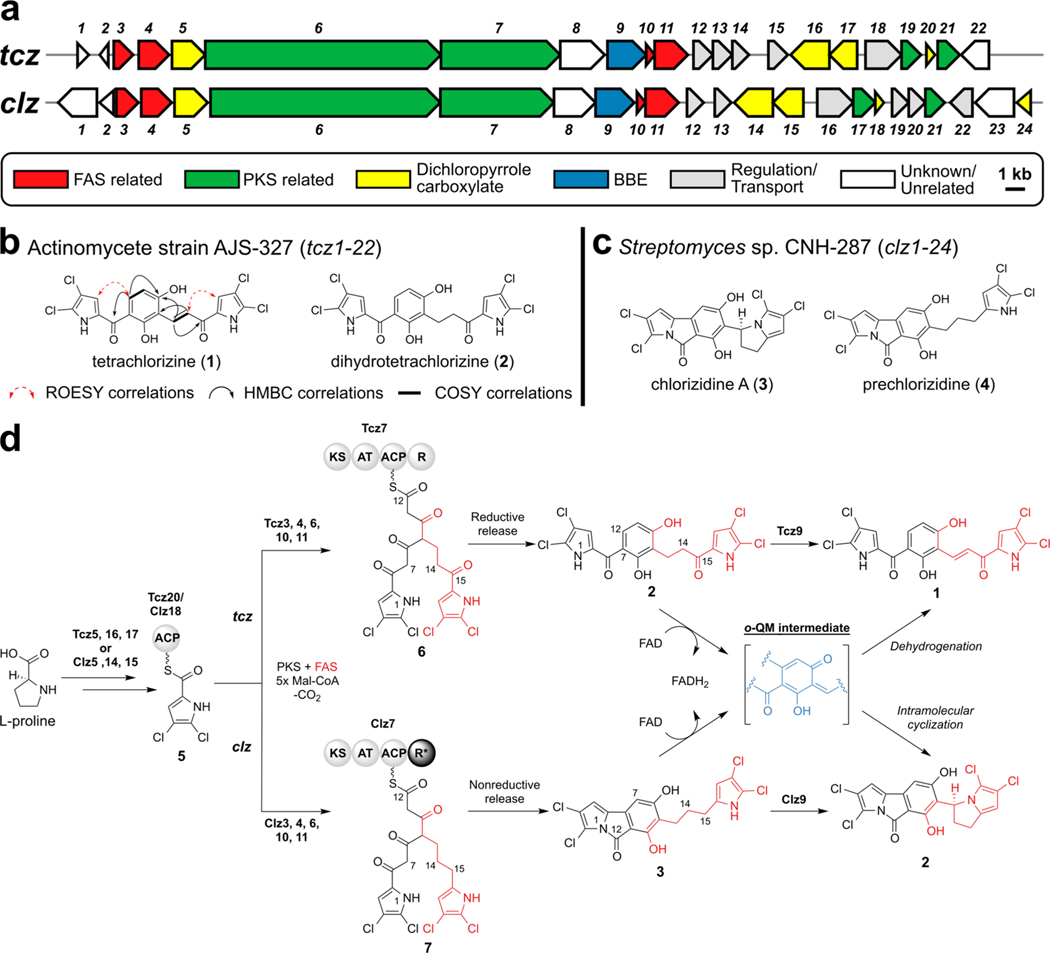
Figure 1.
Molecular basis for tetrachlorizine biosynthesis. (a) Alignment of the tetrachlorizine (tcz) and chlorizidine A (clz) BGCs. Genes are color-coded by associated function. (b) Structures of novel dichloropyrrole-containing natural products, tetrachlorizine (1) and dihydrotetrachlorizine (2). Key 2D NMR correlations of 1 are depicted. (c) Structures of previously characterized chlorizidine A (3) and biosynthetic precursor, prechlorizidine (4). (d) Proposed polyketide biosynthesis of 1, in comparison with the previously reported biosynthesis of 3.17 The rare FAS-derived dichloropyrrolyl extender unit is colored red. The general structure of the o-QM intermediate generated by Tcz9 and Clz9 is colored blue. This o-QM intermediate represents one possible regio- and stereoisomer for both pathways. Abbreviations: ACP, acyl carrier protein; AT, acyl transferase; FAD, flavin adenine dinucleotide; FADH2, reduced FAD (hydroquinone form); FAS, fatty acid synthase; KS, ketoacyl synthase; Mal-CoA, malonyl-coenzyme A; PKS, polyketide synthase; R, thioester reductase; *inactive domain.
To characterize these new tetrachlorinated metabolites that did not match known compounds in various databases, we cultivated 20 L of strain AJS-327 and purified several milligrams of both metabolites. HR-MS data coupled with 13C NMR analysis revealed their molecular formulas as C18H10Cl4N2O4 and C18H12Cl4N2O4, which we have since named tetrachlorizine (1) and dihydrotetrachlorizine (2), respectively (Figure 1b). These molecules differ by one degree of unsaturation, which could be explained by the disappearance of high field methylenic protons in the 1H NMR spectrum of 2 and the appearance of two olefinic doublets in the 1H NMR spectrum of 1. Key HMBC and NOE correlations were essential for determining the full connectivity between each ring system. Both of these metabolites are reminiscent of previously reported dichloropyrrole-containing secondary metabolites, such as chlorizidine A (3) (Figure 1c),17 pyoluteorin,19 the armeniaspirols,20 marinopyrroles,21 pyralomycins,22 and the pyrrolomycins23 (Figure S2). To better understand the biosynthetic relationship of the tetrachlorizines and how they are related to 3, we next turned our attention to the AJS-327 clz-like BGC and the role the putative BBE-like enzyme plays in 1 biosynthesis.
Sequence analysis revealed a contiguous region of 33.7 kb containing 22 open reading frames (ORFs) that displayed extremely high similarity and organization to the chlorizidine A BGC (clz) (Figure 1a).17 Remarkably, a homologue for every gene associated with 3 biosynthesis could be putatively annotated in strain AJS-327.24 Every homologue in the tetrachlorizine BGC (tcz) shares 58−90% amino acid sequence similarity and 46−81% identity with its corresponding gene associated with 3 biosynthesis. As such, we envisioned a highly analogous polyketide synthase (PKS) assembly line pathway for 1 and the previously established 317 with subtle metabolic differences that could repurpose their biosyntheses to achieve different cyclization patterns (Figure 1d).
There are two key structural differences best exemplified in the penultimate pathway intermediates 2 and prechlorizidine (4) that are quite informative. Most significant is the oxidation state and connectivity of the terminal carbon C-12 in both molecules that derives from the fully mature PKS intermediates 6 and 7 (Figure 1d). In the case of 2, 6 is reductively off-loaded by the terminal Tcz7 reductase (R) domain to an aldehyde intermediate that reacts with C-7 to generate the central tetrasubstituted benzene ring. This contrasts 4 in which we earlier envisaged that the PKS substrate 7 is nonreductively off-loaded and that C-12 rather forms an amide bond with N-1.17 The former mechanism is consistent with off-loading of similar PKS-extended, dichloropyrrole-containing secondary metabolites.20,21,25 The second structural difference is the oxidation state of C-15 which maintains the carbonyl of the dichloropyrrolyl-acyl carrier protein (5) substrate in 2 but is reduced to a methylene in 4.
We previously showed that the flavoenzyme Clz9 catalyzes the final reaction in the chlorizidine pathway via an o-QM intermediate to install the dihydropyrrolizine ring (Figure 1d, highlighted in blue).17 Although tetrachlorizine does not have such a structure motif, its BGC encodes the FAD-linked oxidoreductase Tcz9 that shares 69% amino acid sequence similarity and 53% identity with Clz9, suggesting a related enzymatic function. Sequence alignment of Tcz9 with Clz9 and other well characterized BBE family members confirmed the distinguishing active site motifs RSGGH and CxxI/V/LG necessary for bicovalent FAD attachment by the histidine and cysteine residues at positions 67 and 125, respectively (Figure S3).9,12,26 Given the high sequence homology to Clz9 and functional group similarity in 2, we speculated that Tcz9 could generate a similar o-QM intermediate. However, instead of catalyzing nucleophilic attack via the dichloropyrrole-nitrogen as seen in 3 biosynthesis, Tcz9 would facilitate deprotonation at C-14 and subsequent formation of an α,β-unsaturated ketone (Figure 1d). To explore the suspected role of Tcz9 in tetrachlorizine biosynthesis, we overexpressed Tcz9 in Escherichia coli and purified the polyhistidine tagged, recombinant protein fused with a maltose binding protein by affinity chromatography. Upon incubation with 2, we observed the complete conversion to 1 (Figure 2a), thereby confirming the functional role of Tcz9 as a dehydrogenase in tetrachlorizine biosynthesis.
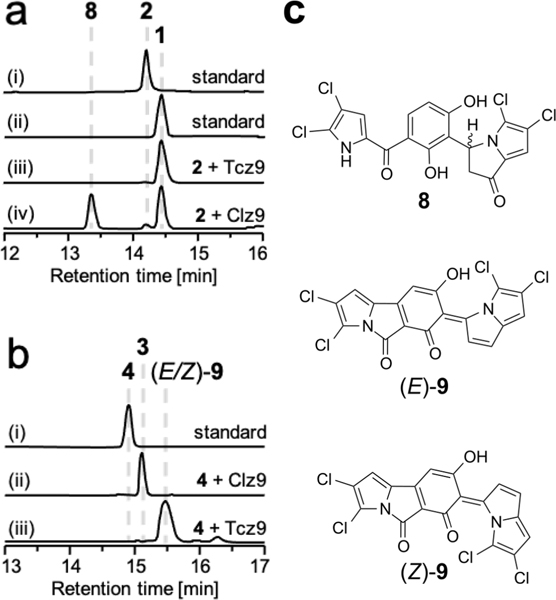
Figure 2.
(a) HPLC analysis of the in vitro conversion with 2 and either Tcz9 or Clz9 after 12 h: UV absorption monitored at 360 nm; (i) purified 2 standard; (ii) purified 1 standard; (iii) 2 + Tcz9, confirming Tcz9’s role in 1 biosynthesis; (iv) 2 + Clz9, yielding the cyclized derivative 8 in addition to 1. (b) HPLC analysis of the in vitro conversion with 4 and either Clz9 or Tcz9 after 12 h: UV absorption monitored at 430 nm; (i) purified 4 standard; (ii) 4 + Clz9, as previously reported;17 (iii) 4 + Tcz9, yielding a mixture of stabilized o-QM isomers, (E)-9 and (Z)-9. (c) Structures of enzymatic derivatives 8, (E)-9 and (Z)-9.
Upon confirming the role of Tcz9 in 1 biosynthesis, we were curious about the divergent reactivities of Clz9 and Tcz9 and whether the conversion of 2 → 1 and 4 → 3 were under enzyme or substrate control. We hypothesized that both BBE-like enzymes catalyzed the oxidative generation of similar o-QM intermediates; however, it was unclear how the C-15 carbonyl in the fatty acid synthase-derived extender unit of 2 influenced the dehydrogenation reaction catalyzed by Tcz9. Upon incubation of 2 with Clz9, we observed a mixture of two products (Figure 2a), one of which was 1. The earlier eluting product exhibited an identical UV spectrum as 2 but the same mass as 1. NMR analysis revealed this product to be a cyclized analog of tetrachlorizine (8), indicating Tcz9 and Clz9 have preferential activity to perform dehydrogenation and cyclization reactions, respectively (Figure 2c). Protein structure models comparing Tcz9 and Clz9 could not unambiguously explain which residues are critical for Tcz9’s dehydrogenation reactions. Future X-ray crystallography and mutagenesis experiments will be required to explain the difference in catalytic activity between Tcz9 and Clz9.
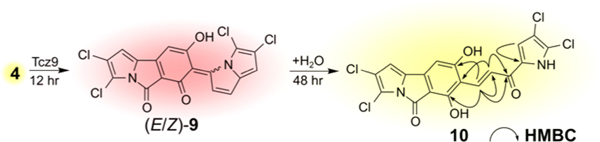
Next, we reversed roles to explore the fate of 4 with the mismatched Tcz9 (Figure 2b). Distinctively, the color of the reaction changed from yellow to a deep red over several hours, indicating the formation of a new product (Video S1). LC-MS analysis clearly showed the consumption of 4 and the appearance of two new products with mass 436.9058 ([M − H]−; C18H6Cl4N2O3) without any formation of 3. NMR analysis revealed this new product to be the cyclized and twice dehydrogenated configurational isomers (E)-9 and (Z)-9 (Figure 2c). This result indicates that Tcz9 not only acts as a dehydrogenase in the absence of the C-15 carbonyl, but it can also act as a cyclase, catalyzing the same intramolecular cyclization reaction as Clz9. Moreover, we were surprised to find that Tcz9 is capable of catalyzing successive dehydrogenation reactions with 4. The isolation of (E)-9 and (Z)-9 confirmed our hypothesis that the oxidation reaction proceeds via an o-QM intermediate. Both of these dearomatized compounds are stabilized via conjugation with the 3H-pyrrolizine ring and are stable at room temperature. However, (E)-9 and (Z)-9 slowly changed from red to yellow following purification, and NMR spectroscopic analysis indicated these two molecules coalesced to the same rearranged, water adduct with a mass of 454.9171 ([M − H]−; C18H8Cl4N2O4). We speculate that (E)-9 and (Z)-9 nonenzymatically react with water by 1,6-conjugate addition to generate the linear α,β-unsaturated ketone product, 10 (Scheme 1; a full proposed mechanism is provided in Scheme S1). Thus, Tcz9 is ultimately capable of dehydrogenation reactions with 2 and 4 that ultimately generate α,β-unsaturated ketones, 1 and 10, but through two distinct mechanisms.
Scheme 1.
Abbreviated Reaction Scheme Depicting the Enzymatic Process for Oxidation of 4 to (E)-9 and (Z)-9 Catalyzed by Tcz9, Which Non-Enzymatically Coalesce to 10 in the Presence of Watera aKey HMBC correlations for assigning the structure of 10 are included.
Numerous biosynthetic proposals feature o-QM intermediates founded on biomimetic total syntheses, nonenzymatic coupling of secondary metabolites, and specific trapping reactions using biosynthetic precurors.27−29 These ephemeral intermediates react with various nucleophiles and can undergo inter- and intramolecular conjugate addition, cycloaddition, and spirolation reactions before they can be isolated and spectroscopically characterized.30−32 Several synthetic approaches have been developed to generate and stabilize natural product-derived o-QMs for spectroscopic analysis, typically by cooling these intermediates to extremely low temperatures.33,34 Here, we have not only demonstrated that Clz9 and Tcz9 catalyze benzylic functionalization reactions via an o-QM intermediate, but we were also able to isolate a stabilized o-QM conjugated with the 3H-pyrrolizine ring system. The unprecedented oxidation of the unactivated propyl chain in 4 to the α,β-unsaturated ketone 10 via a cyclized, twice dehydrogenated, and stabilized o-QM intermediate highlights the versatility and potential of the enzymes to perform challenging chemical transformations.
In conclusion, we established the genetic basis for the biosynthesis of two previously undescribed dichloropyrrole-containing marine natural products, tetrachlorizine and dihydrotetrachlorizine. Bioinformatic analysis revealed a gene cluster with striking similarity to the chlorizidine gene cluster, and further comparative in vitro studies revealed nuanced activity between the corresponding BBE-like tailoring enzymes Tcz9 and Clz9. The benzylic dehydrogenation reactions catalyzed by Tcz9 proceed via an o-QM intermediate. To our knowledge, this is the first example of a dehydrogenation reaction proceeding via an o-QM, expanding the reaction repertoire of these highly reactive intermediates. These findings epitomize the importance of connecting genes to the molecules and understanding the nuanced approaches nature has developed to perform powerful and selective chemical transformations. Further development of BBE-like enzymes for biocatalytic applications may deliver chemoenzymatic solutions to challenging oxidative reactions in total synthesis efforts.
Supplementary Material
ACKNOWLEDGMENTS
We kindly thank Brendan Duggan and Anthony Mrse for NMR assistance, and Alexander Smith for isolating strain AJS-327. Funding was generously provided by the NIH (R01-AI47818 to B.S.M.; R37-CA044848 to W.F.) and the NIH Marine Biotechnology Training Grant Predoctoral Fellowship (T32-GM067550) to T.N.P.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.0c12415.
General procedures, experimental details, spectroscopic spectra (PDF) (MP4)
The authors declare no competing financial interest.
Contributor Information
Trevor N. Purdy, Center for Marine Biotechnology and Biomedicine, Scripps Institution of Oceanography, University of California at San Diego, La Jolla, California 92093, United States
Min Cheol Kim, Center for Marine Biotechnology and Biomedicine, Scripps Institution of Oceanography, University of California at San Diego, La Jolla, California 92093, United States.
Reiko Cullum, Center for Marine Biotechnology and Biomedicine, Scripps Institution of Oceanography, University of California at San Diego, La Jolla, California 92093, United States.
William Fenical, Center for Marine Biotechnology and Biomedicine, Scripps Institution of Oceanography, Moores Comprehensive Cancer Center, and Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California at San Diego, La Jolla, California 92093, United States.
Bradley S. Moore, Center for Marine Biotechnology and Biomedicine, Scripps Institution of Oceanography and Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California at San Diego, La Jolla, California 92093, United States.
REFERENCES
- (1).Doroghazi JR; Albright JC; Goering AW; Ju KS; Haines RR; Tchalukov KA; Labeda DP; Kelleher NL; Metcalf WW A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat. Chem. Biol 2014, 10, 963–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Ziemert N; Alanjary M; Weber T The evolution of genome mining in microbes − a review. Nat. Prod. Rep 2016, 33, 988–1005. [DOI] [PubMed] [Google Scholar]
- (3).Navarro-Muñoz JC; Selem-Mojica N; Mullowney MW; Kautsar SA; Tryon JH; Parkinson EI; De ELC; Santos L; Yeong M; Cruz-Morales P; Abubucker S; Roeters A; Lokhorst W; Fernandez-Guerra A; Dias Cappelini LT; Goering AW; Thomson RJ; Metcalf WW; Kelleher NL; Barona-Gomez F; Medema MH A computational framework to explore large-scale biosynthetic diversity. Nat. Chem. Biol 2020, 16, 60–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Kim E; Moore BS; Yoon YJ Reinvigorating natural product combinatorial biosynthesis with synthetic biology. Nat. Chem. Biol 2015, 11, 649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Donia MS; Fischbach MA Small molecules from the human microbiota. Science 2015, 349, 1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Huffman MA; Fryszkowska A; Alvizo O; Borra-Garske M; Campos KR; Canada KA; Devine PN; Duan D; Forstater JH; Grosser ST; Halsey HM; Hughes GJ; Jo J; Joyce LA; Kolev JN; Liang J; Maloney KM; Mann BF; Marshall NM; McLaughlin M; Moore JC; Murphy GS; Nawrat CC; Nazor J; Novick S; Patel NR; Rodriguez-Granillo A; Robaire SA; Sherer EC; Truppo MD; Whittaker AM; Verma D; Xiao L; Xu Y; Yang H Design of an in vitro biocatalytic cascade for the manufacture of islatravir. Science 2019, 366, 1255–1259. [DOI] [PubMed] [Google Scholar]
- (7).Walsh CT; Wencewicz TA Flavoenzymes: Versatile catalysts in biosynthetic pathways. Nat. Prod. Rep 2013, 30, 175–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Teufel R; Agarwal V; Moore BS Unusual flavoenzyme catalysis in marine bacteria. Curr. Opin. Chem. Biol 2016, 31, 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Huang CH; Lai WL; Lee MH; Chen CJ; Vasella A; Tsai YC; Liaw SH Crystal structure of glucooligosaccharide oxidase from Acremonium strictum: A novel flavinylation of 6-S-cysteinyl, 8α-N1-histidyl FAD. J. Biol. Chem 2005, 280, 38831–38838. [DOI] [PubMed] [Google Scholar]
- (10).Nielsen CA; Folly C; Hatsch A; Molt A; Schröder H; O’Connor SE; Naesby M The important ergot alkaloid intermediate chanoclavine-I produced in the yeast Saccharomyces cerevisiae by the combined action of EasC and EasE from Aspergillus japonicus. Microb. Cell Fact 2014, 13, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Carlson JC; Li S; Gunatilleke SS; Anzai Y; Burr DA; Podust LM; Sherman DH Tirandamycin biosynthesis is mediated by co-dependent oxidative enzymes. Nat. Chem 2011, 3, 628–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Daniel B; Konrad B; Toplak M; Lahham M; Messenlehner J; Winkler A; Macheroux P The family of berberine bridge enzyme-like enzymes: A treasure-trove of oxidative reactions. Arch. Biochem. Biophys 2017, 632, 88–103. [DOI] [PubMed] [Google Scholar]
- (13).Taura F; Morimoto S; Shoyama Y; Mechoulam R First Direct Evidence for the Mechanism of Δ1-Tetrahydrocannabinolic Acid Biosynthesis. J. Am. Chem. Soc 1995, 117 (38), 9766–9767. [Google Scholar]
- (14).Lewis RS; Lopez HO; Bowen SW; Andres KR; Steede WT; Dewey RE Transgenic and Mutation-Based Suppression of a Berberine Bridge Enzyme-Like (BBL) Gene Family Reduces Alkaloid Content in Field-Grown Tobacco. PLoS One 2015, 10, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Shoyama Y; Tamada T; Kurihara K; Takeuchi A; Taura F; Arai S; Blaber M; Shoyama Y; Morimoto S; Kuroki R Structure and Function of Δ1-Tetrahydrocannabinolic Acid (THCA) Synthase, the Enzyme Controlling the Psychoactivity of Cannabis sativa. J. Mol. Biol 2012, 423, 96–105. [DOI] [PubMed] [Google Scholar]
- (16).Kim MC; Cullum R; Machado H; Smith AJ; Yang I; Rodvold JJ; Fenical W Photopiperazines A−D, Photosensitive Interconverting Diketopiperazines with Significant and Selective Activity against U87 Glioblastoma Cells, from a Rare, Marine-Derived Actinomycete of the Family Streptomycetaceae. J. Nat. Prod 2019, 82, 2262–2267. [DOI] [PubMed] [Google Scholar]
- (17).Mantovani SM; Moore BS Flavin-Linked Oxidase Catalyzes Pyrrolizine Formation of Dichloropyrrole-Containing Polyketide Extender Unit in Chlorizidine A. J. Am. Chem. Soc 2013, 135, 18032–18035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Kim MC; Winter JM; Cullum R; Li Z; Fenical W Complementary Genomic, Bioinformatics, and Chemical Approaches Facilitate the Absolute Structure Assignment of Ionostatin, a Linear Polyketide from a Rare Marine-Derived Actinomycete. Chem. Biol 2020, 23, 32. [DOI] [PubMed] [Google Scholar]
- (19).Nowak-Thompson B; Chaney N; Wing JS; Gould SJ; Loper JE Characterization of the Pyoluteorin Biosynthetic Gene Cluster of Pseudomonas fluorescens Pf-5. J. Bacteriol 1999, 181, 2166–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Qiao Y; Yan J; Jia J; Xue J; Qu X; Hu Y; Deng Z; Bi H; Zhu D Characterization of the Biosynthetic Gene Cluster for the Antibiotic Armeniaspirols in Streptomyces armeniacus. J. Nat. Prod 2019, 82, 318–323. [DOI] [PubMed] [Google Scholar]
- (21).Yamanaka K; Ryan KS; Gulder TAM; Hughes CC; Moore BS Flavoenzyme-Catalyzed Atropo-Selective N,C-Bipyrrole Homocoupling in Marinopyrrole Biosynthesis. J. Am. Chem. Soc 2012, 134, 12434–12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Flatt PM; Wu X; Perry S; Mahmud T Genetic Insights into Pyralomicin Biosynthesis in Nonomuraea spiralis IMC A-0156. J. Nat. Prod 2013, 76, 939–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Zhang X; Parry RJ Cloning and Characterization of the Pyrrolomycin Biosynthetic Gene Clusters from Actinosporangium vitaminophilum ATCC 31673 and Streptomyces sp. Strain UC 11065. Antimicrob. Agents Chemother 2007, 51, 946–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).There is one ORF associated with chlorizidine A biosynthesis that is absent in the tetrachlorizine BGC - a flavin mononucleotide reductase gene. Putative FMN reductase genes have been identified elsewhere in the genome.
- (25).Nowak-Thompson B; Gould SJ; Loper JE Identification and sequence analysis of the genes encoding a polyketide synthase required for pyoluteorin biosynthesis in Pseudomonas fluorescens Pf-5. Gene 1997, 204, 17–24. [DOI] [PubMed] [Google Scholar]
- (26).Winkler A; Hartner F; Kutchan TM; Glieder A; Macheroux P Biochemical Evidence That Berberine Bridge Enzyme Belongs to a Novel Family of Flavoproteins Containing a Bi-covalently Attached FAD Cofactor. J. Biol. Chem 2006, 281, 21276–21285. [DOI] [PubMed] [Google Scholar]
- (27).Willis NJ; Bray CD ortho-Quinone Methides in Natural Product Synthesis. Chem. - Eur. J 2012, 18, 9160–9173. [DOI] [PubMed] [Google Scholar]
- (28).Bai WJ; David JG; Feng ZG; Weaver MG; Wu KL; Pettus TRR The Domestication of ortho-Quinone Methides. Acc. Chem. Res 2014, 47, 3655–3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Singh MS; Nagaraju A; Anand N; Chowdhury S ortho-Quinone methide (o-QM): a highly reactive, ephemeral and versatile intermediate in organic synthesis. RSC Adv. 2014, 4, 55924–55959. [Google Scholar]
- (30).Spence JTJ; George JH Total Synthesis of Peniphenones A−D via Biomimetic Reactions of a Common o-Quinone Methide Intermediate. Org. Lett 2015, 17, 5970–5973. [DOI] [PubMed] [Google Scholar]
- (31).Rodriguez R; Adlington RM; Moses JE; Cowley A; Baldwin JE A New and Efficient Method for o-Quinone Methide Intermediate Generation: Application to the Biomimetic Synthesis of (±)-Alboatrin. Org. Lett 2004, 6, 3617–3619. [DOI] [PubMed] [Google Scholar]
- (32).Feng Z-G; Burnett GL; Pettus TRR A Biomimetic Synthesis of des-Hydroxy Paecilospirone. Synlett 2018, 29, 1517–1519. [Google Scholar]
- (33).Cavitt SB; Sarrafizadeh HR; Gardner PD The Structure of o-Quinoxe Methide Trimer. J. Org. Chem 1962, 27, 1211–1216. [Google Scholar]
- (34).Rosenau T; Potthast A; Elder T; Kosma P Stabilization and First Direct Spectroscopic Evidence of the o-Quinone Methide Derived from Vitamin E. Org. Lett 2002, 4, 4285–4288. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.