Abstract
The discovery of clustered regularly interspaced short palindromic repeat (CRISPR)/ CRISPR-associated (Cas) genome editing systems and their applications in human health and medicine has heralded a new era of biotechnology. However, the delivery of CRISPR therapeutics is arguably the most difficult barrier to overcome for translation to in vivo clinical administration. Appropriate delivery methods are required to efficiently and selectively transport all gene editing components to specific target cells and tissues of interest, while minimizing off-target effects. To overcome this challenge, we discuss and critic nanoparticle delivery strategies, focusing on the use of lipid-based and polymeric-based matrices herein.
Graphical Abstract
An Introduction to CRISPR-based Systems
The clustered regularly interspaced short palindromic repeat (CRISPR) locus was originally discovered as part of the adaptive immune system in archaea and bacteria, consisting of spacer sequences that corresponded to extrachromosomal DNA derived from phages and plasmids. These spacer sequences essentially confer resistance against those particular microbes from which they are derived from, serving as genomic memory which can identify and disable invading DNA.[1-3] While there are two classes of CRISPR/ CRISPR-associated (Cas) systems (as well as several subtypes), the class 2 type II system, otherwise known as CRISPR/Cas9, is the most widely studied.
The CRISPR-associated Cas9 protein is an endonuclease that relies on a guide sequence within the tracrRNA (trans-activating CRISPR RNA):crRNA (CRISPR RNA) duplex to base pair with target DNA sequences and induce site-specific double-stranded breaks in the genome. The hybrid tracrRNA:crRNA complex was engineered as a single guide RNA (sgRNA) in which a simple change in the 20-nucleotide guide sequence can program Cas9 to target any DNA sequence of interest.
Target DNA recognition requires: (1) site-specific complementarity between the variable 20 nucleotide sgRNA sequence and target DNA sequence, and (2) the presence of the protospacer adjacent motif (PAM) nucleotide sequence 5’-NGG-3’ (usually found 3-4 nucleotides downstream of the cut site).[4] Then, one of two endogenous DNA repair pathways are used to restore the double-stranded breaks in the genome: the error-prone non-homologous end joining (NHEJ) or the template-driven homology-directed repair (HDR) pathways.
In most organisms, the NHEJ pathway is the predominant repair pathway with the goal of simply restoring the integrity of the DNA by non-specifically rejoining the broken ends, inducing frameshift mutations that may lead to the successful knockout of a gene of interest. Alternatively, the HDR pathway relies on exogenous DNA repair templates to enable the insertion of precise genetic modifications by homologous recombination.[5]
CRISPR-based Technologies for Genome Editing
Previous genome-engineering technologies, such as, zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), require meticulous constructing and testing of newly designed proteins for each target DNA sequence, with design challenges surrounding delivery due to the large size and repetitive nature of TALENs or the substantial protein engineering required to construct ZFNs.[6,7] The main advantage of CRISPR technologies, as opposed to either ZFNs or TALENs, is the ease of which the ∼80 nucleotide sgRNAs can be synthesized and modified to direct Cas9 to different target sequences, significantly increasing throughput. CRISPR/Cas systems have shown great promise in identifying essential genes crucial to regulating various biological processes as well as drug targets, modeling disease, and developing novel therapies.[7-9]
In addition to inducing permanent genetic modifications, CRISPR systems can induce non-permanent modifications in the DNA by way of CRISPR interference (CRISPRi) or alternatively, CRISPR activation (CRISPRa). CRISPRi/a rely on a catalytically inactive Cas9 (dCas9) that lacks endonucleolytic activity. CRISPRi/a represses or activates transcription by either directly disrupting or enhancing RNA polymerase binding.[10] Alternatively, fusion of dCas9 to unique transcriptional effector domains can recruit specific proteins to the DNA, and in turn, modulate transcriptional regulation. One such study found that dCas9-KRAB fusion proteins efficiently regulate transcriptional repression by potentially recruiting chromatin-modifying complexes to the DNA site.[11]
Several studies have established the efficacy of CRISPR-based therapies both in vitro and in vivo in model organisms (Table I). Of particular interest are genetic disorders resulting from a loss-of-function mutation in a single gene (monogenic disorders). For example, several groups have focused on permanent and efficient correction of mutations in the endogenous Duchenne muscular dystrophy (DMD) gene implicated in DMD.[12] Other studies have focused on Beta-Thalassemia, Fanconi anemia, cystic fibrosis, lung disease, and even cholesterol metabolism, providing further evidence that CRISPR/Cas9 gene editing can be employed to correct diseased alleles.[13-16]
Table I.
Applications of CRISPR/Cas9 gene editing
| Disease | Model | Mutated Gene(s) |
Editing Strategy | Delivery Strategy | Reference |
|---|---|---|---|---|---|
| Duchenne Muscular Dystrophy | In vivo* | Dmd exon 44 | sgRNA recognition of a PAM sequence in exon 45 and generation of insertions and deletions 7 base pairs downstream of the 5′-AG-3′ splice acceptor site | Intraperitoneal injection of single stranded AAV - packaged SpCas9 nuclease with double-stranded AAV - packaged sgRNA | [12] |
| Beta-Thalassemia/Sickle Cell Anemia | Ex vivo/In vivo** | Hemoglobin β subunit | Genetic editing of autologous CD34+ hematopoietic stem and progenitor cells at the erythroid-specific enhancer region of BCL11A | Electroporation of CD34+ hematopoietic stem and progenitor cells obtained from healthy donors, with CRISPR/Cas9 targeting the BCL11A erythroid-specific enhancer | [16] |
| Fanconi Anemia (FA) | In vitro/In vivo* | FANCA, FANCB, FANCC, FANCD2 | Editing of human FA hematopoietic cells harboring mutations in genes encoding for FA proteins involved in different steps of the FA pathway | Electroporation of a ribonucleoprotein complex composed of the Cas9 protein and either a chemically modified (sgRNA4 containing, 2′-O-methyl 3′phosphorothioate (MS-sgRNA4)) or an in vitro-transcribed sgRNA4 (IVT-RNP4) | [15] |
| Cystic Fibrosis | In vitro | CFTR | Correction of the CFTR locus by homologous recombination (a donor plasmid encoding wild-type CFTR sequences) in cultured intestinal stem cells of CF patients | Lipofectamine-mediated transfection in patient organoids with a plasmid expressing Cas9 and sgRNA | [24] |
| Children’s Interstitial Lung Disease | In vivo* | Sftpc | CRISPR/Cas9–induced excision of the mutant SftpcI73T gene can rescue the lung from toxic accumulation of the disease-associated protein | Intra-amniotic injection of Ad vectors containing SpyCas9 and an sgRNA into the amniotic cavity of gestational fetuses | [14] |
| Cholesterol Metabolism | In vivo* | PCSK9 | Introduction of loss-of-function mutations into the endogenous PCSK9 gene | Adenovirus to express Cas9 and a CRISPR guide RNA targeting Pcsk9in mouse liver, where the gene is specifically expressed | [13] |
| HIV | In vitro | N/A | Simultaneous genome editing of CXCR4 and CCR5 by CRISPR/Cas9 to block HIV-1 infection in primary CD4+ T cells | Lenti-sgR5-Cas9 vector, containing the gRNA targeting CCR5 region, was inserted by the different CXCR4 targeting sgRNAs | [20] |
| Cancer | In vitro/In vivo* | PLK-1 | Therapeutic genome editing of PLK1 to invoke cell cycle arrest and cell death in dividing cells | Intraperitoneal injection of lipid nanoparticles with an antibody to an overexpressed receptor on ovarian cancer cells (EGFR) to cause their selective uptake into ovarian tumors | [25] |
| Type 2 Diabetes Mellitus | In vivo* | N/A | Downregulation of dipeptidyl peptidase-4 (DPP-4) to prevent degradation of glucagon-like peptide-1 (GLP-1) | Delivered ribonucleoprotein designed to edit the DPP-4 gene delivered in a lecithin-based liposomal nanocarrier | [26] |
| Chronic Myeloid Leukemia (CML) | In vitro/In vivo* | Chimeric BCR-ABL | Disruption of the fusion region of the BCR-ABL gene | Delivered cationic lipid-assisted nanoparticles with a plasmid encoding SpCas9 and guide RNA against BCR-ABL fusion gene | [27] |
Mouse Model
Clinical Trial
PAM – Protospacer Adjacent Motif; sgRNA – single guide RNA; AAV – Adeno Associated Virus
The potential of CRISPR gene editing spans past monogenic disorders. The multiplexed capability of CRISPR/Cas systems allows for the robust knockdown and subsequent analysis of several genes that may be genetically linked with a single guide RNA.[17-19] Furthermore, CRISPR systems can also be used to introduce protective mutations into the genome to prevent or overcome diseases that do not have an underlying genetic cause. For instance, simultaneous genome editing of the CXCR4 and CCR5 receptors via CRISPR/Cas9 editing may be used to block HIV-1 infection in primary CD4 T+ cells. The modified cells showed a selective advantage over those that were unmodified in induced HIV-1 infection assays.[20] Ex vivo gene editing provides a flexible alternative to in vivo gene editing, allowing for the analysis and characterization of edited cells before re-introduction into patients. While ex vivo gene editing strategies require more steps (i.e., cell collection, isolation, expansion, editing, characterization, selection, and transplantation), they avoid the delivery and unintended off-target gene editing challenges that often accompany in vivo approaches.[21]
Challenges Associated with CRISPR-based Technologies
While CRISPR/Cas9 systems show great therapeutic potential in the treatment of human diseases, several challenges must be addressed before CRISPR therapeutics can successfully be administered in the clinic.[22] One key challenge relates to the rational engineering of the CRISPR system to minimize unwanted off-target mutations. High fidelity Cas9 variants and optimized sgRNA libraries have been developed to minimize off-target effects. For example, instead of inducing the native Cas9 protein to initiate double stranded breaks in the DNA, nickase versions of Cas9 directed to paired sites may offer improved specificity. One group developed an original two-step approach named ‘verification of in vivo targets’ (VIVO) to predict the sites at which the gRNA will cause off-target effects, providing a general blueprint for (1) defining, and (2) quantifying the off-target effects of gene-editing nucleases in whole organisms.[11]
Other factors that affect the efficacy of the CRISPR/Cas9 gene editing system include optimizing target DNA site selection and sgRNA design, overcoming epigenetic modifications, such as histone acetylation and DNA methylation, to access previously inaccessible regions of DNA (which may contain the gene of interest), and developing strategies to improve the efficacy of homology-directed repair present barriers to the implementation of CRISPR systems.[23]
Current Approaches for the Delivery of CRISPR/Cas9
In present day, the design of appropriate gene delivery platforms are dependent on the selection of two key factors: cargo and delivery vectors. The CRISPR/Cas9 system consists of the endonuclease Cas9, as well as a target-specific guide RNA (gRNA). Currently, three major methods exist to transport the Cas9 protein and gRNA including: (1) a DNA plasmid encoding both Cas9 and the sgRNA, (2) Cas9 mRNA and sgRNA, and (3) the Cas9 protein itself and sgRNA.[7] Regardless of which strategy is utilized, it is important that the delivery vehicle is designed to accommodate the physical properties of the CRISPR/Cas9 cargo without compromising other desired properties of the delivery vehicle itself, such as targeting or transfection efficiency.
Viral Delivery Approaches
Viral vector systems are one of the most extensively characterized methods for the delivery of nucleic acids.[28-32] The five main classes of viral vectors can be broadly grouped into two different categories depending on their ability to integrate within the host genome (such as is the cases of oncoretrovirsuses and lentiviruses) or persist in the nucleus as extrachromosomal episomes (which is the case for adeno-associated viruses (AAVs), adenoviruses, and the herpes virus).[33] Gene therapy via viral vectors is particularly attractive because of their intrinsic ability of to enter and deliver genetic material to cells.
In particular, lentiviral vectors have become increasingly popular in clinical applications because of their ability to accommodate large DNA payloads and sustain robust expression in dividing and non-dividing cells alike.[34] Lentiviral vectors also have the unique ability to translocate across the nuclear pore of an intact nuclear membrane. However, their ability to constitutively express Cas9 and sgRNA may lead to undesirable off-target effects, resulting from non-specific RNA-DNA interactions and off-target DNA cleavages. Moreover, for CRISPR/Cas9 delivery, the integration capacity of retroviral vectors could lead to unwanted off-target insertional mutagenesis.[35] Furthermore, the safety of retroviral vectors for clinical applications remains open: there is potential for retroviral recombination events during large-scale vector manufacturing, resulting in replication-competent viruses.[36]
AAV vectors possess unique features that are attractive for clinical applications including low immunogenicity, lack of integration machinery, and ease of production. However, concerns over the limited packaging capacity, low transduction efficiency, and ability to induce an immune response further, complicates the design of these systems.[37] The discovery of a smaller Cas9 variant derived from Staphylococcus aureus (SaCas9), with editing efficiencies comparable to those of Streptococcus pyogenes Cas9 (SpCas9), has led to the development SaCas9/sgRNA systems that can be packaged in AAV vectors.[38] Another alternative involves using adenoviral vectors, which are nonenveloped, double-stranded DNA vectors that have a larger packaging capacity of approximately 35 kb compared to that of AAV vectors.[39] However, at high dosages, adenoviral vectors are highly immunogenic and can invoke the host innate immune response, producing an inflammatory cytokine response and initiating the differentiation of dendritic cells into antigen-presenting cells.[40]
Physical Delivery Approaches
Additionally, several physical approaches have been repurposed to deliver the Cas9/sgRNA system including microinjection, electroporation, hydrodynamic injection, and laser irradiation;[41] however, physical delivery methods are largely restricted to both in vitro and ex vivo systems.[42] Physical gene delivery methods avoid the immunogenic complications inherent to viral vectors by directly delivering cargo molecules to the cytoplasm or nucleus and bypassing complications that are associated with targeting, internalization via endocytotic pathways, and immunogenicity.[43]
Two broad categories pertaining to physical delivery exist, namely, methods that directly insert cargo into the cytoplasm or nucleus, and methods that use a physical field to disrupt the cell membrane. While microinjection is conceptually the most direct delivery method enabling delivery of cargo into the host cell nucleus, the slow rate of delivery has limited the applicability of microinjection techniques to low-thruput processes such as, in vitro fertilization and production of transgenic animals. Although automated systems have increased thruput significantly, thruput still lags compared to other physical techniques.[44]
Electroporation is a particularly useful technique in cells that are difficult to transfect by other methods. This method relies on the transient increase in cell membrane permeability upon exposure to an electric field pulse. The successful electro-transfer of nucleic acids into cells depends on: (1) how the cell membrane is permeabilized and (2) DNA-cell membrane interactions and internalization.[45] However, at high electric field strengths, electroporation often leads to high post-transfection mortality. The development of novel electroporation devices that result in more even voltage distributions and homogenous electric field distributions has demonstrated high transfection efficiencies and low cytotoxicity in a variety of cell types.[46]
Non-viral Delivery Approaches
The development of non-viral delivery vectors to deliver CRISPR cargo in vivo, particularly the development of nanoparticles (NPs), has the potential to address several of the aforementioned limitations of the use of viral vectors and physical delivery methods. Specifically, non-viral vectors are compatible with the simultaneous delivery of multiple guide RNAs, can be easily functionalized with targeting ligands to enhance the delivery of CRISPR cargo to specific tissues and cells, and can be designed to accommodate large payloads.
NPs employ endocytic pathways to achieve intracellular localization, most notably via receptor dependent clathrin-mediated or caveolin-mediated endocytosis. Additionally, clathrin- and caveolae-independent endocytosis occurs in cells that lack clathrin and caveolin. NPs can also be internalized nonspecifically via pinocytosis or even phagocytosis.[47] The physicochemical properties of the NPs such as size, shape, surface charge, and polarity directly affect the transfection efficiency, cellular uptake mechanism, and cytotoxicity of the NPs.[48] Upon cellular internalization, the NPs are located within endosomes (pH 6.8-6.5), which later develop into late endosomes (pH 6.2-5.2). Late endosomes fuse with lysosomes (pH 5.2-4.5) to destroy the engulfed material, aided by hydrolytic enzymes and an acidic environment. It is essential that the NPs escape from the endosomes prior to lysosomal digestion. In fact, several cationic nanocarriers, such as those that incorporate polyamidoamine (PAMAM), polyethyleneimine (PEI), or chitosan, have been proposed to induce endosomal escape using a proton sponge mechanism.[49] This mechanism relies on the protonation of the cationic moiety resulting from the acidic endosomal environment, which leads to the methodical influx of ions, swelling, and eventual rupture of the endosome.[50,51]
Nanoparticle Delivery Systems
Important Properties of Carrier Systems
In order to achieve effective delivery of CRISPR/Cas components, drug delivery systems must be stable in circulation and designed with ideal characteristics, such as choice of drug delivery carrier, size, shape, and surface properties (Figure 1) being the key determinants that impact factors such as the cellular uptake, in vivo toxicity, immunogenicity, and overall efficacy of the drug delivery carriers.[52] Once in the bloodstream, conventional nanoparticles with no surface modifications can be opsonized and rapidly cleared by phagocytic cells through the reticuloendothelial system (RES). This clearance system hinders active targeting because of its ability to easily recognize nanoparticle systems and remove them from systemic circulation so particles are not delivered to the targeted cells. Functionalizing the surface of nanoparticles with a hydrophilic chain of linear polymers, such as poly(ethylene glycol) (PEG), results in a hydrating layer that hinders protein absorption and clearance of nanoparticle systems.[53]
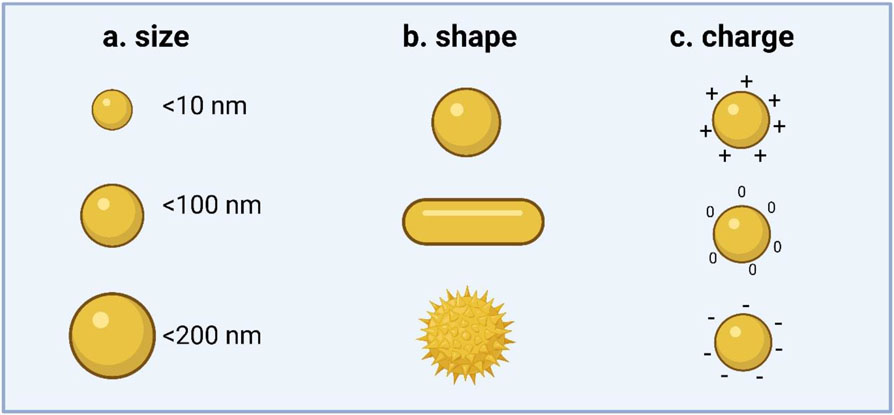
Figure 1. Physicochemical properties of carrier systems.
Size, shape, and charge of nanoparticle systems are the key determinants which impact cellular uptake, cell toxicity, immunogenicity, and the efficacy of the delivered cargo. a.) Carrier size plays a key role in their diffusion profile, adhesion properties, and velocity distribution, all which contribute to the uptake efficiency of particles. b.) Carrier shape effects transport, degradation, internalization, clearance, and even targeting. Most nanoparticles that are designed for intravenous delivery exhibit spherical geometry 10-100 nm in diameter. Particles > 200 nm accumulate in the liver and spleen, and are subsequently cleared by reticuloendothelial system. c.) The surface charge of delivery systems also influences circulation stability and uptake mechanisms by having effects on the molecular interactions between carriers and their target cells, as well as subsequent downstream intracellular events.
Carrier size (Figure 1a) plays a key role in determining the vascular flow dynamics of the carrier in vivo, including their diffusion profile, adhesion properties, and velocity distribution, all which contribute to the uptake efficiency of particles.[54,55] Carrier shape (Figure 1b) also has important consequences in biological applications, with effects on vascular transport, degradation, internalization, clearance, and even targeting.[56,57] Most nanoparticles that are designed for intravenous delivery exhibit spherical geometry and are between 10 and 100 nm in diameter. Particles larger than 200 nm accumulate in the liver and spleen, and are subsequently cleared by RES. Recent research efforts have focused on developing nanoparticles with non-spherical geometries to increase interactions with vessel walls and extravasation sites to develop innovative means to amass nanoparticles at site-specific tissues.
Moreover, the surface properties of delivery systems can also determine stability in circulation and uptake mechanisms. For example, the surface charge (Figure 1c) of carrier systems, namely in non-viral delivery systems, has critical effects on the molecular interactions between carriers and their target cells, as well as subsequent downstream intracellular events. Mukherjee et al., investigated the role of nanoparticle surface charge (cationic, anionic, neutral, and zwitterionic) on the cellular membrane potential and uptake of gold nanoparticles.[58] They found that the internalization of positively charged gold nanoparticles depolarized the membrane potential in a concentration-dependent manner by rapidly increasing the intracellular [Ca2+] concentration by stimulating both plasma membrane [Ca2+] influx and [Ca2+] release from the endoplasmic reticulum. While cationic NPs can disrupt the lipid bilayer and thereby enhance cellular uptake by modulating cell-membrane potential, increases in [Ca2+] can decrease cell proliferation and result in apoptosis. Therefore, these seemingly opposing mechanisms must be reconciled in the design of delivery systems.
Lipid-based Delivery Systems
Lipid-based delivery systems are composed of physiological lipids. Liposomes, which are spherical vesicles enclosed by a lipid bilayer membrane (Figure 2), are the traditional paradigm of lipid-based formulations, capable of encapsulating both hydrophilic and hydrophobic molecules. Despite liposomes being entirely biocompatible and biodegradable, many of their translational applications are limited due to short shelf life, poor stability, low encapsulation efficiency, rapid clearance by the reticuloendothelial system (RES), and intermembrane transfer.[59]
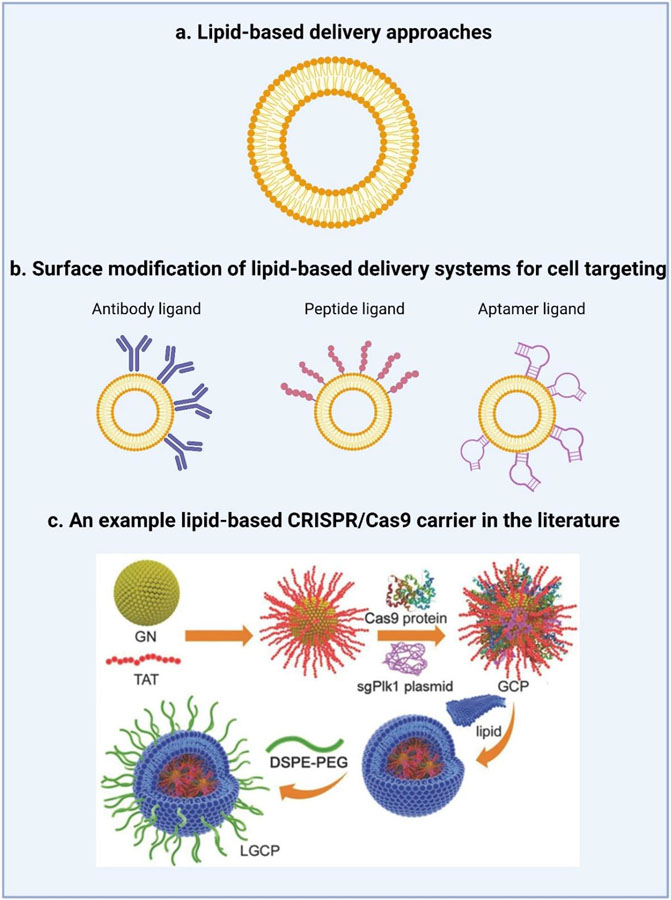
Figure 2. Lipid-based strategies for the delivery of CRISPR/Cas9.
a.) Liposomes are the traditional lipid-based formulations capable of encapsulating both hydrophilic and hydrophobic molecules. b.) To promote cell targeting, the lipid surface can be modified with a ligand, such as an antibody, peptide, or aptamer to interact with the receptors of target cells. c.) From the literature, a schematic diagram of the synthesis of lipid-coated gold nanoclusters (LGCP) adapted from Wang et al.[67] Specifically, the HIV-1-transactivator of transcription (TAT) peptide was used to modify the surface of cationic gold nanoclusters (GNs). The positively charged TAT-GNs and the negatively charged Cas9 proteins and sgRNA plasmids were mixed to form a ternary complex (GCP). GCP was further encapsulated in an anionic lipid shell (1,2-dioleoyl-3-trimethylammoniumpropane (DOTAP)/dioleoyl-phosphatidylethanol-amine (DOPE)/cholesterol, followed by post-modification with polyethylene glycol-phospholipids (DSPE-PEG).[67]
Solid lipid nanoparticles (SLNs) and nanostructured lipid carriers (NLCs) are two major classes of lipid-based nanoparticles that have been developed to overcome some of the aforementioned limitations of liposomes, boasting advantages such as a sustained release profile, targeted drug delivery, and superior physical stability. SLNs are composed of a solid lipid core, surrounded by a layer of surfactants. SLNs are generally smaller than 500 nm in diameter, with diameters between 80 and 120 nm ideal for in vivo delivery.[60]
Nucleic acids can form strong electrostatic attractions to cationic SLNs, facilitating the transport of genetic material and protecting them from enzymatic degradation. The features of the vector are ultimately dictated by the ratio between the particle and nucleic acid, optimized to maximize the condensation capacity of the vehicle and later release of its contents at the appropriate site.[61] An alternative to SLNs are NLCs, with improved drug loading capacities and release properties, composed of a mixture of solid and liquid lipids. NLCs are unique in their ability to co-deliver nucleic acids as well as lipophilic drugs.[62] Some of the commonly used methods for the synthesis of LNPs include high pressure homogenization (solvent emulsification, evaporation, or diffusion), supercritical fluid extraction of emulsions, ultrasonication, and spray drying.[59]
Over the years, several commercial, non-viral transfection reagents have been developed for the transport of nucleic acids. The most commonly used commercially available lipid nanoparticle systems are Lipofectamine-based reagents. Lipofectamine is a cationic liposome formulation that can be complexed with negatively charged nucleic acid molecules via electrostatic interactions. One group conducted a comparative analysis of 5 common transfection reagents including Lipofectamine 3000, Lipofectamine 2000, Fugene, RNAiMAX, and Lipofectin against 10 different cell lines. They found that although transfection efficiency and transfection reagent toxicity were dependent on cell type, generally, the toxicity of the reagents displayed a positive correlation with their transfection efficacy. They found that Lipofectamine 3000, Fugene, and RNAiMAX had the highest transfection efficiencies, and that the transfection efficiency of Lipofectamine 2000 was compromised due to its high toxicity.[63]
As an alternative to using commercially available lipid-based transfection reagents, a wide range of lipid-based nanocarriers have been synthesized in the laboratory and extensively tested for their ability to be used as gene delivery systems (Table II). Traditional, non-viral, cationic carriers have been widely used for the delivery of nucleic acid cargo by taking advantage of electrostatic interactions between the cargo and vehicle. Zhang and colleagues constructed polyethylene glycol phospholipid-modified cationic lipid nanoparticles (PLNP).[64] The principle of this strategy took advantage of the electrostatic interactions between a negatively charged Cas9/sgRNA-fused plasmid DNA/Chondroitin sulfate complex and the positively charged protamine solution at an optimized ratio, resulting in a tightly packed core. The resulting core is encapsulated with cationic lipids composed of DOTAP, DOPE, and cholesterol, post-modified with polyethylene glycol phospholipid (DSPE-PEG) to yield nanoparticles with increased stability, low toxicity, and decreased clearance and immunogenicity.
Table II.
Nanoparticle systems used for the delivery of CRISPR/Cas9
| Nanoparticle Formulation |
Material Details | Cargo | In Vitro Results | In Vivo Results | Reference |
|---|---|---|---|---|---|
| Biodegradable Lipid Nanoparticle | The particles contain: a biodegradable, ionizable lipid—termed “LP01” with an approximate pKa of 6.1— and PEG-DMG | Cas9 mRNA/chemically modified gRNA | N/A | >97% knockdown of the mouse TTR protein (transthyretin (Ttr) gene, the homolog of a target for therapeutic gene editing to treat amyloidosis in humans) | [66] |
| Cationic Lipid Nanoparticle | The particles consist of a negative core of protamine/Cas9-sgPLK-1 plasmid DNA and a cationic lipid shell. The complex is postmodified with DSPE-PEG | CRISPR/Cas9 plasmid | 47.4% successful transfection of Cas9/sgPLK-1 plasmids in A375 cells. | Intratumor injection of Cas9/sgPLK-1 plasmids into melanoma tumor-bearing mice resulted in downregulation of Polo-like kinase 1 (PLK-1) protein and suppression of tumor growth (>67%) | [64] |
| RNA Aptamer Liposome | Chemically synthesized stearyl-polyethylenimine (stPEI)/ CRISPR/Cas9 plasmid /HAS (human serum albumin) nanoparticles were made by double emulsion | CRISPR/Cas9 plasmid | Codelivery of CRISPR/Cas9 and siRNA showed a high-transfection efficiency with a synergistic effect on inhibition of PD-L1 expression by 21.95% compared to about 13% for plasmid-loaded Lipofectamine in the CT26 cell line. | N/A | [67] |
| Bioreducible Lipid Nanoparticle | BAMEA-O16B, a lipid nanoparticle integrated with disulfide bonds for delivery of Cas9 mRNA and sgRNA | Cas9 mRNA/sgRNA | Simultaneous delivery of Cas9 mRNA and sgRNA using BAMEA-O16B knocks out green fluorescent protein expression in human embryonic kidney cells with efficiency up to 90% | Intravenous injection of BAMEA-O16B/Cas9 mRNA/sgRNA nanoparticles effectively accumulate in hepatocytes and knocks down PCSK9 levels in mouse serum down to 20% | [68] |
| Lipid/Gold Nanocluster (GNs ) | Positively charged TAT-GNs were mixed with negatively charged Cas9 proteins and sgRNA, yielding a ternary complex (TAT-GNs/Cas9 protein/sgRNA plasmid) formed via electrostatic interactions. The complex is encapsulated in an anionic lipid shell, and post-modified with DSPE-PEG | Cas9 protein/sgRNA plasmid | Led to >70% downregulation of Plk1 protein expression in A375 cells. | Intratumoral injection suppressed melanoma progression by 75% in Balb/c nude mice (melanoma model mice). | [69] |
| Polymeric and Hybrid silicon dioxide (SiO2) - coated capsules | (PARG/DEXS)3 capsules were prepared using layer-by-layer assembly by alternative deposition of PARG and DEXS on calcium carbonate particles and encapsulated by a silica shell | Cas9/gRNA plasmid | Delivery of CRISPR–Cas9 components using capsules to dTomato-expressing HEK293T cells. Transfection of indicator cells translated in high-level dTomato knockout in approx. 70% of transfected cells | N/A | [70] |
| Liposome-Templated Hydrogel Nanoparticles (LHNPs) | DOTAP liposomes were used as a template. Hydrogel was then formed by crosslinking cyclodextrin (CD)-engrafted PEI (PEI-CD) with adamantine (AD)-engrafted PEI (PEI-AD) | Cas9/minicircle-sgRNA | Cas9/mini-sgPLK1-2-encapsulated LHNPs inhibited cell growth by 79.3% and 80.2% for U87 cells and GS5 cells | LHNPs, which included iRGD and LEX for tumor targeting and autocatalysis, were evaluated in mice bearing intracranial U87 tumors. Compared to the control group, the expression of PLK1 in the nanoparticle treatment group was inhibited by 60.4% | [71] |
DSPE-PEG – 1, 2-Distearoyl-sn-glycero-3-phosphoethanolamine-Poly(ethylene glycol); stPEI – Stearyl-polyethylenimine; HAS – Human serum albumin ; sgRNA – Single guide RNA; TAT-GNs –Transactivating transcriptional activator peptide functionalized gold nanoclusters; CD – Cyclodextrin; PEI – Polyethylenimine; AD – Adamantine
While the dense core serves to minimize the size of the CRISPR system, the cationic shell facilitates nanoparticle-cell interactions and the subsequent internalization of the particles. Another class of lipids, suitable for negatively charged nucleic acid delivery, are ionizable lipids. Ionizable lipids carry the added feature of pH-dependent charge changes, bearing a positive charge in an acidic environment to enable nucleic acid encapsulation, and a neutral charge at physiological pH. Lipid nanoparticles utilizing ionizable lipids usually have additional standard components, such as cholesterol for particle stability, a helper phospholipid for structural stability, and a PEG-derivative for physiological stability.[65,66]
In order to meet the demands of in vivo gene therapy, the cationic lipid system must have the ability to survive in circulation for extended periods of time, increasing the time and likelihood that the carriers reach the intended target tissue. Even for neutral nanoparticles, PEGylation is an essential strategy to minimize nonspecific interactions with serum proteins in the bloodstream and avoid aggregation and subsequent clearance by immune system components.[72]
While several advances have been made for the delivery of short RNAs and plasmids by lipid-based nanoparticles, an ideal formulation and composition has yet to be determined for the delivery of longer RNAs characteristic of the CRISPR/Cas9 system, such as the sgRNA or Cas9 mRNA. Miller and colleagues developed zwitterionic amino lipids (ZALs) to uniquely co-deliver long RNAs including Cas9 mRNA and sgRNAs, reducing protein expression by >90% in cells.[73] Traditionally, highly effective LNPs are composed of a cationic lipid, zwitterionic phospholipid, cholesterol, and PEG. High cationic lipid densities may minimize the stabilization interactions between phospholipids and longer RNAs, and even hinder inclusion of long RNAs. Therefore, the group combined zwitterionic and cationic lipids to increase molecular interactions within the LNP, thereby improving the delivery of long RNA molecules.
The targeting specificity of nanoparticles can be improved when a ligand, such as an antibody, protein, peptide, or aptamer are incorporated and interact with the receptors of target cells (Figure 2b). Zhen et al. designed a flexible RNA aptamer A10-liposome-CRISPR/Cas9 chimera that specifically binds prostate cancer cells expressing a prostate-specific membrane antigen as the ligand. During transcription, they modified the aptamer with a 2’- F and 3’-NH2 to improve its stability and provide a convenient linkage. Their results showed that binding of the RNA-aptamer is more competitive when modified with DSPE-PEG 2000, suggesting that appropriate modifications can promote aptamer function without affecting targeting or gene knockdown.[74] Moreover, target specificity can be enhanced by ensuring that the response of the ligand-receptor pair is particularly strong, and that the receptor for the ligand be overexpressed on target cells rather than normal cells. For instance, tumor cells express high levels of the albumin-binding glycoprotein (Gp60) and secreted protein acidic and rich in cysteine (SPARC) receptors, which are albumin receptors present on the cell surface that increase membrane permeability for receptor-mediated uptake of circulating proteins. Therefore, albumin-based NP formulations may facilitate cellular uptake of CRISPR/Cas9 components to preferentially edit cancer cells.[67] The delivery of nanoparticles to tumors can also be improved through the conjugation of cancer-targeting ligands such as internalizing RGD (iRGD).[71]
In order to create a permanent modification to the genome, NPs must enter the nucleus. Nuclear localization of NPs has been achieved using the inclusion of nuclear localization sequences, cell penetrating peptides, and additional nuclear membrane targeting ligands.[75] Furthermore, effective cellular internalization is a critical process for successful clinical application of nanoparticles for gene delivery. Liu et al. designed bioreducible lipid nanoparticles containing disulfide bonds for the Cas9 mRNA/sgRNA complex.[68] They chose the leading lipid, BAMEA-O16B, which is a general nanocarrier for mRNA delivery to encapsulate the mRNA/sgRNA complex via electrostatic interactions and assemble into nanoparticles. In the intracellular environment, reductive signals release the mRNA through a disulfide bond exchange mechanism. The group found that the BAMEA-O16B/Cas9/mRNA/sgRNA knocked out GFP expression in human embryonic kidney cells with efficiencies as high as 90%. Moreover, the group demonstrated through intravenous injection that the particles could knock down mouse PCSK9, an important mediator of lipid metabolism in the liver, by 80% in vivo. Additionally, Wang and colleagues developed cationic bioreducible lipid nanoparticles for the delivery of super negatively charged Cre protein (to drive electrostatic self-assembly with cationic lipids to form a nanocomplex) and Cas9::sgRNA, enabling gene recombination and gene editing efficiencies greater than 70% in cultured HeLa cells.[76] The group hypothesized that the bioreduction of these lipid/protein nanocarriers inside cells in response to the reductive intracellular environment may facilitate endosomal escape, enabling the protein cargo to enter the nucleus.
Lipids, and in particular phospholipids, have emerged as particularly versatile building blocks for the production of NP coatings that can improve the stability and biocompatibility of NPs synthesized from inorganic materials, which are unstable in aqueous suspensions and can give rise to cytotoxic effects.[77] Several inorganic materials such as calcium phosphate, gold, carbon materials, silicon oxide, iron oxide, and layered double hydroxide (LDH), have been studied in detail.[78] Wang et al. developed a vehicle based on lipid/gold nanoclusters (GNs) for the delivery of both the Cas9 protein and sgRNA plasmid to tumor cells (Figure 2c). To improve nuclear targeting of the cationic GNs, the HIV-1-transactivator of transcription (TAT) peptide was used to modify the GNs. Positively charged TAT-GNs were mixed with negatively charged Cas9 proteins and sgRNA, yielding a ternary complex (TAT-GNs/Cas9 protein/sgRNA plasmid, abbreviated as “GCP”) formed via electrostatic interactions. Furthermore, this complex was encapsulated in an anionic lipid shell (1,2-dioleoyl-3-trimethylammoniumpropane (DOTAP)/dioleoyl-phosphatidyl ethanol-amine (DOPE)/cholesterol, 0.8/1/0.5), followed by post-modification with polyethylene glycol-phospholipids (DSPE-PEG) on the surface.[69]
Polymeric-based Delivery Systems
Polymers are extensively used for drug delivery applications. Several types of polymeric drug delivery vehicles exist including polyplexes, nanoconjugates, micelles, nanocapsules, dendrimers, and nanoparticles (Figure 3a).[79] Similar to cationic liposomes, cationic polymeric vectors are advantageous for gene delivery applications because they often contain ionizable amino groups that can easily be complexed with negatively charged nucleic acids via electrostatic attractions.
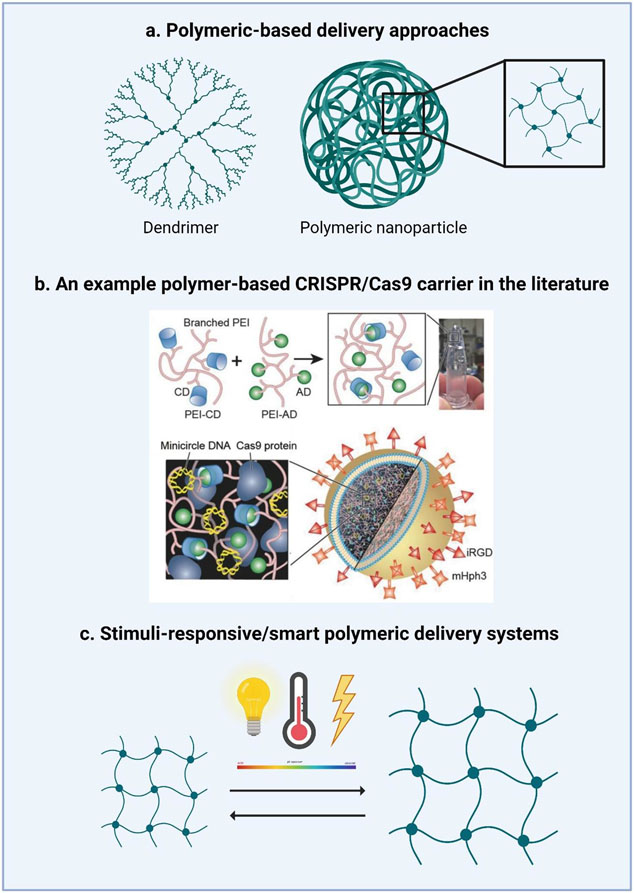
Figure 3. Polymeric-based strategies for the delivery of CRISPR/Cas9.
a.) Dendrimers and polymeric nanoparticles are the most common polymeric systems used to deliver CRISPR/Cas9. b.) An example of the fabrication of liposome-templated hydrogel nanoparticles (LHNPs) from the literature. Reproduced with permission from Chen et al.[69] The core of LHNPs was formed by a polyethylenimine (PEI) hydrogel crosslinked by cyclodextrin (CD)-engrafted PEI and adamantine (AD)-engrafted PEI. The shell consisted of cationic 1,2-dioleoyl-3-trimethylammonium-propane chloride salt (DOTAP) lipids that were selected for the delivery of genetic materials (i.e., minicircle DNA and Cas9 protein). To improve cell uptake, LHNPs were conjugated with internalizing RGD (iRGD) and mHph3targeting ligands.[69] c.) Another class of polymeric systems are those which are stimuli-responsive or “smart” which respond by either swelling or collapsing in response to pH, temperature, electric fields, pressure, or specific analytes/compounds.
The polyethylenimine (PEI) polymer family consists of a variety of linear and branched polymers that are often used in gene and drug delivery. Branched PEI has a high charge density, facilitating efficient plasmid packaging by compacting negatively charged nucleic acids into polyplexes that shield nucleic acids from degradation by nucleases. The high charge density of branched PEI also imparts pH-buffering abilities to the nanocarrier, enabling intracellular endosomal escape. However, these benefits are offset by more alarming characteristics of branched PEI namely, it is not biodegradable and is extremely cytotoxic. This can therefore cause extensive damage to cellular membranes and mitochondrial dysfunction, inhibiting ATP synthesis and activating cellular apoptosis and necrosis. In fact, the effectiveness of PEI transfection is dependent on both architecture and molecular weight, demonstrating a direct correlation with the positive charge of these carriers. While higher molecular weight PEIs demonstrate superior transfection efficiencies in vivo, their net positive charge is unfortunately correlated with greater cytotoxicity.[80]
Zhang and colleagues explored the use of cationic polyethyleneimine-β-cyclodextrin (PC) as a delivery vector a Cas9-sgRNA encoded plasmid.[81] Compared to polyethyleneimine alone, PC is less cytotoxic, allowing for transfection at either high doses, or in repeated numbers. Further modification of the polymer can impart multifunctionalities, such as enhanced targeting capabilities to improve its delivery efficiency. The authors found that PC encapsulation of the Cas9-sgRNA plasmid was dependent on the polymer to DNA (N/P) ratio, with increased encapsulation efficiencies obtained at higher N/P ratios and the greatest transfection (34%) was achieved at an optimal N/P ratio of 60. However, they did observe that increasing the N/P ratio to 80 significantly increased the cytotoxicity of the nanoparticles (since the zeta potential of the complexes becomes strongly positively with increasing N/P ratios). Furthermore, they hypothesized that negatively charged macromolecules in cells would trigger the intracellular release of pDNA from the complexes.
The use of naturally derived, biodegradable polymers may address the inherent cytotoxicity of synthetic polymers such as PEI, PC, and PAMAM. Several natural polymers including chitosan, collagen derivatives, hyaluronic acid, dextran, and β-cyclodextrin (β-CD) have been extensively characterized for nucleic acid delivery. One group developed a novel gene-chemo synergistic treatment nanosystem composed of chitosan (CS) loaded with Paclitaxel (an FDA approved chemical drug which has been clinically used for breast cancer, ovarian cancer, and liver cancer) and sg-VEGFR2/Cas9 plasmid for combination cancer therapy. In addition to excellent biocompatibility and biodegradability, CS use provides the nanoparticles with a pH-responsive effect and the ability to obtain sustained drug release profiles. In addition, -galactose-carrying lactobionic acid (LA) was conjugated to CS to specifically target hepatocellular carcinoma (HCC) cells that expressed asialoglyco protein receptors (ASGPR) on their membrane surface, increasing uptake of agents to tumor cells preferentially. In vivo studies in mice demonstrated that the nanocomplex suppressed >60% VEGFR2 on HepG2 cells and inhibited HCC tumor progression by 70%.[82]
Complex Polymer Systems
Apart from polymeric nanoparticles, polymeric capsules have been leveraged for the delivery of genetic material. Polymeric capsules are synthesized by a multilayer assembly of oppositely charged polyelectrolytes and can be functionalized with antibodies, drugs, analyte sensitive fluorophores, etc. at two distinct positions, their wall and cavity, allowing for the creation of a multifunctional delivery system that possesses tunable physiochemical properties.[83] Compared to traditional nanoparticle systems, capsules are internalized by mammalian cells via phagocytosis and lipid-raft-mediated macropinocytosis. One general obstacle is that upon internalization, the capsules are located in the lysosome and therefore, the delivered cargo is not available for cytosolic targets. Biodegradable capsules provide a solution to translocate capsule-delivered cargo from the lysosomal compartment to the cytosol.[84,85]
Timin and colleagues explored the use of polymeric and hybrid silicon dioxide (SiO2) -coated capsules to deliver CRISPR/Cas9 components. The carriers are composed of biodegradable polymers, such as polysaccharides and polypeptides, modified with a biocompatible silica shell, which endows them with enhanced mechanical strength. A layer-by-layer assembly by alternating deposition of poly-l-arginine hydrochloride (PARG) and dextran sulfate (DEXS) on calcium carbonate particles, followed by the removal of the calcium carbonate cores was used to form the hollow microcapsules. The group showed that in addition to high internalization and low toxicity profiles in vitro, the capsules can be degraded upon cell uptake, resulting in cargo release into the cellular microenvironment. Direct quantitative comparison revealed that the capsules achieved a transfection efficiency greater than 70% for eGFP-mRNA loaded capsules compared to the less than 50% efficiency achieved using a commercial liposome-based transfection reagent (MF PRO) in the HEK293T cell line.[70]
Complexing lipid nanoparticles with polymers such as PEI, PAMAM, and poly(lactic-co-glycolic acid) (PLGA) can be a useful strategy to improve intracellular transfection efficiencies while minimizing cytotoxicity. Xu et al. developed cationic lipid-assisted nanoparticles (CLAN), a type of PEG-b-PLGA based nanoparticle assisted by cationic lipid N,N-bis(2-hydroxyethyl)-N-methyl-N-(2-cholesteryloxycarbonyl aminoethyl)ammonium bromide (BHEM-Chol) for nucleic acid delivery, to deliver Cas9 mRNA and gRNA targeting the NLRP3 inflammasome in macrophages, subsequently mitigating their inflammatory response.[86] The group showed that optimizing the ratio between the surface charge and PEG density of the polymeric nanoparticles could enhance their internalization in macrophages. Another group used a cationic lipid assisted nanoparticle (CLAN ) system to specifically disrupt the NTn1 gene (a potential therapeutic target in macrophages for the treatment of type 2 diabetes) in macrophages and their precursor monocytes in vivo to reduce expression of netrin -1(protein encoded by Ntn1) and improve type 2 diabetes (T2D) symptoms in the T2D mouse model (C57BL/6 mice on a high fat diet).[87] Furthermore, the group found that the CD68 promoter-driven Cas9 plasmids could drive the specific expression of Cas9 in macrophages, thereby allowing macrophage-specific gene editing without disrupting surrounding cells. Another group optimized the CLAN formulation to encapsulate pCas9/gBCR-ABL into chronic myeloid leukemia (CML) cells in vitro and in vivo.[27] CML is caused by the chromosomal translocation of the BCR gene on chromosome 22 and its subsequent fusion with the ABL gene on chromosome 9, resulting in a chimeric BCR-ABL protein with constitutive tyrosine kinase activity. After intravenous injection of CLANpCas9/gBCR-ABL, the group specifically disrupted the fusion region of the BCR-ABL gene in CML mice, selectively sparing the expression of either gene alone in normal cells. Chen and colleagues synthesized liposome-templated hydrogel nanoparticles (LHNPs) using 1,2-dioleoyl-3-trimethylammonium-propane chloride salt (DOTAP) lipids for the codelivery of protein and nucleic acids (Figure 3b).[71] While the DOTAP liposome has demonstrated high safety for clinical use, it is limited by its low encapsulation efficiency of the Cas9 protein at 6.3%. To improve its encapsulation efficiency, the cationic DOTAP liposome was used as a template. By crosslinking cyclodextrin (CD)-engrafted PEI with adamantine (AD)-engrafted PEI, the authors were able to create a non-covalently crosslinked hydrogel complexed with both the Cas9 protein and sgRNA at high encapsulation and delivery efficiencies without the addition of initiators and UV radiation, conditions which are often required for hydrogel synthesis, but hinder protein stability and activity. Furthermore, conjugation of the cell penetrating peptide, mHph3, on the surface of the hydrogel liposomes increased the delivery efficiency of the particles to 1.3 times more than the commercial agent Lipofectamine 2000. The group synthesized LHNPs that encapsulated the Cas9 protein and minicircle sgRNA against polo-like kinase 1 (PLK1), a regulator of mitosis which is overexpressed in various tumor cells, inhibiting cell growth by 79.3% and 80.2% in human brain cancer stem cell lines (specifically, U87 and GS5).[69]
Two or more polymeric materials can be used in the synthesis of a single carrier to form hybrid nanomaterials with interesting features. Gao et al. developed a PAMAM - poly(β-amino ester) (PBAE) hyperbranched copolymer for the delivery of CRISPR/Cas9 to specifically cleave HPV E7 oncogene in HPV-positive cervical cancer cells. Cationic PBAE can efficiently condense the plasmids in nano-sized composite particles, deliver the plasmid into the cell, and transfer the plasmids into the cytoplasm via endosomal escape. Previous work has reported that hyperbranched polymers (with the same molecular weight) showed higher gene transfection efficiencies compared to linear polymers synthesized using the same monomers. Therefore, the group chose PAMAM-G0 as the branching unit. While dendrimers, such as PAMAM, have an abundance of charged coupling sites that allow for the efficient loading of negatively charged nucleic acid molecules, their high cation charge density can lead to non-specific cellular uptake and systemic toxicity in vivo.[88,89] Moreover, PAMAM can only achieve high transfection efficiencies in high generation, which is both difficult to synthesize and expensive. As a result, the group utilized PAMAM-G0 and PBAE to develop hyperbranched copolymers similar in structure to higher generation PAMAM, with good biocompatibility, biodegradability, and high transfection efficiencies.[90]
Semiconducting polymers (SPs) have attracted considerable attention in molecular imaging, photothermal bioactivation, and phototheranostics. One group designed a semiconducting polymer brush and demonstrated its application in fluorescence image-guided, light triggered remote control of CRISPR/Cas9 gene editing, useful for visible genome editing in vivo.[91] A combination of alkyl side chains and fluorinated PEI was responsible for the formation of a hydrophobic core that could bind the CRISPR/Cas9 cassettes via electrostatic interactions. Furthermore, the glucocorticoid Dexamethasone (Dex), which can dilate nuclear pores when bound to the nuclear glucocorticoid receptor, was incorporated into the hydrophobic core. The backbone of the SP brush serves as the photothermal transducer. After administration, NIR-II fluorescence imaging is used to track the in vivo distribution of the NPs. Once the NPs accumulate inside the target tissues and cells, laser irradiation is applied to promote the photothermal conversion of the nanoparticles, facilitating endosomal escape, triggering Dex release which can further promote entry of the gene payloads into the nucleus.
Stimuli-responsive smart materials
Hydrogels are appealing drug delivery systems due to their high tunability and capability to retain large amounts of water (70-99%).[92] Stimuli responsive hydrogels are particularly unique in that they can be engineered to allow the polymeric network to transition from a collapsed to swollen state, or vice versa, in response to a wide variety of environmental cues such as pH, temperature, electric field, pressure, as well as other stimuli (Figure 3c). Stimuli responsive materials allow for the integration of completely different capabilities into a single delivery system. Liu et al., synthesized a multistage delivery nanoparticle (MDNP) system with the goal of modulating the surface properties of the vehicle depending on the stage of delivery from circulation in the bloodstream to entry into cancer cells.[93]
The complex has a core-shell structure, containing a cationic polyplex core consisting of CRISPR/dCas9 plasmid DNA, phenylboronic acid (PBA)-modified low molecular weight polyethyleneimine (PEI–PBA). The shell consists of 2,3-dimethylmaleic anhydride (DMMA)-modified poly(ethylene glycol)-b-polylysine (mPEG113-b-PLys100/DMMA). To overcome the various physiological barriers in the delivery of CRISPR cargo from bloodstream to tumor cells, MDNP exhibits corresponding surface properties suitable for each stage in delivery. The MDNP system is designed to maintain the core-shell structure in the blood stream, with the negatively charged polymer shell reducing the likelihood of immune clearance. Upon entering tumor tissues, the acidic microenvironment induces decomposition of DMMA groups in the polymer shell, thereby leading to the rapid conversion of mPEG113-b-PLys100/DMMA from an anionic polymer to a cationic polymer (mPEG113-b-PLys100). Due to electrostatic repulsion, the polymer shell effectively detaches from the MDNP and exposes the cationic polyplex core, which enhances tumor accumulation and cell internalization. Upon internalization, the PEI groups in the polyplex triggers endosomal disruption and release of CRISPR cargo.
DNA-responsive hydrogels that can interface with synthetic DNA constructs or naturally occurring extracellular DNA have emerged as useful biomaterials, specifically for the delivery of CRISPR/Cas9. However, current DNA-responsive hydrogels require high concentrations of DNA triggers for actuation. Moreover, adapting such DNA hydrogels for the activation by new trigger sequences via modification of nucleic acid components can hinder the structural constraints of the materials, thus limiting the programmability of these systems. To overcome the aforementioned challenges, one group developed a programmable, CRISPR-based hydrogel using ssDNA sequences as both key structural elements and linkers to the cargo. Following introduction of an external DNA stimuli, a Cas enzyme (specifically Cas12a) and a gRNA complex cleaves the ssDNA sequences in the hydrogel, restructuring the hydrogel, releasing the bound cargo, or a combination of the two.[94] Importantly, this hydrogel system eliminates the need to encode target-sequence specificity into the carrier structure. Instead, replacement of the gRNA sequence can easily modulate the specificity of the Cas enzyme. Furthermore, they demonstrated the versatility of this nanoparticle system for the controlled release of various payloads including, but not limited to, branched PEG hydrogels releasing DNA anchored fluorescent molecules and active enzymes, as well as degradable polyacrylamide-DNA hydrogels encapsulating nanoparticles and live cells at low concentrations of DNA stimuli. Furthermore, this system can operate as a stimuli-responsive electric fuse, as well as a controllable valve in fluidic devices.
Cell-Specific Targeting to Improve Translation Outcomes
The formulation of delivery systems with cell specific targeting abilities is unquestionably a critical step in the future translation of gene editing technologies. Active targeting is the most straightforward strategy and involves conjugating ligands that specifically bind to an overexpressed receptor on target cells on the surface of nanoparticles. The ligand-receptor interaction induces the nanoparticles to be internalized by target cells via receptor-mediated endocytosis, thereby minimizing nonspecific interactions with normal cells, which can be of particular interest in anticancer drug delivery applications. The targeting efficiency of nanocarriers is generally a function of the binding affinity of the selected ligand for the target cell. However, it has been suggested that modulating the affinity properties of the nanoparticle system may be counterproductive in the case of solid tumors; high affinity interactions between antibodies and tumor antigens hinder efficient tumor penetration of the nanoparticle systems. [95]
Naoparticles have been functionalized to a diverse range of ligands including antibodies, aptamers, carbohydrates, enzymes, folate, peptides (lectins and transferrin), and vitamins.[74,96] Rosenblum et al. constructed cell targeted CRISPR lipid nanoparticles functionalized with EGFR (epidermal growth factor receptor), an antibody to an overexpressed receptor on ovarian cancer cells, which achieved up to 80% PLK1 gene editing in vivo.[97] Several chemical strategies exist to conjugate antibodies to drug delivery systems, including: (1) the modification of existing functional groups (disulfide, amine, carboxyl, and carbohydrate groups) in antibodies with crosslinking reagents with reactive functionalities, (2) the use of free functional groups present in the antibody molecules, and (3) the use of functionalized PEG derivative which can act as a linking agent between the antibody and drug delivery carrier.[98] However, the presence of targeting ligands does not guarantee in vivo success; it has been shown that the targeting ability of functionalized nanoparticles may disappear when they are placed in a complex, biological environment in which interactions with other proteins in the medium may shield the targeting ligands, resulting in a loss of specificity.[99]
Biomimetic nanoparticles are an emerging class of materials based on the paradigm that natural biomaterials, such as cell membranes, can be interfaced with synthetic nanoparticle systems to enhance drug delivery systems for therapeutic applications. Particles can be fabricated by fusing natural cell membranes onto synthetic cores thus, retaining biological properties such as membrane composition, membrane fluidity, and antigenic profile of source cells. Consequently, such systems may demonstrate superior homotypic targeting, complex antigenic profile, and low intrinsic immunogenicity, which are cornerstones of personalized medicine.[100]
Alyami et al. synthesized cancer cell coated zeolitic imidazolate frameworks encapsulating CRISPR/Cas9 machinery which showed increased selectivity for and accumulation in tumor cells with negligible uptake by healthy cells.[101] Another approach to constructing biomimetic nanoparticles involves the construction of exosome-nanoparticle hybrids. Exosomes are nanoscale vesicles that are secreted by almost all kinds of cells and can be loaded with a diverse range of biomolecules, including proteins, small molecules, and nucleic acids, making them a particularly useful drug delivery vehicles.[102] Compared to synthetic nanoparticles, exosomes possess a number of desirable attributes such as, excellent biocompatibility (since exosomes are produced by cells), long-term, stable circulation from CD47-mediated protection of exosomes from phagocytosis, and ability to cross through various biological barriers, such as the blood-brain barrier.[103] Additionally, they retain the membrane surface composition of progenitor cells, containing membrane proteins and lipids that may facilitate direct contact between exosomes and target cells.
Moreover, exosomes can be engineered to express specific surface proteins that enhance their targeting specificity. One group modified the surface of macrophage-derived exosome-coated poly(lactic-co-glycolic acid) nanoparticles to express a peptide to target the mesenchymal-epithelial transition factor (c-MET), overexpressed by triple-negative breast cancer cells.[104] However, issues such as low production and isolation yield, heterogeneity of isolated exosomes, and complexities in drug loading make it difficult to effectively scale-up exosome production and thus, effect the translational potential of these drug delivery vehicles.[105] To address these issues while leveraging the smart-targeting abilities of exosomes, several groups have developed exosome-liposome hybrid nanoparticles.[106,107] Lin and colleagues developed hybrid nanoparticles by incubating cell-derived exosomes with liposomes that successfully loaded large nucleic acids (such as CRISPR/Cas9 expression vectors) and transfected MSCs.[108] By including cell-targeting moieties to nanoparticle drug delivery vehicles, the future translation of gene editing technologies can be greatly influenced.
Conclusions and Future Outlook
CRISPR allows researchers to direct development and disease using a precise, simple modification of the genome. The simple two-component system has been engineered to harness the potent Cas9 nuclease and its variable sgRNA counterpart to exemplify some of the most basic biological concepts, primarily DNA base pairing. The advent of CRISPR also highlights the utility of understanding, characterizing, and studying the mechanisms that govern development and normal physiology in organisms other than humans.
While CRISPR holds great promise in transforming the advent of genome editing, its entire potential cannot be fully harnessed until we develop the appropriate vehicles to efficiently deliver CRISPR-related cargo. This review has highlighted various nanoparticle-mediated delivery methods that have been developed to transport CRISPR components. The variety in approaches developed serves to demonstrate there is not one singular approach to achieve optimal delivery. Instead, each delivery method comes with its own set of advantages and disadvantages; each must be modified and adapted for in vivo delivery applications. Delivery vehicles must be developed so that they are biocompatible, safe, and efficient in transporting CRISPR components to target cells. Our understanding of the circulation of these particles in the body, their pharmacokinetic profiles in vivo, and their systemic gene editing capabilities is limited, and must be further characterized to develop more efficient vehicles that address potential limitations of current nano-sized carriers.
The intensity and pace of innovation for the development of CRISPR/Cas9 as a gene editing tool is quickly gaining momentum, bringing together some of the brightest minds to understand and unleash the full potential of this gene-editing platform. CRISPR is still in its initial stages and its full capacity remains unknown, but the only way to explore this uncharted territory is through properly addressing potential limitations in the development of this technology. CRISPR may be gaining traction for completely re-envisioning genome engineering; however, its true authenticity lies in its ability to demonstrate the limitless possibilities of unrestrained curiosity, scientific collaboration, and simply put, science.
Acknowledgements
N.A.P. acknowledges support from the National Institutes of Health (R01EB022025), the Cockrell Family Chair Foundation, the office of the Dean of the Cockrell School of Engineering at the University of Texas at Austin (UT) for the Institute for Biomaterials, Drug Delivery, and Regenerative Medicine, and the UT-Portugal Collaborative Research Program. M.E.W acknowledges support from the University of Texas at San Antonio. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the funding agencies. All figures were creating using BioRender.com.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest: none
REFERENCES
- [1].Mojica FJM, Díez-Villaseñor C, García-Martínez J, and Soria E, “Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements,” Journal of Molecular Evolution, vol. 60, no. 2, pp. 174–182, 2005, doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- [2].Pourcel C, Salvignol G, and Vergnaud G, “CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies,” Microbiology, vol. 151, no. 3, pp. 653–663, 2005, doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- [3].Bolotin A, Quinquis B, Sorokin A, and Dusko Ehrlich S, “Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin,” Microbiology, vol. 151, no. 8, pp. 2551–2561, 2005, doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- [4].Doudna JA and Charpentier E, “The new frontier of genome engineering with CRISPR-Cas9,” Science, vol. 346, no. 6213, pp. 19–20, 2014, doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- [5].Chu VT et al. , “Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells,” Nature Biotechnology, vol. 33, no. 5, pp. 543–548, 2015, doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- [6].Chandrasegaran S, “Recent advances in the use of ZFN-mediated gene editing for human gene therapy,” Cell and Gene Therapy Insights, vol. 3, no. 1, pp. 33–41, February. 2017, doi: 10.18609/cgti.2017.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lino CA, Harper JC, Carney JP, and Timlin JA, “Delivering crispr: A review of the challenges and approaches,” Drug Delivery, vol. 25, no. 1, pp. 1234–1257, 2018, doi: 10.1080/10717544.2018.1474964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Barrangou R and Doudna JA, “Applications of CRISPR technologies in research and beyond,” Nature Biotechnology, vol. 34, no. 9, pp. 933–941, 2016, doi: 10.1038/nbt.3659. [DOI] [PubMed] [Google Scholar]
- [9].Szlachta K et al. , “CRISPR knockout screening identifies combinatorial drug targets in pancreatic cancer and models cellular drug response,” Nature Communications, vol. 9, no. 1, 2018, doi: 10.1038/s41467-018-06676-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gilbert LA et al. , “Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation,” Cell, vol. 159, no. 3, pp. 647–661, 2014, doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gilbert LA et al. , “XCRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes,” Cell, vol. 154, no. 2, p. 442, July. 2013, doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhang Y et al. , “Enhanced CRISPR-Cas9 correction of Duchenne muscular dystrophy in mice by a self-complementary AAV delivery system,” Science Advances, vol. 6, no. 8, pp. 1–12, 2020, doi: 10.1126/sciadv.aay6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Qiurong D et al. , “Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing,” Circulation Research, vol. 115, no. 5, pp. 488–492, 2014, doi: 10.1161/CIRCRESAHA.115.304351.Permanent. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Alapati D et al. , “In utero gene editing for monogenic lung disease,” Science Translational Medicine, vol. 11, no. 488, pp. 1–14, 2019, doi: 10.1126/scitranslmed.aav8375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Román-Rodríguez FJ et al. , “NHEJ-Mediated Repair of CRISPR-Cas9-Induced DNA Breaks Efficiently Corrects Mutations in HSPCs from Patients with Fanconi Anemia,” Cell Stem Cell, vol. 25, no. 5, pp. 607–621.e7, November. 2019, doi: 10.1016/j.stem.2019.08.016. [DOI] [PubMed] [Google Scholar]
- [16].Frangoul H et al. , “CRISPR-Cas9 Gene Editing for Sickle Cell Disease and β-Thalassemia,” New England Journal of Medicine, vol. 384, no. 3, pp. 252–260, 2021, doi: 10.1056/nejmoa2031054. [DOI] [PubMed] [Google Scholar]
- [17].Dow LE, “Modeling disease in vivo with CRISPR/Cas9 Lukas,” Trends Mol Med, vol. 21, no. 10, pp. 609–621, 2016, doi: 10.1016/j.molmed.2015.07.006.Modeling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sakuma T, Nishikawa A, Kume S, Chayama K, and Yamamoto T, “Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system,” Scientific Reports, vol. 4, pp. 4–9, 2014, doi: 10.1038/srep05400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Cong L et al. , “Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, … Zhang F (2013). Multiplex Genome Engineering Using CRISPR/Cas Systems. Science (New York, N.Y.),” Science (New York, N.Y.), vol. 339, no. 6121, pp. 819–823, 2013, doi: 10.1126/science.1231143.Multiplex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Liu Z et al. , “Genome editing of the HIV co-receptors CCR5 and CXCR4 by CRISPR-Cas9 protects CD4+ T cells from HIV-1 infection,” Cell and Bioscience, vol. 7, no. 1, pp. 1–15, 2017, doi: 10.1186/s13578-017-0174-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Li Y, Glass Z, Huang M, Chen ZY, and Xu Q, “Ex vivo cell-based CRISPR/Cas9 genome editing for therapeutic applications,” Biomaterials, vol. 234, no. January, p. 119711, 2020, doi: 10.1016/j.biomaterials.2019.119711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Huang D, Miller M, Ashok B, Jain S, and Peppas NA, “CRISPR/Cas systems to overcome challenges in developing the next generation of T cells for cancer therapy,” Advanced Drug Delivery Reviews, vol. 158. Elsevier B.V., pp. 17–35, January. 01, 2020, doi: 10.1016/j.addr.2020.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kleinstiver BP et al. , “High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects,” Nature, vol. 529, no. 7587, pp. 490–495, 2016, doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Schwank G et al. , “Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients,” Cell Stem Cell, vol. 13, no. 6, pp. 653–658, December. 2013, doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- [25].Rosenblum D et al. , “CRISPR-Cas9 genome editing using targeted lipid nanoparticles for cancer therapy,” 2020. [Online]. Available: http://advances.sciencemag.org/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cho EY et al. , “Lecithin nano-liposomal particle as a CRISPR/Cas9 complex delivery system for treating type 2 diabetes,” Journal of Nanobiotechnology, vol. 17, no. 1, January. 2019, doi: 10.1186/s12951-019-0452-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Liu Y, Zhao G, Xu CF, Luo YL, Lu ZD, and Wang J, “Systemic delivery of CRISPR/Cas9 with PEG-PLGA nanoparticles for chronic myeloid leukemia targeted therapy,” Biomaterials Science, vol. 6, no. 6, pp. 1592–1603, 2018, doi: 10.1039/c8bm00263k. [DOI] [PubMed] [Google Scholar]
- [28].Chew WL et al. , “A multifunctional AAV-CRISPR-Cas9 and its host response,” Nature Methods, vol. 13, no. 10, pp. 868–874, 2016, doi: 10.1038/nmeth.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Scobie L, Denner J, and Schuurman HJ, “Inactivation of porcine endogenous retrovirus in pigs using CRISPR-Cas9, editorial commentary,” Xenotransplantation, vol. 24, no. 6, pp. 1303–1307, 2017, doi: 10.1111/xen.12363. [DOI] [PubMed] [Google Scholar]
- [30].Wang X et al. , “Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors,” Nature Biotechnology, vol. 33, no. 2, pp. 175–179, 2015, doi: 10.1038/nbt.3127. [DOI] [PubMed] [Google Scholar]
- [31].Kabadi AM, Ousterout DG, Hilton IB, and Gersbach CA, “Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector,” Nucleic Acids Research, vol. 42, no. 19, pp. 1–11, 2014, doi: 10.1093/nar/gku749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Stephens CJ, Lauron EJ, Kashentseva E, Lu ZH, Yokoyama WM, and Curiel DT, “Long-term correction of hemophilia B using adenoviral delivery of CRISPR/Cas9,” Journal of Controlled Release, vol. 298, no. January, pp. 128–141, 2019, doi: 10.1016/j.jconrel.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Thomas CE, Ehrhardt A, and Kay MA, “Progress and problems with the use of viral vectors for gene therapy,” Nature Reviews Genetics, vol. 4, no. 5, pp. 346–358, 2003, doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- [34].Ortinski PI, O’Donovan B, Dong X, and Kantor B, “Integrase-Deficient Lentiviral Vector as an All-in-One Platform for Highly Efficient CRISPR/Cas9-Mediated Gene Editing,” Molecular Therapy - Methods and Clinical Development, vol. 5, no. June, pp. 153–164, 2017, doi: 10.1016/j.omtm.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chen X and Gonçalves MAFV, “Engineered viruses as genome editing devices,” Molecular Therapy, vol. 24, no. 3, pp. 447–457, 2016, doi: 10.1038/mt.2015.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bear AS et al. , “Replication-competent retroviruses in gene-modified T cells used in clinical trials: Is it time to revise the testing requirements?,” Molecular Therapy, vol. 20, no. 2, pp. 246–249, 2012, doi: 10.1038/mt.2011.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Li C and Samulski RJ, “Engineering adeno-associated virus vectors for gene therapy,” Nature Reviews Genetics, vol. 21, no. 4, pp. 255–272, 2020, doi: 10.1038/s41576-019-0205-4. [DOI] [PubMed] [Google Scholar]
- [38].Ran FA et al. , “In vivo genome editing using Staphylococcus aureus Cas9,” Nature, vol. 520, no. 7546, pp. 186–191, 2015, doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kaspar BK, “Gene therapy: Direct viral delivery,” Encyclopedia of Neuroscience, pp. 633–639, 2009, doi: 10.1016/B978-008045046-9.00012-7. [DOI] [Google Scholar]
- [40].Ghosh SS, Gopinath P, and Ramesh A, “Adenoviral vectors: A promising tool for gene therapy,” Applied Biochemistry and Biotechnology, vol. 133, no. 1, pp. 9–29, 2006, doi: 10.1385/ABAB:133:1:9. [DOI] [PubMed] [Google Scholar]
- [41].Mout R, Ray M, Lee YW, Scaletti F, and Rotello VM, “In Vivo Delivery of CRISPR/Cas9 for Therapeutic Gene Editing: Progress and Challenges,” Bioconjugate Chemistry, vol. 28, no. 4. American Chemical Society, pp. 880–884, April. 19, 2017, doi: 10.1021/acs.bioconjchem.7b00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Yip BH, “Recent advances in CRISPR/Cas9 delivery strategies,” Biomolecules, vol. 10, no. 6, 2020, doi: 10.3390/biom10060839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Mellott AJ, Forrest ML, and Detamore MS, “Physical non-viral gene delivery methods for tissue engineering,” Annals of Biomedical Engineering, vol. 41, no. 3, pp. 446–468, 2013, doi: 10.1007/s10439-012-0678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Meacham JM, Durvasula K, Degertekin FL, and Fedorov AG, “Physical Methods for Intracellular Delivery: Practical Aspects from Laboratory Use to Industrial-Scale Processing,” Journal of Laboratory Automation, vol. 19, no. 1, pp. 1–18, 2014, doi: 10.1177/2211068213494388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Escoffre JM, Portet T, Wasungu L, Teissié J, Dean D, and Rols MP, “What is (Still not) known of the mechanism by which electroporation mediates gene transfer and expression in cells and tissues,” Molecular Biotechnology, vol. 41, no. 3, pp. 286–295, 2009, doi: 10.1007/s12033-008-9121-0. [DOI] [PubMed] [Google Scholar]
- [46].Xu X et al. , “Efficient homology-directed gene editing by CRISPR/Cas9 in human stem and primary cells using tube electroporation,” Scientific Reports, vol. 8, no. 1, pp. 1–12, 2018, doi: 10.1038/s41598-018-30227-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Foroozandeh P and Aziz AA, “Insight into Cellular Uptake and Intracellular Trafficking of Nanoparticles,” Nanoscale Research Letters, vol. 13, 2018, doi: 10.1186/s11671-018-2728-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Mitchell MJ, Billingsley RM, Haley RM, Wechsler ME, Peppas NA, and Langer R, “Engineering precision nanoparticles for drug delivery,” Nature Reviews Drug Discovery, vol 20, no. 2. Nature Research, pp. 101–124, February. 01, 2021, doi: 10.1038/s41573-020-0090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Givens BE, Naguib YW, Geary SM, Devor EJ, and Salem AK, “Nanoparticle-Based Delivery of CRISPR/Cas9 Genome-Editing Therapeutics,” AAPS Journal, vol. 20, no. 6. Springer New York LLC, November. 01, 2018, doi: 10.1208/s12248-018-0267-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Akinc A, Thomas M, Klibanov AM, and Langer R, “Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis,” Journal of Gene Medicine, vol. 7, no. 5, pp. 657–663, 2005, doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- [51].Sonawane ND, Szoka FC, and Verkman AS, “Chloride Accumulation and Swelling in Endosomes Enhances DNA Transfer by Polyamine-DNA Polyplexes,” Journal of Biological Chemistry, vol. 278, no. 45, pp. 44826–44831, 2003, doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- [52].Bamrungsap S, et al. , “Nanotechnology in therapeutics: A focus on nanoparticles as a drug delivery system,” Nanomedicine, vol. 7, no. 8, pp. 1253–1271, 2012, doi: 10.2217/nnm.12.87. [DOI] [PubMed] [Google Scholar]
- [53].Blanco E, Shen H, and Ferrari M, “Principles of nanoparticle design for overcoming biological barriers to drug delivery,” Nature Biotechnology, vol. 33, no. 9, pp. 941–951, 2015, doi: 10.1038/nbt.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Shinde Patil VR, Campbell CJ, Yun YH, Slack SM, and Goetz DJ, “Particle diameter influences adhesion under flow,” Biophysical Journal, vol. 80, no. 4, pp. 1733–1743, 2001, doi: 10.1016/s0006-3495(01)76144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Zauner W, Farrow NA, and Haines AMR, “In vitro uptake of polystyrene microspheres: Effect of particle size, cell line and cell density,” Journal of Controlled Release, vol. 71, no. 1, pp. 39–51, 2001, doi: 10.1016/S0168-3659(00)00358-8. [DOI] [PubMed] [Google Scholar]
- [56].Champion JA, Katare YK, and Mitragotri S, “Particle shape: A new design parameter for micro- and nanoscale drug delivery carriers,” Journal of Controlled Release, vol. 121, no. 1–2, pp. 3–9, 2007, doi: 10.1016/j.jconrel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Banerjee A, Qi J, Gogoi R, Wong J, and Mitragotri S, “Role of nanoparticle size, shape and surface chemistry in oral drug delivery,” Journal of Controlled Release, vol. 238, pp. 176–185, 2016, doi: 10.1016/j.jconrel.2016.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Arvizo RR et al. , “Effect of nanoparticle surface charge at the plasma membrane and beyond,” Nano Letters, vol. 10, no. 7, pp. 2543–2548, July. 2010, doi: 10.1021/nl101140t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Naseri N, Valizadeh H, and Zakeri-Milani P, “Solid lipid nanoparticles and nanostructured lipid carriers: Structure preparation and application,” Advanced Pharmaceutical Bulletin, vol. 5, no. 3, pp. 305–313, 2015, doi: 10.15171/apb.2015.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Rehman ZU, Zuhom IS, and Hoekstra D, “How cationic lipids transfer nucleic acids into cells and across cellular membranes: Recent advances,” Journal of Controlled Release, vol. 166, no. 1, pp. 46–56, 2013, doi: 10.1016/j.jconrel.2012.12.014. [DOI] [PubMed] [Google Scholar]
- [61].del Pozo-Rodríguez A, Solinís MÁ, and Rodríguez-Gascón A, “Applications of lipid nanoparticles in gene therapy,” European Journal of Pharmaceutics and Biopharmaceutics, vol. 109, pp. 184–193, 2016, doi: 10.1016/j.ejpb.2016.10.016. [DOI] [PubMed] [Google Scholar]
- [62].Han Y, Zhang Y, Li D, Chen Y, Sun J, and Kong F, “Transferrin-modified nanostructured lipid carriers as multifunctional nanomedicine for codelivery of DNA and doxorubicin,” International Journal of Nanomedicine, vol. 9, no. 1, pp. 4107–4116, 2014, doi: 10.2147/IJN.S67770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Wang T, Larcher LM, Ma L, and Veedu RN, “Systematic screening of commonly used commercial transfection reagents towards efficient transfection of single-stranded oligonucleotides,” Molecules, vol. 23, no. 10, 2018, doi: 10.3390/molecules23102564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Zhang L et al. , “Lipid nanoparticle-mediated efficient delivery of CRISPR/Cas9 for tumor therapy,” NPG Asia Materials, vol. 9, no. 10, pp. e441–e441, 2017, doi: 10.1038/am.2017.185. [DOI] [Google Scholar]
- [65].Granot Y and Peer D, “Delivering the right message: Challenges and opportunities in lipid nanoparticles-mediated modified mRNA therapeutics—An innate immune system standpoint,” Seminars in Immunology, vol. 34, no. July, pp. 68–77, 2017, doi: 10.1016/j.smim.2017.08.015. [DOI] [PubMed] [Google Scholar]
- [66].Finn JD et al. , “A Single Administration of CRISPR/Cas9 Lipid Nanoparticles Achieves Robust and Persistent In Vivo Genome Editing,” Cell Reports, vol. 22, no. 9, pp. 2227–2235, 2018, doi: 10.1016/j.celrep.2018.02.014. [DOI] [PubMed] [Google Scholar]
- [67].Cheng WJ, Chen LC, Ho HO, Lin HL, and Sheu MT, “Stearyl polyethylenimine complexed with plasmids as the core of human serum albumin nanoparticles noncovalently bound to CRISPR/Cas9 plasmids or siRNA for disrupting or silencing PD-L1 expression for immunotherapy,” International Journal of Nanomedicine, vol. 13, pp. 7079–7094, 2018, doi: 10.2147/IJN.S181440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Liu J et al. , “Fast and Efficient CRISPR/Cas9 Genome Editing In Vivo Enabled by Bioreducible Lipid and Messenger RNA Nanoparticles,” Advanced Materials, vol. 31, no. 33, pp. 1–7, 2019, doi: 10.1002/adma.201902575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Wang P et al. , “Genome Editing for Cancer Therapy: Delivery of Cas9 Protein/sgRNA Plasmid via a Gold Nanocluster/Lipid Core–Shell Nanocarrier,” Advanced Science, vol. 4, no. 11, 2017, doi: 10.1002/advs.201700175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Timin AS et al. , “Efficient gene editing via non-viral delivery of CRISPR–Cas9 system using polymeric and hybrid microcarriers,” Nanomedicine: Nanotechnology, Biology, and Medicine, vol. 14, no. 1, pp. 97–108, 2018, doi: 10.1016/j.nano.2017.09.001. [DOI] [PubMed] [Google Scholar]
- [71].Chen Z et al. , “Targeted Delivery of CRISPR/Cas9-Mediated Cancer Gene Therapy via Liposome-Templated Hydrogel Nanoparticles,” Advanced Functional Materials, vol. 27, no. 46, December. 2017, doi: 10.1002/adfm.201703036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Mishra S, Webster P, and Davis ME, “PEGylation significantly affects cellular uptake and intracellular trafficking of non-viral gene delivery particles,” European Journal of Cell Biology, vol. 83, no. 3, pp. 97–111, 2004, doi: 10.1078/0171-9335-00363. [DOI] [PubMed] [Google Scholar]
- [73].Miller JB et al. , “Non-viral CRISPR/Cas gene editing in vitro and in vivo enabled by synthetic nanoparticle co-delivery of Cas9 mRNA and sgRNA HHS Public Access,” Angew Chem Int Ed Engl, vol. 56, no. 4, pp. 1059–1063, 2017, doi: 10.1002/anie.20X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Zhen S, Takahashi Y, Narita S, Yang Y-C, and Li X, “Oncotarget 9375 www.impactjournals.com/oncotarget Targeted delivery of CRISPR/Cas9 to prostate cancer by modified gRNA using a flexible aptamer-cationic liposome,” Oncotarget, vol. 8, no. 6, pp. 9375–9387, 2017, [Online]. Available: www.impactjournals.com/oncotarget/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Xiao Y, Shi K, Qu Y, Chu B, and Qian Z, “Engineering Nanoparticles for Targeted Delivery of Nucleic Acid Therapeutics in Tumor,” Molecular Therapy - Methods and Clinical Development, vol. 12, no. March, pp. 1–18, 2019, doi: 10.1016/j.omtm.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Wang M et al. , “Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles,” Proceedings of the National Academy of Sciences of the United States of America, vol. 113, no. 11, pp. 2868–2873, 2016, doi: 10.1073/pnas.1520244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Luchini A and Vitiello G, “Understanding the nano-bio interfaces: Lipid-coatings for inorganic nanoparticles as promising strategy for biomedical applications,” Frontiers in Chemistry, vol. 7, no. May, pp. 1–16, 2019, doi: 10.3389/fchem.2019.00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Xu ZP, Zeng QH, Lu GQ, and Yu AB, “Inorganic nanoparticles as carriers for efficient cellular delivery,” Chemical Engineering Science, vol. 61, no. 3, pp. 1027–1040, 2006, doi: 10.1016/j.ces.2005.06.019. [DOI] [Google Scholar]
- [79].Piotrowski-Daspit AS, Kauffman AC, Bracaglia LG, and Saltzman WM, “Polymeric vehicles for nucleic acid delivery,” Advanced Drug Delivery Reviews, vol. 156, pp. 119–132, 2020, doi: 10.1016/j.addr.2020.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Hall A, Lächelt U, Bartek J, Wagner E, and Moghimi SM, “Polyplex Evolution: Understanding Biology, Optimizing Performance,” Molecular Therapy, vol. 25, no. 7, pp. 1476–1490, 2017, doi: 10.1016/j.ymthe.2017.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Zhang Z et al. , “Cationic Polymer-Mediated CRISPR/Cas9 Plasmid Delivery for Genome Editing,” Macromolecular Rapid Communications, vol. 40, no. 5, pp. 1–8, 2019, doi: 10.1002/marc.201800068. [DOI] [PubMed] [Google Scholar]
- [82].Zhang BC et al. , “Efficient CRISPR/Cas9 gene-chemo synergistic cancer therapy via a stimuli-responsive chitosan-based nanocomplex elicits anti-tumorigenic pathway effect,” Chemical Engineering Journal, vol. 393, no. January, p. 124688, 2020, doi: 10.1016/j.cej.2020.124688. [DOI] [Google Scholar]
- [83].Ganas C et al. , “Biodegradable capsules as non-viral vectors for in vitro delivery of PEI/siRNA polyplexes for efficient gene silencing,” Journal of controlled release : official journal of the Controlled Release Society, vol. 196, pp. 132–138, 2014, doi: 10.1016/j.jconrel.2014.10.006. [DOI] [PubMed] [Google Scholar]
- [84].Gil PR, de Koker S, de Geest BG, and Parak WJ, “Intracellular processing of proteins mediated by biodegradable polyelectrolyte capsules,” European Cells and Materials, vol. 20, no. SUPPL.3, p. 213, 2010. [DOI] [PubMed] [Google Scholar]
- [85].Gao H, Goriacheva OA, Tarakina N. v., and Sukhorukov GB, “Intracellularly Biodegradable Polyelectrolyte/Silica Composite Microcapsules as Carriers for Small Molecules,” ACS Applied Materials and Interfaces, vol. 8, no. 15, pp. 9651–9661, 2016, doi: 10.1021/acsami.6b01921. [DOI] [PubMed] [Google Scholar]
- [86].Xu C et al. , “Targeting of NLRP3 inflammasome with gene editing for the amelioration of inflammatory diseases,” Nature Communications, vol. 9, no. 1, 2018, doi: 10.1038/s41467-018-06522-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Luo YL et al. , “Macrophage-Specific in Vivo Gene Editing Using Cationic Lipid-Assisted Polymeric Nanoparticles,” ACS Nano, vol. 12, no. 2, pp. 994–1005, 2018, doi: 10.1021/acsnano.7b07874. [DOI] [PubMed] [Google Scholar]
- [88].Jevprasesphant R, Penny J, Jalal R, Attwood D, McKeown NB, and D’Emanuele A, “The influence of surface modification on the cytotoxicity of PAMAM dendrimers,” International Journal of Pharmaceutics, vol. 252, no. 1–2, pp. 263–266, 2003, doi: 10.1016/S0378-5173(02)00623-3. [DOI] [PubMed] [Google Scholar]
- [89].Malik N et al. , “Dendrimers: Relationship between structure and biocompatibility in vitro, and preliminary studies on the biodistribution of 125I-labelled polyamidoamine dendrimers in vivo,” Journal of Controlled Release, vol. 65, no. 1–2, pp. 133–148, 2000, doi: 10.1016/S0168-3659(99)00246-1. [DOI] [PubMed] [Google Scholar]
- [90].Gao X et al. , “Hyperbranched poly(β-amino ester) based polyplex nanopaticles for delivery of CRISPR/Cas9 system and treatment of HPV infection associated cervical cancer,” Journal of Controlled Release, vol. 321, no. February, pp. 654–668, 2020, doi: 10.1016/j.jconrel.2020.02.045. [DOI] [PubMed] [Google Scholar]
- [91].Li L et al. , “A Rationally Designed Semiconducting Polymer Brush for NIR-II Imaging-Guided Light-Triggered Remote Control of CRISPR/Cas9 Genome Editing,” Advanced Materials, vol. 31, no. 21, pp. 1–9, 2019, doi: 10.1002/adma.201901187. [DOI] [PubMed] [Google Scholar]
- [92].Li J and Mooney DJ, “Designing hydrogels for controlled drug delivery,” Nature Reviews Materials, vol. 1, no. 12, 2016, doi: 10.1038/natrevmats.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Liu Q et al. , “Multistage Delivery Nanoparticle Facilitates Efficient CRISPR/dCas9 Activation and Tumor Growth Suppression In Vivo,” Advanced Science, vol. 6, no. 1, January. 2019, doi: 10.1002/advs.201801423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].English MA et al. , “Programmable CRISPR-responsive smart materials,” vol. 785, no. August, pp. 780–785, 2019. [DOI] [PubMed] [Google Scholar]
- [95].Adams GP et al. , “High affinity restricts the localization and tumor penetration of single-chain Fv antibody molecules,” Cancer Research, vol. 61, no. 12, pp. 4750–4755, 2001. [PubMed] [Google Scholar]
- [96].Marques AC, Costa PJ, Velho S, and Amaral MH, “Functionalizing nanoparticles with cancer-targeting antibodies: A comparison of strategies,” Journal of Controlled Release, vol. 320, no. January, pp. 180–200, 2020, doi: 10.1016/j.jconrel.2020.01.035. [DOI] [PubMed] [Google Scholar]
- [97].Liang C et al. , “Tumor cell-targeted delivery of CRISPR/Cas9 by aptamer-functionalized lipopolymer for therapeutic genome editing of VEGFA in osteosarcoma,” Biomaterials, vol. 147, pp. 68–85, 2017, doi: 10.1016/j.biomaterials.2017.09.015. [DOI] [PubMed] [Google Scholar]
- [98].Manjappa AS et al. , “Antibody derivatization and conjugation strategies: Application in preparation of stealth immunoliposome to target chemotherapeutics to tumor,” Journal of Controlled Release, vol. 150, no. 1, pp. 2–22, 2011, doi: 10.1016/j.jconrel.2010.11.002. [DOI] [PubMed] [Google Scholar]
- [99].Salvati A et al. , “Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface,” Nature Nanotechnology, vol. 8, no. 2, pp. 137–143, 2013, doi: 10.1038/nnano.2012.237. [DOI] [PubMed] [Google Scholar]
- [100].Fang RH et al. , “Cancer cell membrane-coated nanoparticles for anticancer vaccination and drug delivery,” Nano Letters, vol. 14, no. 4, pp. 2181–2188, 2014, doi: 10.1021/nl500618u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Alyami MZ et al. , “Cell-Type-Specific CRISPR/Cas9 Delivery by Biomimetic Metal Organic Frameworks,” Journal of the American Chemical Society, vol. 142, no. 4, pp. 1715–1720, 2020, doi: 10.1021/jacs.9b11638. [DOI] [PubMed] [Google Scholar]
- [102].Elkhoury K, Koçak P, Kang A, Arab-Tehrany E, Ward JE, and Shin SR, “Engineering smart targeting nanovesicles and their combination with hydrogels for controlled drug delivery,” Pharmaceutics, vol. 12, no. 9, pp. 1–24, 2020, doi: 10.3390/pharmaceutics12090849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Kamerkar S et al. , “Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer,” Nature, vol. 546, no. 7659, pp. 498–503, 2017, doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Li S et al. , “Engineering macrophage-derived exosomes for targeted chemotherapy of triple-negative breast cancer,” Nanoscale, vol. 12, no. 19, pp. 10854–10862, 2020, doi: 10.1039/d0nr00523a. [DOI] [PubMed] [Google Scholar]
- [105].Lu M and Huang Y, “Bioinspired exosome-like therapeutics and delivery nanoplatforms,” Biomaterials, vol. 242, no. February, p. 119925, 2020, doi: 10.1016/j.biomaterials.2020.119925. [DOI] [PubMed] [Google Scholar]
- [106].Lv Q et al. , “Thermosensitive Exosome–Liposome Hybrid Nanoparticle-Mediated Chemoimmunotherapy for Improved Treatment of Metastatic Peritoneal Cancer,” Advanced Science, vol. 7, no. 18, pp. 1–13, 2020, doi: 10.1002/advs.202000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Sato YT et al. , “Engineering hybrid exosomes by membrane fusion with liposomes,” Scientific Reports, vol. 6, no. February, pp. 1–11, 2016, doi: 10.1038/srep21933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Lin Y et al. , “Exosome–Liposome Hybrid Nanoparticles Deliver CRISPR/Cas9 System in MSCs,” Advanced Science, vol. 5, no. 4, pp. 1–9, 2018, doi: 10.1002/advs.201700611. [DOI] [PMC free article] [PubMed] [Google Scholar]