Abstract
Environmental programs in the United States face technical challenges that inhibit the ability to use diatoms in water quality monitoring and assessment projects. Specifically, inconsistent taxonomy can obscure diatom responses to environmental variables. Problems are the result of (1) limited access to a common set of taxonomic references, especially those that are geographically relevant, (2) inefficient enumeration protocols, (3) lack of complete and transparent documentation of taxa, and (4) limited opportunities for continued education, training, and knowledge sharing. However, robust resources and practices are available to improve diatom data quality and interpretation. Several resources improve diatom data quality, including a publicly accessible taxonomic reference (diatoms.org) and recommended practices. These practices include adoption of the voucher floras, random sample assignment, replicate microscope slides, and improved quality control. Finally, the Society for Freshwater Science Diatom Taxonomic Certification Committee is developing educational materials and certification exams to support practitioner training and to increase the diatom research knowledge base. The resources and practices in this article are broadly applicable to improving basic and applied research on diatoms worldwide.
Diatoms as measures of aquatic health
Diatoms are single-celled microalgae with cell walls composed of silica (Fig. 1). These organisms are the most species-rich protists on earth. Estimates of the number of diatom species are thought to range from 20,000 to 2 million and scientists continue to discover new species. Diatoms live in a wide variety of semiaquatic and aquatic habitats, including moist soils, wetlands, rivers, lakes, and estuaries. Some diatoms can thrive as free-floating cells in the plankton, while others live attached to surfaces within biofilms. Diatoms also play a pivotal role in the earth’s ecosystems as they are important components of the carbon and oxygen cycles and aquatic food webs (https://diatoms.org; Spaulding et al. 2020). Each diatom species has a range of preferred habitat and optimal conditions for growth. As a result, species exhibit distinct tolerance ranges of pH, salinity, and other environmental variables, such as nutrients, metals, and suspended sediments. Diatoms also have traits related to resource acquisition and resilience to various disturbances, so the composition and relative abundance of diatom species paired with trait information can inform environmental assessments. Diatoms are also preserved well in sediments, and therefore aid in reconstruction of past human and natural disturbances (Stoermer and Smol 2010).
FIG 1.
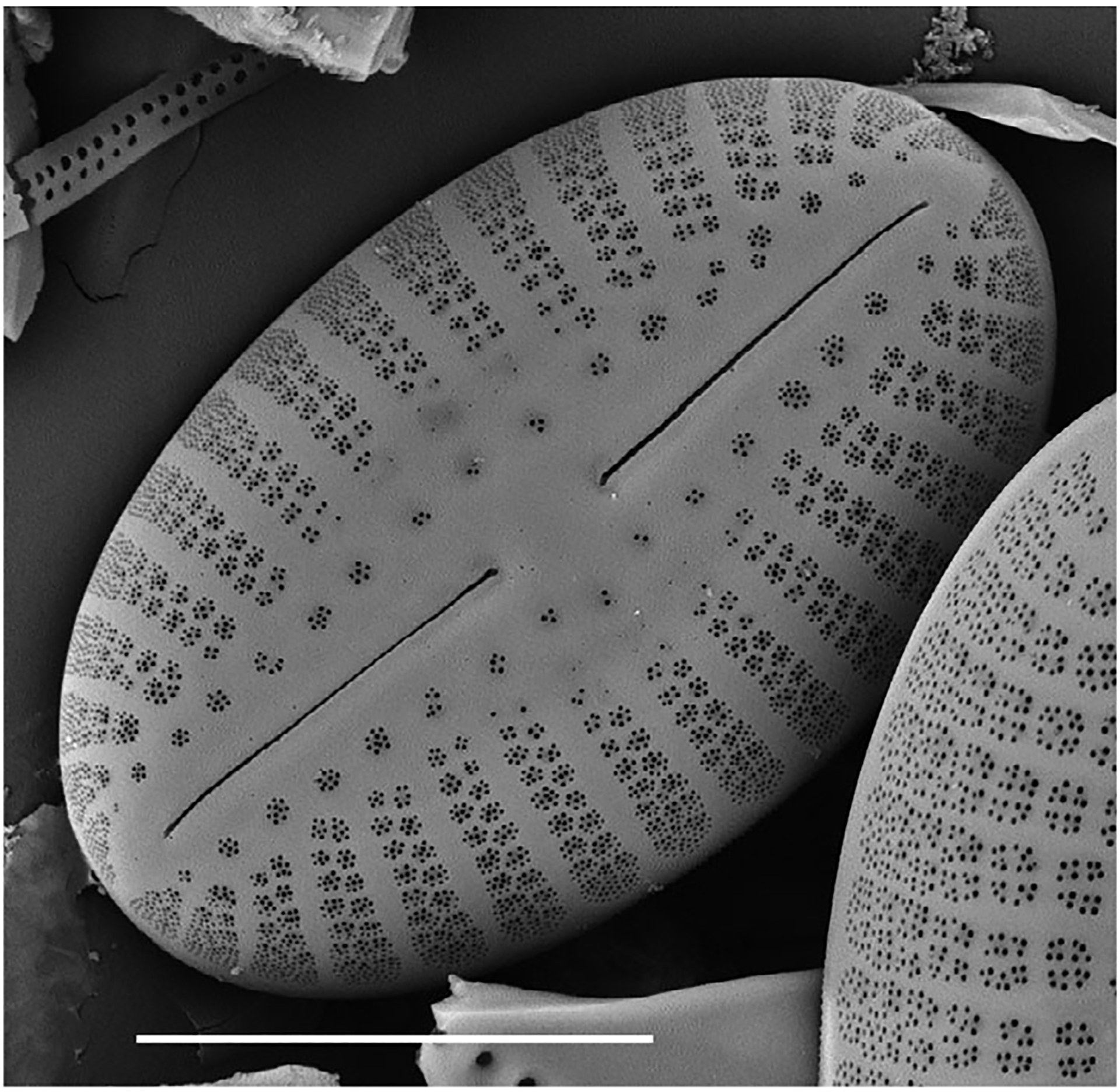
Scanning electron micrograph of Diploneis puella (Schumann) Cleve 1894 (see https://diatoms.org/species/diploneis-puella), a freshwater diatom that is rare in North America. The intricate ornamentation of the silica cell wall aids identification of diatom species. Scale bar equal to 10 μm.
Diatoms in bioassessment
Many state environmental agencies in the United States incorporate diatoms in assessments of biotic integrity to reflect impacts of wastewater, agriculture, urbanization, and atmospheric deposition on water quality. However, the effective use of diatoms in assessments has met challenges because of taxonomic inconsistencies that result in analyst bias in datasets (Kahlert et al. 2016; Werner et al. 2016; Tyree et al. 2020a). Datasets collected by different analysts and/or laboratories may be incompatible and difficult to merge. Taxonomic inconsistencies reduce confidence in the ability to accurately, transparently, and defensibly report biological responses to the environment determined from diatom assemblages (Kahlert et al. 2016).
There are at least four reasons for inconsistent taxonomy in North American datasets. These include (1) reliance on European floras (Kociolek and Spaulding 2000), (2) inefficient enumeration protocols (Bishop et al. 2017), (3) use of different taxonomic references (Tyree et al. 2020a), and finally, (4) a large number of inadequately described or undescribed species (Kociolek and Spaulding 2000; Tyree et al. 2020a). First, in the absence of complete North American references, analysts have relied on European floras to identify North American diatoms. While some North American and European species are shared, many are not. When analysts are faced with an unidentified taxon, it is possible to “force” a name from a reference onto a specimen. Second, analysts are typically required to work under time constraints with limited resources to make species identifications and enumerations. Analysts often lack the opportunity to gain a reasonable understanding of the morphological variation and diversity within samples before making a species identification. Third, different analysts and laboratories often reference a different set of taxonomic resources that range from comprehensive original literature to a small number of European keys, which increases the likelihood of applying different names to the same taxa. Finally, many taxa in North America remain to be described, with 20–25% of species still lacking formal scientific description (Potapova and Charles 2003; Bishop et al. 2017). Many of these taxa have geographic distributions that are regional or even more restricted. Furthermore, these very taxa are often those most sensitive to human disturbance and biodiversity loss (Tyree et al. 2020b). All these factors increase the potential for incorrect identifications and inconsistent taxonomy.
Phosphorus is an important nutrient for diatom growth and influences species composition; however, inconsistent taxonomy can obscure the relationship between diatoms and total phosphorus concentrations (Lee et al. 2019), reducing the value of diatoms as biological indicators and assessment endpoints for water quality (Cao et al. 2007; Werner et al. 2016). Protection of aquatic resources can be vulnerable to contention if there are known errors in the data underlying scientific conclusions.
During the past 10 yr, alternatives to microscopic identification of taxa, such as DNA metabarcoding, have been pursued (Hering et al. 2018; Bailet et al. 2020). Efforts are underway to compare morphological and molecular data to determine the most informative and cost-effective methods for assessments. However, significant challenges remain to improve the cost and reproducibility of metabarcoding outputs (Bailet et al. 2020).
Diatoms of North America
The Diatoms of North America (DONA) website (https://diatoms.org; Spaulding et al. 2020) is a publicly accessible taxonomic reference that represents a collaborative effort to document and provide accurate information about North American diatoms. An increasing number of analysts rely on DONA to identify diatom species and it is required for use in state and federal task agreements. The web resource is based on content provided by over 100 contributors, managed by an Editorial Review Board.
The DONA website provides practitioners with access to information about diatom morphology, nomenclature, ecology, and geographic distribution. It supports accurate identification of diatoms by providing users with an easily accessible, user-friendly, and geographically relevant taxonomic source. Unlike dichotomous keys, such as those for macroinvertebrates and other organisms, DONA uses a multichotomous visual key. Whereas a dichotomous key uses a fixed sequence of identification steps, a multichotomous key presents the user with several characteristics to evaluate in any sequence. For example, many popular bird guides present multiple features of each species as a multichotomous key.
DONA’s visual key allows users to quickly find taxa by their shape (Fig. 2). Users less familiar with diatoms can begin the identification process with the nine basic morphological groups, then the subpages of genera within each group, and continue toward finer-scale taxonomy by studying individual taxon pages. Tools (slider bars) are available to filter subsets of taxa by size and/or stria density. To help practitioners distinguish between closely related species, each taxon page has a “compare” feature. This feature presents species side by side at a common scale of magnification and aids in reaching a correct identification.
FIG 2.
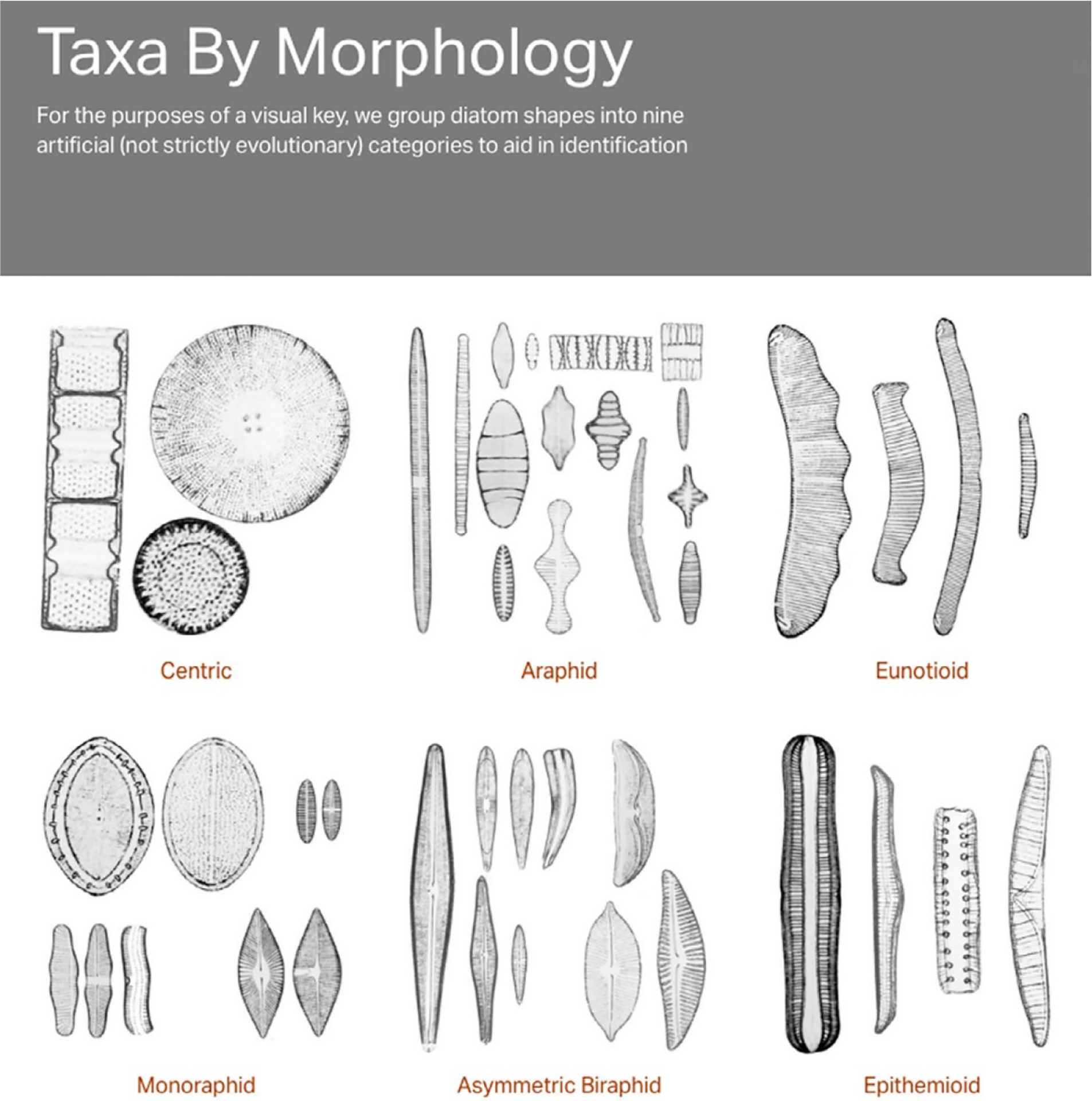
The Diatoms of North America (DONA) website allows practitioners to use a visual key to morphology (https://diatoms.org/morphology). Here, six of the nine possible morphologies are illustrated. Taxonomists can work their way to an identification of a group, genus, or species by following the image and text features.
Each taxon page has images that capture a species’ range of morphological variation, cell wall structures, size range, autecological information, original descriptions, and relevant citations and links. The autecological information includes species traits, such as motility, attachment, growth form (i.e., single or colony), preferred habitats and waterbodies, eco/physiological features (e.g., nitrogen fixer, sensitivity, or tolerance to stressors), and geographic distribution.
In addition, the website contains resources to aid users of differing skill levels. Taxon Updates feature the most recent pages released to the public. Resources for Practitioners provide descriptive techniques for sampling, slide preparation, and enumeration of samples. This section features articles on how to perform sample digestion, prepare slides, and measure diatom features. News informs users about recent or upcoming events, such as online presentations by the Diatom Web Academy, which are held twice a month, and in 2020, gained over 3000 views.
Voucher floras
While DONA is a valuable taxonomic resource, it does not on its own ensure consistent taxonomic identification. It is important to have documented and transparent taxonomic decisions so that multiple datasets can be combined and shared across analysts, temporal ranges, and spatial scales (e.g., state boundaries). The process used to develop project voucher floras meets this need by providing analysts a common voucher or morphological guide to use during diatom enumeration. Multi-analyst teams using a voucher flora approach are more likely to create datasets without analyst bias.
A voucher flora is a collection of diatom images, grouped by species, organized into “plates” (Fig. 3), and labeled by codes (Bishop et al. 2017). These codes are known as operational taxonomic units (OTUs) and are used to group diatoms based on their morphology. Voucher floras are developed for a project, typically for a specific geographic region and habitat type (e.g., southeast U.S. rivers). Even for skilled analysts, the numerous diatom species described in books, journal articles, databases, and other references can be overwhelming when working with diatoms from an unfamiliar or large geographic area. Voucher floras help analysts focus on a set of OTUs they are likely to encounter as part of a project by providing examples of the morphological variation within a given taxon and the necessary visual clues for taxon identification and distinction from morphologically similar taxa. This helps eliminate force-fitting and encourages more careful consideration of taxa with limited or poorly defined geographic distributions. The use of OTUs during enumeration streamlines the process by delaying assignment of scientific names until after enumeration is completed.
FIG 3.
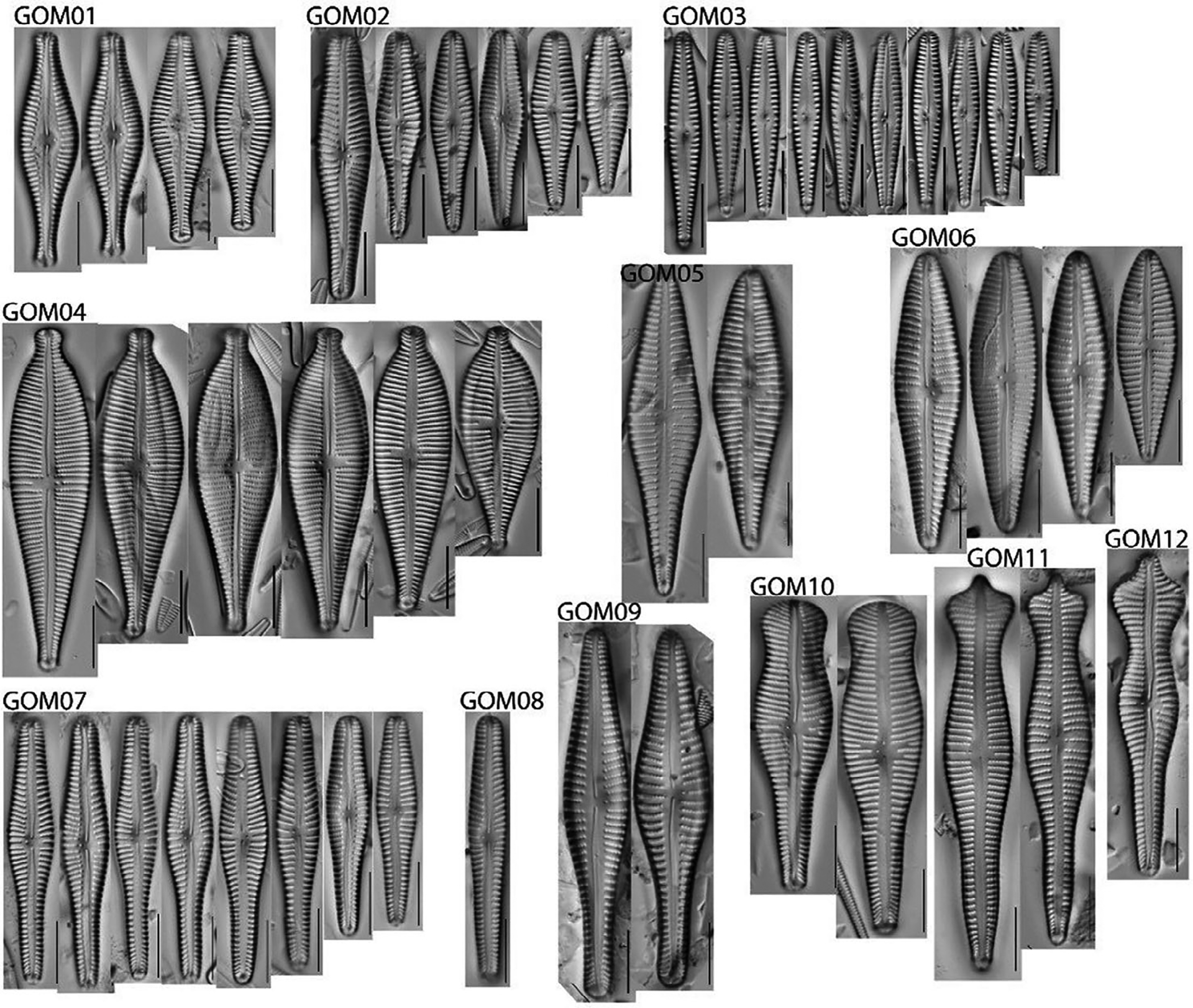
An example of OTUs in the genus Gomphonema in a voucher flora that can be used during enumeration of diatoms to keep track of taxa encountered, facilitate communication among analysts, and document taxonomy in a project. Assignment of scientific names is done after enumeration is finalized.
Shared voucher floras help reduce analyst bias and are composed of key steps. First, a lead analyst conducts an initial survey of samples and creates a precount voucher flora by collecting and organizing specimen images into morphological OTUs. Then, each OTU is assigned a provisional name (e.g., GOM01, GOM02). Analysts further develop the voucher flora during enumeration by adding additional images when they encounter new OTUs and revising OTUs as needed. As a final step in the analysis, scientific names are identified and assigned to OTUs. The voucher serves as a permanent record of the study. Finally, and importantly, the slides are deposited in a public herbarium and can be related back to the voucher flora by future investigators.
The main role of voucher floras is to document OTUs and serve as a common taxonomic guide in a project, whether the project team consists of one or multiple analysts. However, the process of creating and using a voucher flora can especially benefit the ability of a multi-analyst team to work efficiently and maintain taxonomic consistency during enumeration. In addition to collaboratively developing the voucher flora, a multi-analyst team can find inconsistent identifications and correct these while it is still feasible to make revisions and proceed with greater accuracy. This kind of communication and harmonization is even more valuable than analyst experience for producing accurate and consistent data (Kahlert et al. 2009).
A voucher flora increases the transparency of datasets by presenting the range of morphological variation within each taxon’s morphological species concept used in a project. The voucher flora itself allows for taxonomic revisions and/or updates to be made in a systematic and reproducible manner in later projects, if needed. In addition, voucher floras support the use and combination of data from different projects for new statistical analyses; exploration of metrics, indices, and threshold development; and assessment of biological condition at different spatial scales. Voucher floras can be useful for updating nomenclature in existing datasets and for examining trends over time or large spatial scales because the type of documentation compiled during their development allows for traceable accounting of taxonomic discrepancies among datasets.
In summary, using a voucher flora is critical for analysts and data users (e.g., bioassessment programs) because it allows analysts to postpone the process of species identification, minimizing incorrect identification. Moreover, use of voucher floras streamlines the process of species identification, increases analyst efficiency, facilitates taxonomic consistency, and makes taxonomic practice transparent. Several voucher floras from the United States are available for download at: https://diatoms.org/practitioners/what-is-a-voucher-flora.
Other practical ways to improve data quality
In addition to sharing voucher floras, analysts can adopt additional practices to improve data quality. These practices include using Battarbee chambers for slide preparation, randomly assigning samples to analysts, and implementing a multiparty quality control (QC) system (Bishop et al. 2017; Tyree et al. 2020a).
Battarbee chambers are evaporative settling chambers that produce a more homogenous distribution of diatoms on coverslips (Fig. 4, Battarbee 1973). Traditional strewn slides are vulnerable to an uneven diatom distribution because of edge and vibration effects. As a result, strewn slides are more likely to have clumps of valves that are difficult to count, develop an “X” pattern on the coverslip, and have other confounding effects that can introduce errors in quantitative analyses. Because analysts enumerate diatoms from a small portion of the slide (e.g., along a transect or random fields of view), it is important that diatoms are evenly distributed on the slide so that representative subsamples for quantitative analyses are obtained. Moreover, Battarbee chambers facilitate QC by producing four replicate slides that can be shared with other analysts. Projects with a large geographic scope and multi-analyst teams can assign samples to analysts on a random basis. In the past, programs assigned samples from a particular geographic area to project analysts with experience in diatom taxonomy from that geographic area. As a result, it was difficult to untangle the interaction between analyst bias and regional geographic distribution of species. While randomization of samples across analysts does not directly affect analyst bias, it allows clear detection of variation in data due to analysts.
FIG 4.
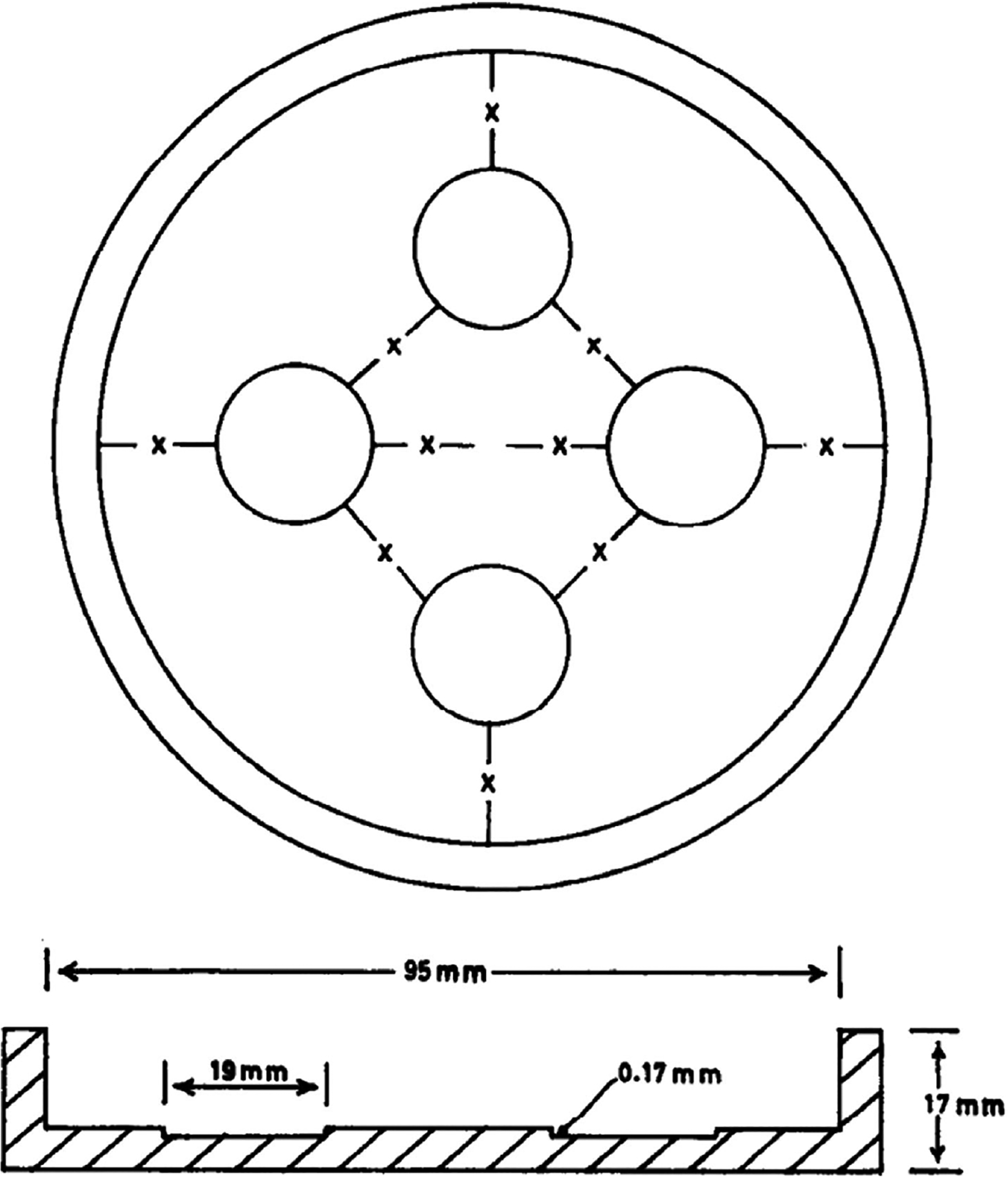
Schematic illustration of a Battarbee chamber (Battarbee 1973). The chamber design allows a sample slurry to settle equally onto four coverslips. Chamber construction has a constant surface area to allow quantitative analyses of species composition. Used with permission.
Implementation of multiparty QC involves assigning analysts 10% of their samples as “self-QC counts,” and 10% of samples from other analysts as “cross-QC counts.” This approach helps assess variation within and among project analysts (i.e., how consistent they are), which are crucial aspects of the confidence and defensibility of datasets. Use of this QC approach also allows separation of analyst taxonomic consistency from the effects of taxon richness.
Examples of improved taxonomic consistency
Two recent projects provide examples of coordination of analysts across the country that produced consistent datasets using voucher floras: the 2014 U.S. Geological Survey Southeast Stream Quality Assessment (USGS SESQA; Bishop et al. 2017) and the 2017 Northeast Lake Sediment Diatom Voucher Flora project (EPA NE Lakes unpubl.). Both studies implemented the previously described protocols (i.e., voucher floras, Battarbee chambers, sample randomization, and multiparty QC), facilitated collaboration among analysts, and, in doing so, arrived at consistent taxonomy. This increased enumeration efficiency and improved data quality and transparency. These efforts minimized errors (Bishop et al. 2017; Tyree et al. 2020a), improved detection of diatom species, and established confidence in the utility of diatom data for bioassessment.
The SESQA voucher flora was developed as part of an assessment of biotic condition across an urban, agricultural, and hydrologic gradient. Algal samples from 108 streams within the Piedmont and southern Appalachian Mountains were examined to create a voucher flora, with over 375 diatom taxa included in the voucher flora (Bishop et al. 2017). The purpose of the NE Lakes voucher flora was to document the diatom taxa within lake sediments of the northeast United States. This voucher flora showed remarkable species richness, as it contained over 1200 OTUs.
For both projects, two or three analysts identified and counted diatom slides using a precount voucher flora that contained an initial set of digital images that represented the taxonomic diversity and morphological range of the species in each region. Analysts met regularly to discuss the criteria used to differentiate morphologically similar taxa and updated OTUs in the voucher flora. After enumerating most of the samples, analysts participated in an expert workshop to finalize the OTUs for the project, assign scientific names to as many OTUs as possible, and discuss the challenges encountered during the process and ideas for improvement. The resulting voucher floras for these projects are compilations of images with their associated OTUs, taxon names, references, and project R scripts that are publicly available for use as a taxonomic resource for species identifications or to compare and/or merge with future voucher floras (https://instaar.colorado.edu/research/labs-groups/diatom-laboratory/research-detail/). Transparent documentation of these project floras will facilitate the incorporation of taxonomic updates, as needed, in the future.
Results from regional projects using the voucher flora approach show evidence of increased taxonomic consistency. Tyree et al. (2020a) showed that in the Midwest Stream Quality Assessment (MSQA), a survey that used traditional taxonomic practices, 14% of the variation in diatom assemblage composition was explained by analyst. In contrast, for SESQA and NE Lakes, which used the proposed protocols, the variation explained by analyst was substantially less: 1% and 4%, respectively (Fig. 5). Despite consistent taxonomic identifications among analysts (i.e., no difference between self- and cross-QC counts), NE Lakes showed an overall low index of taxon similarity in QC samples because of high species richness in the project (Tyree et al. 2020a), as the NE Lakes project included over 1200 taxa. Yet, the multiparty QC system allowed the team to evaluate the analyst performance against sample richness. Analysts reported that using the protocols above made their microscopic tasks straightforward and more enjoyable. Analysts accomplished project goals, appreciated a collaborative environment, and felt rewarded for their expertise.
FIG 5.
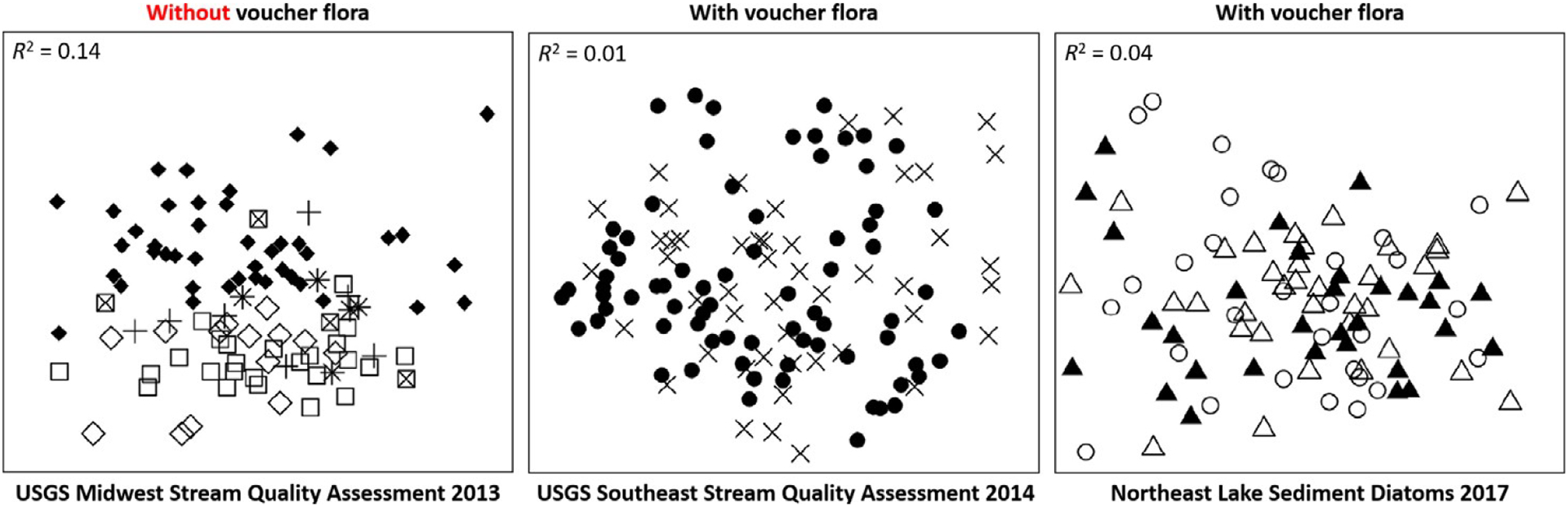
Ordination of diatom species data in nonmetric multidimensional scaling for sites in three regional surveys: Midwest (MSQA), southeast (SESQA), and Northeast Lakes (panels from left to right). Symbols represent analysts. The R2 values represent the amount of variation in the dataset attributable to analyst bias. In MSQA where more variation is explained by analyst, symbols for analysts are clustered. In the SESQA and NE Lakes studies, the symbols distribute more evenly in ordination space because differences in sites is not attributed to analyst. Modified from Tyree et al. (2020a).
Diatom taxonomic certification program
As more agencies adopt the use of diatoms for assessment, it has become desirable to have the ability to certify analysts with expertise needed for this work. A national certification program is supported by the Society for Freshwater Science (SFS) and administered by the Diatom Taxonomic Certification Committee (DTCC). The certification program is designed to encourage the production of high-quality diatom data and taxonomic consistency in large datasets. The objectives of the certification program are to: (1) develop taxonomic expertise of taxonomists through training, (2) encourage new taxonomic experts through professional support, (3) further the technical proficiency of taxonomists, and (4) provide recognition of senior taxonomists committed to teaching and transferring their knowledge to others. A Level 1 certification is available and indicates that an individual has a current working knowledge of the diagnostic characteristics of North American freshwater genera. A Level 2 certification, for species level proficiency, will be announced in 2021. The DTCC regularly holds workshops at the SFS Annual Meeting and online. The DTCC is currently also developing additional training opportunities in the service of accurate and consistent data for assessments and subsequent management decisions.
Practitioners seeking Level 1 certification (genus level) can access study materials: https://diatoms.org/practitioners/diatom-taxonomic-certification.
Practitioners can schedule the Level 1 Certification Exam via the Stroud Water Research Center: https://stroudcenter.org/sfstcp/exam/.
Online training, directed at a range of audiences, is available through Diatom Web Academy: https://diatoms.org/news/diatom-web-academy-1.
For additional information on the Diatom Taxonomic Certification Program, contact the Diatom Taxonomic Certification Committee: diatomtcc@gmail.com.
Summary
Several resources facilitate a high level of diatom data quality and transparency. Academics and managers can sustain the progress achieved to date by encouraging adoption of the activities outlined in this note, including: (1) use of common taxonomic references such as Diatoms of North America (diatoms.org); (2) documentation of taxonomy with a project voucher flora; (3) adoptions of methods for random distribution of diatom cells on microscope slides; (4) implementation of multi-party QC; and (5) participation in professional development and taxonomic certification.
Acknowledgments
This material is based upon work supported in part by the U.S. Geological Survey under Cooperative Agreement G15AC00104. We thank Luisa Riato, Galen Kaufman, and Susan K. Jackson for providing technical reviews. We also thank Meredith Tyree for her contributions to an earlier version of this manuscript.
Footnotes
Publisher's Disclaimer: Disclaimer
Publisher's Disclaimer: The views expressed in this article are those of the authors and do not necessarily reflect the views or policies of the U.S. Environmental Protection Agency. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Conflict of interest
None declared.
Contributor Information
Janice Alers-García, U.S. Environmental Protection Agency, Office of Water, Office of Science and Technology, Washington, DC;.
Sylvia S. Lee, U.S. Environmental Protection Agency, Office of Research and Development, Center for Public Health and Environmental Assessment, Washington, DC;
Sarah A. Spaulding, U.S. Geological Survey, Institute of Alpine and Arctic Research, University of Colorado, Boulder, CO;
References
- Bailet B, Apothéloz-Perret-Gentil L, Baričević A, and others. 2020. Diatom DNA metabarcoding for ecological assessment: Comparison among bioinformatics pipelines used in six European countries reveals the need for standardization. Sci. Total Environ 745: 140948 doi: 10.1016/j.scitotenv.2020.140948. [DOI] [PubMed] [Google Scholar]
- Battarbee RW 1973. A new method for the estimation of absolute microfossil numbers, with reference especially to diatoms. Limnol. Oceanogr 18: 647–653. [Google Scholar]
- Bishop IW, Esposito RM, Tyree M, and Spaulding SA. 2017. A diatom voucher flora from selected southeast rivers. Phytotaxa 332: 101–140. doi: 10.11646/phytotaxa.332.2.1. [DOI] [Google Scholar]
- Cao Y, Hawkins CP., Olson J, and Kosterman MA. 2007. Modeling natural environmental gradients improves the accuracy and precision of diatom-based indicators. J. North Am. Benthol. Soc 26: 566–585. doi: 10.1899/06-078.1. [DOI] [Google Scholar]
- Hering D, Borja A, Iwan Jones J, and others. 2018. Implementation options for DNA-based identification into ecological status assessment under the European Water Framework Directive. Water Res. 138: 192–205. doi: 10.1016/j.watres.2018.03.003. [DOI] [PubMed] [Google Scholar]
- Kahlert M, Albert RL, Anttila EL, and others. 2009. Harmonization is more important than experience—results of the first Nordic–Baltic diatom intercalibration exercise 2007 (stream monitoring). J. Appl. Phycol 21: 471–482. doi: 10.1007/s10811-008-9394-5. [DOI] [Google Scholar]
- Kahlert M, Ács É, Almeida SFP and others. 2016. Quality assurance of diatom counts in Europe: towards harmonized datasets. Hydrobiologia 772: 1–14. doi: 10.1007/s10750-016-2651-8. [DOI] [Google Scholar]
- Kociolek JP and Spaulding SA. 2000. Freshwater diatom biogeography. Nova Hedwigia 71: 223–241. [Google Scholar]
- Lee SS, Bishop IW, Spaulding SA, Mitchell RM, and Yuan LL. 2019. Taxonomic harmonization may reveal a stronger association between diatom assemblages and total phosphorus in large datasets. Ecol. Indic 102: 166–174. doi: 10.1016/j.ecolind.2019.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potapova M and Charles DF. 2003. Distribution of benthic diatoms in U.S. rivers in relation to conductivity and ionic composition. Freshwater Biol. 48: 1311–1328. doi: 10.1046/j.1365-2427.2003.01080.x. [DOI] [Google Scholar]
- Spaulding SA, Bishop IW, Edlund MB, Lee SS, Furey P, Jovanovska E, and Potapova M. 2020. Diatoms of North America. https://diatoms.org/ [Accessed 30 November 2020].
- Stoermer EF, and Smol JP. 2010. The diatoms: applications for the environmental and earth sciences, 2nd Edition. [Google Scholar]
- Tyree MA, Bishop IW, Hawkins CP, Mitchell RM, and Spaulding SA. 2020a. Reduction of taxonomic bias in diatom species data. Limnol. Oceanogr.: Methods 1–9. doi: 10.1002/lom3.10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyree MA, Carlisle DM, and Spaulding SA. 2020b. Improving diatom enumeration methods for use in predictive bioassessment models. Freshw. Sci 39: 183–195. doi: 10.1086/707725. [DOI] [Google Scholar]
- Werner P, Adler S, and Dreßler M. 2016. Effects of counting variances on water quality assessments: implications from four benthic diatom samples, each counted by 40 diatomists. J. Appl. Phycol 28: 2287–2297. doi: 10.1007/s10811-015-0760-9. [DOI] [Google Scholar]


