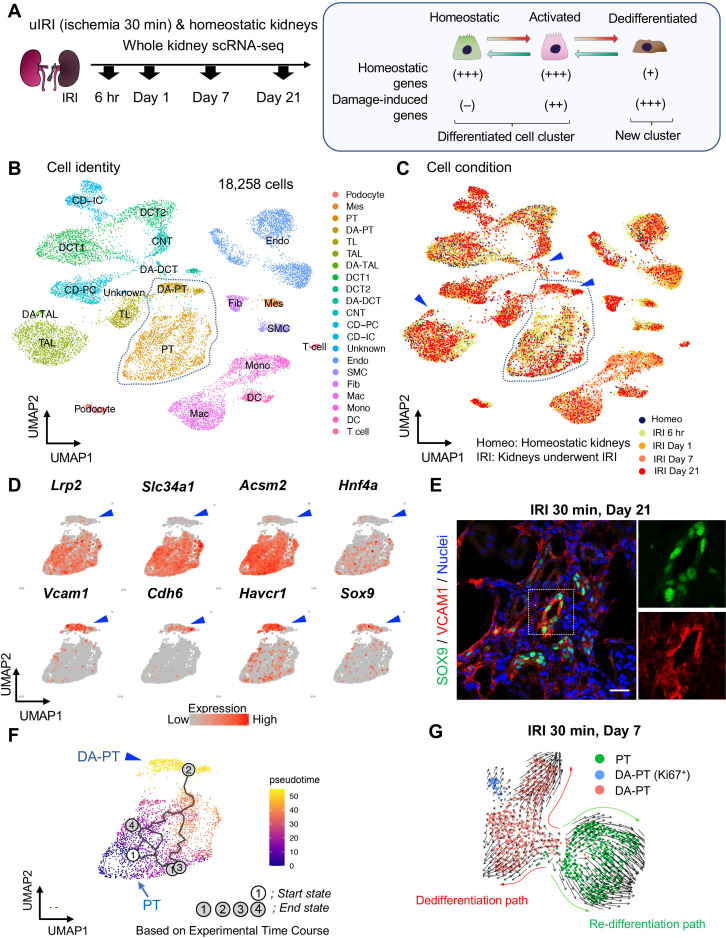
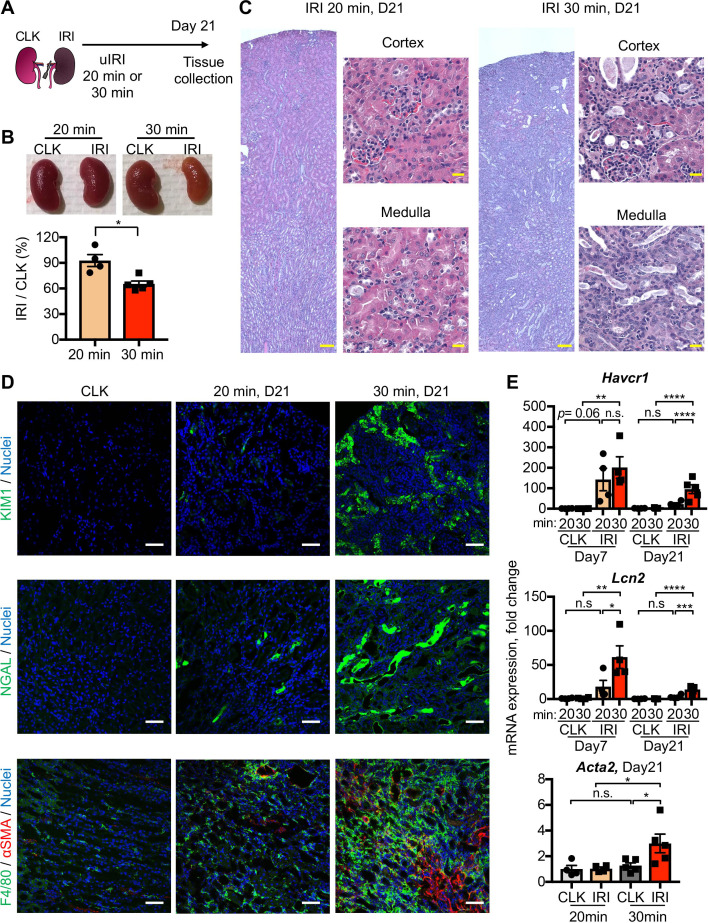
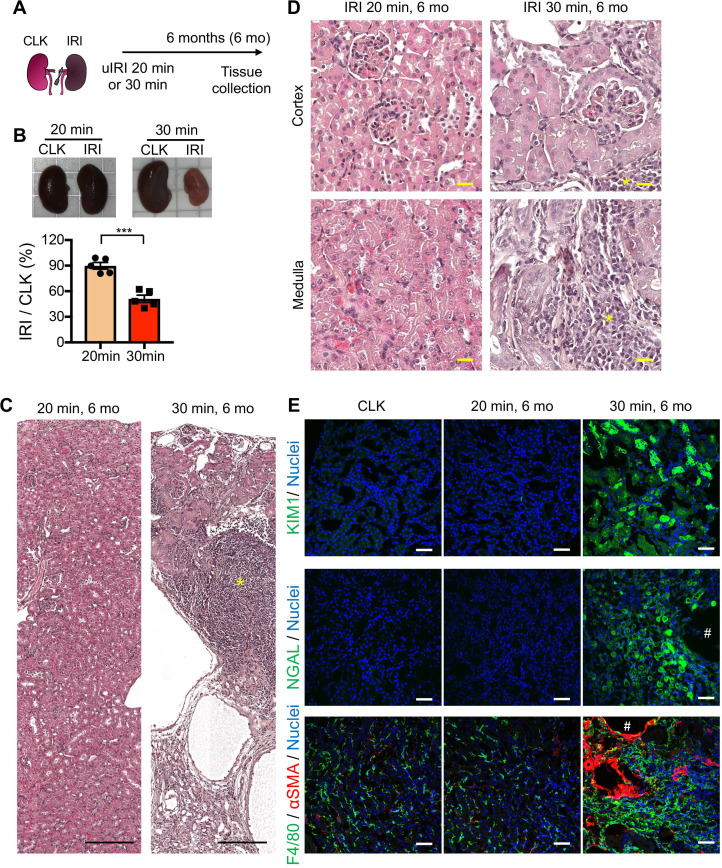
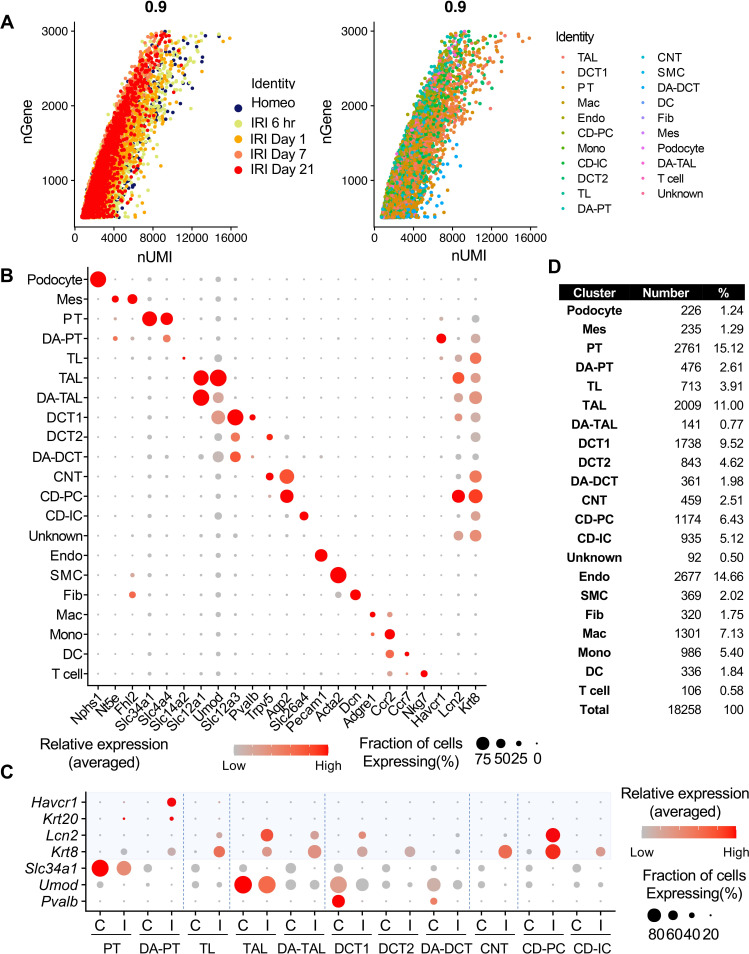
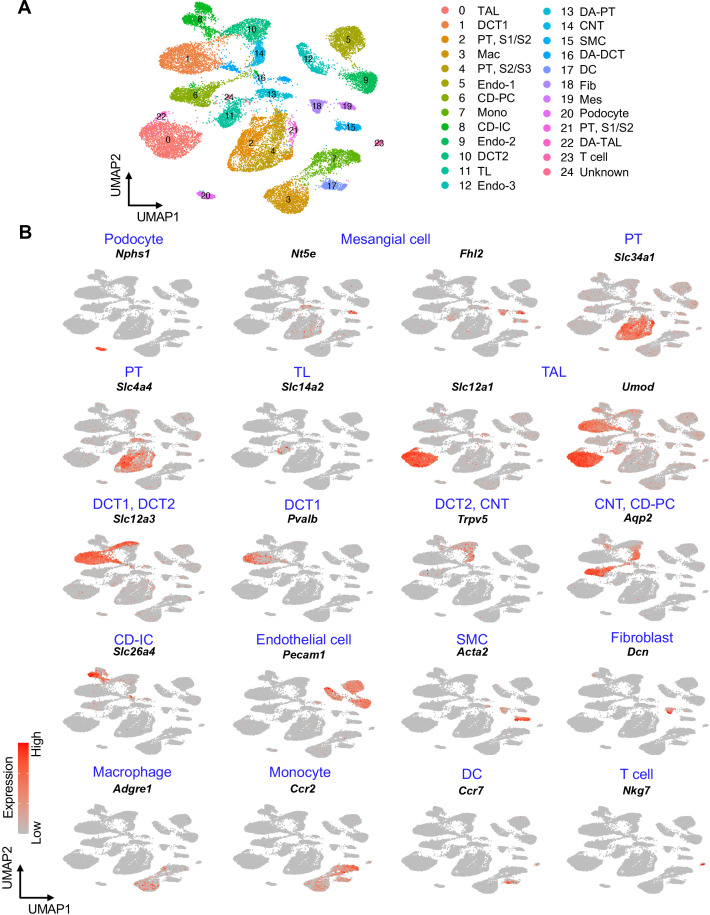
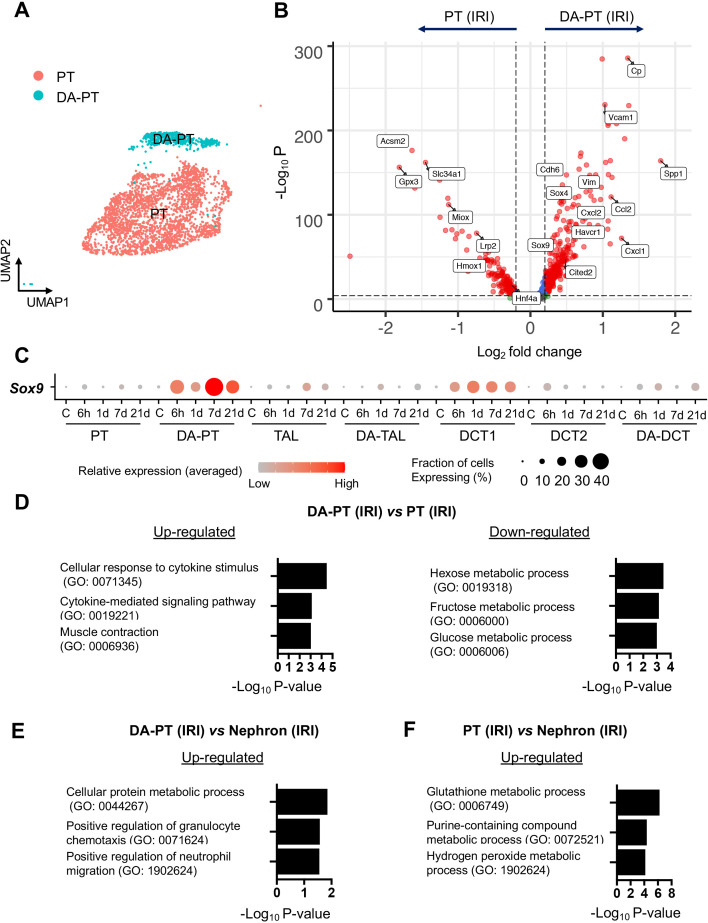
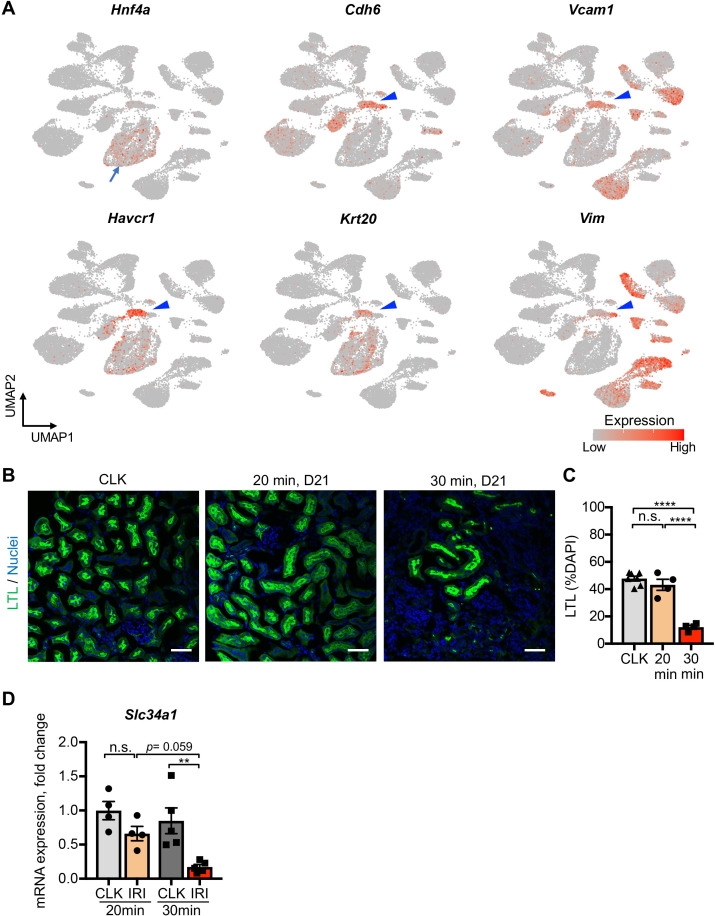
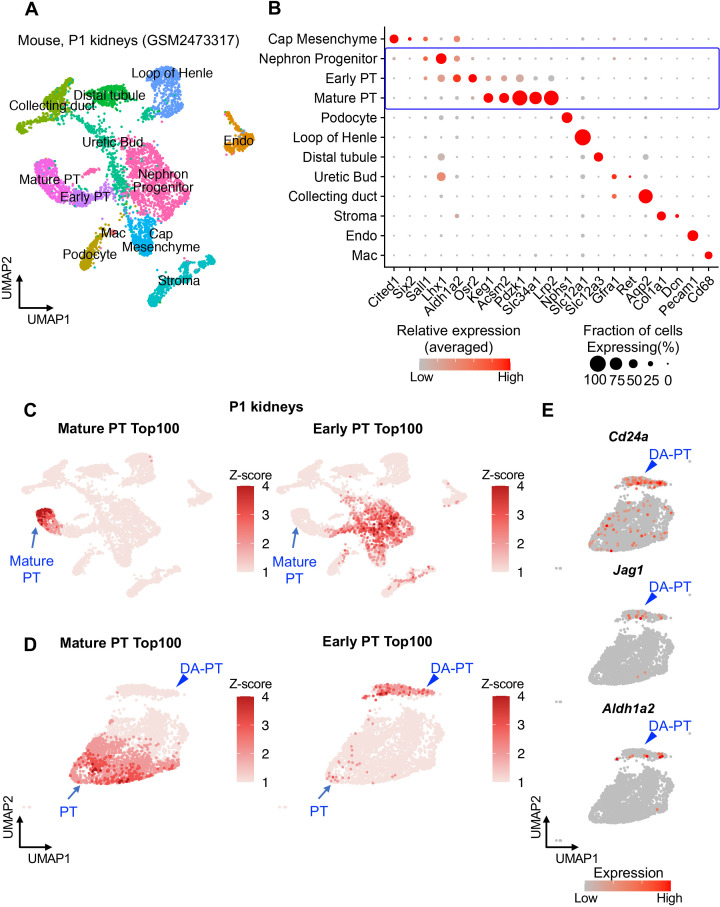
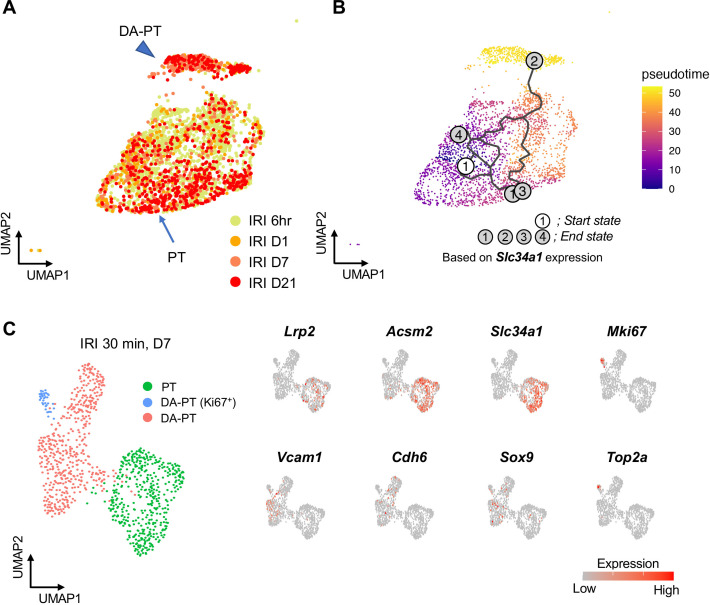
Figure 1. Single-cell RNA sequencing (scRNA-seq) identifies dynamic cellular state transitions of tubular epithelial cells after severe IRI.
(A) Drop-seq strategy. uIRI, unilateral IRI. A schematic illustration of epithelial cell states is shown. (B) and (C) Integrated single-cell transcriptome map. Unsupervised clustering identified 21 distinct clusters in the UMAP plot. Arrowheads indicate damage-associated tubular epithelial cells. The dotted area (PT cell clusters; PT and DA-PT) was used for the downstream analyses in (D)–(G). (D) UMAP plots showing the expression of indicated genes in PT cell clusters (PT and DA-PT in (B)). Differentiated/mature PT cell markers: Lrp2 (megalin), Slc34a1 (sodium-dependent phosphate transporter 2a, NaPi2a), Acsm2 (acyl-coenzyme A synthetase), and Hnf4a (hepatocyte nuclear factor 4α); and damage-induced genes: Vcam1 (vascular adhesion molecule 1), Cdh6 (cadherin 6), Havcr1 (kidney injury molecule-1, KIM1), Sox9 (Sry-box 9). Arrowheads; DA-PT. (E) Immunostaining for SOX9 and VCAM1 using post-severe IRI kidneys on day 21. Scale bar: 20 μm. (F) Pseudotime trajectory analysis of proximal tubular cells (PT and DA-PT clusters) that underwent IRI. A region occupied with cells from 6 hr after post-IRI was set as a starting state. (G) RNA velocity analysis of PT clusters (PT and DA-PT) from post-IRI kidneys on day 7. Cells in PT clusters from IRI day 7 dataset was extracted for the analysis. The arrows indicate predicted lineage trajectories. PT, proximal tubule; DA-PT, damage-associated PT; TL, thin limb; TAL, thick ascending limb; DA-TAL, damage-associated TAL; DCT, distal convoluted tubule; DA-DCT, damage-associated DCT; CNT, connecting tubule; CD, collecting duct (P, principal cells, IC, intercalated cells); Mes, mesangial cells; Endo, endothelial cells; SMC, smooth muscle cells; Fib, fibroblasts; Mac, macrophages; Mono, monocytes; DC, dendritic cells.