Abstract
Bacterial cytochrome P450 (P450) 101A1 (P450cam) has served as a prototype among the P450 enzymes and has high catalytic activity towards its cognate substrate, camphor. X-ray crystallography and NMR and IR spectroscopy have demonstrated the existence of multiple conformations of many P450s, including P450cam. Kinetic studies have indicated that substrate binding to several P450s is dominated by a conformational selection process, in which the substrate binds an individual conformer(s) of the unliganded enzyme. P450cam was found to differ in that binding of the substrate camphor is dominated by an induced fit mechanism, in which the enzyme binds camphor and then changes conformation, as evidenced by the equivalence of binding eigenvalues observed when varying both camphor and P450cam concentrations. The accessory protein putidaredoxin had no effect on substrate binding. Estimation of the rate of dissociation of the P450cam·camphor complex (15 s−1) and fitting of the data yield a minimal kinetic mechanism in which camphor binds (1.5 × 107 M−1 s−1) and the initial P450cam•camphor complex undergoes a reversible equilibrium (kforward 112 s−1, kreverse 28 s−1) to a final complex. This induced fit mechanism differs from those reported for several mammalian P450s and bacterial P450BM-3, indicative of the diversity of how P450s recognize multiple substrates. However, similar behavior was not observed with the alternate substrates (+)-α-pinene and 2-adamantanone, which probably utilize a conformational selection process.
Keywords: camphor, cytochrome P450, conformational selection, enzyme kinetics, enzyme mechanisms, induced fit, P450cam, substrate binding
Graphical Abstract
INTRODUCTION
Cytochrome P450 (P450, CYP) enzymes catalyze the most oxidation-reduction reactions.1 This dominance is due to two major features of this group of > 400,000 enzymes: (i) their collective roles in the oxidations of a great variety of drugs, steroids, vitamins, natural products, industrial chemicals, and other substrates, due in part to spacious and malleable active sites,2 and (ii) multiple types of oxidations and rearrangements that can develop from an almost universally common reactive intermediate, Compound I (FeO3+).3-5 Further, Arnold and others have used P450s as a scaffold to catalyze unnatural reactions outside the normal repertoire of oxidation and reduction.6-13
Even with a single P450, e.g. human P450 3A4, thousands of chemicals are substrates14 but remarkable regio- and enantioselectivity are commonly seen.2, 15 How do P450s achieve such reaction diversity? The question is a part of the more general issue of how enzymes can achieve both catalytic diversity and selectivity as well as remarkable rate enhancement, a biochemical conundrum that is decades old.16-17 One of the concepts that is widely accepted today is the conformational flexibility of enzymes, generating “landscapes” of reaction profiles.18 Conformational flexibility is harnessed into catalytic efficiency and reaction selectivity, and two extremes in the mechanism of achieving optimal enzyme-substrate complementarity are conformational selection and induced fit (Scheme 1).19-23
Scheme 1. Conformational Selection and Induced Fit Mechanisms for Binding Enzyme and Substrate.
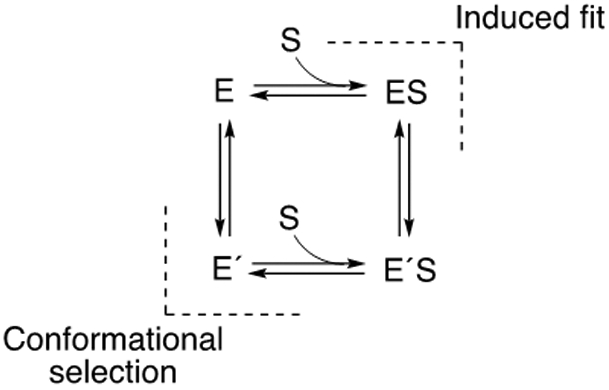
Briefly, conformational selection involves the existence of multiple forms of an enzyme, and one (or more) is complementary to a ligand, which is then bound (in a productive pose).19, 22 With an induced fit mechanism, the binding of a ligand to an enzyme promotes a change in the conformation to a productive mode.24 These two mechanisms can both be operative for a single enzyme. Which of these mechanisms is dominant must be studied kinetically, in that thermodynamics alone does not distinguish which of two paths is followed if the free energy difference in going from E to E´S is identical (Scheme 1).20
Recent work in this laboratory has used kinetic approaches to the problem of conformational dynamics20-21 with P450 enzymes. A number of human P450s (2C8, 2D6, 3A4, 4A11, 17A1, 46A1) exhibited kinetic behavior indicative of a dominant conformational selection process.25-27 The work was extended to a well-characterized soluble bacterial P450, P450BM-3 (CYP102A1), which also exhibited conformational selection behavior in its binding of the substrates myristic acid and dodecyl sulfate.28 In light of these findings, we considered the hypothesis that all P450s might utilize a conformational selection process.
P450cam (CYP101A1) is a bacterial P450, encoded by a plasmid in the bacterium Pseudomonas putida, that was discovered by Gunsalus and associates in 1965.29 Its high level of expression, high catalytic activity, and solubility proved to be major advantages in studying P450 enzymatic mechanisms and biophysical properties, and this was the first P450 to be purified, sequenced, and crystallized.29-35 An October 2020 SciFinder search yielded 1,418 articles published with the terms “P450cam or P-450cam or CYP101.” The enzyme catalyzes the 5-exo hydroxylation of the substrate d-(+)-camphor at high rates for a P450 (e.g., >10 s−1).35-36 However, the enzyme can also utilize some alternate substrates (e.g., norcamphor, (+)-α-pinene, 2-adamantanone), and Wong and others have used site-directed mutagenesis to further expand the catalytic versatility of P450cam.37-39 Thus, questions about conformational dynamics and catalytic specificity are in order even for this relatively well-characterized enzyme.
The first X-ray crystal structure of P450cam was solved with camphor in the active site.34 Since then, at least 158 structures for P450cam and site-directed mutants have been deposited in the Protein Data Bank (https://www.rcsb.org), including those with different oxidation-reduction states and ligands. Although some of the early considerations about P450cam were based on the view of a “closed” state with or without substrate bound,34 today there is a collection of structures of open, closed, and more states, not strictly related to ligand occupancy.40-46 NMR and IR spectroscopy have also been used to indicate the existence of multiple conformations of P450cam in the absence of substrate.47-49
A number of unanswered questions exist about these conformations and related issues. Evidence has been presented for binding of a second molecule of camphor in P450cam,50-51 although this has apparently not been observed in any crystals to date. Another issue involves the accessory redox protein putidaredoxin (Pdx) and when and how it binds in the catalytic cycle.40, 43, 46 Somewhat surprisingly, the effect of Pdx on substrate binding to P450cam has not been reported to our knowledge.
In light of general questions about P450 flexibility and catalysis, we addressed some of the issues. Our kinetic results are consistent with a dominant induced fit mechanism in the binding of camphor, instead of conformational selection. However, this conclusion does not apply to all substrates.
RESULTS
Steady-State Binding of Camphor to P450cam.
Titrations with the substrate camphor were done in the presence of potassium (200 mM KCl), long known to enhance binding.52 The ΔA390-A420 data were used to estimate a Kd value of 1.1 ± 0.2 μM in the presence of 200 mM KCl (Figure 1), similar to values reported earlier.37, 49, 52-57 This value is important later in considering the kinetic mechanism.
Figure 1.
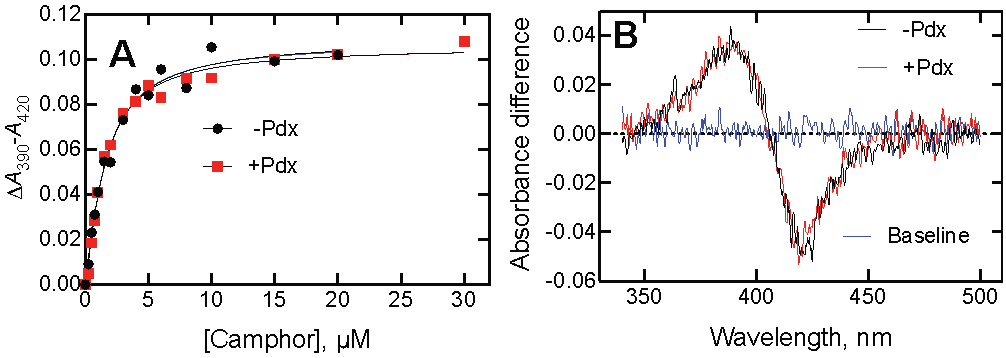
Lack of effect of Pdx on binding of P450cam and camphor. Spectra were recorded using 1.0 μM enzyme concentrations. (A) The estimated Kd values were 1.10 ± 0.20 μM in the absence of Pdx (black, ●) and 0.95 ± 0.12 μM in the presence of Pdx (red, ■). (B) The similarity of the difference spectra in the absence (black) and presence (red) of Pdx is shown for a concentration of 2.5 μM camphor.
A Kd of 0.95 ± 0.12 μM was obtained when the titration was repeated in the presence of Pdx, a study which had apparently not been reported before (Figure 1A). The similarity of the data is shown by the imposition of the difference spectra obtained with 2.5 μM camphor present (Figure 1B).
The possibility was considered that the binding of Pdx may be higher at low ionic strength and alter the binding of camphor under these conditions. Using the work of Peterson52 as a guide, we repeated the binding assays in 10 mM Tris•HCl buffer (pH 7.4) in the absence of inorganic cations and with 15 mM KCl, conditions used in that reference. With only the Tris buffer, the Kd was 23 ± 2 μM without Pdx and 21 ± 2 μM with Pdx (2-fold molar excess) (Figures S4 and S5). With 15 mM KCl present , the Kd was 9.0 ± 0.3 μM without Pdx and 9.8 ± 0.8 μM with Pdx (2-fold molar excess) (Figures S5 and S5).
At least two reports suggest that P450cam can bind a second molecule of camphor and reverse the iron atom back to the low-spin state.50-51 These reports may be related to specific experimental conditions, but no reversal of the high-spin spectral change was observed up to 2 mM camphor (in the presence of 200 mM KCl) in this work (Figure 2).
Figure 2.
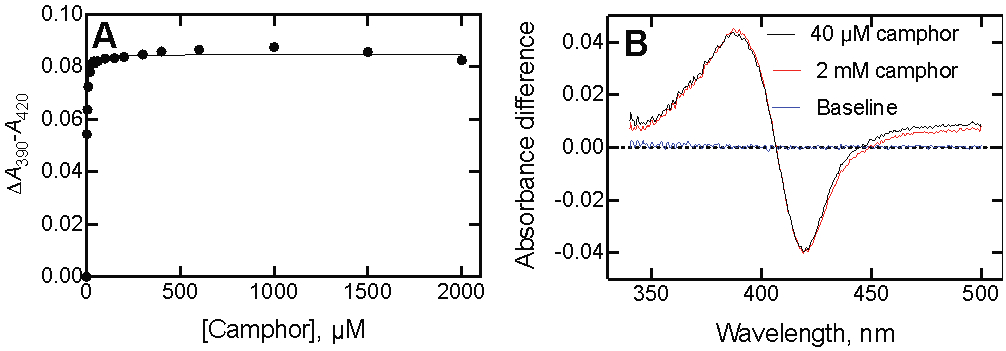
Lack of reversibility of P450cam binding spectra at high concentrations of camphor. (A) ΔA390-A420 as a function of camphor concentration. (B) Difference spectra of P450cam obtained with 40 μM (red) and 2 mM camphor (black). The P450cam concentration was 1.0 μM. The data points were not corrected for dilution.
Kinetics of Camphor Binding to P450cam.
One of the means of discerning the kinetics of conformational changes is the examination of rates of binding at varying concentrations of the ligand and enzyme.20-21, 23, 58 Although rate constants for camphor binding to P450cam have been reported,53, 57, 59 all appear to have been derived from estimates of first-order rates measured only at a single camphor concentration, with that observed rate divided by the substrate concentration to give an apparent second-order rate constant.
The first-order (kobs) rates, or eigenvalues (λ),20,21 of the spectral change upon binding (ΔA390-A420) were measured with a 1 μM final concentration of P450cam and varying concentrations of camphor (Figure 3) (this is λ2 in the convention of Vogt and di Cera20). The eigenvalues increased with increasing concentrations of camphor, a pattern that can be indicative of a simple 2-state system, conformational selection, or an induced fit model.20, 21 As in the case of steady-state binding (Figure 1), the presence of Pdx did not alter rates of binding.
Figure 3.
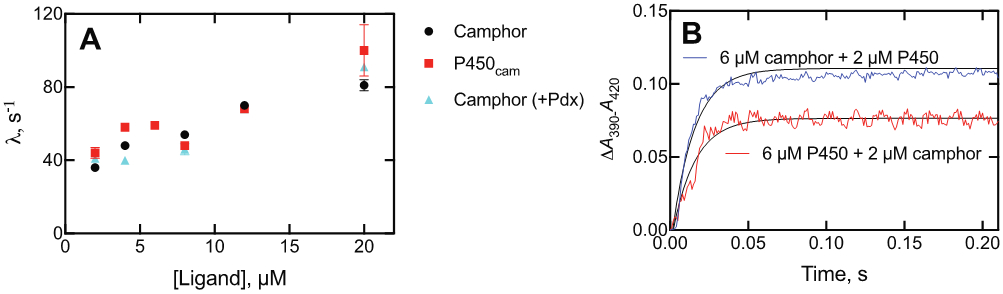
(A) Eigenvalues (λ) for binding of camphor to P450cam. λ (ΔA390-A420, single-exponential) is plotted vs. concentration: variation of the concentration of camphor (final concentration of P450cam was 2 μM) (black, ●); variation of the concentration of camphor, with 2 μM P450cam plus 2 μM Pdx (blue, ▲); variation of P450cam as the ligand in the presence of a final concentration of 2 μM camphor (red, ■). (B) Traces of reactions of varying mixtures: 2 μM P450cam mixed with 6 μM camphor (blue trace, λ 70 ± 2 s−1); 6 μM P450cam mixed with 2 μM camphor (red trace, λ 69 ± 2 s−1).
When a fixed concentration of camphor was used and the P450cam concentration was varied, the plot was nearly superimposable upon the points obtained in the opposite situation (Figure 3A).21 The similarity of eigenvalues for opposite mixtures is shown in Figure 3B.
Analysis of Kinetics of Camphor Binding to P450cam.
Simple linear regression analysis of the eigenvalues for formation of a P450cam•camphor complex as a function of camphor concentration (Figures 3,4) yielded a slope of 2.4 (± 0.3) × 106 M−1 s−1 (a second-order “kon” rate constant for a 2-state system) and an intercept of 35 ± 4 s−1, which would be koff for the complex. However, (i) the plot showed curvature (Figure 4) and (ii) in a simple 2-state system Kd = koff/kon = 35 s−1/2.4 × 106 M−1 s−1 ≅ 15 μM, which is inconsistent with the steady-state Kd value of ~1 μM (Figure 1A), a value rather consistently been reported in earlier literature.37, 52-57
Figure 4.
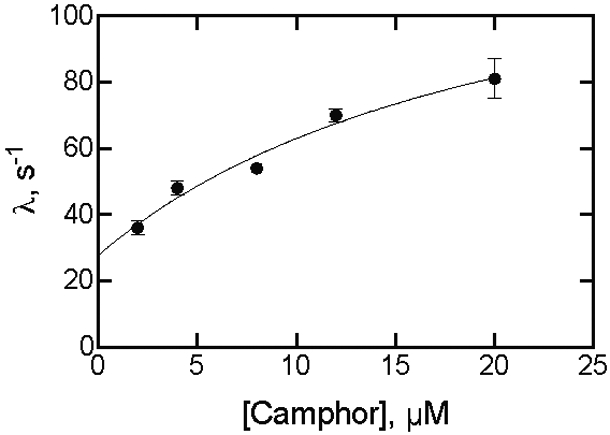
Fitting a plot of eigenvalues (kobs) for binding of camphor to P450cam as a function of camphor concentration. Points are from Figure 3A. The final P450cam concentration was 2 μM. The fit was to the equation λ = kf + kr·S/(Kd + S),20 with S being the camphor concentration, kf the forward rate constant following camphor binding, and kr the rate constant for the reverse reaction (Y= kf + kr*X/(Kd+X) in GraphPad Prism software). The fit is shown for kf = 112 ± 49 s−1 and kr = 28 ± 7 s−1. A linear regression fit yielded a slope (kon) of 2.4 (± 0.3) M−1 s−1 and y-intercept (koff) of 35 ± 4 s−1 (Kd = koff/kon = 15 μM) (not shown).
The data were fit according to Vogt and DiCera,20 using the equation λ = kf + kr·S/(Kd + S), where kf and kr are the forward and reverse rate constants for equilibration following P450cam·camphor complex formation in an induced fit model (Scheme 1). The values kf = 112 ± 49 s−1 and kr = 28 ± 7 s−1 were estimated (the error in the Kd was too high for that parameter to be useful).
Estimation of koff for the P450cam·Camphor Complex.
Peterson and Griffin52-53 used the approach of complexing P450cam with a small excess of camphor, mixing this with a large excess of metyrapone, and measuring the rate of formation of the P450cam•metyrapone complex. The basis of the experiment is the different spectra of the complexes (Figure 5A). In the previous work52 the binding of (1 mM) metyrapone was assumed to be very fast, so that in such a trap experiment the rate of formation of the P450cam•metyrapone complex could be used as the rate constant for koff. However, a control experiment done here by mixing P450cam with a (final) concentration of 2 mM metyrapone yielded a first-order rate of 18.9 s−1, which cannot be neglected. That rate was used in calculations with a simple model with the P450cam and a slight excess of camphor, and fitting in KinTek Explorer® yielded a koff value of 15 s−1 (Figure 5B).
where E is P450cam, S is camphor, I is the inhibitor metyrapone, k2 is 18.9 s−1, and Kd = k-1/k1 is 1 μM (Figure 1A). In Figure 5B, the best fit to the to the data points was obtained with a value of k-1 = 15 s−1.
Figure 5.
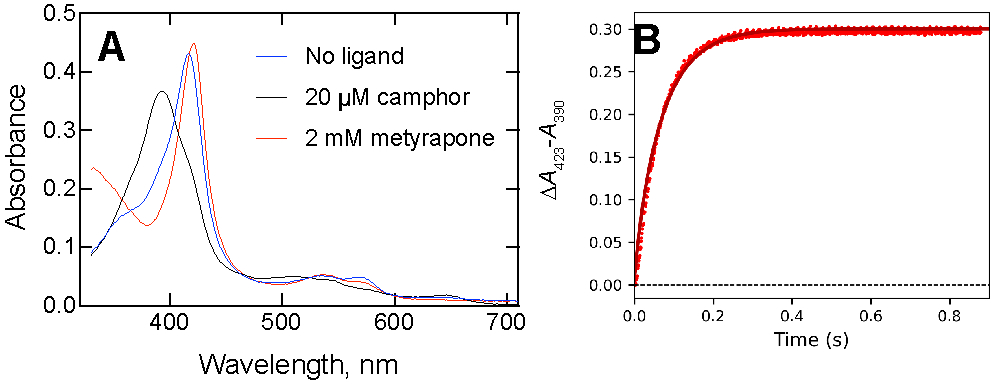
Estimation of koff for the P450cam·camphor complex. (A) Spectra of P450cam (4 μM) without ligand (blue line, λmax 418 nm), with 20 μM camphor (black line, λmax 390 nm, and with 2 mM metyrapone (red line, λmax 423 nm)). (B) A mixture of 2 μM P450cam and 10 μM camphor was mixed with 2 mM metyrapone (all final concentrations). A separate assay yielded a value of 18.9 s−1 for the binding of metyrapone to ligand-free P450cam under the same conditions. The trace was fit to the model (in KinTek Explorer ® software)
Kinetic Fitting to a Minimal Induced Fit Model.
The values of koff = 15 s−1, kf = 112 s−1, and kr = 28 s−1 (vide supra) were used in a minimal kinetic model (Scheme 2) in KinTek Explorer® software. With koff = 15 s−1 (Figure 5B) and Kd = 1 μM (Figure 1A), kon was fit to 1.5 × 107 M−1 s−1.
Scheme 2. Minimal Model for Binding of Substrate to P450cam.
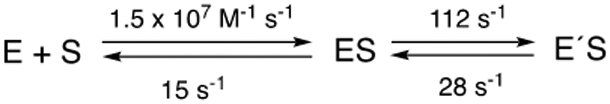
The adequacy of this model was tested by fitting the kinetic courses of multiple experiments with relatively lower concentrations of reagents used in Figure 3A (Figure 6), with reasonable fits for multiple conditions.
Figure 6.
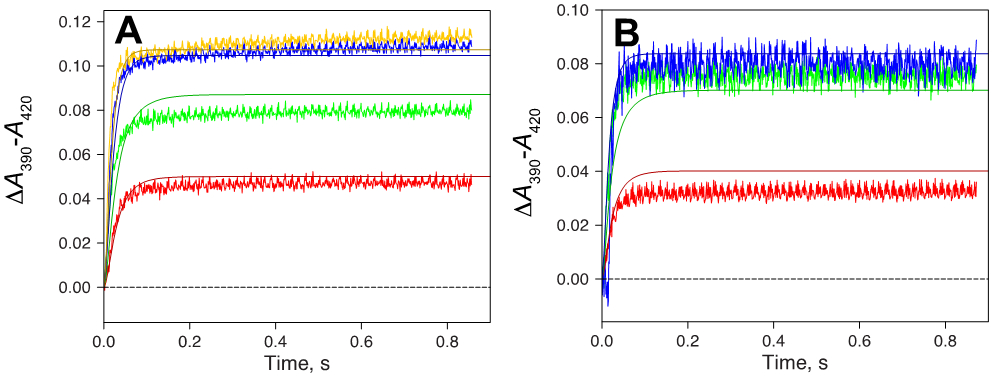
Kinetic modeling of some binding data with parameters from the minimal model of Scheme 2. (A) Fixed final concentration of 1 μM P450cam. Traces are shown for 1 μM (red), 2 μM (green), 4 μM (blue), and 8 μM (yellow) camphor, plus fits (solid lines). (B) Fixed final concentration of 2 μM camphor. Traces are shown for 1 μM (red), 2 μM (green), and 4 μM (blue) P450cam, plus fits (solid lines). The rate constants are from Scheme 2.
Thus, the model and rate constants in Scheme 2 can be used to describe an induced fit mechanism for the binding of camphor to P450cam. The model (Scheme 2) has only the formation of E´S being observed in Figure 6 (if ES were identical to E´S, then the second set of rate constants could be any values and E´S would be meaningless). Again, this is intended to be a minimal model even though the system is known to be more complex, e.g. at least two conformational states of unbound E exist47-48 and there is structural evidence for at least three ES (P450cam•camphor) complexes,41 (Scheme 3).
Scheme 3. Expanded Model for Binding of Substrate to P450cam.
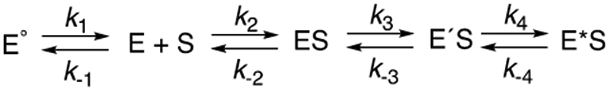
Steady-State Binding and Oxidation of Alternate Substrates for P450cam.
P450cam has ligands and substrates other than camphor.35, 37, 54-56, 60-61 Four were examined: (+)-α-pinene, norcamphor, 2-admantanone, and 3,3,5,5-tetramethylcyclohexanone. Under the conditions used here (i.e., 20 mM potassium MOPS (pH 7.4) and 200 mM KCl), the respective Kd values measured using the partial conversion to the high-spin iron state were 3.4 ± 0.3 μM (Figure 7), 250 ± 8 μM (Figure S6A), 2.7 ± 0.3 μM (Figure 8), and 5.0 ± 0.5 μM (Figure S6C) for (+)-α-pinene, norcamphor, 2-adamantanone, and 3,3,5,5-tetramethylcycolohexanone (Table 1).
Figure 7.
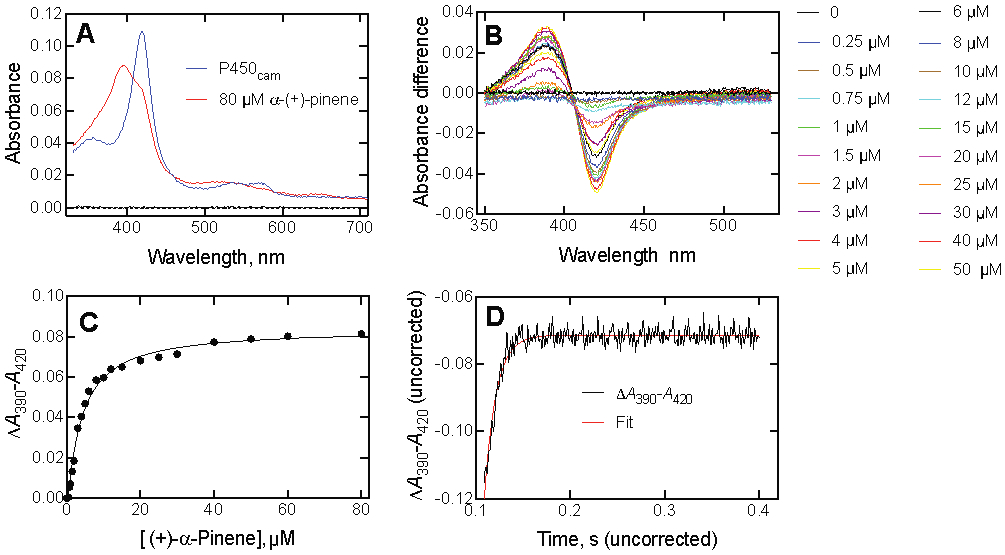
Binding of (+)-α-pinene to P450cam. (A) Absolute spectra of P450cam (1.0 μM) in the absence and presence of 80 μM (+)-α-pinene. (B) Titration of P450cam with increasing concentrations of (+)-α-pinene. The concentrations are listed to the right of the graph. (C) Plot of ΔA390-A420 data from Part B vs. (+)-α-pinene concentration. (D) Kinetics of binding of 2 μM P450cam and 30 μM (+)-α-pinene (final concentrations in the stopped-flow spectrophotometer cell). kobs 81 ± 3 s−1.
Figure 8.
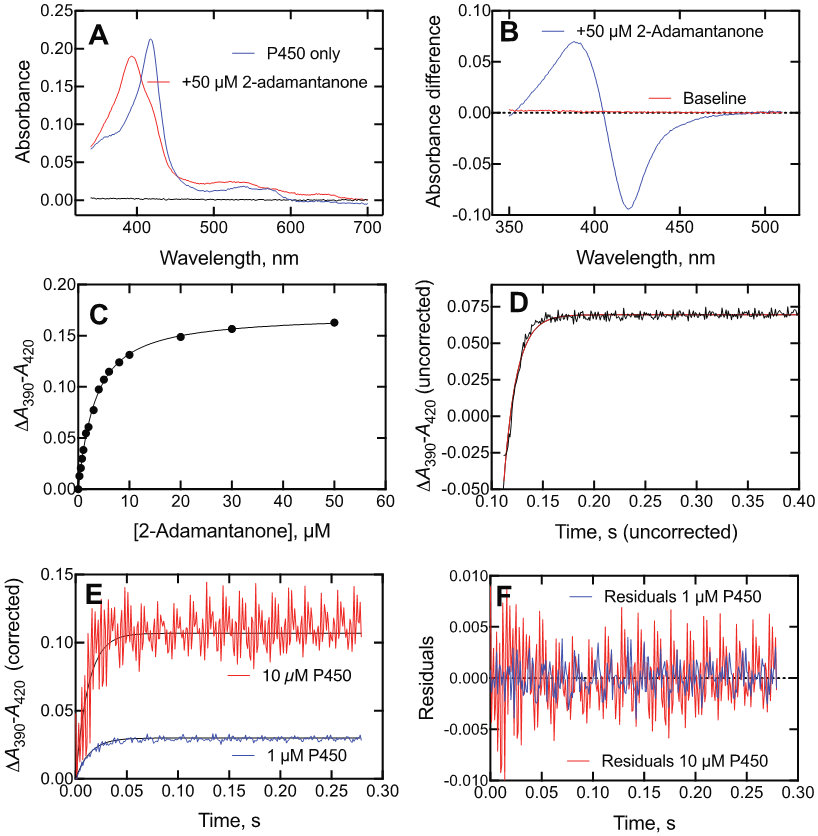
Binding of 2-adamantanone to P450cam. (A) Absolute spectra of P450cam (2.0 μM) in the absence (blue) and presence (red) of 50 μM 2-adamantanone (black line: baseline). (B) Difference spectrum of P450cam with 50 μM 2-adamantanone. (C) Plot of ΔA390-A420 data (i.e., Part B) vs. 2-adamantanone concentration. (D) Kinetics of binding of 2 μM P450cam and 4 μM 2-adamantanone (final concentrations) (axes are uncorrected). kobs 78 ± 2 s−1. (E) Kinetics of binding of 2 μM 2-adamantanone and either 1 μM (blue line) or 10 μM (red line) P450cam. A 20 mm flow cell was used for 1 μM P450cam and a 4 mm cell for 10 μM P450cam. The respective exponentials were 72 and 82 s−1. (F) Residuals plots for the fits in Part E (1 μM P450cam–blue, 10 μM P450cam–red).
Table 1.
Rates of NADH oxidation by P450cam with camphor and alternate substrates
| NADH oxidation | |||||
|---|---|---|---|---|---|
| Substrate | Rate, s−1 |
Substrate concentration, μM |
Kd, μM | Rate/Kd, M−1 s−1 |
% relative to camphor |
| Camphor | 7.7 | 250 | 1.1 | 7.0 × 106 | 100 |
| Norcamphor | 1.75 | 3000 | 250 | 7.0 × 104 | 1 |
| (+)-α-Pinene | 1.1 | 80 | 3.4 | 3.2 × 105 | 5 |
| 2-Adamantanone | 3.6 | 50 | 2.7 | 1.3 × 106 | 19 |
| 3,3,5,5-Tetramethylcyclohexanone | 0.2 | 70 | 5.0 | 4.0 × 104 | 0.6 |
These Kd estimates (Table 1) are similar to the respective values of 1.1 μM,37 350 μM,55, 61 34 and 45 μM,55, 56 and 3.5 μM54 reported previously for (+)-α-pinene, norcamphor, 2-adamantanone, and 3,3,5,5-tetramethylcycolohexanone, with the exception being 3,3,5,5-tetramethylcycolohexanone. 2-Adamantanone is oxidized by P450cam to a single product, the 5-alcohol,54 and norcamphor is oxidized to three products, the 5-exo-, 6-exo, and 3-exo alcohols.60 (+)-α-Pinene is oxidized to at least six products.37 No products have been reported for 3,3,5,5-tetramethylcyclohexanone to our knowledge.
Rates for catalytic activity for P450cam can vary considerably in the literature, due in part to the design of experiments in terms of which enzyme component is limiting.36, 62 The NADH oxidation rate increased with the concentration of NADH-Pdx reductase (PdR). The basic system of Holden et al.62 was used with 5 μM Pdx and 0.1 μM P450cam, and the increased NADH oxidation rates observed above 2 μM PdR were not due to the P450cam reaction (Supporting Information Fig. S3). Few efforts have been reported to measure kcat and Km for camphor with P450cam (presumably due to the issues of rapid hydroxylation, low Kd, and substrate depletion at low substrate concentrations), with the exception of a Km based on O2 as a co-substrate.36 Those values for kcat (55-66 s−1)36 are surprisingly much higher than typically reported here or anywhere else for P450cam and camphor, usually done on the basis of NADH oxidation, e.g. 7-14 s−1.57 One approach to estimating specificity constants (usually kcat/Km)58 is to use a rate measured at a saturating substrate concentration divided by Kd, as proposed for P450cam by Mueller et al.59 To our knowledge, a classical kcat and Km based on the hydroxylation of the substrate camphor has never been reported. The specificity constant defined by rate (at optimal substrate concentration) divided by Kd was 7.0 × 106 M−1 s−1 under our conditions (Table 1), which is intermediate between values of 1.6 × 107 M−1 s−1 57 and 3.4 × 105 M−1 s−1 54 reported by others for camphor, and used for reference for other substrates (Table 1). Our estimated specificity constant (rate/Kd) for (+)-α-pinene (3.2 × 105 M−1 s−1, Table 1) was very similar to the value of 3 × 105 M−1 s−1 reported for product formation by Bell et al.37 although less than their 1.3 × 106 M−1 s−1 based on NADH oxidation. White et al.54 reported a rate/Kd value of 2.4 × 105 M−1 s−1 for 2-adamantanone, which can be compared with our value of 1.3 × 106 M−1 s−1 (Table 1).
The rate/Kd values in Table 1 were all based on NADH oxidation rates (ΔA340 measurements) and may not reflect actual coupling to the formation of oxygenated products. In the case of camphor, the efficiency is generally considered high (but note the potential for NADH oxidation by Pdr/Pdx alone, vide supra36). NADH coupling for P450cam has been reported under specified conditions in the cases of (+)-α-pinene (23%)37 and norcamphor (15%)60 but not 2-adamantanone.54 Low coupling efficiency will reduce the actually rate/Kd estimates of specificity constants even further for alternative substrates to camphor (Table 1).
Assays were done at 23 °C with 0.1 μM P450cam, 5 μM Pdx, 2 μM PdR, the indicated concentration of substrate, and 0.17 μM NADH, used to start the reaction. A blank rate of 0.9 s−1 was obtained in the absence of substrate and the same value was observed without P450cam or substrate; this value was subtracted before calculating the rates.
Pre-Steady-State Binding of Alternate Substrates with P450cam.
(+)-α-Pinene, reported to be oxidized at a rate of 1.1 s−1 in our work (Table 1) or 0.3 s−1 by Bell et al.,37 bound fairly tightly to P450cam (Figure 7C, Table 1) and rates of binding could be measured (Figure 7D). However, the eigenvalues did not change appreciably with the concentration of (+)-α-pinene (Figure 9). This behavior is consistent with a conformational selection mechanism but not induced fit.20-21
Figure 9.
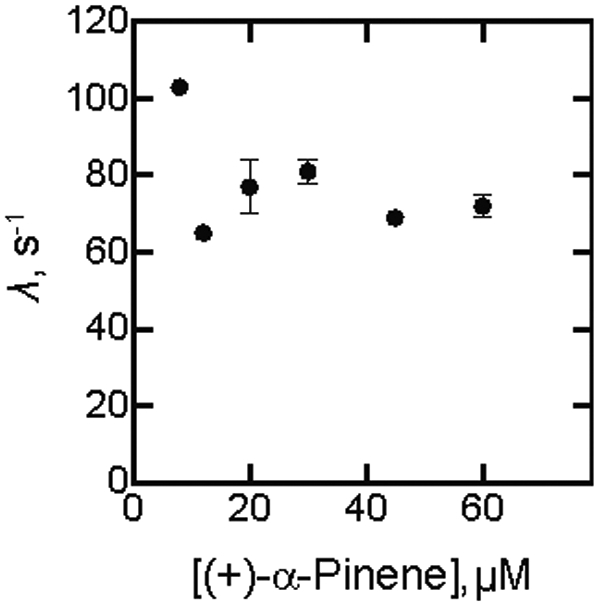
Kinetics of binding of (+)-α-pinene to P450cam as a function of concentration of (+)-α-pinene. The λ values shown (± SD) are from the averaged kinetic traces.
2-Adamantone, oxidized by P450cam to a single product at a rate of 1 s−1 by White at al.54 or 3.6 s−1 in our work (Table 1), yielded a partial shift to the high-spin iron state (Figure 8A), with saturable binding (Figure 8B, 8C). The change in ΔA390-A420 is shown as a function of time in Figure 8D, with final concentrations of 2 μM P450cam and 4 μM 2-adamantanone. The similarity of the eigenvalue patterns of varying both ligand and P450cam was not seen (Figure 10), indicating that an induced fit mechanism observed with the substrate camphor (Figure 3A) is probably not dominant here either,21 as observed with the substrate (+)-α-pinene (Figure 9).
Figure 10.
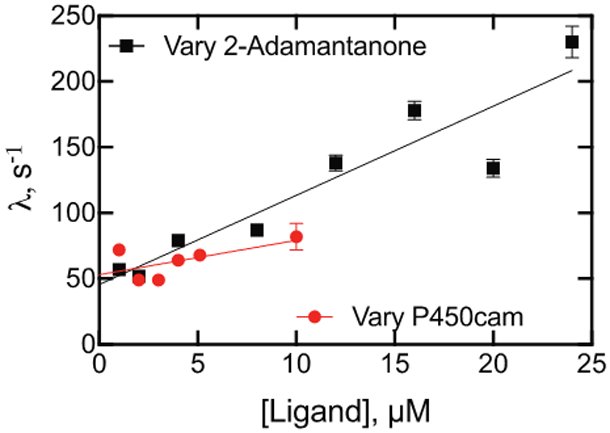
Kinetics of binding of 2-adamantanone and P450cam as a function of concentration, varying either 2-adamantanone (black, ■) or P450cam (red, ●) in the presence of a 2 μM (final) concentration of the opposite component (i.e., P450cam or 2-adamantanone). Linear regression fits are shown to denote trends, although this fit does not have a physical basis.
Attempts to measure binding rates were also made with the ligands norcamphor and 3,3,5,5-tetramethylcyclohexanone. The partial high-spin spectra of each were observed in the stopped-flow spectrophotometer cell following mixing but the changes could not be observed (ΔA390-A420), presumably because the rates are too fast. The actual kon rates are unknown and could be faster than for camphor and 2-adamantone. In the case of norcamphor, the Kd is much higher (> 100-fold, vide supra) and since kobs = kon + koff and Kd = koff/kon in a simple 2-state system,58 then a kon of ~ 107 M−1 s−1 and Kd of 250 μM would yield (at 10 μM norcamphor) kobs = 100 s−1 + 2,500 s−1 = 2,600 s−1, i.e. t1/2 ~ 0.3 ms, an order of magnitude less than the deadtime of the stopped-flow spectrophotometer. Although the Kd for binding 3,3,5,5,-tetramethylcyclohexanone was 5 μM (Figure S6B), the binding step was also too fast to measure.
DISCUSSION
Considerable evidence has accumulated that P450cam can exist in a number of conformational states both in the absence and presence of its cognate substrate, camphor.40-43, 45-49, 63 As stated by Basom et al.,48 based on IR spectroscopy results, “…the local energy landscape within each state appears to be independent of the bound substrate, and is rather a function of which conformation is populated, with the role of the substrate binding only to favor the population of one of the other state…” and “…the current data are strongly indicative of a conformational selection or induced fit mechanism of substrate recognition.” Later, the authors proposed a conformational selection mechanism based on static IR studies with P450cam, site-directed mutants, and substrate analogs.49 In the present work, we examined several aspects of the binding of several substrates to P450cam using established approaches20,21 and concluded that the evidence favors the dominance of an induced fit mechanism, as opposed to conformational selection (Scheme 1), in the binding of the prototypic substrate camphor. The strongest evidence is the similarity of the dependence of the observed increasing binding eigenvalues measured when both the substrate and P450cam were varied (Figure 3A). However, this phenomenon was not observed with the alternate substrates (+)-α-pinene and 2-adamantanone (Figures 9, 10), and those binding mechanisms are considered to involve domination by conformational selection (Scheme 1).
The distinction between conformational selection and induced fit mechanisms (Scheme 1) may seem subtle but has practical relevance in terms of rational design of site-directed mutants and in discovery of enzyme inhibitors.19 A conformational selection mechanism is implicated if the eigenvalue λ (i.e., kobs) decreases or if there is no change as the ligand concentration is increased. Conformational selection can also be associated with an increase in the eigenvalue λ with ligand concentration, depending on the rates of koff, kf, and kr,20 but induced fit can only be associated with the overlap of the plots of varying both ligand and protein in the presence of a fixed concentration of the other component.21 One simplified way of looking at this phenomenon is that the formation of the initial ES complex is related to both concentrations of both E and S (first order in each) but then the induced fit conformational change “takes over” further kinetics. The approach of varying the protein concentration is expensive, in terms of protein; in the case of spectral assays such as these done here reducing the absorbance (of the protein) was also necessary (using a shorter pathlength in the spectrophotometer).
Initial studies indicated that the accessory protein Pdx did not appear to alter camphor binding to P450cam, as judged by spectral assays (Figures 1 S4, S5). To our knowledge, this point had never been addressed and is relevant in light of the substantial effects of another ferredoxin, adrenodoxin, on the binding of substrates to mitochondrial P450s (e.g., 11B2, 24A1).64,65 Hollingsworth et al.46 did report that the presence of camphor increased the Kd of a P450cam·Pdx complex 2-fold, which should change the Kd for camphor when Pdx is present (2-fold) in consideration of a thermodynamic “box” analysis,66 but even this difference was not detected (Figures 1, S4, S5). Accordingly, all of our P450cam measurements were made in the absence of Pdx. To our knowledge there is no report of complexation of PdR with P450cam, and no binding measurements included this protein.
Some evidence has been presented for the binding of a second molecule of camphor in P450cam and that this reverses this high-spin state,50,51 and such binding has support from molecular dynamics.45 However, a titration of up to 2 mM camphor under our conditions (20 mM potassium MOPS buffer (pH 7.4) and 200 mM KCl) did not show any evidence for reversal (Figure 2). To our knowledge, no X-ray crystal structure has shown occupancy of a second molecule of camphor. However, one of the alternate substrate/ligands considered, 3,3,5,6-tetramethylcyclohexanone, did reverse the spin state change at higher concentrations (Figure S6C).
Only two spectral states of P450cam were observed, the classic low-spin and high-spin iron forms (e.g., Figures 5A, 7A). Singular value decomposition (SVD) analysis (OLIS GlobalWorks) did not reveal any others (Figure S7). However, as mentioned earlier, more than two states do exist (at least five), with the “open” and “closed” forms being two extremes (Scheme 3).41-42, 45, 47, 55 Fisher and Sligar55 used temperature jump spectroscopy to measure rates of spin changes related to the binding of camphor and other ligands. These measurements involve the iron spin state in the protein-ligand complex, i.e. low-spin P450cam·ligand ⇄ high-spin P450cam·ligand, and are much faster than rates measured here (kobs 890-2300 s−1), with the forward and reverse rates also being much faster than we report or would be able to detect (e.g., Figures 3B, 7D, 8D). Exactly how the forms examined in that work and their kinetics relate to our minimal model (Scheme 2) or the expanded one (Scheme 3) is unknown. An expanded version of the system (five conformations), which includes the structural work41, 42, 47 as well as the temperature jump work55 is shown in Scheme 3, although individual rate constants and conformations cannot be assigned.
With regard to the relevance of kinetic evidence for an induced fit mechanism, consideration was given to known alternate substrates, particularly those with relaxed catalytic regioselectivity (Table 1). Norcamphor (devoid of three methyl groups) is a substrate, produces a spin shift, and yields three products,60 but it has weak affinity (Table 1, Fig. S6A)55 and apparently the high koff rates made kinetic analysis of binding impossible using stopped-flow methods, at least with our system. 3,3,5,5-Tetramethylcyclohexanone is a known ligand55-56, 67 with relatively high affinity (Kd 5 μM, Figure S6B, Table 1) but no products have been reported and this ligand proved not to be useful in kinetic studies.
(+)-α-Pinene has also been established as a substrate and is oxidized to four known products, plus at least two others.37 It also binds tightly to P450cam (Figure 7)37 and was useful in kinetic studies (Figures 7, 9). The eigenvalues did not vary with the substrate concentration (Figure 9), a result consistent with conformational selection and not induced fit.20 2-Adamantanone is a known substrate, with a single product, 5-hydroxy-2-adamantanone.54 It is bound tightly54 (Figure 8C), and White et al.54 reported an oxidation rate of ~ 1 s−1 with P450cam, almost as high as for camphor in that particular study. This proved to be a useful ligand in terms of the kinetics of binding (Figures 8, 10). In contrast to camphor, a conformational selection mechanism appears to be more prominent than induced fit (Scheme 1), as seen in the differences in the eigenvalues when concentrations of both 2-adamantanone and P450cam were varied (Figure 10).
We conclude that the binding of camphor to P450cam fits the criteria supporting a series of conformational states in which an induced fit mechanism is dominant (Scheme 3). This kinetic behavior may be related to the high regio- and stereo-selectivity of camphor hydroxylation. To date most mammalian P450s seem to be dominated by conformational selection, not induced fit, 25-27 and it can be speculated that induced fit is linked to higher rates of catalysis and regioselectivity in P450cam. The P450cam alternate substrate 2-adamantanone, with high reported regioselectivity,54 was concluded to be involved in conformational selection with the enzyme, not induced fit (Figure 8). (+)-α-Pinene, with multiple products,37 also fits the criteria for a conformational selection process being dominant (Figure 9).20
Recent work shows that P450cam samples a variety of stable conformational states with some involving rather large motions and rearrangement of active site residues.40-49 Even in the substrate-free form, P450cam samples a broad range of conformational space with some having been captured in crystal structures. One or a few of the many conformational states of the enzyme favors camphor binding; these are depicted collectively as E° in Scheme 3 (vide supra).47, 48 Thus, there is an element of conformational selectivity but once bound, the enzyme closes down to a single conformation. In the case of camphor binding, the progression of multiple states of the enzyme substrate complex constitutes an induced fit mechanism because this process is dominant in driving the kinetics. With the alternate substrates (+)-α-pinene and 2-adamantanone, the evidence appears to favor the conformational selection process over induced fit. This is further evidence for the existence of multiple and functionally relevant conformations of substrate-free P450cam, which must be present in the camphor binding mechanisms (Scheme 3), even though it appears to be dominated by the induced fit process. Thus, for all of the substrates there is a combination of induced fit and conformational selectivity and one or the other is dominant for each substrate, even though both are operative.
The question can be raised as to whether an induced fit mechanism drives the high regio- and stereoselectivity and relatively high (for P450s) specificity constant seen in the oxidation of camphor by P450cam. Clearly the alternate reactions catalyzed by this enzyme have lower specificity constants (Table 1), even considering the parameters based on NADH oxidation. An interesting comparison is bacterial P450BM-3 (CYP102A1), with a kinetically defined conformational selection mechanism and oxidation rates as high or higher than P450cam/camphor and low Kd values.28 A specificity constant of P450BM-3 with the substrate tetradecyl sulfate of 3 × 106 M−1 s−1 (or rate/Kd approximation of 30 s−1/1.4 μM = 2 × 107 M−1 s−1)28 compares favorably with P450cam/camphor (Table 1). However, oxidation of fatty acids and alkyls sulfates by P450BM-3 is known not to be completely regioselective, with ω−1, ω−2, and ω−3 products all being formed.28 It is possible that an induced fit mechanism is associated with enhanced selectivity and high sensitivity, as concluded by Johnson and others for DNA polymerases,68-71 but whether there is a general pattern with P450s will require analysis of more examples.
Recently, Murarka et al.72 characterized another P450 enzyme, P450tcu, from the bacterium Pseudomonas sp. strain TCU-HL1 that also catalyzes the 5-exo hydroxylation of camphor with a kcat as high as P450cam (~10 s−1). The addition of camphor induces a transition to high-spin iron but, in contrast to P450cam, the binding was enthalpically-driven and very slow (t1/2 25 min). Exactly how the high-spin transition relates to catalytic activity in this enzyme is unclear.
In conclusion, P450s can utilize both conformational selection and induced fit mechanisms (Scheme 1) to achieve catalytic specificity. Rates of catalysis are probably not strictly linked to which mechanism is used. Although P450cam (using induced fit) has a high rate of catalysis with camphor, so does P450BM-3 (which has the ability to use multiple substrates).28 Thus, P450s provide an example of an enzyme family that uses multiple conformations to achieve catalytic selectivity in different ways, and even a single P450 (i.e., P450cam) can have different mechanisms predominating with individual substrates.
EXPERIMENTAL SECTION
Chemicals.
d-(+)-Camphor was purchased from Aldrich and recrystallized from hot C2H5OH (mp 179-179.5 °C, uncorr, lit 179.8 °C73.) Metyrapone (Aldrich) was recrystallized from (C2H5)2O/petroleum ether (l:l, v/v)74 (mp 50-51 °C, lit 50-51 °C73). Both compounds were dissolved in H2O or buffer, except when camphor stocks were prepared at > 8 mM (solubility limit73) in which case C2H5OH was used. 2-Adamantanone and 3,3,5,5-tetramethylcyclohexanone (both SigmaAldrich) were dissolved in H2O at concentrations up to 2 mM, and norcamphor was soluble in H2O up to at least 50 mM. (+)-α-Pinene is reported not to be soluble in H2O at concentrations ≥18 μM (PubChem) and was dissolved in C2H5OH and diluted into aqueous solutions (≤1% C2H5OH, v/v) except in cases in which low concentrations were used without C2H5OH (<18 μM). In the substrate binding titrations (Figure 7C), the C2H5OH concentration reached 2% (v/v) and C2H5OH aliquots were also used in the reference cuvette.
Enzymes.
P450cam, Pdx, and PdR were expressed in Escherichia coli from plasmids provided by S-W. Chuo and D. B. Goodin (Univ. California, Davis) and purified using the general methods previously described by that group.40 The purity of the three enzymes was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Figure S1). P450cam generated a typical Fe2+•CO vs. Fe2+ difference spectrum, devoid of inactive cytochrome P420 (Figure S2). The enzymes were stored frozen at −80 °C. P450cam (~ 100 μM) was stored in the presence of 1 mM camphor, which was removed prior to use by gel filtration chromatography (Sephadex G-10, 1.2 cm × 85 cm, 4 °C) using 20 mM Tris-acetate buffer (pH 7.4), containing 0.1 mM EDTA, to reduce the camphor affinity by excluding monovalent cations.
Catalytic Activity.
Assays were done at 23 °C with 0.1 μM P450cam, 5 μM Pdx, 2 μM PdR, the indicated concentration of substrate, and 0.17 μM NADH, used to start the reaction (total volume 0.33 mL in black self-masking cuvettes).62 The decrease in A340 was monitored for 2 min in an Aminco DW2-OLIS spectrophotometer (On-Line Instrument Systems, Athens, GA), and linear data (30-120 s) were used to calculate rates using OLIS GlobalWorks software. A blank rate of 0.9 s−1 was obtained under these conditions in the absence of substrate (and the same value without P450cam or substrate); this value was subtracted before calculating the rates (Figure S3).
Substrate Binding Titrations.
P450cam (1.0 μM) was diluted in 20 mM potassium MOPS buffer (pH 7.4) containing 200 mM KCl. In the experiments described in Figure S4, the buffer was 10 mM Tris•HCl (pH 7.4), with or without the addition of 15 mM KCl. Two 1.0-mL cuvettes were used in either an OLIS-Cary14 or an OLIS-Aminco DW2 spectrophotometer and a baseline was established (23 °C). Increasing μL amounts of camphor, (+)-α-pinene, 2-adamantanone, norcamphor, or 3,3,5,5-tetramethylcyclohexanone were added to the sample cuvette using a cuvette mixer (Bel-Art, Wayne, NJ) (from 0.5-5 mM aqueous solutions or, at high concentrations, a 100 mM ethanolic solution). Spectra were recorded, and the difference ΔA390-A420 at each concentration was used to estimate Kd with a quadratic fitting equation in GraphPad Prism software: Y = B + (A/2)*(1/E)*((Kd+E+X)=sqrt((Kd+E+X)∧2-(4*E*X))), with E fixed.
Ligand Binding Kinetics.
All measurements were made at 23 °C in 20 mM potassium MOPS buffer (pH 7.4) containing 200 mM KCl, using an OLIS RSM-1000 stopped-flow spectrophotometer equipped with a 16 × 0.2 mm spinning disk, acquiring 4000 scans over 4 seconds. The slits were both 1.24 mm and the 400 L/mm, 500 nm> gratings were used, covering a 332-565 nm wavelength range. Equal volumes of buffer solutions of P450cam and ligand were mixed, with a nominal dead time of 2 ms. From the accumulated spectra from each shot (5-10 shots per concentration), the A420 data were subtracted from the A390 data points and the difference (ΔA390-A420 vs. time) traces were averaged using the OLIS GlobalWorks® software and fit to first-order fits (or used in SVD analysis, Figure S7). The ΔA390-A420 data sets were saved as Excel files, transferred to an Apple Mac OS 10.15.6 system, saved as txt files, and in some cases used in KinTek Explorer® software (KinTek, Snow Shoe, PA).
The first-order rates, as a function of substrate concentration, were fit in GraphPad Prism as described in individual cases.
Supplementary Material
ACKNOWLEDGMENTS
We thank S.-W. Chu and D. B. Goodin for supplying the expression plasmids and K. Trisler for assistance in preparation of the manuscript. This work was supported by National Institutes of Health Grant R01 GM118122. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supporting Information
Purity of enzymes; Fe2+-CO vs. Fe2+ difference spectrum of purified P450cam; optimization of PdR concentrations for catalytic assays; titrations of P450cam with some alternate ligands; SVD analysis of representative kinetic binding data; titrations of P450cam and Kd estimates with camphor at low concentrations of monovalent ions in the absence and presence of Pdx.
The authors declare no competing financial interest.
REFERENCES
- 1.Rendic S; Guengerich FP, Survey of Human Oxidoreductases and Cytochrome P450 Enzymes Involved in the Metabolism of Xenobiotic and Natural Chemicals, Chem. Res. Toxicol 2015, 28, 38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ortiz de Montellano PR, Ed., Cytochrome P450: Structure, Mechanism, and Biochemistry. 4th ed.; Springer: New York, 2015. [Google Scholar]
- 3.Guengerich FP, Common and Uncommon Cytochrome P450 Reactions Related to Metabolism and Chemical Toxicity, Chem. Res. Toxicol 2001, 14, 611–650. [DOI] [PubMed] [Google Scholar]
- 4.Ortiz de Montellano PR Substrate Oxidation by Cytochrome P450 Enzymes. In Cytochrome P450: Structure, Mechanism, and Biochemistry, 4th ed.; Ortiz de Montellano PR, Ed. Springer: New York, 2015; pp 111–176. [Google Scholar]
- 5.Guengerich FP, Perspective: Mechanisms of Cytochrome P450-Catalyzed Oxidations, ACS Catalysis 2018, 8, 10964–10976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen K; Arnold FH, Engineering Cytochrome P450s for Enantioselective Cyclopropenation of Internal Alkynes, J. Am. Chem. Soc 2020, 142, 6891–6895. [DOI] [PubMed] [Google Scholar]
- 7.Brandenberg OF; Chen K; Arnold FH, Directed Evolution of a Cytochrome P450 Carbene Transferase for Selective Functionalization of Cyclic Compounds, J. Am. Chem. Soc 2019, 141, 8989–8995. [DOI] [PubMed] [Google Scholar]
- 8.Sawayama AM; Chen MM; Kulanthaivel P; Kuo MS; Hemmerle H; Arnold FH, A Panel of Cytochrome P450BM3 Variants to Produce Drug Metabolites and Diversify Lead Compounds, Chemistry 2009, 15, 11723–11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coelho PS; Wang ZJ; Ener ME; Baril SA; Kannan A; Arnold FH; Brustad EM, A Serine-substituted P450 Catalyzes Highly Efficient Carbene Transfer to Olefins In Vivo, Nat. Chem. Biol 2013, 9, 485–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farinas ET; Schwaneberg U; Glieder A; Arnold FH, Directed Evolution of a Cytochrome P450 Monoxygenase for Alkane Oxidation, Adv. Synth. Catal 2001, 343, 601–606. [Google Scholar]
- 11.Fasan R; Meharenna YT; Snow CD; Poulos TL; Arnold FH, Evolutionary History of a Specialized P450 Propane Monooxygenase, J. Mol. Biol 2008, 383, 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh R; Bordeaux M; Fasan R, P450-Catalyzed Intramolecular sp3 C-H Amination with Arylsulfonyl Azide Substrates, ACS Catalyis 2014, 4, 546–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fasan R, Enzymatic Catalysis: New Functional Twists for P450s, Nat. Chem 2017, 9, 609–611. [DOI] [PubMed] [Google Scholar]
- 14.Guengerich FP Human Cytochromes P450. In Cytochrome P450: Structure, Mechanism, and Biochemistry, 4th ed.; Ortiz de Montellano PR, Ed. Springer: New York, 2015; Vol. 2, pp 523–785. [Google Scholar]
- 15.Krauser JA; Guengerich FP, Cytochrome P450 3A4-catalyzed Testosterone 6β-Hydroxylation. Stereochemistry, Kinetic Deuterium Isotope Effects, and Rate-Limiting Steps, J. Biol. Chem 2005, 280, 19496–19506. [DOI] [PubMed] [Google Scholar]
- 16.Dixon M; Webb EC, Enzymes. 2nd ed.; Longman's, Green and Co.: London, 1964. [Google Scholar]
- 17.Jencks WP, Catalysis in Chemistry and Enzymology. McGraw-Hill: New York, 1969. [Google Scholar]
- 18.Benkovic SJ; Hammes GG; Hammes-Schiffer S, Free-energy Landscape of Enzyme Catalysis, Biochemistry 2008, 47, 3317–3321. [DOI] [PubMed] [Google Scholar]
- 19.Changeux JP; Edelstein S, Conformational Selection or Induced Fit? 50 Years of Debate Resolved, F1000 Biol. Rep 2011, 3, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vogt AD; Di Cera E, Conformational Selection or Induced Fit? A Critical Appraisal of the Kinetic Mechanism, Biochemistry 2012, 51, 5894–5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gianni S; Dogan J; Jemth P, Distinguishing Induced Fit from Conformational Selection, Biophys. Chem 2014, 189, 33–39. [DOI] [PubMed] [Google Scholar]
- 22.Chakraborty P; Di Cera E, Induced Fit Is a Special Case of Conformational Selection, Biochemistry 2017, 56, 2853–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agafonov RV; Wilson C; Otten R; Buosi V; Kern D, Energetic Dissection of Gleevec's Selectivity Toward Human Tyrosine Kinases, Nat. Struct. Mol. Biol 2014, 21, 848–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koshland DE Jr.; Nemethy G; Filmer D, Comparison of Experimental Binding Data and Theoretical Models in Proteins Containing Subunits, Biochemistry 1966, 5, 365–385. [DOI] [PubMed] [Google Scholar]
- 25.Guengerich FP; Wilkey CJ; Phan TTN, Human Cytochrome P450 Enzymes Bind Drugs and Other Substrates Mainly Through Conformational-Selection Modes, J. Biol. Chem 2019, 294, 10928–10941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guengerich FP; Wilkey CJ; Glass SM; Reddish MJ, Conformational Selection Dominates Binding of Steroids to Human Cytochrome P450 17A1, J. Biol. Chem 2019, 294, 10028–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mast N; Verwilst P; Wilkey CJ; Guengerich FP; Pikuleva IA, In Vitro Activation of Cytochrome P450 46A1 (CYP46A1) by Efavirenz-Related Compounds, J. Med. Chem 2020, 63, 6477–6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guengerich FP; Fekry MI, Methylene Oxidation of Alkyl Sulfates by Cytochrome P450BM-3 and a Role for Conformational Selection in Substrate Recognition, ACS Catalysis 2020, 10, 5008–5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hedegaard J; Gunsalus IC, Mixed Function Oxidation. IV. An Induced Methylene Hydroxylase in Camphor Oxidation, J. Biol. Chem 1965, 240, 4038–4043. [PubMed] [Google Scholar]
- 30.Yu C-A; Gunsalus IC; Katagiri M; Suhara K; Takemori S, Cytochrome P-450cam. I. Crystallization and Properties, J. Biol. Chem 1974, 249, 94–101. [PubMed] [Google Scholar]
- 31.Haniu M; Armes LG; Tanaka M; Yasunobu KT; Shastry BS; Wagner GC; Gunsalus IC, The Primary Structure of the Monoxygenase Cytochrome P450CAM, Biochem. Biophys. Res. Commun 1982, 105, 889–894. [DOI] [PubMed] [Google Scholar]
- 32.Haniu M; Armes LG; Yasunobu KT; Shastry BA; Gunsalus IC, Amino Acid Sequence of the Pseudomonas putida Cytochrome P-450. II. Cyanogen Bromide Peptides, Acid Cleavage Peptides, and the Complete Sequence, J. Biol. Chem 1982, 257, 12664–12671. [PubMed] [Google Scholar]
- 33.Poulos TL; Finzel BC; Gunsalus IC; Wagner GC; Kraut J, The 2.6-Å Crystal Structure of Pseudomonas putida Cytochrome P-450, J. Biol. Chem 1985, 260, 16122–16130. [PubMed] [Google Scholar]
- 34.Poulos TL; Finzel BC; Howard AJ, Crystal Structure of Substrate-free Pseudomonas putida Cytochrome P-450, Biochemistry 1986, 25, 5314–5322. [DOI] [PubMed] [Google Scholar]
- 35.Mueller EJ; Loida PJ; Sligar SG Twenty-five Years of P450cam Research-Mechanistic Insights into Oxygenase Catalysis. In Cytochrome P450–Structure, Mechanism, and Biochemistry, 2nd ed., Ortiz de Montellano PR, Ed. Plenum Press: New York, 1995; pp 83–124. [Google Scholar]
- 36.Purdy MM; Koo LS; Ortiz de Montellano PR; Klinman JP, Steady-state Kinetic Investigation of Cytochrome P450cam: Interaction with Redox Partners and Reaction with Molecular Oxygen, Biochemistry 2004, 43, 271–281. [DOI] [PubMed] [Google Scholar]
- 37.Bell SG; Chen X; Sowden RJ; Xu F; Williams JN; Wong LL; Rao Z, Molecular Recognition in (+)-α-Pinene Oxidation by Cytochrome P450cam, J. Am. Chem. Soc 2003, 125, 705–714. [DOI] [PubMed] [Google Scholar]
- 38.Bell SG; Harford-Cross CF; Wong LL, Engineering the CYP101 System for In Vivo Oxidation of Unnatural Substrates, Protein Eng. 2001, 14, 797–802. [DOI] [PubMed] [Google Scholar]
- 39.Jones JP; O'Hare EJ; Wong LL, Oxidation of Polychlorinated Benzenes by Genetically Engineered CYP101 (Cytochrome P450cam), Eur. J. Biochem 2001, 268, 1460–1467. [DOI] [PubMed] [Google Scholar]
- 40.Liou SH; Myers WK; Oswald JD; Britt RD; Goodin DB, Putidaredoxin Binds to the Same Site on Cytochrome P450cam in the Open and Closed Conformation, Biochemistry 2017, 56, 4371–4378. [DOI] [PubMed] [Google Scholar]
- 41.Lee YT; Glazer EC; Wilson RF; Stout CD; Goodin DB, Three Clusters of Conformational States in P450cam Reveal a Multistep Pathway for Closing of the Substrate Access Channel, Biochemistry 2011, 50, 693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee YT; Wilson RF; Rupniewski I; Goodin DB, P450cam Visits an Open Conformation in the Absence of Substrate, Biochemistry 2010, 49, 3412–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skinner SP; Liu W-M; Hiruma Y; Timmer M; Blok A; Hass MAS; Ubbink M, Delicate Conformational Balance of the Redox Enzyme Cytochrome P450cam, Proc. Natl. Acad. Sci. U. S. A 2015, 112, 9022–9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Follmer AH; Tripathi S; Poulos TL, Ligand and Redox Partner Binding Generates a New Conformational State in Cytochrome P450cam (CYP101A1), J. Am. Chem. Soc 2019, 141, 2678–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Follmer AH; Mahomed M; Goodin DB; Poulos TL, Substrate-Dependent Allosteric Regulation in Cytochrome P450cam (CYP101A1), J. Am. Chem. Soc 2018, 140, 16222–16228. [DOI] [PubMed] [Google Scholar]
- 46.Hollingsworth SA; Batabyal D; Nguyen BD; Poulos TL, Conformational Selectivity in Cytochrome P450 Redox Partner Interactions, Proc. Natl. Acad. Sci. U. S. A 2016, 113, 8723–8728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Asciutto EK; Young MJ; Madura J; Pochapsky SS; Pochapsky TC, Solution Structural Ensembles of Substrate-Free Cytochrome P450cam, Biochemistry 2012, 51, 3383–3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Basom EJ; Spearman JW; Thielges MC, Conformational Landscape and the Selectivity of Cytochrome P450cam, J. Phys. Chem. B 2015, 119, 6620–6627. [DOI] [PubMed] [Google Scholar]
- 49.Basom EJ; Manifold BA; Thielges MC, Conformational Heterogeneity and the Affinity of Substrate Molecular Recognition by Cytochrome P450cam, Biochemistry 2017, 56, 3248–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marden MC; Hui Bon Hoa G, P-450 Binding to Substrates Camphor and Linalool Versus Pressure, Arch. Biochem. Biophys 1987, 253, 100–107. [DOI] [PubMed] [Google Scholar]
- 51.Yao H; McCullough CR; Costache AD; Pullela PK; Sem DS, Structural Evidence for a Functionally Relevant Second Camphor Binding Site in P450cam: Model for Substrate Entry into a P450 Active Site, Proteins: Struct., Funct., Bioinf 2007, 69, 125–138. [DOI] [PubMed] [Google Scholar]
- 52.Peterson JA, Camphor Binding by Pseudomonas putida Cytochrome P-450, Arch. Biochem. Biophys 1971, 144, 678–693. [DOI] [PubMed] [Google Scholar]
- 53.Griffin BW; Peterson JA, Camphor Binding by Pseudomonas putida Cytochrome P-450. Kinetics and Thermodynamics of the Reaction, Biochemistry 1972, 11, 4740–4746. [DOI] [PubMed] [Google Scholar]
- 54.White RE; McCarthy MB; Egeberg KD; Sligar SG, Regioselectivity in the Cytochromes P-450: Control by Protein Constraints and by Chemical Reactivities, Arch. Biochem. Biophys 1984, 228, 493–502. [DOI] [PubMed] [Google Scholar]
- 55.Fisher MT; Sligar SG, Temperature Jump Relaxation Kinetics of the P-450cam Spin Equilibrium, Biochemistry 1987, 26, 4797–4803. [DOI] [PubMed] [Google Scholar]
- 56.Di Primo C; Hui Bon Hoa G; Douzou P; Sligar SG, Heme-Pocket-Hydration Change During the Inactivation of Cytochrome P-450camphor by Hydrostatic Pressure, Eur. J. Biochem 1992, 209, 583–588. [DOI] [PubMed] [Google Scholar]
- 57.Gerber NC; Sligar SG, Catalytic Mechanism of Cytochrome P-450: Evidence for a Distal Charge Relay, J. Am. Chem. Soc 1992, 114, 8742–8743. [Google Scholar]
- 58.Johnson KA, Kinetic Anaylsis for the New Enzymology. 1st ed.; KinTek: Austin, TX, 2019; p 361–383. [Google Scholar]
- 59.Atkins WM; Sligar SG, Metabolic Switching in Cytochrome P-450cam: Deuterium Isotope Effects on Regiospecificity and the Monooxygenase/Oxidase Ratio, J. Am. Chem. Soc 1987, 109, 3754–3760. [Google Scholar]
- 60.Schulze H; Hui Bon Hoa G; Jung C, Mobility of Norbornane-type Substrates and Water Accessibility in Cytochrome P-450cam, Biochim. Biophys. Acta 1997, 1338, 77–92. [DOI] [PubMed] [Google Scholar]
- 61.Holden M; Mayhew M; Bunk D; Roitberg A; Vilker V, Probing the Interactions of Putidaredoxin with Redox Partners in Camphor P450 5-Monooxygenase by Mutagenesis of Surface Residues, J. Biol. Chem 1997, 272, 21720–21725. [DOI] [PubMed] [Google Scholar]
- 62.Follmer AH; Tripathi S; Poulos TL, Ligand and Redox Partner Binding Generates a New Conformational State in Cytochrome P450cam (CYP101A1), 2019, 141, 2678–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reddish MJ; Guengerich FP, Human Cytochrome P450 11B2 Produces Aldosterone by a Processive Mechanism Due to the Lactol Form of the Intermediate 18-Hydroxycorticosterone, J. Biol. Chem 2019, 294, 12975–12991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hartfield KA; Stout CD; Annalora AJ, The Novel Purification and Biochemical Characterization of a Reversible CYP24A1:Adrenodoxin Complex, J. Steroid Biochem. Mol. Biol 2013, 136, 47–53. [DOI] [PubMed] [Google Scholar]
- 65.Kyte J, Mechanism in Protein Chemistry. 1st ed.; Garland: New York, 1995. [Google Scholar]
- 66.Fisher MT; Sligar SG, Control of Heme Protein Redox Potential and Reduction Rate: Linear Free Energy Relation Between Potential and Ferric Spin State Equilibrium, J. Am. Chem. Soc 1985, 107, 5018–5019. [Google Scholar]
- 67.Wong I; Patel SS; Johnson KA, An Induced-Fit Kinetic Mechanism for DNA Replication Fidelity: Direct Measurement by Single-Turnover Kinetics, Biochemistry 1991, 30, 526–537. [DOI] [PubMed] [Google Scholar]
- 68.Johnson KA, Role of Induced Fit in Enzyme Specificity: A Molecular Forward/Reverse Switch, J. Biol. Chem 2008, 283, 26297–26301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Beckman JW; Wang Q; Guengerich FP, Kinetic Analysis of Correct Nucleotide Insertion by a Y-Family DNA Polymerase Reveals Conformational Changes Both Prior To and Following Phosphodiester Bond Formation as Detected by Tryptophan Fluorescence, J. Biol. Chem 2008, 283, 36711–36723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kellinger MW; Johnson KA, Role of Induced Fit in Limiting Discrimination Against AZT by HIV Reverse Transcriptase, Biochemistry 2011, 50, 5008–5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dangerfield TL; Johnson KA, Optimized Incorporation of an Unnatural Fluorescent Amino Acid Affords Measurement of Conformational Dynamics Governing High-Fidelity DNA Replication, J. Biol. Chem 2020, 295, 17265–17280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Murarka VC; Batabyal D ; Amaya JA; Sevrioukova IF ; Poulos TL Unexpected Differences between Two Closely Related Bacterial P450 Camphor Monooxygenase. Biochemistry 2020, 59, 2743–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Budavari S, in Merck Index. 12th ed.; Merck Research Laboratories: Whitehouse Station, NJ, 2000. [Google Scholar]
- 74.Rossi M, Structural Studies of Metyrapone: A Potent Inhibitor of Cytochrome P-450, 1983, 26, 1246–1252. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


