Abstract
The rapid emergence of multidrug resistant microorganisms has become one of the most critical threats to public health. A decrease in the effectiveness of available antibiotics has led to the failure of infection control, resulting in a high risk of death. Among several alternatives, antimicrobial peptides (AMPs) serve as potential alternatives to antibiotics to resolve the emergence and spread of multidrug-resistant pathogens. These small proteins exhibit potent antimicrobial activity and are also an essential component of the immune system. Although several AMPs have been reported and characterized, studies associated with their potential medical applications are limited. This review highlights the novel sources of AMPs with high antimicrobial activities, including the entomopathogenic nematode/bacterium (EPN/EPB) symbiotic complex. Additionally, the AMPs derived from insects, nematodes, and marine organisms and the design of peptidomimetic antimicrobial agents that can complement the defects of therapeutic peptides have been used as a template.
Keywords: antimicrobial peptides, multidrug-resistant pathogens, insects, nematodes, marine
Introduction
Antimicrobial peptides (AMPs) are small molecules that generally consist of 10–50 amino acids and are highly conserved in a wide range of species, including insects, nematodes, microbes, and mammals. AMPs serve as an essential component of the body’s immune system and defend against exogenous pathogens. They possess significant structural variations in the α-helices, β-strands with one or more disulfide bridges, loop, and extended structures associated with their broad-spectrum activities (Hancock, 2001; Pushpanathan et al., 2013). Other important factors associated with the functional activities of AMPs are size, hydrophobicity, charge, amphipathic stereo-geometry, and peptide self-association to the biological membrane (Nissen-Meyer and Nes, 1997; Marcos and Gandía, 2009; Pushpanathan et al., 2013). AMPs can be considered potential drug candidates to treat pathogenic microorganisms due to their broad-spectrum activity, lesser toxicity, decreased resistance development by the target cells, and capability to modulate the host immune response (Hancock and Patrzykat, 2002; Xu et al., 2019). AMPs can ameliorate the drug-resistant crisis and associated toxicity with conventional AMP drugs and also can be employed as an alternative to antibiotics (Lewies et al., 2019). They exhibit several similarities to antibiotics, such as killing microbial cells and targeting a broad spectrum of pathogens, including antibiotic resistance.
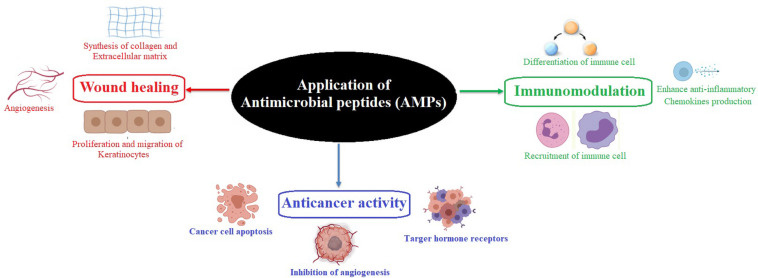
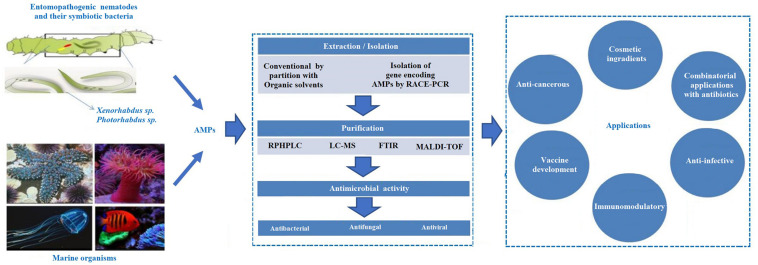
Moreover, compared to antibiotics, AMPs have unique epitopes that serve as protease recognition sites, thereby less likely to be targeted by the protease (Zasloff, 2002; Lai and Gallo, 2009). Different mechanisms, such as inhibition of gene expression or protein synthesis, inhibition of cell wall synthesis, or delocalization of bacterial cell surface proteins are commonly employed by the AMPs (Baltzer and Brown, 2011). Most of the AMPs are cationic and capable of adapting to amphipathic conformations. This helps them interact with the negatively charged bacterial cell wall and integrate it into the lipid bilayers (Haney et al., 2017; Zharkova et al., 2019). The success of AMPs against multidrug-resistant pathogens is due to the widescale multitargeted action (Zharkova et al., 2019). They are also active at lower minimum inhibitory concentrations (MICs) as compared to antibiotics. AMPs demonstrate higher killing effects and show a narrower mutation-selection window, accounting for the less likely development of resistance to AMP (Fantner et al., 2010; Yu et al., 2018). They are also active against biofilm-producing antibiotic-resistant microbes and induce non-opsonic phagocytosis. However, the combined use of AMPs with other antimicrobial compounds such as specific antibiotics may play a vital role against multidrug-resistant pathogens and associated adverse health conditions. In addition, some AMPs have been identified to exhibit antiviral activities (Chia et al., 2010; Van Der Does et al., 2010; Chung and Kocks, 2011; Steckbeck et al., 2014; Elnagdy and AlKhazindar, 2020). The AMPs play an essential role in modulating immunogenic activities, improving wound healing, enhancing chemokine production, exhibiting anti-inflammatory properties, regulating epithelial cell differentiation, and modulating angiogenesis (Koczulla and Bals, 2007; Mahlapuu et al., 2016; Otvos, 2016; Patrulea et al., 2020; Figure 1). Nowadays, scientists are participating in developing enhanced AMPs with novel modes of actions to replace or complement traditional antibiotics to treat various diseases (Morikawa et al., 1992; Wang et al., 2016). So far, 3257 AMPs have been reported from six kingdoms (bacteria, archaea, fungi, protists, plants, and animals) in the Antimicrobial Peptide Database1 (Wang et al., 2016). This review provides insights into developing different AMPs from novel sources and their multifunctional properties and elaborates their future prospects (Figure 2). Particular focus has been given to the AMPs in bacteria that form a symbiotic relationship with the entomopathogenic nematodes (EPNs), displaying varied modes of actions.
FIGURE 1.
Role of an antimicrobial peptide in immunomodulation, wound healing, and anticancer activity.
FIGURE 2.
Schematic representation of the antimicrobial peptides (AMPs) extracted from the different source and their applications.
AMPs in Insects
Insects represent the largest class in the animal kingdom and are found in most of the biological niches. One of the critical features of their successful adaptation is their resistance to various pathogens. The AMPs play a critical role in innate immunity against insect pathogens (Bulet et al., 1999). They produce a large number of AMPs that varies between species, ranging from 50 (Harmonia axyridis) to 0 (Hermetia illucens) (Gerardo et al., 2010; Vilcinskas, 2013; Vogel et al., 2018). Cecropin, the first insect AMP, was isolated and characterized from Hyalophora cecropia (Steiner et al., 1981). Since then, many insect AMPs have been reported, which are mainly classified into three groups based on the sequence and structural features, i.e., linear peptides with α-helices that lack cysteine residues and cyclic peptides containing cysteine residues and peptides with an overexpression of proline and glycine residues (Hetru, 1998; Bulet et al., 1999). The most explored insect AMPs are defensin, cecropin, drosocin, attacin, diptericin, ponericin, drosomycin, and metchnikowin. However, it is expected that insects may have more AMPs with novel modes of action (Mylonakis et al., 2016).
Cecropins are small peptides that destroy bacterial cell membranes, inhibit proline uptake, and cause leaky membranes (Moore et al., 1996). It has also been reported that cecropin A (CecA) destroys uropathogenic Escherichia coli (UPEC) cells, alone or in combination with nalidixic acid (NAL), and could be a practical approach to treat antibiotic-resistant UPEC infections (Kalsy et al., 2020). CecA from H. cecropia exhibits only antibacterial activity, whereas CecA from Anopheles gambiae exhibits antibacterial and antifungal activities (Bulet et al., 2004). BR003-CecA from Aedes aegypti actively inhibits multiple species of Gram-negative bacteria (GNB), including A. baumannii (Jayamani et al., 2015). Cec D from Galleria mellonella exhibits vigorous activity against Gram-positive bacterium (GPB) Listeria monocytogenes (Mukherjee et al., 2011). Defensins are the second primary class of inducible insect AMPs active against GPB, including Staphylococcus aureus, but are less active against GNB (Hetru et al., 2003; Gomes and Fernandes, 2010). Few defensins also possess antifungal activities against filamentous fungi, e.g., gallerimycin from the greater wax moth G. mellonella (Langen et al., 2006). Insect defensin-like peptides are found in Leiurus quinquestriatus and Androctonus australis (Cociancich et al., 1993; Ehret-Sabatier et al., 1996). Defensin-like peptide 4 (DLP4) reported from the black soldier fly is active against GPB (Park et al., 2015).
The AMP drosocin, isolated from Drosophila melanogaster, is a 19-residue peptide containing six proline and four arginine residues (McManus et al., 1999). Glycosylated drosocin is active against E. coli and fungi (Imler and Bulet, 2005). These O-glycosylated AMPs are also found in other insects such as Pyrrhocoris apterus (pyrrhocoricin), Bombyx mori (lebocins), and Myrmecia gulosa (formations) (Cociancich et al., 1994; Hara and Yamakawa, 1995; Mackintosh et al., 1998; Wu et al., 2018).
Attacins, glycine-rich AMP, were first discovered in H. cecropia and is active against GNB (Hultmark et al., 1983; Carlsson et al., 1991). Attacins from Spodoptera exigua exhibit activity against E. coli, Pseudomonas cichorii, Bacillus subtilis, L. monocytogenes, Trypanosoma brucei, Citrobacter freundii, and Candida albicans (Hu and Aksoy, 2005; Kwon et al., 2008; Bang et al., 2012). Attacins and attacin-related proteins are also isolated from B. mori, Heliothis virescens, Trichoplusia ni, Samia cynthia ricini, and Musca domestica (Dushay et al., 2000; Geng et al., 2004).
Diptericin (9 kDa), found in D. melanogaster, Sarcophaga peregrina, and Mayetiola destructor, is active against GNB such as E. coli, Erwinia herbicola T, and E. carotovora (Keppi et al., 1989; Ishikawa et al., 1992; Reichhart et al., 1992).
However, limited reports are available on antifungal peptides in insects such as drosomycin from D. melanogaster, termicin from termites, heliomicin from H. virescens, and gallerimycin peptide from G. mellonella (Fehlbaum et al., 1994; Da Silva et al., 2003; Schuhmann et al., 2003). The antifungal peptide drosomycin is active against fungal pathogens, whereas thanatin is effective against a broad range of β-lactamase-producing E. coli (Bulet et al., 1999; Hou et al., 2011).
Xu et al. (2019) reported a novel Moricin (Px-Mor) from the diamondback moth that showed a broad-spectrum activity against GPB, GNB, and fungi, including the opportunistic human pathogen Aureobasidium pullulans. They suggested that Px-Mor can be used as a potential topical antimicrobial agent (Xu et al., 2019). These results indicate the importance of insect-derived AMPs against pathogens and could be further employed against multidrug-resistant pathogens or in combination with existing antibiotics (Table 1).
TABLE 1.
Recently identified insect AMPs with their mechanism of action.
| Name of AMP | Type of AMP | Source | 3D structure | Pathogenic species | Molecular mechanism | Inhibitory concentration | References |
| ETD151 (Heliomicin) | Defensin | Heliothis virescens | Combine helix and beta structure | Botrytis cinerea | Interact with glucosylceramides of the fungal membrane | IC50 = 0.59 μM | Aumer et al., 2020 |
| Holosins | Ixodes holocyclus | Cys-stabilized α/β-fold | Staphylococcus aureus, Listeria grayi, F. graminearum, and C. albicans | Peptide–lipid interactions result in the formation of membrane penetrating pores | MIC = 8 μM MIC = 5 μM | Cabezas-Cruz et al., 2019 | |
| Oxysterlins | Cecropin | Oxysternon conspicillatum | Linear α-helix | Staphylococcus saprophyticus, Klebsiella pneumoniae, and Pseudomonas aeruginosa | Membrane lysis due to formation of pores | MIC = 12.5 μg/ml MIC = 3.12 μg/ml MIC = 12.5 μg/ml | Toro Segovia et al., 2017 |
| Cecropin D | Galleria mellonella | α-Helix | K. pneumoniae (MDR), P. aeruginosa (MDR) | MIC = 256 μg/ml MIC = 32 μg/ml | Ocampo-Ibáñez et al., 2020 | ||
| Cecropin B | Antheraea pernyi | P. aeruginosa | MIC = 0.4 μg/ML | Wu et al., 2012; Yang et al., 2018; Gholizadeh and Moradi, 2020 | |||
| Cecropin AD | Hyalophora cecropia | Staphylococcus aureus | MIC = 0.2 μg/ml | ||||
| HI-attacin | Attacin | Hermetia illucens | Unknown | E. coli and methicillin-resistant Staphylococcus aureus | Blocking the synthesis of the major outer membrane proteins, thus disturbing the integrity of the cell wall | NS | Shin and Park, 2019 |
| Prolixicin | Rhodnius prolixus | E. coli, Citrobacter freundii, Enterobacter aerogenes, and Bacillus coagulans | MIC = 1.6 μM MIC = 12.5 μM | Ursic-Bedoya et al., 2011 | |||
| SlLeb-1 | Lebocin | Spodoptera litura | Rich | E. coli and B. subtilis | Disrupt cell membrane and cause cell elongation | MIC = 50 μM | Yang et al., 2018; Yang et al., 2020 |
| Apidaecin IB | Drosocin | Apis cerana | Rich | Escherichia coli and Klebsiella pneumoniae | Binds to the substrate binding site of E. coli DnaK to compete with natural substrates | NS | Berthold and Hoffmann, 2014; Feng et al., 2020 |
| Api795 | Apidaecin | P. aeruginosa | Insert into bacterial mimic membranes and initiates a structural change leading to a thicker and more rigid membrane layer | MIC = 8 μg/ml | Bluhm et al., 2016 | ||
| EtDip | Diptericin | Eristalis tenax | Unknown | Candida albicans FH2173 and Mycobacterium smegmatis ATCC 607 | MIC > 1024 μg/ml MIC = 64 μg/ml | Hirsch et al., 2020 | |
| Mtk | Metchnikowin | Drosophila melanogaster | Rich | Fusarium graminearum | Interacts with the fungal enzyme β(1,3)-glucanosyltransferase Gel1 (FgBGT), which is one of the enzymes responsible for fungal cell wall synthesis | NS | Moghaddam et al., 2017 |
| Ponericin-Q42 | Ponericins | Ectatomma quadridens | α-Helical folds | Arthrobacter globiformis, B. subtilis, E. coli MH1, and P. aeruginosa PAO1 | Membrane blebbing, formation of swollen cells and finally membrane destruction and cell death | MIC = 0.2 μM MIC = 0.6 μM MIC = 10 μM | Pluzhnikov et al., 2014 |
| Jelleine-I | Jelleines | Apis mellifera | Unknown | Candida glabrata and C. albicans | Increase the production of cellular ROS and bind with genome DNA | MIC and MFC = 30 μg/ml MIC and MFC = 61 μg/ml | Jia et al., 2018 |
| Pyrrhocoricin | Pyrrhocoricin | Pyrrhocoris apterus | Non-helix beta | Cell-free E. coli system and Cryptosporidium parvum | Inhibit the protein folding activity of the ATP-dependent DnaK/DnaJ molecular chaperone system | IC for transcription = 427 μM | Boxell et al., 2008; Taniguchi et al., 2016 |
| Melittin | Melittin | Apis mellifera | Helix | Lactobacillus casei and Streptococcus mutans | Interact with bacterial membrane | MIC = 4 μg/ml MIC = 40 μg/ml | Leandro et al., 2015 |
AMPs in Nematode
Antimicrobial peptides are produced by microorganisms associated with insect symbioses and play a significant role in maintaining the symbiotic microbe in specific anatomical compartments (Ovchinnikova et al., 2004; Tasiemski et al., 2015). A lot of the literature have highlighted the EPN/entomopathogenic bacterium (EPB) symbioses. Nematodes are specialized organisms with the ability to adapt both free-living and parasitic lifestyle in different environments. Nematodes also serve as a novel invertebrate model to study innate immunity and host–pathogen interactions (Kurz and Tan, 2004; Nicholas and Hodgkin, 2004). EPNs and their associated bacteria have evolved with several defense mechanisms to elude and counteract the host (insect) immune responses (Brivio et al., 2018; Brivio and Mastore, 2020). The nematode Steinernema carpocapsae can produce proteolytic secretions that can interfere with the host immune system in S. feltiae, S. glaseri, and G. mellonella (Balasubramanian et al., 2010; Chaubey and Garg, 2019). Similarly, the surface proteins of S. glaseri protect from encapsulation by the host immune system of Popillia japonica (Wang and Gaugler, 1999; Chaubey and Garg, 2019). It has also been reported that symbiotic bacteria and nematode cooperate to overcome the host immune response. Many defense molecules are produced as immune effectors against various microbial infections (Bulet et al., 2004). The AMP cecropins are found in the worm Ascaris suum (cecropin-P1, cecropin-P2, cecropin-P3, and cecropin-P4), a parasite inhabiting the intestine of pigs. These short AMPs, rich in serine residues, are stabilized by the disulfide bonds and contain potential antimicrobial activities against BPB (S. aureus, B. subtilis, Micrococcus luteus) and GNB (Pseudomonas aeruginosa, Salmonella typhimurium, Serratia marcescens, and E. coli) and are less effective against fungi (Saccharomyces cerevisiae, C. albicans) (Andrä et al., 2001; Andersson et al., 2003; Pillai et al., 2005; Bruno et al., 2019). Cecropin P1-like sequences were also identified in two other species, i.e., Ascaris lumbricoides and Toxocara canis (Pillai et al., 2005).
Another group of AMPs called the caenopores belong to the saposin-like protein (SAPLIP) superfamily detected in Caenorhabditis elegans. It contains conserved positions of six cysteine residues. Caenopore-1 (SPP-1), caenopore-5 (SPP-5), and caenopore-12 (SPP-12) exhibit antimicrobial activity against B. megaterium, E. coli, and SPP-12 Bacillus thuringiensis (Roeder et al., 2010; Hoeckendorf et al., 2012).
Defensins are the most studied AMPs in nematodes. Ascaris suum antibacterial factors (ASABFs) was the first nematode defensin identified in C. elegans. They are short AMPs with eight cysteine residues that form four disulfide bonds except for ASABF-6Cys-α (Minaba et al., 2009; Tarr, 2012). These peptides are primarily active against GPB, especially the common pathogen S. aureus. However, it is less effective against GNB and yeast (Tarr, 2012). A recent study by Lim et al. (2016) reported two novel C. elegans AMPs (NLP-31 and Y43C5A.3) that exhibit antimicrobial activity against Burkholderia pseudomallei, the causative agent of melioidosis, by interfering with DNA synthesis. They also revealed that these AMPs might act by modulating host cytokine production to interfere with the inflammatory response, and modifications could enhance anti-B. pseudomallei activities (Lim et al., 2016).
AMPs Linked With EPN/EPB Symbiotic Complex
Several bacterial genera belonging to the Enterobacteriaceae family are mutualistically associated with the EPNs (Boemare, 2002). These EPNs, with their symbiotic bacteria, are lethal to many soil insects, as they synthesize diverse secondary metabolites, including small AMPs. These nematode-associated microbes exist in two distinct phases: phase 1, where they are generally associated with the nematodes, and phase 2, where they may also colonize with the nematode. However, they have never been reported to be associated with the naturally occurring nematodes. Both the phases have distinguished physiological, biochemical, and behavioral features; also phase 1 is considered more virulent than phase 2 (Akhurst et al., 1990; Volgyi et al., 1998; Abdel-Razek, 2002; Sugar et al., 2012). During the infective juvenile (IJ) stage, the nematodes enter inside the insects by piercing the body wall or via natural openings and releasing these bacteria inside the hemocoel. They reproduce exponentially, producing bioactive compounds with broad-spectrum antimicrobial activities (Sanda et al., 2018). They provide nutrients to the nematodes and protect them from environmental predators such as bacteria and fungi. They also compete for nutrition with other microbes, including the saprophytic soil microbes and the bacteria present in the insect gut or cuticle of the nematode. The elimination of the competitors is facilitated by the production of colicin E3-type killer proteins, insect toxin complexes, phage-derived bacteriocins, and several secondary metabolites (Thaler et al., 1995; Ffrench-Constant and Waterfield, 2006; Singh and Banerjee, 2008; Bode, 2009; Piel, 2009). The presence of high content of non-ribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) genes facilitates them to produce novel and new bioactive molecules (Tobias et al., 2017). These bioactive molecules disrupt the insect’s metabolic and functional properties, leading to septicemia (Khandelwal and Banerjee-Bhatnagar, 2003; Tran and Goodrich-Blair, 2009; Ellis and Kuehn, 2010; Brivio et al., 2018). Nematodes also play a significant role in the pathogenicity of the nemato-bacterial complex (Han and Ehlers, 2000; Chang et al., 2019). The EPNs, along with the mutualistic bacteria, kill their host within 48−72 h (Forst and Nealson, 1996). These features are now being exploited for the biological control of pests (Brivio and Mastore, 2018).
The bacterial genus Xenorhabdus is often found in close association with EPNs of the family Steinernematidae (Webster et al., 2002). Xenorhabdus synthesizes and releases antibiotic compounds in the host hemocoel that suppresses the microbial competitors, thereby manipulating the environment to promote growth, proliferation, and nematode development (Vallet-Gely et al., 2008; Richards and Goodrich-Blair, 2009; Gaugler, 2018). The antimicrobial compounds produced by these bacterial genera are highly toxic to the insect but not toxic to the nematodes. Various surface structures such as pili/fimbriae, flagella, and the outer membrane vesicles (OMVs) present in the Xenorhabdus interact with the host and promote adhesion and invasion of the host tissues. They also promote larvicidal activity by releasing proteases, lytic factors, and phospholipase C (Brivio et al., 2018). Ribosomal-encoded bacteriocins (xenorhabdicins) are found in Xenorhabdus nematophilus. These AMPs compete against more closely related bacteria, such as other Xenorhabdus and Photorhabdus strains (Thaler et al., 1995). The indole-containing Xenematide from Xenorhabdus nematophila exhibits moderate antibacterial and insecticidal activities (Lang et al., 2008). Two novel depsipeptides, xenematides F and G, were isolated from Xenorhabdus budapestensis SN84 with high antibacterial activity (Xi et al., 2019).
The cyclic peptide-antimicrobial-Xenorhabdus (PAX) lipopeptides, obtained by the fermentation of the X. nematophila F1 strain, exhibit significant activity against plants and human fungal pathogens and moderately effective against a few bacteria and yeast (Gualtieri et al., 2009). Two novel AMPs GP-19 and EP-20 from the bacterial strain X. budapestensis NMC-10. GP-19 exhibited inhibitory activity mainly against bacteria, while EP-20 was highly effective against plant pathogens. The synthetic GP-19 and EP-20 peptide exhibited inhibitory activities against the fungal pathogen Verticillium dahlia and Phytophthora capsici with EC50 values of 17.54 and 3.14 μg/ml, respectively (Xiao et al., 2012).
The AMPs xenocoumacin 1 (XCN 1) and 2 (XCN 2), from the bacterium X. nematophila, is effective against GPB and fungi. This peptide is synthesized by the PKS/NRPS multienzyme (xcnA-N) (McInerney et al., 1991; Reimer, 2013). Six novel linear peptides (rhabdopeptides) in X. nematophila and two other rhabdopeptide derivatives by X. cabanillasii were also identified (Reimer et al., 2013).
Nematophins, from X. nematophila YL001, inhibit mycelial growth of Rhizoctonia solani and Phytophthora infestans with an EC50 value of 40.00 and 51.25 μg/ml, respectively, and can be employed as a potential biopesticide in the agriculture sector (Zhang et al., 2019). Similarly, the novel peptide, xenoamicin, tridecadepsipeptides with hydrophobic amino acids, from the entomopathogenic X. doucetiae DSM 17909 and X. mauleonii DSM 17908 was effective against Plasmodium falciparum (Zhou et al., 2013).
Another dipeptide xenobactin was isolated from Xenorhabdus sp., strain PB30.3, and szentiamide from X. szentirmaii. Both AMPs are active against P. falciparum and have moderately effective against T. brucei rhodesiense and Trypanosoma cruzi (Nollmann et al., 2012; Grundmann et al., 2013). Similarly, the depsipentapeptide chaiyaphumine A from Xenorhabdus sp. PB61.4 was effective against P. falciparum (IC50 of 0.61 μM) and other protozoal tropical disease-causing agents (Grundmann et al., 2014). Xenorhabdus indica can produce depsipeptides and lipodepsipeptides with an additional fatty acid chain linked to one of the amino acids, also called taxlllaids (A–G), and exhibits antiprotozoal activity (Kronenwerth et al., 2014). Taken together, these reports suggest that the mutualistic association between Xenorhabdus and Steinernematidae could serve as a potential source for novel AMPs against bacteria, fungi, and protozoal disease-causing agents.
The bacterium Photorhabdus spp. forms a symbiotic relationship with the EPNs of the genus Heterorhabditis (Gerrard et al., 2006). They cause pathogenicity in most insects post invading the hemolymph (Boemare et al., 1997). Genomic analysis of Photorhabdus can interpret the relation between pathogenesis and symbiosis, thereby providing vital information for the development of biocontrol agents. The genomic sequence of P. luminescens subsp. laumondii strain TT01 revealed several genes that encode toxins, hemolysins, adhesins, hemolysins, proteases, lipases, and a wide array of antibiotics. Two identified protein, PirA and PirB, exhibit similarity to both δ-endotoxins (B. thuringiensis) and a developmentally regulated protein from a beetle (Leptinotarsa decemlineata) (Duchaud et al., 2003; Waterfield et al., 2005). Research on the larvicidal activity of Photorhabdus sp. showed that Photorhabdus insect-related (Pir) protein is associated with high toxicity against the primary vector of dengue virus A. aegypti and Aedes albopictus (Ahantarig et al., 2009). These novel insecticidal proteins could further be exploited to develop alternative agents to control insect pests. Genomic analysis of P. luminescens subsp. laumondii strain TT01 indicates the presence of several enzymes associated with the secondary metabolite biosynthesis. The genomic sequence analysis identifies biosynthetic gene clusters associated with the synthesis of linear or cyclized peptides, lipopeptides, or depsipeptides; NRPS; unusual fatty acid synthase or a FAS/PKS hybrid; and siderophore biosynthesis (Bode, 2009). Photorhabdus sp. also produces numerous antimicrobials such as isopropyl stilbene, ethylstilbenes, anthraquinones (AQs) photobactin, ethyl stilbene, epoxystilbene, and ulbactin E (Li et al., 1995; Webster et al., 2002; Bode, 2009). The bioactive compounds exhibit a broad range of antimicrobial activities. Photorhabdus antibacterial compounds include trans-stilbenes and anthraquinone pigments (Boemare and Akhurst, 2002) that have enthralled substantial interest in the agronomic and pharmaceutical sectors (Webster et al., 2002; Hazir et al., 2016). Phthalic acid or 1,2-benzene dicarboxylic acid purified from Photorhabdus temperata M1021 exhibits an antibacterial activity with MIC values of 0.1 and 0.5 M (Ullah et al., 2014), benzaldehyde exhibits an antibacterial activity with MIC values of 6 and 10 mM, and antifungal activity with MIC values between 8 and 10 mM (Ullah et al., 2015). P. temperata subsp. temperata inhibits the growth of 10 strains of drug-resistant bacteria (Muangpat et al., 2017), including S. typhimurium KCTC 1926 and M. luteus KACC 10488 (Jang et al., 2012). Therefore, Photorhabdus spp. can be a suitable biocontrol agent in drug industries. AMPs from nemato-bacterial complexes with their inhibitory concentrations are shown in Table 2.
TABLE 2.
Antimicrobial peptides from nematobacterial complexes with their inhibitory concentrations.
| Name of AMP | Source | Pathogenic species | Inhibitory concentration | References |
| Xenematide C | Xenorhabdus budapestensis SN19 | Botrytis cinerea | EC50 = 22.71 μg/ml | Xing-zhong et al., 2016 |
| Xenematides F | Xenorhabdus budapestensis SN84 | P. aeruginosa | MIC = 32 μg/ml | Xi et al., 2019 |
| Xenematides G | B. subtilis | MIC = 16 μg/ml | ||
| PAX lipopeptides Xenocoumacin 2 | Xenorhabdus khoisanae SB10 | B. subtilis subsp. subtilis Escherichia coli Candida albicans | NS | Dreyer et al., 2019 |
| Diketopiperazines | EPN Rhabditis sp. | Penicillium expansum | MIC = 2 μg/ml | Kumar et al., 2013 |
| Nematophin | Xenorhabdus nematophila YL001 | Rhizoctonia solani Phytophthora infestans | EC50 = 40.00 μg/ml EC50 = 51.25 μg/ml | Zhang et al., 2019 |
| Nematophin | Xenorhabdus PB62.4 | Staphylococcus aureus | MIC = 0.7 μg/ml | Cai et al., 2017 |
| GP-19 EP-20 | Xenorhabdus budapestensis NMC-10 | Verticillium dahlia Phytophthora capsici | EC50 = 17.54 μg/ml EC50 = 3.14 μg/ml | Xiao et al., 2012 |
| Threonine–glutamine dipeptide) domain containing protein | Bacillus cereus | E. coli, S. aureus, and B. subtilis | MIC = 62.55 μg/ml MIC = 125 μg/ml MIC = 250 μg/ml | Anju et al., 2015 |
| Xenocoumacin 1 Xenocoumacin 2 | Xenorhabdus nematophila | Botrytis cinerea | Inhibition rate of 100 ml/L cell-free filtrate on the mycelial growth of the pathogens is 100% | Guo et al., 2017 |
| Nematophins, Xenocoumacins and Xenorhabdins | Xenorhabdus assam-isolate (SG as1) | Macrophomina phaseolina | EC50 = 55.98 μg/ml | Sharma et al., 2016 |
| Cabanillasin | Xenorhabdus cabanillasii | Fusarium oxysporum | IC50 = 6.25 μg/ml | Houard et al., 2013 |
| Xenobactin | Xenorhabdus sp. PB30.3 | Micrococcus luteus Plasmodium falciparum NF 54 Trypanosoma brucei rhodesiense STIB900 Trypanosoma cruzi Tulahuen C4 | MIC = 64 μg/ml IC50 = 12.45 μg/ml IC50 = 12.45 μg/ml IC50 = 67.03 μg/ml | Grundmann et al., 2013 |
| Xenortide D | Xenorhabdus nematophila | Plasmodium falciparum Trypanosoma brucei | NS | Reimer et al., 2014 |
| Taxlllaids | Xenorhabdus indica (DSM 17382) | Plasmodium falciparum | NS | Kronenwerth et al., 2014 |
| Phototemtide A | Photorhabdus temperata Meg1 | Plasmodium falciparum Trypanosoma brucei rhodesiense | IC50 = 9.8 μM IC50 = 62 μM | Zhao L. et al., 2020 |
The Targets and Mechanism of Action of AMPs Derived From Nematobacterial Complexes
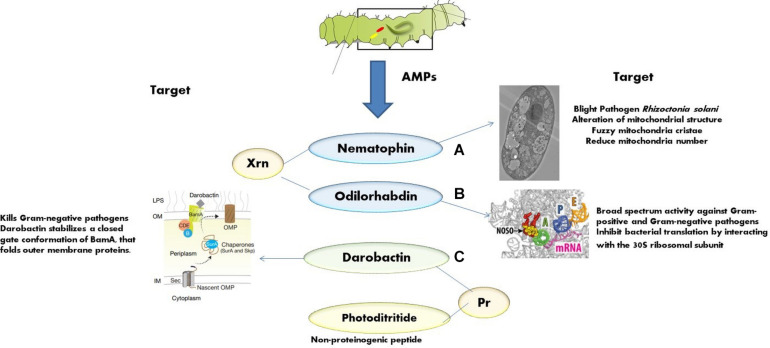
The AMPs from Xenorhabdus spp. and Photorhabdus spp. are non-lethal to nematode but toxic to insect pathogens and other opportunistic microorganisms with unique targets and modes of action. This section highlights some of the recently identified AMPs from EPB with novel modes of action, namely, nematophin, odilorhabdin, darobactin, and photoditritide (Figure 3).
FIGURE 3.
Mode of action of various AMPs: (A) nematophins act on mitochondrial structure of Rhizoctonia solani [adapted from Zhang et al. (2019)]; (B) odilorhabdins act on the ribosome and inhibit protein synthesis in Gram-negative bacteria [adapted from Pantel et al. (2018)]; (C) darobactins act on outer membrane chaperone [adapted from Imai et al. (2019)].
Nematophin
First isolated from X. nematophilus strain BC1 (Li et al., 1997), it contains 3-indoleethyl (3′-methyl-2′-oxo) pentanamide with an N-terminal α-keto group and a C-terminal tryptamine residue. Recently, few novel nematophin analogs were identified from Xenorhabdus strains (Cai et al., 2017). Nematophin is effective against S. aureus (MIC = 0.125 μg/ml) (Li et al., 1997), methicillin-resistant S. aureus (MRSA) (MIC = 1.5 μg/ml), and fungal pathogens, Botrytis cinerea (MIC = 12 μg/ml) (Li et al., 1997) and R. solani (MIC = 40 μg/ml) (Zhang et al., 2019). The synthetic nematophin analog with N-methyl substitution exhibits nanomolar activity toward S. aureus (15 ng/ml), S. intermedius 9503 (50 ng/ml) (Himmler et al., 1998), S. hyicus (60 ng/ml), MRSA ATCC 43300 (31 ng/ml), and methicillin-susceptible S. aureus ATCC 29213 (125 ng/ml) (Wesche et al., 2019). Recent studies indicate that nematophin is a potent biopesticide against a necrotrophic fungal pathogen R. solani. It interferes with the sclerotial development and hyphal morphology of R. solani at 40.00 μg/ml and germination at 15.00 μg/ml. The ultrastructure shows that the hyphae becomes twisted, shriveled, and deformed at the growing points after the exposure to nematophin at 40.00 μg/ml, and the mitochondrial structural abnormalities such as reduction in number, vacuolar degeneration, and fuzzy cristae are also observed (Figure 3A).
Odilorhabdin
This is a new class of AMP with broad-spectrum activity encoded by the enzymes (of NRPS gene cluster) of X. nematophila. This peptide binds to the decoding center of the small ribosomal subunit, leading to faulty coding procedure and prohibits non-cognate aminoacyl-tRNAs binding (Pantel et al., 2018). Odilorhabdin can directly bind with the new site on 16S rRNA (Figure 3B) and with the anticodon loop of the A-site aminoacyl-tRNA concurrently, resulting in the precision of translation decreased. At very high concentrations, odilorhabdin hinders the ribosome movement on mRNA (Pantel et al., 2018). Studies reported that odilorhabdin acts against Gram-negative and Gram-positive bacterial pathogens, including carbapenem-resistant Enterobacteriaceae, which strain especially shows resistance toward many classes of available antibiotics and causes severe infections with a 50% mortality rate (van Duin et al., 2013).
Darobactin
It is a novel peptide antibiotic produced by Photorhabdus khanii HGB1456 (Imai et al., 2019) that is effective against several Gram-negative drug-resistant pathogens. Instead of targeting the enzymes, darobactin targets outer membrane chaperone BamA (Figure 3C), catalyzing the insertion and folding of β-barrel outer membrane proteins in many Gram-negative pathogens. As the target of darobactin is a cell surface protein, there is no permeability obstacle encountered (Imai et al., 2019). No antibiotics were reported to act on the two surface proteins, namely, BamA and LptD, present on the GNB; therefore, darobactin could act as a potential drug candidate due to its distinctive sizeable molecular structure fused rings and unusual cell surface target (Konovalova et al., 2017).
Photoditritide
Photoditritide is the first non-proteinogenic peptide reported from P. temperata Meg1 through promoter exchange (Maglangit et al., 2021). Photoditritide 19 consists of two tyrosines, two homo-arginines, and two tryptophans (Bode et al., 2015). It is effective against E. coli (MIC = 24 μM), M. luteus (MIC = 3.0 μM), and antiprotozoal activity against P. falciparum (IC50 = 27 μM), T. cruzi (IC50 = 71 μM), and T. brucei rhodesiense (IC50 = 13 μM) (Bode et al., 2015).
The increasing evidence of antibiotic resistance is a serious issue. Drug-resistant pathogens develop new resistance mechanisms and interfere in the treatment of common infections. Moreover, multidrug resistance pathogenic strains have developed tolerance against most of the available antibiotics. Researchers searching for novel sources of antimicrobial agents through synthetic compound library screening have mostly failed to get efficient antimicrobial agents (Payne et al., 2007). Therefore, exploiting new natural antimicrobial sources to fill the research gap in antimicrobial drug discovery is a promising approach. Most of the antibiotics used to date belong to soil actinomycetes. The present review aims to compile novel natural sources, highlighting the unnoticed and ignored sources to identify new AMPs with a unique mode of action. The marine ecosystem presents a vast repository of microorganisms, invertebrates, and vertebrates that produce various natural products and AMPs with the perspective of treating several infectious diseases (Bertrand and Munoz-Garay, 2019).
Marine-Derived Antimicrobial Peptides
The marine ecosystem encompasses an unprecedented variety of organisms that have shown remarkable contribution in discovering and developing novel biomolecules, nutraceuticals, and secondary metabolites that pave the way to produce antimicrobial agents (Malve, 2016; Sekurova et al., 2019; Figure 1). AMPs derived from marine sources are novel and revolutionary therapeutic agents with distinctive pharmacological properties such as antimicrobial, antiproliferative, antioxidant, anticoagulant, antihypertensive, antidiabetic, and antiobesity properties (Jo et al., 2017).
Antimicrobial Peptides Derived From Marine Invertebrates
Marine invertebrates produce AMPs to activate innate immune machinery to recognize, neutralize, and eliminate invading pathogens (Loker et al., 2004). A wide variety of corals produce structurally unique bioactive metabolites that can serve as significant novel compounds in drug development against various human diseases. For example, the marine fungus Simplicillium sp. associated with soft coral Sinularia sp. synthesizes five new peptides, including Sinularia peptides A–E. These bioactive AMPs exhibit significant antimicrobial activity against Mycobacterium tuberculosis, Colletotrichum asianum, and Pyricularia oryzae Cav. Mollusks such as Mytilus edulis, Ruditapes decussatus, and oyster Mytilus galloprovincialis produce AMPs such as myticins and mytilin. A cyclic hexapeptide, cyclo-(Gly-Leu-Val-IIe-Ala-Phe), bacicyclin isolated from Bacillus sp. associated with M. edulis, exhibits antibacterial activities against clinically relevant bacterial strains such as S. aureus and Enterococcus faecalis (Wiese et al., 2018; Zanjani et al., 2018). AMPs derived from marine invertebrates can modulate the lifecycle of bacterial biofilm and also inhibit biofilm formation. Crustin, an antibacterial protein, consists of alanine or threonine, glycine, and glutamine residues at their cleavage site and is derived from the hemolymph of crustaceans (Destoumieux-Garzón et al., 2016). It effectively inhibits biofilm formation of various antibiotic-resistant bacterial strains, including B. pumilis and B. thuringiensis and also is effective against Aeromonas hydrophila and E. coli (Rekha et al., 2018; Sivakamavalli et al., 2020). A novel antibacterial peptide named PcnAMP, extracted from Procambarus clarkia (Pcn) (a red swamp crayfish), exhibits a significant inhibitory effect against Gram-positive and GNB strains such as S. aureus and M. luteus (Zhao B. R. et al., 2020). AMPs from ascidian Didemnum sp. exhibit an antibacterial effect against human pathogens E. faecalis, S. marcescens, S. typhimurium, and S. aureus at MICs of 2.30, 2.17, 2.05, and 1.95 μg/ml, respectively (Arumugam et al., 2020). The AMPs halocyntin and papillosin from tunicate H. papillosa exhibit antibacterial activity against M. luteus and E. coli (Palanisamy et al., 2017). A novel AMP myticusin-beta isolated from the mantle of Mytilus coruscus exhibits a broad range of antibacterial activity and acts as a substitute to antibiotics (Oh et al., 2020). Therefore, the diverse forms of marine invertebrates act as natural reservoirs for novel AMPs, which can be exploited for the treatment of various microbial infections (Thoms et al., 2007; Destoumieux-Garzón et al., 2016; Table 3).
TABLE 3.
Antimicrobial peptides from marine organisms.
| Name of the peptide | Source of peptide | Mode of action | Inhibitory concentration | References |
| An antimicrobial peptide from marine invertebrates | ||||
| Sinulariapeptides A–E | Coral Sinularia sp. | Inhibitory effects against protein tyrosine phosphatases of Mycobacterium tuberculosis (MptpA and MptpB) | IC50 values of 35.0 and 25.9 μM against MptpA and MptpB | Dai et al., 2018 |
| Bacicyclin | Mytilus edulis | Cell membrane damage of Enterococcus faecalis and Staphylococcus aureus | MIC values of Enterococcus faecalis and Staphylococcus aureus was noted to be 8 and 12 mM, respectively | Wiese et al., 2018 |
| Crustin | Portunus pelagicus | The growth reduction and biofilm inhibition potential of on Gram-positive bacteria and Gram-negative bacteria | MIC of both Gram-positive and Gram-negative bacteria was noted to be 30 and 20 μg/ml, respectively | Rekha et al., 2018 |
| An antimicrobial peptide from marine microorganisms | ||||
| Hyporientalin A | Trichoderma orientale | Growth inhibitory effects toward clinical isolates like Candida albicans | MICs of Candida albicans species (247FN and 098 VC) was noted to be 2.55–4.92 μM, respectively | Touati et al., 2018 |
| Fengycins | Bacillus subtilis | Inducing the mitochondrial membrane potential (MMP), reactive oxygen species (ROS), downregulate the ROS-scavenging enzymes and chromatin condensation in plant-pathogenic fungus Magnaporthe grisea | Zhang and Sun, 2018 | |
| EeCentrocin 1 | Echinus esculentus | Cell membrane damage | MIC of Corynebacterium glutamicum and S. aureus (MIC = 0.78 μM) | Solstad et al., 2019 |
| Tetrapeptides 1 | Streptomyces sp. | Growth inhibition of Burkholderia gladioli and Burkholderia glumae | MIC was noted to be 0.068 and 1.1 mM in Burkholderia gladioli and Burkholderia glumae | Betancur et al., 2019 |
| Thr-Pro-Asp-Ser -Glu-Ala-Leu (TPDSEAL) | Porphyra yezoensis | The surface of S. aureus became blurred, loose, irregular, and cell wall damage | Jiao et al., 2019 | |
| An antimicrobial peptide from marine vertebrates | ||||
| Epinecidin-1 | Epinephelus coioides | Disrupted the membrane of metronidazole-resistant Trichomonas vaginalis and completely killed the pathogen | Minimal Epi-1 concentration was noted to be 62.5 μg/ml to produce 100% growth inhibition of Trichomonas vaginalis | Huang et al., 2019 |
| Tissue factor pathway inhibitor 1 (TFPI-2) | Sciaenops ocellatus | TFPI-2 destroying cell membrane integrity, penetrating the cytoplasm and inducing degradation of genomic DNA and total RNA | MICs of TFPI-2 against M. luteus, S. aureus, V. litoralis, V. ichthyoenteri, V. vulnificus, and V. scophthalmi were 3, 6, 11, 85, 170, and 340 μM, respectively | He et al., 2018 |
| Caspian trout (CtHep) | Salmo caspius | The growth inhibition of infectious bacteria | MICs concentration was noted to be 50 and 12.5 μM for Aeromonas hydrophila and Bacillus subtilis | Shirdel et al., 2019 |
Antimicrobial Peptides From Marine Microorganisms
Marine microbial systems are the significant resources of AMPs with unique pharmacological features, including antimicrobial, cytostatic, animal growth, immunosuppressant, antiviral, antimalarial, antiparasitic, promoters, and insecticides activities (Semreen et al., 2018). AMPs extracted from symbiotic marine microorganisms exhibit enhanced broad-spectrum antimicrobial activity. These natural compounds are now being exploited to resolve the microbial drug-resistance problem. Hyporientalin A, an anti-Candida peptaibol, a moronecidin-like peptide from Trichoderma orientale strains, symbiotic fungi of Mediterranean marine sponge Cymbaxinella damicornis, inhibits the growth of clinical isolates of C. albicans, Gram-positive and Gram-negative bacteria (Touati et al., 2018). Cyclic lipopeptide Fengycins from marine bacterium B. subtilis (BS155) is effective against the plant-pathogenic fungus Magnaporthe grisea. Host-dependent marine microbes are excellent sources of many active antimicrobial cyclic peptides (e.g., the cyclolipopeptides cyclodysidins A–D). These peptides, secondary metabolites of Streptomyces sp. associated with sponge Dysidea tupha, exhibit broad-spectrum antimicrobial activities (Indraningrat et al., 2016). Different marine gamma-proteobacteria associated with seaweeds, particularly, Pseudomonas sp., are the primary sources in cyclotetrapeptide cyclo-(isoleucyl-prolyl-leucyl-alanyl), cyclic heptapeptide, scopularides A and B, and ogipeptin A–C. These peptides exhibit intense antimicrobial and anthelmintic activities. Ogipeptin is a powerful agent suppressing the immunostimulatory role of lipopolysaccharides present in the cell wall of GNB (Betancur et al., 2019). Similarly, the marine sponge Tethya aurantium associated with fungus Scopulariopsis brevicaulis synthesizes cyclodepsipeptides scopularides A and B that exhibit effective cytotoxic activity against pathogens (Agrawal et al., 2017). New cyclic lipopeptides maribasins A and B from the broth culture of marine microorganism B. marinus exhibit broad-spectrum activities against phytopathogens such as Fusarium oxysporum, Fusarium graminearum, Verticillium alboatrum, Alternaria solani, and R. solani with the MICs of 25–200 mg/ml (Zhang et al., 2010). Additionally, the two new cyclic tetrapeptides, from the marine strain Streptomyces sp., are effective against Burkholderia gladioli and Burkholderia glumae at MIC of 0.068 and 1.1 mM, respectively. Furthermore, tetrapeptide-2 is effective against B. glumae (MIC = 1.1 mM) and fungal phytopathogens (Betancur et al., 2019). Hence, the diversified marine microorganisms prove to be an effective substitute to the existing antibiotics, thereby reducing the probability of antibiotic-resistant pathogens (Table 3).
Antimicrobial Peptides From Marine Vertebrates
Antimicrobial peptides in marine vertebrates are mainly localized in body fluids, mucous layers, and epithelial surfaces (Edilia Avila, 2017). AMPs participate in body defense mechanisms to eliminate the invading pathogens and enhance physiological and metabolic processes such as toxin neutralization, wound healing, angiogenesis, and iron metabolism. For instance, epinecidin-1 (Epi-1) disrupts the cell membrane of metronidazole-resistant Trichomonas vaginalis and terminates the pathogen with a minimal dose of 62.5 μg/ml. T. vaginalis treated with different concentrations of Epi-1 (62.5, 125, 250, or 500 μg/ml) exhibits 100% growth inhibition (Huang et al., 2019). 3C-terminal peptide tissue factor pathway inhibitor 1 (TFPI-1) from Cyprinus carpio (common carp) exhibits bactericidal effects against M. luteus, S. aureus, and Vibrio vulnificus (Su et al., 2020). Orange-spotted grouper (Epinephelus coioides) derived from AMP EPI is effective against GPB (Su and Chen, 2020). Cysteine-rich Hepcidins (CtHep) from vertebrates such as fish, reptiles, and amphibians can significantly inhibit Streptococcus iniae and A. hydrophila (Shirdel et al., 2019). Marine betta fish Betta splendens produce four families of AMPs, including defensins, piscidins, hepcidins, and LEAP-2, which vigorously suppress the growth of fungi, bacteria, virus, and parasites (Amparyup et al., 2020). A short novel peptide synthesized from the core region of the LCNKL2 of a marine fish Larimichthys crocea inhibits S. aureus and Vibrio harveyi (Zhou et al., 2019). Antibacterial activity of piscidin-5 like AMP has been reported from L. crocea (Pan et al., 2019). Therefore, AMPs are essential to induce adaptive response and participate in a vertebrate’s metabolic and reproductive processes (Table 3).
Conclusion
The exponentially increasing cases of antibiotic resistance requires the introduction of novel and alternative drug molecules. Insects, nematodes, insect–nematode–bacterial associations and marine organisms could be promising sources for natural AMPs to address the challenges of multidrug-resistant infections. The conventional method of overmining natural antibiotic sources has failed to develop new drugs to overcome drug resistance. Genomic analysis indicates the presence of several gene clusters for the novel secondary metabolite biosynthesis. The exploitation of these secondary metabolites might lead to the discovery of potential antimicrobial compounds. This review thereby highlights the symbiotic bacteria–EPN complexes as prospective antimicrobial peptide sources and opens the window to new sources of intervention and invention of natural bioactive compounds to combat antimicrobial resistance. Further research is required to understand the metabolic pathways to optimize the conditions for large-scale production and commercialization of these drug molecules as adequate substitutes.
Author Contributions
SD, FJ, and XX conceptualized the manuscript. SD, AP, and CM drafted the manuscript. AP was responsible for preparing the figures in the manuscript. FJ, NS, SD, AP, and CM assisted in revising the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the reviewers for their invaluable comments and suggestions.
Funding. This work was supported by a grant from the National Natural Science Foundation of China (31972345) and Natural Science Foundation of Guangdong Province of China (2019A151501122).
References
- Abdel-Razek A. (2002). Pathogenicity of bacteria symbiotically associated with insect pathogenic nematodes against the greater wax moth, Galleria mellonella (L.). Arch. Phytopathol. Plant Protection 35 53–60. 10.1080/0323540021000009579 [DOI] [Google Scholar]
- Agrawal S., Acharya D., Adholeya A., Barrow C. J., Deshmukh S. K. (2017). Nonribosomal peptides from marine microbes and their antimicrobial and anticancer potential. Front. Pharmacol. 8:828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahantarig A., Chantawat N., Waterfield N. R., Kittayapong P. J. A., Microbiology E. (2009). PirAB toxin from Photorhabdus asymbiotica as a larvicide against dengue vectors. Appl. Environ. Microbiol. 75 4627–4629. 10.1128/aem.00221-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhurst R., Boemare N., Gaugler R., Kaya H. (1990). Entomopathogenic Nematodes in Biological Control. Boca Raton, Fla: CRC Press. [Google Scholar]
- Amparyup P., Charoensapsri W., Samaluka N., Chumtong P., Yocawibun P., Imjongjirak C. (2020). Transcriptome analysis identifies immune-related genes and antimicrobial peptides in Siamese fighting fish (Betta splendens). Fish Shellfish Immunol. 99 403–413. 10.1016/j.fsi.2020.02.030 [DOI] [PubMed] [Google Scholar]
- Andersson M., Boman A., Boman H. (2003). Ascaris nematodes from pig and human make three anti-bacterial peptides: isolation of cecropin P1 and two ASABF peptides. Cell. Mol. Life Sci. 60 599–606. 10.1007/s000180300051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrä J., Berninghausen O., Leippe M. (2001). Cecropins, antibacterial peptides from insects and mammals, are potently fungicidal against Candida albicans. Med. Microbiol. Immunol. 189 169–173. 10.1007/s430-001-8025-x [DOI] [PubMed] [Google Scholar]
- Anju K., Archana M., Mohandas C., Nambisan B. J. (2015). Purification and identification of an antibacterial protein from the symbiotic bacteria associated with novel entomopathogenic nematode, Rhabditis (Oscheius) sp. World J. Microbiol. Biotechnol. 31 621–632. 10.1007/s11274-015-1816-3 [DOI] [PubMed] [Google Scholar]
- Arumugam V., Venkatesan M., Ramachandran K., Ramachandran S., Palanisamy S. K., Sundaresan U. (2020). Purification, characterization and antibacterial properties of peptide from marine ascidian Didemnum sp. Int. J. Pept. Res. Ther. 26, 201–208. 10.1007/s10989-019-09829-z [DOI] [Google Scholar]
- Aumer T., Voisin S. B. N., Knobloch T., Landon C. L., Bulet P. J. (2020). Impact of an antifungal insect defensin on the proteome of the phytopathogenic fungus Botrytis cinerea. ACS Publications 19 1131–1146. 10.1021/acs.jproteome.9b00638 [DOI] [PubMed] [Google Scholar]
- Balasubramanian N., Toubarro D., Simoes N. (2010). Biochemical study and in vitro insect immune suppression by a trypsin-like secreted protease from the nematode Steinernema carpocapsae. Parasite Immunol. 32 165–175. 10.1111/j.1365-3024.2009.01172.x [DOI] [PubMed] [Google Scholar]
- Baltzer S. A., Brown M. H. (2011). Antimicrobial peptides–promising alternatives to conventional antibiotics. J. Mol. Microbiol. Biotechnol. 20 228–235. 10.1159/000331009 [DOI] [PubMed] [Google Scholar]
- Bang K., Park S., Yoo J. Y., Cho S. (2012). Characterization and expression of attacin, an antibacterial protein-encoding gene, from the beet armyworm, Spodoptera exigua (Hübner)(Insecta: Lepidoptera: Noctuidae). Mol. Biol. Rep. 39 5151–5159. 10.1007/s11033-011-1311-3 [DOI] [PubMed] [Google Scholar]
- Berthold N., Hoffmann R. J. P. (2014). Cellular uptake of apidaecin 1b and related analogs in Gram-negative bacteria reveals novel antibacterial mechanism for proline-rich antimicrobial peptides. Protein Pept. Lett. 21 391–398. 10.2174/09298665113206660104 [DOI] [PubMed] [Google Scholar]
- Bertrand B., Munoz-Garay C. (2019). Marine antimicrobial peptides: a promising source of new generation antibiotics and other bio-active molecules. J. Int. J. Peptide Res. Therapeutics 25 1441–1450. 10.1007/s10989-018-9789-3 [DOI] [Google Scholar]
- Betancur L. A., Forero A. M., Romero-Otero A., Sepúlveda L. Y., Moreno-Sarmiento N. C., Castellanos L., et al. (2019). Cyclic tetrapeptides from the marine strain Streptomyces sp. PNM-161a with activity against rice and yam phytopathogens. J. Antibiotics 72 744–751. 10.1038/s41429-019-0201-0 [DOI] [PubMed] [Google Scholar]
- Bluhm M. E., Schneider V. A., Schäfer I., Piantavigna S., Goldbach T., Knappe D., et al. (2016). N-terminal Ile-Orn-and Trp-Orn-motif repeats enhance membrane interaction and increase the antimicrobial activity of apidaecins against Pseudomonas aeruginosa. Front. Cell Dev. Biol. 4:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode E., Brachmann A. O., Kegler C., Simsek R., Dauth C., Zhou Q., et al. (2015). Simple “on-demand” production of bioactive natural products. J. Chem. Bio Chem. 16 1115–1119. 10.1002/cbic.201500094 [DOI] [PubMed] [Google Scholar]
- Bode H. B. (2009). Entomopathogenic bacteria as a source of secondary metabolites. Curr. Opin. Chem. Biol. 13 224–230. 10.1016/j.cbpa.2009.02.037 [DOI] [PubMed] [Google Scholar]
- Boemare N. (2002). Systematics of Photorhabdus and Xenorhabdus. Entomopathogenic Nematol. 60(Pt 8), 1921–1937. [Google Scholar]
- Boemare N., Akhurst R. (2002). Genera Photorhabdus and Xenorhabdus. Berlin: Springer. [Google Scholar]
- Boemare N., Givaudan A., Brehélin M., Laumond C. J. S. (1997). Symbiosis and pathogenicty of nematode-bacterium complexes. Biol. Bull. 223 85–102. [Google Scholar]
- Boxell A., Lee S. H. C., Jefferies R., Watt P., Hopkins R., Reid S., et al. (2008). Pyrrhocoricin as a potential drug delivery vehicle for Cryptosporidium parvum. Exp. Parasitol. 119 301–303. 10.1016/j.exppara.2008.02.005 [DOI] [PubMed] [Google Scholar]
- Brivio M. F., Mastore M. (2018). Nematobacterial complexes and insect hosts: different weapons for the same war. Insects 9:117. 10.3390/insects9030117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brivio M. F., Mastore M. J. I. (2020). When appearance misleads: the role of the entomopathogen surface in the relationship with its host. Insects 11:387. 10.3390/insects11060387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brivio M. F., Toscano A., De Pasquale S. M., De Lerma Barbaro A., Giovannardi S., Finzi G., et al. (2018). Surface protein components from entomopathogenic nematodes and their symbiotic bacteria: effects on immune responses of the greater wax moth, Galleria mellonella (Lepidoptera: Pyralidae). Pest. Manag. Sci. 74 2089–2099. 10.1002/ps.4905 [DOI] [PubMed] [Google Scholar]
- Bruno R., Maresca M., Canaan S., Cavalier J.-F., Mabrouk K., Boidin-Wichlacz C., et al. (2019). Worms’ antimicrobial peptides. Mar. Drugs 17:512. 10.3390/md17090512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulet P., Hetru C., Dimarcq J.-L., Hoffmann D. (1999). Antimicrobial peptides in insects; structure and function. Dev. Compar. Immunol. 23 329–344. [DOI] [PubMed] [Google Scholar]
- Bulet P., Stöcklin R., Menin L. (2004). Anti-microbial peptides: from invertebrates to vertebrates. Immunol. Rev. 198 169–184. 10.1111/j.0105-2896.2004.0124.x [DOI] [PubMed] [Google Scholar]
- Cabezas-Cruz A., Tonk M., Bleackley M. R., Valdés J. J., Barrero R. A., Hernández-Jarguín A., et al. (2019). Antibacterial and antifungal activity of defensins from the Australian paralysis tick, Ixodes holocyclus. Ticks Tick Borne Dis. 10:101269. 10.1016/j.ttbdis.2019.101269 [DOI] [PubMed] [Google Scholar]
- Cai X., Challinor V. L., Zhao L., Reimer D., Adihou H. L. N., Grün P., et al. (2017). Biosynthesis of the antibiotic nematophin and its elongated derivatives in entomopathogenic bacteria. J. Org. Lett. 19 806–809. 10.1021/acs.orglett.6b03796 [DOI] [PubMed] [Google Scholar]
- Carlsson A., Engström P., Palva E. T., Bennich H. (1991). Attacin, an antibacterial protein from Hyalophora cecropia, inhibits synthesis of outer membrane proteins in Escherichia coli by interfering with omp gene transcription. Infect. Immun. 59 3040–3045. 10.1128/iai.59.9.3040-3045.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D. Z., Serra L., Lu D., Mortazavi A., Dillman A. R. (2019). A core set of venom proteins is released by entomopathogenic nematodes in the genus Steinernema. PLoS Pathog. 15:e1007626. 10.1371/journal.ppat.1007626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaubey A. K., Garg A. P. (2019). “Entomopathogenic nematodes in the biological control of insect pests with reference to insect immunity,” in Plant Biotic Interactions, eds Varma A., Tripathi S., Prasad R. (Berlin: Springer; ). [Google Scholar]
- Chia T.-J., Wu Y.-C., Chen J.-Y., Chi S.-C. (2010). Antimicrobial peptides (AMP) with antiviral activity against fish nodavirus. J. Fish Shellfish Immunol. 28 434–439. 10.1016/j.fsi.2009.11.020 [DOI] [PubMed] [Google Scholar]
- Chung Y.-S. A., Kocks C. J. (2011). Recognition of pathogenic microbes by the Drosophila phagocytic pattern recognition receptor eater. J. Biol. Chem. 286 26524–26532. 10.1074/jbc.m110.214007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cociancich S., Dupont A., Hegy G., Lanot R., Holder F., Hetru C., et al. (1994). Novel inducible antibacterial peptides from a hemipteran insect, the sap-sucking bug Pyrrhocoris apterus. Biochem. J. 300 567–575. 10.1042/bj3000567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cociancich S., Goyffon M., Bontems F., Bulet P., Bouet F., Menez A., et al. (1993). Purification and characterization of a scorpion defensin, a 4kDa antibacterial peptide presenting structural similarities with insect defensins and scorpion toxins. Biochem. Biophys. Res. Commun. 194 17–22. 10.1006/bbrc.1993.1778 [DOI] [PubMed] [Google Scholar]
- Da Silva P., Jouvensal L., Lamberty M., Bulet P., Caille A., Vovelle F. (2003). Solution structure of termicin, an antimicrobial peptide from the termite Pseudacanthotermes spiniger. Protein Sci. 12 438–446. 10.1110/ps.0228303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y., Lin Y., Pang X., Luo X., Wang J., Zhou X., et al. (2018). Peptides from the soft coral-associated fungus Simplicillium sp. SCSIO41209. J Phytochem. 154 56–62. 10.1016/j.phytochem.2018.06.014 [DOI] [PubMed] [Google Scholar]
- Destoumieux-Garzón D., Rosa R. D., Schmitt P., Barreto C., Vidal-Dupiol J., Mitta G., et al. (2016). Antimicrobial peptides in marine invertebrate health and disease. Philos. Trans. R. Soc. B Biol. Sci. 371:20150300. 10.1098/rstb.2015.0300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer J., Rautenbach M., Booysen E., van Staden A., Deane S., Dicks L. J. (2019). Xenorhabdus khoisanae SB10 produces Lys-rich PAX lipopeptides and a Xenocoumacin in its antimicrobial complex. BMC Microbiol. 19:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaud E., Rusniok C., Frangeul L., Buchrieser C., Givaudan A., Taourit S., et al. (2003). The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 21 1307–1313. [DOI] [PubMed] [Google Scholar]
- Dushay M. S., Roethele J. B., Chaverri J. M., Dulek D. E., Syed S. K., Kitami T., et al. (2000). Two attacin antibacterial genes of Drosophila melanogaster. Gene 246 49–57. 10.1016/s0378-1119(00)00041-x [DOI] [PubMed] [Google Scholar]
- Edilia Avila E. (2017). Functions of antimicrobial peptides in vertebrates. Curr. Protein Pept. Sci. 18 1098–1119. [DOI] [PubMed] [Google Scholar]
- Ehret-Sabatier L., Loew D., Goyffon M., Fehlbaum P., Hoffmann J. A., van Dorsselaer A., et al. (1996). Characterization of novel cysteine-rich antimicrobial peptides from scorpion blood. J. Biol. Chem. 271 29537–29544. 10.1074/jbc.271.47.29537 [DOI] [PubMed] [Google Scholar]
- Ellis T. N., Kuehn M. J. (2010). Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol. Mol. Biol. Rev. 74 81–94. 10.1128/mmbr.00031-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elnagdy S., AlKhazindar M. J. (2020). The potential of antimicrobial peptides as an antiviral therapy against COVID-19. ACS Pharmacol. Transl. Sci. 3 780–782. 10.1021/acsptsci.0c00059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantner G. E., Barbero R. J., Gray D. S., Belcher A. M. (2010). Kinetics of antimicrobial peptide activity measured on individual bacterial cells using high-speed atomic force microscopy. Nat. Nanotechnol. 5 280–285. 10.1038/nnano.2010.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehlbaum P., Bulet P., Michaut L., Lagueux M., Broekaert W. F., Hetru C., et al. (1994). Insect immunity. septic injury of Drosophila induces the synthesis of a potent antifungal peptide with sequence homology to plant antifungal peptides. J. Biol. Chem. 269 33159–33163. 10.1016/s0021-9258(20)30111-3 [DOI] [PubMed] [Google Scholar]
- Feng M., Fei S., Xia J., Labropoulou V., Swevers L., Sun J. J. (2020). Antimicrobial peptides as potential antiviral factors in insect antiviral immune response. Front. Immunol. 11:2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ffrench-Constant R., Waterfield N. J. (2006). An ABC guide to the bacterial toxin, complexes. Adv. Appl. Microbiol. 58 169–183. 10.1016/s0065-2164(05)58005-5 [DOI] [PubMed] [Google Scholar]
- Forst S., Nealson K. J. (1996). Molecular biology of the symbiotic-pathogenic bacteria Xenorhabdus spp. and Photorhabdus spp. Microbiol. Rev. 60:21. 10.1128/mr.60.1.21-43.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaugler R. (2018). Entomopathogenic Nematodes in Biological Control. Boca Raton: CRC press. [Google Scholar]
- Geng H., An C.-J., Hao Y.-J., Li D.-S., Du R.-Q. (2004). Molecular cloning and expression of attacin from housefly (Musca domestica). Acta Genet. Sin. 31 1344–1350. [PubMed] [Google Scholar]
- Gerardo N. M., Altincicek B., Anselme C., Atamian H., Barribeau S. M., De Vos M., et al. (2010). Immunity and other defenses in pea aphids, Acyrthosiphon pisum. Genome Biol. 11:R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrard J. G., Joyce S. A., Clarke D. J. (2006). Nematode symbiont for Photorhabdus asymbiotica. Emerg. Infect. Dis. 12:1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gholizadeh A., Moradi B. J. (2020). Cecropins activity against bacterial pathogens. Infect. Dis. Clin. Pract. 29 e6–e12. [Google Scholar]
- Gomes P. D. S., Fernandes M. H. (2010). Defensins in the oral cavity: distribution and biological role. J. Oral Pathol. Med. 39 1–9. 10.1111/j.1600-0714.2009.00832.x [DOI] [PubMed] [Google Scholar]
- Grundmann F., Kaiser M., Kurz M., Schiell M., Batzer A., Bode H. B. (2013). Structure determination of the bioactive depsipeptide xenobactin from Xenorhabdus sp. PB30. 3. RSC Adv. 3 22072–22077. 10.1039/c3ra44721a [DOI] [Google Scholar]
- Grundmann F., Kaiser M., Schiell M., Batzer A., Kurz M., Thanwisai A., et al. (2014). Antiparasitic Chaiyaphumines from entomopathogenic Xenorhabdus sp. PB61. 4. J. Nat. Prod. 77 779–783. 10.1021/np4007525 [DOI] [PubMed] [Google Scholar]
- Gualtieri M., Aumelas A., Thaler J.-O. (2009). Identification of a new antimicrobial lysine-rich cyclolipopeptide family from Xenorhabdus nematophila. J. Antibiot. 62 295–302. 10.1038/ja.2009.31 [DOI] [PubMed] [Google Scholar]
- Guo S., Zhang S., Fang X., Liu Q., Gao J., Bilal M., et al. (2017). Regulation of antimicrobial activity and xenocoumacins biosynthesis by pH in Xenorhabdus nematophila. Microbiol. Cell Factories 16:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han R., Ehlers R.-U. (2000). Pathogenicity, development, and reproduction of Heterorhabditis bacteriophora and Steinernema carpocapsae under axenic in vivo conditions. J. Invertebr. Pathol. 75 55–58. 10.1006/jipa.1999.4900 [DOI] [PubMed] [Google Scholar]
- Hancock R. E. (2001). Cationic peptides: effectors in innate immunity and novel antimicrobials. Lancet Infect. Dis. 1 156–164. 10.1016/s1473-3099(01)00092-5 [DOI] [PubMed] [Google Scholar]
- Hancock R., Patrzykat A. (2002). Clinical development of cationic antimicrobial peptides: from natural to novel antibiotics. Curr. Drug Targets Infect. Disord. 2 79–83. 10.2174/1568005024605855 [DOI] [PubMed] [Google Scholar]
- Haney E. F., Mansour S. C., Hancock R. E. (2017). Antimicrobial peptides: an introduction. J. Antimicrobial Peptides 1548 3–22. 10.1007/978-1-4939-6737-7_1 [DOI] [PubMed] [Google Scholar]
- Hara S., Yamakawa M. (1995). A novel antibacterial peptide family isolated from the silkworm, Bombyx mori. Biochem. J. 310 651–656. 10.1042/bj3100651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazir S., Shapiro-Ilan D. I., Bock C. H., Hazir C., Leite L. G., Hotchkiss M. W. (2016). Relative potency of culture supernatants of Xenorhabdus and Photorhabdus spp. on growth of some fungal phytopathogens. Eur. J. Plant Pathol. 146 369–381. 10.1007/s10658-016-0923-9 [DOI] [Google Scholar]
- He S.-W., Wang J.-J., Du X., Yue B., Wang G.-H., Zhou S., et al. (2018). A teleost TFPI-2 peptide that possesses a broad antibacterial spectrum and immune-stimulatory properties. Fish Shellfish Immunol. 82 469–475. 10.1016/j.fsi.2018.08.051 [DOI] [PubMed] [Google Scholar]
- Hetru C. (1998). Antimicrobial peptides from insects. Molecular mechanisms of immune responses in insects. Dev. Comp. Immunol. 23 329–344. [DOI] [PubMed] [Google Scholar]
- Hetru C., Troxler L., Hoffmann J. A. (2003). Drosophila melanogaster antimicrobial defense. J. Infect. Dis. 187 S327–S334. [DOI] [PubMed] [Google Scholar]
- Himmler T., Pirro F., Schmeer N. (1998). Synthesis and antibacterial in vitro activity of novel analogues of nematophin. J. Bioorganic Med. Chem. Lett. 8 2045–2050. 10.1016/s0960-894x(98)00358-8 [DOI] [PubMed] [Google Scholar]
- Hirsch R., Wiesner J., Bauer A., Marker A., Vogel H., Hammann P. E., et al. (2020). Antimicrobial peptides from rat-tailed maggots of the drone fly eristalis tenax show potent activity against multidrug-resistant gram-negative bacteria. Microorganisms 8:626. 10.3390/microorganisms8050626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeckendorf A., Stanisak M., Leippe M. (2012). The saposin-like protein SPP-12 is an antimicrobial polypeptide in the pharyngeal neurons of Caenorhabditis elegans and participates in defence against a natural bacterial pathogen. Biochem. J. 445 205–212. 10.1042/bj20112102 [DOI] [PubMed] [Google Scholar]
- Hou Z., Lu J., Fang C., Zhou Y., Bai H., Zhang X., et al. (2011). Underlying mechanism of in vivo and in vitro activity of C-terminal–amidated thanatin against clinical isolates of extended-spectrum β-lactamase–producing Escherichia coli. J. Infect. Dis. 203 273–282. 10.1093/infdis/jiq029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houard J., Aumelas A., Noël T., Pages S., Givaudan A., Fitton-Ouhabi V., et al. (2013). Cabanillasin, a new antifungal metabolite, produced by entomopathogenic Xenorhabdus cabanillasii JM26. J. Antibiotics 66 617–620. 10.1038/ja.2013.58 [DOI] [PubMed] [Google Scholar]
- Hu Y., Aksoy S. (2005). An antimicrobial peptide with trypanocidal activity characterized from Glossina morsitans morsitans. Insect Biochem. Mol. Biol. 35 105–115. 10.1016/j.ibmb.2004.10.007 [DOI] [PubMed] [Google Scholar]
- Huang H.-N., Chuang C.-M., Chen J.-Y., Chieh-Yu P. (2019). Epinecidin-1: a marine fish antimicrobial peptide with therapeutic potential against trichomonas vaginalis infection in mice. Peptides 112 139–148. 10.1016/j.peptides.2018.12.004 [DOI] [PubMed] [Google Scholar]
- Hultmark D., Engström A., Andersson K., Steiner H., Bennich H., Boman H. (1983). Insect immunity. Attacins, a family of antibacterial proteins from Hyalophora cecropia. EMBO J. 2 571–576. 10.1002/j.1460-2075.1983.tb01465.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y., Meyer K. J., Iinishi A., Favre-Godal Q., Green R., Manuse S., et al. (2019). A new antibiotic selectively kills Gram-negative pathogens. Nature 576 459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imler J.-L., Bulet P. (2005). “Antimicrobial peptides in Drosophila: structures, activities and gene regulation,” in Mechanisms of Epithelial Defense, eds Kabelit D., Schroder J. M. (Switzerland: Karger Publishers; ). [DOI] [PubMed] [Google Scholar]
- Indraningrat A. A. G., Smidt H., Sipkema D. (2016). Bioprospecting sponge-associated microbes for antimicrobial compounds. Mar. Drugs 14:87. 10.3390/md14050087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa M., Kubo T., Natori S. (1992). Purification and characterization of a diptericin homologue from Sarcophaga peregrina (flesh fly). Biochem. J. 287 573–578. 10.1042/bj2870573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang E.-K., Ullah I., Lim J.-H., Lee I.-J., Kim J.-G., Shin J.-H. (2012). Physiological and molecular characterization of a newly identified entomopathogenic bacteria, Photorhabdus temperata M1021. J. Microbiol. Biotechnol. 22 1605–1612. 10.4014/jmb.1203.03068 [DOI] [PubMed] [Google Scholar]
- Jayamani E., Rajamuthiah R., Larkins-Ford J., Fuchs B. B., Conery A. L., Vilcinskas A., et al. (2015). Insect-derived cecropins display activity against Acinetobacter baumannii in a whole-animal high-throughput Caenorhabditis elegans model. Antimicrob. Agents Chemother. 59 1728–1737. 10.1128/aac.04198-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia F., Wang J., Peng J., Zhao P., Kong Z., Wang K., et al. (2018). The in vitro, in vivo antifungal activity and the action mode of Jelleine-I against Candida species. Amino Acids 50 229–239. 10.1007/s00726-017-2507-1 [DOI] [PubMed] [Google Scholar]
- Jiao K., Gao J., Zhou T., Yu J., Song H., Wei Y., et al. (2019). Isolation and purification of a novel antimicrobial peptide from Porphyra yezoensis. J. Food Biochem. 43:e12864. [DOI] [PubMed] [Google Scholar]
- Jo C., Khan F. F., Khan M. I., Iqbal J. (2017). Marine bioactive peptides: types, structures, and physiological functions. Food Rev. Intl. 33 44–61. 10.1080/87559129.2015.1137311 [DOI] [Google Scholar]
- Kalsy M., Tonk M., Hardt M., Dobrindt U., Zdybicka-Barabas A., Cytrynska M., et al. (2020). The insect antimicrobial peptide cecropin a disrupts uropathogenic Escherichia coli biofilms. npj Biofilms Microbiomes 6:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppi E., Pugsley A. P., Lambert J., Wicker C., Dimarcq J. L., Hoffmann J. A., et al. (1989). Mode of action of diptericin A, a bactericidal peptide induced in the hemolymph of Phormia terranovae larvae. Arch. Insect. Biochem. Physiol. 10 229–239. 10.1002/arch.940100306 [DOI] [Google Scholar]
- Khandelwal P., Banerjee-Bhatnagar N. (2003). Insecticidal activity associated with the outer membrane vesicles of Xenorhabdus nematophilus. Appl. Environ. Microbiol. 69 2032–2037. 10.1128/aem.69.4.2032-2037.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koczulla R., Bals R. (2007). “Cathelicidin antimicrobial peptides modulate angiogenesis,” in Therapeutic Neovascularization-Quo Vadis?, eds Deindl E., Kupatt C. (Netherlands: Springer: ), 191–196. 10.1007/1-4020-5955-8_10 [DOI] [Google Scholar]
- Konovalova A., Kahne D. E., Silhavy T. J. (2017). Outer membrane biogenesis. J. Annu. Rev. Microbiol. 71 539–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenwerth M., Bozhüyük K. A., Kahnt A. S., Steinhilber D., Gaudriault S., Kaiser M., et al. (2014). Characterisation of Taxlllaids A–G; natural products from Xenorhabdus indica. Chem. A Eur. J. 20 17478–17487. 10.1002/chem.201403979 [DOI] [PubMed] [Google Scholar]
- Kumar N., Mohandas C., Nambisan B., Kumar D. S., Lankalapalli R. S. J. (2013). Isolation of proline-based cyclic dipeptides from Bacillus sp. N strain associated with rhabitid entomopathogenic nematode and its antimicrobial properties. Biotechnology 29 355–364. 10.1007/s11274-012-1189-9 [DOI] [PubMed] [Google Scholar]
- Kurz C. L., Tan M. W. (2004). Regulation of aging and innate immunity in C. elegans. Aging Cell 3 185–193. 10.1111/j.1474-9728.2004.00108.x [DOI] [PubMed] [Google Scholar]
- Kwon Y., Kim H., Kim Y., Kang Y., Lee I., Jin B., et al. (2008). Comparative analysis of two attacin genes from Hyphantria cunea. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 151 213–220. 10.1016/j.cbpb.2008.07.002 [DOI] [PubMed] [Google Scholar]
- Lai Y., Gallo R. L. (2009). AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. J. Trends Immunol. 30 131–141. 10.1016/j.it.2008.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G., Kalvelage T., Peters A., Wiese J., Imhoff J. F. (2008). Linear and cyclic peptides from the entomopathogenic bacterium Xenorhabdus nematophilus. J. Nat. Prod. 71 1074–1077. 10.1021/np800053n [DOI] [PubMed] [Google Scholar]
- Langen G., Imani J., Altincicek B., Kieseritzky G., Kogel K.-H., Vilcinskas A. (2006). Transgenic expression of gallerimycin, a novel antifungal insect defensin from the greater wax moth Galleria mellonella, confers resistance to pathogenic fungi in tobacco. Biol. Chem. 387 549–557. 10.1515/bc.2006.071 [DOI] [PubMed] [Google Scholar]
- Leandro L. F., Mendes C. A., Casemiro L. A., Vinholis A. H., Cunha W. R., Almeida R. D., et al. (2015). Antimicrobial activity of apitoxin, melittin and phospholipase A2 of honey bee (Apis mellifera) venom against oral pathogens. An. Acad. Bras. Cienc. 87 147–155. 10.1590/0001-3765201520130511 [DOI] [PubMed] [Google Scholar]
- Lewies A., Du Plessis L. H., Wentzel J. F. (2019). Antimicrobial peptides: the Achilles’ heel of antibiotic resistance? J. Probiotics Antimicrob. Proteins 11 370–381. 10.1007/s12602-018-9465-0 [DOI] [PubMed] [Google Scholar]
- Li J., Chen G., Webster J. M. (1997). Nematophin, a novel antimicrobial substance produced by Xenorhabdus nematophilus (Enterobactereaceae). Can. J. Microbiol. 43 770–773. 10.1139/m97-110 [DOI] [PubMed] [Google Scholar]
- Li J., Chen G., Wu H., Webster J. M. (1995). Identification of two pigments and a hydroxystilbene antibiotic from Photorhabdus luminescens. Appl. Environ. Microbiol. 61 4329–4333. 10.1128/aem.61.12.4329-4333.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim M.-P., Firdaus-Raih M., Nathan S. (2016). Nematode peptides with host-directed anti-inflammatory activity rescue Caenorhabditis elegans from a Burkholderia pseudomallei infection. Front. Microbiol. 7:1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loker E. S., Adema C. M., Zhang S. M., Kepler T. B. (2004). Invertebrate immune systems–not homogeneous, not simple, not well understood. Immunol. Rev. 198 10–24. 10.1111/j.0105-2896.2004.0117.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackintosh J. A., Veal D. A., Beattie A. J., Gooley A. A. (1998). Isolation from an ant Myrmecia gulosa of two inducible O-glycosylated proline-rich antibacterial peptides. J. Biol. Chem. 273 6139–6143. 10.1074/jbc.273.11.6139 [DOI] [PubMed] [Google Scholar]
- Maglangit F., Yu Y., Deng H. (2021). Bacterial pathogens: threat or treat (a review on bioactive natural products from bacterial pathogens). J. Nat. Product Rep. 38 782–821. 10.1039/d0np00061b [DOI] [PubMed] [Google Scholar]
- Mahlapuu M., Håkansson J., Ringstad L., Björn C. (2016). Antimicrobial peptides: an emerging category of therapeutic agents. J. Front. Cell. Infect. Microbiol. 6:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malve H. (2016). Exploring the ocean for new drug developments: marine pharmacology. J. Pharm. Bioallied Sci. 8 83–91. 10.4103/0975-7406.171700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcos J. F., Gandía M. (2009). Antimicrobial peptides: to membranes and beyond. J. Expert Opin. Drug Discov. 4 659–671. 10.1517/17460440902992888 [DOI] [PubMed] [Google Scholar]
- McInerney B. V., Taylor W. C., Lacey M. J., Akhurst R. J., Gregson R. P. (1991). Biologically active metabolites from Xenorhabdus spp., Part 2. Benzopyran-1-one derivatives with gastroprotective activity. J. Nat. Prod. 54 785–795. 10.1021/np50075a006 [DOI] [PubMed] [Google Scholar]
- McManus A. M., Otvos L., Hoffmann R., Craik D. J. (1999). Conformational studies by NMR of the antimicrobial peptide, drosocin, and its non-glycosylated derivative: effects of glycosylation on solution conformation. Biochemistry 38 705–714. 10.1021/bi981956d [DOI] [PubMed] [Google Scholar]
- Minaba M., Ueno S., Pillai A., Kato Y. (2009). Evolution of ASABF (Ascaris suum antibacterial factor)-type antimicrobial peptides in nematodes: Putative rearrangement of disulfide bonding patterns. Dev. Comp. Immunol. 33 1147–1150. 10.1016/j.dci.2009.06.011 [DOI] [PubMed] [Google Scholar]
- Moghaddam M.-R. B., Vilcinskas A., Rahnamaeian M. J. (2017). The insect-derived antimicrobial peptide metchnikowin targets Fusarium graminearum β (1, 3) glucanosyltransferase Gel1, which is required for the maintenance of cell wall integrity. Biol. Chem. 398 491–498. 10.1515/hsz-2016-0295 [DOI] [PubMed] [Google Scholar]
- Moore A. J., Beazley W. D., Bibby M. C., Devine D. A. (1996). Antimicrobial activity of cecropins. J. Antimicrob. Chemother. 37 1077–1089. 10.1093/jac/37.6.1077 [DOI] [PubMed] [Google Scholar]
- Morikawa N., Hagiwara K. I., Nakajima T. (1992). Brevinin-1 and-2, unique antimicrobial peptides from the skin of the frog, Rana brevipoda porsa. Biochem. Biophys. Res. Commun. 189 184–190. 10.1016/0006-291x(92)91542-x [DOI] [PubMed] [Google Scholar]
- Muangpat P., Yooyangket T., Fukruksa C., Suwannaroj M., Yimthin T., Sitthisak S., et al. (2017). Screening of the antimicrobial activity against drug resistant bacteria of Photorhabdus and Xenorhabdus associated with entomopathogenic nematodes from Mae Wong National Park, Thailand. Front. Microbiol. 8:1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee K., Mraheil M. A., Silva S., Müller D., Cemic F., Hemberger J., et al. (2011). Anti-Listeria activities of Galleria mellonella hemolymph proteins. Appl. Environ. Microbiol. 77 4237–4240. 10.1128/aem.02435-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylonakis E., Podsiadlowski L., Muhammed M., Vilcinskas A. (2016). Diversity, evolution and medical applications of insect antimicrobial peptides. Philos. Trans. R. Soc. B Biol. Sci. 371:20150290. 10.1098/rstb.2015.0290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas H. R., Hodgkin J. (2004). Responses to infection and possible recognition strategies in the innate immune system of Caenorhabditis elegans. Mol. Immunol. 41 479–493. 10.1016/j.molimm.2004.03.037 [DOI] [PubMed] [Google Scholar]
- Nissen-Meyer J., Nes I. F. (1997). Ribosomally synthesized antimicrobial peptides: their function, structure, biogenesis, and mechanism of action. Arch. Microbiol. 167 67–77. 10.1007/s002030050418 [DOI] [PubMed] [Google Scholar]
- Nollmann F. I., Dowling A., Kaiser M., Deckmann K., Grösch S., Bode H. B. (2012). Synthesis of szentiamide, a depsipeptide from entomopathogenic Xenorhabdus szentirmaii with activity against Plasmodium falciparum. Beilstein J. Org. Chem. 8 528–533. 10.3762/bjoc.8.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocampo-Ibáñez I. D., Liscano Y., Rivera-Sánchez S. P., Oñate-Garzón J., Lugo-Guevara A. D., Flórez-Elvira L. J., et al. (2020). A novel cecropin D-Derived short cationic antimicrobial peptide exhibits antibacterial activity against wild-type and multidrug-resistant strains of Klebsiella pneumoniae and Pseudomonas aeruginosa. Evol. Bioinform Online 16:1176934320936266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh R., Lee M. J., Kim Y.-O., Nam B.-H., Kong H. J., Kim J.-W., et al. (2020). Myticusin-beta, antimicrobial peptide from the marine bivalve, Mytilus coruscus. Fish Shellfish Immunol. 99 342–352. 10.1016/j.fsi.2020.02.020 [DOI] [PubMed] [Google Scholar]
- Otvos L. (2016). Immunomodulatory effects of anti-microbial peptides. J. Acta Microbiol. Immunol. Hungarica 63 257–277. 10.1556/030.63.2016.005 [DOI] [PubMed] [Google Scholar]
- Ovchinnikova T. V., Aleshina G. M., Balandin S. V., Krasnosdembskaya A. D., Markelov M. L., Frolova E. I., et al. (2004). Purification and primary structure of two isoforms of arenicin, a novel antimicrobial peptide from marine polychaeta Arenicola marina. FEBS Lett. 577 209–214. 10.1016/j.febslet.2004.10.012 [DOI] [PubMed] [Google Scholar]
- Palanisamy S. K., Rajendran N., Marino A. (2017). Natural products diversity of marine ascidians (tunicates; ascidiacea) and successful drugs in clinical development. Nat. Products Bioprospecting 7 1–111. 10.1007/s13659-016-0115-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Zheng L.-B., Mao Y., Wang J., Lin L.-S., Su Y.-Q., et al. (2019). The antibacterial activity and mechanism analysis of piscidin 5 like from Larimichthys crocea. Dev. Comp. Immunol. 92 43–49. 10.1016/j.dci.2018.10.008 [DOI] [PubMed] [Google Scholar]
- Pantel L., Florin T., Dobosz-Bartoszek M., Racine E., Sarciaux M., Serri M., et al. (2018). Odilorhabdins, antibacterial agents that cause miscoding by binding at a new ribosomal site. J. Mol. Cell 70 83–94.e87. [DOI] [PubMed] [Google Scholar]
- Park S.-I., Kim J.-W., Yoe S. M. (2015). Purification and characterization of a novel antibacterial peptide from black soldier fly (Hermetia illucens) larvae. Dev. Comp. Immunol. 52 98–106. 10.1016/j.dci.2015.04.018 [DOI] [PubMed] [Google Scholar]
- Patrulea V., Borchard G., Jordan O. (2020). An update on antimicrobial peptides (AMPs) and their delivery strategies for wound infections. J Pharmaceutics 12:840. 10.3390/pharmaceutics12090840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne D. J., Gwynn M. N., Holmes D. J., Pompliano D. L. (2007). Drugs for bad bugs: confronting the challenges of antibacterial discovery. J Nat. Rev. Drug Discov. 6 29–40. 10.1038/nrd2201 [DOI] [PubMed] [Google Scholar]
- Piel J. (2009). Metabolites from symbiotic bacteria. Nat. Prod. Rep. 26 338–362. 10.1039/b703499g [DOI] [PubMed] [Google Scholar]
- Pillai A., Ueno S., Zhang H., Lee J. M., Kato Y. (2005). Cecropin P1 and novel nematode cecropins: a bacteria-inducible antimicrobial peptide family in the nematode Ascaris suum. Biochem. J. 390 207–214. 10.1042/bj20050218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluzhnikov K. A., Kozlov S. A., Vassilevski A. A., Vorontsova O. V., Feofanov A. V., Grishin E. (2014). Linear antimicrobial peptides from Ectatomma quadridens ant venom. Biochimie 107 211–215. 10.1016/j.biochi.2014.09.012 [DOI] [PubMed] [Google Scholar]
- Pushpanathan M., Gunasekaran P., Rajendhran J. (2013). Antimicrobial peptides: versatile biological properties. J. Int. J. Peptides 2013:675391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichhart J.-M., Meister M., Dimarcq J.-L., Zachary D., Hoffmann D., Ruiz C., et al. (1992). Insect immunity: developmental and inducible activity of the Drosophila diptericin promoter. EMBO J. 11 1469–1477. 10.1002/j.1460-2075.1992.tb05191.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer D. (2013). Identification and Characterization of Selected Secondary Metabolite Biosynthetic Pathways from XENORHABDUS Nematophila. Ph. D. thesis. Frankfurt: Johann Wolfgang Groethe-Universität. [Google Scholar]
- Reimer D., Cowles K. N., Proschak A., Nollmann F. I., Dowling A. J., Kaiser M., et al. (2013). Rhabdopeptides as insect-specific virulence factors from entomopathogenic bacteria. Chembiochem 14 1991–1997. 10.1002/cbic.201300205 [DOI] [PubMed] [Google Scholar]
- Reimer D., Nollmann F. I., Schultz K., Kaiser M., Bode H. (2014). Xenortide biosynthesis by entomopathogenic Xenorhabdus nematophila. J. Nat. Prod. 77 1976–1980. 10.1021/np500390b [DOI] [PubMed] [Google Scholar]
- Rekha R., Vaseeharan B., Ishwarya R., Anjugam M., Alharbi N. S., Kadaikunnan S., et al. (2018). Searching for crab-borne antimicrobial peptides: crustin from Portunus pelagicus triggers biofilm inhibition and immune responses of Artemia salina against GFP tagged Vibrio parahaemolyticus Dahv2. Mol. Immunol. 101 396–408. 10.1016/j.molimm.2018.07.024 [DOI] [PubMed] [Google Scholar]
- Richards G. R., Goodrich-Blair H. (2009). Masters of conquest and pillage: Xenorhabdus nematophila global regulators control transitions from virulence to nutrient acquisition. Cell Microbiol. 11 1025–1033. 10.1111/j.1462-5822.2009.01322.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder T., Stanisak M., Gelhaus C., Bruchhaus I., Grötzinger J., Leippe M. (2010). Caenopores are antimicrobial peptides in the nematode Caenorhabditis elegans instrumental in nutrition and immunity. Dev. Comp. Immunol. 34 203–209. 10.1016/j.dci.2009.09.010 [DOI] [PubMed] [Google Scholar]
- Sanda N. B., Muhammad A., Ali H., Hou Y. (2018). Entomopathogenic nematode Steinernema carpocapsae surpasses the cellular immune responses of the hispid beetle, Octodonta nipae (Coleoptera: Chrysomelidae). Microb. Pathog. 124 337–345. 10.1016/j.micpath.2018.08.063 [DOI] [PubMed] [Google Scholar]
- Schuhmann B., Seitz V., Vilcinskas A., Podsiadlowski L. (2003). Cloning and expression of gallerimycin, an antifungal peptide expressed in immune response of greater wax moth larvae, Galleria mellonella. Arch. Insect Biochem. Physiol. 53 125–133. [DOI] [PubMed] [Google Scholar]
- Sekurova O. N., Schneider O., Zotchev S. B. (2019). Novel bioactive natural products from bacteria via bioprospecting, genome mining and metabolic engineering. Microbial Biotechnol. 12 828–844. 10.1111/1751-7915.13398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semreen M. H., El-Gamal M. I., Abdin S., Alkhazraji H., Kamal L., Hammad S., et al. (2018). Recent updates of marine antimicrobial peptides. Saudi Pharmaceutical J. 26 396–409. 10.1016/j.jsps.2018.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K., Walia S., Ganguli S., Kundu A. (2016). Analytical characterization of secondary metabolites from Indian Xenorhabdus species the symbiotic bacteria of entomopatathogenic nematode (Steinernema spp.) as Antifungal Agent. Natl. Acad. Sci. Lett. 39 175–180. 10.1007/s40009-016-0453-1 [DOI] [Google Scholar]
- Shin H. S., Park S. I. (2019). Novel attacin from Hermetia illucens: cDNA cloning, characterization, and antibacterial properties. Prep. Biochem. Biotechnol. 49 279–285. 10.1080/10826068.2018.1541807 [DOI] [PubMed] [Google Scholar]
- Shirdel I., Kalbassi M. R., Hosseinkhani S., Paknejad H., Wink M. (2019). Cloning, characterization and tissue-specific expression of the antimicrobial peptide hepcidin from caspian trout (Salmo caspius) and the antibacterial activity of the synthetic peptide. Fish Shellfish Immunol. 90 288–296. 10.1016/j.fsi.2019.05.010 [DOI] [PubMed] [Google Scholar]
- Singh J., Banerjee N. (2008). Transcriptional analysis and functional characterization of a gene pair encoding iron-regulated xenocin and immunity proteins of Xenorhabdus nematophila. J. Bacteriol. 190 3877–3885. 10.1128/jb.00209-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakamavalli J., James R. A., Park K., Kwak I.-S., Vaseeharan B. (2020). Purification of WAP domain-containing antimicrobial peptides from green tiger shrimp Peaneaus semisulcatus. Microb. Pathog. 140:103920. 10.1016/j.micpath.2019.103920 [DOI] [PubMed] [Google Scholar]
- Solstad R. G., Johansen C., Stensvåg K., Strøm M. B., Haug T. (2019). Structure-activity relationship studies of shortened analogues of the antimicrobial peptide EeCentrocin 1 from the sea urchin Echinus esculentus. J. Pept. Sci. 26:e3233. [DOI] [PubMed] [Google Scholar]
- Steckbeck J. D., Deslouches B., Montelaro R. C. (2014). Antimicrobial peptides: new drugs for bad bugs? Expert Opin. Biol. Ther. 14 11–14. 10.1517/14712598.2013.844227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner H., Hultmark D., Engström Å, Bennich H., Boman H. G. (1981). Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature 292 246–248. 10.1038/292246a0 [DOI] [PubMed] [Google Scholar]
- Su B.-C., Chen J.-Y. (2020). Epinecidin-1: an orange-spotted grouper antimicrobial peptide that modulates Staphylococcus aureus lipoteichoic acid-induced inflammation in macrophage cells. Fish Shellfish Immunol. 99 362–367. 10.1016/j.fsi.2020.02.036 [DOI] [PubMed] [Google Scholar]
- Su Y.-L., Wang G.-H., Wang J.-J., Xie B., Gu Q.-Q., Hao D.-F., et al. (2020). TC26, a teleost TFPI-1 derived antibacterial peptide that induces degradation of bacterial nucleic acids and inhibits bacterial infection in vivo. Fish Shellfish Immunol. 98 508–514. 10.1016/j.fsi.2020.01.057 [DOI] [PubMed] [Google Scholar]
- Sugar D. R., Murfin K. E., Chaston J. M., Andersen A. W., Richards G. R., deLéon L., et al. (2012). Phenotypic variation and host interactions of Xenorhabdus bovienii SS-2004, the entomopathogenic symbiont of Steinernema jollieti nematodes. Environ. Microbiol. 14 924–939. 10.1111/j.1462-2920.2011.02663.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi M., Ochiai A., Kondo H., Fukuda S., Ishiyama Y., Saitoh E., et al. (2016). Pyrrhocoricin, a proline-rich antimicrobial peptide derived from insect, inhibits the translation process in the cell-free Escherichia coli protein synthesis system. J. Biosci. Bioeng. 121 591–598. 10.1016/j.jbiosc.2015.09.002 [DOI] [PubMed] [Google Scholar]
- Tarr D. (2012). Nematode antimicrobial peptides. Invertebrate Survival J. 9 122–133. [Google Scholar]
- Tasiemski A., Massol F., Cuvillier-Hot V., Boidin-Wichlacz C., Roger E., Rodet F., et al. (2015). Reciprocal immune benefit based on complementary production of antibiotics by the leech Hirudo verbana and its gut symbiont Aeromonas veronii. Sci. Rep. 5:17498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler J.-O., Baghdiguian S., Boemare N. (1995). Purification and characterization of xenorhabdicin, a phage tail-like bacteriocin, from the lysogenic strain F1 of Xenorhabdus nematophilus. Appl. Environ. Microbiol. 61 2049–2052. 10.1128/aem.61.5.2049-2052.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms C., Schupp P. J., Custódio M., Lôbo-Hajdu G., Hajdu E., Muricy G. (2007). Chemical defense strategies in sponges: a review. Porifera Res. Biodivers. Innov. Sustainabil. 28 627–637. [Google Scholar]
- Tobias N. J., Wolff H., Djahanschiri B., Grundmann F., Kronenwerth M., Shi Y.-M., et al. (2017). Natural product diversity associated with the nematode symbionts Photorhabdus and Xenorhabdus. Nat. Microbiol. 2 1676–1685. 10.1038/s41564-017-0039-9 [DOI] [PubMed] [Google Scholar]
- Toro Segovia L. J., Téllez Ramírez G. A., Henao Arias D. C., Rivera Duran J. D., Bedoya J. P., Castaño Osorio J. C. (2017). Identification and characterization of novel cecropins from the Oxysternon conspicillatum neotropic dung beetle. PLoS One 12:e0187914. 10.1371/journal.pone.0187914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touati I., Ruiz N., Thomas O., Druzhinina I. S., Atanasova L., Tabbene O., et al. (2018). Hyporientalin A, an anti-Candida peptaibol from a marine Trichoderma orientale. World J. Microbiol. Biotechnol. 34:98. [DOI] [PubMed] [Google Scholar]
- Tran E. E. H., Goodrich-Blair H. (2009). CpxRA contributes to Xenorhabdus nematophila virulence through regulation of lrhA and modulation of insect immunity. Appl. Environ. Microbiol. 75 3998–4006. 10.1128/aem.02657-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah I., Khan A. L., Ali L., Khan A. R., Waqas M., Hussain J., et al. (2015). Benzaldehyde as an insecticidal, antimicrobial, and antioxidant compound produced by Photorhabdus temperata M1021. J. Microbiol. 53 127–133. 10.1007/s12275-015-4632-4 [DOI] [PubMed] [Google Scholar]
- Ullah I., Khan A. L., Ali L., Khan A. R., Waqas M., Lee I.-J., et al. (2014). An insecticidal compound produced by an insect-pathogenic bacterium suppresses host defenses through phenoloxidase inhibition. Molecules 19 20913–20928. 10.3390/molecules191220913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursic-Bedoya R., Buchhop J., Joy J., Durvasula R., Lowenberger C. (2011). Prolixicin: a novel antimicrobial peptide isolated from Rhodnius prolixus with differential activity against bacteria and Trypanosoma cruzi. Insect Mol. Biol. 20 775–786. 10.1111/j.1365-2583.2011.01107.x [DOI] [PubMed] [Google Scholar]
- Vallet-Gely I., Lemaitre B., Boccard F. (2008). Bacterial strategies to overcome insect defences. Nat. Rev. Microbiol. 6 302–313. 10.1038/nrmicro1870 [DOI] [PubMed] [Google Scholar]
- Van Der Does A. M., Bogaards S. J., Ravensbergen B., Beekhuizen H., Van Dissel J. T., Nibbering P. H. (2010). Antimicrobial peptide hLF1-11 directs granulocyte-macrophage colony-stimulating factor-driven monocyte differentiation toward macrophages with enhanced recognition and clearance of pathogens. J. Antimicrobial Agents 54 811–816. 10.1128/aac.00652-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duin D., Kaye K. S., Neuner E. A., Bonomo R. A. (2013). Carbapenem-resistant Enterobacteriaceae: a review of treatment and outcomes. J. Diagn. Microbiol. Infect. Dis. 75 115–120. 10.1016/j.diagmicrobio.2012.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilcinskas A. (2013). Evolutionary plasticity of insect immunity. J. Insect Physiol. 59 123–129. 10.1016/j.jinsphys.2012.08.018 [DOI] [PubMed] [Google Scholar]
- Vogel H., Müller A., Heckel D. G., Gutzeit H., Vilcinskas A. (2018). Nutritional immunology: diversification and diet-dependent expression of antimicrobial peptides in the black soldier fly Hermetia illucens. Dev. Comp. Immunol. 78 141–148. 10.1016/j.dci.2017.09.008 [DOI] [PubMed] [Google Scholar]
- Volgyi A., Fodor A., Szentirmai A., Forst S. (1998). Phase variation in Xenorhabdus nematophilus. Appl. Environ. Microbiol. 64 1188–1193. 10.1128/aem.64.4.1188-1193.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Li X., Wang Z. (2016). APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 44 D1087–D1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Gaugler R. (1999). Steinernema glaseriSurface Coat protein suppresses the immune response of popillia japonica (Coleoptera: Scarabaeidae) Larvae. Biol. Control 14 45–50. 10.1006/bcon.1998.0672 [DOI] [Google Scholar]
- Waterfield N., George Kamita S., Hammock B. D., Ffrench-Constant R. (2005). The Photorhabdus Pir toxins are similar to a developmentally regulated insect protein but show no juvenile hormone esterase activity. FEMS Microbiol. Lett. 245 47–52. 10.1016/j.femsle.2005.02.018 [DOI] [PubMed] [Google Scholar]
- Webster J., Chen G., Hu K., Li J. (2002). Entomopathogenic Nematology. Wallingfort: CABI. [Google Scholar]
- Wesche F., Adihou H., Wichelhaus T. A., Bode H. B. (2019). Synthesis and SAR of the antistaphylococcal natural product nematophin from Xenorhabdus nematophila. Beilstein J. Org. Chem. 15 535–541. 10.3762/bjoc.15.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese J., Abdelmohsen U. R., Motiei A., Humeida U. H., Imhoff J. F. (2018). Bacicyclin, a new antibacterial cyclic hexapeptide from Bacillus sp. strain BC028 isolated from Mytilus edulis. Bioorg. Med. Chem. Lett. 28 558–561. 10.1016/j.bmcl.2018.01.062 [DOI] [PubMed] [Google Scholar]
- Wu Q., Patoc̆ka J., Kuc̆a K. (2018). Insect antimicrobial peptides, a mini review. Toxins 10:461. 10.3390/toxins10110461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Zhang F., Huang Z., Liu H., Xie C., Zhang J., et al. (2012). Effects of the antimicrobial peptide cecropin AD on performance and intestinal health in weaned piglets challenged with Escherichia coli. Peptides 35 225–230. 10.1016/j.peptides.2012.03.030 [DOI] [PubMed] [Google Scholar]
- Xi X., Lu X., Zhang X., Bi Y., Li X., Yu Z. (2019). Two novel cyclic depsipeptides Xenematides F and G from the entomopathogenic bacterium Xenorhabdus budapestensis. J. Antibiot. 72 736–743. 10.1038/s41429-019-0203-y [DOI] [PubMed] [Google Scholar]
- Xiao Y., Meng F., Qiu D., Yang X. (2012). Two novel antimicrobial peptides purified from the symbiotic bacteria Xenorhabdus budapestensis NMC-10. Peptides 35 253–260. 10.1016/j.peptides.2012.03.027 [DOI] [PubMed] [Google Scholar]
- Xing-zhong L. U., Dan-shu S., Chun-zhi G., Xiao-mei T., Yu-hui B. (2016). Isolation and identification of secondary metabolites from Xenorhabdus budapestensis. Nat. Prod. Res. Dev. 28 828–832. [Google Scholar]
- Xu X., Zhong A., Wang Y., Lin B., Li P., Ju W., et al. (2019). Molecular identification of a moricin family antimicrobial Peptide (Px-Mor) from Plutella xylostella with activities against the opportunistic human pathogen Aureobasidium pullulans. Front. Microbiol. 10:2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L. L., Zhan M. Y., Zhuo Y. L., Dang X. L., Li M. Y., Xu Y., et al. (2020). Characterization of the active fragments of Spodoptera litura Lebocin-1. Insect Biochem. Physiol. 103:e21626. [DOI] [PubMed] [Google Scholar]
- Yang L.-L., Zhan M.-Y., Zhuo Y.-L., Pan Y.-M., Xu Y., Zhou X.-H., et al. (2018). Antimicrobial activities of a proline-rich proprotein from Spodoptera litura. Dev. Comp. Immunol. 87 137–146. [DOI] [PubMed] [Google Scholar]
- Yu G., Baeder D. Y., Regoes R. R., Rolff J. (2018). Predicting drug resistance evolution: insights from antimicrobial peptides and antibiotics. J. Proc. R. Soc. B Biol. Sci. 285:20172687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanjani N. T., Miranda-Saksena M., Cunningham A. L., Dehghani F. (2018). Antimicrobial peptides of marine crustaceans: the potential and challenges of developing therapeutic agents. Curr. Med. Chem. 25 2245–2259. [DOI] [PubMed] [Google Scholar]
- Zasloff M. (2002). Antimicrobial peptides of multicellular organisms. Nature 415 389–395. [DOI] [PubMed] [Google Scholar]
- Zhang D.-J., Liu R.-F., Li Y.-G., Tao L.-M., Tian L. (2010). Two new antifungal cyclic lipopeptides from Bacillus marinus B-9987. Chem. Pharm. Bull. 58 1630–1634. [DOI] [PubMed] [Google Scholar]
- Zhang L., Sun C. (2018). Fengycins, cyclic lipopeptides from marine Bacillus subtilis strains, kill the plant-pathogenic fungus Magnaporthe grisea by inducing reactive oxygen species production and chromatin condensation. Appl. Environ. Microbiol. 84:e0445-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Liu Q., Han Y., Han J., Yan Z., Wang Y., et al. (2019). Nematophin, an antimicrobial dipeptide compound from Xenorhabdus nematophila YL001 as a potent biopesticide for Rhizoctonia solani control. Front. Microbiol. 10:1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B.-R., Zheng Y., Gao J., Wang X.-W. (2020). Maturation of an antimicrobial peptide inhibits aeromonas hydrophila infection in crayfish. J. Immunol. 204 487–497. [DOI] [PubMed] [Google Scholar]
- Zhao L., Vo T. D., Kaiser M., Bode H. (2020). Phototemtide A, a cyclic lipopeptide heterologously expressed from photorhabdus temperata Meg1, shows selective antiprotozoal activity. Chembiochem 21 1288–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zharkova M. S., Orlov D. S., Golubeva O. Y., Chakchir O. B., Eliseev I. E., Grinchuk T. M., et al. (2019). Application of antimicrobial peptides of the innate immune system in combination with conventional antibiotics—a novel way to combat antibiotic resistance? J. Front. Cell. Infect. Microbiol. 9:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Grundmann F., Kaiser M., Schiell M., Gaudriault S., Batzer A. (2013). Structure and biosynthesis of xenoamicins from entomopathogenic Xenorhabdus. Chem. A Eur. J. 19 16772–16779. [DOI] [PubMed] [Google Scholar]
- Zhou Q.-J., Wang J., Mao Y., Liu M., Su Y.-Q., Ke Q.-Z., et al. (2019). Molecular structure, expression and antibacterial characterization of a novel antimicrobial peptide NK-lysin from the large yellow croaker Larimichthys crocea. Aquaculture 500 315–321. [Google Scholar]