Abstract
Background
Elevated Interleukin-6 (IL-6) may play an important role in the pathophysiology of COVID-19 yet attenuated response is not seen across all severe patients. We aimed to determine the effect of IL-6 baseline levels and other clinical variables on mortality and outcomes in hospitalized COVID-19 patients as well as to explore genetic variants associated with attenuated IL-6 response.
Methods
Baseline IL-6 cytokine levels were measured in hospitalized patients participating in ongoing ODYSSEY phase 3 randomized study of tradipitant and placebo in hospitalized patients with severe COVID-19 who are receiving supplemental oxygen support. Furthermore blood samples for whole genome sequencing analysis were collected from 150 participants.
Results
We report significantly elevated IL-6 in COVID-19 infected hospitalized patients, n = 100 (p-value < 0.0001) when compared to controls n = 324. We also report a significantly increased level of IL-6 (p-value < 0.01) between the severe and mild COVID-19 patients with severity defined on a WHO scale. Excessive IL-6 plasma levels correlate with higher mortality (p-value 0.001). Additionally, based on our classification analysis, combination of IL-6 elevation and high levels of serum glucose can identify highest risk-group of COVID19 patients.
Furthermore, we explore the role of genetic regulatory variants affecting baseline IL-6 levels specifically in COVID-19 patients. We have directly tested the association between variants in the IL6 and IL6R gene region and IL6 plasma levels. We provide results for a common IL-6 variant previously associated with pneumonia, rs1800795, and rs2228145 that was previously shown to affect IL-6 plasma levels, as well as report on novel variants associated with IL-6 plasma levels detected in our study patients.
Conclusions
While it is unlikely that “cytokine storm” is the norm in severe COVID19, baseline elevations above 150 pg/ml may be associated with worst outcomes and as such may warrant treatment considerations. So far no clinical studies used IL-6 baseline assessment to stratify the patient population participating in clinical studies. We believe that careful examination and interpretation of the IL-6 levels and genetic variants can help to determine a patient population with a potentially very robust clinical response to IL-6 inhibition.
Trial Registration: Clinicaltrials.gov: NCT04326426.
1. Background
Elevated IL-6 may play an important biomarker in the body’s response to and course of coronavirus disease 2019 (COVID-19) [1]. In Covid-19 positive patients, a high number of T lymphocytes and mononuclear macrophages are activated, resulting in increased production of IL-6, binding the IL-6 receptor ultimately resulting in cytokine storm [2]. Interleukin 6 is a potent pleiotropic cytokine that regulates cell growth and differentiation and plays an important role in the immune response. High IL-6 serum levels have been associated with various attenuated immune responses, associated with asthma, atopic dermatitis and recently in COVID-19. IL-6 and TNF-∝ serum levels are associated with early death in community-acquired pneumonia patients [3]. Recently, Kox et al., and other studies reported cytokine levels in critically ill patients with COVID-19 and compared those levels in patients with other critical illnesses [4], [5]. We believe we are able to contribute to the current knowledge on cytokines response as we have conducted a controlled clinical study on hospitalized COVID-19 patients. We have used a panel consisting of 13 cytokines measuring baseline cytokine levels in all our clinical study participants. Our primary focus was studying the distribution of IL-6 at baseline.
Beyond exploring the plasma cytokine levels at baseline we further explore genetic variants in the IL-6 and IL6R gene region to see if certain variants act as risk loci in terms of IL-6 response. We have conducted whole genome sequencing analysis of the samples collected to explore the potential role of genetic variants affecting plasma IL-6 levels [6].
2. Results
2.1. IL-6
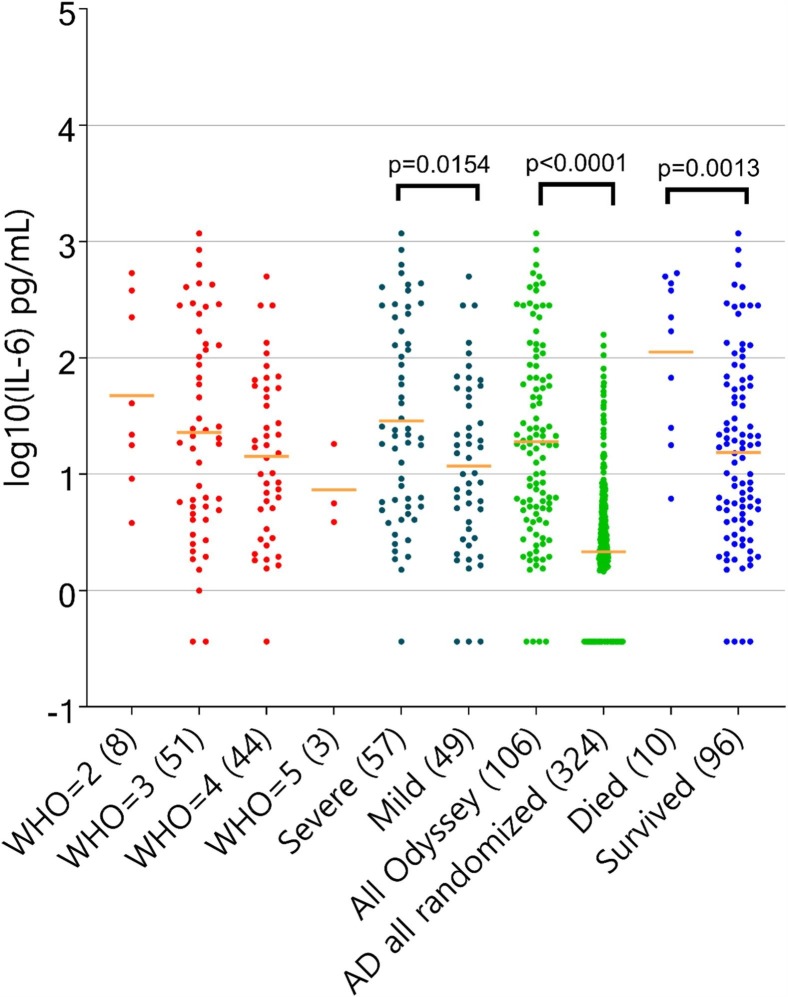
As part of explorative tests on patients participating in our trial we measured baseline plasma cytokine levels of IL-6. We report baseline demographic summary as follows: sex (male) 65.2%, mean age 62.6, mean BMI 32.1. Fig. 1 shows IL-6 distribution based on baseline samples obtained from our study cohort. We report significantly elevated IL-6 in COVID-19 infected hospitalized patients, n = 106 (p-value < 0.0001) when compared to controls n = 324. IL-6 levels above 300 pg/mL were associated with elevated mortality when compared to the rest of the COVID-19 patient cohort (55.5% vs 5.2%, p-value < 0.001). Additionally IL-6 levels were higher among advanced clinical patients (baseline, WHO2/3 vs WHO4/5, p-value < 0.015. COVID-19 hospitalized patients had significantly higher levels of IL-6 compared to Atopic dermatitis patients. Our clinical samples also seem to have a higher IL-6 baseline, average values than those recently reported COVID-19 ARDS cases in Kox et al [4]. We additionally assessed the effects of sex, age, and BMI (Supplementary Fig. 1). In this cohort, sex and BMI did not have an effect (no statistically significant difference reported across BMI nor between males and females). The effect of age was trending with older individuals (>=65 years old) having higher IL6 levels (result did not attain statistical significance p-value = 0.06).
Fig. 1.
Distribution of IL-6 levels across multiple subsets grouped by WHO scale (red), severe versus mild (black), Odyssey patients versus controls (green), died versus survived (blue).
For all the patients treated with Tocilizumab (TZM), observed mortality was higher than rest of the cohort 5/13 (38.5%) vs 4/46 (8.7%). Out of 13 patients treated with TZM 4 patients had IL-6 baseline levels above 300 pg/mL – 3 out of those 4 patients died on TZM treatment. For patients treated with TZM and with IL-6 baseline levels smaller than 300 pg/mL mortality was 22.2% (2/9), well above mortality for the rest of the cohort. This observation limited by sample size warrants further confirmation. In a classification using combination of all baseline clinical variables and cytokines, the majority of the mortality can be captured by a combination of baseline elevated plasma glucose (>127 mg/dL) and elevated IL-6 (>150 pg/mL). This high risk group consists of only 8 patients but accounts for ~66% of mortality for the cohort.
2.2. Genetic modifiers of IL-6 levels
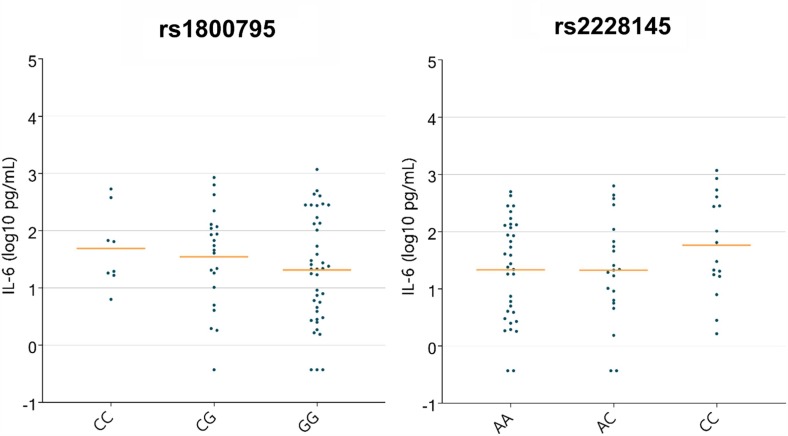
Several genetic variants have been reported in the past with respect to IL-6 serum levels [7], [8], [6]. We evaluate such pre-identified associations as well as explore new ones in the current whole genome sequencing data analyses. IL-6-174 G/C (rs1800795) is a variant in the promoter of the IL-6 gene that was shown to affect IL-6 serum levels. It has been previously reported in relation to serum IL6 in Egyptian children with community-acquired pneumonia [8]. Interestingly, in our study the minor allele seems to correlate with mortality. It is a common variant with a global MAF of 0.68 (higher in males and large allelic differences between East Asian (1) and European (0.55). Looking at mortality 6 of the 30 allele carriers died, in comparison to 3 out of 41 as shown in Table 1 . Fig. 2 A displays IL6 levels across rs1800795 genotypes (log scale). In the current study cohort, the minor allele trend is consistent with literature reports [8], but significance is not reached (linear regression model). This locus is a significant (p-value 8.2e-22) eQTL in the lung for IL6-AS1 as reported by GTEX. Minor allele, and CC genotype is reported to confer higher expression levels. The minor allele for this variant if further replicated in other studies could then be a risk locus associated with higher mortality and perhaps a biomarker of earlier intervention.
Table 1.
Mortality in carriers of the IL-6 rs1800795 minor allele.
| GG (n = 41) |
CG (n = 22) |
CC (n = 8) |
||||
|---|---|---|---|---|---|---|
| N | % | N | % | N | % | |
| Hospital mortality | 3 | (7.3) | 4 | (18.1) | 2 | (25.0) |
Fig. 2.
Distribution of IL-6 across genotypes for the two variants a. rs1800795 and b. 2228145.
We have also inspected WGS samples from our COVID-19 patients for the presence of loss-of-function IL-6 variants – we do not report any such variants in this study cohort. Furthermore, we conducted analysis of modifiers of plasma IL-6 level on this set of sample WGS samples obtained from study patients. We have directly tested the association between variants in the IL6 and IL6R region and IL-6 plasma levels. Linear regression corrected for PCs and covariates identified a strong significant signal within the IL6R region, IL6R Asp358Ala variant (rs2228145) is displayed on Fig. 2B. The minor allele is associated with higher plasma IL-6 levels (linear regression assuming additive effects p-value < 0.03). This variant was previously reported to be associated with aberrant IL-6 levels in a cohort of 1674 individuals (P < 2.0 × 10−9 for IL-6) in a non COVID-19 background [9]. It is a significant eQTL for expression of TDRD10 in the lung tissue, a gene associated previously with IL6R level and for IL6R in whole blood [10]. All other variants identified in this analysis with significant association with IL-6 serum levels are listed in Supplementary Table 1 and include rs13306435 – a variant furthermore associated with different profiles of IL-6 response to immunization and rs12132326, an upstream IL6R variant.
We were able to obtain viral sequencing as described in the Methods section, for 77 patients. We had only two cases with the A lineage infection and 75 individuals infected by the B lineage (B.1, B.1.2, B.1.3, B.1.1.7). We did not observe a significant association between IL and 6 levels and variant lineage – however this warrants further replication on a larger cohort.
3. Methods
3.1. Participants
ODYSSEY is a double-blinded Phase 3 study with a planned randomization of a total of 300 hospitalized severely ill COVID-19 patients (Clinicaltrials.gov: NCT04326426). Inclusion criteria for the study comprised of: 1. Adults aged 18–90; 2. confirmed laboratory COVID-19 infection; 3. confirmed pneumonia by chest radiograph or computed tomography; 4. fever defined as temperature ≥36.6 °C armpit, ≥37.2C oral, or ≥37.8 °C rectal since admission or the use of antipyretics; 5. PaO2/FiO2 ≤ 300; 6. In patient hospitalization. Patients were to be followed for up to 28 days to record clinical outcomes. Patients’ clinical progress was recorded on a 7 point clinical status ordinal scale defined as follows: 1- Death; 2- Hospitalized on mechanical ventilation or ECMO; 3- Hospitalized on non invasive ventilation or high-flow oxygen supplementation; 4- Hospitalized requiring supplemental oxygen; 5- Hospitalized not requiring supplemental oxygen, requiring continued medical care; 6- Hospitalized not requiring supplemental oxygen, not requiring continued medical care; 7- Not hospitalized. Main Exclusion Criteria included: 1. Inability to provide informed consent or to have an authorized relative or designated person provide informed consent, or to comply with the protocol requirements; 2. Known allergy to tradipitant or other neurokinin-1 antagonists; 3. Pregnancy; 4. Uncontrolled HIV, HBV, or HCV infection; 5. Other uncontrolled medically significant diseases; 6. Enrollment in another clinical trial of an investigational therapy; 7. Alanine aminotransferase >5X Upper Limit of Normal or Creatinine clearance <50 m 174 l/min; 8. Requiring mechanical ventilation for >72 h.
3.2. Cytokines
Quantitative determination of a multiplex bead-based method (Luminex) for the detection of cytokines (CM-CSF, IFN-g, IL-12(p70), IL-1B, IL-4, IL-5, IL-6, IL-8, IL-10, IL-13, IL-23, TNF-a and IL-17A) in Human plasma was performed at Charles River Laboratories (Montreal, Canada). The Human Cytokine panel - catalogue # HSTCMAG28SK from Millipore was used in this study.
3.3. Viral sequencing and genome assembly
Sequencing was attempted on all samples with a positive RT-PCR assay result that had Ct < 32 using either a metagenomic approach described previously, via IDT probe-capture, or by amplicon sequencing with SWIFT library preparation [11], [12], [13]. Libraries were sequenced on Illumina MiSeq or NextSeq instruments using 1 × 150 or 1 × 75 runs respectively. Consensus sequences were assembled using a custom bioinformatics pipeline [https://github.com/proychou/hCoV19] adapted for SARS-CoV-2 from previous work. Briefly, raw reads were trimmed to remove adapters and low quality regions using BBDuk and a k-mer based filter was used to pull out reads matching the reference sequence NC_045512. Filtered reads were de novo assembled using SPAdes and contigs were ordered against the reference using BWA-MEM [14]. Gaps were filled by remapping reads against the assembled scaffold and a consensus sequence was called from this alignment using a custom script in R/Bioconductor.
3.4. Genetic analysis: DNA quantification
Incoming nucleic acid samples are quantified using fluorescent-based assays (PicoGreen) to accurately determine whether sufficient material is available for library preparation and sequencing. DNA sample size distributions are profiled by a Fragment Analyzer (Advanced Analytics) or BioAnalyzer (Agilent Technologies), to assess sample quality and integrity. HumanCoreExome 24v1.3 array was performed on all human DNA samples sequenced. Whole genome sequencing (WGS) libraries were prepared using the Truseq DNA PCR-free Library Preparation Kit. Whole Genome data were processed on NYGC automated pipeline. Paired-end 150 bp reads were aligned to the GRCh37 human reference (BWA-MEM v0.7.8) and processed with GATK best-practices workflow (GATK v3.4.0). The mean coverage was 35.8, it reflects the samples average. All high quality variants obtained from GATK were annotated for functional effects (intronic, intergenic, splicing, nonsynonymous, stopgain and frameshifts) based on RefSeq transcripts using Annovar31. Additionally, Annovar was used to match general population frequencies from public databases (Exac, gnomAD, ESP6500, 1000 g) and to prioritize rare, loss-of-function variants. Linear models adjusted for PC, age and sex were conducted in PLINK.
3.5. Statistical analysis and machine learning
WEKA package was used for classification and regression trees, specifically J48. All additional Statistical analysis were done in Graphpad and Octave.
4. Discussion
Recently several trials based on an IL6 hypothesis were conducted with mixed results thus far. A randomized clinical study using anti-IL-6 monoclonal antibody Actemra (tocilizumab) failed to show improvement in response (COVACTA, NCT04320615) [15]. The same was true of another anti-IL-6 mAbs Kevzara (sarilumab) from Regeneron Pharmaceuticals Inc. and Sanofi. On the other hand a small cohort, open-label study from China treatment of with tocilizumab has been reported preliminary data showing tocilizumab improved the clinical outcomes severe COVID-19 patients [2].This group reported 0% mortality when treated with tocilizumab on 21 patients that is strikingly different than other studies where mortality is ~20% Covacta, and observational, retrospective study reported mortality of ~49% in another large cohort. This is in line with previous studies reporting tocilizumab effective in decreasing mortality in community acquired pneumonia. Unfortunately, many large controlled trials have not reported IL-6 baseline levels; having the information regarding cytokine levels would likely be very informative. Our results similarly to Kox et al., do not lend universal support but suggest that for certain patients IL-6 inhibition could be a reasonable treatment option. Our data emphasize the importance of measuring IL-6 levels, especially given the heterogeneous levels reported across COVID-19 infected patients. Our data also emphasize the need for further investigations of IL-6 levels pre and post treatments. Furthermore, the detected gene variants within IL6 and IL6R in this study, could be considered risk loci biomarkers indicative of higher IL-6 plasma levels. If confirmed with further replication studies, such loci could serve as biomarkers of greater risk of higher IL-6 response, and hence could suggest altered treatment regimens for carriers, tailored to their IL-6 levels.
The present study is limited by sample size and warrants further replication on larger sets especially in terms of replicating the novel variants reported in this study. Although most patients were infected by the same viral lineage, there are couple individuals infected with variants carrying novel variants. Such a situation that may produce non-uniform clinical outcomes among the patients including in this study, affecting the production of IL-6. In future follow up analyses, it will be informative to discern the potential association between the infection with specific viral variant categories and the induction of IL-6 in infected patients.
5. Conclusion
In a severe COVID19 cohort, IL-6 levels were on average elevated but not to levels described before in ARDS cohorts or in cohorts with a cytokine storm. Patients with significantly increased levels of over 150 pg/ml showed a trend towards increased mortality. Furthermore we identify potential novel variants as well as confirm known risk loci for attenuated IL-6 response. Using combination of all baseline labs and all cytokines, a majority of the mortality can be captured by a combination of baseline elevated serum glucose and elevated IL-6. While it is unlikely that “cytokine storm” is the norm in severe COVID19, baseline elevations above 150 pg/ml may be associated with worse outcomes and as such may warrant treatment considerations. Genetic variants may help discern risk for more severe COVID-19 infection.
Declarations
Ethics approval and consent to participate.
IRB approval status: Reviewed and approved by Advarra IRB; Pro00043096.
Consent for publication
All authors consent.
Availability of data and material
Vanda shares data related to this trial in peer-reviewed journals, medical conferences, and ClinicalTrials.gov. Individual patient data are not publicly available. Patient level clinical and genetic data are presented in this paper and are freely available upon request
Funding
Vanda Pharmaceuticals Inc.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to thank all our investigators, contributors and foremost study participants.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cyto.2021.155662.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
Legend: Linear model for IL6 levels.
References
- 1.Liu B.M., Martins T.B., Peterson L.K., Hill H.R. Clinical significance of measuring serum cytokine levels as inflammatory biomarkers in adult and pediatric COVID-19 cases: a review. Cytokine. 2021;142:155478. doi: 10.1016/j.cyto.2021.155478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu X., Han M., Li T., Sun W., Wang D., Fu B., Zhou Y., Zheng X., Yang Y., Li X., Zhang X., Pan A., Wei H. Effective treatment of severe COVID-19 patients with tocilizumab. PNAS. 2020;117(20):10970–10975. doi: 10.1073/pnas.2005615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bacci M.R., Leme R.C.P., Zing N.P.C., Murad N., Adami F., Hinnig P.F., Feder D., Chagas A.C.P., Fonseca F.L.A. IL-6 and TNF-α serum levels are associated with early death in community-acquired pneumonia patients. Braz. J. Med. Biol. Res. 2015;48(5):427–432. doi: 10.1590/1414-431X20144402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kox M., Waalders N.J.B., Kooistra E.J., Gerretsen J., Pickkers P. Cytokine levels in critically ill patients with COVID-19 and other conditions. JAMA. 2020;324(15):1565. doi: 10.1001/jama.2020.17052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sinha P., Matthay M.A., Calfee C.S. Is a ‘cytokine Storm’ relevant to COVID-19? JAMA Int. Med. 2020;180:E1–E3. doi: 10.1001/jamainternmed.2020.3313. [DOI] [PubMed] [Google Scholar]
- 6.J. Van Dongen et al., The contribution of the functional IL6R polymorphism rs2228145, eQTLs and other genome-wide SNPs to the heritability of plasma sIL-6R levels. [DOI] [PMC free article] [PubMed]
- 7.Shah T., et al. Gene-centric analysis identifies variants associated with interleukin-6 levels and shared pathways with other inflammation markers. Circ.: Cardiovascular Gen. 2013:163–170. doi: 10.1161/CIRCGENETICS.112.964254. [DOI] [PubMed] [Google Scholar]
- 8.Zidan H.E., Elbehedy R.M., Azab S.F. IL6-174 G/C gene polymorphism and its relation to serum IL6 in Egyptian children with community-acquired pneumonia. Cytokine. 2014;67(2):60–64. doi: 10.1016/j.cyto.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 9.Reich D., Patterson N., Ramesh V., De Jager P.L., McDonald G.J., Tandon A., Choy E., Hu D., Tamraz B., Pawlikowska L., Wassel-Fyr C., Huntsman S., Waliszewska A., Rossin E., Li R., Garcia M., Reiner A., Ferrell R., Cummings S., Kwok P.-Y., Harris T., Zmuda J.M., Ziv E. Admixture mapping of an allele affecting interleukin 6 soluble receptor and interleukin 6 levels. Am. J. Hum. Genet. 2007;80(4):716–726. doi: 10.1086/513206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., Hasz R., Walters G., Garcia F., Young N., Foster B., Moser M., Karasik E., Gillard B., Ramsey K., Sullivan S., Bridge J., Magazine H., Syron J., Fleming J., Siminoff L., Traino H., Mosavel M., Barker L., Jewell S., Rohrer D., Maxim D., Filkins D., Harbach P., Cortadillo E., Berghuis B., Turner L., Hudson E., Feenstra K., Sobin L., Robb J., Branton P., Korzeniewski G., Shive C., Tabor D., Qi L., Groch K., Nampally S., Buia S., Zimmerman A., Smith A., Burges R., Robinson K., Valentino K., Bradbury D., Cosentino M., Diaz-Mayoral N., Kennedy M., Engel T., Williams P., Erickson K., Ardlie K., Winckler W., Getz G., DeLuca D., MacArthur D., Kellis M., Thomson A., Young T., Gelfand E., Donovan M., Meng Y., Grant G., Mash D., Marcus Y., Basile M., Liu J., Zhu J., Tu Z., Cox N.J., Nicolae D.L., Gamazon E.R., Im H.K., Konkashbaev A., Pritchard J., Stevens M., Flutre T., Wen X., Dermitzakis E.T., Lappalainen T., Guigo R., Monlong J., Sammeth M., Koller D., Battle A., Mostafavi S., McCarthy M., Rivas M., Maller J., Rusyn I., Nobel A., Wright F., Shabalin A., Feolo M., Sharopova N., Sturcke A., Paschal J., Anderson J.M., Wilder E.L., Derr L.K., Green E.D., Struewing J.P., Temple G., Volpi S., Boyer J.T., Thomson E.J., Guyer M.S., Ng C., Abdallah A., Colantuoni D., Insel T.R., Koester S.E., Little A.R., Bender P.K., Lehner T., Yao Y., Compton C.C., Vaught J.B., Sawyer S., Lockhart N.C., Demchok J., Moore H.F. The genotype-tissue expression (GTEx) project. Nat. Genet. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greninger A.L., Zerr D.M., Qin X., Adler A.L., Sampoleo R., Kuypers J.M., Englund J.A., Jerome K.R., McAdam A.J. Rapid metagenomic next-generation sequencing during an investigation of hospital-acquired human parainfluenza virus 3 infections. J. Clin. Microbiol. 2017;55(1):177–182. doi: 10.1128/JCM.01881-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greninger A.L., Roychoudhury P., Xie H., Casto A., Cent A., Pepper G., Koelle D.M., Huang M.-L., Wald A., Johnston C., Jerome K.R., Fernandez-Sesma A., Houldcroft C., Cohrs R. Ultrasensitive capture of human herpes simplex virus genomes directly from clinical samples reveals extraordinarily limited evolution in cell culture. mSphere. 2018;3(3) doi: 10.1128/mSphereDirect.00283-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greninger A.L., et al. Proteomic reannotation of human herpesvirus. BMC Genomics. 2018;6:1–17. doi: 10.1186/s12864-018-4604-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., Pyshkin A.V., Sirotkin A.V., Vyahhi N., Tesler G., Alekseyev M.A., Pevzner P.A. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campochiaro C., Dagna L. The conundrum of interleukin-6 blockade in COVID-19. Lancet Rheumatol. 2020;2(10):e579–e580. doi: 10.1016/S2665-9913(20)30287-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Legend: Linear model for IL6 levels.
Data Availability Statement
Vanda shares data related to this trial in peer-reviewed journals, medical conferences, and ClinicalTrials.gov. Individual patient data are not publicly available. Patient level clinical and genetic data are presented in this paper and are freely available upon request