Abstract
Aims
Stem cell therapy has shown promise for treating myocardial infarction via re-muscularization and paracrine signalling in both small and large animals. Non-human primates (NHPs), such as rhesus macaques (Macaca mulatta), are primarily utilized in preclinical trials due to their similarity to humans, both genetically and physiologically. Currently, induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) are delivered into the infarcted myocardium by either direct cell injection or an engineered tissue patch. Although both approaches have advantages in terms of sample preparation, cell–host interaction, and engraftment, how the iPSC-CMs respond to ischaemic conditions in the infarcted heart under these two different delivery approaches remains unclear. Here, we aim to gain a better understanding of the effects of hypoxia on iPSC-CMs at the transcriptome level.
Methods and results
NHP iPSC-CMs in both monolayer culture (2D) and engineered heart tissue (EHT) (3D) format were exposed to hypoxic conditions to serve as surrogates of direct cell injection and tissue implantation in vivo, respectively. Outcomes were compared at the transcriptome level. We found the 3D EHT model was more sensitive to ischaemic conditions and similar to the native in vivo myocardium in terms of cell–extracellular matrix/cell–cell interactions, energy metabolism, and paracrine signalling.
Conclusion
By exposing NHP iPSC-CMs to different culture conditions, transcriptome profiling improves our understanding of the mechanism of ischaemic injury.
Keywords: Non-human primate, Induced pluripotent stem cells, Cardiomyocytes, Hypoxia, Engineered heart tissue, Transcriptome
1. Introduction
Excessive loss of cardiomyocytes (CMs) in the heart resulting from myocardial infarction (MI) can give rise to heart failure.1–3 In comparison to conventional medical treatment, cell therapy has the potential to restore heart function by regenerating the damaged myocardium through the reintroduction of endogenous/exogenous CMs.4 Pluripotent stem cell-derived cardiomyocytes (PSC-CMs) are a potential cell source to improve cardiac structure and function and remuscularize the damaged myocardium resulting from a MI.5–9 A number of preclinical studies using direct human embryonic stem cell-derived cardiomyocyte (hESC-CM) injections into the infarcted myocardium of non-human primates (NHPs) have shown integration of the cells and regeneration of the infarcted myocardium.5,6,10 Moreover, allogeneic induced pluripotent stem cell-derived cardiomyocyte (iPSC-CM) transplantation was also performed in cynomolgus monkeys.6 NHPs are utilized in preclinical studies, primarily because of their physiologic, anatomic, and genetic similarity to humans. Cardioprotective effects using directly injected NHP (rhesus macaques) or human iPSC-CMs for the treatment of MI in athymic nude rat model have also been reported.11 Collectively, these studies support the notion that NHPs can be used to recapitulate human pathophysiological conditions.11,12
Generally, two approaches have been developed for in vivo cell delivery in the infarcted myocardium: direct cell injection and cell patch transplantation.13 Both methods have their individual advantages and disadvantages. For example, direct cell injection has few cell culture and technical obstacles, and this approach allows cells to have prolonged contact with the host tissue; however, the resulting cell engraftment rate is relatively low.14 In contrast, the preparation of a cell patch is time-consuming and requires pre-culturing in special engineering platforms. However, when patching from outside of the myocardium, cell patches enhance the survival rate post-implantation with less ‘washing-away’ of cells,15,16 and can also mechanically support myocardial contractions.17,18 Overall, both approaches have been shown to improve cardiac function, but the manner in which these two delivery methods respond to ischaemic conditions in the infarcted heart before and after transplantation remains unclear.
Here, we used iPSC-CMs from rhesus macaques (Macaca mulatta) in both 2D monolayer and 3D engineered heart tissue (EHT) cultures to model direct cell injection vs. tissue patch approaches, with hypoxic conditioning included to recapitulate the ischaemic milieu effects on transplanted CMs. This allowed for the isolation of two key factors: the dimension outcome (2D vs. 3D) of cell transplantation and the impact of ischaemia. We compared 2D NHP iPSC-CM monolayer culture and 3D NHP EHTs using an equivalent cell differentiation protocol. These in vitro models were then pre-conditioned in hypoxic conditions to simulate the ischaemia associated with MI. After the hypoxic treatment, the differential transcriptome of all four groups (2D-Normoxia, 2D-Hypoxia, 3D-Normoxia, and 3D-Hypoxia) were further analysed and profiled. The transcriptional profiling approach allowed a better understanding of the effects of NHP iPSC-CMs under ischaemic conditions and in different culture dimensions, which may help improve future delivery approaches for regenerative medicine.
2. Methods
2.1 Fabrication and characterization of EHT
2.1.1 Master mold design and 3D printing
The master mold was designed by AutoCAD (Autodesk Inc.), and then fabricated using a 3D printer with a proprietary ‘acrylic’ type material (3D Systems, Inc.). The final products were cleaned and heated to 65°C for polymerization deactivation.
2.1.2 Polydimethylsiloxane mold fabrication
The polydimethylsiloxane (PDMS) mold was cast from the acrylic master mold after PDMS mixing (Dow Corning Sylgard 184), degassing, and polymerization. The PDMS mold for EHT formation consisted of three reservoirs. Each reservoir was 4 mm wide, 12 mm long, and 6 mm deep, and had two posts at either end for EHT attachments that were 1 mm thick, 2 mm wide, and 6 mm apart. The centre-to-centre distance between the two reservoirs was 8 mm.
2.1.3 EHT formation
The NHP EHT was fabricated with the same method as the human EHT as previously reported.19 Briefly, 200 µL of 10 g/mL collagen type I, 21 µL of 10× PBS (Life Technologies, 70011-044), 4 µL of 1 N NaOH, 67.4 µL of CM media (RPMI+B27), and 100 µL of Matrigel were pre-mixed on ice, then added into the pellet of NHP iPSC-CMs at 2 million per 275 µL. Afterwards, the cell and hydrogel mixture were carefully loaded into each reservoir. The entire plate was placed in a cell culture incubator at 37°C for 1 h to allow for hydrogel solidification. Then 4 mL of media was added to the wells for further EHT culture. On Day 45, EHT was used for the following experiments.
2.1.4 Immunofluorescent staining of EHT
EHT was fixed in 4% paraformaldehyde in PBS (PFA) for 15 min and then washed with PBS for 10 min three times. EHT was equilibrated in 30% sucrose (filter-sterilized) overnight, embedded in OCT on dry ice, and frozen at ≤ −80°C. The samples were cryosectioned into 10 µm sections and mounted onto Superfrost Plus slides (Thermo Fisher Scientific). The slides were permeabilized with 0.1% Triton X-100 for 30 min at room temperature (RT) and then blocked with 10% normal goat serum in PBS for 25 min at RT. Then the primary antibodies (anti-alpha-actinin from mouse, 1:200 (Sigma) and anti-human Troponin T from rabbit, 1:200 (Abcam)) were applied at 4°C overnight. Secondary antibodies (Goat-anti-mouse-Alexa Fluor 488 and Goat-anti-rabbit-Alexa Fluor 633, 1:200, Thermo Fisher) were applied for 1 h at RT and followed by 5 min washes (three times) in wash buffer. Finally, the slides were sealed in a mounting solution with DAPI co-staining (ProLong Gold Antifade Mountant with DAPI).
2.2 Hypoxic conditioning
Three individual EHT samples were placed and sealed in three respective anaerobic pouches (GasPak EZ Anaerobe Pouch System, Fisher Scientific, MA) with fresh culture medium in a cell culture incubator at 37°C and 5% CO2. The other three individual EHT samples served as normoxic controls at 37°C and 5% CO2. EHTs were further incubated for another 24 h, then the conditioned culture medium from each EHT sample was collected for secreted growth factor measurement, and the EHT was collected for RNA extraction and RNA sequencing. The conditioned medium and RNA from the remaining three EHT samples under normoxic condition were also collected at the same time and served as controls.
2.3 Proteomic angiogenesis assay
A similar procedure was described previously.11 In brief, cytokines (VEGF, IL-8, and MCP-1) in the conditioned medium collected from the four groups were detected by the reconstituted antibody cocktail. After incubating with blotting membrane, 2 mL of diluted streptavidin-HRP was added into each well of the 4-well multi-dish for chemical conjugation. The membrane was then developed with the Chemi Reagent Mix for imaging with the Chemidoc Imaging system (Bio-Rad). The intensities of VEGF, IL-8, and MCP-1 blots were processed and analysed with ImageJ (NIH).
2.4 RNA-sequencing and analysis
RNA was extracted using the RNeasy Mini Kit (Qiagen, Germany). Briefly, EHTs were homogenized in Trizol (Roche, Switzerland), and chloroform was further added. After vortexing and incubating at RT for 10 min, samples were centrifuged at 16000g at 4°C for 30 min. The upper, transparent supernatant was collected, and an equal volume of 70% ethanol was added and mixed thoroughly, and then the mixture was loaded into an isolation column. Following the steps of column washes, RNA was eluted and collected from the column with nuclease-free water. The concentration of RNA was measured by the NanoDrop (2000/2000c Spectrophotometers, Thermo Fisher), and the purity of RNA was determined by the ratio of A260/280. All the RNAs used in this study were A260/280 = 1.9 ∼ 2.1.
Kits used for RNA-sequencing (RNA-seq) were purchased from ThermoFisher. Total RNA was mixed with ERCC RNA Spike-In Mix 1, and polyadenylated mRNA was isolated using the Dynabeads mRNA DIRECT™ Micro Purification Kit. A cDNA library was prepared using the Ion Total RNA-Seq Kit v2 and sequenced by the Ion Proton method using Ion PI Hi-Q Sequencing kit. Unaligned BAM files were generated by the Ion Torrent Suite 5.0.4 and converted into FASTQ files. The raw reads were aligned to the rhesus monkey genome rheMac2 by Bowtie2 and Tophat2, and then annotated with ENSEMBL Release 84 by HT-seq.20 The differentially expressed genes (DEGs) were detected by DESeq2,21 and the adjusted P-value cutoff of DEGs was 0.001. The total reads of each sample were normalized by the ERCC reads. The likelihood ratio test was performed as the statistical inference to estimate DEGs. The variances stabilizing transformed values generated by DESeq2 were used as input for hierarchical clustering and further analyses. The line plots of gene expression levels were generated based on clustering by R package ggplot2 (https://cran.r-project.org/web/packages/ggplot2/index.html). Functional enrichment analysis was implemented by the Bioconductor package GeneAnswers (https://www.bioconductor.org/ packages/release/bioc/html/GeneAnswers.html).
2.5 Analysis of microarray data
The public microarray data of heart failure patients with post-MI (GSE59867, 10 patients and 10 healthy controls) were extracted by GEO2R from NCBI. Downstream analysis was the same as described RNA-seq analysis.
2.6 Statistical analysis
Data are presented as mean±SEM. Statistical significance was determined using either the Student’s t-test or one-way ANOVA. P < 0.05 was considered statistically significant.
Animal experiments and procedures were approved by the Stanford Administrative Panel on Laboratory Animal Care (APLAC) guidelines and policies, following the NIH Guide for the Care and Use of Laboratory Animals. Mice were anesthetized with 1–5% isoflurane for NHP iPSCs (1–5 million cells in 50 µL Matrigel) injection and were euthanized with saturated KCl after being anesthetized with 4% isoflurane. NHP fibroblasts were obtained from a 1-year-old male rhesus monkey small skin biopsy according to a University of California Davis Institutional Animal Care and Use Committee (IACUC) approved protocol.
3. Results
3.1 Establishment of NHP iPSC line
The overall experimental design is shown in Figure 1, including NHP iPSC reprogramming, NHP iPSC-CM differentiation and characterization, 2D monolayer culture, 3D EHT fabrication and culture, and RNA-seq analysis of NHP iPSC-CMs in 2D monolayer and 3D EHT cultures under normoxic or hypoxic conditions. The reprogramming strategy for generating NHP iPSCs from skin fibroblasts of a male rhesus monkey is shown in Supplementary material online, Figure S1A. The NHP iPSC colony was formed after reprogramming as shown in Supplementary material online, Figure S1B. NHP iPSCs exhibited pluripotency markers (Oct3/4, Sox2, SSEA-4, and TRA-4) which are shown in Supplementary material online, Figure S1C. After reprogramming and passaging for 30 times, the NHP iPSCs maintained a normal karyotype with 20 pairs of autosomal chromosomes, as well as X and Y sex chromosomes (Supplementary material online, Figure S1D). Moreover, teratoma were formed from subcutaneous injection in rats after 1–2 months, with formation of three germ layers as identified by haematoxylin and eosin (H&E) staining. Supplementary material online, Figure S1E shows the ectodermal lineage with primitive neuroectodermal development with rosettes and mesodermal and endodermal lineages with cartilage and endodermal gland.
Figure 1.
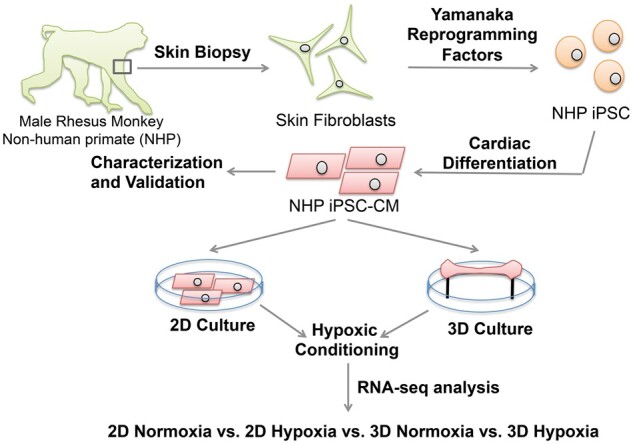
Diagram of experimental design. Skin fibroblasts were grown from a 1-year-old male rhesus monkey skin biopsy. This was followed by generating iPSCs which were differentiated using a cardiac differentiation protocol to derive the NHP iPSC-CMs for generation of the 2D monolayer and 3D EHT. Both culture conditions (2D, 3D) were then exposed to hypoxic conditions to allow transcriptional analysis by RNA-seq.
3.2 Cardiac differentiation of NHP iPSCs
As shown in the protocol illustrated in Supplementary material online, Figure S2A, NHP iPSCs were successfully differentiated into beating NHP iPSC-CMs (Supplementary material online, Video S1). The confocal images of NHP iPSC-CMs immunostained with cardiac troponin T (cTnT) and alpha-actinin show the characteristic striated sarcomeric structure (Supplementary material online, Figure S2B). The purity of iPSC-CMs was ∼80% of cTnT and alpha-actinin double-positive cells as measured by FACS (Supplementary material online, Figure S2C and D). From the single-cell RT–qPCR experiment, the ∼20% non-CMs in the differentiated NHP iPSC-CMs were THY-1 positive fibroblasts (Supplementary material online, Figure S2G). The pluripotency genes (SOX2, NANOG, POU5F1, and LIN28) were not detected in the NHP iPSC-CMs or the whole heart RNA from NHP left ventricle (Zyagen, rhesus monkey) in comparison to the NHP iPSCs (Supplementary material online, Figure S2F). Cardiac-related genes were also examined, and NHP iPSC-CMs (∼80% CMs and 20% non-CMs, such as fibroblasts) were found to have relatively higher expression of NKX2.5 and relatively lower expression of TNNT2 and MYH7 than the RNA from NHP left ventricle (Supplementary material online, Figure S2F). The derived NHP iPSC-CMs include all three CM action potential (AP) subtype morphologies: ventricular-, atrial-, and nodal-like, presenting similar action potential patterns to human iPSC-CM, as noted in Supplementary material online, Figure S3A and B. The detailed parameters of AP of NHP iPSC-CMs and human iPSC-CMs are summarized in Supplementary material online, Table S1. Calcium imaging suggests that NHP iPSC-CMs exhibit decreased transient amplitude, and slower calcium transient dynamics compared to human iPSC-CMs (Supplementary material online, Figure S3C).
3.3 3D culture of NHP EHT
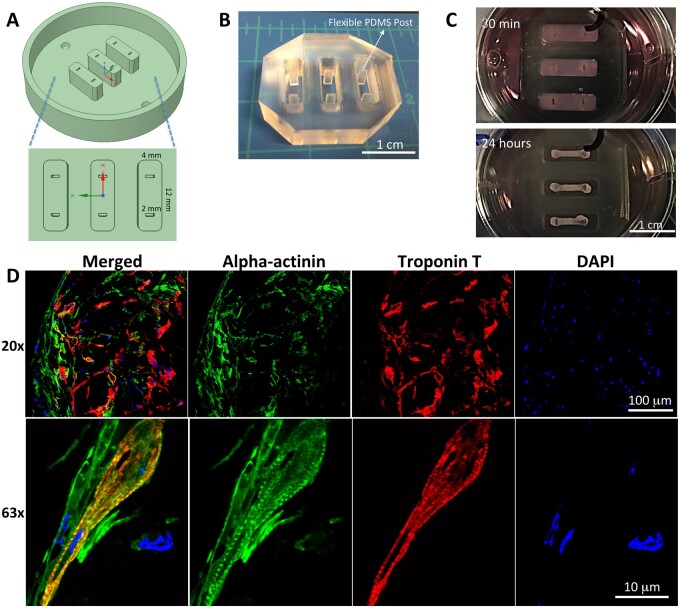
Our previous approach for EHT fabrication was used to make the NHP EHT.19 First, the master mold shown in Figure 2A was designed, 3D-printed with polyacrylic, and polished. Then the silicone mold with two flexible and separated posts, as shown in Figure 2B, was cast from the master mold with PDMS. After filling the reservoirs of the PDMS mold, the hydrogel/cell mixture was solidified (Figure 2C). The elongated 3D EHT was formed after 24 h upon the stimulation of passive force from separated flexible posts (Figure 2C). The EHT was cultured for four additional weeks. The anisotropic contraction is shown in Supplementary material online, Video S2. The sarcomeric structure of NHP iPSC-CMs inside the EHT was examined by fluorescent immunostaining of EHT sections with anti-alpha actinin and cTnT. The results are shown in Figure 2D and Supplementary material online, Figure S4, revealing a well-distributed and interconnected NHP iPSC-CM network (10× and 20×) with striated sarcomeres (63×).
Figure 2.
Design and fabrication of the master mold, PDMS mold, and NHP EHTs. (A) Master mold designed by AutoCAD and fabricated by 3D printing, and sizes in three dimensions are shown. (B) PDMS mold with flexible posts. (C) EHT formation in the PDMS mold at 30 min and 24 h. (D) Immunofluorescence staining of NHP EHTs at Day 50 with alpha-actinin (green), troponin T (red), and DAPI (blue) showing striated sarcomeres.
3.4 Distinct transcriptional program of NHP iPSC-CMs in 2D and 3D hypoxic cultures
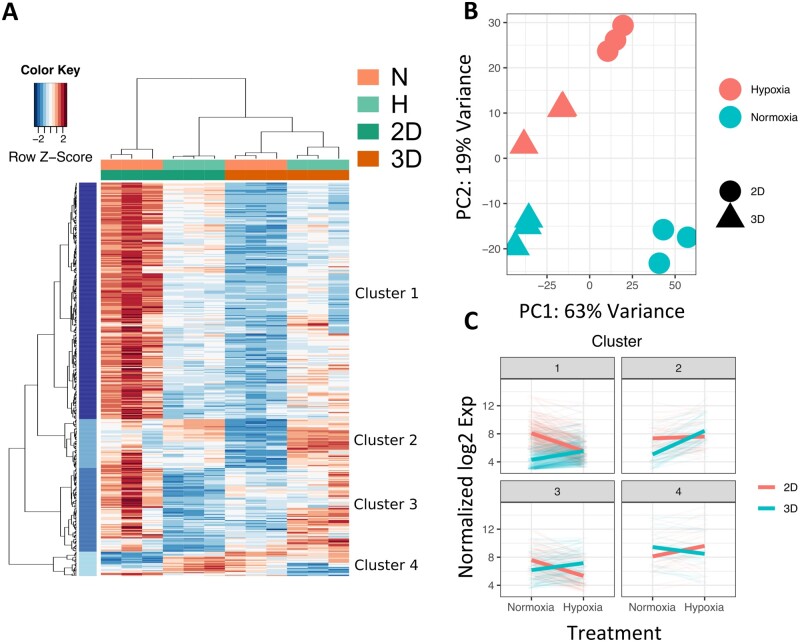
To determine the microenvironment-specific transcriptional program deployed in NHP iPSC-CMs, we performed RNA-seq on the following four groups: 2D-Normoxia, 2D-Hypoxia, 3D-Normoxia, and 3D-Hypoxia. Thousands of differentially regulated genes were identified within each microenvironment condition, with the clear majority of differences accounted for by the culture dimensional variance and ischaemia conditioning (Figure 3A). The four clusters were labelled as Clusters 1, 2, 3, and 4 as shown in Figure 3A.
Figure 3.
Differential transcriptome of NHP iPSC-CMs in 2D and 3D culture models under normoxic or hypoxic conditions by RNA-seq analysis. (A) Heatmap of four groups: 2D-Normoxia, 2D-Hypoxia, 3D-Normoxia, and 3D-Hypoxia, and with four clusters segregated, N = 3 biological samples in each group. (B) PCA for each group: red represents hypoxia, green represents normoxia, a solid circle represents 2D, and solid triangle represents 3D. (C) Overall gene expression trends of four clusters in 2D and 3D culture models under normoxic vs. hypoxic conditions. N = 3 biological samples in each group. The detailed RNA-seq and statistical analysis are in the Methods section.
To visualize the relative transcriptional distances of each RNA-seq sample, we employed principal component analysis (PCA). Each microenvironment condition within the RNA-seq dataset was grouped into a defined cluster in the PCA plot (Figure 3B). PC1 contributed 63% variance and clearly indicated the difference between 2D and 3D cultures, whereas PC2 contributed 19% variance and indicated the difference between normoxia (N) and hypoxia (H).
We further performed Pathway Enrichment Analysis on the 2D and 3D culture samples in response to the hypoxic treatment. In Figure 3C, the weighted trends of enriched pathways in 2D and 3D cultures in the four clusters were compared between normoxia and hypoxia conditions. In general, the gene expression of the enriched pathways in Clusters 1, 2, and 3 were up-regulated in the 3D culture, whereas the gene expression in Cluster 4 was up-regulated in the 2D culture.
3.5 Differential gene expression and related pathways in four clusters
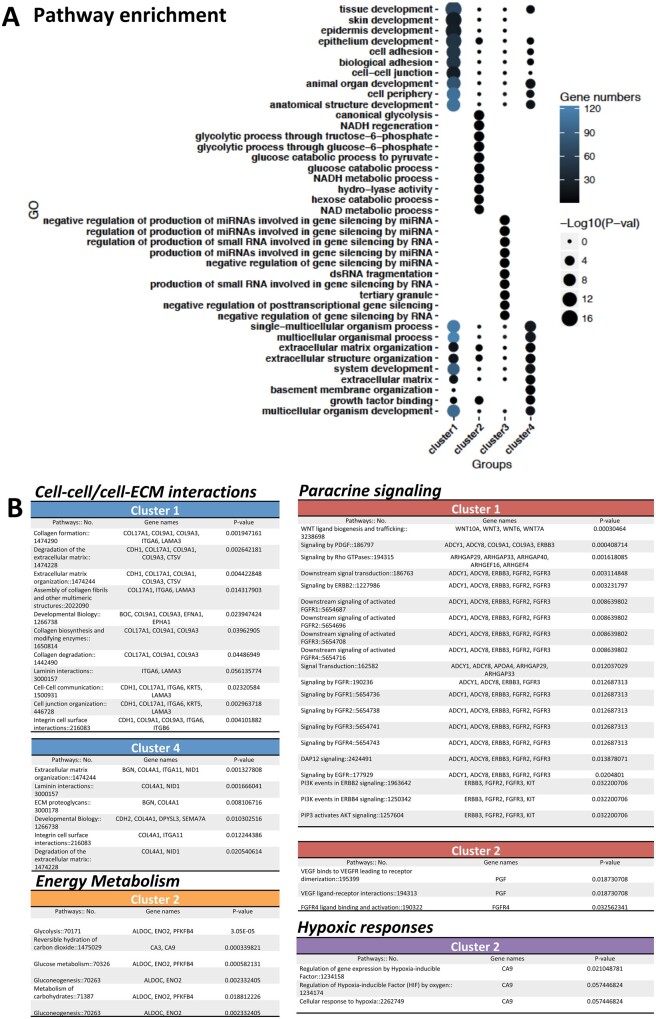
We further performed Pathway Enrichment Analysis on the DEGs of 2D and 3D culture samples in response to the hypoxic treatment. The top 10 enriched pathways in each cluster and the corresponding gene numbers and P-values are shown in Figure 4A. Detailed information on pathways and genes is summarized in Figure 4B, which shows that four key pathways in each cluster were significantly regulated in response to the culture model and hypoxia: cell–extracellular matrix (ECM)/cell–cell interactions, energy metabolism, hypoxic response, and paracrine signalling.
Figure 4.
Enrichment analysis in the groups of 2D-Normoxia, 2D-Hypoxia, 3D-Normoxia, and 3D-Hypoxia. (A) Top 10 pathways in four clusters by GO enrichment analysis. (B) Corresponding genes of the activated pathways with GO numbers in the following four categories: cell–cell/cell–ECM interactions, energy metabolism, hypoxic responses, and paracrine signalling.
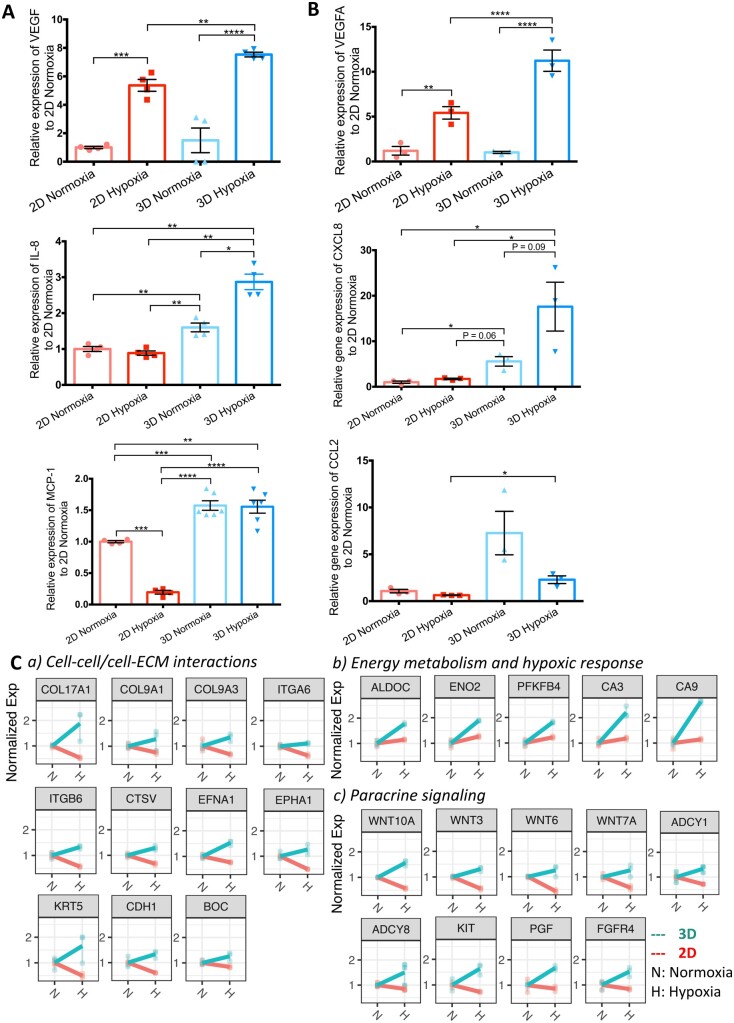
Differential enrichment was found when comparing 2D and 3D culture conditions as shown in Figure 4A and B by applying gene ontology (GO) and reactome enrichment analysis (Supplementary material online, Figure S5). In comparison to the 2D culture, iPSC-CMs interacted more with the microenvironment via ECM in the 3D culture via collagen formation, degradation of the ECM, ECM organization, integrin cell surface interactions, cell–cell communications, and cell junction organizations in Cluster 1. A number of these hub-genes are highlighted in Figure 4B: COL17A1, COL9A1, COL9A3, ITGA6, LAMA3, and CDH1.
In Cluster 2 (Figure 4B), a number of energy metabolism-related pathways were up-regulated in the 3D culture, including glycolysis, glucose metabolism, and metabolism of carbonates. We also found the ischaemia-induced hypoxic responses were differentially modulated between the 2D and 3D cultures: regulation of gene expression by a hypoxia-inducible factor, and cellular response to hypoxia. The genes involved in energy metabolism were ALDOC, ENO2, and PFKFB4, whereas CA9 appeared in both energy metabolism and the hypoxic response.
Upon exposure to hypoxia, the NHP iPSC-CMs in 3D culture were more likely to engage in paracrine signalling than those in 2D culture; such as signalling by PDGF, as well as Wnt ligand biogenesis and Wnt trafficking. Signalling of FGFR, ERBB2, EGFR, and PI3K was enriched in Cluster 1 (Figure 4B). The genes included ADCY1, ADCY8, ERBB3, FGFR2, and FGFR3. In Cluster 2, the VEGF ligand–receptor interaction pathway was up-regulated in the 3D culture, with the pivotal gene, PGF, playing a significant role.
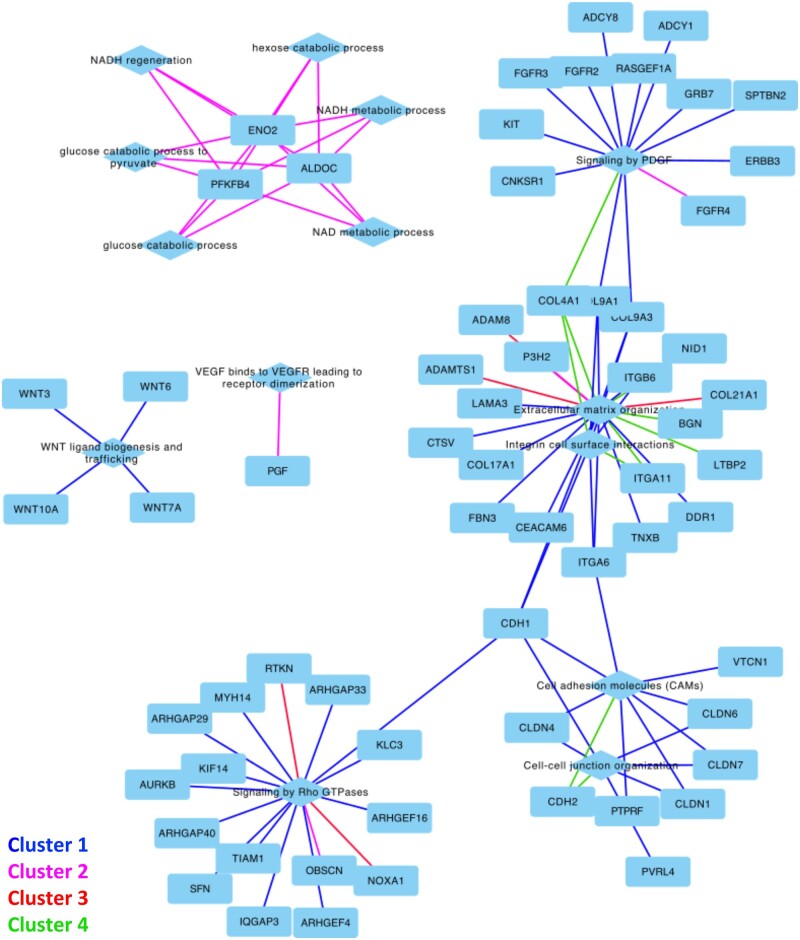
3.6 Gene networks and validation of representative genes in corresponding pathways
The corresponding gene networks in the four clusters are shown in Figure 5 and annotated with corresponding pathways. Specifically, the ECM organization, integrin cell surface interactions, cell adhesion molecules, and cell–cell junction organization in the four clusters bridged the paracrine signalling of PDGF and Rho GTPases. These processes of energy metabolism were enriched in Cluster 2 of glucose catabolic process and NADH regeneration. The WNT ligand biogenesis and trafficking, as well as VEGF binding to VEGFR that led to receptor dimerization, were also significantly represented in Clusters 1 and 2, respectively.
Figure 5.
Networks of selective genes and pathways. The corresponding four clusters by the RNA-seq enrichment analysis interconnect genes and pathways belonging to the cell–cell/cell–ECM interactions, energy metabolism, hypoxic responses, and paracrine signalling.
Moreover, the secreted VEGF, IL-8, and MCP-1 proteins were examined before and after hypoxic treatment in both 2D and 3D, along with the corresponding VEGFA, CXCL8, and CCL2 gene expressions under normoxic and hypoxic conditions. As demonstrated in Figures 6A and B, the expression of VEGF and IL-8 protein and VEGFA and CXCL8 increased under hypoxic treatment in both 2D and 3D cultures in comparison to normoxia; however, the 3D culture had significantly higher expression of VEGF and VEGFA and IL-8 and CXCL8 in response to hypoxia compared with the 2D culture (P < 0.01). The decreases of MCP-1 and CCL2 were found in the 2D hypoxia condition, while relatively fewer decreases were noticed in the 3D hypoxia condition.
Figure 6.
Validation of gene and protein expressions in four groups: 2D-Normoxia, 2D-Hypoxia, 3D-Normoxia, and 3D-Hypoxia. (A) Relative VEGF, IL-8, and MCP-1 secretions. N = 4∼6 biological samples in each group, Student’s t-test. (B) Relative VEGFA, CXCL8, and CCL2 expressions. N = 3 biological samples in each group, Student’s t-test. (C) Profile of key genes expressed in 2D and 3D culture models under normoxic or hypoxic conditions to validate the enrichment analysis in the categories: cell–cell/cell–ECM interactions, energy metabolism, hypoxic responses, and paracrine signalling, N = 3 biological samples in each group.
The corresponding key genes and their relative expression are further noted in Figure 6C. Cell–cell/cell–ECM interaction-related pathways, as well as the expression of COL17A1, COL9A1, COL9A3, ITGA6, LAMA3, CDH1, CTSV, BOC, EFNA1, EPHA1, ITGB6, and KRT5 were significantly increased in the 3D culture under hypoxia compared to 2D conditions, whereas the expression of BGN, COL4A1, ITGA11, NID1, CDH2, DPYSL3, and SEMA7A had no significant changes when the 2D and 3D cultures were compared under ischaemic conditions. Energy metabolism and hypoxic response-related pathways, as well as the expression of ALDOC, ENO2, PFKFB4, CA3, and CA9, were significantly up-regulated in 3D culture compared to 2D hypoxic conditions. Paracrine signalling-related pathways such as WNT10A, WNT3, WNT6, WNT7A, ADCY1, ADCY8, ERBB3, ADCY1, ADCY8, ERBB3, FGFR2, FGFR3, KIT, PGF, and FGFR4 were also up-regulated under the 3D hypoxic conditions.
We further validated our results by comparing them with public microarray data from heart failure patients’ post-MI. The genes from four clusters were analysed and are summarized in Supplementary material online, Figure S6. We found a similar trend of higher DEGs in the MI group in comparison to healthy controls (Supplementary material online, Figure S6A). The same set of genes from Figure 6C, related to the pathways of cell–ECM/cell–cell interactions, energy metabolism/hypoxia, and paracrine signalling were selected and compared, suggesting higher gene expressions in MI patients, as demonstrated in the 3D hypoxic group shown in Supplementary material online, Figure S6B–D.
4. Discussion
Recently, iPSC-CMs have been extensively employed in cell therapy-based small and large animal (e.g. NHP) preclinical trials to treat MI.6,8,9 Prior to in vivo cell delivery, iPSC-CMs are prepared either in 2D monolayer or as a 3D bioengineered construct.6,8,9 How the iPSC-CMs under the different culture models respond to the ischaemic microenvironment at the transcriptional level remains unclear. In this study, we established a simple and direct comparison of NHP iPSC-CMs within the 2D monolayer and 3D EHT models, focusing on the transcriptional signatures of the ischaemic niche. Based on our results, the NHP iPSC-CMs cultured in the different culture conditions responded differently to hypoxia. In general, the responses can be summarized into the following categories: cell–cell/cell–ECM interactions, energy metabolism, hypoxic responses, and paracrine signalling.
4.1 Cell–cell/cell–ECM interactions
Cell–cell/cell–ECM interactions were one of the primary responses to the cell culture model and hypoxia. We found that COL9A1, ITGB6, CTSV, and EPHA1 exhibited higher expression in the 3D than the 2D model under hypoxic conditions. Recently, COL9A1 was found to be one of the key markers in response to low oxygen in ischaemic cardiomyopathy.22 Similarly, ITGB6 was also up-regulated in the hypoxia-related pathways as an adhesion receptor for signalling from ECM to cells.23–25 Both COL9A1 and ITGB6 were up-regulated in the 3D model but down-regulated in 2D, suggesting that the 3D culture better recapitulates the host myocardium than the 2D culture in terms of cell–ECM interactions in ischaemia. The up-regulation of EPHA1 in our 3D model was also observed in the 3D culture of melanoma cells, which has a higher EPHA1 expression level when compared to the 2D culture in response to hypoxia.26 Additionally, the expression of Epha1 in the murine heart was also shown to be enhanced post-MI.27 EPHA1 belongs to the ephrinA ligand family and plays a key role in cell–cell interactions via the ephrin–eph signalling.28 This finding suggests that 3D culture better recapitulates the host myocardium in terms of cell–cell interactions in response to ischaemia.
4.2 Energy metabolism and hypoxic responses
When the myocardium suffers from ischaemia, the oxygen consumption and ATP production in the heart are greatly reduced; to compensate for this, glycolysis and lactate production are accelerated.29 In our in vitro models, we showed that the relevant key genes (ALDOC, ENO2, PFKFB4, CA3, and CA9) were up-regulated in both 2D and 3D cultures. These are significant hypoxic regulators in energy metabolism,30–32 indicating NHP iPSC-CMs in both culture models displayed the same trend of response to hypoxia. In other studies, when rodent CMs were exposed to hypoxic conditions for 24 h, the expression of ALDOC was augmented, and the increased glycolytic flux further demanded an increased expression of genes involved in glycolysis.33 When the metabolic pathways of the neonatal mouse heart at Days 1 and 7 were compared, ENO2 was found to be more highly expressed in the glycolysis-dominated heart.34 The expression of the enzyme carbonic anhydrase family is also significantly increased in human heart failure and cardiac hypertrophy patients.35 Interestingly, the NHP iPSC-CMs in 3D culture had greater gene expression fold-changes than those in 2D culture, an outcome that was also found in the melanoma cells cultured in 3D when compared to 2D.26 Moreover, earlier mesoderm formation and faster cardiac maturation were realized in the 3D iPSC cardiac differentiation conditions when compared to the 2D differentiation. In addition, the RNA-seq comparison between 3D and 2D cardiac differentiation suggested that the activation of hypoxia/energy metabolism-related pathways might be responsible for these findings.36 Overall, the 3D culture more efficiently recapitulates the hypoxic responses and energy metabolism when compared to 2D culture.
4.3 Paracrine signalling
In our system, we also highlight the top enriched genes/pathways of paracrine signalling, including WNT, PDGF, FGFR, EGFR, PI3K, and VEGF. These pathways showed opposite responses between 2D and 3D cultures in response to hypoxia. The WNT pathway is known to play a key role in cardiac development and cardiomyopathy.37,38 Canonical Wnt signalling actively contributes to cardiac myogenesis via the Wnt/β-catenin pathways,39,40 and further promotes left ventricular growth.41 The recapitulation of Wnt up-regulation in heart failure patients by our 3D hypoxic culture provides a practical in vitro model for studying the role of the Wnt pathway on pathological remodelling of the infarcted heart.38 Another key paracrine signalling family, FGF/FGFR, also has been shown to play a central role in cardiac development: FGFR4 is exclusively activated by FGF23 on CMs, whereas FGFR2 and the augmented expression of FGFR4 are associated with left ventricular hypertrophy.42,43 Typically, FGF21 efficiently binds and activates FGFR1c, FGFR2c, and FGFR3c with βKlotho to prevent cardiac hypertrophy in a paracrine manner.44 Importantly, we found the up-regulation of PGF in the 3D hypoxic culture serves as a paracrine mediator of cardiac protection under stress.45PGF has also been shown to promote cardiac growth and angiogenesis, further correlating with elevated VEGF secretion during hypoxia.26,45,46 Enhanced angiogenesis is also reflected by the increase of IL-8 secretion and CXCL8 expression.47,48
Previous studies have shown that transplantation of a 3D cardiac patch typically results in a higher cell engraftment rate over a relatively long period of time when compared to direct injection,8,9,49 suggesting the CMs in the 3D patch can better adapt to the ischaemic microenvironment. In our study, we found that NHP iPSC-CMs in the 3D culture model demonstrated similar responses to the host myocardium under hypoxic conditioning with respect to the cell–cell/cell–ECM interactions, energy metabolism, paracrine signalling, and hypoxic response to promote cardiac development and growth. In addition, the enhanced cardiac protection, cardiac growth, and angiogenesis may be better achieved by iPSC-CMs in the 3D culture model. The goal of in vitro hypoxic conditioning of the iPSC-CMs in this study was to recapitulate the in vivo ischaemia milieu of post-MI hearts. However, we acknowledge limitations such as the lack of cells of the immune system, intercellular crosstalk, and cell–host interactions that occur in vivo.
5. Conclusions
In this study, we compared two independent cell culture systems: a 2D monolayer and 3D EHT using NHP iPSC-CMs. Modulating exposure to hypoxia was used to recapitulate the in vivo hypoxic setting of transplanted NHP iPSC-CMs in the infarcted myocardium. Through systematic comparisons at the transcriptome level, we found that NHP iPSC-CMs in the 3D EHT had similar hypoxic responses to in vivo ischaemia in terms of cell–ECM/cell–cell interaction, energy metabolism, and paracrine signalling, with elevation of key gene expression and corresponding pathway activation. This transcriptional profiling of NHP iPSC-CMs in response to different culture conditions enhances our understanding of the mechanism of ischaemic injury, which may prove useful for advancing cell therapy delivery approaches in the future.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Authors’ contributions
H.Y. and O.J.A. fabricated the EHTs. H.Y. conducted the hypoxic treatment. N.S. performed the transcriptome analysis. H.C., X.Z., and M.Z. prepared the RNA-seq sample. H.C. performed the RT–qPCR. A.H., M.A., T.C., and N.J.C. performed the NHP iPSC reprogramming and iPSC-CM differentiation. H.Y. and C.T. performed cell and EHT immunostaining and confocal imaging. H.W. and I.I. recorded and analysed the electrophysiological signals of iPSC-CMs. A.F.T. provided rhesus monkey fibroblasts and edited the manuscript drafts. H.Y., N.S., and J.C.W wrote the article. J.C.W. provided funding support. All the authors have reviewed the manuscript.
Supplementary Material
Acknowledgements
The authors thank Dr Andrew Olsen from Stanford Neuroscience Microscopy Service for the support of confocal imaging of iPSC-CMs and EHTs. This publication was supported in part by the American Heart Association (AHA) Postdoctoral Fellowship Award 18POST34030106 (H.Y.), Steven M. Gootter Foundation seed grant (O.J.A. and H.Y.), National Institutes of Health (NIH) K01 HL130608 (O.J.A.), AHA 18CDA34110293 (M.Z.), Center for Fetal Monkey Gene Transfer for Heart, Lung, and Blood Diseases NIH R24 HL085794 (A.F.T.), California National Primate Research Center base operating grant NIH P51-OD011107 (A.F.T.), NIH R01 HL141371 (J.C.W.), NIH R01 HL141851 (J.C.W.), NIH R01 HL145676 (J.C.W.), NIH UG3 TR002588 (J.C.W.), and California Institute of Regenerative Medicine RT3-07798 (J.C.W.).
Conflict of interest: J.C.W. is a co-founder of Khloris Biosciences but has no competing interests, as the work presented here is completely independent.
Data availability
The data underlying this article are available in the Gene Expression Omnibus (GEO) Database at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc=GSE156237 and can be accessed with GSE156237.
Translational perspective
Stem cell therapy has shown promise for treating ischaemic heart tissue. However, how stem cells respond following different delivery method is unclear. Here, hypoxic conditioning was applied to non-human primate iPSC-CMs in 2D monolayer culture and 3D engineered heart tissue to model cell injection vs. patch implantation, respectively, in an ischaemic milieu. The differential transcriptome of hypoxic effects on iPSC-CMs show up-regulation of ECM–cell/cell–cell interactions (COL9A1, ITGB6, CTSV, and EPHA1), energy metabolism/hypoxia (ALDOC, ENO2, PFKFB4, CA3, and CA9), and paracrine signalling (WNT, PDGF, FGFR, EGFR, PI3K, and VEGF) in the 3D format, which suggest engineered heart tissue as more suitable model for evaluating cardiac regenerative therapy.
References
- 1. Anderson JL, Morrow DA.. Acute myocardial infarction. N Engl J Med 2017;376:2053–2064. [DOI] [PubMed] [Google Scholar]
- 2. Benjamin EJ, Blaha MJ, Chiuve SE, Cushman M, Das SR, Deo R, de Ferranti SD, Floyd J, Fornage M, Gillespie C, Isasi CR, Jimenez MC, Jordan LC, Judd SE, Lackland D, Lichtman JH, Lisabeth L, Liu S, Longenecker CT, Mackey RH, Matsushita K, Mozaffarian D, Mussolino ME, Nasir K, Neumar RW, Palaniappan L, Pandey DK, Thiagarajan RR, Reeves MJ, Ritchey M, Rodriguez CJ, Roth GA, Rosamond WD, Sasson C, Towfighi A, Tsao CW, Turner MB, Virani SS, Voeks JH, Willey JZ, Wilkins JT, Wu JH, Alger HM, Wong SS, Muntner P.. American Heart Association Statistics C and Stroke Statistics S. Heart disease and stroke statistics-2017 update: a report from the American Heart Association. Circulation 2017;135:e146–e603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Taqueti VR, Solomon SD, Shah AM, Desai AS, Groarke JD, Osborne MT, Hainer J, Bibbo CF, Dorbala S, Blankstein R, Di Carli MF.. Coronary microvascular dysfunction and future risk of heart failure with preserved ejection fraction. Eur Heart J 2018;39:840–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Doppler SA, Deutsch MA, Serpooshan V, Li G, Dzilic E, Lange R, Krane M, Wu SM.. Mammalian heart regeneration: the race to the finish line. Circ Res 2017;120:630–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chong JJ, Yang X, Don CW, Minami E, Liu YW, Weyers JJ, Mahoney WM, Van Biber B, Cook SM, Palpant NJ, Gantz JA, Fugate JA, Muskheli V, Gough GM, Vogel KW, Astley CA, Hotchkiss CE, Baldessari A, Pabon L, Reinecke H, Gill EA, Nelson V, Kiem HP, Laflamme MA, Murry CE.. Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature 2014;510:273–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shiba Y, Gomibuchi T, Seto T, Wada Y, Ichimura H, Tanaka Y, Ogasawara T, Okada K, Shiba N, Sakamoto K, Ido D, Shiina T, Ohkura M, Nakai J, Uno N, Kazuki Y, Oshimura M, Minami I, Ikeda U.. Allogeneic transplantation of iPS cell-derived cardiomyocytes regenerates primate hearts. Nature 2016;538:388–391. [DOI] [PubMed] [Google Scholar]
- 7., Zhu K, Wu Q, Ni C, Zhang P, Zhong Z, Wu Y, Wang Y, Xu Y, Kong M, Cheng H, Tao Z, Yang Q, Liang H, Jiang Y, Li Q, Zhao J, Huang J, Zhang F, Chen Q, Li Y, Chen J, Zhu W, Yu H, Zhang J, Yang HT, Hu X, Wang J. Lack of remuscularization following transplantation of human embryonic stem cell-derived cardiovascular progenitor cells in infarcted nonhuman primates. Circ Res 2018;122:958–969., , , [DOI] [PubMed] [Google Scholar]
- 8. Tiburcy M, Hudson JE, Balfanz P, Schlick S, Meyer T, Chang Liao ML, Levent E, Raad F, Zeidler S, Wingender E, Riegler J, Wang M, Gold JD, Kehat I, Wettwer E, Ravens U, Dierickx P, van Laake LW, Goumans MJ, Khadjeh S, Toischer K, Hasenfuss G, Couture LA, Unger A, Linke WA, Araki T, Neel B, Keller G, Gepstein L, Wu JC, Zimmermann WH.. Defined engineered human myocardium with advanced maturation for applications in heart failure modeling and repair. Circulation 2017;135:1832–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weinberger F, Breckwoldt K, Pecha S, Kelly A, Geertz B, Starbatty J, Yorgan T, Cheng KH, Lessmann K, Stolen T, Scherrer-Crosbie M, Smith G, Reichenspurner H, Hansen A, Eschenhagen T.. Cardiac repair in guinea pigs with human engineered heart tissue from induced pluripotent stem cells. Sci Transl Med 2016;8:363ra148. [DOI] [PubMed] [Google Scholar]
- 10. Liu YW, Chen B, Yang X, Fugate JA, Kalucki FA, Futakuchi-Tsuchida A, Couture L, Vogel KW, Astley CA, Baldessari A, Ogle J, Don CW, Steinberg ZL, Seslar SP, Tuck SA, Tsuchida H, Naumova AV, Dupras SK, Lyu MS, Lee J, Hailey DW, Reinecke H, Pabon L, Fryer BH, MacLellan WR, Thies RS, Murry CE.. Human embryonic stem cell-derived cardiomyocytes restore function in infarcted hearts of non-human primates. Nat Biotechnol 2018;36:597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhao X, Chen H, Xiao D, Yang H, Itzhaki I, Qin X, Chour T, Aguirre A, Lehmann K, Kim Y, Shukla P, Holmstrom A, Zhang JZ, Zhuge Y, Ndoye BC, Zhao M, Neofytou E, Zimmermann WH, Jain M, Wu JC.. Comparison of non-human primate versus human induced pluripotent stem cell-derived cardiomyocytes for treatment of myocardial infarction. Stem Cell Rep 2018;10:422–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rogers J, Gibbs RA.. Comparative primate genomics: emerging patterns of genome content and dynamics. Nat Rev Genet 2014;15:347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Haraguchi Y, Shimizu T, Yamato M, Okano T.. Concise review: cell therapy and tissue engineering for cardiovascular disease. Stem Cells Transl Med 2012;1:136–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14., Nguyen PK, Rhee JW, Wu JC. Adult stem cell therapy and heart failure, 2000 to 2016: a systematic review. JAMA Cardiol 2016;1:831–841., , [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15., Liu C, Oikonomopoulos A, Sayed N, Wu JC. Modeling human diseases with induced pluripotent stem cells: from 2D to 3D and beyond. Development 2018;145:dev156166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. , Huang K, Ozpinar EW, , Su T, Tang J, Shen D, Qiao L, Hu S, Li Z, Liang H, Mathews K, Scharf V, Freytes DO, Cheng K.. An off-the-shelf artificial cardiac patch improves cardiac repair after myocardial infarction in rats and pigs. Sci Transl Med 2020;12. eaat9683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Menasche P, Vanneaux V, Hagege A, Bel A, Cholley B, Cacciapuoti I, Parouchev A, Benhamouda N, Tachdjian G, Tosca L, Trouvin JH, Fabreguettes JR, Bellamy V, Guillemain R, Suberbielle Boissel C, Tartour E, Desnos M, Larghero J.. Human embryonic stem cell-derived cardiac progenitors for severe heart failure treatment: first clinical case report. Eur Heart J 2015;36:2011–2017. [DOI] [PubMed] [Google Scholar]
- 18. Cambria E, Pasqualini FS, Wolint P, Gunter J, Steiger J, Bopp A, Hoerstrup SP, Emmert MY.. Translational cardiac stem cell therapy: advancing from first-generation to next-generation cell types. NPJ Regen Med 2017;2:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Abilez OJ, Tzatzalos E, Yang H, Zhao MT, Jung G, Zollner AM, Tiburcy M, Riegler J, Matsa E, Shukla P, Zhuge Y, Chour T, Chen VC, Burridge PW, Karakikes I, Kuhl E, Bernstein D, Couture LA, Gold JD, Zimmermann WH, Wu JC.. Passive stretch induces structural and functional maturation of engineered heart muscle as predicted by computational modeling. Stem Cells 2018;36:265–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Anders S, McCarthy DJ, Chen YS, Okoniewski M, Smyth GK, Huber W, Robinson MD.. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc 2013;8:1765–1786. [DOI] [PubMed] [Google Scholar]
- 21. Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gil-Cayuela C, Rivera M, Ortega A, Tarazón E, Triviño JC, Lago F, González-Juanatey JR, Almenar L, Martínez-Dolz L, Portolés M.. RNA sequencing analysis identifies new human collagen genes involved in cardiac remodeling. J Am Coll Cardiol 2015;65:1265–1267. [DOI] [PubMed] [Google Scholar]
- 23. Zhuang Y, Gong YJ, Zhong BF, Zhou Y, Gong L.. Bioinformatics method identifies potential biomarkers of dilated cardiomyopathy in a human induced pluripotent stem cell-derived cardiomyocyte model. Exp Ther Med. 2017;14:2771–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Diamanti D, Mori E, Incarnato D, Malusa F, Fondelli C, Magnoni L, Pollio G.. Whole gene expression profile in blood reveals multiple pathways deregulation in R6/2 mouse model. Biomark Res 2013;1:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chakraborty D, Cui W, Rosario GX, Scott RL, Dhakal P, Renaud SJ, Tachibana M, Rumi MA, Mason CW, Krieg AJ, Soares MJ.. HIF-KDM3A-MMP12 regulatory circuit ensures trophoblast plasticity and placental adaptations to hypoxia. Proc Natl Acad Sci U S A 2016;113:E7212–E7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ghosh S, Spagnoli GC, Martin I, Ploegert S, Demougin P, Heberer M, Reschner A.. Three-dimensional culture of melanoma cells profoundly affects gene expression profile: a high density oligonucleotide array study. J Cell Physiol 2005;204:522–531. [DOI] [PubMed] [Google Scholar]
- 27. DuSablon A, Parks J, Whitehurst K, Estes H, Chase R, Vlahos E, Sharma U, Wert D, Virag J.. EphrinA1-Fc attenuates myocardial ischemia/reperfusion injury in mice. PLoS One 2017;12:e0189307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dries JL, Kent SD, Virag JA.. Intramyocardial administration of chimeric ephrinA1-Fc promotes tissue salvage following myocardial infarction in mice. J Physiol 2011;589:1725–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Opie LH. Myocardial ischemia–metabolic pathways and implications of increased glycolysis. Cardiovasc Drug Ther 1990;4:777–790. [DOI] [PubMed] [Google Scholar]
- 30. Staudacher JJ, Naarmann-de Vries IS, Ujvari SJ, Klinger B, Kasim M, Benko E, Ostareck-Lederer A, Ostareck DH, Bondke Persson A, Lorenzen S, Meier JC, Blüthgen N, Persson PB, Henrion-Caude A, Mrowka R, Fähling M.. Hypoxia-induced gene expression results from selective mRNA partitioning to the endoplasmic reticulum. Nucleic Acids Res 2015;43:3219–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pegoraro C, Figueiredo AL, Maczkowiak F, Pouponnot C, Eychene A, Monsoro-Burq AH.. PFKFB4 controls embryonic patterning via Akt signalling independently of glycolysis. Nat Commun 2015;6:5953. [DOI] [PubMed] [Google Scholar]
- 32. Mirtschink P, Krek W.. Hypoxia-driven glycolytic and fructolytic metabolic programs: pivotal to hypertrophic heart disease. Biochim Biophys Acta 2016;1863:1822–1828. [DOI] [PubMed] [Google Scholar]
- 33. Jian B, Wang D, Chen D, Voss J, Chaudry I, Raju R.. Hypoxia-induced alteration of mitochondrial genes in cardiomyocytes: role of Bnip3 and Pdk1. Shock 2010;34:169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lalowski MM, Bjork S, Finckenberg P, Soliymani R, Tarkia M, Calza G, Blokhina D, Tulokas S, Kankainen M, Lakkisto P, Baumann M, Kankuri E, Mervaala E.. Characterizing the key metabolic pathways of the neonatal mouse heart using a quantitative combinatorial omics approach. Front Physiol 2018;9:365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Alvarez BV, Quon AL, Mullen J, Casey JR.. Quantification of carbonic anhydrase gene expression in ventricle of hypertrophic and failing human heart. BMC Cardiovasc Disord 2013;13:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Branco MA, Cotovio JP, Rodrigues CAV, Vaz SH, Fernandes TG, Moreira LM, Cabral JMS, Diogo MM.. Transcriptomic analysis of 3D cardiac differentiation of human induced pluripotent stem cells reveals faster cardiomyocyte maturation compared to 2D culture. Sci Rep 2019;9:9229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stylianidis V, Hermans KCM, Blankesteijn WM.. Wnt signaling in cardiac remodeling and heart failure. Handb Exp Pharmacol 2017;243:371–393. [DOI] [PubMed] [Google Scholar]
- 38. Moon J, Zhou H, Zhang LS, Tan W, Liu Y, Zhang S, Morlock LK, Bao X, Palecek SP, Feng JQ, Williams NS, Amatruda JF, Olson EN, Bassel-Duby R, Lum L.. Blockade to pathological remodeling of infarcted heart tissue using a porcupine antagonist. Proc Natl Acad Sci U S A 2017;114:1649–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pahnke A, Conant G, Huyer LD, Zhao Y, Feric N, Radisic M.. The role of Wnt regulation in heart development, cardiac repair and disease: a tissue engineering perspective. Biochem Biophys Res Commun 2016;473:698–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nakamura T, Sano M, Songyang Z, Schneider MD.. A Wnt- and beta-catenin-dependent pathway for mammalian cardiac myogenesis. Proc Natl Acad Sci U S A 2003;100:5834–5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ai D, Fu X, Wang J, Lu MF, Chen L, Baldini A, Klein WH, Martin JF.. Canonical Wnt signaling functions in second heart field to promote right ventricular growth. Proc Natl Acad Sci U S A 2007;104:9319–9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Itoh N, Ohta H, Nakayama Y, Konishi M.. Roles of FGF signals in heart development, health, and disease. Front Cell Dev Biol 2016;4:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Grabner A, Amaral AP, Schramm K, Singh S, Sloan A, Yanucil C, Li J, Shehadeh LA, Hare JM, David V, Martin A, Fornoni A, Di Marco GS, Kentrup D, Reuter S, Mayer AB, Pavenstadt H, Stypmann J, Kuhn C, Hille S, Frey N, Leifheit-Nestler M, Richter B, Haffner D, Abraham R, Bange J, Sperl B, Ullrich A, Brand M, Wolf M, Faul C.. Activation of cardiac fibroblast growth factor receptor 4 causes left ventricular hypertrophy. Cell Metab 2015;22:1020–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Itoh N, Ohta H.. Pathophysiological roles of FGF signaling in the heart. Front Physiol 2013;4:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Accornero F, van Berlo JH, Benard MJ, Lorenz JN, Carmeliet P, Molkentin JD.. Placental growth factor regulates cardiac adaptation and hypertrophy through a paracrine mechanism. Circ Res 2011;109:272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Klapperich CM, Bertozzi CR.. Global gene expression of cells attached to a tissue engineering scaffold. Biomaterials 2004;25:5631–5641. [DOI] [PubMed] [Google Scholar]
- 47. Khurana R, Simons M, Martin JF, Zachary IC.. Role of angiogenesis in cardiovascular disease: a critical appraisal. Circulation 2005;112:1813–1824. [DOI] [PubMed] [Google Scholar]
- 48. Li A, Dubey S, Varney ML, Dave BJ, Singh RK.. IL-8 directly enhanced endothelial cell survival, proliferation, and matrix metalloproteinases production and regulated angiogenesis. J Immunol 2003;170:3369–3376. [DOI] [PubMed] [Google Scholar]
- 49. Riegler J, Tiburcy M, Ebert A, Tzatzalos E, Raaz U, Abilez OJ, Shen Q, Kooreman NG, Neofytou E, Chen VC, Wang M, Meyer T, Tsao PS, Connolly AJ, Couture LA, Gold JD, Zimmermann WH, Wu JC.. Human engineered heart muscles engraft and survive long term in a rodent myocardial infarction model. Circ Res 2015;117:720–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the Gene Expression Omnibus (GEO) Database at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc=GSE156237 and can be accessed with GSE156237.