Arabidopsis ANT and AIL6 directly regulate genes involved in different aspects of early flower development including genes that specify floral organ identity and those that regulate growth and vascular development.
Keywords: AINTEGUMENTA (ANT), AINTEGUMENTA-LIKE6 (AIL6), Arabidopsis thaliana, ChIP-Seq, floral homeotic genes, floral organ identity, flower development, organ growth, organ size, vascular development
Abstract
Arabidopsis flower primordia give rise to organ primordia in stereotypical positions within four concentric whorls. Floral organ primordia in each whorl undergo distinct developmental programs to become one of four organ types (sepals, petals, stamens, and carpels). The Arabidopsis transcription factors AINTEGUMENTA (ANT) and AINTEGUMENTA-LIKE6 (AIL6) are required for correct positioning of floral organ initiation, contribute to the specification of floral organ identity, and regulate the growth and morphogenesis of developing floral organs. To gain insight into the molecular mechanisms by which ANT and AIL6 contribute to floral organogenesis, we identified the genome-wide binding sites of both ANT and AIL6 in stage 3 flower primordia, the developmental stage at which sepal primordia become visible and class B and C floral homeotic genes are first expressed. AIL6 binds to a subset of ANT sites, suggesting that AIL6 regulates some but not all of the same target genes as ANT. ANT- and AIL6-binding sites are associated with genes involved in many biological processes related to meristem and flower organ development. Comparison of genes associated with both ANT and AIL6 ChIP-Seq peaks and those differentially expressed after perturbation of ANT and/or AIL6 activity identified likely direct targets of ANT and AIL6 regulation. These include class B and C floral homeotic genes, growth regulatory genes, and genes involved in vascular development.
Introduction
Flowers have long fascinated humans for both their beauty and their morphological diversity. In addition, flowers are of practical importance since they contribute to human nutrition in the form of fruits, seeds, and grains. In Arabidopsis thaliana, flowers arise iteratively from the periphery of the inflorescence meristem, a dome-shaped structure at the apex of the plant. Auxin accumulation within these cells activates MONOPTEROS/AUXIN RESPONSE FACTOR5 (MP/ARF5), a transcription factor that up-regulates expression of the floral meristem identity gene LEAFY (LFY) as well as two AINTEGUMENTA-LIKE/PLETHORA (AIL/PLT) genes, AINTEGUMENTA (ANT) and AIL6/PLT3, to promote flower primordia identity and outgrowth (Yamaguchi et al., 2013). ANT and AIL6, two members of the larger APETALA2/ETHYLENE RESPONSE FACTOR (AP2/ERF) transcription factor family, and LFY, a plant-specific transcription factor, act early to establish flower primordia and later to promote their continued development with the specification and elaboration of different floral organ types (Weigel et al., 1992; Weigel and Meyerowitz, 1993; Elliott et al., 1996; Klucher et al., 1996; Krizek, 2009; Yamaguchi et al., 2016).
Floral meristems give rise to floral organ primordia at precise positions within four concentric whorls. Floral organ primordia adopt one of four fates (sepal, petal, stamen, or carpel) based on their relative position within the developing flower. Positional information within floral primordia is conveyed through distinct combinations of floral organ identity gene activities in each whorl of the flower, as summarized in the ABCE model (reviewed in Wellmer et al., 2014). In the outermost whorl one, the class A [APETALA1 (AP1) and APETALA2 (AP2)] and E [SEPALLATA1–4 (SEP1–SEP4)] genes specify sepal identity. In whorl two, the class A, B [APETALA3 (AP3) and PISTILLATA (PI)], and E genes specify petal identity. In whorl three, class B, C [AGAMOUS (AG)], and E genes specify stamen identity, and in whorl four, class C and E genes specify carpel identity. Mutations in class A, B, and C genes result in homeotic changes in floral organ identity in two adjacent whorls of the flower. Thus, these genes are also referred to as floral homeotic genes. All of the class A, B, C, and E genes, except the class A gene AP2, encode MADS domain transcription factors (Yanofsky et al., 1990; Jack et al., 1992; Mandel et al., 1992; Goto and Meyerowitz, 1994; Pelaz et al., 2000; Ditta et al., 2004). AP2 is a member of the AP2/ERF transcription factor family which also includes the AIL/PLT proteins ANT and AIL6 (Jofuku et al., 1994).
Both lfy single mutants and ant ail6 double mutants display loss of floral organ identities but do not show homeotic transformations in organ identity as described for mutations in the class A, B, and C genes (Weigel et al., 1992; Krizek, 2009). lfy flowers consist primarily of leaf-like and carpel-like organs, and exhibit reduced expression of class B and C genes (Weigel and Meyerowitz, 1993). Broadly expressed LFY acts with region-specific cofactors to directly activate expression of class B and C genes in stage 3 flowers (Lee et al., 1997; Busch et al., 1999; Lenhard et al., 2001; Lohmann et al., 2001; Lamb et al., 2002; Liu et al., 2009). ant ail6 double mutants produce flowers with sepals, filaments, stamenoid organs, unfused carpel valves, and structures that do not resemble any wild-type floral organs (Krizek, 2009). ANT and AIL6 act redundantly to promote petal, stamen, and carpel identity as these floral organs are present in both ant and ail6 single mutants (Elliott et al., 1996; Klucher et al., 1996; Krizek, 2009). Previous work has shown that expression of the class B genes AP3 and PI, and the class C gene AG is reduced in ant ail6 flowers, but it was not known whether ANT and AIL6 directly activate these genes in stage 3 flowers (Krizek, 2009; Krizek et al., 2016).
In addition to defects in floral organ identity, ant ail6 flowers make fewer and smaller floral organs that do not arise with regular phyllotaxy (Krizek, 2009). ANT plays a more important role than AIL6 in promoting floral organ growth as ant floral organs are reduced in size while ail6 flowers are normal in appearance (Elliott et al., 1996; Klucher et al., 1996; Krizek, 2009). However, the enhanced growth defects in ant ail6 double mutant flowers indicate that AIL6 also contributes to organ growth (Krizek, 2009). Overexpression of either ANT or AIL6 can result in larger flowers, indicating that both genes are sufficient for floral organ growth (Krizek, 1999; Mizukami and Fischer, 2000; Krizek and Eaddy, 2012; Han and Krizek, 2016).
Here we used ChIP in combination with next-generation sequencing (ChIP-Seq) to investigate the molecular means by which ANT and AIL6 regulate early events in flower development. We identified genome-wide binding sites for both ANT and AIL6 in stage 3 flowers, the stage at which sepal primordia are first visible and class B and C gene expression is initiated. Our work reveals that the partial redundancy of ANT and AIL6 results from AIL6 binding a subset of the genomic regions bound by ANT. Both ANT and AIL6 bind genomic loci associated with genes regulating many different developmental processes including radial patterning, polarity specification, boundary formation, floral meristem determinacy, and floral organ morphogenesis. To identify the most likely direct targets of ANT and AIL6 regulation, we compared our ChIP-Seq data with genes previously identified as differentially expressed in either ant ail6 double mutant inflorescences compared with the wild type or those differentially expressed after induction of ANT activity in steroid-treated 35S:ANT-GR inflorescences (Krizek et al., 2016, 2020). Furthermore, we investigated how the expression of some of these candidate direct target genes responds to inducible down-regulation of AIL6 in the ant mutant background or inducible down-regulation of ANT alone. Our data support a direct role for ANT and AIL6 in activating the floral homeotic genes AP3 and AG in stage 3 flowers, in regulating the expression of four growth regulatory genes: BIG BROTHER (BB), ROTUNDIFOLIA 3 (ROT3), ANGUSTIFOLIA3/GRF-INTERACTING FACTOR 1 (AN3/GIF1), and XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE9 (XTH9), and in regulating genes associated with vascular development.
Materials and methods
Plant materials and growth conditions
ANT:ANT-VENUS ant-4 AP1:AP1-GR ap1 cal plants and AIL6:AIL6-VENUS plants were described previously (Han and Krizek, 2016; Krizek et al., 2020). AIL6:AIL6-VENUS plants were crossed to AP1:AP1-GR ap1cal plants. AP1:AP1-GR ap1 cal, ANT:ANT-VENUS ant-4 AP1:AP1-GR ap1 cal, and AIL6:AIL6-VENUS AP1:AP1-GR ap1 cal inflorescences were grown on a soil mixture of Fafard 4P:perlite:vermiculite (8:1:1) in 24 h days at a light intensity of ~160 μmol m–2 s–1 at 20 °C. An artificial miRNA (amiRNA) that targets AIL6 was designed using http://wmd3.weigelworld.org/cgi-bin/webapp.cgi. A gene fragment containing this AIL6-amiR within the miR319a precursor was synthesized by IDT and cloned into the EcoRI/BamHI sites of BJ36_AlcA to create AlcA:AIL6-amiR/BJ36. AlcA:AIL6-amiR was subcloned into the NotI site of pMLBart_AlcR to create 35S:ALCR/AlcA:AIL6-amiR/pMLBart. 35S:ALCR/AlcA:AIL6-amiR/pMLBart was transformed into Agrobacterium strain ASE, which was then used to transform ant-4/+ plants. 35S:ALCR/AlcA:AIL6-amiR transgenic plants were selected for Basta resistance. The first 736 nucleotides of the ANT coding region were cloned in the sense and antisense directions in pHannibal using ANTIR-5 and ANTIR-6 (Supplementary Table S1). The ANT-IR fragment was digested from pHannibal with BamHI and cloned into BJ36_AlcA to create AlcA:ANT-IR. AlcA:ANT-IR was subcloned into the NotI site of pMLBart_AlcR to create 35S:ALCR/AlcA:ANT-IR/pMLBart. 35S:ALCR/AlcA:ANT-IR/pMLBart was transformed into Agrobacterium strain ASE, which was then used to transform Ler plants. 35S:ALCR/AlcA:ANT-IR transgenic plants were selected for Basta resistance. 35S:ALCR/AlcA:AIL6-amiR ant-4 and 35S:ALCR/AlcA:ANT-IR plants were grown on a soil mixture of Fafard 4P:perlite:vermiculite (8:1:1) in 16 h days at a light intensity of ~160 μ mol m–2 s–1 at 22 °C .
ChIP-Seq and ChIP-qPCR
Plants for ChIP-Seq and ChIP-qPCR were treated by pipetting a dex (10 μ M dexamethasone+0.015% Silwet) solution onto the inflorescences. ChIP was performed as described previously except that inflorescences were collected 2 d after dex treatment when the tissue consists of stage 3 flowers (Krizek et al., 2020). Primers for ChIP-qPCR are listed in Supplementary Table S1. Fold enrichment was determined relative to a negative control, the transposon TA3. Sequencing libraries were prepared from two biological replicates of input and ChIP DNA for AP1:AP1-GR ap1 cal, ANT:ANT-VENUS ant-4 AP1:AP1-GR ap1 cal, and AIL6:AIL6-VENUS AP1:AP1-GR ap1 cal as described previously (Krizek et al., 2020). Sequencing was performed on an Illumina HiSeq 2500 producing 150 base paired-end reads. Sequence reads were aligned to the reference A. thaliana genome (version TAIR9, released June 2009) using bowtie2. Examination by eye of the coverage graphs for each chromosome revealed high reproducibility between the two ChIP-Seq replicates. In addition, the input samples closely resembled the control untagged AP1:AP1-GR ap1 cal samples. ANT and AIL6 binding peaks were identified using a visual analytics approach within the Integrated Genome Brower (IGB) (Freese et al., 2016). Specifically, coverage graphs were generated for the combined data from the two replicates and normalized. A difference coverage graph was generated by subtracting coverage graphs of the untagged sample (AP1:AP1-GR ap1 cal) from the coverage graphs for the tagged samples (ANT:ANT-VENUS ant-4 AP1:AP1-GR ap1 cal and AIL6:AIL6-VENUS AP1:AP1-GR ap1 cal). Peaks were defined using the thresholding feature. A thresholding value of 2.5 identified 595 peaks for AIL6 and 2631 peaks for ANT in stage 3 flowers. For each identified peak, ChIPpeakAnno was used to identify the gene with the closest transcription start site (TSS) (Zhu et al., 2010). In some cases, a peak located within a gene is associated with two genes if the TSS of an adjacent gene is closer than the TSS of the gene overlapping the peak. Gene Ontology (GO) analyses were performed with AmiGO 2 (http://amigo.geneontology.org/amigo). Enriched GO terms were identified using a Fisher’s exact test of contingency tables followed by Bonferroni correction for multiple hypothesis testing. BEDtools intersect was used to identify overlapping ANT and AIL6 peaks in stage 3 flowers. De novo motif discovery was performed with MEME-ChIP (Machanick and Bailey, 2011) searching for five MEME motifs using the Arabidopsis DAP motifs (O’Malley et al., 2016). FIMO was used to locate individual occurrences of MEME-identified motifs within common ANT- and AIL6-binding sites (Grant et al., 2011). The relative position of identified motifs was calculated using the Distance program. Source codes for bioinformatic analyses including the Distance program are available in the project ‘git’ repository (https://bitbucket.org/krizeklab/antail6stage3chipseq/). Venn diagrams were created using Venn Diagram Plotter (https://omics.pnl.gov) created by the Pacific Northwest National Lab.
RT-qPCR
35S:AlcR/AlcA:AIL6-amiR ant-4 and 35S:ALCR/AlcA:ANT-IR plants were mock treated or treated with ethanol vapor by placing 2 ml of water or 2 ml of 100% ethanol in 2 ml centrifuge tubes in half of the pots in a tray. The tray was covered with a plastic dome. Inflorescences were collected at the end of an 8 h (35S:AlcR/AlcA:AIL6-amiR ant-4) or 24 h (35S:ALCR/AlcA:ANT-IR) treatment. RNA was isolated using an RNeasy Plant Mini Kit (Qiagen) or TRIzol (Life Technologies). Samples isolated with TRIzol were further purified on an RNeasy column (Qiagen) and treated with DNase while on the column. First-strand cDNA synthesis was performed using Quanta qScript cDNA SuperMix (Quanta BioSciences) following the manufacturer’s instructions. Quantitative PCR (qPCR) was performed on a BioRad CFX96 or CFX Connect real-time PCR system using PerfeCTa SYBR Green FastMix for iQ (Quanta BioSciences) and primers listed in Supplementary Table S1. Data analyses were carried out as described previously (Krizek and Eaddy, 2012). At least two biological replicates were analyzed for each experiment.
Gel mobility shift assays
The gel mobility shift assays were performed as described previously (Nole-Wilson and Krizek, 2000).
Yeast strain and β-galactosidase assays
A reporter plasmid was made by cloning 76 bp of AG intron sequence upstream of the lacZ gene in the vector pLacZi (Clontech). The yeast reporter strain was made by integration of the linearized AG intron reporter plasmid into the yeast strain YM4271. This new reporter strain was transformed with the previously described ANT/pGAD424 lacking the GAL4 activation domain (Krizek, 2003). Transformants were selected on plates containing synthetic medium lacking leucine. β-Galactosidase assays were performed as described previously (Krizek, 2003).
Petal size measurements
Petal measurements for mock- and ethanol-treated Ler and 35S:ALCR/AlcA:ANTIR were performed on 10–20 petals from different flowers at positions 1–10 on an inflorescence. Petal measurements were performed as described previously (Trost et al., 2014). Petals from stage 13 flowers were removed with forceps and placed on Sellotape. The tape was adhered to a piece of black plexiglass and scanned at a resolution of 3600 dpi in 8-bit grayscale. Petal area, length, and width were determined using Image J software.
Results
ChIP-Seq identifies many more ANT-binding peaks than AIL6-binding peaks
To begin to understand the overlapping functions of ANT and AIL6 in early stages of flower development, we performed ChIP-Seq using previously described ANT–VENUS and AIL6–VENUS fusion lines expressed under their respective endogenous promoters in the AP1:AP1-GR ap1 cal synchronized floral induction system (O’Maoiléidigh et al., 2015; Han and Krizek, 2016; Krizek et al., 2020). The AP1:AP1-GR ap1 cal system allows for the collection of inflorescences composed of flowers of a single stage of flower development (O’Maoiléidigh et al., 2015). Inflorescences from AP1:AP1-GR ap1 cal (no tag), ANT:ANT-VENUS ant AP1:AP1-GR ap1 cal (ANT–VENUS), and AIL6:AIL6-VENUS AP1:AP1-GR ap1 cal (AIL6–VENUS) plants were collected 2 d after dex treatment when the inflorescences are composed of stage 3 flowers. Peaks were identified using a visual analytic method in IGB (Freese et al., 2016) in which signal from the no tag sample was subtracted from the tagged sample and signals above a threshold value were identified. This method revealed 2631 peaks for ANT–VENUS and 595 peaks for AIL6–VENUS. Visual inspection of the data reveals many overlapping ANT and AIL6 peaks. However, there are many more ANT peaks than AIL6 peaks, and ANT peaks are higher in signal than AIL6 peaks. This suggests that ANT regulates more genes than AIL6 and that there is generally lower occupancy of genomic regions by AIL6 as compared with ANT in the collected tissue. This is consistent with the more important role of ANT as compared with AIL6 in floral organ development (Krizek, 2009). Loss of ANT function results in smaller floral organs and slight reductions in floral organ number, while loss of AIL6 function has no phenotypic consequence (Elliott et al., 1996; Klucher et al., 1996; Krizek, 2009). The lower signal of the majority of AIL6-binding peaks as compared with corresponding ANT peaks may be a consequence of lower levels of AIL6 protein as compared with ANT in developing flowers or from AIL6 having lower affinity for these genomic regions as compared with ANT. In addition, competition between the endogenous AIL6 protein and the transgenic AIL6–VENUS protein may have lowered the ChIP-Seq signal in this experiment compared with that for ANT–VENUS. For the ANT–VENUS line, we used an ant mutant background in which there is probably no endogenous ANT protein produced.
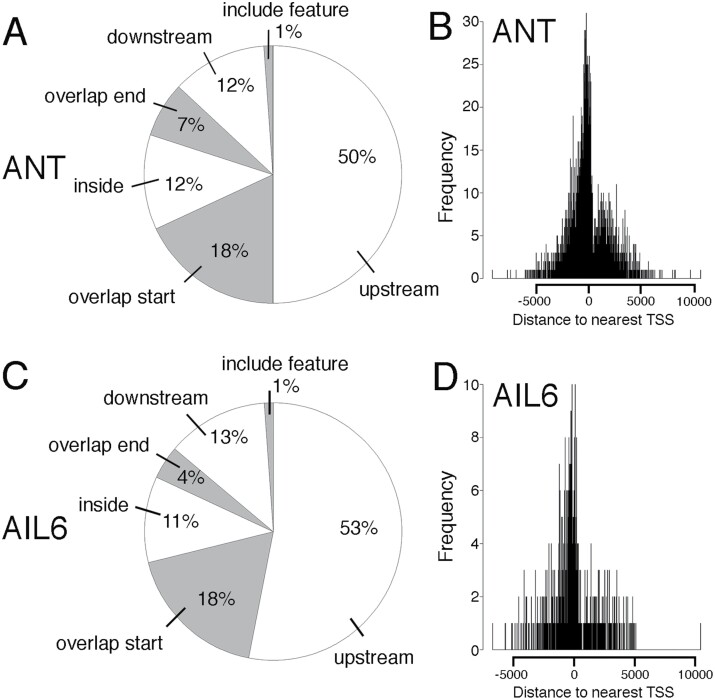
We used ChIPpeakAnno to identify genes associated with ANT- and AIL6-binding peaks (Zhu et al., 2010). ANT peaks were associated with 2318 unique genes while AIL6 peaks were associated with 592 unique genes (Supplementary Dataset S1). ANT and AIL6 peaks showed similar distributions relative to the positions of genes. For ANT, half of the peaks (50%) are present upstream of the gene, with the remaining peaks either overlapping the start of transcription (18%), within the gene (12%), overlapping the end of transcription (7%), downstream of the gene (12%), or encompassing the gene (1%) (Fig. 1A). For AIL6, over half of the peaks (53%) are present upstream of the gene with the remaining peaks either overlapping the start of transcription (18%), within the gene (11%), overlapping the end of transcription (4%), downstream of the gene (13%), or encompassing the gene (1%) (Fig. 1C). For both ANT and AIL6, the majority of peaks are located within 2.5 kb of the TSS (Fig. 1B, D). The average distance upstream from the TSS was 37 bp for ANT and 90 bp for AIL6.
Fig. 1.
Position of ANT and AIL6 ChIP-Seq peaks relative to the closest gene. (A) Pie chart showing the position of ANT ChIP-Seq binding peaks relative to the closest gene. Approximately half of the peaks are upstream of the closest gene (50%). The remaining peaks either overlap the start of the gene (18.0%), are within the gene (12%), overlap the end of the gene (7%), are downstream of the gene (12%), or overlap the entire gene (1%). (B) Position of ANT-binding peaks relative to the transcriptional start site (TSS) of the closest gene. (C) Pie chart showing the position of AIL6 ChIP-Seq binding peaks relative to the closest gene. Approximately half of the peaks are upstream of the closest gene (53%). The remaining peaks either overlap the start of the gene (18.0%), are within the gene (11%), overlap the end of the gene (4%), are downstream of the gene (13%), or overlap the entire gene (1%). (D) Position of AIL6-binding peaks relative to the TSS of the closest gene.
GO term enrichment analyses link ANT and AIL6 function with meristem and flower development, hormone physiology, and transcriptional regulation
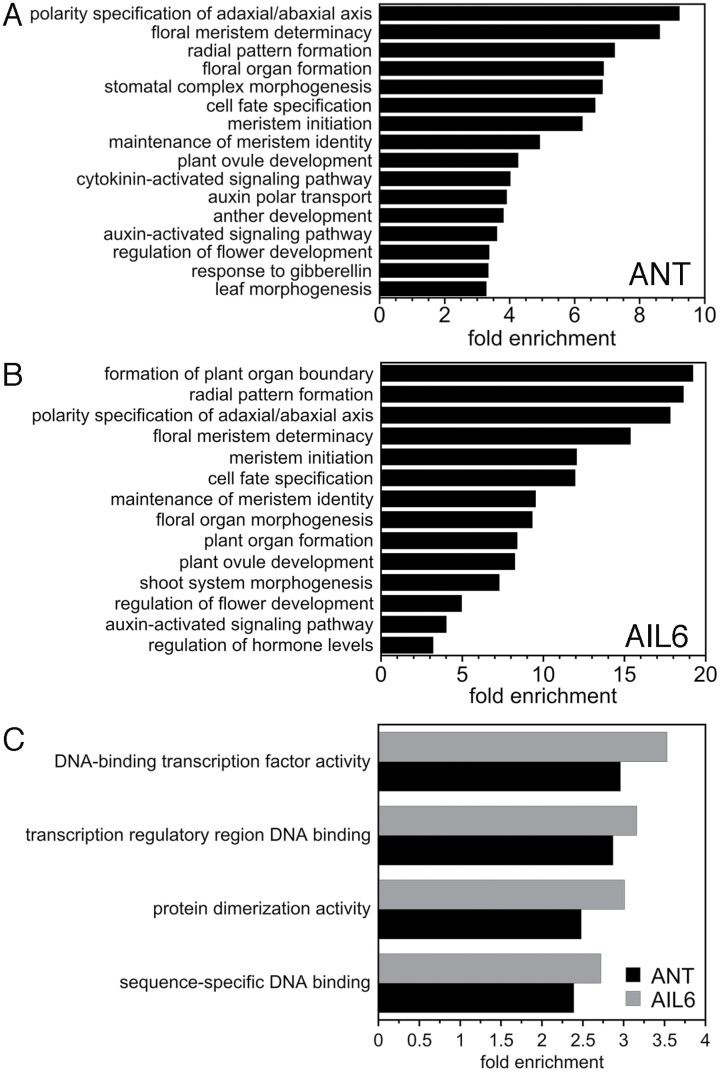
GO enrichment analyses identified a number of over-represented terms in the set of genes associated with either ANT or AIL6 peaks (Fig. 2; Supplementary Datasets S2, S3). Over-represented GO biological process terms that were identified for both ANT and AIL6 include many associated with meristem and lateral organ development including: polarity specification of the adaxial/abaxial axis (GO:0009944), floral meristem determinacy (GO:0010582), radial pattern formation (GO:0009956), cell fate specification (GO:0001708), meristem initiation (GO:0010014), maintenance of meristem identity (GO:0010074), plant ovule development (GO:0048481), and regulation of flower development (GO:0009909) (Fig. 2A, B). Other over-represented biological process development-related GO terms for genes associated with ANT-binding peaks include: floral organ formation (GO:0048449), stomatal complex morphogenesis (GO:0010103), anther development (GO:0048653), and leaf morphogenesis (GO:0009965) (Fig. 2A). Additional over-represented GO terms for genes associated with AIL6-binding peaks were: formation of plant organ boundary (GO:0090691), floral organ morphogenesis (GO:0048444), plant organ formation (GO:1905393), and shoot system morphogenesis (GO:0010016) (Fig. 2B).
Fig. 2.
GO enrichment analyses on genes associated with ANT and AIL6 ChIP-Seq binding peaks. (A) Biological process GO terms enriched in genes associated with ANT-binding peaks. (B) Biological process GO terms enriched in genes associated with AIL6-binding peaks. (C) Molecular function GO terms enriched in genes associated with ANT- and AIL6-binding peaks.
One over-represented biological process hormone-related GO term associated with both ANT- and AIL6-binding peaks was the auxin-activated signaling pathway (GO:0009734) (Fig. 2A, B). In addition, other over-represented hormone terms for genes associated with ANT peaks include cytokinin-activated signaling pathway (GO:0009736), auxin polar transport (GO:0009926), response to gibberellin (GO:0009739), ethylene-activated signaling pathway (GO:0009873), and response to abscisic acid (GO:0009737) (Fig. 2A, B; Supplementary Dataset S2). These results suggest that both ANT and AIL6 regulate auxin signaling while ANT plays a more general role in mediating multiple hormone signaling pathways and responses.
Several over-represented molecular function GO terms for both ANT and AIL6 are associated with transcriptional regulation. These include DNA-binding transcription factor activity (GO:0003700), transcription regulatory region DNA binding (GO:0044212), protein dimerization activity (GO:0046983), and sequence-specific DNA binding (GO:0043565) (Fig. 2C; Supplementary Datasets S2, S3). ANT and AIL6 bind to numerous genomic regions associated with transcription factors that regulate development.
AIL6-binding peaks correspond to a subset of ANT peaks
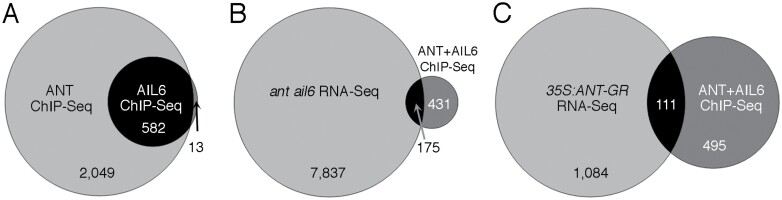
We determined the degree of overlap between ANT- and AIL6-binding peaks. A total of 582 out of the 595 AIL6 peaks (~98%) overlap at least 50% with an ANT peak (Fig. 3A). For the 13 AIL6 peaks that did not overlap at least 50% with an ANT peak, this was because there was no nearby ANT peak (three), the AIL6 peak was wider with a summit shifted compared with an ANT peak in the same region (three), or there was an overlapping ANT peak but its height was below the 2.5 threshold value used for peak identification (seven) (Supplementary Dataset S4). These results indicate that AIL6 binds almost exclusively to a subset of regions also bound by ANT in stage 3 flowers. This is consistent with AIL6 providing some but not all of the same functions as ANT (Krizek, 2009). A total of 606 unique genes are associated with the 582 overlapping peaks between ANT and AIL6 (Supplementary Dataset S5). Some of the developmental genes bound by both ANT and AIL6 are shown in Table 1.
Fig. 3.
Venn diagrams showing the overlap of ANT and AIL6 ChIP-Seq data and the overlap of genes that are bound by both ANT and AIL6 and differentially expressed in ant ail6 or 35S:ANT-GR RNA-Seq experiments. (A) Venn diagram showing the overlap of ANT and AIL6 ChIP-Seq binding peaks. (B) Venn diagram showing the overlap of genes associated with both ANT and AIL6 ChIP-Seq peaks and genes differentially expressed in ant ail6 compared with Ler (RNA-Seq). (C) Venn diagram showing the overlap of genes associated with both ANT and AIL6 ChIP-Seq peaks and genes differentially expressed in dex-treated 35S:ANT-GR compared with mock-treated 35S:ANT-GR (RNA-Seq).
Table 1.
Developmental genes associated with both ANT and AIL6 ChIP-Seq peaks
| Gene identifier | Gene name | ChIP-Seq binding peak position | Log2 fold change (ant ail6 or 35S:ANT-GR) |
|---|---|---|---|
| Boundary genes | |||
| AT2G31160 | LSH3/OBO1 | Upstream; downstream | 0.90 (ant ail6) |
| AT3G23290 | LSH4 | Upstream | 0.85 (ant ail6) |
| AT4G32980 | ATH1 | Overlap start | 1.12 (ant ail6) –0.238 (4 h 35S:ANT-GR) –0.258 (4 h 35S:ANT-GR) |
| AT3G15170 | CUC1 | Overlap start | – |
| AT1G76420 | CUC3 | Overlap start | – |
| AT1G26780 | LOF1 | Upstream | – |
| Vascular genes | |||
| AT2G37590 | DOF2.4/PEAR1 | Upstream; downstream | – |
| AT5G60200 | DOF5.3/TMO6 | Upstream | 0.253 (4 h 35S:ANT-GR) 0.247 (8 h 35S:ANT-GR) |
| AT1G07640 | OBP2 | Downstream | 1.15 (ant ail6) |
| AT2G34925 | CLE42 | Upstream | |
| AT2G27230 | LHW | Overlap end | |
| AT1G26600 | CLE9 | Overlap start | |
| AT5G61480 | PXY | Upstream | 0.67 (ant ail6) |
| AT5G65700 | BAM1 | Downstream | |
| AT1G19850 | MP | Upstream | –0.59 (ant ail6) |
| AT5G60690 | REV | Overlap start | |
| AT1G19050 | ARR7 | Overlap start | 1.25 (ant ail6) –0.541 (4 h 35S:ANT-GR) –0.477 (8 h 35S:ANT-GR) |
| AT5G62230 | ERL1 | Upstream; overlap start | –0.76 (ant ail6) |
| Polarity genes | |||
| AT5G60690 | REV | Overlap start | – |
| AT2G37630 | AS1 | Upstream | –0.235 (4 h 35S:ANT-GR) –0.175 (8 h 35S:ANT-GR) |
| AT3G57130 | BOP1 | Upstream | –0.449 (4 h 35S:ANT-GR) |
| AT2G41370 | BOP2 | Upstream | 0.65 (ant ail6) |
| AT2G45190 | FIL/YAB1 | Upstream | –0.47 (ant ail6) |
| AT5G16560 | KAN1 | Upstream; inside | – |
| AT1G32240 | KAN2 | Inside | 0.227 (4 h 35S:ANT-GR) 0.255 (8 h 35S:ANT-GR) |
| Floral organ morphogenesis genes | |||
| AT3G54340 | AP3 | Overlap start | –2.38 (ant ail6) |
| AT5G20240 | PI | Overlap start | –2.29 (ant ail6) |
| AT4G18960 | AG | Inside | –1.26 (ant ail6) |
| AT1G24260 | SEP3 | Inside | 0.175 (8 h 35S:ANT-GR) |
| AT5G67060 | HEC1 | Upstream | – |
| AT3G50330 | HEC2 | Upstream | –2.24 (ant ail6) |
| AT2G01940 | SGR5/IDD15 | Downstream | 0.320 (4 h 35S:ANT-GR) 0.497 (8 h 35S:ANT-GR) |
| AT1G25250 | IDD16 | Downstream | – |
ANT and AIL6 ChIP-Seq peaks contain DNA-binding motifs for AIL/PLT, BPC, and bHLH transcription factors
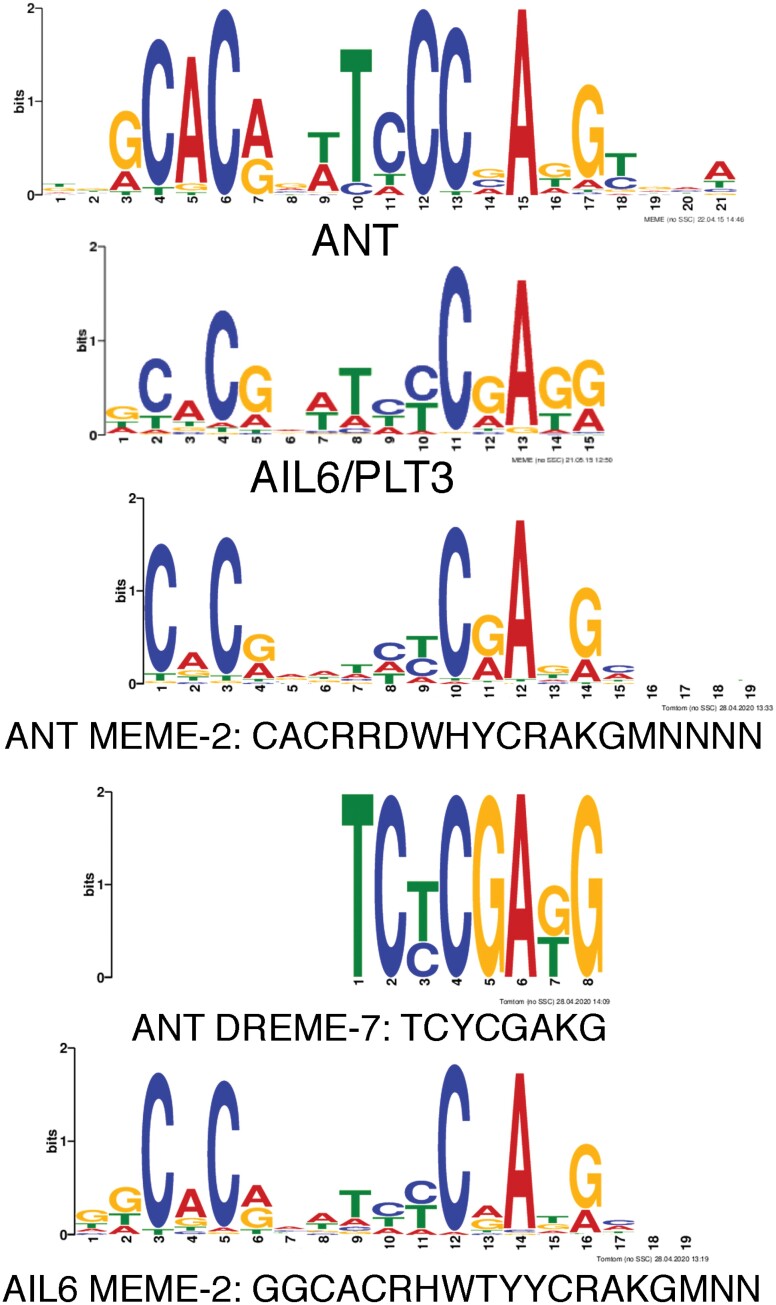
MEME-ChIP from the MEME Suite was used to perform de novo motif discovery on the binding peaks for both ANT and AIL6 (Machanick and Bailey, 2011). This analysis used the DAP-Seq database for motif discovery which contains motifs for several AIL/PLT transcription factors including AIL6 but not ANT (O’Malley et al., 2016). AIL/PLT-binding motifs are fairly long, with a few conserved residues on both ends of the motif and somewhat fewer conserved nucleotides in the center, as shown for AIL6 (Fig. 4). This is less true for the in vitro determined ANT-binding motif which consists of more conserved residues throughout the motif (Fig. 4) (Nole-Wilson and Krizek, 2000). For both ANT and AIL6 ChIP-Seq binding peaks, motifs with similarity to those bound by AIL/PLT transcription factors were identified (Table 2). For ANT, these motifs are MEME-2 (CACRRDWHYCRAKGMNNNN) and DREME-7 (TCYCGAKG), and for AIL6 the single motif MEME-2 (GGCACRHWTYYCRAKGMNN) was identified (Table 2; Fig. 4). The ANT MEME-2 and AIL6 MEME-2 motifs are very similar and contain conserved nucleotides toward both ends of the AIL/PLT-like-binding motif, while the DREME-7 motif identified in ANT-binding peaks has similarity to the right half of AIL/PLT-like motifs (Fig. 4).
Fig. 4.
DNA motifs with similarity to ANT- and AIL6-binding motifs are over-represented in ANT and AIL6 ChIP-Seq binding peaks. Top: sequence logos for ANT- and AIL6-binding motifs. Middle: two motifs over-represented in ANT-binding peaks with similarity to the ANT-binding motif are ANT MEME-2 and ANT DREME-7. Bottom: one motif over-represented in AIL6-binding peaks with similarity to the AIL6-binding motif is AIL6 MEME-2.
Table 2.
MEME-ChIP analysis of ANT and AIL6 ChIP-Seq peaks
| ANT motifs | Motif ID | Width | Sites | e-value | Most similar DAP-Seq motif |
|---|---|---|---|---|---|
| RRRRARARARARARARARARR | MEME-1 | 21 | 689 | 5.70E-194 | BPC5 (BBR/BPC) |
| CACRRDWHYCRAKGMNNNN | MEME-2 | 19 | 355 | 8.00E-45 | AIL7 (AP2/ERF) |
| AGAGAGM | DREME-1 | 7 | 437 | 1.20E-29 | BPC1 (BBR/BPC) |
| CACRTS | DREME-2 | 6 | 686 | 8.10E-22 | AREB3 (bZIP) |
| CACGTGDCAYDYRCR | MEME-4 | 15 | 88 | 1.50E-11 | BIM2 (bHLH) |
| TCYCGAKG | DREME-7 | 8 | 57 | 1.40E-08 | AIL6 (AP2/ERF) |
| AIL6 motifs | Motif ID | Width | Sites | e-value | Most similar DAP-Seq motif |
| GGCACRHWTYYCRAKGMNN | MEME-2 | 19 | 219 | 3.20E-186 | PLT1 (AP2/ERF) |
| RRRARARARAGARARAGARG | MEME-1 | 20 | 198 | 1.40E-118 | BPC5 (BBR/BPC) |
| CACGTGDCKTBYKC | MEME-3 | 14 | 50 | 6.00E-13 | bHLH74 (bHLH) |
| AGAGAVA | DREME-1 | 7 | 161 | 3.80E-10 | BPC1 (BBR/BPC) |
| CACGHG | DREME-2 | 6 | 103 | 8.20E-10 | bHLH74 (bHLH) |
FIMO (Find Individual Motif Occurrences) identified 224 ANT MEME-2 motifs and 298 AIL6 MEME-2 motifs present in ANT/AIL6 overlapping binding peaks (Grant et al., 2011). A total of 185 of the 224 ANT MEME-2 motifs map to the same location as an AIL6 MEME-2 motif with an offset of two nucleotides, because the ANT MEME-2 site lacks the GG nucleotides at positions one and two of the AIL6 MEME-2 motif. This suggests that either ANT or AIL6 can bind to a single DNA sequence that matches both the ANT MEME-2 and AIL6 MEME-2 motifs present within these peaks. The remaining ANT MEME-2 motifs were not separated with any conseved distance from an AIL6 MEME-2 motif.
BARLEY B-RECOMBINANT/BASIC PENTACYSTEINE (BBR/BPC) and basic helix–loop–helix (bHLH) transcription factor-binding motifs were over-represented in both ANT and AIL6 stage 3 binding peaks (Table 2; Supplementary Fig. S1). Previously, we found that binding motifs for these two families of transcription factors were over-represented in ANT-binding peaks identified in stage 6 and 7 flowers (Krizek et al., 2020). BBR/BPC are broadly expressed transcription factors involved in many developmental processes (Monfared et al., 2011). They mediate gene silencing by recruiting Polycomb repressive complexes or other regulatory proteins to GAGA motifs (Simonini et al., 2012; Hecker et al., 2015; Xiao et al., 2017). Binding motifs for basic leucine zipper transcription factors (bZIPs) were also over-represented in ANT-binding peaks (Table 2). These other transcription factors may act in combination with ANT and AIL6 in regulating transcription of target genes.
Identification of genes bound by ANT and AIL6 and differentially expressed in ant ail6 or 35S:ANT-GR inflorescences
Direct targets of a transcription factor are typically defined as genes whose regulatory region is bound by the transcription factor and whose expression is altered in response to changes in the activity of the transcription factor. While ChIP-Seq can identify many hundreds or thousands of transcription factor-binding sites, not all of the genes associated with these sites may be transcriptionally regulated by these binding events. Jointly analyzing ChIP-Seq and RNA-Seq offers the best approach to identify direct target genes. A comparison of the set of genes bound in stage 3 flowers by both ANT and AIL6 (606 genes) with the set of genes that are differentially expressed in ant ail6 inflorescences (8012 genes) identified an overlap of 175 genes (Fig. 4B) (Krizek et al., 2016).
A second comparison of the genes bound in stage 3 flowers by both ANT and AIL6 (606 genes) with genes that are differentially expressed after induction of ANT activity in 35S:ANT-GR inflorescences (1195 genes) (Krizek et al., 2020) identified 111 genes that may be direct targets of ANT and AIL6 regulation (Fig. 4C). Within this set of 111 genes, 29 genes are differentially expressed in both the ant ail6 and 35S:ANT-GR RNA-Seq datasets (Supplementary Dataset S6). Of these 29 genes, 15 show opposite regulation in 35S:ANT-GR and ant ail6 inflorescences (i.e. down-regulated in 35S:ANT-GR and up-regulated in ant ail6 or, vice versa, up-regulated in 35S:ANT-GR and down-regulated in ant ail6).
ANT and AIL6 activate class B and C homeotic genes in stage 3 flowers
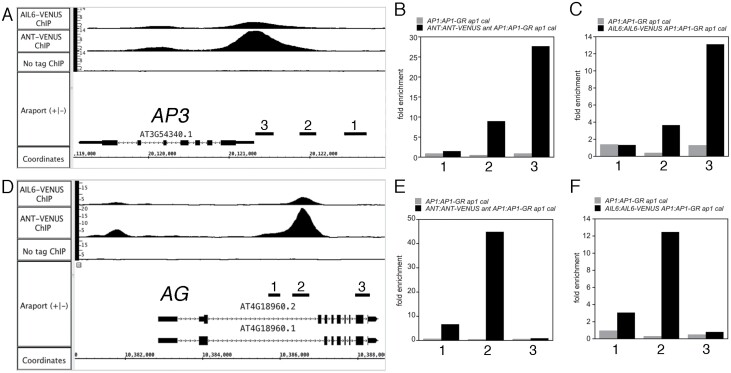
Included in the set of 175 genes that are both bound by ANT and AIL6 and differentially expressed in ant ail6 inflorescences are the class B floral homeotic genes AP3 and PI, and the class C floral homeotic gene AG (Fig. 5A, D; Table 1; Supplementary Fig. S2A). We confirmed the ChIP-Seq results for AP3, PI, and AG using ChIP-qPCR (Fig. 5B, C, E, F; Supplementary Fig. S2B, C). Expression of these three genes is down-regulated in ant ail6 inflorescences (Krizek et al., 2016). In addition, in situ hybridization had shown previously that AP3 and AG were expressed in fewer cells of stage 3 floral primordia (Krizek, 2009). These results are consistent with the loss of petal, stamen, and carpel identities in ant ail6 flowers (Krizek, 2009). These data suggest that ANT and AIL6 might directly activate the expression of class B and C floral homeotic genes in young flowers.
Fig. 5.
ANT and AIL6 bind directly to regulatory regions associated with the floral organ identity genes AP3 and AG. (A) ChIP-Seq of ANT and AIL6 binding to the AP3 genomic region. 1, 2, and 3 are the genomic regions tested for binding using ChIP-qPCR in (B) and (C). (B) ChIP-qPCR of ANT binding to AP3 genomic region 3. (C) ChIP-qPCR of AIL6 binding to AP3 genomic region 3. (D) ChIP-Seq of ANT and AIL6 binding to the AG genomic region. 1, 2, and 3 are the genomic regions tested for binding using ChIP-qPCR in (E) and (F). (E) ChIP-qPCR of ANT binding to AG genomic region 2. (F) ChIP-qPCR of AIL6 binding to AG genomic region 2.
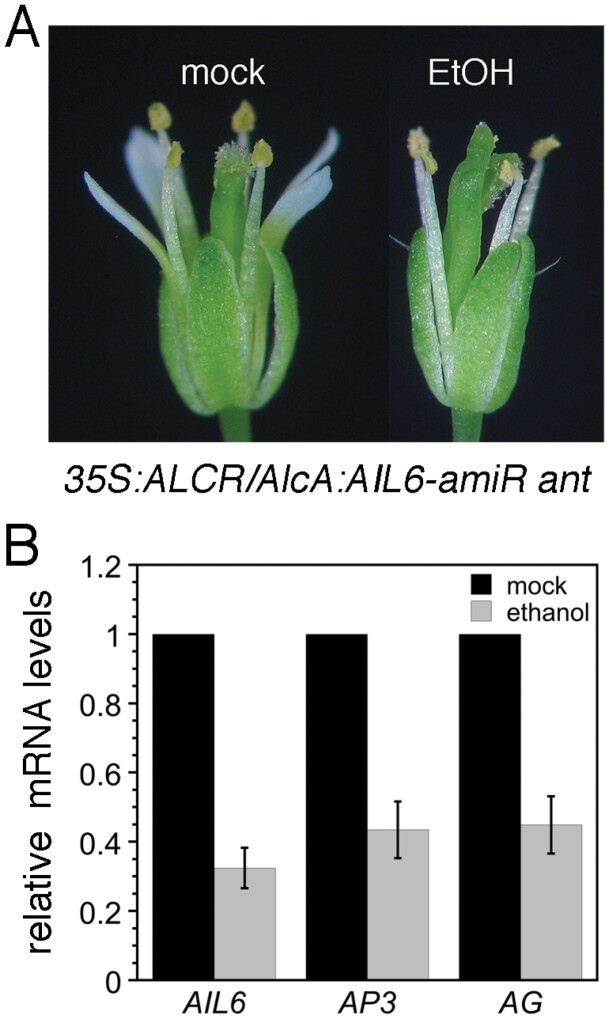
To investigate whether ANT and AIL6 directly control transcription of AP3, PI, and AG, we examined whether class B and C gene expression responds quickly to changes in ANT and AIL6 activity. We used an ethanol-inducible transgenic line in which AIL6 expression is down-regulated by an amiRNA in the ant mutant background (35S:ALCR/AlcA:AIL6-amiR ant; i.e. AIL6-amiR ant). After an 8 h ethanol treatment, AIL6-amiR ant flowers exhibit a phenotype more severe than ant, with loss of petals and partially unfused carpels, suggesting that AIL6 activity is reduced (Fig. 6A). Mock-treated AIL6-amiR ant flowers resemble ant (Fig. 6A). AIL6 mRNA levels in the ethanol-treated inflorescences are ~30% of those in mock-treated inflorescences, indicating that the AIL6-amiR knocks down AIL6 expression (Fig. 6B). AP3 and AG mRNA levels but not PI mRNA levels are reduced after this 8 h ethanol treatment (Fig. 6B; Supplementary Fig. S2D). These data further support the conclusion that ANT and AIL6 directly activate expression of the class B gene AP3 and the class C gene AG in stage 3 flowers.
Fig. 6.
AP3 and AG expression is reduced soon after down-regulation of AIL6 expression in 35S:ALCR/AlcA:AIL6-amiR ant inflorescences. (A) Mock- and ethanol (EtOH)-treated 35S:ALCR/AlcA:AIL6-amiR ant flowers. (B) Expression of AIL6, AP3, and AG is reduced in 35S:ALCR/AlcA:AIL6-amiR ant inflorescences after an 8 h EtOH treatment. Error bars show the SD.
No DNA sequences with obvious similarity to ANT- or AIL6-binding motifs are present in AP3 genomic regions (Nole-Wilson and Krizek, 2000; O’Malley et al., 2016; Krizek et al., 2020). The ANT MEME-2 motif and the AIL6 MEME-2 motif identified here were also not present in these peaks. Thus, is it not clear what DNA sequence is bound by ANT and AIL6 within the AP3 genomic region. The ANT- and AIL6-binding peaks upstream of AP3 do overlap the defined PEE core element that is required for early stage 3–5 expression (Lamb et al., 2002). Two partially overlapping sequences with weak similarity to the ANT-binding motif are present near the summit of the ANT and AIL6 ChIP-Seq binding peaks within the AG intron (Supplementary Fig. S3A). ANT can bind specifically to this region of the AG intron in vitro and can activate transcription in yeast through this sequence (Supplementary Fig. S3B, C). For both AP3 and AG, the ANT- and AIL6-binding peaks overlap those of other known regulators of AP3, PI, and AG, including LFY, AP1, AP2, AP3, PI, AG, and SEP3 (Kaufmann et al., 2009, 2010; Yant et al., 2010; Winter et al., 2011; Wuest et al., 2012; Ó’Maoiléidigh et al., 2013) (Supplementary Fig. S4A, B).
Cross-regulation of AIL expression may involve direct repression by ANT and AIL6
Previously, we found evidence of cross-regulation among AIL gene expression (Krizek et al., 2016). Specifically, we found that AIL6 mRNA levels are increased in ant mutants and that ANT, AIL6, and AIL7 mRNA levels are increased in ant ail6 double mutants (Krizek et al., 2016). Our ChIP-Seq results show that both ANT and AIL6 bind the regulatory regions of ANT, AIL5, and AIL6; ANT also binds to the regulatory region of AIL7 (Supplementary Fig. S5). This suggests that the observed repression of AIL expression by ANT and AIL6 may be mediated by direct binding of ANT and AIL6 to AIL regulatory regions.
As seen with the floral homeotic genes, ANT- and AIL6-binding peaks in these regulatory regions overlap those of other floral regulators, including LFY, AP1, AP3, PI, and AG (Kaufmann et al., 2010; Winter et al., 2011; Wuest et al., 2012; Ó’Maoiléidigh et al., 2013) (Supplementary Fig. S5). ANT and AIL6 expression in floral anlagen occurs with similar timing to LFY expression, and LFY was previously shown to bind to ANT and AIL6 genomic regions (Winter et al., 2011). Thus, LFY may contribute to ANT and AIL6 expression in floral primordia. In contrast, AP1, AP3, PI, and AG are expressed later in flower development than ANT and AIL6 and thus would only contribute to maintenance or refinement of ANT and AIL6 expression at later stages of development, if they play a role at all.
ANT and AIL6 directly regulate organ growth genes
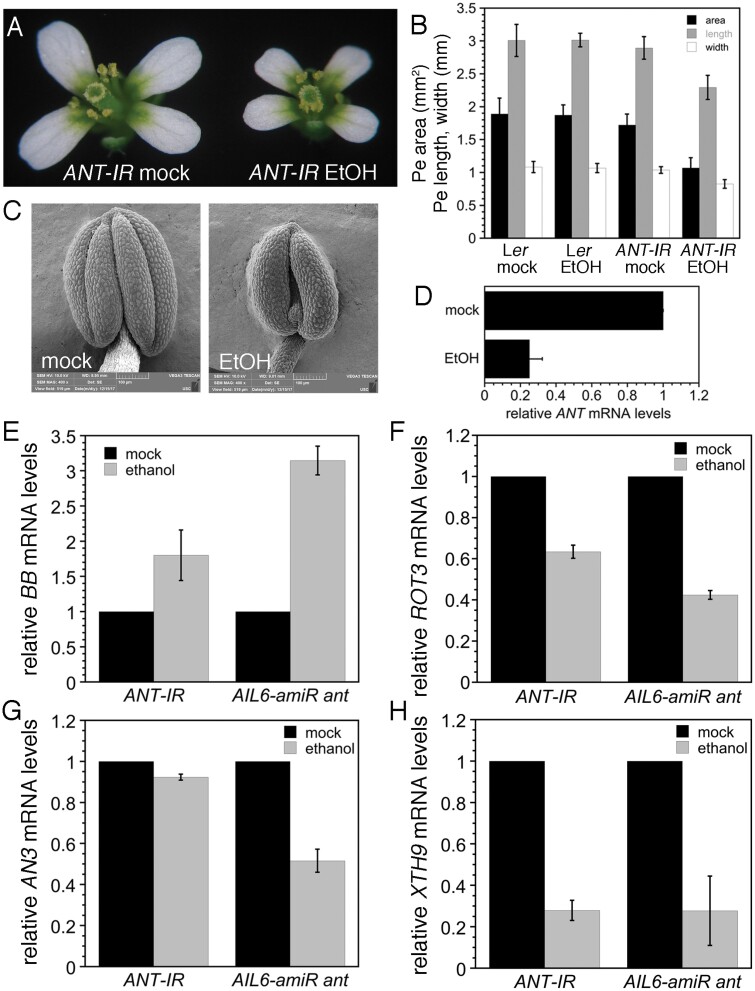
ANT and AIL6 both positively contribute to floral organ growth, although AIL6 cannot completely substitute for ANT in this role as ant single mutants produce smaller floral organs (Elliott et al., 1996; Klucher et al., 1996). Several growth-regulating genes are bound by both ANT and AIL6 in our ChIP-Seq experiments and exhibit differential expression in either ant ail6 double mutants or dex-treated 35S:ANT-GR inflorescences. These include the growth repressor BIG BROTHER (BB), and the growth-promoting genes ROTUNDIFOLIA3 (ROT3), ANGUSTIFOLIA3/GRF-INTERACTING FACTOR 1 (AN3/GIF1), and XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE9 (XTH9) (Supplementary Fig. S6). BB expression was elevated in ant ail6 double mutants, ROT3 expression was reduced in ant ail6 double mutants, and AN3/GIF1 and XTH9 expression was up-regulated by induction of ANT-GR activity (Krizek et al., 2016, 2020). AN3/GIF1 and XTH9 were previously identified as likely direct targets of ANT regulation as their regulatory regions are bound by ANT in stage 6/7 flowers (Krizek et al., 2020).
We further investigated whether these genes are likely to be direct targets of ANT and AIL6 regulation by examining their expression in response to down-regulation of AIL6 in the ant mutant background and down-regulation of ANT alone. To down-regulate ANT expression, we generated an RNAi line that utilizes an ethanol-inducible inverted repeat transgene (35S:ALCR/AlcA:ANT-IR; i.e. ANT-IR). After ethanol treatment, ANT-IR flowers produce smaller petals and anthers with two locules similar to ant mutants (Fig. 7A–C). After a 24 h ethanol treatment, ANT mRNA levels are reduced to ~25% of that of the mock-treated plants (Fig. 7D). BB expression was increased 1.8-fold in ethanol-treated ANT-IR and 3.1-fold in ethanol-treated AIL6-amiR ant inflorescences, suggesting that both ANT and AIL6 contribute to repression of BB expression (Fig. 7E). After ethanol induction, ROT3 expression levels were 63% of the levels seen in mock-treated ANT-IR and 42% of the levels seen in mock-treated AIL6-amiR inflorescences (Fig. 7F). AN3/GIF1 expression was slightly reduced in ANT-IR inflorescences and reduced ~2-fold in AIL6-amiR inflorescences (Fig. 7G). This suggests that for AN3/GIF1 regulation, AIL6 can largely compensate for loss of ANT. After ethanol induction, XTH9 expression was reduced ~3-fold in both ANT-IR and AIL6-amiRNA ant inflorescences (Fig. 7H).
Fig. 7.
Expression of growth-regulatory genes is altered after down-regulation of ANT expression in 35S:ALCR/AlcA:ANT-IR inflorescences and after down-regulation of AIL6 expression in 35S:ALCR/AlcA:AIL6-amiR ant inflorescences. (A) Mock- and ethanol (EtOH)-treated 35S:ALCR/AlcA:ANT-IR flowers. (B) Petal area, length, and width measurements for mock- and EtOH-treated Ler and 35S:ALCR/AlcA:ANT-IR flowers. Between 10 and 20 petals from different flowers were measured for each genotype and treatment. (C) Stamen anther of a mock-treated (left) and ethanol-treated (right) 35S:ALCR/AlcA:ANT-IR flower. (D) Expression of ANT is reduced in 35S:ALCR/AlcA:ANT-IR inflorescences after a 24 h EtOH treatment. (E) BB expression is up-regulated in 35S:ALCR/AlcA:ANT-IR and 35S:ALCR/AlcA:AIL6-amiR ant inflorescences after ethanol treatment. (F) ROT3 expression is down-regulated in 35S:ALCR/AlcA:ANT-IR and 35S:ALCR/AlcA:AIL6-amiR ant inflorescences after ethanol treatment. (G) AN3 expression is slightly down-regulated in 35S:ALCR/AlcA:ANT-IR inflorescences and more severely down-regulated in 35S:ALCR/AlcA:AIL6-amiR ant inflorescences. (H) XTH9 expression is down-regulated in 35S:ALCR/AlcA:ANT-IR and 35S:ALCR/AlcA:AIL6-amiR ant inflorescences after ethanol treatment. Error bars in all panels show the SD.
ANT and AIL6 directly regulate genes with roles in vascular development
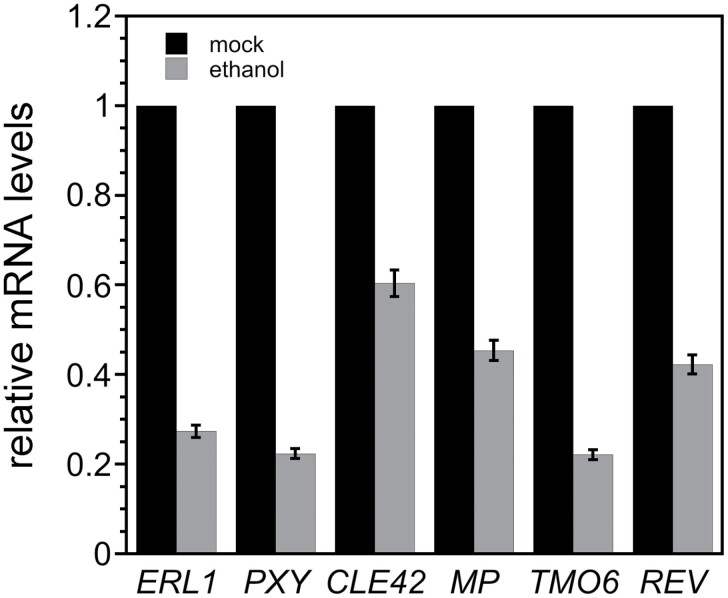
A number of genes that function in vascular development were present among the radial patterning genes bound by both ANT and AIL6 (Table 1; Supplementary Fig. S7). Because ANT and AIL6 are expressed in the procambium of young inflorescence tissue and flowers, it is possible that some of these genes are direct targets of ANT and AIL6 regulation (Elliott et al., 1996; Krizek, 2015). ant mutants in both Arabidopsis and maize show defects in leaf vascular pattern (Kang et al., 2007; Liu et al., 2020). In addition, ant ail6 double mutants show more severe reductions in leaf vein density and complexity than ant single mutants (Krizek, 2009). ERECTA-LIKE1 (ERL1), PHLOEM INTERCALATED WITH XYLEM/TDIF RECEPTOR (PXY/TDR), and CLAVATA3/ESR-RELATED 42 (CLE42) mRNA levels were reduced to 27, 22, and 60% of the mock-treated levels (Fig. 8). MP, TARGET OF MONOPTEROS (TMO6), and REVOLUTA (REV) mRNA levels were reduced to 45, 27, and 42% of the mock-treated levels (Fig. 8).
Fig. 8.
Vascular development genes are expressed at reduced levels after down-regulation of AIL6 expression in 35S:ALCR/AlcA:AIL6-amiR ant inflorescences. ERL1, PXY, CLE42, MP, TMO6, and REV mRNA levels are reduced after a 8 h ethanol treatment of 35S:ALCR/AlcA:AIL6-amiR inflorescences.
Discussion
Our data indicate that the partially overlapping roles of ANT and AIL6 in flower development are a consequence of these transcription factors regulating many of the same target genes. The more important role of ANT in floral organogenesis as compared with AIL6 appears to result from ANT regulating additional genes that are not targets of AIL6 regulation. In addition, at most of these genomic sites, ANT exhibits higher occupancy than AIL6, which may lead to a more significant effect on transcriptional regulation at these sites. Higher occupancy by ANT may be a consequence of greater amounts of ANT protein in these stage 3 flowers as compared with AIL6 rather than intrinsic differences in the DNA binding affinities of ANT and AIL6. ANT mRNA levels in wild-type inflorescences are ~8-fold higher than those of AIL6 (Han and Krizek, 2016). In addition, endogenous AIL6 protein may have competed with AIL6–VENUS for binding to genomic sites in the ChIP-Seq experiment. Previous work has shown that AIL6 can compensate for loss of ANT function when AIL6 is expressed under the control of the ANT promoter at levels similar to ANT mRNA levels (Han and Krizek, 2016).
GO analyses on genes associated with ANT and AIL6 ChIP-Seq peaks suggest that these transcription factors regulate a number of different processes during early stages of flower development. In particular, terms associated with floral meristem development (meristem initiation, maintenance of meristem identity, and floral meristem determinacy) and floral organ development (polarity specification, formation of plant organ boundary, radial pattern formation, and floral organ morphogenesis) were identified (Fig. 2). Many of the GO terms identified here match those identified in our earlier ANT ChIP-Seq experiment using stage 6/7 flowers (Krizek et al., 2020). However, several more terms associated with the initiation and patterning of floral organ primordia were identified here, including floral organ formation, formation of plant organ boundary, radial pattern formation, and floral organ morphogenesis (Fig. 2). This suggests additional roles for ANT and AIL6 in boundary specification and radial pattern formation during floral organogenesis.
We show that ANT and AIL6 directly activate the class B gene AP3 and the class C gene AG in stage 3 flowers. ANT and AIL6 bind to AP3 and AG regulatory regions (Fig. 5), expression of AP3 and AG is reduced soon after down-regulation of AIL6 activity in the ant mutant background (Fig. 6), and AP3 and AG are expressed in fewer cells in stage 3 flowers (Krizek, 2009). The regulation of AG expression by ANT appears to be complex as ANT acts with APETALA2 (AP2) to repress AG expression in second whorl cells, although it is not known if this regulation is direct (Krizek et al., 2000). Thus, ANT and AIL6 may directly activate AG expression in third and fourth whorl cells while repressing (directly or indirectly) AG expression in second whorl cells. How broadly expressed ANT and AIL6 activate AP3 specifically in second and third whorl cells and AG specifically in third and fourth whorl cells is not clear. It is possible that ANT and AIL6 act with region-specific cofactors as has been shown previously for LFY (Lee et al., 1997; Busch et al., 1999; Lenhard et al., 2001; Lohmann et al., 2001; Lamb et al., 2002; Liu et al., 2009). Another question still to be answered is what is the DNA sequence bound by ANT and AIL6 within the AP3 promoter. No obvious AIL/PLT-like-binding motif was identified within the ChIP-Seq binding peaks. Two overlapping sequences with weak similarity to the ANT-binding motif are present within the AG intron and bound by ANT in vitro (Supplementary Fig. S3). This weak similarity suggests that there may be flexibility in the DNA sequences recognized by ANT. These sequences are located near the LFY- and WUSCHEL (WUS)-binding sites within the AG intron (Busch et al., 1999; Lohmann et al., 2001). The ANT and AIL6 ChIP-Seq binding peaks within both AP3 and AG overlap with those of other floral regulatory genes (Supplementary Fig. S4). Thus, these genomic regions appear to contain binding sites for multiple transcription factors that may act cooperatively to regulate transcription. In particular, ANT and AIL6 may act in combination with other transcriptional regulators such as LFY, SEP3, AP1, and AP2 to control the spatial and temporal expression of AP3 and AG.
ANT and AIL6 contribute to other aspects of floral organogenesis including growth; this appears to involve the regulation of both growth-promoting and growth-repressing genes. ANT and AIL6 directly bind to a region near the TSS of the growth repressor BB (Supplementary Fig. S6A), and BB expression is increased in ant ail6 double mutant inflorescences (Fig. 7E) (Krizek et al., 2016). bb mutants produce larger flowers than the wild type, while overexpression of BB results in smaller flowers (Disch et al., 2006). BB is expressed throughout young stage 1–4 flowers, similar to ANT expression in young flowers and overlapping with AIL6 (Elliott et al., 1996; Nole-Wilson et al., 2005; Disch et al., 2006). Previous work has shown that transgenic bb plants containing a genomic copy of BB with a deletion in an upstream region corresponding to the location of the ANT and AIL6 ChIP-Seq binding peaks produced smaller flowers than the wild type (Breuninger and Lenhard, 2012). The reduced size of these flowers suggests that these transgenic plants have higher levels of BB expression than the wild type and that this region contains a negative cis-regulatory element bound by ANT and AIL6 that represses transcription of BB. BB is an E3 ubiquitin ligase that ubiquitinates and activates the protease DA1 and is subsequently cleaved by DA1 (Disch et al., 2006; Dong et al., 2017). DA1 limits the duration of the cell proliferation phase of organ growth by cleaving the deubiquitylase UBP15 and two TEOSINTE BRANCHED 1/CYCLOIDEA/PCF transcription factors, TCP15 and TCP22 (Dong et al., 2017). While BB protein levels are regulated by DA1, ANT and AIL6 are the first identified potential regulators of BB transcription.
ANT and AIL6 also bind to regions associated with three growth-promoting genes, ROT3, AN3, and XTH9 (Supplementary Fig. S6B–D). Previously we showed that ANT binds to the regulatory regions of AN3 and XTH9 in stage 6/7 flowers and that both of these genes are activated upon dex induction of ANT-GR activity (Krizek et al., 2020). Here we show that ANT and AIL6 bind to these genomic regions in stage 3 flowers. Furthermore, reduced expression of ROT3, AN3, and XTH9 upon down-regulation of AIL6 in the ant mutant background provides additional evidence that ANT and AIL6 directly regulate the expression of these genes (Fig. 7F–H). This suggests that ANT and AIL6 regulate a variety of growth-regulating pathways as ROT3 is a cytochrome P450 acting in brassinosteroid biosynthesis, AN3 is a transcriptional co-activator that works with GROWTH REGULATING FACTOR (GRF) transcription factors, and XTH enzymes modify xyloglucan in the cell wall (Kim et al., 1998; Rose et al., 2002; Kim and Kende, 2004).
Several of the radial patterning genes bound by ANT and AIL6 are associated with vascular development (Supplementary Fig. S7). ANT and AIL6 are expressed in procambial cells of the young inflorescence stem and developing flowers, although their function in these cells is not known (Elliott et al., 1996; Krizek, 2015). Transcriptional profiling of inflorescence stems detected the highest expression of AIL6 and ANT in the cambium, although there is expression of each gene in other vascular cell types (Shi, 2020). To further investigate the potential role of ANT and AIL6 in development of the procambium in young floral buds, we measured the expression of six genes with known roles in regulating cell division of procambial cells. ERL1, PXY, CLE42, MP, TMO6, and REV RNA levels were reduced in ethanol-treated 35S:ALCR/AlcA:AIL6-amiR ant inflorescences, suggesting that these genes might be directly regulated by ANT and AIL6 (Fig. 8). ERL1 and PXY are leucine rich-repeat receptor-like kinases (LRR-RLKs) that act in parallel signaling pathways to maintain procambial cell identity (Fisher and Turner, 2007; Uchida and Tasaka, 2013; Wang et al., 2019). EPIDERMAL PATTERNING FACTOR LIKE (EPFL) peptides perceived by ERL1 in phloem cells promote cell proliferation and/or inhibit differentiation of the adjacent cambial cells (Uchida and Tasaka, 2013). TRACHEARY ELEMENT DIFFERENTIATION INHIBITORY FACTOR (TDIF) peptides, including CLE42, bound by PXY receptors in the procambium promote division of the procambium, inhibit xylem differentiation, and organize vascular patterning (reviewed in Etchells et al., 2016)] In addition, ANT and AIL6 may contribute to the regulation of procambium proliferation through MP. One pathway mediated by MP is through the PEAR transcription factor TMO6 (Schlereth et al., 2010; Miyashima, 2019). TMO6 produced in phloem precursor cells moves into adjacent procambial cells to promote their division (Miyashima, 2019). PEAR activity is restricted to cambial cells on the phloem side by HD-ZIP transcription factors including REV (Miyashima, 2019). Thus, our data suggest that ANT and AIL6 regulate procambium identity and proliferation through multiple pathways.
In summary, our work provides new insights into the roles of ANT and AIL6 in the initiation and development of floral organs from the floral meristem. We have identified direct targets of ANT and AIL6 regulation that mediate their roles in the establishment of floral organ identity, promotion of floral organ growth, and development of the procambium. Additional work is needed to further elaborate the roles of ANT and AIL6 in these processes and to determine whether other genes identified here are direct targets of ANT and AIL6 that may mediate their roles in additional aspects of floral organogenesis.
Supplementary data
The following supplementary data are available at JXB online.
Table S1. Primers used in this study.
Fig. S1. DNA motifs with similarity to BBR/BPC- and bHLH-binding motifs are over-represented in ANT and AIL6 ChIP-Seq binding peaks.
Fig. S2. ANT and AIL6 bind directly to regulatory regions associated with the floral organ identity gene PI, but PI expression is not altered after down-regulation of AIL6 expression in 35S:ALCR/AlcA:AIL6-amiR ant inflorescences.
Fig. S3. ANT binds to the AG intron in vitro and can activate transcription through this binding site in yeast.
Fig. S4. ANT- and AIL6-binding peaks within AP3 and AG regulatory regions overlap those of other floral regulators.
Fig. S5. ANT- and AIL6-binding peaks within ANT, AIL6, and AIL7 regulatory regions overlap those of other floral regulators.
Fig. S6. ANT and AIL6 ChIP-Seq binding peaks in BB, ROT3, AN3, and XTH9 genomic regions.
Fig. S7. ANT and AIL6 ChIP-Seq binding peaks in ERL1, PXY, CLE42, MP, TMO6, and REV genomic regions.
Dataset S1. Genes associated with ANT or AIL6 stage 3 ChIP-Seq binding peaks
Dataset S2. GO terms over-represented in genes associated with ANT stage 3 ChIP-Seq binding peaks
Dataset S3. GO terms over-represented in genes associated with AIL6 stage 3 ChIP-Seq binding peaks
Dataset S4. AIL6 peaks that lack an overlapping ANT peak
Dataset S5. Genes associated with overlapping ANT and AIL6 stage 3 ChIP-Seq binding peaks
Dataset S6. Genes associated with overlapping ANT and AIL6 stage 3 ChIP-Seq binding peaks and differentially expressed in ant ail6 and 35S:ANT-GR inflorescences (2, 4, and 8 h)
Acknowledgements
We thank Detlef Weigel for the BJ36_AlcA and pMLBart_AlcR plasmids, and Kevin Higgins for the Distance program. This work was supported by the National Science Foundation (NSF) grant IOS 1354452. The Integrated Genome Browser software was supported by National Institutes of Health grants R01-GM103463 and R01-GM121927. Data hosting is provided by the SciDas project, funded by NSF award 1659300 to PI Frank (Alex) Feltus.
Author contributions
BAK and AEL: conceptualization and design; BAK, ATB, JMH, and HH: conducting the experiments; BAK, NHF and AEL: performing bioinformatic analyses; BAK: writing—original draft; BAK, ATB, JMH, HH, NHF, and AEL: writing—editing.
Data availability
ChIP-Seq sequences are available from the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) accession number PRJNA685489. Version-controlled source codes used to process and analyze data are available from https://bitbucket.org/krizeklab. Sequence alignments and coverage graphs are available for interactive visualization within the Integrated Genome Browser (Nicol et al., 2009). To view the data in IGB, readers may download and install the software from https://bioviz.org. Once installed, datasets from the study can be opened within IGB by selecting the latest Arabidopsis thaliana genome and then choosing the ChIP-Seq folder within the Available Data Sets section of the Data Access Panel.
References
- Breuninger H, Lenhard M. 2012. Expression of the central growth regulator BIG BROTHER is regulated by multiple cis-elements. BMC Plant Biology 12, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch MA, Bomblies K, Weigel D. 1999. Activation of a floral homeotic gene in Arabidopsis. Science 285, 585–587. [DOI] [PubMed] [Google Scholar]
- Disch S, Anastasiou E, Sharma VK, Laux T, Fletcher JC, Lenhard M. 2006. The E3 ubiquitin ligase BIG BROTHER controls arabidopsis organ size in a dosage-dependent manner. Current Biology 16, 272–279. [DOI] [PubMed] [Google Scholar]
- Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF. 2004. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Current Biology 14, 1935–1940. [DOI] [PubMed] [Google Scholar]
- Dong H, Dumenil J, Lu FH, et al. 2017. Ubiquitylation activates a peptidase that promotes cleavage and destabilization of its activating E3 ligases and diverse growth regulatory proteins to limit cell proliferation in Arabidopsis. Genes & Development 31, 197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQ, Gerentes D, Perez P, Smyth DR. 1996. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. The Plant Cell 8, 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchells JP, Smit ME, Gaudinier A, Williams CJ, Brady SM. 2016. A brief history of the TDIF–PXY signalling module: balancing meristem identity and differentiation during vascular development. New Phytologist 209, 474–484. [DOI] [PubMed] [Google Scholar]
- Fisher K, Turner S. 2007. PXY, a receptor-like kinase essential for maintaining polarity during plant vascular-tissue development. Current Biology 17, 1061–1066. [DOI] [PubMed] [Google Scholar]
- Freese NH, Norris DC, Loraine AE. 2016. Integrated genome browser: visual analytics platform for genomics. Bioinformatics 32, 2089–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K, Meyerowitz EM. 1994. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes & Development 8, 1548–1560. [DOI] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Krizek BA. 2016. AINTEGUMENTA-LIKE6 can functionally replace AINTEGUMENTA but alters Arabidopsis flower development when misexpressed at high levels. Plant Molecular Biology 92, 597–612. [DOI] [PubMed] [Google Scholar]
- Hecker A, Brand LH, Peter S, Simoncello N, Kilian J, Harter K, Gaudin V, Wanke D. 2015. The Arabidopsis GAGA-binding factor BASIC PENTACYSTEINE6 recruits the POLYCOMB-REPRESSIVE COMPLEX1 component LIKE HETEROCHROMATIN PROTEIN1 to GAGA DNA motifs. Plant Physiology 168, 1013–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack T, Brockman LL, Meyerowitz EM. 1992. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68, 683–697. [DOI] [PubMed] [Google Scholar]
- Jofuku KD, den Boer BG, Van Montagu M, Okamuro JK. 1994. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. The Plant Cell 6, 1211–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Mizukami Y, Wang H, Fowke L, Dengler NG. 2007. Modification of cell proliferation patterns alters leaf vein architecture in Arabidopsis thaliana. Planta 226, 1207–1218. [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Muiño JM, Jauregui R, Airoldi CA, Smaczniak C, Krajewski P, Angenent GC. 2009. Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biology 7, e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Wellmer F, Muiño JM, et al. 2010. Orchestration of floral initiation by APETALA1. Science 328, 85–89. [DOI] [PubMed] [Google Scholar]
- Kim GT, Tsukaya H, Uchimiya H. 1998. The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes & Development 12, 2381–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Kende H. 2004. A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proceedings of the National Academy of Sciences, USA 101, 13374–13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klucher KM, Chow H, Reiser L, Fischer RL. 1996. The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. The Plant Cell 8, 137–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek B. 2009. AINTEGUMENTA and AINTEGUMENTA-LIKE6 act redundantly to regulate Arabidopsis floral growth and patterning. Plant Physiology 150, 1916–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA. 1999. Ectopic expression of AINTEGUMENTA in Arabidopsis plants results in increased growth of floral organs. Developmental Genetics 25, 224–236. [DOI] [PubMed] [Google Scholar]
- Krizek BA. 2003. AINTEGUMENTA utilizes a mode of DNA recognition distinct from that used by proteins containing a single AP2 domain. Nucleic Acids Research 31, 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA. 2015. Intronic sequences are required for AINTEGUMENTA-LIKE6 expression in Arabidopsis flowers. BMC Research Notes 8, 556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA, Bequette CJ, Xu K, Blakley IC, Fu ZQ, Stratmann JW, Loraine AE. 2016. RNA-seq links the transcription factors AINTEGUMENTA and AINTEGUMENTA-LIKE6 to cell wall remodeling and plant defense pathways. Plant Physiology 171, 2069–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA, Blakley I, Ho Y-Y, Freese N, Loraine AE. 2020. The Arabidopsis transcription factor AINTEGUMENTA orchestrates patterning genes and auxin signaling in the establishment of floral growth and form. The Plant Journal 103, 752–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA, Eaddy M. 2012. AINTEGUMENTA-LIKE6 regulates cellular differentiation in flowers. Plant Molecular Biology 78, 199–209. [DOI] [PubMed] [Google Scholar]
- Krizek BA, Prost V, Macias A. 2000. AINTEGUMENTA promotes petal identity and acts as a negative regulator of AGAMOUS. The Plant Cell 12, 1357–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb RS, Hill TA, Tan QK-G, Irish VF. 2002. Regulation of APETALA3 floral homeotic gene expression by meristem identity genes. Development 129, 2079–2086. [DOI] [PubMed] [Google Scholar]
- Lee I, Wolfe DS, Nilsson O, Weigel D. 1997. A LEAFY co-regulator encoded by UNUSUAL FLORAL ORGANS. Current Biology 7, 95–104. [DOI] [PubMed] [Google Scholar]
- Lenhard M, Bohnert A, Jürgens G, Laux T. 2001. Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105, 805–814. [DOI] [PubMed] [Google Scholar]
- Liu C, Xi W, Shen L, Tan C, Yu H. 2009. Regulation of floral patterning by flowering time genes. Developmental Cell 16, 711–722. [DOI] [PubMed] [Google Scholar]
- Liu WY, Lin HH, Yu CP, et al. 2020. Maize ANT1 modulates vascular development, chloroplast development, photosynthesis, and plant growth. Proceedings of the National Academy of Sciences, USA 117, 21747–21756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann JU, Hong RL, Hobe M, Busch MA, Parcy F, Simon R, Weigel D. 2001. A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105, 793–803. [DOI] [PubMed] [Google Scholar]
- Machanick P, Bailey TL. 2011. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27, 1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF. 1992. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360, 273–277. [DOI] [PubMed] [Google Scholar]
- Miyashima S, Roszak P, Sevilem I, et al. 2019. Mobile PEAR transcription factors integrate positional cues to prime cambial growth. Nature 565, 490–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami Y, Fischer RL. 2000. Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proceedings of the National Academy of Sciences, USA 97, 942–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monfared MM, Simon MK, Meister RJ, Roig-Villanova I, Kooiker M, Colombo L, Fletcher JC, Gasser CS. 2011. Overlapping and antagonistic activities of BASIC PENTACYSTEINE genes affect a range of developmental processes in Arabidopsis. The Plant Journal 66, 1020–1031. [DOI] [PubMed] [Google Scholar]
- Nicol JW, Helt GA, Blanchard SG Jr, Raja A, Loraine AE. 2009. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25, 2730–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nole-Wilson S, Krizek BA. 2000. DNA binding properties of the Arabidopsis floral development protein AINTEGUMENTA. Nucleic Acids Research 28, 4076–4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nole-Wilson S, Tranby TL, Krizek BA. 2005. AINTEGUMENTA-like (AIL) genes are expressed in young tissues and may specify meristematic or division-competent states. Plant Molecular Biology 57, 613–628. [DOI] [PubMed] [Google Scholar]
- O’Malley RC, Huang SC, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. 2016. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165, 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Maoiléidigh DS, Thomson B, Raganelli A, Wuest SE, Ryan PT, Kwasniewska K, Carles CC, Graciet E, Wellmer F. 2015. Gene network analysis of Arabidopsis thaliana flower development through dynamic gene perturbations. The Plant Journal 83, 344–358. [DOI] [PubMed] [Google Scholar]
- Ó’Maoiléidigh DS, Wuest SE, Rae L, et al. 2013. Control of reproductive floral organ identity specification in Arabidopsis by the C function regulator AGAMOUS. The Plant Cell 25, 2482–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF. 2000. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203. [DOI] [PubMed] [Google Scholar]
- Rose JKC, Braam J, Fry SC, Nishitani K. 2002. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspective and a new unifying nomenclature. Plant & Cell Physiology 43, 1421–1435. [DOI] [PubMed] [Google Scholar]
- Schlereth A, Möller B, Liu W, Kientz M, Flipse J, Rademacher EH, Schmid M, Jürgens G, Weijers D. 2010. MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464, 913–916. [DOI] [PubMed] [Google Scholar]
- Shi D, Jouannet V, Agusti J, Kaul V, Levitsky V, Sanchez P, Mironova VV, Greb T. 2020. Tissue-specific transcriptome profiling of the Arabidopsis inflorescence stem reveals local cellular signatures. The Plant Cell 33, 200–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonini S, Roig-Villanova I, Gregis V, Colombo B, Colombo L, Kater MM. 2012. Basic pentacysteine proteins mediate MADS domain complex binding to the DNA for tissue-specific expression of target genes in Arabidopsis. The Plant Cell 24, 4163–4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trost G, Vi SL, Czesnick H, Lange P, Holton N, Giavalisco P, Zipfel C, Kappel C, Lenhard M. 2014. Arabidopsis poly(A) polymerase PAPS1 limits founder-cell recruitment to organ primordia and suppresses the salicylic acid-independent immune response downstream of EDS1/PAD4. The Plant Journal 77, 688–699. [DOI] [PubMed] [Google Scholar]
- Uchida N, Tasaka M. 2013. Regulation of plant vascular stem cells by endodermis-derived EPFL-family peptide hormones and phloem-expressed ERECTA-family receptor kinases. Journal of Experimental Botany 64, 5335–5343. [DOI] [PubMed] [Google Scholar]
- Wang N, Bagdassarian KS, Doherty RE, Kroon JT, Conner KA, Wang XY, Wang W, Jermyn IH, Turner SR, Etchells JP. 2019. Organ-specific genetic interactions between paralogues of the PXY and ER receptor kinases enforce radial patterning in Arabidopsis vascular tissue. Development 146, dev177105. [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. 1992. LEAFY controls floral meristem identity in Arabidopsis. Cell 69, 843–859. [DOI] [PubMed] [Google Scholar]
- Weigel D, Meyerowitz EM. 1993. Activation of floral homeotic genes in Arabidopsis. Science 261, 1723–1726. [DOI] [PubMed] [Google Scholar]
- Wellmer F, Graciet E, Riechmann JL. 2014. Specification of floral organs in Arabidopsis. Journal of Experimental Botany 65, 1–9. [DOI] [PubMed] [Google Scholar]
- Winter CM, Austin RS, Blanvillain-Baufumé S, et al. 2011. LEAFY target genes reveal floral regulatory logic, cis motifs, and a link to biotic stimulus response. Developmental Cell 20, 430–443. [DOI] [PubMed] [Google Scholar]
- Wuest SE, O’Maoiléidigh DS, Rae L, Kwasniewska K, Raganelli A, Hanczaryk K, Lohan AJ, Loftus B, Graciet E, Wellmer F. 2012. Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proceedings of the National Academy of Sciences, USA 109, 13452–13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Jin R, Yu X, et al. 2017. Cis and trans determinants of epigenetic silencing by Polycomb repressive complex 2 in Arabidopsis. Nature Genetics 49, 1546–1552. [DOI] [PubMed] [Google Scholar]
- Yamaguchi N, Jeong CW, Nole-Wilson S, Krizek BA, Wagner D. 2016. AINTEGUMENTA and AINTEGUMENTA-LIKE6/PLETHORA3 induce LEAFY expression in response to auxin to promote the onset of flower formation in Arabidopsis. Plant Physiology 170, 283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, Wu MF, Winter CM, Berns MC, Nole-Wilson S, Yamaguchi A, Coupland G, Krizek BA, Wagner D. 2013. A molecular framework for auxin-mediated initiation of flower primordia. Developmental Cell 24, 271–282. [DOI] [PubMed] [Google Scholar]
- Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM. 1990. The protein encoded by the Arabidopsis homeotic gene AGAMOUS resembles transcription factors. Nature 346, 35–39. [DOI] [PubMed] [Google Scholar]
- Yant L, Mathieu J, Dinh TT, Ott F, Lanz C, Wollmann H, Chen X, Schmid M. 2010. Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. The Plant Cell 22, 2156–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu LJ, Gazin C, Lawson ND, Pagès H, Lin SM, Lapointe DS, Green MR. 2010. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 11, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-Seq sequences are available from the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) accession number PRJNA685489. Version-controlled source codes used to process and analyze data are available from https://bitbucket.org/krizeklab. Sequence alignments and coverage graphs are available for interactive visualization within the Integrated Genome Browser (Nicol et al., 2009). To view the data in IGB, readers may download and install the software from https://bioviz.org. Once installed, datasets from the study can be opened within IGB by selecting the latest Arabidopsis thaliana genome and then choosing the ChIP-Seq folder within the Available Data Sets section of the Data Access Panel.