Abstract
The spike protein of the SARS-CoV-2 virus is the foremost target for the designing of vaccines and therapeutic antibodies and also acts as a crucial antigen in the assessment of COVID-19 immune responses. The enveloped viruses; such as SARS-CoV-2, Human Immunodeficiency Virus-1 (HIV-1) and influenza, often hijack host-cell glycosylation pathways and influence pathobiology and immune selection. These glycan motifs can lead to either immune evasion or viral neutralization by the production of cross-reactive antibodies that can lead to antibody-dependent enhancement (ADE) of infection. Potential cross-protection from influenza vaccine has also been reported in COVID-19 infected individuals in several epidemiological studies recently; however, the scientific basis for these observations remains elusive. Herein, we show that the anti-SARS-CoV2 antibodies cross-reacts with the Hemagglutinin (HA) protein. This phenomenon is common to both the sera from convalescent SARS-CoV-2 donors and spike immunized mice, although these antibodies were unable to cross-neutralize, suggesting the presence of a non-neutralizing antibody response. Epitope mapping suggests that the cross-reactive antibodies are targeted towards glycan epitopes of the SARS-CoV-2 spike and HA. Overall, our findings address the cross-reactive responses, although non-neutralizing, elicited against RNA viruses and warrant further studies to investigate whether such non-neutralizing antibody responses can contribute to effector functions such as antibody-dependent cellular cytotoxicity (ADCC) or ADE.
Keywords: SARS-CoV2, Influenza, Cross-reactivity, HIV-1, Glycan dependent, Neutralizing antibodies
1. Introduction
The ongoing SARS CoV-2 pandemic is devastating and has spread its grip worldwide. The spread of this disease has brought about a revolution in the field of vaccinology; the early development of diverse vaccines, a number of which are under clinical trials and few of these are available for emergency use [1]. There are only a few studies that have evaluated commonalities in the immune responses elicited against different corona viruses, other common respiratory viruses (influenza or RSV) or similar enveloped RNA viruses like HIV-1 [2], [3]. The tendency to exhibit host-derived glycans is a common feature of class 1 fusion proteins such as SARS-CoV-2 spike, HIV-1 Env glycoprotein (gp160) and Influenza HA [4]. Such host derived glycan motifs can serve as the basis for antibody mediated cross-reactivity, or provide mechanisms for viral escape. Glycan directed cross-reactive antibodies can have significant implications for viral neutralizing activity, ADCC mediated protection or ADE of infection.
Numerous retrospective studies have shown that influenza vaccines may enhance cross-protection and responsiveness to COVID-19 [5], [6]. However, the mechanisms behind these cross-reactive immune responses and their co-relations are poorly understood. Contrastingly, a few studies have shown that influenza vaccines have no synergistic or divergent effects on heterologous diseases, such as non-influenza respiratory virus infection (rhinovirus and coxsackie/echovirus infection) [7], [8], [9].
Both coronaviruses and influenza viruses are single-stranded, enveloped RNA viruses and both are nucleoprotein-encapsulated. However, the genomes of these two viruses vary in polarity and segmentation. The Influenza virus consists of 8 single-stranded, negative, viral RNA segments, whereas, SARS-CoV-2 is a single-stranded, non-segmented, positive-sense, RNA virus. The SARS CoV-2 infection involves a series of conformational changes in the spike (S) protein, which leads to membrane fusion following binding to the host receptor. However, this process requires appropriate activation of the spike protein by host proteases. The furin protease site between S1 and S2 subunits of the SARS-CoV-2 S protein is homologous to the highly pathogenic influenza viruses [10].
The envelope proteins of SARS-CoV2 and influenza have evolved to be extensively glycosylated and these glycans are derived from host-cells. The envelopes of these viruses fuse with the host cell utilizing a Class I fusion mechanism, which does not require any other viral surface proteins for fusion [4]. The glycan shield on these viruses provides diverse structural and functional features which help with the viral life-cycle and immune evasion mechanisms by misdirecting the humoral immune response to target non-neutralizing epitopes. Glycan density is especially high in some of the class I fusion proteins [11], which is consistent with their role in shielding. Differences in the composition, density, and conservation of glycans have been observed across distinctive families of enveloped viruses, e.g. HIV-1, SARS, influenza virus, Lassa, Zika, dengue, and Ebola viruses [12], [13]. However, cross-reactive responses of newly emerged SARS-CoV-2 antibodies and their potential protective or adverse responses are poorly understood.
In this study, we aimed to investigate the reactivity of the SARS-CoV-2 directed antibodies, present in convalescent donor sera and spike immunized mice sera, and investigated whether they confer cross-reactive protection against influenza virus, in terms of neutralization. Our findings highlight that SARS-CoV-2 specific antibodies in the convalescent sera are cross-reactive, although they do not exhibit any potential to cross neutralize, as shown by viral neutralization assays, ELISA and other immune-reactive studies. Further, we characterized the epitopes defining the cross- reactivity among the crucial viral targets studied herein, in an attempt to identify shared epitopes towards immunogen design and therapeutic targets against SARS-CoV-2. We believe our findings will aid in understanding whether the antibodies elicited during natural infection or through active immunization can provide protection against circulating infection or lead to disease enhancement as a long-term response in the event of future re-emergence or co-infections.
2. Material and methods
2.1. Human participant:
A longitudinal cohort of COVID-19 positive patients was enrolled at designated COVID-19 testing centers or hospitals within five days of their positive RT-PCR test. The study protocol was approved by the Institute Ethics Committees of all participating institutions. The eligibility criterion was a confirmed positive RT-PCR test for SARS-CoV-2 using nasopharyngeal swabs. These patients were enrolled after written consent and baseline active phase samples were collected, processed and archived at the NCR Biotech Science Cluster Biorepository for all subsequent analyses. The follow-up visits were designed to capture the clinical outcomes of illness (10–28 days after being diagnosed with SARS-CoV-2 infection), early (6–10 weeks) and late (6 and 12 months) convalescent periods. The duration of illness was defined as the date of onset of symptoms in symptomatic participants and from the date of testing positive for SARS-CoV-2 infection among those who were asymptomatic.
2.2. Expression and purification of recombinant proteins
Recombinant proteins were expressed using the Expi 293F expression system, from a codon-optimized nucleic sequence of RBD-His, Spike-His, HA-His, following the methodology published earlier [14], [15], [16]. Briefly, the culture supernatant was harvested 5–7 days post-transfection and purified by Ni-NTA affinity chromatography followed by dialysis in phosphate buffer saline (pH 7.4) as described in our previous papers [14], [15], [16], [17]. The N protein of SARS-CoV-2 was expressed in the bacterial expression system and was purified by Ni-NTA affinity chromatography.
2.3. Animal immunization and binding reactivity assays
Animal immunizations for RBD and spike proteins were performed on 7–8 week old, C57BL/6 (male) mice inbred in the THSTI small animal facility (SAF) with 5 animals immunized in each group based on prime/boost immunization regimen, as described in the the published literature [18]. For influenza HA and SARS-CoV-2 N protein, 6–8 week old BALB/c (male) mice, inbred in THSTI small animal facility (SAF) were immunized (i.m; intramuscular route) with 30 µg of the purified recombinant protein in combination with AddaVax as adjuvant in a prime/boost immunization regimen, prime and boost immunization was done 21 days apart. Pre-bleed sera was collected on day 0 and sera post immunization were collected 14 days after each immunization. For our studies, we used sera collected 14 days after the booster dose. All experiments were performed in accordance with the Guidelines of the CPCSEA, under the protocol approved by the Institutional Animal Ethics Committee (IAEC Approval number: IAEC/THSTI/53 and IAEC/THSTI/93).
2.4. ELISA binding assays
For ELISA binding assays, NUNC Maxisorp plates (Thermo Scientific) were coated with 100 μL of recombinant soluble proteins (RBD protein, bovine serum albumin, soluble S protein, and N –protein 2 µg mL−1) overnight in coating buffer, 0.1 M Sodium Carbonate, pH 9.5 at 4 °C. Next day, the ELISA plate was washed and blocked with 250 µL of 5% (wt/vol) non-fat milk in PBS (MPBS) for 90 m at room temperature (RT). Serum samples, 100 μL each, of dilution ranging from 150 to 328,050 were used for the ELISA assays. The serum dilutions were prepared in 5% MPBS. HRP conjugated secondary antibodies (anti-Mouse IgG Fc, Code: 115–035-003, Anti-Human HRP, Code: 109–035-003, Jackson Immunoresearch) were used in 1:2000 dilutions for 1 h at RT. Following the primary and secondary antibody steps, the plates were washed six times with 0.1% Tween-20 in PBS. The ELISA reaction was developed with a tetramethylbenzidine substrate (TMB, Thermo Cat. No. N301) for colour reaction. In competition ELISA binding assay, all steps were the same except plates were coated overnight as described above and evaluation of cross-reactivity to HA and spike proteins was assessed in the presence of increasing concentrations of methyl-d-mannopyranoside (Sigma-Aldrich Cat. No. M6882).
2.5. Coating of magnetic beads and depletion flow cytometry assays
Purified soluble H1-HA was covalently coupled to Tosylactivated MyOneDynabeads (Life Technologies Inc. Cat. No 65,501) according to the manufacturer's protocol as described by Patil et al; 2016 [16]. For depletion studies, SARS-CoV-2 infected human serum (neutralization CPEE titre 3200) was diluted to 1:50 in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS). Diluted serum (800 µL) along with magnetic beads were incubated at room temperature for 30–60 mins. Unbound plasma antibodies were separated from bound antibodies to protein-coated beads using a DynaMag 15 magnet as described above. This step was repeated 5 times for the depletion of serum antibodies. As a negative control, serum antibodies were depleted from BSA coated beads in parallel.
2.6. Flow-cytometry based assay for cross-reactive binding
Purified soluble H1-HA was covalently coupled to Tosylactivated MyOneDynabeads (Life Technologies Inc.) according to the manufacturer's protocol as described by Patil et al., 2016 [16]. In the mice study, HA-coated beads were incubated for 1 h at RT with the sera from HA, spike, and SARS-CoV-2 nucleoprotein (N) immunized groups. For control, pre-immune mice sera and beads with a secondary antibody were included. The beads were incubated with anti-mouse secondary antibody labelled with Alexa Fluor 488 (Jackson ImmunoResearch, Code: 115–545-062), post two washes with 1% FACS buffer. Beads were finally washed twice and re-suspended in 1% FACS buffer and data were acquired in BD FACS Canto-II flow-cytometer.
For the human study, HA-coated beads were incubated with sera from SARS-CoV-2 positive patients for 1 h at RT. For control, beads were added with secondary antibody only. The staining procedures followed were the same as described above. The secondary antibody used was anti-human PE (Jackson ImmunoResearch, Code:109–116-097). Flow data were analysed using FlowJo software and statistical analysis was done by applying ‘t-test’ using GraphPad Prism 7 software.
2.7. Protein’s enzymatic de-glycosylation:
Protein de-glycosylation was performed by PNGase F (NEB, Cat. No P0705S) (Non-Denaturing Reaction Conditions) and Endo H (NEB, Cat. No P0702S) following the manufacturer’s protocol. Briefly, 20 µg of purified dialyzed influenza HA and SARS-CoV-2 spike protein was mixed with Glyco Buffer 2 in 20 µL volume of the reaction mixture. 2 µL of PNGase F was added to the final reaction and reaction mixture was incubated at 37 °C for 4 h. As a control, to estimate the extent of de-glycosylation, one protein reaction with Glyco Buffer 2 and without PNGase F was also incubated at 37 °C for the same time period. Both control and enzymatic reaction samples were further run on SDS-PAGE and the extent of deglycosylation was estimated by the shift in mobility of protein bands.
For de-glycosylation, using Endo H, 20 µg of proteins was mixed with Glycoprotein Denaturing Buffer in 10 µL of reaction volume and incubated at 100 °C for 10 mins. The denatured protein was further mixed with Glyco Buffer 3 and 2 µL of Endo H and reaction mix was incubated at 37 °C for 2 h.
2.8. Cytopathic effect (CPE) based SARS-CoV-2 neutralization assay
CPE based neutralization assays were performed as described previously in Parray et al., 2020 [14]. Briefly, 1 × 102 TCID50 isolate; USA-WA1/2020 virus, passaged once in Vero cells, was incubated with serum dilution ranging from 1:20 to 3260 for 90 mins, followed by 1 h of adsorption on the Vero cells. After washing the cells, DMEM supplemented with 2% (vol/vol) FBS was added. The presence of cytopathic effect (CPE) in cells was detected using a microscope after incubation for 4–5 days at 37 °C with 5% CO2. Non-infected VERO E6 cells were used as a positive control, and infected VERO E6 cells were used as a negative control.
2.9. Immunofluorescence (IF) staining of MDCK cells
2.9.1. Cells
Madin-Darby canine kidney (MDCK) cells (ATCC® CCL-34TM) were grown in Advanced minimal essential medium (Advance MEM, Gibco, United States) supplemented with 10% fetal bovine serum (FBS; 10082–147, Gibco, United States), GlutaMAX (Cat No.35050061, Gibco, United States), penicillin G sodium (CAS No. 69–57-8, Sigma-Aldrich) 100 units/ml and streptomycin sulfate (Cat. No. 15140122, Thermo Fisher Scientific)100 μg/ml.
2.9.2. Virus infection
Laboratory strain PR8 virus [A/Puerto Rico/8/34 (H1N1)] was used in this research. The multiplicity of infection (MOI) was calculated using the virus titer determined by a plaque assay. In a 6-well plate, cells were expanded to 80% confluency before being inoculated with influenza virus at 0.002 MOI. After adsorption for 1 h at 37 °C, the inoculum was removed and the cells were incubated in medium supplemented with BSA (0.3%) and TPCK-trypsin (1 µg mL−1).
2.9.3. Immunofluorescence
After 40 h of PR8 infection in MDCK cells, cells were fixed in 4% paraformaldehyde for 15 min, then permeabilized with 0.1% Triton X-100 in PBS for 10 min. Non-specific binding was blocked using 3% goat serum in PBS for 1 h at room temperature. Cells were then probed with primary antibodies; anti-Influenza A virus, PR8 mice sera (1:100) and 313 (1:2000) and incubated at RT for 1 h. After incubation, cells were washed followed by incubation with secondary antibodies, including Alexa488-labeled rabbit anti-mouse IgG (1:1000), and anti-human-PE (1:300) for 1 h at room temperature. The nuclei of cells were counterstained with DAPI (D9542, Sigma-Aldrich, United States) for another 10 min at room temperature. The expression of proteins was observed by fluorescence microscope (IX-71, Olympus) at 60X magnification.
3. Results
3.1. Cross-reactive binding of SARS-CoV-2 antibodies to influenza HA surface glycoproteins
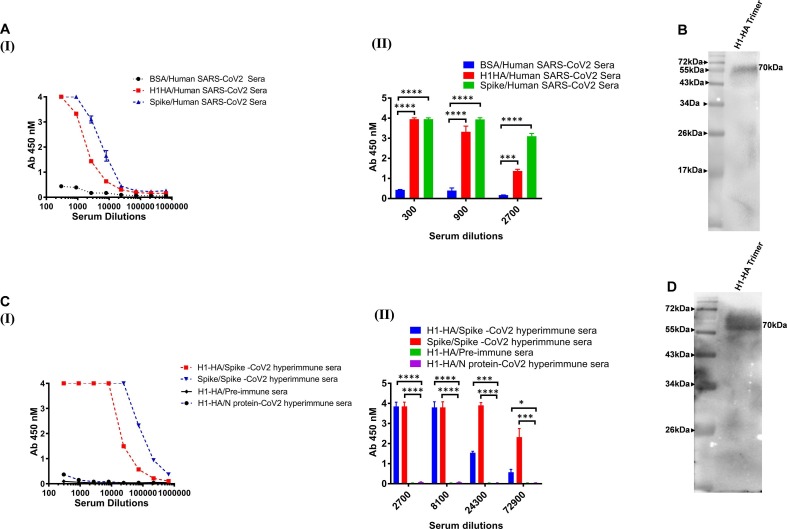
As both SARS-CoV-2 and the influenza virus show similar clinical presentations, we first investigated any possible correlation between the two diseases in terms of reactivity or cross-reactivity. We selected SARS-CoV-2 polyclonal sera obtained from SARS-CoV-2 infected human subjects with high titres of neutralizing antibodies (cytopathogenic effect value, CPE 3200), to evaluate them for cross binding to influenza HA proteins by ELISA. Interestingly, the SARS-CoV-2 human polyclonal sera showed a varied degree of cross-reactivity with influenza HA protein (Fig. 1 A (I) (II)). The cross-reactivity of the polyclonal sera was further confirmed by western blot analysis. Detection of ~ 70 kDa band for HA protein substantiate the cross-reactive binding of the SARS-CoV-2 polyclonal sera with HA proteins (Fig. 1B).
Fig. 1.
Cross-reactive binding of SARS-CoV-2 antibodies to influenza HA surface glycoproteins. A (I and II). Human SARS-CoV-2 infected sera was used in ELISA to assess antibody binding and cross-reactivity with HA proteins. BSA coated wells were used as a negative control in the ELISA binding assay. B). His-tagged HA was detected by Western blot analysis using convalescent sera of a human SARS-CoV-2 infected donor as the source of primary antibody, followed by using an HRP conjugated anti-Fc antibody. C & D). Polyclonal sera from the spike immunized mice was tested for its cross-reactive binding to HA protein in ELISA and Western blot analysis. Nucleoprotein (N) protein of SARS-CoV-2 immunized mice sera and pre-immune sera was used as experimental negative control. The binding and immunoblot assays were repeated at least three times. Data are presented as the mean ± SD, and differences between groups were determined by two-way analysis of variance (ANOVA) followed by Tukey’s post hoc tests using GraphPad Prism 7. Statistical significance between the control and different groups is shown as *P < 0.05, **P < 0.01, ***P < 0.0002, ****P < 0.0001.
To further confirm the above results, we investigated the binding of the immune sera, from SARS-CoV-2 spike protein (prefusion spike trimer S2P) immunized mice, and cross-reactivity with influenza HA proteins (Fig. 1C (I) (II)). The hyperimmune spike sera from mice also showed a similar cross-reactive binding pattern to influenza HA protein both in ELISA and western blot analysis (Fig. 1D). To test the specificity of this cross-reactivity and rule out any pre-existing cross-reactive antibodies in mouse polyclonal sera, pre-immune sera from mice and sera from mice immunized with the SARS-CoV-2 nucleoprotein (N) were used as a negative control. Neither the pre-immune sera nor the sera from the N-immunized mice showed any reactive binding to the tested proteins (Fig. 1C).
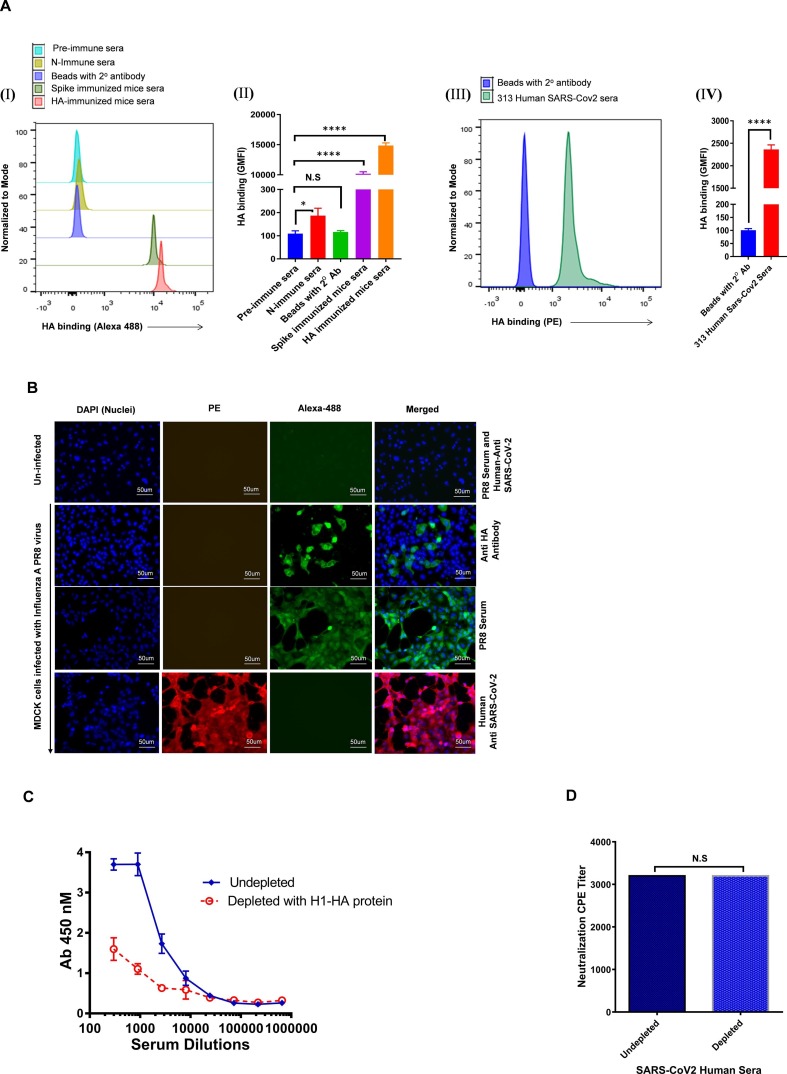
The above findings were confirmed by a flow cytometry-based assay in which purified soluble HA protein was coated on magnetic beads and tested for reactivity against SARS-CoV-2 convalescent sera and spike protein immunized mice sera. Similar to the ELISA results, we found that the HA coated beads produced significant positive signals with both human and mice immunized sera when compared to sera from N immunized mice (used as control), which did not show any reactivity with the coated beads (Fig. 2 A I, II, III, IV & Fig. S1).
Fig. 2.
Cross reactive potential of spike polyclonal antibodies. A). (i) Histogram showing binding of various mice sera with HA-coated beads. (ii) GMFI data for the binding of HA-coated beads with various mice serum samples. (iii) Representative histogram showing binding of human SARS-CoV-2 infected convalescent sera with magnetic beads bound full-length HA protein. (iv) GMFI data for the binding of HA-coated beads with human SARS-CoV-2 serum sample. All the experiments were done in duplicate and repeated at least two times. B). Cross-reactivity of antibody against SARS-CoV-2 to influenza A virus surface proteins. Localization of influenza A virus surface proteins using a panel of anti-influenza antibodies. MDCK cells infected with laboratory Influenza A strain-PR8 after 40 h of infection cells were fixed and permeabilized. Un-infected cells were co-stained with PR8 infected mouse polyclonal serum and anti-SARS-CoV-2 infected human polyclonal serum. PR8 infected cells were stained with anti-HA mouse antibody and PR8 infected mouse polyclonal serum, counterstained with Alexa fluor-488 tagged anti-mouse secondary antibody (green). In another set, PR8 infected MDCK cells were stained with primary 313 anti-SARS-CoV-2 infected human polyclonal serum and counterstained with PE-tagged anti-human secondary antibody. The nuclei were stained with DAPI, 4′,6-diaminidino-2-phenylindole (blue). Scale bar: 50 μm and magnification 60x. C). The extent of binding of the depleted and undepleted Human SARS-CoV2 infected plasma with magnetic beads coated with H1-HA protein was accessed by ELISA. D). Degree of the shift in sensitivity of Wuhan SARS-CoV2 viruses to plasma depleted with HA protein. The value in the graph represents the CPE value. No change in neutralization of the plasma sample was observed in depleted and undepleted samples. The binding assay was done in duplicate and repeated at least three times. Statistical significance was determined using ‘t-test’. *P < 0.05, **P < 0.01, ***P < 0.0002, ****P < 0.0001 and N.S (non-significant).
Further, to assess the cross-reactivity of SARS-CoV-2 serum against Influenza A, we performed immunofluorescence (IF) staining of MDCK cells infected with live Influenza A-PR8 virus. The SARS-CoV-2 convalescent sera showed strong binding reactivity with the Influenza A PR8 virus infected cells. Simultaneously, incubation of the PR8 infected MDCK cells with immune sera from PR8 infected mouse and anti-HA antibody also showed binding reactivity with the structural proteins of the Influenza A virus efficiently. In another set, when we probed un-infected cells with PR8 infected mice serum and SARS-CoV-2 infected human serum, no staining was observed, serving as an experimental negative control (Fig. 2B). Our data suggests that this cross-reactivity of anti-SARS-CoV-2 human sera or hyperimmune mice sera is due to the antibody responses directed towards the spike protein of SARS-CoV-2 and the possible presence of shared epitopes between SARS-CoV-2 and the Influenza A virus.
3.2. Cross-reactive anti-SARS-CoV-2 antibodies do not show cross-neutralization
Next, we investigated whether these anti-SARS-CoV-2 cross-reactive antibodies confer any cross-neutralization. To validate the presence or absence of cross-neutralization potential of the cross-reactive anti-SARS-CoV-2 antibodies, we performed serum depletion assays, where HA cross-reactive binding antibodies were depleted from the convalescent serum of a SARS-CoV-2 infected donor that demonstrated potent anti-SARS-CoV-2 neutralizing activity (Fig. 2C). Depletion was performed using Dynabeads coated with purified H1-HA protein. The depletion of SARS-CoV-2 infected human serum of antibodies directed at HA trimer protein did not show any change in neutralization potency of the polyclonal antibodies towards the SARS-CoV-2 virus, suggesting that cross-reactive binding antibodies to HA proteins do not confer cross-neutralization (Fig. 2D).
3.3. Cross-reactive binding of HA immunized mice sera to SARS-CoV-2 spike proteins
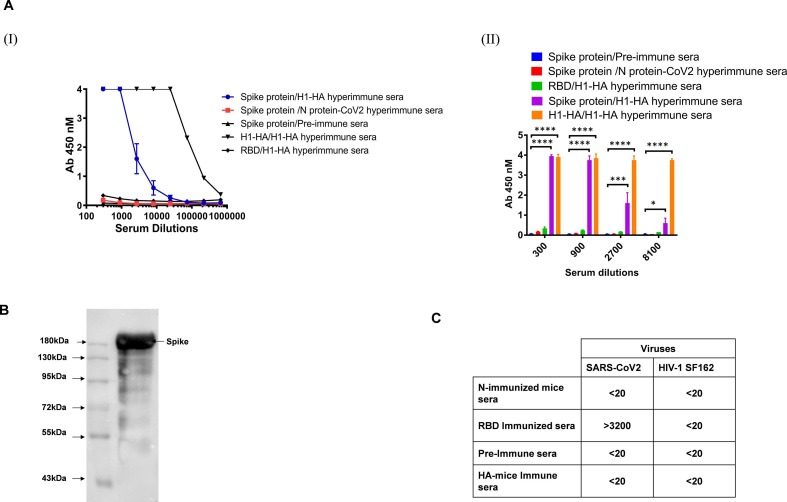
We further investigated the possible bidirectional cross-reactivity of influenza HA immunized sera with SARS-CoV-2 proteins. The mice were immunized with the purified H1-HA protein. We observed that the HA hyperimmune sera showed cross-reactive binding towards the spike protein of SARS-CoV-2 both in ELISA (Fig. 3 A (I) (II)) and in a Western blot (~180 kDa band size) (Fig. 3B). Interestingly, HA polyclonal antibodies did not cross-react with the receptor binding domain (RBD) or the N protein of SARS-CoV-2 as compared to the spike in the ELISA binding assay. These findings support the hypothesis that antibodies in the sera of HA-immunized mice target non-RBD epitopes in the spike protein. The hyperimmune sera of N protein of SARS-CoV-2 immunized mice and sera of pre-immunized mice showed no reactivity to the full length full length spike or RBD proteins. Our experimental findings show that the cross-reactivity in binding is common between SARS-CoV-2 and HA immunized sera and is bidirectional, with no reactivity towards other proteins tested in the present experimental setup, suggesting some commonality of the epitopes between the HA and SARS-CoV-2 spike protein, excluding the RBD region.
Fig. 3.
Cross reactive binding and accessment of cross-neutralization potential of HA polyclonal antibodies. A (I and II) & B). The H1-HA immunized mice polyclonal serum was tested for its cross-reactive binding to Spike proteins in ELISA and Western blot analysis. Nucleoprotein (N) protein of SARS-CoV-2 immunized mice sera and pre-immune sera were used as an experimental negative control. C). Cross neutralization potential of sera was tested in CPE based assay. The value represents the 100% serum neutralization titers. The neutralization assay was done in duplicate and repeated at least three times. Statistical significance between the control and other experimental groups was estimated by two-way analysis of variance (ANOVA) followed by Tukey’s post hoc tests using GraphPad Prism 7. Statistical significance between control and different groups is shown as *P < 0.05, **P < 0.01, ***P < 0.0002, ****P < 0.0001.
3.4. Cross-reactive anti-HA antibodies did not show cross-neutralizing activity against SARS-CoV-2
We next sought to determine the possible cross-neutralization capabilities of these cross-reactive antibodies. HA immunized hyperimmune sera were tested against SARS-CoV-2 virus in a CPE based virus neutralization assay. None of the tested sera showed any reduction in cytopathic effect at 1:20 serum dilution (Fig. 3C).
3.5. Cross-reactive antibodies target glycan epitopes
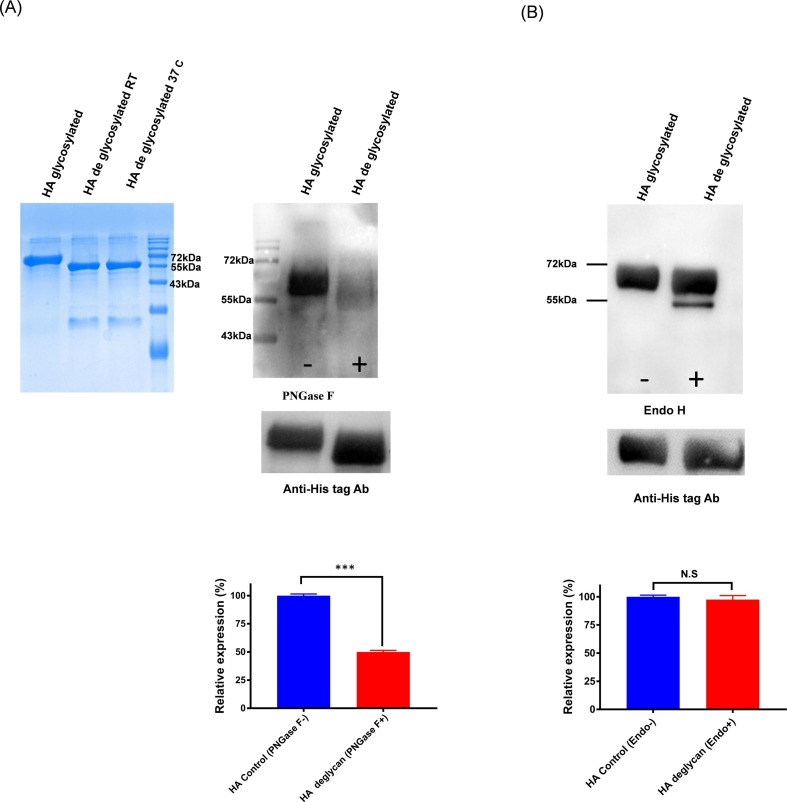
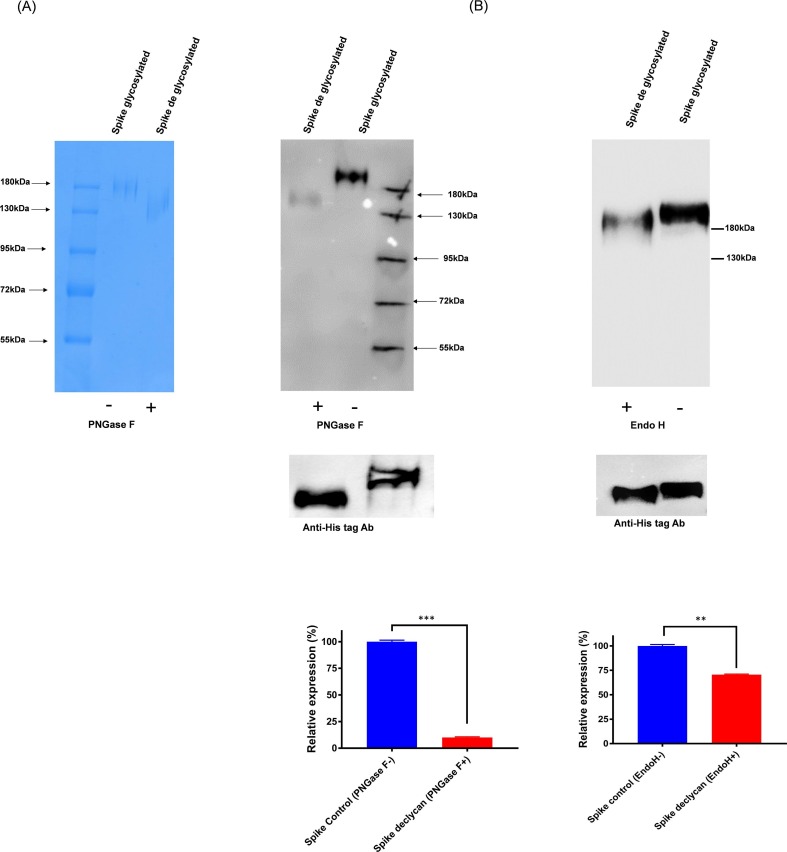
To confirm and investigate the epitopes shared by SARS-CoV-2 and influenza HA proteins, which directing the cross-reactivity among the two group of proteins, we studied the possible contribution of glycosylation. We found that deglycosylation of HA protein shows selectively decreased reactivity towards SARS-CoV-2 polyclonal antibodies in a western blot (Fig. 4 A). Whereas the fully glycosylated HA Envs bind better with the SARS-CoV-2 polyclonal sera than PNGase H deglycosylated Envs. In a similar manner, the spike protein was deglycosylated and tested for HA immune sera cross-reactivity. The HA immune sera showed reduced binding reactivity towards the PNGase H deglycosylated spike protein (Fig. 5 A). This data suggests that cross-reactive binding antibodies present in HA and spike immune sera show bi-directional reactivity and are targeted towards glycans.
Fig. 4.
A). Role of glycosylation in cross-reactive antibody binding. H1-HA purified protein was deglycosylated with PNGase F enzyme and Western blot analysis was performed with the spike immunized mice sera. The spike immunized mice sera preferentially bind with the glycosylated form of HA protein and poorly react with de-glycosylated protein. In parallel, as a control experiment, equal amount of protein was loaded and probed with Anti-His Tag antibody. B). H1-HA purified protein was deglycosylated with endo H enzyme and Western blot analysis was performed with the spike immunized mice sera. The spike immunized mice sera bind with both the glycosylated and endo H de-glycosylated protein. The relative expression/decrease in binding is calculated by densitometry analysis and is represented in bottom panel as bar diagram. Statistical significance was determined using t-test and p < 0.05 was considered significant and N.S (non-significant).
Fig. 5.
Effect of de glycosylation on cross reactive binding A). The spike protein was de-glycosylated with the PNGase F enzyme and Western blot analysis was performed with H1-HA immunized mice sera. The H1-HA mice preferentially bind with the glycosylated form of spike protein and poorly reacts with PNGase F deglycosylated protein. The immunoblot assay was repeated at least two times. As a control experiment same blot/in parrel, equal amount of both proteins were loaded and probed with Anti-His tag antibody, B). In this, the spike protein was de-glycosylated with the endo H enzyme and Western blot analysis was performed with H1-HA immunized mice sera. The H1-HA mice bind with both the glycosylated and endo H deglycosylated proteins. The immunoblot assay was repeated at least two times. As a control experiment same blot/in parrel, equal amount of both proteins were loaded and probed with Anti-His tag antibody. The relative expression/decrease in binding is calculated by densitometry analysis and is represented in bottom panel as bar diagram. Statistical significance was determined using t-test and p < 0.05 was considered significant.
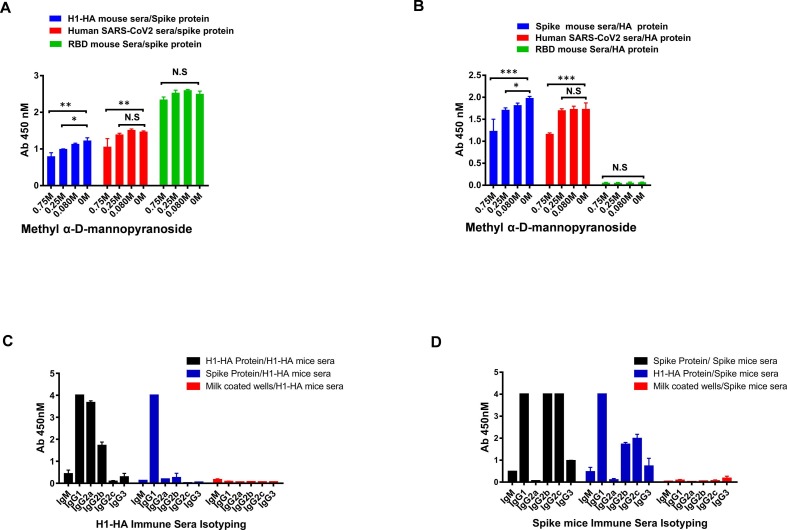
To further elucidate the role and the extent of involvement of complex glycosylations in cross-reactivity, we treated both the HA and spike protein with the Endo H enzyme; that selectively removes high mannose and hybrid glycans, and tested for its bi-directional cross-reactivity. The immune sera shows minimal difference in the recognition of Endo H treated and untreated proteins in the western blots, the Endo H treated HA protein shows significantly less changes in the recognition of spike sera as compared to treated spike protein, which shows slightly reduced reactivity (30%) with the HA sera (Fig. 4B & 5B). However, when tested in a methyl α-d-mannopyranoside (a stabilized mannose analogue) ELISA-based competition assay for the cross-reactive interactions, both the protein shows partial disruption of the binding with the sera at the highest concentrations of methyl-d-mannopyranoside (750 mM), indicating that these interactions are moderately susceptible (~35–40 % inhibition) in the presence of mannose analogue (Fig. 6 A & B). Hence, our results suggest that the cross-reactive antibody responses are preferentially directed towards N-linked complex type glycans with differential involvement of complex glycosylation.
Fig. 6.
Isotyping of cross reactive antibody responses. A & B). Cross-reactivity of binding was assessed in the presence of methyl α-d-mannopyranoside. Plates were coated with SARS-CoV-2 spike and H1-HA protein and incubated with dilutions of methyl α-d-mannopyranoside along with a constant amount of the indicated antibodies. Antibody binding was quantified via ELISA. All the experiments were performed three times separately. Data are presented as the mean ± SD, and differences between groups were determined by two-way analysis of variance (ANOVA) followed by Dunnett post test using GraphPad Prism 7. Statistical significance between control and different groups is shown as P > 0.05 (N.S not significant), *P < 0.05, **P < 0.01, and ***P < 0.0001. C & D). Isotyping of cross-reactive antibodies in the mice immune sera was tested in ELISA binding assay.
3.6. Characterization of IgG subclasses and their cross-reactivity
To investigate if any specific IgG subclasses are contributing to driving the mechanism of cross-reactive response, we evaluated the antibody isotypes and specificities of HA and spike hyperimmune sera against the cross-reactive protein. The IgG1 subclass was found to be the most dominant class of cross-reactive antibodies. The H1-HA mice polyclonal sera cross-reacts with spike protein and the most dominant class of cross-reactive antibodies were IgG1 (Fig. 6C). However, in spike immunized mice sera that cross-reactive antibody isotype with HA proteins was IgG1 followed by IgG2b, IgG2c and IgG3 (Fig. 6D). It will be important to study the antibody isotypes and subclasses of these cross-reactive antibodies generated in response to SARS-CoV-2 in the natural course of infection and their role in COVID-19 pathogenesis.
4. Discussion
Several recent large-cohort studies have shown that receiving an influenza vaccine shot before or shortly after contracting SARS-CoV-2 improved health outcomes and reduced the risk of contracting a severe COVID-19 infection [5], [19], [20]. Our study shows that non-neutralizing SARS-CoV-2 directed polyclonal antibodies (SARS-CoV-2 convalescent sera and spike immunized mice sera) demonstrate cross-reactivity with the HA glycans of influenza virus. However, there is no scientific report so far that elucidates the mechanism behind this possible cross-protection [21]. Our study describes that antibodies elicited against influenza HA protein; a major component of the flu vaccine, cross-react with the spike protein of SARS-CoV-2, though these cross-reactive antibodies do not show any direct protection in terms of neutralization of the SARS-CoV-2 virus. Evaluation of the ADCC activity of these antibodies will reveal if their effector functions can confer any protection, though it has not been addressed herein and is a limitation of the study. Using targeted antibody-depletion experiments, we demonstrated that SARS-CoV-2 antibodies that cross-react with HA antibodies are preferentially non-neutralizing to SARS-CoV-2. We found that these cross-reactive antibodies do not bind or bind poorly to the RBD protein. One possible reason for the non-neutralizing behaviour of these cross-reactive antibodies could be that these antibodies target non-RBD regions, as RBD is a highly immunogenic component of spike protein and is recognised by the majority of nAbs, and is, therefore, a major target of current nAb-based vaccine design efforts [22], [23]. Another possible explanation for the poor binding of these cross-reactive antibodies to the SARS-CoV-2 RBD could be the limited number of glycosylation moieties within the RBD. Any protection that these cross-reactive antibodies may confer, possibly by enhancing innate immune functions by binding to FcRs, including ADCC and complement activation, might help in the clearance of the virus and reduce the severity of the disease. In a study conducted by Zanettini et al, it was observed that the influenza vaccine had a positive effect on COVID-19 mortality in the elderly population and is supportive of our findings [24].
Viral envelope spike and HA proteins are heavily glycosylated with a variable array of host-derived glycans [25]. These glycans play an important role in viral defence mechanisms via epitope occlusion and host immune system evasion [26]. These glycans are immunogenic in nature and are reported to elicit potent neutralizing antibodies in the case of HIV-1 (2G12) [27] and SARS-CoV-2 spike protein (S309) [28]. In SARS-CoV-2, glycosylation of the spike protein is essential for viral infection and is important for viral escape and defence mechanisms [29]. The shedding of viral glycoproteins can redirect the humoral immune response by exposing immunodominant (non-neutralizing) epitopes, not exposed on the functional native closed trimeric conformation of the Env proteins, which plausibly leads to the production of cross-reactive binding antibodies which are, however, non-neutralizing [3], [30].
Studies conducted by Lander et al, have shown that SARS-CoV-2 infection elicits cross-reactive antibody responses towards the FP and HR2 epitopes of endemic CoVs, implying that B cell memory for these epitopes exists in the general population and hypothesised that these antibodies might exhibit cross neutralisation to SARS-CoV-2 [31], [32]. Several epidemiological studies have indicated a substantially higher prevalence of cross-reactive antibodies against SARS-CoV-2 that protect against SARS-CoV-2 infection in sub-Saharan African populations, with a much lower COVID-19-related morbidity and mortality rate, plausibly due to these regions having a higher incidence rate of infectious diseases [33]. In a similar study, it has recently been documented that a large fraction of non-exposed individuals show antibody cross-reactivity and T-cell reactivity to SARS-CoV-2 peptides. Such antibodies, which developed during the pre-pandemic period, are probably those elicited against homologous peptides shared by the endemic HCoVs and related viruses [34], [35]. Future studies need to address the functional implications of these cross-reactive antibody responses to understand how the history of an individual's exposure to the endemic virus can influence their immune response to COVID-19 infection and disease progression.
The findings of our study suggest that most of these cross-reactive antibodies target the glyco-epitopes (influenza HA). In dengue infection, it has been shown that human or hamster polyclonal cross-reactive antibody response against West Nile virus (WNV) is primarily directed against a cross-reactive domain II fusion loop epitope (DII-FL) on the envelope (E) protein; these cross-reactive antibodies were found to be poorly neutralizing. Passive transfer of these purified cross-reactive IgGs from DENV-immunized hamsters protects mice from lethal WNV infection via Fc receptor and complement-dependent effector mechanisms [36].
Furthermore, we discovered that HA immunized mice sera did not cross-react with the RBD of SARS-CoV-2 and could not neutralize the Wuhan live SARS-CoV-2 virus in a CPE assay.
The background signal in SARS-CoV-2 serological assays, leading to a false predictive value between antibody titers and viral neutralisation, disease status, and disease progression, may be attributed, at least partially, to such cross-reactive antibodies. Our results suggest that adding RBD and/or N protein to similar serological assay platforms, with no cross-reactivity, may reduce the number of false positives, improve sensitivity and may be useful signatures for differentiating vaccine responses.
In our analysis, we found that SARS-CoV-2 and HA displayed bi-directional and similar mechanistic (glycan-dependent) cross-reactivity, possibly since both these viruses share the same ecological niche and exhibit a similar mode of propagation and clinical presentation [37], [38]. One possible explanation for the emergence of cross-reactive non-neutralizing antibodies might be deceptive imprinting of the immune system towards non-protecting epitopes [39] and the mechanism that favours viral escape through pre-existing cross-reactivity. The immune response, though cross-reactive with the evading virus is unable to mount a protective neutralizing response. These cross-reactive antibodies can form immune complexes with the virus and can activate systemic immune responses that can indirectly help with viral degradation pathways [40], [41].
To conclude, the cross-reactive responses elicited during natural infection or by vaccines, as observed by us and others, have implications for the development of immune-based therapies and vaccines. Whether these non-neutralizing cross-reactive antibody responses have effector functions with a protective effect or lead to antibody-dependent disease enhancement of SARS-CoV-2 infection remains to be evaluated in further studies.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgment
We thank Dr. Pramod Garg, THSTI for the development of project and Dr. Anna George, for critical inputs. We thank Prof. S Pöhlmann, Infection Biology Unit, Göttingen, Germany for ACE2-Fc plasmids as a kind gift. SARS-CoV-2-S-RBD-Fc was a gift from Erik Procko (Addgene plasmid # 141183). The RBD-His is proprietary reagent with IP No. 202011018845. We thank Dr B Graham (VRC/NIAID/NIH) for providing us with spike constructs (SARS-2-CoV S 2P). We thank St. Jude Children's Research Hospital, Inc., (Drs. Erich Hoffmann and Robert Webster and Dr. Raghavan Varadarajan, IISc India for proving PR8 virus [A/Puerto Rico/8/34 (H1N1) reverse genetics. We thank International Reagent Resource (IRR) established by the Centers for Disease Control and Prevention (CDC) for proving Influenza reagent. The following reagent was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, National Institutes of Health: SARS-related coronavirus 2, Isolate USA-WA1/2020, NR-5228. This research has been conducted with the contribution of NCR Biotech Science Cluster Biorepository and DBT India Consortium.
Funding
This work was supported through the THSTI core grant.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.intimp.2021.108020.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Kumar A., Singh R., Kaur J., Pandey S., Sharma V., Thakur L., Sati S., Mani S., Asthana S., Sharma T.K., Chaudhuri S., Bhattacharyya S., Kumar N. Wuhan to World: The COVID-19 Pandemic. Front. Cell. Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.596201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity | Nature Communications, (n.d.). https://www.nature.com/articles/s41467-020-18450-4 (accessed June 29, 2021). [DOI] [PMC free article] [PubMed]
- 3.Mannar D., Leopold K., Subramaniam S. Glycan reactive anti-HIV-1 antibodies bind the SARS-CoV-2 spike protein but do not block viral entry. Sci. Rep. 2021;11:12448. doi: 10.1038/s41598-021-91746-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Structures and Mechanisms of Viral Membrane Fusion Proteins, (n.d.). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2649671/ (accessed March 17, 2021).
- 5.Fink G., Orlova-Fink N., Schindler T., Grisi S., Ferrer A.P.S., Daubenberger C., Brentani A. Inactivated trivalent influenza vaccination is associated with lower mortality among patients with COVID-19 in Brazil. BMJ Evid. Based Med. 2021;26(4):192–193. doi: 10.1136/bmjebm-2020-111549. [DOI] [PubMed] [Google Scholar]
- 6.Candelli M., Pignataro G., Torelli E., Gullì A., Nista E.C., Petrucci M., Saviano A., Marchesini D., Covino M., Ojetti V., Antonelli M., Gasbarrini A., Franceschi F. Effect of influenza vaccine on COVID-19 mortality: a retrospective study. Intern. Emerg. Med. 2021 doi: 10.1007/s11739-021-02702-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelly H., Jacoby P., Dixon G.A., Carcione D., Williams S., Moore H.C., Smith D.W., Keil A.D., Van Buynder P., Richmond P.C. WAIVE Study Team, Vaccine Effectiveness Against Laboratory-confirmed Influenza in Healthy Young Children: A Case-Control Study. Pediatr. Infect. Dis. J. 2011;30:107–111. doi: 10.1097/INF.0b013e318201811c. [DOI] [PubMed] [Google Scholar]
- 8.Cowling B.J., Fang V.J., Nishiura H., Chan K.-H., Ng S., Ip D.K.M., Chiu S.S., Leung G.M., Peiris J.S.M. Increased risk of noninfluenza respiratory virus infections associated with receipt of inactivated influenza vaccine. Clin. Infect. Dis. 2012;54:1778–1783. doi: 10.1093/cid/cis307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sundaram M.E., McClure D.L., VanWormer J.J., Friedrich T.C., Meece J.K., Belongia E.A. Influenza vaccination is not associated with detection of noninfluenza respiratory viruses in seasonal studies of influenza vaccine effectiveness. Clin. Infect. Dis. 2013;57(6):789–793. doi: 10.1093/cid/cit379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petersen E., Koopmans M., Go U., Hamer D.H., Petrosillo N., Castelli F., Storgaard M., Al Khalili S., Simonsen L. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect. Dis. 2020;20(9):e238–e244. doi: 10.1016/S1473-3099(20)30484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bosch B.J., van der Zee R., de Haan C.A.M., Rottier P.J.M. The Coronavirus Spike Protein Is a Class I Virus Fusion Protein: Structural and Functional Characterization of the Fusion Core Complex. J. Virol. 2003;77(16):8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe Y., Bowden T.A., Wilson I.A., Crispin M. Exploitation of glycosylation in enveloped virus pathobiology. Biochim. Biophys. Acta Gen. Subj. 2019;1863(10):1480–1497. doi: 10.1016/j.bbagen.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao X., Chen H., Wang H. Glycans of SARS-CoV-2 Spike Protein in Virus Infection and Antibody Production. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.629873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parray H.A., Chiranjivi A.K., Asthana S., Yadav N., Shrivastava T., Mani S., Sharma C., Vishwakarma P., Das S., Pindari K., Sinha S., Samal S., Ahmed S., Kumar R. Identification of an anti-SARS-CoV-2 receptor binding domain directed human monoclonal antibody from a naïve semi-synthetic library. J. Biol. Chem. 2020;295(36):12814–12821. doi: 10.1074/jbc.AC120.014918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shrivastava T., Samal S., Tyagi A.K., Goswami S., Kumar N., Ozorowski G., Ward A.B., Chakrabarti B.K. Envelope proteins of two HIV-1 clades induced different epitope-specific antibody response. Vaccine. 2018;36(12):1627–1636. doi: 10.1016/j.vaccine.2018.01.081. [DOI] [PubMed] [Google Scholar]
- 16.Patil S., Kumar R., Deshpande S., Samal S., Shrivastava T., Boliar S., Bansal M., Chaudhary N.K., Srikrishnan A.K., Murugavel K.G., Solomon S., Simek M., Koff W.C., Goyal R., Chakrabarti B.K., Bhattacharya J., Sundquist W.I. Conformational Epitope-Specific Broadly Neutralizing Plasma Antibodies Obtained from an HIV-1 Clade C-Infected Elite Neutralizer Mediate Autologous Virus Escape through Mutations in the V1 Loop. J. Virol. 2016;90(7):3446–3457. doi: 10.1128/JVI.03090-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perween R., Ahmed S., Shrivastava T., Parray H.A., Singh B., Pindari K.S., Sharma C., Shukla S., Sinha S., Panchal A.K., Kumar R. A rapid novel strategy for screening of antibody phage libraries for production, purification, and functional characterization of amber stop codons containing single-chain antibody fragments. Biotechnol. Prog. 2021;37(3) doi: 10.1002/btpr.3136. [DOI] [PubMed] [Google Scholar]
- 18.Shrivastava T., Singh B., Rizvi Z.A., Verma R., Goswami S., Vishwakarma P., Jakhar K., Sonar S., Mani S., Bhattacharyya S., Awasthi A., Surjit M. Comparative Immunomodulatory Evaluation of the Receptor Binding Domain of the SARS-CoV-2 Spike Protein; a Potential Vaccine Candidate Which Imparts Potent Humoral and Th1 Type Immune Response in a Mouse Model. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.641447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maltezou H.C., Theodoridou K., Poland G. Influenza immunization and COVID-19. Vaccine. 2020;38(39):6078–6079. doi: 10.1016/j.vaccine.2020.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jang Y.H., Seong B.L. The Quest for a Truly Universal Influenza Vaccine. Front. Cell. Infect. Microbiol. 2019;9:344. doi: 10.3389/fcimb.2019.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The effect of influenza vaccination on trained immunity: impact on COVID-19 | medRxiv, (n.d.). https://www.medrxiv.org/content/10.1101/2020.10.14.20212498v1 (accessed March 17, 2021).
- 22.Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., Hägglöf T., Oliveira T.Y., Viant C., Hurley A., Hoffmann H.-H., Millard K.G., Kost R.G., Cipolla M., Gordon K., Bianchini F., Chen S.T., Ramos V., Patel R., Dizon J., Shimeliovich I., Mendoza P., Hartweger H., Nogueira L., Pack M., Horowitz J., Schmidt F., Weisblum Y., Michailidis E., Ashbrook A.W., Waltari E., Pak J.E., Huey-Tubman K.E., Koranda N., Hoffman P.R., West A.P., Rice C.M., Hatziioannou T., Bjorkman P.J., Bieniasz P.D., Caskey M., Nussenzweig M.C. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584(7821):437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X.u., Zheng Y., Geng C., Chai X., He R., Li X., Lv Q.i., Zhu H., Deng W., Xu Y., Wang Y., Qiao L., Tan Y., Song L., Wang G., Du X., Gao N., Liu J., Xiao J., Su X.-D., Du Z., Feng Y., Qin C., Qin C., Jin R., Xie X.S. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell. 2020;182(1):73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zanettini C., Omar M., Dinalankara W., Imada E.L., Colantuoni E., Parmigiani G., Marchionni L. Influenza Vaccination and COVID19 Mortality in the USA. MedRxiv. 2020 doi: 10.1101/2020.06.24.20129817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe Y., Berndsen Z.T., Raghwani J., Seabright G.E., Allen J.D., Pybus O.G., McLellan J.S., Wilson I.A., Bowden T.A., Ward A.B., Crispin M. Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nat. Commun. 2020;11:2688. doi: 10.1038/s41467-020-16567-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balzarini J. Targeting the glycans of glycoproteins: a novel paradigm for antiviral therapy. Nat. Rev. Microbiol. 2007;5(8):583–597. doi: 10.1038/nrmicro1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Structure of 2G12 Fab2 in Complex with Soluble and Fully Glycosylated HIV-1 Env by Negative-Stain Single-Particle Electron Microscopy, (n.d.). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4136306/ (accessed March 17, 2021). [DOI] [PMC free article] [PubMed]
- 28.Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., Peter A., Guarino B., Spreafico R., Cameroni E., Case J.B., Chen R.E., Havenar-Daughton C., Snell G., Telenti A., Virgin H.W., Lanzavecchia A., Diamond M.S., Fink K., Veesler D., Corti D. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583(7815):290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 29.Lardone R.D., Garay Y.C., Parodi P., de la Fuente S., Angeloni G., Bravo E.O., Schmider A.K., Irazoqui F.J. How glycobiology can help us treat and beat the COVID-19 pandemic. J. Biol. Chem. 2021;296:100375. doi: 10.1016/j.jbc.2021.100375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore P.L., Crooks E.T., Porter L., Zhu P., Cayanan C.S., Grise H., Corcoran P., Zwick M.B., Franti M., Morris L., Roux K.H., Burton D.R., Binley J.M. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J. Virol. 2006;80(5):2515–2528. doi: 10.1128/JVI.80.5.2515-2528.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ladner J.T., Henson S.N., Boyle A.S., Engelbrektson A.L., Fink Z.W., Rahee F., D’ambrozio J., Schaecher K.E., Stone M., Dong W., Dadwal S., Yu J., Caligiuri M.A., Cieplak P., Bjørås M., Fenstad M.H., Nordbø S.A., Kainov D.E., Muranaka N., Chee M.S., Shiryaev S.A., Altin J.A. Epitope-resolved profiling of the SARS-CoV-2 antibody response identifies cross-reactivity with an endemic human CoV. BioRxiv. 2020 doi: 10.1101/2020.07.27.222943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorse Geoffrey J., Patel Gira B., Vitale Joseph N., O'Connor Theresa Z. Prevalence of antibodies to four human coronaviruses is lower in nasal secretions than in serum. Clin. Vaccine Immunol. 2010;17(12):1875–1880. doi: 10.1128/CVI.00278-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tso F.Y., Lidenge S.J., Peña P.B., Clegg A.A., Ngowi J.R., Mwaiselage J., Ngalamika O., Julius P., West J.T., Wood C. High prevalence of pre-existing serological cross-reactivity against severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) in sub-Saharan Africa. Int J Infect Dis. 2021;102:577–583. doi: 10.1016/j.ijid.2020.10.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ladner Jason T., Henson Sierra N., Boyle Annalee S., Engelbrektson Anna L., Fink Zane W., Rahee Fatima, D’ambrozio Jonathan, Schaecher Kurt E., Stone Mars, Dong Wenjuan, Dadwal Sanjeet, Yu Jianhua, Caligiuri Michael A., Cieplak Piotr, Bjørås Magnar, Fenstad Mona H., Nordbø Svein A., Kainov D.E., Muranaka Norihito, Chee M.S., Shiryaev S.A., Altin J.A. Epitope-resolved profiling of the SARS-CoV-2 antibody response identifies cross-reactivity with endemic human coronaviruses. Cell Rep Med. 2021;2(1):100189. doi: 10.1016/j.xcrm.2020.100189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grifoni Alba, Weiskopf Daniela, Ramirez Sydney I., Mateus Jose, Dan Jennifer M., Moderbacher Carolyn Rydyznski, Rawlings Stephen A., Sutherland Aaron, Premkumar Lakshmanane, Jadi Ramesh S., Marrama Daniel, de Silva Aravinda M., Frazier April, Carlin Aaron F., Greenbaum Jason A., Peters Bjoern, Krammer Florian, Smith Davey M., Crotty Shane, Sette Alessandro. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181(7):1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vogt M.R., Dowd K.A., Engle M., Tesh R.B., Johnson S., Pierson T.C., Diamond M.S. Poorly neutralizing cross-reactive antibodies against the fusion loop of West Nile virus envelope protein protect in vivo via Fcgamma receptor and complement-dependent effector mechanisms. J. Virol. 2011;85:11567–11580. doi: 10.1128/JVI.05859-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Araújo M.B., Naimi B. Spread of SARS-CoV-2 Coronavirus likely constrained by climate. Epidemiology. 2020 doi: 10.1101/2020.03.12.20034728. [DOI] [Google Scholar]
- 38.Zheng J., Perlman S. Immune responses in influenza A virus and human coronavirus infections: an ongoing battle between the virus and host. Curr Opin. Virol. 2018;28:43–52. doi: 10.1016/j.coviro.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kohler Heinz, Nara Peter. A Novel Hypothesis for Original Antigenic Sin in the Severe Disease of SARS-CoV-2 Infection. Monoclon Antib. Immunodiagn. Immunother. 2020;39(4):107–111. doi: 10.1089/mab.2020.0029. [DOI] [PubMed] [Google Scholar]
- 40.Fu Yajing, Cheng Yuanxiong, Wu Yuntao. Understanding SARS-CoV-2-Mediated Inflammatory Responses: From Mechanisms to Potential Therapeutic Tools. Virol Sin. 2020;35(3):266–271. doi: 10.1007/s12250-020-00207-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Is COVID-19 receiving ADE from other coronaviruses? - PubMed, (n.d.). https://pubmed.ncbi.nlm.nih.gov/32092539/ (accessed March 17, 2021).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.