Abstract
Recent advancement in molecular techniques has spurred waves of studies on responses of microorganisms to lead contamination exposure, leveraging detailed phylogenetic analyses and functional gene identification to discern the effects of lead toxicity on microbial communities. This work provides a comprehensive review of recent research on (1) microbial community changes in contaminated aquatic sediments and terrestrial soils; (2) lead resistance mechanisms; and (3) using lead resistance genes for lead biosensor development. Sufficient evidence in the literature, including both in vitro and in situ studies, indicates that exposure to lead contamination inhibits microbial activity resulting in reduced respiration, suppressed metabolism, and reduced biomass as well as altered microbial community structure. Even at sites where microbial communities do not vary compositionally with contamination levels due to extremely long periods of exposure, functional differences between microbial communities are evident, indicating that some microorganisms are susceptible to lead toxicity as others develop resistance mechanisms to survive in lead contaminated environments. The main mechanisms of lead resistance involve extracellular and intracellular biosorption, precipitation, complexation, and/or efflux pumps. These lead resistance mechanisms are associated with suites of genes responsible for specific lead resistance mechanisms and may serving as indicators of lead contamination in association with dominance of certain phyla. This allows for development of several lead biosensors in environmental biotechnology. To promote applications of these advanced understandings, molecular techniques, and lead biosensor technology, perspectives of future work on using microbial indicators for site ecological assessment is presented.
Keywords: Lead contamination, microbial community, lead resistance, lead biosensors, ecological risk assessment
Graphical Abstract
1. Introduction
Environmental and occupational exposure to lead (Pb) continues to cause health effects globally, primarily through exposure to contaminated air, water, food, soils and household dust (Tong et al., 2000; WHO, 2011). Lead reserves are estimated at 7.1 x 107 tons in ores such as galena (PbS), anglesite (PbSO4) and cerussite (PbCO3), and over the past three centuries, largely due to industrialization, environmental lead levels have significantly increased (U.S. DHHS/ATSDR, 2007). Lead is mined worldwide and purified through smelting, which releases lead into the environment in the form of hazardous fumes, fallout, and dust (WHO/IARC, 2006; U.S. DHHS, 2007). Total lead concentrations on contaminated sites can reach up to 10 000 mg/kg, while the average value in natural soils ranges from 10 to 50 mg/kg. Pb can be found as a co-contaminant with other heavy metals such as aresenic (As), cadmium (Cd), cobalt (Co), copper (Cu), nickel (Ni) and zinc (Zn). Aside from mining and smelting activities, sequestered lead is released into the environment from dredging activities and has been associated with oceanic micro- and macro-plastics (Nayar, Goh, & Chou, 2004; Yang et al., 2019). Lead is a component of many commercial products such as automobile batteries, solder, x-ray machine shielding, and corrosion resistant paints. In the USA, it was used as a paint pigment until 1978, and as lead solder for food cans and an “anti-knock” agent in gasoline until 1995 (U.S. DHHS/ATSDR, 2007). Industrial waste can be contaminated with lead and other heavy metals, which in turn, pollute the surrounding environment (Jiang et al., 2019; Kuppusamy et al., 2016; Li et al., 2017; Shi et al., 2002). Banning lead as a fuel additive resulted in decreasing air concentrations. However, because lead does not degrade in the environment, lead based paint and contaminated soils remain public health concerns.
Lead causes neurological, cardiovascular, renal, hematological, gastrointestinal, musculoskeletal, endocrinological, immunological, reproductive and developmental effects and is listed as a probable human carcinogen (U.S. DHHS, 2007; WHO, 2011; WHO/IARC, 2006). Many countries regulate environmental lead (Tong et al., 2000). In the United States, the Environmental Protection Agency (U.S. EPA) stipulates that the National Ambient Air Quality Standard for lead is 0.15 μg/m3 total suspended particles (Fed Register, 2008; Fed Register, 2016) and 15 μg/L in drinking water (U.S. EPA, 2008). The US EPA’s standard for lead in bare soil in play areas is 400 mg/kg and 1200 mg/kg for non-play areas (U.S. EPA, 2001). Lead is frequently found as a co-contaminant at Superfund sites; there is no reference dose (RfD) for lead, and the risk reduction goal used for cleanup is based on the probability that ≤ 5% children will have a blood lead concentration of ≥ 10 μg/dL (U.S. EPA, 2019).
Cleanup of lead contamination at U.S. Superfund sites includes removal of material, groundwater treatment, engineering controls and other approaches focused on reducing the risk of lead exposure, especially to children, consistent with the U.S. Federal Lead Action Plan (U.S. Executive Office of the President, 2018). As part of the risk assessment, lead bioaccessibility, bioavailability, bioaccumulation, and bioconcentration are considered (U.S. EPA, 2007). Currently, the ecological risk assessment considers higher level organism effects and remains silent on microorganisms and microbial processes due to considerable spatial and temporal variation and experimental uncertainties (U.S. EPA, 2003). However, because microorganisms play a critical role in carbon and nitrogen cycling and other fundamental geochemical processes, groups of European scientists advocate for ecological risk assessments protective of microorganisms (Dahlin, Witter, Martensson et al., 1997; de Vries et al., 2007; Giller, Witter, & McGrath, 2009). Therefore, it is important to understand the impact of lead contamination on microbial community.
Lead is not biologically essential and can be toxic to microorganisms at low concentrations. Once exposed to lead contamination, microorganisms develop a variety of resistance mechanisms that are selective in high lead concentration areas, resulting in an increase in lead resistance genes and changes in microbial community activity and composition (Braud et al., 2009; Naik & Dubey, 2011; O’Brien et al., 2014). These resistance genes have been harnessed in lead biosensors to detect bioavailable lead (Hobman, Julian, & Brown, 2012; Zhang et al.,2017). Furthermore, microbial processes can form insoluble lead precipitates, thus reducing lead bioaccessibility and bioavailability. Twenty-five years ago, microbial process studies focused on measuring respiration, biomass, litter decomposition and enzyme synthesis and activity, and using those end points, critical toxicity concentrations of lead and other heavy metals could be determined (de Vries et al., 2007). Microbial community diversity studies, comparing contaminated and uncontaminated soils and water, used microscopy, plate counting and phospholipid fatty acid analysis (Dahlin et al., 1997; Fakruddin & Mannan, 2013). As molecular approaches, such as 16S rRNA analysis, became more commonplace, higher resolution in microbial community composition was possible. While uncertainties including temporal and spatial variation remain, quantifying microbial resistance genes, indicative of increased environmental lead load or changes in microbial diversity using molecular approaches, can serve as indices of lead contamination and bioavailability to inform both human health and ecological risk assessments.
The objective of this work is to provide a critical review of recent research on the impact of lead contamination on microbial community structure and function as well as lead resistance mechanisms and their use for lead biosensor development for potential application in bioavailability assessment of contaminated sites. The specific objectives are as follows: 1) to evaluate the impact of lead contamination on microbial community structure and function in contaminated terrestrial soils and aquatic sediments; 2) to elucidate various lead resistance mechanisms; and 3) to leverage resistance genes for lead biosensor development to detect lead and assess lead bioavailability. Perspectives of future work on using microbial indicators for site ecological risk assessment is presented.
2. Impact of lead contamination on microbial community structure and function
Understanding the effect of lead contamination on microbial communities, including activity and community composition, is important because microorganisms may serve as a direct or indirect indicator of ecosystem changes or effect of perturbations. Microbial lead toxicity involves displacement or substitution of essential elements in nuclear proteins, inhibition of enzyme activity, and damage to cell membrane or DNA structure (Chen et al., 2018; Tipayno et al., 2018). Thus, suppressed microbial activity is typically observed. For example, in lotic sediments contaminated with heavy metals (Cd, Cr, Cu, Ni, Pb, Zn) from mixed sources (e.g. treated and untreated industrial effluents, urban sewage, atmospheric deposition, rain waste waters), microbial respiration and biomass are negatively correlated with heavy metal concentration (Jaiswal & Pandey, 2018). Microbial activity is assayed by measuring fluorescein diacetate hydrolytic activity, a surrogate for lipase, protease, esterase, alkaline phosphatase, and β-D-glucosidase. Except for alkaline phosphatase (due to high concentrations of phosphate), the other parameters are negatively correlated with heavy metal concentration (Green et al., 2006; Jaiswal & Pandey, 2018). Both bioavailability and toxicity of metals are also negatively correlated with organic matter, possibly reflecting complexation of heavy metals by organic materials (ref).
While lead is bacteriostatic or bactericidal to many microorganisms, some have developed resistance mechanisms that enable them to survive or thrive. Comparative studies between varying levels of contamination often reveal changes in microbial community structure and function (e.g., Akmal et al., 2005; Liu, Lin, Dong, Li, & Liu, 2018). Conventionally, alpha and beta diversity, respiration, biomass, or metabolic enzymes are measured to infer microbial community changes. For example, measurement of phospholipid fatty acids (PLFA) has been used to identify microbial community structure alteration due to metal toxicity (Lenart & Wolny-Koladka, 2013; Zhang et al., 2018). However, PLFA analysis does not provide detailed quantitative information about microbial community structure, albeit insights from a few biomarker fatty acids indicative of a broad change can be gained. Recent advancements in molecular techniques (e.g., 16S rRNA gene sequencing, functional gene microarray, metagenomic sequencing) coupled with statistical analyses (e.g., principle component analysis, canonical correspondence analysis) have allowed for detailed phylogenetic analyses, functional gene identification, and discernment of the effects of lead and other heavy metals on microbial communities (Chen et al., 2018). For example, Functional Response Group (FRG) analysis, which is based on RNA and DNA abundance patterns instead of phylogeny, can functionally characterize microbial communities in terms of their metal tolerant nature in a contaminated environment (Jacquiod et al., 2018).
This section is based on a review of available literature relating lead and other heavy metal contamination with microbial community structure and function in aquatic sediments and terrestrial soils (Tables 1–3). While several of the evaluated studies are conducted through short-term in vitro experiments whereby lead exposure is artificially imposed by incubation with lead nitrate in a green house or laboratory setting (Table 1), most studies involve in situ field sampling of contaminated sites that have a contamination history of decades to over one hundred years (Tables 2 & 3). In these contaminated sites, lead typically co-exists with other heavy metals. Thus, most in situ studies on microbial communities reflect a collective effect of a group of heavy metals. Because the concentrations of different heavy metals in these contaminated sites often covary, determination of the effects of any individual heavy metal contaminant on microbial community can be challenging. In spite of limited in situ studies on lead-specific ecological and toxicological effects, the impact of lead exposure can be inferred because lead is one of the major heavy metal contaminants and many of them share the same toxicological effects on microbial community (Xu et al., 2019). The In vitro studies that used lead dosing and incubation help to better isolate lead-specific ecological and toxicological effects.
Table 1.
Lead spiked soil impacts microbial metabolism and community composition.
| Soil Source | Lead treatment | Microbial Effectsa | Reference |
|---|---|---|---|
| Barossa Valley region, South Australia | 5000 (4605; 92.1% recovery) mg/kg soil incubated with Pb(NO3)2 for 49 days | (−) Microbial respiration (CO2 evolution) & biomass carbon (+) Gram negative bacteria & Gram positive bacteria (−) Fungi & Actinomycetes |
Xu et al., 2018 |
| Highly sodic yet low acidity (pH > 5.5) soils in the Barossa Valley region, South Australia | 50 and 5000 mg/kg incubated with Pb(NO3)2 for 7 and 49 days | (−) Microbial activity (basal respiration, microbial biomass carbon, and microbial functional groups) (−) Microbial community compositions based on the microbial phospholipid fatty acids (PLFA) analysis: Greater negative influence on the fungal population than bacteria |
Xu et al., 2019 |
| Resembling Mississippi silty clay soil & Delta topsoil (Yazoo silty clay), Mississippi, USA | 500, 1000, 2000 mg/kg incubated with Pb(NO3)2 at 18-24°C for 18 months | Selected for Pb resistant nitrite reducers (nirK) Shift in microbial community (−) Community diversity |
Sobolev and Begonia, 2008 |
| Uncontaminated soil, Beijing, China | 100 & 500 mg Pb/kg incubated with Pb(NO3)2 for 4 weeks | Dominant phyla (all soils): Bacteroidetes, Proteobacteria, Firmicutes, Actinobacteria On genus levelb: Firmicutes: (+) Bacillus Bacteroidetes: (+) Adhaeribacter, Pontibacter, Flavisolibacter Alphaproteobacteria: (+) Kaistobacter |
Liu et al., 2018 |
| Hangzhou, China | 2200, 400, 600, 800, 1000 mg/kg incubated with Pb(NO3)2 at 25°C for 56 days | (−) Biomass C & N, C mineralization, abundance, diversity (−) Metabolism: (+) L-phenylalanine, D-mannitol, α-ketobutyric acid utilization, and (−) Tween40, pyruvic acid methyl-ester, hydroxy butyric acid, Itaconic acid utilization |
Akmal et al., 2005 |
(+) or (−) indicates positive or negative correlation to lead concentration, respectively.
Phylum classification was provided to each genus.
Table 3.
Lead and heavy metal impact on microbial communities in terrestrial soils.
| Site History | Total Metal Concentration (mg/kg) | Community Effectsa | Reference | |
|---|---|---|---|---|
| Industrial waste (20 years), Dapu Town, Hunan Province, China | As, 10.8-146.3 Cd, 1.09-46.55 Cr, 1.21-36.9 Pb, 33.6-2866 Zn, 57.7-1300 +Ca, Ti, V, Mn, Fe, Co, Ni, Rb, Sr, Y, Zr, Bi, Th, K |
(+)b
Proteobacteria, Crenarchaeota, Euryarchaeota (−) Chloroflexi Cd, As, Zn, Pb (+) Archea abundance; (−) Bacteria abundance |
Li et al., 2017 | |
| Electronic waste & smouldering activities (Cu enrichment) Alaba Internatl Market Lagos State, Nigeria | Cd, 1.2-20.7 Cr, 16.6-187 Cu, 10-21600 Ni, 10-3830 Pb, 10-8670 Zn, 20-3040 |
Dominant (in descending order of abundance in high heavy metal soil): Proteobacteria, Firmicutes, Actinobacteria, Chloroflexi, Acidobacteria, Planctomycetes, Bacteroidetes |
Jiang et al., 2019 | |
| Manufactured gas plant (150-200 years) Multiple cities, Australia | Al, 418-3341 As, 3-177 Cd, 0.04-3 Cr, 15-431 Cu, 10-1388 Ni, 6-107 Pb, 19-1622 Zn, 20-953 +Polycyclic Aromatic Hydrocarbons |
Dominant: Proteobacteria, Gram negative bacteria Lower abundance: Gemmatimonadetes, Bacteriodetes, Chloroflexi |
Kuppusamy et al., 2016 | |
| Au, Pb, & Zn mining, Zhen’an County, Shangluo City, Shaanxi Province, China | As, 19.83-79.81 Cd, 0.36-1.97 Hg, 0.12-2.09 Pb, 30.91-59.86 Zn, 28.63-50.68 |
Dominant (in descending order of abundance): Proteobacteria, Acidobacteria, Bacteroidetes, Actinobacteria, Gemmatimonadetes, Planctomycetes, Firmicutes (+) Acidobacteria, Bacteroidetes, Planctomycetes (−) Proteobacteria, Gemmatimonadetes, Actinobacteria, Firmicutes Gammaroteobacteria Acidobacteria Bacteroidetes Alphaproteobacteria Gammaproteobacteria Gemmatimonadetes Betaproteobacteria Actinobacteria |
Genera: (+) Acidibacter (+) Blastocatella (+) Flavobacterium, Pedobacter (−) Sphingomonas (−) Acinetobacter, Pseudomonas, Mizugakiibacter, Rhodanobacter (−) Gemmatimonas (−)Ralstonia (−) Arthrobacter |
Guo, et al., 2017 |
| Pb & Zn mining (1904-1970) Picher, Ottawa Co., Oklahoma, USA |
Al, 34.8-4040.4 Cd, 0.1-43.6 Mg, 94.0-841.7 Pb, 3.1-1115.2 Zn, 8.1-4486.9 +Te, W, K, Mn, Ni, Fe, V, B, Cu, Co, Cr, Ti, Na |
Pb, Cd, Zn, Mg (−) Bacteria Al, Cd, Pb, and/or Zn (+) or (−) Acidobacteria, Actinobacteria, Bacteriodetes, Chloroflexi, Plantomycetes, Proteobacteria, Verrucomicrobia Actinobacteria Bacteroidetes Alphaproteobacteria Deltaproteobacteria Planctomycetes |
Genera: (+) Streptomyces, Amycolatopsis (+) Flavisolibacter (+) Sphingomonas (+) Geobacter (+) Gemmata, Planctomyces |
Beattie et al, 2018
Beattie et al., 2017 (analytical chemistry) |
| Zn & Pb mining & smelting (since medieval times; industrial, 1967-2013) Miasteczko Śláskie & Olkusz, Poland |
Cd, 3.98-82.6 Cu, 2.25-127 Mn, 57-770 Pb, 298-7200 Zn, 80-4249 |
(+) Gram positive bacteria (−) Gram negative bacteria & fungi |
Azarbad et al., 2013 | |
| Pb & Zn enrichment facility (40 years) & Pb & Zn mining (100 years) Yunnan Province, China | Pb, 23.61-36,586 Zn, 66.99-31,089 |
Pb & Zn (+) Verrucomicrobia Alphaproteobacteria Actinobacteria Betaproteobacteria Alphaproteobacteria Mining (−) Actinobacteria, Chloroflexi Mining (+) Verrucomicrobia Actinobacteria Alphaproteobacteria Betaproteobacteria Alphaproteobacteria |
Genera: Pb & Zn (+) Bradyrhizobium Pb & Zn (−) Nocardioides, Gaiella, Acidimicrobiaceae (Family) uncultured, Actinobacteria norank Pb & Zn (−) Comamonadaceae (Family) unclass Pb & Zn (−) Skermanella Mining (−) Nocardioides, Gaiella, Acidimicrobiaceae uncultured & Actinobacteria norank Mining (−) Skermanella Mining (−)Comamonadaceae (Family) unclass Mining (+) Bradyrhizobium |
Xu et al., 2017 |
| Smelter, (1936-1989) Seocheon city, Chungnam, Republic of Korea |
As, 1.3-16.9 Cd, 1.02-3.2 Cu, 65.33-154.8 Ni, 2.7-3.82 Pb, 53.8-455.3 Zn, 44.6-102.6 |
Dominant (in descending order of abundance): Proteobacteria, Chloroflexi, Acidobacteria, Bacteroidetes Smelting (−) Chloroflexi, Chlorobi Firmucutes Deltaproteobacteria Deltaproteobacteria Firmucutes Deltaproteobacteria Actinobacteria Deltaproteobacteria Deltaproteobacteria Deltaproteobacteria Deltaproteobacteria |
Genera: As(+) Bacillus As(+) Desulfatibacillum As(−) Desulfovirga Cd (+) Bacillus Cd (−) Desulfatibacillum, Desulfoglaeba, Desulfovirga Cu (+) Streptomyces Cu (−) Desulfatibacillum, Desulfococcus,Desulfovirga Pb (+) Desulfatibacillum, Desulfovirga Pb (−) Desulfococcus Ni (+) Desulfovirga |
Tipayno et al., 2017 |
Phylum unless otherwise noted. Proteobacteria are listed by class if associated information is available: Alphaproteobacteria, Betaproteobacteria, Deltaproteobacteria, and Gammaproteobacteria. When a genus is identified, its corresponding phylum is provided for clarity.
(+), positive correlation to heavy metal concentration; (−), negative correlation to heavy metal concentration
Table 2.
Impact of lead and other heavy metal pollution on microbial communities in aquatic sediments
| Site history | Total metal concentration (mg/kg) | Community Effectsa | Reference | |
|---|---|---|---|---|
| Cu Mining, Clark Fork River, Montana, USA | As, 2.01-68.9 Cd, .011-1.84 Cu, 1.14-332 Pb, 2.59-69.8 Zn, BDLc-433 |
(+)b
Gammaproteobacteria (−) Betaproteobacteria (+) prokaryotes (−) eukaryotes & actinomycetes |
Feris et al., 2003 | |
| Sewage outlet of mining and smelting operations over 30 years, Xiangjian river, Hunan Province, China |
As, 73-48 Cd, 3-22.1 Co, 12.1-23.2 Cr, 57-87 Cu, 34-69 Hg, 0.18-0.65 Ni, 30.2-53.7 Mn, 788-2012 Pb, 83-124 Zn, 158-496 |
Betaproteobacteria Gammaproteobacteria Firmicutes Firmicutes Deltaproteobacteria Alphaproteobacteria Betaproteobacteria Actinobacteria Bacteroidetes |
Genera: Dominant: Janthinobacterium, Massilia Acinetobacter Proteiniclasticum (+) Fusibacter, Proteiniclasticum (+) Geobacter (−) Sphingomonas (−) Janthinobacterium (−) Arthrobacter (−) Flavobacterium |
Ren et al., 2016 |
| Baiyin Nonferrous Metal Co., 19 million tons of wastewater discharged during 1960s-1995, Dondagou River, Gansu Province, China | As, 8.8-57.1 Cd, 0.2-5.9 Cr, 32.6-59.1 Cu, 19.7-57 Hg, BDL-1.3 Ni, 14.7-26.2 Pb, 20.1-153.7 Zn, 54.8-330.2 |
(+) viral abundance (+) Proteobacteria, Cyanobacteria, Tenericutes, Firmicutes, Bacteroidetes (−) Verrucomicrobia, Actinobacteria, Chloroflexi Betaproteobacteria Gammaproteobacteria Bacteroidetes Planctomycetes |
Genera: (+) Thauera, Hydrogenophaga, Thiobacillus, Acidovorax, Albidiferax, Ramlibacter, Methyloversatilis (+) Luteibacter (+) Algoriphagus (+) Pirellula |
Chen et al., 2018 |
| MetalEurop Smelter, over 100 years of accidental discharges into river from 1893, River Deûle, France |
MetalEurop Cd, 1.29-38.13 Cu, 13.76-99.99 Pb, 11.58-913.83 Zn, 348.6-3218.3 +Al, As, Co, Cr, Fe, Mn, Ni, V |
Gammaproteobacteria |
Genus: Pb, As, Cd, Co, Cr, Cu, Ni, and Zn (+) Pseudomonas |
Roosa, Wauven et al., 2014 |
| MetalEurop Smelter, Decades of accidental discharges into river from 1893, River Deûle, France Férin, Sansée Canal, France |
MetalEurop Cd, 38.13 Cu, 99.99 Pb, 913.83 Zn, 3218.27 +Al, As, Co, Cr, Fe, Mn, Ni, V Férin Cd, 1.29 Cu, 13.76 Pb, 111.58 Zn, 348.55 +Al, As, Co, Cr, Fe, Mn, Ni, V |
Betaproteobacteria Gammaproteobacteria Alphaproteobacteria Deltaproteobacteria Gammaproteobacteria Betaproteobacteria Betaproteobacteria Deltaproteobacteria Actinobacteria |
Dominant: Burkholderia, Rubrivivax, Leptothrix, Cupriavidus, Methylibium, Variovorax, Thauera, Azoarcus Pseudomonas Methylobacterium Anaeromyxobacter Genera: (+) Pseudomonas, Pseudoxanthomonas, Stenotrophomonas (+) Thiobacillus, Acidovorax, Dechloromonas, Alicycliphilus (−) Rubrivivax, Leptothrix (−) Anaeromyxobacter, Sorangium (−) Mycobacterium, Streptomyces |
Gillan et al., 2015 |
| MetalEurop Smelter, Decades of accidental discharges into river from 1893, River Deûle, France Férin, Sansée canal, France |
MetalEurop Cd, 38.13 Cu, 99.99 Pb, 913.83 Zn, 3218.27 +Al, As, Co, Cr, Fe, Mn, Ni, V Férin Cd, 1.29 Cu, 13.76 Pb, 111.58 Zn, 348.55 +Al, As, Co, Cr, Fe, Mn, Ni, V |
(−) Gammaroteobacteria (−) Firmicutes (−) Alphaproteobacteria (with higher RNA levels (more active) (+) Actinobacteria, with lower RNA levels (sensitivity) |
Orders: Enterobacteriales, Pseudomonadales, Aeromonadales dominant) Clostridiales, Lactobacillales |
Jacquiod et al., 2018 |
| Poyang Lake, China Tungsten & other mining | As, 4.23-10.95 Cd, 0.307-1.022 Pb, 29.40-40.06 +Fe, Li, Cr, Co, Ni, Cu, Zn, W, Tl, Sn, Sb |
(−) Betaproteobacteria, Gammaproteobacteria (+) Acidobacteria, Latescibacteria, Verrucomicrobia, Nitrospirae, Gemmatimonadetes |
Zhang et al., 2018 | |
| Heavy metal spill in April 2016, Xiannvhu Lake, Jiangxi Province, China | As, 098-137.87 Cd, 0.13-137.87 Cu, 5.31-119.32 Pb, 3.75-54.12 Zn, 14.44-205.59 |
(−) Denitrifiers Betaproteobacteria Gammaproteobacteria |
Genera: Primary denitrifiers: Pseudogulbenkiania Pseudomonas + 2 unknown groups |
Guo et al., 2018 |
| Zn & Ti smelters (80 years) Sørfjord, southern Norway | Cd, 0.22-3.76 Cu, 1.32-43.83 Pb, 12.18-259.71 Zn, 18.72-333.37 |
Cu, Pb, Zn (−) Gammaproteobacteria, Cytophaga-Flexibacter-Bacteroides Group (Bacteroidetes) No correlation with Deltaproteobacteria |
Gillan et al., 2005 | |
Phylum unless otherwise noted. Proteobacteria are listed by class if associated information is available: Alphaproteobacteria, Betaproteobacteria, Deltaproteobacteria, and Gammaproteobacteria. When a genus is identified, its corresponding phylum is provided for clarity.
(+), positive correlation to heavy metal concentration; (−), negative correlation to heavy metal concentration
BDL, Below detection limit
2.1. Microbial community under early stage exposure to lead contamination
Several studies have indicated that early stage exposure to lead contamination results in suppressed microbial metabolism, reduced biomass production in association with higher energy demand, and changes in diversity and relative abundance (Table 1; Akmal et al., 2005; Liu et al., 2018; Sobolev & Begonia, 2008; Xu et al., 2018). The immediate inhibitory effects are typically reflected in carbon and nutrient cycling such as effects on the denitrifying microbial community (Sobolev & Begonia, 2008; Xu et al., 2019). Sediments from a lentic system acutely contaminated due to a heavy metal spill (Cd, Zn, Pb, Cu, As; China) showed reduced abundance of the nitrous oxide reductase gene, nosZ, suggesting lower denitrifier community richness (Table 2; Guo et al., 2018). Following the spill, the dominant nosZ-denitrifier genus observed is Pseudogulbenkiania (Betaproteobacteria), whereas the genus Pseudomonas (Gammaproteobacteria) increases following the spill and two other unidentified denitrifier groups decrease (Guo et al., 2018), suggesting that the some members of the microbial community react to contamination of lead and other heavy metals, in part, by developing metal-resistant mechanisms while others are susceptible. The selection of microorganisms for lead-resistant forms of nitrite reductase in early stage exposure to lead contamination is also evidenced by changes in the community harboring nirK in lead nitrate amended soil incubated for 18 months at 18-24°C (Sobolev & Begonia, 2008). This study further notes that lead has detectable effects upon the community diversity at relatively low concentrations, and there are several thresholds as concentrations increase, each causing a shift in microbial diversity. Several other studies have also reported microbial community shifts in lead amended soil (Liu et al., 2018; Xu et al., 2018). In another study with lead nitrate amended soil, incubated for 56 days at 25°C, utilization of L-phenylalanine, D-mannitol, α-ketobutyric acid increased while Tween40, pyruvic acid methyl-ester, hydroxy butyric acid, itaconic acid utilization decreased, suggesting alteration in microbial community-level metabolism and activity (Akmal et al., 2005).
In the presence of lead, resistant microorganisms clearly have a selective advantage due to their ability to persist in a lead contaminated environment which selects against susceptible microbes. Soil incubated with 5000 mg/kg lead nitrate for 49 days results in an increase of Gram-positive bacteria accompanied by a decrease in the abundance of fungi and actinomycetes (Xu et al., 2018). The relative abundance of Gram-negative bacteria is unaffected based on their increased numbers in the lead spiked soil. Upon the addition of macadamia nutshell biochar, which reduces lead bioavailability, fungi and actinomycetes recover slightly and microbial respiration and biomass production increases (Xu et al., 2018). When lead amended soil is acclimated for 4 weeks, the dominant phyla observed are Bacteroidetes, Proteobacteria, Firmicutes, and Actinobacteria. Acidobacteria, Verrucomicrobia, and Chloroflexi are less abundant. However, as the lead concentration decreases during phytoremediation, these latter phyla increased in abundance. At the genus level, Bacillus, Adhaeribacter, Pontibacter, Flavisolibacter, and Kaistobacter are present in the lead amended soil. Following phytoremediation, Flavisolibacter, Kaistobacter, and Pseudomonas increase in abundance with a relative decrease in Bacillus, Adhaeribacter, Pontibacter, and Paenibacillus, thus suggesting reduced lead tolerance. Bioavailability and bioremediation clearly influence the community structure in lead amended soil (Liu et al., 2018; Xu et al., 2018).
2.2. Microbial community under long-term exposure to lead contamination: aquatic sediments
Sediment microbial communities in aquatic systems adjacent to mining and smelting operations are impacted by heavy metal contamination, with lead identified as a primary constituent (Table 2; Feris et al., 2003; Gillan, Danis, Pernet, Joly, & Dubois, 2005; Jie et al., 2016; Roosa, Wattiez, et al., 2014; Zhang, Xu, Zhao, Rong, & Zhang, 2018). In sediments contaminated from copper (USA) and tungsten (China) mining operations which contain significant quantities of lead, dissimilarity in microbial communities along a chemical concentration gradient increases as metal content increased (Feris et al., 2003; Zhang et al., 2018). However, in contaminated sediments due to copper mining activity, biomass is unaffected by metal concentration with no apparent correlation between sediment metal content and diversity or total productivity (Feris et al., 2003), suggesting that other factors controlling biomass outweigh the impact of heavy metal contamination. Rather, the structure of microbial communities is significantly affected. Phospholipid fatty acid analysis indicates that heavy metal concentration positively correlates with prokaryote abundance and eukaryotes and actinomycetes negatively correlate. Further analysis by qPCR using specific primers reveals a positive correlation with Gammaproteobacteria and a negative correlation with Betaproteobacteria abundance (Feris et al., 2003). In the tungsten mining site (China), 16S rRNA gene sequencing identifies Proteobacteria and Actinobacteria as the dominant sediment phyla (Zhang et al., 2018). While all sites with varying concentrations of heavy metals have unique members, they also share many members in common. Zhang et al. (2018) confirms the negative correlation of metal concentration with Betaproteobacteria and Deltaproteobacteria while members of the phyla Acidobacteria, Latescibacteria, Verrucomicrobia, Nitrospirae and Gemmatimonadetes are positively correlated with metal concentration.
Sediments sampled along a gradient from a sewage outfall on the Xiangjian River (China), linked to heavy non-ferrous metal mining and smelting operations over the past 30 years, have been studied extensively to understand the effects of heavy metal stressors on both the microbial communities and functional genes (Table 2). Inductively coupled plasma atomic emission spectroscopy reveals that the sediments are contaminated with Cu, Pb, Zn, As, Cd, Ni, Hg, Cr, Mn, Co, and S, forming a gradient of decreasing concentrations with distance from the sewage outlet (Jie et al., 2016; Ren et al., 2016; Yin et al., 2015). Microbial communities in sediments closest to the outfall have lower Shannon diversity and Pielou eveness indices are dissimilar to other less contaminated sediments (Ren et al., 2016; Yin et al., 2015). Members of the phyla Firmicutes, Chloroflexi and Crenarchaeota are more abundant in the highly contaminated sediments whereas Proteobacteria and Actinobacteria are present but in lower abundance (Yin et al., 2015). Molecular ecological network analysis reveals that Pb, as well as mercury (Hg), Zn and carbon (C), correlate with the network module containing the phyla Bacteroidetes, Chloroflexi, Proteobacteria, Acidobacteria, Firmicutes and Actinobacteria from the highly contaminated sediments, thus suggesting microbial community composition and co-occurrence of phyla, is driven by the heavy metals in the sediment (Yin et al., 2015). Dominant microbial genera in all sediments along the gradient are Fusibacter, Janthinobacterium, Proteiniclasticum, Acinetobacter, and Massilia with ~15-18% unclassified to the genus level. Fusibacter, Geobacter, and Proteiniclasticum are more abundant in sediments with higher heavy metal concentration whereas Janthinobacterium, Arthrobacter, Sphingomonas and Flavobacterium are at higher numbers in the less contaminated samples (Ren et al., 2016). Each sediment sample also had a unique microbial community functional gene structure though all samples along the gradient were ~80-90% similar. Whole microbial community functional gene structure and lead resistant genes abundance correlated with lead concentration across the gradient (Jie et al., 2016). In the most heavily contaminated sediment, genes encoding for heavy metal resistance and carbon cycling pathways, to include degradation of aromatic and nitroaromatic compounds, were more prevalent (Yin et al., 2015).
The Dondagou River (China), a tributary to the Yellow River, is heavily contaminated with Cu, Zn, Cd, Pb, As, Hg, Cr and Ni due to approximately 19 million tons of wastewater discharge from non-ferrous metal mining and processing during the 1960s – 1995 (Table 2; Li et al., 2006; Chen et al., 2018). Lead concentrations are in the same range (153 ppm vs 124 ppm) as those observed in the Xiangjian River site, with other heavy metals present at both sites at comparable levels (Chen et al., 2018; Ren et al., 2016). Microbial community diversity is negatively correlated with heavy metal concentration and carries a higher viral load than sediments from less contaminated sites. The phyla Proteobacteria, Cyanobacteria, Tenericutes, Firmicutes, and Bacteroidetes abundance show a positive correlation with heavy metal contamination while Verrucomicrobia, Actinobacteria and Chloroflexi are less represented (Chen et al., 2018). Proteobacteria, Bacteroidetes, Firmicutes are the most dominant phyla harboring heavy metal resistance and reduction genes However, heavy metal resistance and reduction genes are detected in the Bacteria Gemmatimonadetes, Planctomycetes, Acidobacteria, Tenericutes, Spirochaetes, Chlorobi and Parubacteria and the Archea Euryarchaeota and Thaumarchaeota using quantitative polymerase chain reaction (qPCR) analysis. Within the phylum Proteobacteria, the classes Betaproteobacteria, Gammaproteobacteria, and Alphaproteobacteria harbor the highest number of heavy metal resistance genes, and they are present in a few representatives of the classes Deltaproteobacteria and Zetaproteobacteria (Chen et al., 2018). Genes associated with DNA recombination, DNA damage repair, and heavy metal resistance are more prevalent in the more highly contaminated sediments.
In northern France, accidental discharges of Cd, Cu, Pb, and Zn from a smelter located on the River Deûle occurred over a 100 yr period (Table 2; Roosa et al., 2014; Roosa, Wauven, et al., 2014). Directly adjacent to the smelter, sediment lead concentration averages 913 mg/kg and co-occurs with aluminum (Al), As, Cd, Co, Cr, Cu, iron (Fe), manganese (Mn), Ni, Pb, vanadium (V), and Zn (Gillan et al., 2015). Lead concentrations are higher than in the most contaminated sediments of the Xiangjian River (737 vs 913 mg/kg; Jie et al., 2016). The upstream lead concentrations average 112 mg/kg, comparable to those observed in two of the Xiangjian River studies (Chen et al., 2018; Ren et al., 2016). Sediments from this site, and lessor contaminated sites upstream, have been studied extensively to elucidate the impact of heavy metal contamination on microbial community structure and function and better understand how these communities adapt to the presence of the heavy metals (Roosa, Wauven et al., 2014; Gillan et al., 2015; Jacquiod et al., 2018). At the phylum level, microbial communities in sediments adjacent to the smelter are similar to those in the upstream less contaminated site, possibly reflecting the effect of long-term exposure to contamination. This finding is similar to one from a study of marine sediment associated with 80 years of Zn and Pb smelter operation (Norway), which reports that microbial communities from contaminated sites are similarly diverse due to acclimation over the 80-year period (Gillan et al., 2005). Also note that there is a potential for microorganisms in upstream sediment to continually re-inoculate sediment downstream in a riverine system. While the metal contaminated site and upstream control sediments contain taxonomically similar microbial communities (70% similar), functionally they are different (Gillan et al., 2015). The higher-Pb sediments contain more genes encoding “cell wall and capsule substances”, “virulence, disease and defense mechanisms”, and “phages, prophages, transposable elements and plasmids” (Gillan et al., 2015). For example, Co/Zn/Cd efflux system genes, czcA and czcD, and genes encoding exopolysaccharides are more prevalent in the higher-Pb sediments (Gillan et al., 2015). The change in the community genetic potential clearly shows a lead resistance selective advantage in the contaminated sediment. Betaproteobacteria dominate at both lead contaminated sites, primarily by the genera Burkholderia, Rubrivivax, Leptothrix, and Cupriavidus. Gammaproteobacteria (Pseudomonas) and Alphaproteobacteria (Methylobacterium) also are represented. Pseudomonas, Thiobacillus, Acidovorax, Dechloromonas, Pseudoxanthomonas, Stenotrophomonas and Alicycliphilus are more prevalent in the higher contaminated sediment whereas Rubrivivax, Anaeromyxobacter, Leptothrix, Sorangium, Mycobacterium and Streptomyces are more numerous in the less contaminated, upstream site (Gillan et al., 2015; Shi et al., 2002).
Roosa, Wauven et al. (2014) further noted that metal contaminated sediments contain microorgainsms from the phyla Actinobacteria (Mycobacterium vaccaevaccae, Mycobacterium llatzerense, Rhodococcus erythreus, Streptomyces coelicoflavus), Alphaproteobactera (Sphingomonas xenophaga Methylobacterium extorquens, Methylobacterium radiotolerans), Gammaproteobacteria (Klebsiella oxytoca, Klebsiella ornithinolytica, Enterobacter minipressuralis, Enterobacter mori, Pseudomonas nitroreducens, Pseudomonas monteilli, Pseudomonas putida, Pseudomonas lutea, Pseudomonas putida, Pseudomonas arsenicoxydans, Aeromonas salmonicida) and Betaproteobacteria (Delftia lacustris) which are lead and multiple heavy metal resistant even though they were originally selected on media supplemented with Pb, Cu, Ni, Cd, Co or Zn. The Pseudomonas community is positively correlated with concentrations of Pb as well as Cu, Co, Ni, and Zn, as evidenced by 16S rRNA and qPCR detection of the outer membrane lipoprotein I (oprI) gene, specific for Pseudomonas q. The pbrT gene is present in the high-Pb sediments; however, correlation with Pb concentration is inconclusive (Roosa, Wattiez, et al., 2014). Interestingly, copy numbers of the czcA gene (encodes Co/Zn/Cd efflux pump) show a positive correlation with Pb concentration even though it is not a czcA target metal.
Taking a more precise 16S rRNA gene DNA and 16S rRNA (measure cDNA) sequencing approach, Jacquiod et al., (2018) confirms that the microbial community is dominated by Proteobacteria with Firmicutes the second most prevalent. Alphaproteobacteria and Betaproteobacteria are more numerous in metal contaminated sediments whereas Gammaproteobacteria are more abundant in control sediments. Alphaproteobacteria 16S rRNA levels (cDNA) relative to 16S rRNA gene (DNA) levels (RNA/DNA) are higher in metal contaminated sediment, implying increased activity; Actinobacteria have a lower OTU RNA/DNA ratio, suggesting sensitivity. The Bacteroidetes represent the passive part of community. Ignavibacteriae, Deltaproterobacteria, Gemmatinmonadetes and Verrucomicrobia are present in low abundance in the metal contaminated sediments. However, the three latter groups, as well as Acidobacteria, Alphaproteobacteria, Betaproteobacteria, Nitrospirae, Planctomycetes, have increased RNA/DNA ratios (Jacquiod et al., 2018). The Functional Response Group (FRG) analysis employed in this study reveals that the metal contaminated sediments contain “seed bank” (metal tolerant/slow growing or inactive, e.g. Gammaproteobacteria, Betaproteobacteria, Bacteroidetes), “upcoming bacteria” (metal tolerant, active or passive, e.g. Firmicutes, Proteobacteria), “fecal-related bacteria” (e.g. Clostridium, Enterobactericeae), “dominant metal sensitive bacteria” (lower DNA and RNA signal, e.g. Gammaproteobacteria, Pseudomonas) and “rare metal sensitive bacteria” (significantly impacted by metals, e.g. Bacteriodetes, Acidobacteria, and Deltaproteobacteria) (Jacquiod et al., 2018). This elegant analysis confirms that metal resistance mechanisms and adaptation are at work in these long-term contaminated sediments.
2.2. Microbial community under long-term exposure to lead contamination: terrestrial soils
Lead and zinc mining and lead-containing industrial activities have occurred globally for decades and left an environmental footprint as contaminated soils that influence microbial biomass, activity, diversity, and community structure (Table 3; Azarbad et al., 2013; Beattie et al., 2018; Guo, Kang, & Feng, 2017; Hu, Qi, Zeng, & Zhang, 2007; Xu et al., 2017). For example, twenty years of heavy metal contamination explains the dissimilarity of microbial communities and bacterial abundance between contaminated and uncontaminated control soils (China; Hu et al., 2007). Bacterial abundance is negatively correlated with Pb, Cd, Zn, Mg whereas Archaea are positively correlated with pH, and Al, Pb, Cd, Zn significantly impact community composition (Beattie et al., 2018). Gram-positive bacteria positively correlate to heavy metal contamination and Gram-negative bacteria and fungi show a negative correlation (Azarbad et al., 2013). Organic matter has a stronger correlation than heavy metal content, suggesting a link with heavy metal bioavailability (Azarbad et al., 2013). However, at a site with over 100 years of mining operations and 40 years of lead and zinc enrichment, no apparent heavy metal induced differences in diversity or richness are observed (China; Xu et al., 2017). Biomass and respiration are negatively correlated with levels of nine metals including lead, accounting for differences in microbial community structure (Poland, USA; Azarbad et al., 2013; Beattie et al., 2018; Xu et al., 2017). At a lead battery recycling facility that contaminated soil for 40 years (lead 10,000 mg/kg; 5% water), microbial biomass and respiration are decreased. However, addition of a carbon source stimulates biomass production (Shi et al., 2002). Near a metallurgy plant (in operation since the 1970s), Pb, Cd, Cu and Zn contaminated soil also reduces microbial biomass and respiration as well as dehydrogenase activity (Chen et al., 2014).
Lead and other heavy metals are statistically linked to changes in microbial community structure in heavy metal contaminated soils (Azarbad et al., 2013; Beattie et al., 2018; Guo et al., 2017; Xu et al., 2017). In soil contaminated for twenty years by industrial waste, community structure is statistically linked to heavy metal concentration unlike diversity richness and evenness are not (Xiaoqi Li et al., 2017). Archea abundance is positively correlated to Cd, As, Zn and Pb, and bacteria are negatively correlated to these metals (Li et al., 2017). Acidobacteria, Ascomycota and Chytridiomycota are inhibited in Cd, As, Zn and Pb contaminated soils (Chen et al., 2014). Proteobacter, Crenarchaeota and Euryarchaeota are more abundant in the heavy metal contaminated soil whereas Chloroflexi are more abundant in control soil (Li et al., 2017). Electronic waste and copper enriching smelting activities contaminated surrounding soil with Pb as well as Cu, Zn, Cd, Cr and Ni where Proteobacteria, Firmicutes, Actinobacteria, Chloroflexi, Acidobacteria, Planctomcetes and Bacteroidetes dominate (in descending order; Jiang et al., 2019). Physical and other chemical properties, such as organic matter levels and pH, influence microbial diversity (Jiang et al., 2019). Manufactured gas plants (Australia), in operation for 150-200 years, resulted in polycyclic aromatic hydrocarbon and heavy metal (Al, Cr, Ni, Cu, Zn, As, Cd, Pb) soil contamination which selects for Proteobacteria and other Gram-negative bacteria, Gemmatimonadetes and Bacteriodetes, and influenced Chloroflexi which are present in lower abundance (Kuppusamy et al., 2016). At the site of a large mining operation which ceased in mid-1950s (USA), Al, Cd, Pb, and/or Zn show significant influence on selecting for the phyla Acidobacteria, Actinobacteria, Bacteriodetes, Chloroflexi, Plantomycetes, Proteobacteria and Verrucomicrobia in the soil microbial community (Beattie et al., 2018). The phyla Proteobacteria, Acidobacteria, Bacteroidetes, Actinobacteria, Gemmatimonadetes Planctomycetes and Firmicutes (in descending order of dominance) are associated heavy metal contamination in soils polluted by mining wastes in China (J. Guo et al., 2017). In another study, Actinobacteria and Chloroflexi are more dominant in the mine soil compared to control where Verrucomicrobia are more abundant (Xu et al., 2017). These changes in microbial community structure clearly show the selection of tolerant groups under long-term exposure to heavy metal contamination while sensitive ones are reduced.
The above studies also indicate that most bacteria living in extremely polluted soils belong to the class Proteobacteria. Guo et al., 2017 showed that at the genus level, Sphinogomonas is the most abundant genus in soils polluted by mining wastes in China, and Acinetobacter, Pseudomonas, Gemmatimonas, Ralstonia, Mizugakiibacter, Rhodanobacter, Arthrobacter, Acidobacter, Blastocatella, Flavobacterium and Pedobacter correlate with Cd and Pb levels (p<0.05). Ralstonia and Gemmatimonas negatively correlate with Cd, Pb, As, Hg and pH; Mizugakiibacter and Rhodanobacter negatively correlate with Cd, Pb, As and soluble organic matter; and Blastocatella, an unidentified Nitrospiraceae, and an unidentified Acidobacteria positively correlate with Pb, Cd, Zn, Hg and soluble organic matter (Jing Guo, Yong Kang, & Ying Feng, 2017). In another lead and zinc enrichment site with many years of mining operation in China, Xu et al. (2017) indicate that below the phylum level, Norcardioides, Gaiella, Comamonadaceae, Acidimicrobiaceae, Actinobacteria and Skermanella are negatively correlated with metal concentrations and present in lesser numbers than in control samples. In contrast, abundances of Verrucomicrobia and Bradyrhizobium show positive correlations with Pb and Zn levels.
Approximately fifty years of nonferrous smelter operation, which ceased in 1989 (South Korea), resulted in enduring soil contamination by Pb and other heavy metals (As, Cd, Cu, Ni, Pb and Zn; Tipayno et al., 2018). In these soils, lead significantly correlates to the soil bacterial community composition at the phylum level, with relative abundance linked to pH. At the genus level, lead positively correlates with Desulfatibacillum and Desulfovirga; and negatively correlates with Desulfococcus (Tipayno et al., 2018). Analysis of community functional profiles (“Pathway abundance profiles”) reveals an increase in genes encoding enzymes associated with DNA replication and repair, translation, transcription, and nucleotide metabolism pathways in metal contaminated soil, whereas genes encoding enzymes associated with amino acid, lipid, and energy metabolism and biodegradation potential of xenobiotics are less abundant (Tipayno et al., 2018). As, Cd and Pb levels are positively correlated with enzymes associated with cell growth/death, transcription, signaling molecules, and interaction pathways; enzymes associated with transport, catabolism and metabolism of terpenoids and polyketides are negatively correlated.
3. Lead resistance mechanisms
Microorganisms have developed extracellular and intracellular strategies to persist in lead contaminated environments (reviewed by Jaroslawiecka & Piotrowska-Seget, 2014; Naik & Dubey, 2013; Pan et al., 2017). Lead can be sequestered extracellularly in exopolysaccharide matrices (De, Ramaiah, & Vardanyan, 2008; Macaskie & Dean, 1987; Naik, Pandey, & Dubey, 2012a; Nelson, Lo, Lion, Shuler, & Ghiorse, 1995; Roane, 1999) or scavenged by excreted siderophores (Naik & Dubey, 2011; O’Brien, Hodgson, & Buckling, 2014), ultimately resulting in lead precipitates, such as lead phosphates through phosphatase action. Surface biosorption (lipopolysaccharide, Gram negative; peptidoglycan, Gram positive) also excludes lead from the cell (Chang, Law, & Chang, 1997; Karimpour et al., 2018). Lead can enter the cell and either bind with metallothionein (Murthy, 2011; Naik, Shamim, & Dubey, 2012c), or, through an efflux mechanism, be transported by P-type ATPase to the periplasm where phosphatase release of pyrophosphate results in lead precipitation (Borremans, Hobman,& Provoost et al., 2001; Hynninen, Touze, & Pitkanen, et al., 2009; Murthy, 2011; Naik, Shamim, et al., 2012c; Rensing, et al., 1998). These lead resistance mechanisms are schematically shown in Figure 1. Some of the extracellular and intracellular strategies are described in detail below.
Figure 1.
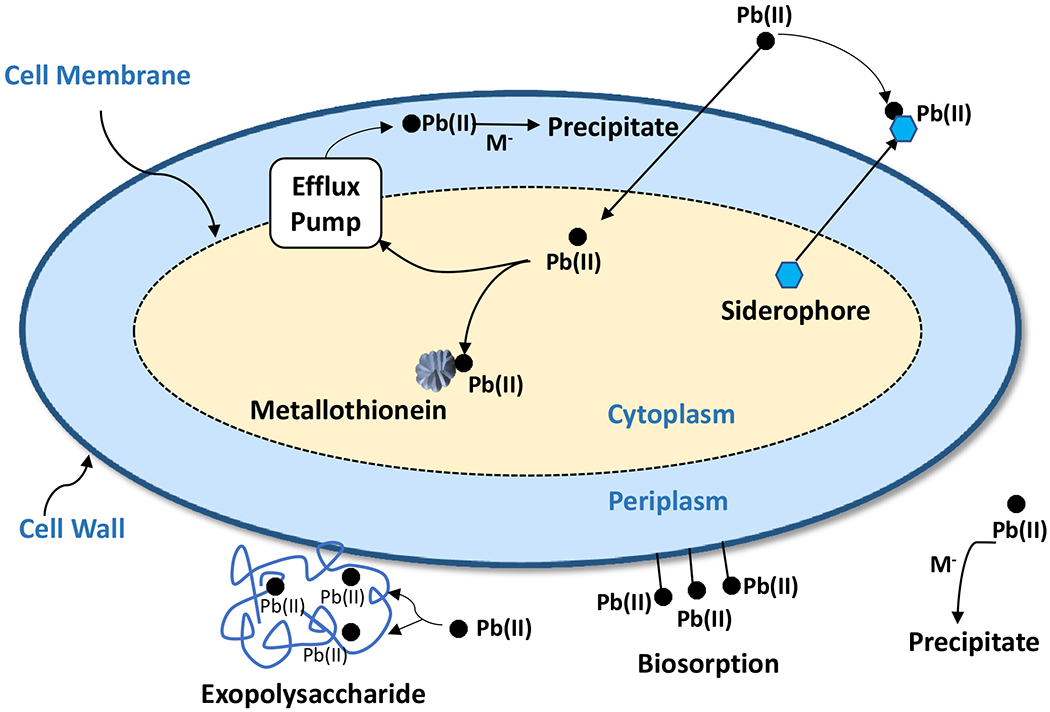
Schematic representation of lead resistance mechanisms operational in extracellular and intracellular spaces of a bacteria cell. “M−” represents anions such as phosphate that precipitate lead ions, Pb(II).
3.1. Extracellular immobilization
Selected microbial species with extracellular biosorption functions reported in the literature are listed in Table 4. When exposed to lead contamination, microorganisms will firstly invoke extracellular immobilization strategies to limit the entry of lead ion [Pb(II)] into the cell envelop to maintain metal homeostasis. In general, extracellular immobilization is accomplished mainly through biosorption and precipitation. While extracellular precipitation of lead resembles intracellular precipitation, which is described in the next section, biosorptive binding of lead involves a series of polymers or compounds produced by microorganisms including exopolysaccharides, siderophores, and various functional groups on the cell wall (Figure 1).
Table 4.
Bacteria species with extracellular immobilization functions involving exopolysaccharide (EPS), siderophores and cell surface biosorption.
| Isolation source | Pb(II) media | Phyluma | Genus/Species | Reference |
|---|---|---|---|---|
| Exopolysacchride (EPS) production | ||||
| Coast | Pb(CH3COO)2 100ppm | Gammaproteobacteria | Pseudomonas aeruginosa | De et al., 2008 |
| Lead battery manufacturing plant | Pb(NO3)2 1.6 mM |
Gammaproteobacteria | Enterobacter cloacae | Naik et al., 2012 |
| - | Pb(C2H3O2)2 1.3 – 13 mg/L |
Gammaproteobacteria | Marinobacter sp. | Bhaskar & Bhosle et al., 2006 |
| - | - |
Betaproteobacteria |
Burkholderia cepacia (formerly Pseudomonas cepacia) | Nelson et al., 1995 |
| - | Pb(NO3)2 1mM |
Gammaproteobacteria |
Citrobacter sp. |
Macaskie and Dean, 1987 |
| Metal mine | Pb(NO3)2 2.5 mM |
Gammaproteobacteria | Pseudomonas marginalis | Roane, 1999 |
| Industrial waste water | Pb(NO3)2 | Firmicutes |
Bacillus anthracis
|
El-Shanshoury et al., 2012 |
| Siderophore production | ||||
| Car battery waste | Pb(NO3)20.5mM | Gammaproteobacteria | Pseudomonas aeruginosa | Naik and Duby, 2011 |
| - | Pb(NO3)2 | Gammaproteobacteria | Pseudomonas aeruginosa | O’Brien et al., 2014 |
| Surface biosorption | ||||
| Hospital sewage | PbCI2 | Gammaproteobacteria | Pseudomonas aeruginosa | Chang et al., 1997 |
| Soil | - | Gammaproteobacteria | Pseudomonas aeruginosa | Karimpour et al., 2018 |
Proteobacteria are listed by class if available
Exopolysaccharides (EPS) are high molecular weight polyanionic polymers secreted by microorganisms into their environment. These extracellular polysaccharides protect the cell from lead and other heavy metals by preventing their entry into the cell and providing an advantageous environmental niche for the EPS-coated cell and its sensitive neighbors that become enveloped with the EPS (Bitton & Freihofer, 1978; Roane, 1999). Transmission electron microscopy reveals lead accumulation in Pseudomonas marginalis, resisting 2.5 mM lead nitrate (Roane, 1999), and Enterobacter cloacae strain P2B, resisting 1.6 mM lead nitrate (Naik, Pandey et al., 2012a), is through binding to carboxyl, hydroxyl, and amide functional groups and glucuronic acid in EPS polymer chains. Glucuronic acid EPS is produced constitutively, such as in Pseudomonas marginalis (Roane, 1999) or induced in the presence of metals, including lead, in Pseudomonas aeruginosa, Bacillus amyloliquefaciens, Bacillus subtilis subsp. subtilis, Enterobacter cloacae strain P2B and other Enterobacter spp. (Bhaskar & Bhosle, 2006; Chowdhury et al., 2008; El-Shanshoury et al., 2012; Naik, Pandey et al., 2012a). A Bacillus anthracis isolated from industrial waste water excretes EPS and precipitates lead sulfide (PbS) extracellularly (El-Shanshoury et al., 2012). Purified EPS from Marinobacter sp. unexposed to lead, binds lead and copper in vitro (Bhaskar & Bhosle, 2006). Immobilized biofilms, which contain polysaccharides and other biopolymers (reviewed by Marvasi et al., 2010), accumulate lead, a phenomenon enhanced by the addition of iron in Burkholderia cepacia (formerly Pseudomonas cepacia) (Macaskie & Dean, 1987; Nelson et al., 1995). Inactivated Pseudomonas aeruginosa cells have been shown to bioabsorb lead, possibly due to interactions with EPS (Chang et al., 1997; Karimpour et al., 2018).
Siderophores (Greek: “iron carrier”) are small, high-affinity iron-chelating compounds secreted by microorganisms such as bacteria and fungi. Microorganisms excrete siderophores to chelate iron and transport it back across the cell membrane (Neilands, 1995). However, these same relatively low molecular weight compounds also can bind heavy metals, such as lead, and reduce toxicity (O’Brien et al., 2014). In Pseudomonas aeruginosa PAO1, lead binds to the siderophore pyoverdine but is not transported into the cell (Braud et al., 2009). Some heavy metals, such as Cd, Co, gallium (Ga), Hg, Mn and Zn, can inhibit iron uptake, however lead does not (Braud et al., 2009). Furthermore, it does not inhibit pyoverdine production (Braud et al., 2009). A lead resistant strain of Pseudomonas aeruginosa (strain 4EA), isolated from soil contaminated with lead battery waste, increases pyochelin and pyoverdine production coupled with reduced cell size in the presence of lead (Naik & Dubey, 2011). Scanning electron microscopy and energy dispersive X-ray spectroscopy reveal biosorption of lead (Naik & Dubey, 2011). Excreted siderophores also scavenge lead, resulting in the production of lead precipitates through phosphatase action and formation of lead phosphates (Naik & Dubey, 2011; O’Brien et al., 2014).
Organic functional groups such as hydroxyl, carboxyl, and nitrogen-, sulfur-, and phosphorus-containing groups on the cell wall can sorb Pb(II) onto the cell surface. This may occur either in living or in inactivated cells. Chang et al. (1997) noted that both inactivated cells and resting cells of Pseudomonas aeruginosa, isolated from sewage, can adsorb Pb (II) with high capacities. This is further confirmed by Karimpour et al. (2018) wth Pseudomonas aeruginosa isolated from contaminated soil. Scanning electron microscopy and energy dispersive X-ray spectroscopy reveal biosorption of lead onto cell surface of Pseudomonas aeruginosa, isolated from soil contaminated with lead battery waste (Naik & Dubey, 2011). Therefore, these lead resistance bacterial strains may potentially serve as biosorbents for lead remediation.
3.2. Intracellular accumulation
When Pb(II) enters the cell under exposure to high concentrations, intracellular resistance strategies are triggered, first through an efflux pump, or metal transporting ATPases, to transport lead outside the cell membrane to the periplasm where lead can form insoluble precipitates lead through oxidation or reduction, thereby sequestering the lead and protecting the cell from its toxic effects. Meanwhile, metallothioneins can also bind lead in the cytoplasm (Figure 1). Selected microbial species with intracellular accumulation through precipitation and binding with metallothioneins reported in the literature are listed in Table 5.
Table 5.
Bacteria species with intracellular Pb accumulation: precipitation or metallothionein binding
| Isolation source | Pb (II) media | Phyluma | Species | Precipitate/Metallo-thionein gene | Reference |
|---|---|---|---|---|---|
| Precipitation | |||||
| Coast | Pb(CH3COO)2 100ppm |
Actinobacteria Firmicutes |
Brevibacterium iodinum
Bacillus pumilus |
PbS | De et al., 2008 |
| Sewage treatment plant | Pb(NO3)2 300 mg/L | Firmicutes | Desulfotomaculum sp. | PbS | Gong et al., 2007 |
| - | Pb(NO3)2 2.5 mM |
Gammaproteobacteria | Vibrio harveyi | Pb(PO4)6 | Mire et al., 2004 |
| Sewage treatment wetland | Pb(NO3)2 0.1mM |
Gammaproteobacteria Firmicutes Actinobacteria |
Pseudomonas aeruginosa Bacillus amyloliquefaciens Bacillus subtilis subsp. subtilis Microbacterium luteolum |
Lead nano-particles (mineral not identified) | Chowdhury et al., 2008; 2011 |
| Leather workshop | Pb(NO3)2 | Gammaproteobacteria | Enterobacter sp. | PbO | El-Shanshoury et al., 2012 |
| Industrial waste water | Pb(NO3)2 | Firmicutes | Bacillus anthracis | PbS | El-Shanshoury et al., 2012 |
| - | Pb(NO3)2 6 mM | Gammaproteobacteria | Escherichia coli | Lead particles | Essa et al., 2017 |
| - | Pb(NO3)2 4 mM |
Firmicutes | Staphylococcus aureus | Lead particles | Essa et al., 2018 |
| Sludge | Pb(NO3)2 | Gammaproteobacteria | Escherichia coli | PbS & PbCO3 | Essa et al., 2018 |
| Metal mine | PbCl2 100 mM |
Gammaproteobacteria | Enterobacter cloacae | PbCO3 | Kang et al., 2015 |
| Oil field injection water | Pb(NO3)2 150 mg/L | Proteobacteria, alpha | Rhodobacter sphaeroides | PbS & PbSO4 | Li et al., 2016 |
| Metal mine soil | Pb(NO3)2 0.1 mM | Firmicutes | Bacillus megaterium | Mineral unidentified | Roane, 1998 |
| Battery waste | Pb(NO3)2 3.0 mM | Gammaproteobacteria | Providencia vermicola | PbSO3 | Sharma et al., 2017 |
| Metallothionein (intracellular accumulation) | |||||
| Soil, car battery waste | Pb(NO3)2 0.2-0.4 mM |
Gammaproteobacteria Gammaproteobacteria |
Salmonella enterica (formerly Salmonella choleraesuis) Proteus penneri |
smtAB | Naik et al., 2018 |
| Estuary | Pb(NO3)2 0.6 mM | Gammaproteobacteria | Pseudomonas sp. | bmtA | Naik et al., 2012 |
| Industrial effluent | Pb(NO3)2 0-500 mg/L | Firmicutes | Bacillus cereus | Murthy et al., 2011 | |
Proteobacteria are listed by class if available
Precipitates identified in lead-exposed microbes include lead(II) oxide (PbO; El-Shanshoury et al., 2012), lead(II) sulfide (PbS; De et al., 2008; El-Shanshoury et al., 2012; Essa, Al Abboud, & Khatib, 2018; Gong, Zhang, Bai, & Yang, 2007; X. Li et al., 2016), lead(II) sulfite (PbSO3; Sharma et al., 2017), lead(II)sulfate (PbSO4; X. Li et al., 2016), lead(II) carbonate (PbCO3; Essa et al., 2018; Kang et al., 2015), lead(II) phosphate (Pb(PO4)2; Borremans et al., 2001; Hynninen et al., 2009), and an unusual lead phosphate salt, Pb9(PO4)6, precipitated by Vibrio harveyi (Mire et al., 2004). Lead precipitates are typically formed in the periplasm, a concentrated gel-like matrix in the space between the inner cytoplasmic membrane and the bacterial outer membrane. Intracellular lead oxide (PbO) precipitation has been reported to occur in the periplasm of an Enterobacter sp. (El-Shanshoury et al., 2012). Sharma et al. (2017) reports periplasmic lead sulfite accumulation in Providencia vermicola strain SJ2A, isolated from a battery manufacturing plant. Lead sulfide (PbS) has been associated with lead exposed Brevibacterium iodinium GP13, Bacillus pumilus S3, Escherichia coli Z3, and Rhodobacter sphaeroides (De et al., 2008; Essa et al., 2018; Li et al., 2016) and accumulates in the periplasm of Desulfotomaculum sp. (Gong et al., 2007) and Bacillus megaterium, isolated from a silver mining region (Roane, 1999). Accumulation of lead phosphate in the periplasm, by Cupriavidus metallidurans strain CH34 (formerly Alcaligenes eutrophus, Wautersia metallidurans, and Ralstonia metallidurans) also has been noted (Borremans et al., 2001; Hynninen et al., 2009). Lead is exported to the periplasm by an efflux mechanism with a PIB family P-type ATPase from where it combines with pyrophosphate. P-type ATPase efflux has been associated with lead resistance in Escherichia coli (zntA; Beard et al., 1997; Rensing et al., 1997; Rensing et al., 1998), Staphylococcus aureus (cadA), and Pseudomonas putida (cadA2) (Hynninen et al., 2010). Pseudomonas aeruginosa strains C1 and C2, Bacillus amyloliquefaciens strains F1, Bacillus subtilis subsp. subtilis strain F3, and Microbacterium luteolum strain GZ, isolated from a wetland inundated by sewage contaminated with toxic metals, harbor lead containing nanoparticles intracellularly (Chowdhury et al., 2008; Chowdhury et al., 2011). These strains have been used to develop a bioremediation package for removal of lead from lead contaminated wastewater.
Intracellular accumulation of lead precipitates has been associated with cell morphological changes such as shortening and thickening cells of P. aeruginosa (Chowdhury et al., 2008), spheroidal cells of Desulfotomaculum sp. (Gong et al., 2007), and interconnected filaments formed from Providencia vermicola strain SJ2A rod-shaped cells (Sharma et al., 2017). Enterobacter sp. (El-Shanshoury et al., 2012), Pseudmononas aeruiginosa, Bacillus subtilis subsp. subtilis and Bacillus amyloliquerfaciens (Chowdhury et al., 2008; Chowdhury et al., 2011) have increased exopolysaccharide secretion associated with lead precipitation, thus providing a two-layer defense against lead toxicity.
Metallothioneins are known to bind divalent metals and contribute to cellular metal homeostasis (Robinson et al., 1990). These cysteine-rich proteins are involved in zinc and cadmium resistance (Turner et al., 1993; Naz et al., 2005) and the literature suggests that lead may also be sequestered intracellularly by a metallothionein (Roane, 1999). Roane (1999) observed cytoplasmic and periplasmic lead accumulation in Bacillus megaterium by transmission electron microscopy and postulated both efflux and metallothionein involvement in lead resistance. Cadmium localization is similar in Desulfivibrio desulfuricans DSM 1926 and Desulfococcus multivorans DSM 2059. Cytoplasmic accumulation has been linked to a Synechococcus PCC 7942 metallothionein smtAB gene homolog involved in zinc resistance, providing more supporting evidence of metallothionein involvement (Robinson et al., 1990; Naz et al., 2005). Cadmium, zinc and copper treatment increase smtA and smtB transcript abundance and deletion mutations in the smtA gene, which is known to bind zinc and cadmium, results in decreased zinc resistance (Robinson et al., 1990; Turner et al., 1993). Increased metallothionein production has been correlated to lead exposure in Bacillus cereus (Murthy et al, 2011). Salmonella choleraesuis strain 4A and Proteus penneri strain GM10 harbor the smtA gene and bioaccumulate lead, suggesting probable metallothionein involvement in lead sequestration in these species (Naik, Shamim, et al., 2012c). The gene bmtA, encoding a related bacterial metallothionein, is detected in Pseudomonas aeruginosa strain WI-1, accompanied by induction of a probable bmtA gene product (metallothionein protein) and intracellular sequestration of lead (Naik et al., 2012b). The bmtA gene has been described in Pseudomonas aeruginosa, Pseudomonas putida and Anabaena PCC 7120 and binds zinc (Blindauer et al., 2002).
4. Leveraging lead resistance genes for lead biosensor development
4.1. lead resistance genes
Metal resistance genes are chromosomally- and plasmid-linked (Janssen et al., 2010) in the lead resistant C. metallidurans strain CH34 (Borremans et al., 2001; Hynninen et al., 2010; Mergeay et al., 1985). Resistance is ascribed to plasmid pMOL30, which also conveys resistance to Ag(I), Cd(II), Co(II), Hg(II), and Zn(II) (Mergeay et al., 1985; Monchy et al., 2007). The functional aspects of the lead resistance operon, pbrUTRABCD, have been elegantly described (Borremans et al., 2001; Chen et al., 2007; Hynninen et al., 2009; Monchy et al., 2007). The operon confers uptake, efflux and accumulation of Pb(II).
The Pb(II) uptake permease protein, pbrT, transports Pb(II) into the cell (Borremans et al., 2001; Jencova et al., 2008). Pb(II) binds to the MerR family pbrR regulator, which, in turn, induces transcription of pbrABCD from the pbrA promoter (Borremans et al., 2001; Hobman, Julian, & Brown, 2012). An intracellular lead chaperone protein (Taghavi et al., 2009), the pbrD gene product, binds lead and transfers it to the cell membrane where the pbrA gene product, a P1B-type ATPase efflux protein actively exports Pb(II) to the periplasm (Rensing et al., 1998). The pbrB gene product, an undecaprenyl pyrophosphate phosphatase (C55-PP phosphatase), produces inorganic phosphate (from the cell membrane) which combines with Pb(II) to form lead phosphate and is sequestered in the periplasm. This leads to discontinued expression of the pbr operon; however, pbrC and pbrD gene product synthesis is initiated (Hynninen et al., 2009). PbrD may also accumulate Pb(II) and prevent increased uptake of Pb(II) into the cell (Taghavi et al., 2009). The pbrC gene product is a lipoprotein signal peptidase that interacts with pbrB gene product (Taghavi et al., 2009). The pbrU gene product may be a major facilitator superfamily (MFS) membrane bound permease however pbrU is inactivated in C. metallidurans (Taghavi et al., 2009; Van Houdt, Monchy, Leys, & Mergeay, 2009). PbrA and pbrB are required for lead resistance; pbrT, pbrC, pbrD and pbrU are not (Hynninen et al., 2009; Taghavi et al., 2009).
C. metallidurans strain CH34 harbors additional genes that confer lead resistance. The pbrR2 (Rmet_2302), cadA, pbrC2 operon is located on chromosome 1 in a genomic island (CMGI-1) and may maintain low cellular Pb(II) concentration (Taghavi et al., 2009). The zntA gene (Zn(II) efflux protein), which is induced by Pb(II) (a Staphylococcus aureus CadA (Cd(II) efflux protein homolog in Escherichia coli induced by Zn(II)/Cd(II)/Pb(II); Beard et al., 1997; Rensing et al., 1998), is a P-Type ATPase located on chromosome 2. The pbrR3 gene (Rmet_3456; pbrR691) which preferentially binds Pb(II), is located on chromosome 1 (Monsieurs et al., 2011; Taghavi et al., 2009). These genes can rescue mutations in complementary genes in the C. metallidurans strain CH34 primary lead resistance operon, pbrUTRABCD (Taghavi et al., 2009).
Like the pbrR2, cadA, and pbrC2 operons (Rmet_2302), lead resistance genes are associated with genomic islands flanked by mobile genetic elements (Monchy et al., 2007; Taghavi et al., 2009; Van Houdt et al., 2009). For example, in Cupriavidus metallidurans, Pb(II) induces genes involved in transposition (tnpA, tnpR, and orf-2) and open reading frames (ORFs) from truncated insertion sequence (IS) elements (orf-102 and orf-103), and the lead resistance pbr operon is flanked by TN4380, a mercury transposon, suggesting potential for mobilization in the presence of lead (Monchy et al., 2007). Delftia acidovorans strain SPH-1 harbors pbr genes in a chromosomally-linked genomic island with other metal resistance determinants (Van Houdt et al., 2009). Interestingly, antibiotic resistance, which also can be linked to mobile genetic elements, has been reported to co-occur with heavy metal (including lead) resistance in many bacteria (Bharagava et al., 2014; El-Sayed, 2016; Hu and Chen, 2016; Koc, Kabatas, & Icgen, 2013; Learman et al., 2018; Matyar, 2012; Matyar et al., 2014; Pirela et al., 2014; Tomova et al., 2015). Antimicrobial resistance genes show correlation to heavy metal contamination in sediments (Ohore et al., 2018). Table 6 shows examples of multi-antibiotic & multi-heavy metal resistance bacteria such as Bacillus subtilis, Bacillus cereus, Bacillus pumilus, Frankia sp., Acinetobacter baumanni, Raoultella planticola, Microbacterium sp., Vibrio parahaemolyticus, Burkholderia sordidcola, and Pantoea sp. The fact that these bacteria are resistant to both antibiotics and heavy metals suggests that they have the ability to accumulate a suite of resistance genes. Each of these genes may encode a unique resistance functionality to a particular antibiotic or heavy metal and some of them may possess resistance mechanisms, such as efflux pumps, that provide resistance to several pollutants.
Table 6.
Examples of multi-antibiotic and multi-heavy metal resistance in bacteria.
| Source | Resistance | Phylum | Species | Reference |
|---|---|---|---|---|
| Industrial waste water | 14 antibiotics & 5 heavy metals | Gammaproteobacteria | Acinetobacter baumanni | El-Sayad, 2016 |
| Surface water | 15 antibiotics & 11 heavy metals | Gammaproteobacteria | Raoultella planticola | Koc et al., 2013 |
| Soil | 3 antibiotics & 7 heavy metalsb | Actinobacteria | Microbacterium sp. | Learman et al., 2018 |
| Coast | 16 antibiotics & 5 heavy metals | - | Gram negative bacteria | Matyar, 2012 |
| River | 4 antibiotics & 5 heavy metals | - | Gram negative bacteria | Matyar et al., 2014 |
4.2. Lead biosensors
Biological sensors are biologically active organisms that can detect the substrate, transport it into the cell or bind it on the cell surface through resistance mechanisms, and produce a rapid easy to measure response. Biosensors have been developed to detect lead by incorporating lead resistance genes, such as pbrR and pbrA and the luciferase reporter (luxCDABE) into a bacterial strain, such as Cupriavidus metallidurans AE2448 (formally Alcaligenes eutrophus; also referred to as BIOMET® Pb Biosensor or strain AE2450 (Table 7; (Geebelen et al., 2003; van der Lelie, Tibarzawa, & Corbisier, 2000). Lead specific biosensors are rare. C. metallidurans AE2448, which detects 0.5 μM lead did not detect any other heavy metals tested (Zn, Cu, Cd, Hg, Bi, Tl, Au; (Corbisier et al., 1999). Lead specific BIOMET® (Cupriavidus metallidurans AE2448/AE2450) has successfully detected 7 – 404 mg/kg Pb (both Pb spiked and unspiked) in environmental soil samples contaminated with lead and other heavy metals, demonstrating its utility in quantifying bioavailable lead (Geebelen et al., 2003; van der Lelie et al., 2000). In addition to lead, metal specific BIOMET® biosensor constructs using Cuprividus metallidurans CH4 have been developed for Zn, Cd, Cr and Ni and Cu in Cupriavidus silverii DS185 (formally Ralstonia silverii) (Corbisier et al., 1994; Corbisier et al., 1999; van der Lelie et al., 2000a; Van der Lelie et al., 2000b). When pbrR and pbrA are inserted into a Pseudomonas fluorescens plasmid (level of detection (LOD) 0.2 μM Pb) or the chromosome (LOD 0.9 μM Pb) with the lux reporter, Hg, Cd, and Zn also are detected (Corbisier et al., 1999). While the biosensors use the same genetic elements, host strains and constructs are not identical, and this may account for differences in specificity.
Table 7.
Selected lead bioreporters and host strains in in vitro experiments and natural environmental samples.
| Bacterial Host Strain | Sensor-Reporter Element | Pb2+ Level of Detection (Sensitivity) μM | Other Heavy Metals Detected (Specificity) | Reference |
|---|---|---|---|---|
| In vitro | ||||
| Alcaligenes eutrophusa AE2448 | pbrRPOΔpbrA::luxd | 0.5 | N.D.c | Corbisier et al., 1999 |
| Pseudomonas fluorescens OS8 | pDNpbrRPpbrAlux | 0.9 | Hg, Cd, Zn | Ivask et al., 2009 |
| Pseudomonas fluorescens OS8 | KnpbrRPpbrAlux | 0.2 | Hg, Cd, Zn | Ivask et al., 2009 |
| Pseudomonas fluorescens OS8 | pDNzntRPzntAlux | 0.4 | Hg, Cd, Zn | Ivask et al., 2009 |
| Pseudomonas fluorescens OS8 | pDNcadRPcadAlux | 0.4 | Hg, Cd, Zn | Ivask et al., 2009 |
| Staphyloccus aureus RN4220 | pTO024f | 0.033 | Cd, Sb Antimony | Tauriainen et al., 1998 |
| Pseudomonas putida KT2440 | pDNPczc1lux | 0.9 | Zn, Cd | Hynninen et al., 2010 |
| Pseudomonas putida KT2440.2431g | pDNPczc1lux | 0.02 | Zn, Cd | Hynninen et al., 2010 |
| Escherichia coli MG1655 | pZNTlux | 0.03 | Cd, Zn, Hg, Co, Ni, Sb, Cr | Riether et al., 2001 |
| Environmental samples | ||||
| BIOMET® Pb biosensor | pbrRPpbrAlux | 7 – 404 mg/kg Soil +/− Pb2+ amendmenth |
Geebelen et al., 2003 | |
| Cupriavidus metallidurans | zntAPlux | 1.2 – 4 ng/L Water |
N.D. | Zhang et al., 2017 |
| Alcaligenes eutrophusa AE1433 | pMOL30::Tn4431b | 4036-10469 mg/kg Incinerator fly-ash |
Zn, Cd, Co | Corbisier et al., 1996 |
Cupriavidus metallidurans (Betaproteobacteria); formally Alcaligenes eutrophus, Wautersia metallidurans, Ralstonia metallidurans)
Tn4431 inserted 1.4 kb downstream of czc region in cupS gene
N.D., not detected
Promoter/Operator region of pbrR. Also referenced as AE2450 or BIOMET® (Pb(II); Van der Lelie et al., 2007;)
N.A., not available
cadA
disrupted chromosomal P-type ATPase (cadA1, cadA2) and CBA transporter (czcCBA1, czcCBA2). Note that Pb2+ did not induce luciferase production in strains with pDNPcadA1lux.
BIOMET assay determined Pb(II) concentration
P1B-type ATPase efflux pumps, such as the pbrA gene product in Cupriavidus metallidurans, can confer Pb, Zn, and Cd resistance (Lee, Glickmann, & Cooksey, 2001; J. Liu, Dutta, Stemmler, & Mitra, 2006; Rensing et al., 1998). The Staphylococcus aureus and Pseudomonas putida efflux pump is encoded by cadA; its homolog in Escherichia coli is zntA (Lee et al., 2001; Liu et al., 2006; Rensing et al., 1997; Rensing et al., 1998). Staphylococcus aureus cadA confers lead resistance; however results are mixed in Pseudomonas putida (Lee et al., 2001; Leedjarv, Ivask, & Virta, 2008). Several Pseudomonas fluorescens heavy metal biosensors that harbor cadR (receptor) and cadA (P-type ATPase) inducible by Pb have been developed. However, they also detect Hg, Cd, and Zn (Ivask, Rolova, & Kahru, 2009). CadA Pseudmonas putida biosensors are not inducible by lead; yet they can detect Zn (Hynninen et al., 2010). Chromosomal insertion slightly improves lead detection performance (LOD 0.3 μM versus 0.4 μM). Similar performance (Pb LOD 0.33 μM) is observed in Staphylococcus aureus RN4220 harboring a cadAlux constructed plasmid that also detects Cd and antimony (Sb) (Tauriainen, Karp, Chang, & Virta, 1998).
Another biosensor approach harnesses zntA and zntR (receptor), the Zn/Cd/Pb/Hg transporting ATPase, involved in heavy metal resistance (Ivask et al., 2009). This construct shows good sensitivity to lead (LOD 0.4 μM, Pseudomonas fluorescens) and also detects Hg, Cd, and Zn (Ivask et al., 2009). In Escherichia coli MG1655 harboring plasmid pZNTlux, lead detection improves 10 fold (LOD 0.03 μM); however, this biosensor also detects Cd, Zn, Hg, Co, Ni, Sb, and Cr (Riether, Dollard, & Billard, 2001; Reither et al., 2001). Zhang et al. (2017) have developed a zntAlux biosensor that detects 1.2-4 ng/L of lead in environmental water samples, however specificity is not tested.
The czc1 gene product, Co/Zn/Cd efflux permease (CBA transporter), when incorporated into a plasmid with lux operon in Pseudmonas putida KT2440, detects Pb (LOD 0.9 μM), Zn, and Cd (Hynninen et al., 2010). Interestingly, in the cadA1, cadA2, czcCBA1, and czcCBA2 deletion strain Pseudomonas putida KT2440.2431, lead sensor sensitivity improves 45 fold (LOD 0.02 μM), possibly because chromosomal genes are not binding available lead or, because czcCBA1 may be involved in lead export, more lead remains in the cell (Hynninen et al., 2010; Leedjarv et al., 2008). Cuprividus metallidurans AE1433, which has a transposon (Tn4431, promoterless lux operon) inserted 1.4 kb downstream of the czc region in the cupS gene, can detect 4036-10469 mg/kg Pb in incinerator fly-ash as well as Zn, Cd, and Co (Corbisier, Thiry, & Diels, 1996).
5. Conclusions and future perspectives
While it has long been recognized that exposure to lead contamination is a critical stressor to microorganisms, the spatiotemporal variability of the microbial community and experimental uncertainties associated with the measurement of lead in the laboratory are the main reasons for the recommendation to the US EPA that microorganisms not be considered as an ecological endpoint in the screening level ecological risk assessment for Superfund sites (U.S. EPA, 2003). This recommendation partly inspired this literature review of relative research conducted over the past decades. Upon conclusion of this review, evidence supports that revisiting the recommendation with a focus on microbial indicators of lead contamination, could lead to the development of a framework to inform the ecological risk assessment for a contamination site (e.g., Superfund sites in the USA). To that end, a few points concluded from this review along with future research are worth discussing.
First, the current literature, including both in vitro and in situ studies, indicates that exposure to lead contamination inhibits microbial activity resulting in reduced respiration, suppressed metabolism, and reduced biomass as well as altered microbial community structure (Table 2; Table 3; de Vries et al., 2007; Xu et al., 2018; Xu et al., 2019). While most in vitro lead incubation studies are not long enough to reach equilibrium and represent field conditions, findings from them are generally in agreement with conclusions from long term in situ studies of lead and other heavy metals in terms of lead toxicity and impacts on microbial community structure and activities (Table 1). Although other physical, chemical, and biological factors in the field such as temperature, moisture, other concomitant heavy metals, pH, soil or sediment particle size distribution, organic carbon content, and vegetation types could mask microbial community shifts associated with lead contamination, the latest advancements in molecular techniques (e.g. 16S rRNA gene sequencing, functional gene microarray; metagenomic sequencing) coupled with statistical and network analyses have allowed for detailed phylogenetic analyses, functional gene identification, and discernment of the effects of lead and other heavy metals on microbial communities (Table 2; Table 3; Jacquiod et al., 2018). Even at sites where microbial communities are compositionally similar, long term heavy metal exposure results in functional differences regardless of the contamination level. Gene transfer and DNA recombination are more prevalent, thus providing a selective advantage at heavily contaminated sites more so than at less contaminated sites. While genomic approaches will continue to draw research interest in the scientific community, an emphasis of future research for consideration is standardizing the methodology for potential application in ecological risk assessments of contaminated sites. To that end, a unified molecular methodology for microbial community structure analysis should be developed and tested with samples representing varying spatiotemporal scales along with determination of total and bioavailable lead. Interlaboratory comparison of the resulting methodology can inform reduction of measurement uncertainties. The resulting microbial community shifts can serve as indices of lead contamination and bioavailability and inform both human health and ecological risk assessments.
Second, in apparent response to varying environmental levels of lead stress, microorganisms can initiate a series of lead resistance and adaptation mechanisms, even at a very early stage of exposure in the presence of low lead concentrations. It is well known that microbial lead resistance can be accomplished through intracellular and/or extracellular precipitation, adsorption, complexation and efflux pumps (Table 4; Table 5). Although identifying a unified set of lead resistant microorganisms among different sites can be challenging due to variable site conditions, some lead resistant bacteria such as the Gram negative Proteobacteria and Gram positive Firmicutes, Bacteroidetes, and Actinobacteria are common and dominant in many contaminated sites. These phyla also are found in piping biofilms where microorganisms are exposed to lead contamination due to corrosion of lead pipes (White et al., 2011). Thus, another important future research area is to examine lead resistant bacteria as benchmark indictors for evaluation of lead contamination and toxicity, because microbial lead toxicity is directly dependent upon lead bioavailability. Varying levels of bioavailable lead could trigger different resistance and adaptation strategies possessed by different bacterial strains in the community. In other words, the genes bacteria use to become adapted to a specific level or range of environmentally bioavailable lead offer a means to evaluate the ecotoxicological effects at contaminated sites. Thus, quantifying the relationship between the bioavailable portion of total lead and microbial ecotoxicity and lead resistance of benchmark bacteria can be very useful for site assessment. Carefully designed in vitro studies with varying magnitude and duration of exposure can be conducted in the laboratory to help achieve this goal.
Last but not least, the latest molecular understanding of lead resistance mechanisms has identified suites of functional genes responsible for specific lead resistance mechanisms. These resistance genes may serve as indicators of lead contamination in association with dominance of certain microbial phyla. This advanced understanding has led to the development of several lead biosensors in the realm of environmental biotechnology (Table 7). Using biosensors containing lead resistance genes may provide an indication of lead bioavailability because the microbes can sense biologically available lead (as opposed to unavailable, bound lead). In deciding which sensor to use, specificity is a key consideration because some sensors can detect several metals while a few are specific for lead. Nevertheless, the biosensor approach can be less costly to use as a screening assay and has application in lead bioavailability for ecological risk assessment. Future research should be directed to validate the effectiveness and reliability of these sensors with conventional methods for bioavailable lead detection.
In conclusion, microorganisms play an important role in lead biogeochemistry and have adapted to populate niches left available by lead sensitive microbes following contamination events. Understanding the dynamics of lead induced community shifts, to include prevalent resistance mechanisms, gives better insight into the toxicological effects of lead. Harnessing these organisms at the molecular, population, and community levels has the potential to better predict bioavailable lead levels, thereby strengthening both human and ecological risk assessments.
Figure 2.
Bacterial phyla in selected in situ studies of contaminated soils (light brown) and aquatic sediments (light blue): x denotes dominant and + or − indicates positive or negative correlation with levels of Pb and other heavy metals.
Acknowledgements
We would like to thank Drs. Richard Devereux and David Thomas for valuable reviews of earlier versions of this manuscript and Ms. Deborah Vivian for creating the figure. The views expressed in this chapter are those of the authors and do not necessarily reflect the views or policies of the U.S. Environmental Protection Agency. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Footnotes
Potential submission to Critical Reviews in Environmental Science and Technology (https://www.tandfonline.com/toc/best20/current).
References
- Akmal M, Wang H, Wu J, Xu J, & Xu D (2005). Changes in enzymes activity, substrate utilization pattern and diversity of soil microbial communities under cadmium pollution. J Environ Sci, 17, 802–807. [PubMed] [Google Scholar]
- Azarbad H, Niklinska M, van Gestel CA, van Straalen NM, Roling WF, & Laskowski R (2013). Microbial community structure and functioning along metal pollution gradients. Environ Toxicol Chem, 32(9), 1992–2002. doi: 10.1002/etc.2269 [DOI] [PubMed] [Google Scholar]
- Bharagava RN, Yadav S, & Chandra R (2014). Antibiotic and heavy metal resistance properties of bacteria isolated from the aeration lagoons of common effluent treatment plant (CETP) of tannery industries (Unnao, India). Indian Journal of Biotechnology, 13, 514–519. [Google Scholar]
- Beard SJ, Hashim R, Membrillo-Hernández J, Hughes MN, & Poole RK (1997). Zinc(II) tolerance inEscherichia coliK-12: evidence that thezntAgene (o732) encodes a cation transport ATPase. Molecular Microbiology, 25(05), 883–891. doi: 10.1111/j.1365-2958.1997.mmi518.x [DOI] [PubMed] [Google Scholar]
- Beattie RE, Henke W, Campa MF, Hazen TC, McAliley LR, & Campbell JH (2018). Variation in microbial community structure correlates with heavy-metal contamination in soils decades after mining ceased. Soil Biology and Biochemistry, 126, 57–63. doi: 10.1016/j.soilbio.2018.08.011 [DOI] [Google Scholar]
- Bhaskar PV, & Bhosle NB (2006). Bacterial extracellular polymeric substance (EPS): a carrier of heavy metals in the marine food-chain. Environ Int, 32(2), 191–198. doi: 10.1016/j.envint.2005.08.010 [DOI] [PubMed] [Google Scholar]
- Bitton G, & Freihofer V (1978). Influence of Extracellular Polysaccharides on the Toxicity of Copper and Cadmium toward Klebsiella aerogenes. Microbial Ecology, 4, 119–125. [DOI] [PubMed] [Google Scholar]
- Borremans B, Hobman JL, Provoost A, Brown NL, & van Der Lelie D (2001). Cloning and functional analysis of the pbr lead resistance determinant of Ralstonia metallidurans CH34. J Bacteriol, 183(19), 5651–5658. doi: 10.1128/JB.183.19.5651-5658.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braud A, Hoegy F, Jezequel K, Lebeau T, & Schalk IJ (2009). New insights into the metal specificity of the Pseudomonas aeruginosa pyoverdine-iron uptake pathway. Environ Microbiol, 11(5), 1079–1091. doi: 10.1111/j.1462-2920.2008.01838.x [DOI] [PubMed] [Google Scholar]
- Chang J-S, Law R, & Chang C-C (1997). Biosorption of lead, copper and cadmium by biomass of Pseudomonas aeruginosa PU21. Water Research, 31(7), 1651–1658. doi: 10.1016/S0043-1354(97)00008-0 [DOI] [Google Scholar]
- Chen J, He F, Zhang X, Sun X, Zheng J, & Zheng J (2014). Heavy metal pollution decreases microbial abundance, diversity and activity within particle-size fractions of a paddy soil. FEMS Microbiol Ecol, 87(1), 164–181. doi: 10.1111/1574-6941.12212 [DOI] [PubMed] [Google Scholar]
- Chen PR, Wasinger EC, Zhao J, van der Lelie D, Chen LX, & He C (2007). Spectroscopic insights into lead(II) coordination by the selective lead(II)-binding protein PbrR691. J Am Chem Soc, 129(41), 12350–12351. doi: 10.1021/ja0733890 [DOI] [PubMed] [Google Scholar]
- Chen Y, Jiang Y, Huang H, Mou L, Ru J, Zhao J, & Xiao S (2018). Long-term and high-concentration heavy-metal contamination strongly influences the microbiome and functional genes in Yellow River sediments. Sci Total Environ, 637-638, 1400–1412. doi: 10.1016/j.scitotenv.2018.05.109 [DOI] [PubMed] [Google Scholar]
- Chowdhury S, Thakur AR and Chaudhuri SR (2011). Novel Microbial Consortium for Laboratory Scale Lead Removal from City Effluent. J Environ Sci Tech, 4, 41–54 [Google Scholar]
- Chowdhury S, Mishra M, Adarsh VK, Mukherjee A, Thakur AR, & R. CS. (2008). Novel metal Accumulator and Protease Secretor Microbes from East Calcutta Wetland. American Journal of Biochem Biotech, 4(3), 255–264. [Google Scholar]
- Chu D, 2018. Effects of heavy metals on soil microbial community. IOP Conf. Series: Earth Environ Sci, 113, 012009, doi: 10.1088/1755-1315/113/1/012009. [DOI] [Google Scholar]
- Corbisier P, Thiry E, & Diels L (1996). Bacterial biosensors for the toxicity assessment of solid waste. Environ Toxicol Water Quality, 11(3), 171–177. doi:Doi [DOI] [Google Scholar]
- Corbisier P, Thiry E, Masolijn A & Diels L (1994) Construction and development of metal ion biosensors, in “Bioluminescence and Chemiluminsescence: Fundamentals and Applied Aspects” (pp. 1.50–155). Campbell AK, Kricka LJ and Stanley PE (Eds.), John Wiley & Sons, Chichester, New York, Brisbane, Toronto, Singapore. [Google Scholar]
- Corbisier P, van der Lelie D, Borremans B, Provoost A, de Lorenzo V, Brown NL, Lloyd JR, Hobman JL, Csöregi E, Johansson G and Mattiasson B (1999). Whole cell- and protein-based biosensors for the detection of bioavailable heavy metals in environmental samples. Anal Chem Acta 387, 235–244. doi:Doi 10.1016/S0003-2670(98)00725-9 [DOI] [Google Scholar]
- Dahlin S, Witter E, Martensson A, Turner A, & Baath E (1997). Where’s the Limit? Changes in the Microbiological Properties of Agricultural Soils at Low Levels of Metal Contamination. Soil. Bio. Biochem, 29(9), 1405–1415. [Google Scholar]
- De J, Ramaiah N, & Vardanyan L (2008). Detoxification of toxic heavy metals by marine bacteria highly resistant to mercury. Mar Biotechnol (NY), 10(4), 471–477. doi: 10.1007/s10126-008-9083-z [DOI] [PubMed] [Google Scholar]
- de Vries W, Lofts S, Tipping E, Meili M, Groenenberg JE, & Schutze G (2007). Impact of soil properties on critical concentrations of cadmium, lead, copper, zinc, and mercury in soil and soil solution in view of ecotoxicological effects. Rev Environ Contam Toxicol, 191, 47–89. [DOI] [PubMed] [Google Scholar]
- El-Sayed MH (2016). Multiple Heavy Metala and Antibiotic Resistance of Acinetobacter Baumannii Strain HAF - 13 Isolated from Industrial Effluents. Amer J Microbiol Res, 4(1), 26–36. doi: 10.12691/ajmr-4-1-3 [DOI] [Google Scholar]
- El-Shanshoury AE-RR, Elsilk SE, Ateya PS, & Ebeid EM (2012). Synthesis of lead nanoparticles by Enterobacter sp. and avirulent Bacillus anthracis PS2010. Ann Microbiol, 62(4), 1803–1810. doi: 10.1007/s13213-012-0438-3 [DOI] [Google Scholar]
- Essa AMM, Al Abboud MA, & Khatib SI (2018). Metal transformation as a strategy for bacterial detoxification of heavy metals. J Basic Microbiol, 58(1), 17–29. doi: 10.1002/jobm.201700143 [DOI] [PubMed] [Google Scholar]
- Fakruddin M, & Mannan KSB (2013). Methods for Analyzing Diversity of Microbial Communities in Natural Environments. Ceylon J Sci (Bio. Sci), 42(1), 19–33. doi: 10.4038/cjsbs.v42il.5896 [DOI] [Google Scholar]
- Federal Register, 2008. Rules and Regulations, National Ambient Air Quality Standards for Lead. 73, 66964–67062. [Google Scholar]
- Federal Register, 2016. Rules and Regulations, Review of the National Ambient Air Quality Standards for Lead. 81, 71906–81943. [Google Scholar]
- Feris K, Ramsey P, Frazar C, Moore JN, Gannon JE, & Holben WE (2003). Differences in Hyporheic-Zone Microbial Community Structure along a Heavy-Metal Contamination Gradient. Appl Environ Microbiol, 69(9), 5563–5573. doi: 10.1128/aem.69.9.5563-5573.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geebelen W, Adriano DC, van der Lelie D, Mench M, Carleer R, Clijsters H, & Vangronsveld J (2003). Selected bioavailability assays to test the efficacy of amendment-induced immobilization of lead in soils. Plant Soil, 249(1), 217–228. doi:Doi 10.1023/A:1022534524063 [DOI] [Google Scholar]
- Gillan DC, Danis B, Pernet P, Joly G, & Dubois P (2005). Structure of sediment-associated microbial communities along a heavy-metal contamination gradient in the marine environment. Appl Environ Microbiol, 71(2), 679–690. doi: 10.1128/AEM.71.2.679-690.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillan DC, Roosa S, Kunath B, Billon G, & Wattiez R (2015). The long-term adaptation of bacterial communities in metal-contaminated sediments: a metaproteogenomic study. Environ Microbiol, 17(6), 1991–2005. doi: 10.1111/1462-2920.12627 [DOI] [PubMed] [Google Scholar]
- Giller KE, Witter E, & McGrath SP (2009). Heavy metals and soil microbes. Soil Biol Biochem, 41(10), 2031–2037. doi: 10.1016/j.soilbio.2009.04.026 [DOI] [Google Scholar]
- Green VS, Stottb DE, & Diacka M (2006). Assay for fluorescein diacetate hydrolytic activity: Optimization for soil samples. Soil Biol Biochem 38, 693–701. [Google Scholar]
- Gong J, Zhang Z, Bai H, & Yang G (2007). Microbiological synthesis of nanophase PbS by Desulfotomaculum sp. Science in China Series E: Technological Sciences, 50(3), 302–307. doi: 10.1007/s11431-007-0045-x [DOI] [Google Scholar]
- Guo J, Kang Y, & Feng Y (2017). Bioassessment of heavy metal toxicity and enhancement of heavy metal removal by sulfate-reducing bacteria in the presence of zero valent iron. J Environ Manage, 203(Pt 1), 278–285. doi: 10.1016/j.jenvman.2017.07.075 [DOI] [PubMed] [Google Scholar]
- Guo J, Kang Y, & Feng Y (2017). Bioassessment of heavy metal toxicity and enhancement of heavy metal removal by sulfate-reducing bacteria in the presence of zero valent iron. Journal of Environmental Management, 203, 278–285. doi: 10.1016/j.jenvman.2017.07.075 [DOI] [PubMed] [Google Scholar]
- Guo Q, Li N, Bing Y, Chen S, Zhang Z, Chang S, … Xie S (2018). Denitrifier communities impacted by heavy metal contamination in freshwater sediment. Environ Poll, 242, 426–432. doi: 10.1016/j.envpol.2018.07.020 [DOI] [PubMed] [Google Scholar]
- Hobman JL, Julian DJ, & Brown NL (2012). Cysteine coordination of Pb(II) is involved in the PbrR-dependent activation of the lead-resistance promoter, PpbrA, from Cupriavidus metallidurans CH34. BMC Microbiol, 12, 109. doi: 10.1186/1471-2180-12-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, & Chen L (2016). Virulence and antibiotic and heavy metal resistance of Vibrio parahaemolyticus isolated from crustaceans and shellfish in Shangahai, China. J Food Protect, 79(8), 1371–1377. doi: 10.4315/0362-028X.JFP-16-031 [DOI] [PubMed] [Google Scholar]
- Hu Q, Qi H.-y., Zeng J.-h., & Zhang H.-x. (2007). Bacterial diversity in soils around a lead and zinc mine. Journal of Environmental Sciences, 19(1), 74–79. doi: 10.1016/S1001-0742(07)60012-6 [DOI] [PubMed] [Google Scholar]
- Hynninen A, Tonismann K, & Virta M (2010). Improving the sensitivity of bacterial bioreporters for heavy metals. Bioeng Bugs, 1(2), 132–138. doi: 10.4161/bbug.1.2.10902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynninen A, Touze T, Pitkanen L, Mengin-Lecreulx D, & Virta M (2009). An efflux transporter PbrA and a phosphatase PbrB cooperate in a lead-resistance mechanism in bacteria. Mol Microbiol, 74(2), 384–394. doi: 10.1111/j.1365-2958.2009.06868.x [DOI] [PubMed] [Google Scholar]
- Ivask A, Rolova T, & Kahru A (2009). A suite of recombinant luminescent bacterial strains for the quantification of bioavailable heavy metals and toxicity testing. BMC Biotechnol, 9, 41. doi: 10.1186/1472-6750-9-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquiod S, Cyriaque V, Riber L, Al-Soud WA, Gillan DC, Wattiez R, & Sorensen SJ (2018). Long-term industrial metal contamination unexpectedly shaped diversity and activity response of sediment microbiome. J Hazard Mater, 344, 299–307. doi: 10.1016/j.jhazmat.2017.09.046 [DOI] [PubMed] [Google Scholar]
- Jaiswal D, & Pandey J (2018). Impact of heavy metal on activity of some microbial enzymes in the riverbed sediments: Ecotoxicological implications in the Ganga River (India). Ecotoxicology and Environmental Safety, 150, 104–115. doi: 10.1016/j.ecoenv.2017.12.015 [DOI] [PubMed] [Google Scholar]
- Jaroslawiecka A, & Piotrowska-Seget Z (2014). Lead resistance in micro-organisms. Microbiology, 160(Pt 1), 12–25. doi: 10.1099/mic.0.070284-0 [DOI] [PubMed] [Google Scholar]
- Jencova V, Strnad H, Chodora Z, Ulbrich P, Vlcek C, Hickey WJ, & Paces V (2008). Nucleotide sequence, organization and characterization of the (halo)aromatic acid catabolic plasmid pA81 from Achromobacter xylosoxidans A8. Res Microbiol, 159(2), 118–127. doi: 10.1016/j.resmic.2007.11.018 [DOI] [PubMed] [Google Scholar]
- Jiang B, Adebayo A, Jia J, Xing Y, Deng S, Guo L, … Zhang D (2019). Impacts of heavy metals and soil properties at a Nigerian e-waste site on soil microbial community. Journal of Hazardous Materials, 362, 187–195. doi: 10.1016/j.jhazmat.2018.08.060 [DOI] [PubMed] [Google Scholar]
- Jie S, Li M, Gan M, Zhu J, Yin H, & Liu X (2016). Microbial functional genes enriched in the Xiangjiang River sediments with heavy metal contamination. BMC Microbiol, 16(1), 179. doi: 10.1186/s12866-016-0800-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C-H, Oh SJ, Shin Y, Han S-H, Nam I-H, & So J-S (2015). Bioremediation of lead by ureolytic bacteria isolated from soil at abandoned metal mines in South Korea. Ecological Engineering, 74, 402–407. doi: 10.1016/j.ecoleng.2014.10.009 [DOI] [Google Scholar]
- Karimpour M, Ashrafi SD, Taghavi K, Mojtahedi A, Roohbakhsh E, & Naghipour D (2018). Adsorption of cadmium and lead onto live and dead cell mass of Pseudomonas aeruginosa: A dataset. Data Brief, 18, 1185–1192. doi: 10.1016/j.dib.2018.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koc S, Kabatas B, & Icgen B (2013). Multidrug and heavy metal-resistant Raoultella planticola isolated from surface water. Bull Environ Contam Toxicol, 91(2), 177–183. doi: 10.1007/s00128-013-1031-6 [DOI] [PubMed] [Google Scholar]
- Kuppusamy S, Thavamani P, Megharaj M, Venkateswarlu K, Lee YB, & Naidu R (2016). Pyrosequencing analysis of bacterial diversity in soils contaminated long-term with PAHs and heavy metals: Implications to bioremediation. J Hazard Mater, 317, 169–179. doi: 10.1016/j.jhazmat.2016.05.066 [DOI] [PubMed] [Google Scholar]
- Lee SW, Glickmann E, & Cooksey DA (2001). Chromosomal locus for cadmium resistance in Pseudomonas putida consisting of a cadmium-transporting ATPase and a MerR family response regulator. Appl Environ Microbiol, 67(4), 1437–1444. doi: 10.1128/AEM.67.4.1437-1444.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leedjarv A, Ivask A, & Virta M (2008). Interplay of different transporters in the mediation of divalent heavy metal resistance in Pseudomonas putida KT2440. J Bacteriol, 190(8), 2680–2689. doi: 10.1128/JB.01494-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenart A, & Wolny-Koladka K (2013). The effect of heavy metal concentration and soil pH on the abundance of selected microbial groups within ArcelorMittal Poland steelworks in Cracow. Bull Environ Contam Toxicol, 90(1), 85–90. doi: 10.1007/s00128-012-0869-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Meng D, Li J, Yin H, Liu H, Liu X, … Yan M (2017). Response of soil microbial communities and microbial interactions to long-term heavy metal contamination. Environmental Pollution, 231, 908–917. doi: 10.1016/j.envpol.2017.08.057 [DOI] [PubMed] [Google Scholar]
- Li X, Peng W, Jia Y, Lu L, & Fan W (2016). Bioremediation of lead contaminated soil with Rhodobacter sphaeroides. Chemosphere, 156, 228–235. doi: 10.1016/j.chemosphere.2016.04.098 [DOI] [PubMed] [Google Scholar]
- Li Y, Wang Y, Gou X, Su Y, & Wang G (2006). Risk assessment of heavy metals in soils and vegetables around non-ferrous metals mining and smelting sites, Baiyin, China. Journal of Environmental Sciences, 18(6), 1124–1134. [DOI] [PubMed] [Google Scholar]
- Liu C, Lin H, Dong Y, Li B, & Liu Y (2018). Investigation on microbial community in remediation of lead-contaminated soil by Trifolium repensL. Ecotoxicol Environ Saf, 165, 52–60. doi: 10.1016/j.ecoenv.2018.08.054 [DOI] [PubMed] [Google Scholar]
- Liu J, Dutta SJ, Stemmler AJ, & Mitra B (2006). Metal-binding affinity of the transmembrane site in ZntA: implications for metal selectivity. Biochemistry, 45(3), 763–772. doi: 10.1021/bi051836n [DOI] [PubMed] [Google Scholar]
- Macaskie LE, & Dean ACR (1987). Use of Immobilized Biofilm of Citrobacter Sp for the Removal of Uranium and Lead from Aqueous Flows. Enzyme and Microbial Technology, 9(1), 2–4. doi:Doi 10.1016/0141-0229(87)90042-1 [DOI] [Google Scholar]
- Macaskie LE, & Dean ACR (1987). Use of immobilized biofilm of Citrobacter sp. for the removal of uranium and lead from aqueous flows. Enzyme and Microbial Technology, 9(1), 2–4. doi: 10.1016/0141-0229(87)90042-1 [DOI] [Google Scholar]
- Marvasi M, Visscher PT, & Casillas Martinez L (2010). Exopolymeric substances (EPS) from Bacillus subtilis: polymers and genes encoding their synthesis. FEMS Microbiol Lett, 313(1), 1–9. doi: 10.1111/j.1574-6968.2010.02085.x [DOI] [PubMed] [Google Scholar]
- Mergeay M, Nies D, Schlegel HG, Gerits J, Charles P, & Vangijsegem F (1985). Alcaligenes-Eutrophus Ch34 Is a Facultative Chemolithotroph with Plasmid-Bound Resistance to Heavy-Metals. Journal of Bacteriology, 162(1), 328–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mire CE, Tourjee JA, O’Brien WF, Ramanujachary KV, & Hecht GB (2004). Lead Precipitation by Vibrio harveyi: Evidence for Novel Quorum-Sensing Interactions. Applied and Environmental Microbiology, 70(2), 855–864. doi: 10.1128/aem.70.2.855-864.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monchy S, Benotmane MA, Janssen P, Vallaeys T, Taghavi S, van der Lelie D, & Mergeay M (2007). Plasmids pMOL28 and pMOL30 of Cupriavidus metallidurans are specialized in the maximal viable response to heavy metals. J Bacteriol, 189(20), 7417–7425. doi: 10.1128/JB.00375-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsieurs P, Moors H, Van Houdt R, Janssen PJ, Janssen A, Coninx I, … Leys N (2011). Heavy metal resistance in Cupriavidus metallidurans CH34 is governed by an intricate transcriptional network. Biometals, 24(6), 1133–1151. doi: 10.1007/s10534-011-9473-y [DOI] [PubMed] [Google Scholar]
- Murthy. (2011). Effect of lead on metallothionein concentration in lead-resistant bacteria Bacillus cereus isolated from industrial effluent. African Journal of Biotechnology, 10(71), 15966–15972. doi: 10.5897/ajb11.1645 [DOI] [Google Scholar]
- Naik MM, & Dubey SK (2011). Lead-enhanced siderophore production and alteration in cell morphology in a Pb-resistant Pseudomonas aeruginosa strain 4EA. Curr Microbiol, 62(2), 409–414. doi: 10.1007/s00284-010-9722-2 [DOI] [PubMed] [Google Scholar]
- Naik MM, & Dubey SK (2013). Lead resistant bacteria: lead resistance mechanisms, their applications in lead bioremediation and biomonitoring. Ecotoxicol Environ Saf, 98, 1–7. doi: 10.1016/j.ecoenv.2013.09.039 [DOI] [PubMed] [Google Scholar]
- Naik MM, Pandey A, & Dubey SK (2012a). Biological characterization of lead-enhanced exopolysaccharide produced by a lead resistant Enterobacter cloacae strain P2B. Biodegradation, 23(5), 775–783. doi: 10.1007/s10532-012-9552-y [DOI] [PubMed] [Google Scholar]
- Naik MM, Pandey A, & Dubey SK (2012b). Pseudomonas aeruginosa strain WI-1 from Mandovi estuary possesses metallothionein to alleviate lead toxicity and promotes plant growth. Ecotoxicol Environ Saf, 79, 129–133 [DOI] [PubMed] [Google Scholar]
- Naik MM, Shamim K, & Dubey SK (2012c). Biological characterization of lead-resistant bacteria to explore role of bacterial metallothionein in lead resistance. Current Science, 103(4), 426–429. [Google Scholar]
- National Research Council of the National Academies (2003). Bioavailability of Contaminants in Soils and Sediments: Processes, Tools, and Applications. The National Academies Press, Washington, D.C. pp 1–420. [Google Scholar]
- Nayar S, Goh BP, & Chou LM (2004). Environmental impact of heavy metals from dredged and resuspended sediments on phytoplankton and bacteria assessed in in situ mesocosms. Ecotoxicol Environ Saf, 59(3), 349–369. doi: 10.1016/j.ecoenv.2003.08.015 [DOI] [PubMed] [Google Scholar]
- Naz N, Young HK, Ahmed N, & Gadd GM (2005). Cadmium accumulation and DNA homology with metal resistance genes in sulfate-reducing bacteria. Appl Environ Microbiol, 71(8), 4610–4618. doi: 10.1128/AEM.71.8.4610-4618.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilands JB (1995). Siderophores: Structure and Function of Microbial Iron Transport Compounds. Journal of Biological Chemistry, 270(45), 26723–26726. doi: 10.1074/jbc.270.45.26723 [DOI] [PubMed] [Google Scholar]
- Nelson YM, Lo W, Lion LW, Shuler ML, & Ghiorse WC (1995). Lead distribution in a simulated aquatic environment: Effects of bacterial biofilms and iron oxide. Water Research, 29(8), 1934–1944. doi: 10.1016/0043-1354(94)00351-7 [DOI] [Google Scholar]
- O’Brien S, Hodgson DJ, & Buckling A (2014). Social evolution of toxic metal bioremediation in Pseudomonas aeruginosa. Proc Biol Sci, 281(1787). doi: 10.1098/rspb.2014.0858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohore OE, Addo FG, Zhang S, Han N, & Anim-Larbi K (2018). Distribution and relationship between antimicrobial resistance genes and heavy metals in surface sediments of Taihu Lake, China. Journal of Environmental Sciences. doi: 10.1016/j.jes.2018.09.004 [DOI] [PubMed] [Google Scholar]
- Pal C, Asiani K, Arya S, Rensing C, Stekel DJ, Larsson DGJ, & Hobman JL (2017). Metal resistance and its association with antibiotic resistance. Adv Microb Physiol, 70, 261–313. doi: 10.1016/bs.ampbs.2017.02.001 [DOI] [PubMed] [Google Scholar]
- Pan X, Chen Z, Li L, Rao W, Xu Z, & Guan X (2017). Microbial strategy for potential lead remediation: a review study. World J Microbiol Biotechnol, 33(2), 35. doi: 10.1007/s11274-017-2211-z [DOI] [PubMed] [Google Scholar]
- Pirela MLR, Suárez WAB, & Vargas MMB (2014). Antibiotic- and heavy-metal resistance in bacteria isolated from deep subsurface in El Callao region, Venezuela. Rev Colomb Biotecnol, 16(2), 141–149. [Google Scholar]
- Ren Y, Niu J, Huang W, Peng D, Xiao Y, Zhang X, … Yin H (2016). Comparison of microbial taxonomic and functional shift pattern along contamination gradient. BMC Microbiol, 16(1), 110. doi: 10.1186/s12866-016-0731-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing C, Mitra B, & Rosen B (1997). The zntA gene of Escherichia coli encodes a Zn(II)-translocating P-type ATPase. Proc. Natl. Acad. Sci. USA, 94, 14326–14331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing C, Sun Y, Mitra B, & Rosen BP (1998). Pb(II)-translocating P-type ATPases. Journal of Biological Chemistry, 273(49), 32614–32617. doi: 10.1074/jbc.273.49.32614 [DOI] [PubMed] [Google Scholar]
- Riether K, Dollard MA, & Billard P (2001). Assessment of heavy metal bioavailability using Escherichia coli zntAp::lux and copAp::lux-based biosensors. Applied Microbiology and Biotechnology, 57(5-6), 712–716. doi: 10.1007/s00253-001-0852-0 [DOI] [PubMed] [Google Scholar]
- Roane TM (1999). Lead Resistance in Two Bacterial Isolates from Heavy Metal–Contaminated Soils. Microbial Ecology, 37(3), 218–224. doi: 10.1007/s002489900145 [DOI] [PubMed] [Google Scholar]
- Robinson NJ, Gupta A, Fordham-Skelton AP, Croy RRD, Whitton BA, & Huckle JW (1990). Prokaryotic metallothionein gene characterization and expression: chromosome crawling by ligation-mediated PCR. 10.1098/rspb.1990.0130 [DOI] [PubMed] [Google Scholar]
- Roosa S, Wattiez R, Prygiel E, Lesven L, Billon G, & Gillan DC (2014). Bacterial metal resistance genes and metal bioavailability in contaminated sediments. Environ Pollut, 189, 143–151. doi: 10.1016/j.envpol.2014.02.031 [DOI] [PubMed] [Google Scholar]
- Roosa S, Wauven CV, Billon G, Matthijs S, Wattiez R, & Gillan DC (2014). The Pseudomonas community in metal-contaminated sediments as revealed by quantitative PCR: a link with metal bioavailability. Res Microbiol, 165(8), 647–656. doi: 10.1016/j.resmic.2014.07.011 [DOI] [PubMed] [Google Scholar]
- Seiler S, & Berendonk TU (2012). Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Frontiers in Microbiology, 3(Article 399), 1–9. doi: 10.3389/fmicb.2012.00399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma J, Shamim K, Dubey SK, & Meena RM (2017). Metallothionein assisted periplasmic lead sequestration as lead sulfite by Providencia vermicola strain SJ2A. Sci Total Environ, 579, 359–365. doi: 10.1016/j.scitotenv.2016.11.089 [DOI] [PubMed] [Google Scholar]
- Shi W, Bischoff M, Turco R, & Konopka A (2002). Long-term effects of chromium and lead upon the activity of soil microbial communities. Applied Soil Ecology, 21(2), 169–177. doi 10.1016/S0929-1393(02)00062-8 [DOI] [Google Scholar]
- Sobolev D, & Begonia MFT (2008). Effects of heavy Metal Contamination upon Soil Microbes: Lead-induced Changes in General and Dneitrifying Microbial Communities as Evidenced by Molecular Markers. International Journal of Environmental Research and Public Health, 5(5), 450–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghavi S, Lesaulnier C, Monchy S, Wattiez R, Mergeay M, & van der Lelie D (2009). Lead(II) resistance in Cupriavidus metallidurans CH34: interplay between plasmid and chromosomally-located functions. Antonie Van Leeuwenhoek, 96(2), 171–182. doi: 10.1007/s10482-008-9289-0 [DOI] [PubMed] [Google Scholar]
- Tauriainen S, Karp M, Chang W, & Virta M (1998). Luminescent bacterial sensor for cadmium and lead. Biosensors and Bioelectronics, 13(9), 931–938. doi: 10.1016/s0956-5663(98)00027-x [DOI] [PubMed] [Google Scholar]
- Tipayno SC, Truu J, Samaddar S, Truu M, Preem JK, Oopkaup K, … Sa T (2018). The bacterial community structure and functional profile in the heavy metal contaminated paddy soils, surrounding a nonferrous smelter in South Korea. Ecol Evol, 8(12), 6157–6168. doi: 10.1002/ece3.4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomova I, Stoilova-Disheva M, Lazarkevich I, & Vasileva-Tnkova E (2015). Antimicrobial activity and resistance to heavy metals and antibiotics of heterotophic bacteria isolated from sediment and soil samples collected from two Antarctic islands. Frontiers in Life Science, 8(4), 348–357. doi: 10.1080/21553769.2015.1044130 [DOI] [Google Scholar]
- Tong S, von Schirnding YE, and Prapamontol T. 2000. Environmental lead exposure: a public health problem of global dimensions. Bulletin of the World Health Organization 78: 1068–1077. [PMC free article] [PubMed] [Google Scholar]
- Turner JS, Morby AP, A. WB, Gupta A, & Robinson NJ (1993). Construction of Zn2+/Cd2+Hypersensitive Cyanobacterial Mutants Lacking a Functional Metallothioinein Locus. The Journal of Biological Chemistry, 268(6), 4494–4498. [PubMed] [Google Scholar]
- U.S. Department of Health and Human Services, Public Health Service, Agency for Toxic Substances and Disease Registry (2007). Toxicology Profile for Lead. https://www.atsdr.cdc.gov/toxprofiles/tp13.pdf, retrieved 04/15/2019.
- U.S. Environmental Protection Agency, 2001. Hazard standards for lead in paint, dust and soil (TSCA Section 403).
- U.S. Environmental Protection Agency, 2003. Attachment 1-2, Guidance for Developing Ecological Soil Screening Levels (Eco-SSLs): Assessment of Whether to Develop Ecological Soil Screening Levels for Microbes and Microbial Processes. OSWER Directive 92857–55. [Google Scholar]
- U.S. Environmental Protection Agency, 2007. Framework for the Metals Risk Assessment. EPA 120/R-07/001. [Google Scholar]
- U.S. Environmental Protection Agency, 2007. Guidance for Evaluating the Oral Bioavailability of Metals in Soils for Use in Human Health Risk Assessment. OSWER 9285.7-80.
- U.S. Environmental Protection Agency, 2008. Lead and Copper Rule: A Quick Reference Guide. EPA; 816-F-08-018. [Google Scholar]
- U.S. Environmental Protection Agency (2019a). Lead at Superfund Sites: Risk Assessment. https://www.epa.gov/superfund/lead-superfund-sites-risk-assessment, retrieved 2/14/2019.
- U.S. Executive Office of the President, 2018. President’s Task Force on Environmental Health Risks and Safety Risks to Children. Federal Action Plan to Reduce Childhood Lead Exposures and Associated Health Impacts.
- World Health Organization (2011). Lead in Drinking-water: Background document for development of WHO Guidelines for Drinking-water Quality, WHO/SDE/WSH/03.04/09/Rev/1. [Google Scholar]
- World Health Organization, International Agency for Research on Cancer (2006). Inorganic and Organic Lead Compounds, IARC Working Group on the Evaluation of Carcinogenic Risks to Humans (2004: Lyon, France: ), volume 87, 1–506. [PMC free article] [PubMed] [Google Scholar]
- Van Der Lelie D, Tibarzawa C, & Corbisier P, Vangronsveld J & Mench M (2000a) Bacterial Biosensors to quanitify bioavailable concentratoin of heavy metals in polluted soils and to predict their risk of transfer to the food chain. 25th Int Conf Heavy Metals in the Environment, Bioadsorption and biomonitoring, University of Michigan, Ann Arbor, MI. [Google Scholar]
- Van Der Lelie D, Verschaeve L, Regniers L, Corbisier P (2000b) Use of bacterial tests (the VITOTOX® genotoxicity test and the BIOMET heavy metal test) to analyze chemicals and environmental samples. In: Persoone G, Janssen C, De Coen W (eds) New Microbiotests for Routine Toxicity Screening and Biomonitoring. Springer, Boston, MA. doi: 10.1007/978-1-4615-4289-6_20 [DOI] [Google Scholar]
- Van Houdt R, Monchy S, Leys N, & Mergeay M (2009). New mobile genetic elements in Cupriavidus metallidurans CH34, their possible roles and occurrence in other bacteria. Antonie Van Leeuwenhoek, 96(2), 205–226. doi: 10.1007/s10482-009-9345-4 [DOI] [PubMed] [Google Scholar]
- Xu X, Zhang Z, Hu S, Ruan Z, Jiang J, Chen C, & Shen Z (2017). Response of soil bacterial communities to lead and zinc pollution revealed by Illumina MiSeq sequencing investigation. Environ Sci Pollut Res Int, 24(1), 666–675. doi: 10.1007/s11356-016-7826-3 [DOI] [PubMed] [Google Scholar]
- Xu Y, Seshadri B, Bolan N, Sarkar B, Ok YS, Zhang W, … Dong Z (2019). Microbial functional diversity and carbon use feedback in soils as affected by heavy metals. Environ Int, 125, 478–488. doi: 10.1016/j.envint.2019.01.071 [DOI] [PubMed] [Google Scholar]
- Xu Y, Seshadri B, Sarkar B, Wang H, Rumpel C, Sparks D, … Bolan N (2018). Biochar modulates heavy metal toxicity and improves microbial carbon use efficiency in soil. Science of The Total Environment, 621, 148–159. doi: 10.1016/j.scitotenv.2017.11.214 [DOI] [PubMed] [Google Scholar]
- Yang Y, Liu G, Song W, Ye C, Lin H, Li Z, & Liu W (2019). Plastics in the marine environment are reservoirs for antibiotic and metal resistance genes. Environment International, 123, 79–86. doi: 10.1016/j.envint.2018.11.061 [DOI] [PubMed] [Google Scholar]
- Yin H, Niu J, Ren Y, Cong J, Zhang X, Fan F, … Liu X (2015). An integrated insight into the response of sedimentary microbial communities to heavy metal contamination. Sci Rep, 5, 14266. doi: 10.1038/srep14266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Xu Y, Zhao M, Rong H, & Zhang K (2018). Influence of inoculating white-rot fungi on organic matter transformations and mobility of heavy metals in sewage sludge based composting. J Hazard Mater, 344, 163–168. doi: 10.1016/j.jhazmat.2017.10.017 [DOI] [PubMed] [Google Scholar]
- Zhang H, Wan Z, Ding M, Wang P, Xu X, & Jiang Y (2018). Inherent bacterial community response to multiple heavy metals in sediment from river-lake systems in the Poyang Lake, China. Ecotoxicology and Environmental Safety, 165, 314–324. doi: 10.1016/j.ecoenv.2018.09.010 [DOI] [PubMed] [Google Scholar]
- Zheng H, Chen L, Li N, Liu B, Meng N, Wang M, & Chen S.-b. (2017). Toxicity threshold of lead (Pb) to nitrifying microorganisms in soils determined by substrate-induced nitrification assay and prediction model. Journal of Integrative Agriculture, 16(8), 1832–1840. doi: 10.1016/s2095-3119(16)61586-1 [DOI] [Google Scholar]


