Abstract
Background
Inclusion body hepatitis (IBH) is an economically important viral disease primarily affecting broiler and breeder chickens. All 12 serotypes of fowl adenovirus (FAdV) can cause IBH.
Objectives
To characterize FAdV isolates based on phylogenetic analysis, and to study the pathogenicity of FAdV-8b in specific-pathogen-free (SPF) chickens following virus inoculation via oral and intramuscular (IM) routes.
Methods
Suspected organ samples were subjected to virus isolation and polymerase chain reaction (PCR) for FAdV detection. Hexon gene sequencing and phylogenetic analysis were performed on FAdV-positive samples for serotype identification. One FAdV-8b isolate, UPM/FAdV/420/2017, was selected for fiber gene characterization and pathogenicity study and was inoculated in SPF chickens via oral and IM routes.
Results
The hexon gene phylogenetic analysis revealed that all isolates belonged to FAdV-8b. The fiber gene-based phylogenetic analysis of isolate UPM/FAdV/420/2017 supported the grouping of that isolate into FAdV species E. Pathogenicity study revealed that, chickens infected with UPM/FAdV/420/2017 via the IM route had higher clinical score values, higher percent mortality, higher degree of the liver lesions, higher antibody response (p < 0.05), and higher virus shedding amounts (p < 0.05) than those infected via the oral route. The highest virus copy numbers were detected in liver and gizzard.
Conclusions
FAdV-8b is the dominant FAdV serotype in Malaysia, and pathogenicity study of the FAdV-8b isolate UPM/FAdV/420/2017 indicated its ability to induce IBH in young SPF chickens when infected via oral or IM routes.
Keywords: Fowl adenovirus, hexon, inclusion body hepatitis, pathogenicity, phylogenetic analysis
INTRODUCTION
Fowl adenoviruses (FAdVs) are non-enveloped double-stranded DNA viruses belonging to the genus Aviadenovirus within the family Adenoviridae [1]. Based principally on serology analyses and genome characterization, FAdVs have been classified into 12 different serotypes (FAdV-1 to -8a and FAdV-8b to -11) within five different species (FAdV-A to FAdV-E) [2]. Globally, the circulation of FAdV is widespread, covering most geographical locations, and it appears to infect not only chickens but also various other avian species [3]. A feature of FAdV epidemiology is the unusually large number of serotypes that can be isolated on a single farm [4]. It is also common to isolate multiple FAdV serotypes from the same diseased bird, suggesting a lack of cross-protection among serotypes [3].
The pathogenic role of FAdV under field conditions remains unclear since the virus is ubiquitous and can be isolated in both sick and healthy birds [5]. Heterogeneity of FAdV serotypes has led to discrepancies regarding disease behavior and the producibility of clinical signs among different serotypes or different isolates of similar serotypes [6]. Previously, FAdV was considered a secondary agent because it was mostly retrieved from birds that succumbed due to immunosuppression [7,8,9,10]. However, later reports have substantiated that FAdV has emerging roles in eliciting disease symptoms in the absence of immunosuppression as a co-factor [11,12,13]. Nevertheless, the underlying factors that bring about synergistic effects on the severity of the disease that are due to infections with mixed serotypes are yet to be fully described [14].
The FAdV has been isolated in various clinical conditions, and they are most notable as primary pathogens of inclusion body hepatitis (IBH), hepatitis-hydropericardium syndrome (HHS), and avian gizzard erosion (AGE) [15]. Serotypes of FAdVs isolated from these cases are varied, and certain serotypes are predisposed to certain clinical conditions and geographical locations [5]. For example, IBH outbreaks in New Zealand are associated with serotypes 1, 8, and 12, while in Korea, the viruses are mostly of serotypes 3, 9, and 11 [16]. Meanwhile, HHS is mainly associated with the serotype 4 FAdV, and a hydropericarditis outbreak was first reported in Angara Goth near Karachi, Pakistan [17].
In Malaysia, FAdVs infections have been associated with sporadic IBH outbreaks in broiler farms, and those outbreaks are characterized by a rapid onset of mortality, peaking at days 3 - 5 post infection [4]. FAdV infections often result in a low degree of morbidity, but mortality rates typically range between 5% to 10%, although in severe cases, mortality may be as high as 30% [18]. High mortality accompanied by poor growth performance of surviving chickens in the infected chicken house has a direct effect on the profit and loss of the poultry industry. It is vital to identify the serotypes and pathogenic behavior of the FAdVs involved in field outbreaks, especially when vaccination is implemented as the main preventive approach. Therefore, the study aimed to identify the FAdV serotypes isolated from chickens suspected to have IBH by undertaking molecular characterization and determining FAdV pathogenicity in specific-pathogen-free (SPF) chickens.
MATERIALS AND METHODS
Virus isolation and identification
Between 2016 and 2017, 55 organ samples were obtained from broiler chickens suspected for IBH and all the organ tissue samples were included in this study. Individual tissues were homogenized up to 30% (w/v) in sterilized 0.01 M PBS (pH 7.4; Merck, Germany) and frozen at - 20°C. For use, the homogenates were thawed and centrifuged for 10 min at 8,000 r/min at 4°C. The collected supernatant was filtered through a 0.45 μm syringe filter (Sartorius, Germany) before inoculation into 10-day-old embryonated SPF chicken eggs (Malaysian Vaccines and Pharmaceuticals, Puchong, Malaysia) via the chorioallantoic membrane (CAM) inoculation route. After 7 days of incubation at 37°C, the livers and CAMs of the inoculated embryos were collected for viral DNA extraction using DNeasy Blood and Tissue Kit (Qiagen, Germany) following the manufacturer's instructions. Hexon-based PCR was performed to confirm the presence of FAdV in the samples and used a previously described primer pair [1] to amplify 1219-bp of hexon gene fragment of FAdV. The PCR reaction was prepared in a final volume of 25 µL which consisted of 12.5 µL TopTaq Master Mix Kit (Qiagen), 5 µL Coral Load PCR enhancer (Qiagen), 1 µL (10 µM) of each primer, 4 µL of DNA and 1.5 µL nuclease-free water. The amplification was carried out using a C1000 Touch Thermal Cycler (Bio-Rad, USA) under the following conditions: a thermal cycle of 94°C for 3 min, followed by 30 cycles of 3 cycling steps, denaturation at 94°C for 30 sec, annealing at 60°C for 30 sec, and product extension at 72°C for 1 min, with a final elongation at 72°C for 10 min. The amplified PCR product was analyzed by performing 1.5% agarose gel electrophoresis at 80V for 45 min before staining with RedSafe DNA Stain (Chembio, UK) and visualizing by using the Gel Doc XR+ System (Bio-Rad).
Hexon gene analysis and phylogenetic tree
The PCR products were submitted to a commercial analytical company (Apical Scientific, Malaysia) for sequencing using the Sanger method in both forward and reverse directions. The raw sequences from both strands were trimmed, edited, and assembled into a single consensus sequence using BioEdit software. The generated sequence was then compared with other FAdV reference sequences by utilizing the Basic Local Alignment Search Tool (BLAST; NCBI) platform to identify gene homologs. Hexon nucleotide sequences of Malaysian FAdV isolates were aligned with FAdV reference strains representative of all 12 FAdV serotypes (1-8a and 8b-11) retrieved from the GenBank database by applying the ClustalW method as provided in BioEdit software. Construction of the phylogenetic tree was based on the L1 loop of the hexon gene sequence of 16 Malaysian isolates and reference strains, obtained using the maximum-likelihood method within the Kimura-2 parameter model with 1000 bootstrap replicate values and using MEGA v7.0 software. Pairwise evolutionary distances between the nucleotide sequences of the Malaysian FAdV isolates and the representative strains of the different FAdV serotypes were computed using the p-distance method included in MEGA v7.0 software.
Fiber gene analysis and phylogenetic tree
The isolate designated as UPM/FAdV/420/2017, previously confirmed as FAdV serotype 8b based on hexon gene sequence analysis, was chosen for further analysis based on the fiber gene. The pooled CAMs and liver homogenates of UPM/FAdV/420/2017 were subjected to DNA extraction using the commercially available DNeasy Blood and Tissue Kit (Qiagen) and following the manufacturer's instruction. Conventional PCR was performed using PCR master mix (Qiagen) according to the manufacturer's protocol. A previously published primer pair [19] was used to amplify 1124 bp of the FAdV fiber gene. The amplified fiber gene product was processed using 1.5% agarose gel electrophoresis and sent for Sanger sequencing in both forward and reverse directions at Apical Scientific, Malaysia. The sequence was compared with other FAdV fiber sequences available in the NCBI GenBank database by using the BLAST program. Multiple sequence alignments of the fiber gene of UPM/FAdV/420/2017 with reference FAdV isolates of species A to E were performed using the ClustalW method in BioEdit software. A phylogenetic tree of the FAdV fiber nucleotide sequences was constructed using the maximum-likelihood method and the Hasegawa-Kishono-Yano parameter model (1000 bootstrap replicates) provided in MEGA v7.0 software. Computation of pairwise evolutionary distances, based on the UPM/FAdV/420/2017 and 12 serotypes of reference FAdV fiber gene, was performed using the p-distance method provided in MEGA v7.0.
Animals, housing, and ethics statement
Eleven-day-old SPF chickens were used for pathogenicity assessment of the UPM/FAdV/420/2017 FAdV isolate. The SPF chickens were hatched from White Leghorn SPF chicken eggs (Malaysian Vaccines and Pharmaceuticals, Puchong, Malaysia) at their hatching facility in the Institute of Bioscience, Universiti Putra Malaysia (UPM). At 9 days of age, the FAdV seronegative status of the SPF chickens was confirmed by ELISA for the detection of antibodies against group I avian adenovirus (BioChek, USA). All chickens were reared in stainless steel cages at the Animal Research Facility, Faculty of Veterinary Medicine, UPM. The rearing facilities and experimental procedures were approved by the UPM Institutional Animal Care and Use Committee (IACUC) under reference number UPM/IACUC/AUP-R027/2018.
Pathogenicity assessment of FAdV serotype 8b
Eighty 11-day-old SPF chickens of mixed sex were divided randomly into four groups. Chickens in group 1 and group 2 comprised 24 chickens inoculated with 108.53 TCID50 of UPM/FAdV/420/2017 per chick via oral or intramuscular (IM) routes, respectively. Chickens in groups 3 and 4, each with 16 birds per group, were inoculated with PBS via oral or IM routes, respectively, and regarded as control groups. After virus inoculation, all birds were observed daily for clinical signs until 28 days-post-inoculation (dpi). At 1, 3, 5, 7, 10, 14, 21, and 28 dpi, three chickens from the FAdV-inoculated groups and two chickens from the mock-infected groups were euthanized for post-mortem examination. Cloacal swabs and tissue samples such as heart, liver, spleen, kidney, bursa, cecal tonsil, gizzard, and thymus were collected for viral load determination via the quantitative PCR (qPCR) method. The swabs and tissue samples were processed for DNA extraction using a DNeasy Blood and Tissue Kit (Qiagen) and following the manufacturer's instructions. Liver tissues were fixed in 10% formalin and processed by performing hematoxylin and eosin staining at the Histopathology Unit, Faculty of Veterinary Medicine, UPM. Histologic slides were examined for lesions and scored for severity as 0 for normal, 1 for minimal severity, 2 for mild, 3 for moderate, 4 for marked, and 5 for severe. Lesion scoring was performed by a certified pathologist located at Veterinary Diagnostic Pathology, Fort Valley, Virginia, 22652 USA. Blood serum samples were collected at 7, 14, 21, and 28 dpi from surviving chickens in each group for antibody testing against FAdV (BioChek, USA).
Quantitative PCR detection of viral DNA
Viral quantification of tissues and cloacal swabs were determined by measuring DNA copy number of FAdV by applying the standard curve method in quantitative PCR (qPCR). A new primer-probe set that could specifically amplify a 141-bp fragment of the hexon gene was designed based on the nucleotide sequences of UPM/FAdV/420/2017 (Table 1). Amplification of DNA was performed in a 20 µL PCR reaction mix containing 10 µL of SensiFAST Probe No-ROX Kit (Bioline, UK), 0.8 µL of primer pair, 0.2 µL probe, 4.2 µL of nuclease-free water, and 4 µL of the template. The qPCR steps were conducted in a CFX96 Real-Time PCR Detection System (Bio-Rad). The qPCR conditions included initial denaturation at 95°C for 5 min, 40 cycles of denaturation at 95°C for 10 sec, and an extension at 60°C for 40 sec. The positive control DNA used to determine the standard curve was amplified through a series of ten-fold dilutions starting at 100 ng/µL and increasing by 0.001 ng/µL per reaction. For each dilution, the reaction was prepared in triplicate per run.
Table 1. Primer-probe sequences used for quantitative polymerase chain reaction detection of viral DNA from organ samples and cloacal swabs.
| ID | Sequences |
|---|---|
| FAdV420_For | 5′-ACACCACCGCACAGAAATAC-3′ |
| FAdV420_Rev | 5′-TGGGTCTGTTGGCTTTCATC-3′ |
| FAdV420_Probe | 5′-/56-FAM/CCCTCCTTC/ZEN/TGAGTACAGAGCGGTTA/3IABkFQ/-3′ |
FAdV, fowl adenovirus.
Statistical analysis
All statistical analyses were performed using SPSS Statistics 23 (IBM, USA). Comparison of the means of the experimental group parameters was performed by using independent t-tests and two-way ANOVA (SPSS v23.0). A p-value of < 0.05 was considered statistically significant. Results are expressed as mean and standard error values.
RESULTS
Virus isolation and identification
Sixteen liver samples from IBH-suspected chickens were observed to be FAdV positive. These samples were collected from commercial broiler farms with apparent IBH outbreaks. The farms were located in three different provinces of Peninsular Malaysia and one province, Sabah, of East Malaysia (Table 2). All positive samples exhibited common gross lesions of an IBH infection.
Table 2. Description of Malaysian FAdVs isolated in this study.
| No | Sample ID | Location | Case history/necropsy finding |
|---|---|---|---|
| 1 | UPM/FAdV/344A/2016 | Negeri Sembilan | Enlarged liver |
| 2 | UPM/FAdV/350A/2016 | Melaka | Pale liver |
| 3 | UPM/FAdV/350B/2016 | Melaka | Pale liver |
| 4 | UPM/FAdV/392A/2017 | Not available | High mortality and pale liver |
| 5 | UPM/FAdV/392B/2017 | Not available | High mortality and pale liver |
| 6 | UPM/FAdV/420/2017 | Melaka | High mortality with a typical lesion of hepatitis |
| 7 | UPM/FAdV/424/2017 | Pahang | High mortality with a typical lesion of hepatitis |
| 8 | UPM/FAdV/428/2017 | Sabah | A typical lesion of hepatitis |
| 9 | UPM/FAdV/440/2017 | Sabah | Pale liver |
| 10 | UPM/FAdV/520/2017 | Not available | Pale liver |
| 11 | UPM/FAdV/522A/2017 | Melaka | Pale liver |
| 12 | UPM/FAdV/522B/2017 | Melaka | Pale liver |
| 13 | UPM/FAdV/522C/2017 | Melaka | Pale liver |
| 14 | UPM/FAdV/522D/2017 | Melaka | Pale liver |
| 15 | UPM/FAdV/540A/2017 | Melaka | Pale liver |
| 16 | UPM/FAdV/540B/2017 | Melaka | Pale liver |
FAdV, fowl adenovirus.
Molecular characterization of the hexon gene
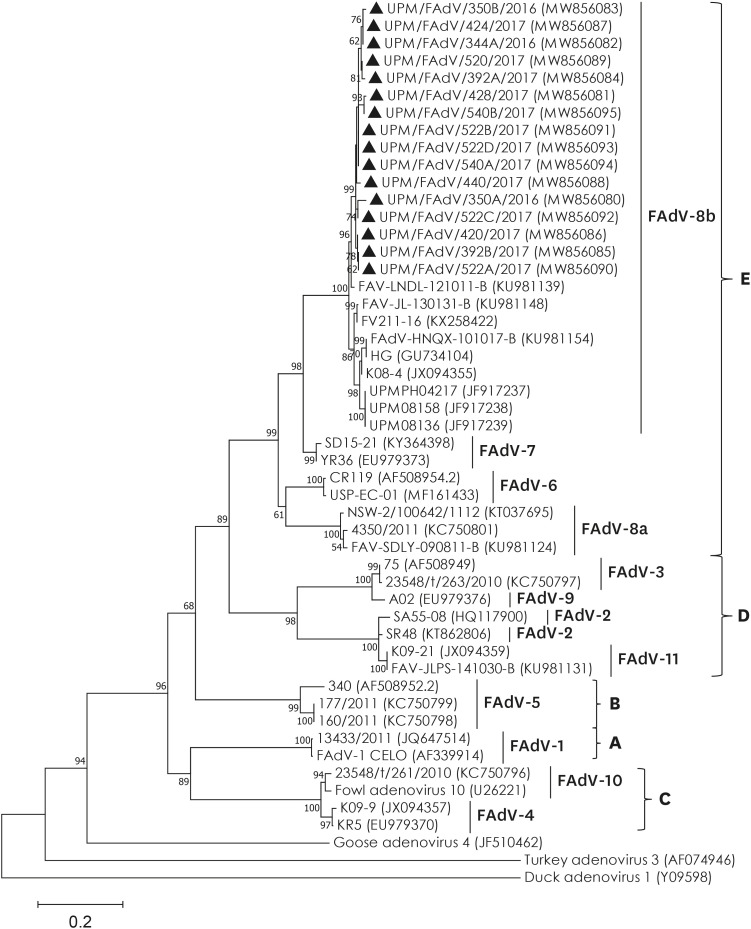
Based on the sequence alignment in GenBank (NCBI) of the PCR-amplified hexon fragment, all 16 Malaysian FAdV isolates were closely related to reference FAdV species E, serotype 8b, which was isolated previously (> 95.3% identity). The phylogenetic tree was generated based on a 593 bp region of the L1 protein of a hexon gene segment corresponding to nt200 to nt792 of the FAdV-A CELO hexon sequence (accession no. AF339914). The phylogenetic analysis results showed that all 16 Malaysian isolates, together with other previously isolated strains, were clustered within a single genotype of FAdV-E serotype 8b (Fig. 1). Intertypic sequence comparison of the studied isolates showed that the highest sequence divergence was with FAdV species A serotype 1 with similar percentage ranges between 57.9%–58.9% (nt) and 46.4%–53.6% (aa) (data not shown). In addition, FAdV-E shared the highest genetic homology with FAdV species D, which comprised serotypes 2, 3, 9, and 11 and had similarity percentage ranges (65.0%–68.3% nt and 56.4%–74.3% aa).
Fig. 1. Phylogenetic analysis based on 593 bp hexon gene fragment containing the L1 region from 16 Malaysian FAdV isolates collected in this study (marked with ▲) and 35 previously characterized isolates representative of all FAdV serotypes and outgroups. Numbers in parenthesis correspond to GenBank accession number. Letter A to E represents FAdV species. The tree was inferred using the maximum-likelihood method based on the Kimura-2 parameter model (1000 bootstrap replicates) included in MEGA v7.0 software.
FAdV, fowl adenovirus.
Phylogenetic analysis based on fiber gene
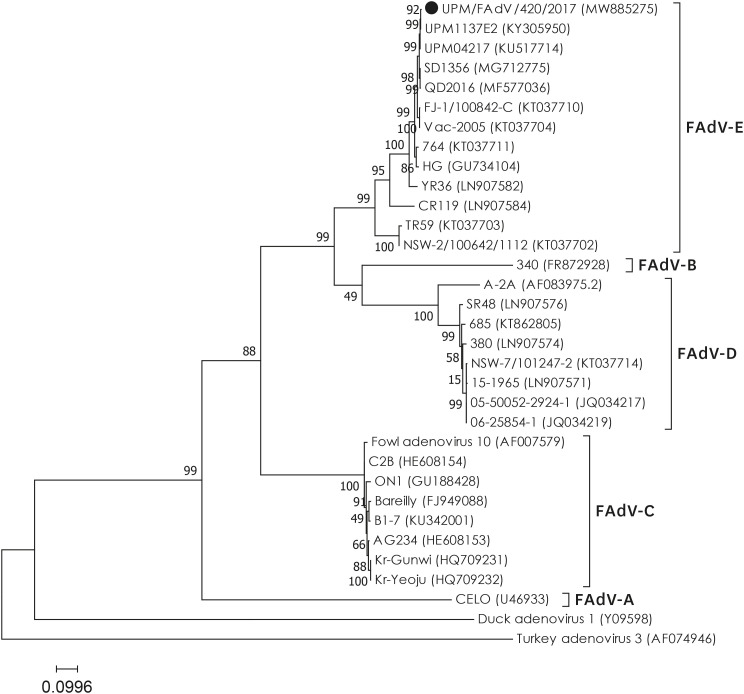
A 1094 bp of the fiber nucleotide sequence corresponding to the fiber tail (1–66 nt), shaft (67–907 nt) and knob (946–1094 nt) regions, was aligned using NCBI's BLAST platform, and the result was consistent with that of FAdV species E serotype 8b, exhibiting a more than 95.1% nucleotide similarity. As expected, high nucleotide identities (> 99.0%) were recorded for previously isolated Malaysian FAdV isolates, UPM04217 (GenBank Accession No. KU517714) and UPM1137E2 (GenBank Accession No. KF866370), that were isolated in 2004 and 2011, respectively. A phylogenetic tree was constructed based on 1094 bp of UPM/FAdV/420/2017 and 32 reference strains retrieved from GenBank (Fig. 2). The resulting dendrogram showed a distinct division of the reference strains into five different FAdV species (A to E). Based on the tree, the isolate UPM/FAdV/420/2017 was shown to be closely related with the FAdV-8b reference strains, and it formed a cluster with FAdV species E serotype 8a (strain TR59 and strain NSW-2/100642/1112), serotype 7 (YR36), and serotype 6 (CR119). In addition, the fiber gene sequence of UPM/FAdV/420/2017 was compared with 12 reference strains, each representative of the 12 different FAdV serotypes, for computation of pairwise similarity matrix values based on the p-distance method (data not shown).
Fig. 2. Phylogenetic tree based on 1094 bp of fiber gene comprising the tail, shaft and knob regions. The isolate UPM/FAdV/420/2017 (marked with ●) was clustered with FAdV species E. Numbers in parenthesis correspond to GenBank accession number. Letters A to E represents FAdV species. The tree was inferred using the maximum-likelihood method based on the Hasegawa-Kishono-Yano parameter model (1000 bootstrap replicates) included in MEGA v7.0 software.
FAdV, fowl adenovirus.
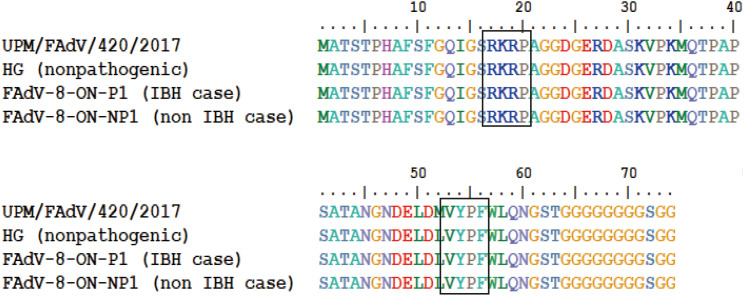
The isolate UPM/FAdV/420/2017 had the highest percentage similarity at both the nucleotide (nt) and amino acid (aa) levels of 96.0% and 90.6%, respectively, when compared to the reference FAdV-8b. Meanwhile, the lowest identities were 43.1% (nt) and 39.0% (aa) with reference FAdV-1. For comparison, an aa pairwise alignment was also performed to compare the fiber tail sequence of isolate UPM/FAdV/420/2017 with the non-pathogenic FAdV-8b (strain HG), a pathogenic FAdV-8b from an IBH case (strain FAdV8-ON-P1), and a non-pathogenic FAdV-8b from a non-IBH case (strain FAdV8-ON-NP1) [20]. The result shows that the first 63 aa residues of the fiber gene sequences are highly conserved within the group (Fig. 3). Inside the tail region, two conserved motifs are present: The identical “RKRP” motif at amino acid position 17 to 20 is equivalent to the conserved “KRAR” motif shared among human adenovirus. The second conserved motif is “VYPF” presented at 53-56 aa and is equivalent to the mammalian “VYPY” motif sequence. A conserved poly-G stretch is also presented at the carboxy end of the tail domain, starting at amino acid position 64 and stretching from 6 residues to up to 10 residues. This stretch indicates the separation between the tail and the shaft region.
Fig. 3. The amino acid pairwise alignment of the fiber tail region as computed by ClustalW for comparison between studied isolate with reference isolate of non-pathogenic FAdV-8b and reference isolates from IBH and non-IBH cases [11]. The boxes indicate the presence of conserved sequence motifs found in the tail region of FAdV-8b.
FAdV, fowl adenovirus; IBH, inclusion body hepatitis.
Pathogenicity assessment of FAdV serotype 8b isolate UPM/FAdV/420/2017
Clinical signs and gross lesions
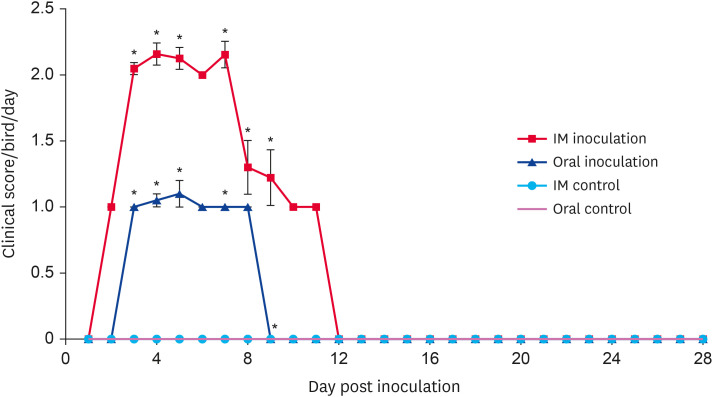
Chickens infected with UPM/FAdV/420/2017 via the oral or IM routes showed clinical signs commonly seen in IBH cases, namely depression, ruffled feathers, huddling in corners, inappetence, and diarrhea. The daily clinical scores for chickens orally inoculated with FAdV were significantly less than those chickens infected via the IM route at 3, 4, 5, 7, and 9 dpi (p < 0.05) (Fig. 4). Orally inoculated chickens started to show clinical signs at 3 dpi; signs peaked at 5 dpi and gradually subsided at 8 dpi. From 9 dpi onward, no overt clinical signs were detected; however, all chickens showed reduced growth at the end of the observation period compared with that of control chickens. In the IM-inoculated group, clinical signs in chickens appeared as early as day 2 pi; signs peaked between days 3 and 7 before reduced steadily until 11 dpi. From 12 dpi onward, no overt clinical signs were observed, and by the end of the trial, the IM-inoculated chickens were distinctively smaller in body size compared to control chickens. One chicken (3.8%) died because of the oral virus inoculation, and 10 chickens (40%) died in the group of IM-inoculated chickens. All control chickens were clinically healthy throughout the experiment. A swollen, friable liver with pale to yellowish discoloration and the presence of necrotic foci were noticed upon necropsy (data not shown). The moribund diseased chickens that died between experimental time points showed pathological lesions similar to those of the euthanized chickens. No gross lesions were seen in tissues of mock-infected chickens.
Fig. 4. Clinical scores of chickens after inoculation with UPM/FAdV/420/2017 via the oral and intramuscular route of inoculation. Values indicate the mean clinical score per group per day. The error bars indicate standard error. Asterisk (*) mark a significant difference in score between groups (p < 0.05). FAdV, fowl adenovirus; IM, intramuscular.
Histopathology
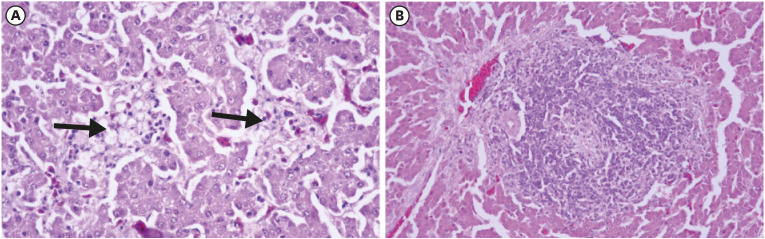
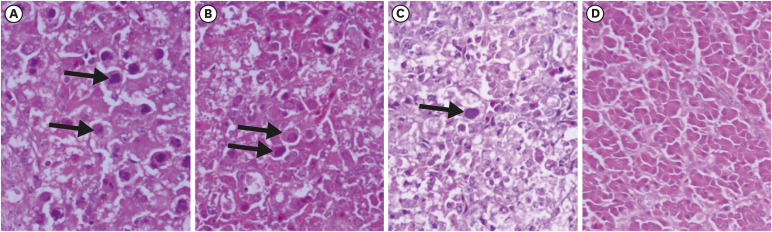
Chickens inoculated with isolate UPM/FAdV/420/2017 via oral and IM routes presented classical IBH histopathological lesions of intranuclear inclusion bodies (INIB) in hepatocytes. The oral-infected group showed milder histopathological liver lesions than the IM group as indicated by minimal multifocal hepatitis with a small collection of heterophils and histiocytes in the hepatocellular parenchyma at 7 dpi and minimal cholangiohepatitis at 10 dpi (Fig. 5). Meanwhile, the IM-infected group presented minimal multifocal hepatitis as early as 1 dpi. This was followed by severe diffuse necrotizing hepatitis with fatty degeneration of hepatocytes and intralesional basophilic inclusion bodies in degenerating hepatocytes at 3, 5, and 7 dpi (Fig. 6). At day 10 pi, moderate periportal lymphohistiocytic hepatitis and mild to moderate bile duct proliferation were observed in the liver of IM-inoculated chickens. Histologic liver lesions were no longer detected at and after 14 dpi in both groups of chickens. No significant histologic lesions were observed in control chickens.
Fig. 5. Histopathology in liver tissues of orally inoculated chickens (hematoxylin and eosin); (A) liver at 7 dpi showing mild histiocytic hepatitis with the fatty change, and (B) liver at 10 dpi showing minimal cholangiohepatitis.
Fig. 6. Histopathology in liver tissues of intramuscularly inoculated chickens (H & E) at 3 dpi (A), 5 dpi (B) and 7 dpi (C) showing intranuclear inclusion bodies in comparison with normal liver (D).
dpi, day post infection.
Antibody response
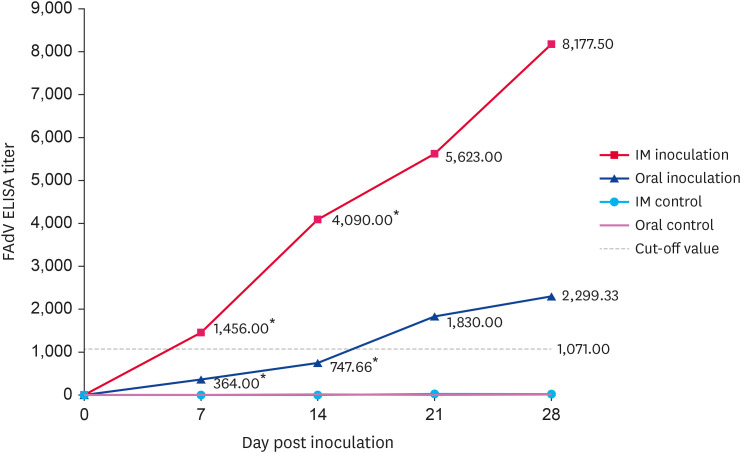
FAdV-specific antibodies were not detected in any of the birds prior to virus inoculation. The antibody response was initially detected at 7 dpi and the response steadily increased until 28 dpi. The difference in antibody titer between oral and IM-inoculated birds differed significantly at 7 and 14 dpi (p < 0.05). At all observation times, IM-inoculated birds had higher antibody titers than orally inoculated birds (Fig. 7).
Fig. 7. The ELISA antibody titer in chickens inoculated with isolate UPM/FAdV/420/2017. Serum samples were collected at 7, 14, 21 and 28-day post-inoculation. Antibodies in control groups were negatively detected at all time points. Asterisks (*) mark the significant difference between groups.
FAdV, fowl adenovirus; ELISA, enzyme-linked immunosorbent assay; IM, intramuscular.
Viral copy number in tissues
Viral DNA levels in the liver, gizzard, cecal tonsil, bursa, spleen, kidney, and thymus at 5 dpi were measured by qPCR method, and the results are summarized in Table 3. Viral DNA was not detected in tissues of control chickens or in chickens before virus inoculation. In both inoculation route groups, the liver was the tissue with the highest amount of viral copies, followed by gizzard tissue. In contrast, the thymus was the tissue with the lowest DNA copies regardless of the inoculation route. Compared to the oral-inoculated birds, the IM-inoculated birds presented significantly higher DNA copies in all sampled tissues (p < 0.05).
Table 3. FAdV copy numbers in tissues of chickens infected with UPM/FAdV/420/2017 at 5 day post infection via oral and IM route of inoculation.
| Tissue | Viral copy numbers, log 10 ± SEM | |
|---|---|---|
| Group | ||
| Infected oral | Infected IM | |
| Liver | 9.85 ± 0.23* | 12.51 ± 0.24† |
| Gizzard | 9.26 ± 0.51* | 11.35 ± 0.19† |
| Cecal tonsil | 8.71 ± 0.21* | 10.19 ± 0.11† |
| Bursa | 7.95 ± 0.6* | 9.90 ± 0.36† |
| Kidney | 7.67 ± 0.12* | 10.64 ± 0.13† |
| Spleen | 7.43 ± 0.14* | 10.50 ± 0.26† |
| Thymus | 7.23 ± 0.05* | 9.20 ± 0.35† |
Values with different superscripts differ significantly (p < 0.05).
No FAdV was detected from tissue samples collected from the control groups inoculated with PBS.
FAdV, fowl adenovirus; IM, intramuscular.
Virus shedding
The viral copy numbers in cloacal swabs of chickens inoculated with UPM/FAdV/420/2017 are shown in Table 4. No virus was detected in chickens before inoculation or in the mock-infected groups. Irrespective of the inoculation route, the highest amount of virus shed by the chickens was at 3 dpi. However, the chickens in the IM-inoculated group continued to shed the virus until the end of the experimental period (28 dpi), whereas virus shedding by the oral-inoculated birds ceased at 14 dpi. The difference in the amount of virus shed between the inoculated groups of chickens was statistically significant at 3, 5, and 7 dpi, with IM-inoculated birds having higher levels of virus shedding (p < 0.05).
Table 4. FAdV copy numbers in cloacal swabs of chickens infected with UPM/FAdV/420/2017 via oral and IM route of inoculation collected at different time points.
| dpi | Viral copy numbers, log 10 ± SEM | |
|---|---|---|
| Group | ||
| Infected oral | Infected IM | |
| 3 | 7.40 ± 0.10* | 10.24 ± 0.40† |
| 5 | 7.54 ± 0.29* | 9.35 ± 0.56† |
| 7 | 7.30 ± 0.20* | 8.45 ± 0.35† |
| 10 | 6.89 ± 0.05 | 6.87 ± 0.01 |
| 14 | ND | 6.85 ± 0.13 |
| 21 | ND | 6.68 ± 0.10 |
| 28 | ND | 6.31 ± 0.05 |
Values with different superscripts differ significantly (p < 0.05).
No FAdV was detected from tissue samples collected from the control groups inoculated with PBS.
FAdV, fowl adenovirus; IM, intramuscular; dpi, day post infection; ND, not detected.
DISCUSSION
Although all 12 serotypes of FAdV have been linked to outbreaks of IBH, species FAdV-D and FAdV-E are the most likely causative agent of the disease [21,22]. To control the further spread of FAdV, it is necessary to identify the prevalent serotypes present in the field, especially if vaccination is the principal method of control and induction of serotype-specific protection. In this study, 16 FAdV strains were isolated from liver tissues of IBH-infected chickens received from commercial broiler farms between 2016 and 2017. Sequence analysis based on 563 bp of hexon nucleotide containing the L1 fragment and the adjacent pedestal region showed clustering of all the isolates into a single serotype of FAdV-8b, species E. The phylogenetic analysis based on a derived amino acid sequence of the hexon gene region showed results consistent with the sequence results, thus supporting the clustering of isolates within serotype FAdV-8b. Therefore, it can be suggested that FAdV-8b is the major serotype responsible for IBH outbreaks in Malaysian poultry. A similar finding was reported in previous Malaysian FAdV studies based on limited numbers of field cases [23,24]. So far, there are no reports on the isolation of FAdV serotypes other than serotype 8b in Malaysia. This lack of serotype diversity in the Malaysian field may be due to this country being a self-sufficient chicken producer and not heavily dependent on cross-country chicken trade, thereby limiting the chance of serotype transfer and mixing. In Indonesia, IBH was associated with FAdV-8a, -8b, and -11, while Thailand reported FAdV-2 as the main agent for IBH [25,26]. In this study, all the IBH positive cases were associated with FAdV-8b; no other FAdV serotypes were detected.
The role of FAdV-8b as a causative agent of IBH has also been reported in other countries, indicating the 8b serotype is a common agent for IBH. A study in Iran observed serotypes 8b and -11 in IBH outbreaks, observations that are similar to those in reports from Korea and Australia [19,21,27]. Meanwhile, in China, recent reports have indicated that serotypes 8a, -8b, and -11 are the predominant agents of IBH [14,28]. The serotypes isolated from IBH cases in Canada include type FAdV-2, -8a, and -11 [22], in South Africa FAdV-2 and 8b [29], and in Slovenia, only serotype 8b has been isolated from IBH cases [30].
Isolation of multiple FAdV serotypes in the same bird has been previously reported, and it appears that mixed infections are not limited to FAdV serotypes of the same species but also FAdV serotypes of different species [31,32]. In Asia, it is not uncommon to isolate FAdV-8 and FAdV-4 together [14]. In the European, North American, and South African countries, mixed infections of FAdV-8 with serotypes FAdV-2, -7, and -11 have been reported [29,33]. So far, no report has addressed the synergistic effect of mixed FAdV infections on the severity of the disease. Moreover, control measures often involve treatment against the predominant serotype, ignoring the possible infectivity and severity effects of mixed FAdV infections [14,28].
In the present study, the 1094 bp of the fiber sequence was compared with fiber reference strains representative of all FAdV serotypes to reconstruct the virus's phylogeny. Based on the resulting dendrogram, the grouping of the isolates was consistent with the phylogenic grouping based on L1-hexon, indicating the possible use of fiber-based molecular typing to identify FAdV serotypes. Previous studies have also suggested the potential use of the fiber gene for FAdV typing as alternatives to hexon gene typing [8,33]. Although grouping and assignment of species were consistent with the hexon L1-based typing, the pairwise similarity matrix showed a higher degree of intertypic nucleotide and amino acid divergence. The hexon-based pairwise calculation showed that the highest difference between the studied isolates was for FAdV-1 strain CELO at 58.9% (nt) and 53.6% (aa), whereas the fiber-based pairwise calculation showed far greater divergence at 43.1% (nt) and 39.0% (aa). This finding has been reported previously in FAdV-E and in human adenovirus implying independent evolutionary rates for hexon and fiber [33].
An association of tissue tropism with adenovirus virulence has been addressed previously [34]. The association implies that the role of the fiber protein in virus attachment may be affected by virus virulence, which subsequently causes different tissue tropism to be expressed by the same virus with a different virulence capacity. In addition, the previous study demonstrated that changes in the aa of the FAdV fiber knob are linked with attenuation of FAdV-8b virulence after multiple passages in embryonated SPF eggs [35]. In this study, a direct sequence comparison of the fiber gene between pathogenic and non-pathogenic FAdV-8b strains could not distinguish between different FAdV pathotypes due to a lack of virulence markers in the sequence. It seems that, despite molecular changes in the fiber gene, it may work as an indicator of attenuation and infectivity of the virus. However, to define the virulence of the FAdV solely based on a fiber gene sequence is impossible because a relevant sequence marker is unavailable. These findings suggest that other genetic factors have possible roles in FAdV virulence [20].
The pathogenicity of isolate UPM/FAdV/420/2017 has shown that the isolate can produce IBH in young SPF chickens when inoculated intramuscularly and via the natural oral route. The observed disease symptoms of IBH were a swollen pale liver with necrotic foci in both groups of inoculated chickens. However, the degree of pathogenicity differed with IM-infected chickens demonstrating higher mortality and clinical scores (p < 0.05) than those of the oral-infected group. This finding is consistent with a previous report that FAdV pathogenicity is dependent on the route of inoculation and not solely influenced by the viral serotype [36].
Results of the histopathologic examination indicated the presence of INIB in hepatocytes. In the field, inclusion bodies are common histologic features, in addition to the presence of degenerative or necrotic cells and lymphoid infiltration, observed in moribund IBH chickens [37]. In this study, prominent basophilic inclusion bodies were spotted in the liver of IM-infected birds at 3, 5, and 7 dpi. Presentation of prominent basophilic inclusion bodies in hepatocytes, along with highest FAdV copy numbers in tissues, indicated the liver was a major replication site and the tissue tropism for the tested isolate. Furthermore, the INIB in the liver occurred in the presence of histologically normal bursa and thymus, indicating the development of IBH was solely due to the virulence of the isolate, without immunosuppression as a co-factor. Although previous studies have reported a close association of IBH-inducing FAdV with an immunosuppressive role [19,37,38], the results of this study suggest that the ability of FAdV to cause immunosuppression may vary from strain to strain. Nonetheless, further study is required to elucidate the mechanism of FAdV-induced immunosuppression.
Pathogenicity study of young SPF chickens showed the production of IBH after FAdV infection via oral and IM routes. No lesions were observed in the gizzard, although the presence of a high virus copy number in gizzard tissue suggests the isolate is not associated with gizzard erosion (GE) but rather, the gizzard is a viral reservoir that is associated with consistent shedding of the virus. Previous studies have indicated that GE is associated primarily with FAdV-1 (species FAdV-A) infection [37,39].
Overall, it is almost impossible to establish a clear-cut indicator of the connection between FAdV and its pathogenicity without deciphering the virulence determinants of the virus. In addition, many factors may have influenced the pathogenic properties of FAdV, which include infection route, individual virulence of FAdV strain, chicken susceptibility, and experimental viral dosage [35,40].
This study confirmed FAdV-8b as the dominant serotype causing IBH in commercial broiler chickens in Malaysia. The FAdV-8b isolate UPM/FAdV/420/2017 produced obvious pathogenicity in SPF chickens when infected via oral or intramuscular routes. Since the development of a commercial vaccine against IBH in Malaysia is currently underway, information from this study has clarified the epidemiological status of Malaysian FAdV, which should help establish effective vaccination strategies against FAdV infection.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Siti Nor Azizah Mahamud for assistance with the experimental animal procedures and other staff of the Laboratory of Vaccines and Immunotherapeutics, Institute of Bioscience, Universiti Putra Malaysia for their helpfulness in the collection of FAdV isolates.
Footnotes
Funding: This research was supported by Boehringer Ingelheim Animal Health (Study No. 2018070) and Higher Institution Centre of Excellence (HICoE), Ministry of Higher Education, Government of Malaysia (Grant no. 6369101).
Conflict of Interest: The authors declare no conflicts of interest.
- Conceptualization: Sabarudin NS, Omar AR.
- Data curation: Sabarudin NS, Omar AR.
- Formal analysis: Sabarudin NS.
- Funding acquisition: Phang YF, Omar AR.
- Investigation: Sabarudin NS, Tan SW, Phang YF.
- Methodology: Sabarudin NS, Tan SW.
- Project administration: Sabarudin NS, Omar AR.
- Resources: Sabarudin NS, Tan SW, Phang YF, Omar AR.
- Software: Sabarudin NS, Tan SW.
- Supervision: Omar AR.
- Validation: Tan SW, Omar AR.
- Visualization: Sabarudin NS.
- Writing - original draft: Sabarudin NS.
- Writing - review & editing: Omar AR.
References
- 1.Hess M. Detection and differentiation of avian adenoviruses: a review. Avian Pathol. 2000;29(3):195–206. doi: 10.1080/03079450050045440. [DOI] [PubMed] [Google Scholar]
- 2.Harrach B, Benko M, Both GW, Brown M, Davison AJ, Echavarria M, et al. Family adenoviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Taxonomy: Classification and Nomenclature of Viruses. Ninth Report of the International Committee on Taxonomy of Viruses. San Diego: Elsevier; 2011. pp. 125–141. [Google Scholar]
- 3.Adair BM, Fitzgerald S. Group I adenovirus infections. In: Saif YM, Fadly AM, Glisson JR, McDougald LR, Nolan LK, Swayne DE, editors. Diseases of Poultry. 12th ed. Iowa: Blackwell Publishing Professional; 2008. pp. 251–266. [Google Scholar]
- 4.Pereira CG, Marin SY, Santos BM, Resende JS, Resende M, Gomes AM, et al. Occurrence of aviadenovirus in chickens from the poultry industry of Minas Gerais. Arq Bras Med Vet Zootec. 2014;66(3):801–808. [Google Scholar]
- 5.McFerran JB, Smyth JA. Avian adenoviruses. Rev Sci Tech. 2000;19(2):589–601. [PubMed] [Google Scholar]
- 6.Gaba A, Pal JK, Parmar H, Prajapati KS. Isolation, identification and molecular characterization of inclusion body hepatitis virus. Vet World. 2010;9:415–417. [Google Scholar]
- 7.Fadly AM, Winterfield RW, Olander HJ. Role of the bursa of Fabricius in the pathogenicity of inclusion body hepatitis and infectious bursal disease viruses. Avian Dis. 1976;20(3):467–477. [PubMed] [Google Scholar]
- 8.Marek A, Nolte V, Schachner A, Berger E, Schlötterer C, Hess M. Two fiber genes of nearly equal lengths are a common and distinctive feature of Fowl adenovirus C members. Vet Microbiol. 2012;156(3-4):411–417. doi: 10.1016/j.vetmic.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 9.McFerran JB, McCracken RM, Connor TJ, Evans RT. Isolation of viruses from clinical outbreaks of inclusion body hepatitis. Avian Pathol. 1976;5(4):315–324. doi: 10.1080/03079457608418201. [DOI] [PubMed] [Google Scholar]
- 10.Saifuddin M, Wilks CR. Effects of fowl adenovirus infection on the immune system of chickens. J Comp Pathol. 1992;107(3):285–294. doi: 10.1016/0021-9975(92)90004-e. [DOI] [PubMed] [Google Scholar]
- 11.Erny KM, Barr DA, Fahey KJ. Molecular characterization of highly virulent fowl adenoviruses associated with outbreaks of inclusion body hepatitis. Avian Pathol. 1991;20(4):597–606. doi: 10.1080/03079459108418799. [DOI] [PubMed] [Google Scholar]
- 12.Gomis S, Goodhope AR, Ojkic AD, Willson P. Inclusion body hepatitis as a primary disease in broilers in Saskatchewan, Canada. Avian Dis. 2006;50(4):550–555. doi: 10.1637/7577-040106R.1. [DOI] [PubMed] [Google Scholar]
- 13.Reece RL, Barr DA, Grix DC, Forsyth WM, Condron RJ, Hindmarsh M. Observations on naturally occurring inclusion body hepatitis in Victorian chickens. Aust Vet J. 1986;63(6):201–202. doi: 10.1111/j.1751-0813.1986.tb02981.x. [DOI] [PubMed] [Google Scholar]
- 14.Changjing L, Haiying L, Dongdong W, Jingjing W, Youming W, Shouchun W, et al. Characterization of fowl adenoviruses isolated between 2007 and 2014 in China. Vet Microbiol. 2016;197:62–67. doi: 10.1016/j.vetmic.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 15.Hess M. Aviadenovirus infections. In: Swayne DE, Glisson JR, McDougald LR, Nolan LK, Suarez DL, Nair VL, editors. Diseases of Poultry. 13th ed. Ames: Wiley-Blackwell; 2013. pp. 290–300. [Google Scholar]
- 16.Lim TH, Lee HJ, Lee DH, Lee YN, Park JK, Youn HN, et al. Identification and virulence characterization of fowl adenoviruses in Korea. Avian Dis. 2011;55(4):554–560. doi: 10.1637/9730-032011-Reg.1. [DOI] [PubMed] [Google Scholar]
- 17.Anjum AD, Sabri MA, Iqbal Z. Hydropericarditis syndrome in broiler chickens in Pakistan. Vet Rec. 1989;124(10):247–248. doi: 10.1136/vr.124.10.247. [DOI] [PubMed] [Google Scholar]
- 18.Smyth JA, McNulty M. Adenoviridae. In: Pattison M, McMullin PF, Bradbury JM, Alexander DJ, editors. Poultry Diseases. 6th ed. UK: Butterworth Heinemann; 2008. pp. 367–381. [Google Scholar]
- 19.Steer PA, O'Rourke D, Ghorashi SA, Noormohammadi AH. Application of high-resolution melting curve analysis for typing of fowl adenoviruses in field cases of inclusion body hepatitis. Aust Vet J. 2011;89(5):184–192. doi: 10.1111/j.1751-0813.2011.00695.x. [DOI] [PubMed] [Google Scholar]
- 20.Grgić H, Krell PJ, Nagy E. Comparison of fiber gene sequences of inclusion body hepatitis (IBH) and non-IBH strains of serotype 8 and 11 fowl adenoviruses. Virus Genes. 2014;48(1):74–80. doi: 10.1007/s11262-013-0995-y. [DOI] [PubMed] [Google Scholar]
- 21.Morshed R, Hosseini H, Langeroudi AG, Fard MH, Charkhkar S. Fowl adenoviruses D and E cause inclusion body hepatitis outbreaks in broiler and broiler breeder pullet flocks. Avian Dis. 2017;61(2):205–210. doi: 10.1637/11551-120516-Reg.1. [DOI] [PubMed] [Google Scholar]
- 22.Ojkic D, Martin E, Swinton J, Vaillancourt JP, Boulianne M, Gomis S. Genotyping of Canadian isolates of fowl adenoviruses. Avian Pathol. 2008;37(1):95–100. doi: 10.1080/03079450701805324. [DOI] [PubMed] [Google Scholar]
- 23.Juliana MA, Nurulfiza I, Hair-Bejo M, Omar AR, Aini I. Molecular characterization of fowl adenoviruses isolated from inclusion body hepatitis outbreaks in commercial broiler chickens in Malaysia. Pertanika J Trop Agric Sci. 2014;37:483–497. [Google Scholar]
- 24.Norina L, Norsharina A, Nurnadiah AH, Redzuan I, Ardy A, Nor-Ismaliza I. Avian adenovirus isolated from broiler affected with inclusion body hepatitis. Malaysian J Vet Res. 2016;7:121–126. [Google Scholar]
- 25.Teguh YP, Khrisna P, Wayan YW, Inna H. The rise of fowl adenovirus-associated diseases in Asia. Int Hatch Pract. 2019;33(7):13–16. [Google Scholar]
- 26.Junnu S, Lertwatcharasarakul P, Jala S, Phattanakulanan S, Monkong A, Kulprasertsri S, et al. An inactivated vaccine for prevention and control of inclusion body hepatitis in broiler breeders. Wetchasan Sattawaphaet. 2015;45(1):55–62. [Google Scholar]
- 27.Choi KS, Kye SJ, Kim JY, Jeon WJ, Lee EK, Park KY, et al. Epidemiological investigation of outbreaks of fowl adenovirus infection in commercial chickens in Korea. Poult Sci. 2012;91(10):2502–2506. doi: 10.3382/ps.2012-02296. [DOI] [PubMed] [Google Scholar]
- 28.Chen L, Yin L, Zhou Q, Peng P, Du Y, Liu L, et al. Epidemiological investigation of fowl adenovirus infections in poultry in China during 2015–2018. BMC Vet Res. 2019;15(1):271–277. doi: 10.1186/s12917-019-1969-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maartens LH, Joubert HW, Aitchison H, Venter EH. Inclusion body hepatitis associated with an outbreak of fowl adenovirus type 2 and type 8b in broiler flocks in South Africa. J S Afr Vet Assoc. 2014;85(1):e1–e5. doi: 10.4102/jsava.v85i1.1146. [DOI] [PubMed] [Google Scholar]
- 30.Zadravec M, Slavec B, Krapež U, Kaján GL, Račnik J, Juntes P, et al. Inclusion body hepatitis associated with fowl adenovirus type 8b in broiler flock in Slovenia: a case report. Slovenian Vet Res. 2011;48(3-4):107–113. [Google Scholar]
- 31.Kaján GL, Kecskeméti S, Harrach B, Benkő M. Molecular typing of fowl adenoviruses, isolated in Hungary recently, reveals high diversity. Vet Microbiol. 2013;167(3-4):357–363. doi: 10.1016/j.vetmic.2013.09.025. [DOI] [PubMed] [Google Scholar]
- 32.Niu Y, Sun Q, Zhang G, Sun W, Liu X, Xiao Y, et al. Epidemiological investigation of outbreaks of fowl adenovirus infections in commercial chickens in China. Transbound Emerg Dis. 2018;65(1):e121–e126. doi: 10.1111/tbed.12691. [DOI] [PubMed] [Google Scholar]
- 33.Schachner A, Marek A, Grafl B, Hess M. Detailed molecular analyses of the hexon loop-1 and fibers of fowl aviadenoviruses reveal new insights into the antigenic relationship and confirm that specific genotypes are involved in field outbreaks of inclusion body hepatitis. Vet Microbiol. 2016;186:13–20. doi: 10.1016/j.vetmic.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 34.Rasmussen UB, Schlesinger Y, Pavirani A, Mehtali M. Sequence analysis of the canine adenovirus 2 fiber-encoding gene. Gene. 1995;159(2):279–280. doi: 10.1016/0378-1119(95)00110-r. [DOI] [PubMed] [Google Scholar]
- 35.Sohaimi NM, Bejo MH, Omar AR, Ideris A, Isa NM. Hexon and fiber gene changes in an attenuated fowl adenovirus isolate from Malaysia in embryonated chicken eggs and its infectivity in chickens. J Vet Sci. 2018;19(6):759–770. doi: 10.4142/jvs.2018.19.6.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cook JK. Fowl adenoviruses: studies on aspects of the pathogenicity of six strains for 1-day-old chicks. Avian Pathol. 1983;12(1):35–43. doi: 10.1080/03079458308436147. [DOI] [PubMed] [Google Scholar]
- 37.Schachner A, Matos M, Grafl B, Hess M. Fowl adenovirus-induced diseases and strategies for their control - a review on the current global situation. Avian Pathol. 2018;47(2):111–126. doi: 10.1080/03079457.2017.1385724. [DOI] [PubMed] [Google Scholar]
- 38.Wang J, Zaheer I, Saleemi MK, Qi X, Gao Y, Cui H, et al. The first complete genome sequence and pathogenicity characterization of fowl adenovirus 11 from chickens with inclusion body hepatitis in Pakistan. Vet Microbiol. 2020;244:108670. doi: 10.1016/j.vetmic.2020.108670. [DOI] [PubMed] [Google Scholar]
- 39.Marek A, Kaján GL, Kosiol C, Benkő M, Schachner A, Hess M. Genetic diversity of species Fowl aviadenovirus D and Fowl aviadenovirus E. J Gen Virol. 2016;97(9):2323–2332. doi: 10.1099/jgv.0.000519. [DOI] [PubMed] [Google Scholar]
- 40.Cowen B, Lu H, Weinstock D, Castro E. Proceeding International Symposium on Adenovirus and Reovirus Infections in Poultry. Germany: Rauischholzhausen; 1996. Pathogenicity studies of fowl adenovirus isolated in several region in the world; pp. 79–84. [Google Scholar]