Genetically diverse IgM+ peripheral B cells reside in the lung microvasculature and dampen neutrophil-mediated lung inflammation via lipoxins. Podstawka et al. reveal that transitional B cells marginate in the lung capillaries, where they dampen neutrophil inflammation. These discoveries help elucidate regulatory mechanisms that attenuate acute lung inflammation, which, when unchecked, can lead to acute respiratory distress syndrome and pulmonary failure.
Abstract
Pulmonary innate immunity is required for host defense; however, excessive neutrophil inflammation can cause life-threatening acute lung injury. B lymphocytes can be regulatory, yet little is known about peripheral transitional IgM+ B cells in terms of regulatory properties. Using single-cell RNA sequencing, we discovered eight IgM+ B cell subsets with unique gene regulatory networks in the lung circulation dominated by transitional type 1 B and type 2 B (T2B) cells. Lung intravital confocal microscopy revealed that T2B cells marginate in the pulmonary capillaries via CD49e and require CXCL13 and CXCR5. During lung inflammation, marginated T2B cells dampened excessive neutrophil vascular inflammation via the specialized proresolving molecule lipoxin A4 (LXA4). Exogenous CXCL13 dampened excessive neutrophilic inflammation by increasing marginated B cells, and LXA4 recapitulated neutrophil regulation in B cell–deficient mice during inflammation and fungal pneumonia. Thus, the lung microvasculature is enriched in multiple IgM+ B cell subsets with marginating capillary T2B cells that dampen neutrophil responses.
Introduction
The lungs are constantly bombarded by a broad array of infectious and noninfectious stimuli that can lead to inappropriate or excessive neutrophil inflammatory responses, resulting in severe acute lung injury and acute respiratory distress syndrome (ARDS; Lloyd and Marsland, 2017; Matthay et al., 2019; Williams and Chambers, 2014). Therefore, identifying regulatory systems that mitigate excessive lung inflammation has clinical importance. Many pathogens, including viruses, bacteria, and fungi, can induce excessive neutrophilic inflammation causing ARDS, and these pathogens include novel viruses such as SARS-CoV-2, where lung injury can be mediated by neutrophil inflammation (Carvelli et al., 2020; Radermecker et al., 2020; Barnes et al., 2020; Zuo et al., 2020). Independent of the inciting pathogen, neutrophils are the primary cell involved in all types of lung injury leading to ARDS, which highlights the importance of studying the regulatory mechanisms governing neutrophil dynamics during lung inflammation and injury. Cross-talk between neutrophils and other leukocytes has been proposed to reciprocally mediate regulation, but there are limited examples of this in vivo. We previously discovered that aged neutrophils have a propensity to marginate in the lung capillaries and that older neutrophils have a greater tendency for activation and inflammation (Kim et al., 2018; Uhl et al., 2016; Zhang et al., 2015); thus, local lung-specific regulatory mechanisms to restrain neutrophils are critical to the avoidance of acute lung injury and syndromes such as ARDS.
B cells are essential effectors of the adaptive immune response owing to their role in humoral immunity (Cyster and Allen, 2019; LeBien and Tedder, 2008). Conventional B cells develop within the bone marrow into immature cells that enter the peripheral circulation. Maturing naive B cells recirculate between blood and spleen as IgM+ IgD+ transitional cells (type 1 B [T1B] and type 2 B [T2B]) before homing to germinal centers within secondary lymphoid tissues to undergo somatic hypermutation, Ig class switching, and differentiation into plasma or memory cells (Victora and Nussenzweig, 2012; Boothby et al., 2019; Su et al., 2004). Limited information exists to characterize these transitional B cells in the peripheral blood. B cells, including T2B cells, have regulatory capacities (Rosser and Mauri, 2015; Rosser et al., 2014; Mauri and Bosma, 2012; Candando et al., 2014); yet, these aspects of B cell function and behavior remain ill defined. Recent advances in single-cell genomics and in vivo imaging have accelerated our understanding of immune cells, including B cells, in vivo (Papalexi and Satija, 2018; Scharer et al., 2020; Nehar-Belaid et al., 2020; Laidlaw et al., 2020). Additionally, intravital imaging has revealed new understandings of leukocyte behaviors, both in the bloodstream and within tissues, including lung and lymphoid tissues (Tas et al., 2016; Beck et al., 2014; Cinamon et al., 2004).
Combining single-cell RNA sequencing (scRNAseq) and confocal lung intravital microscopy, we genetically defined IgM+ B cell states and their gene regulatory pathways and discovered that T2B cells marginate within the lung capillaries via CD49e and C-X-C motif chemokine receptor 5 (CXCR5)/C-X-C motif chemokine ligand 13 (CXCL13). Moreover, marginated B cells dampen neutrophilic lung inflammation via the resolving molecule lipoxin A4 (LXA4). Additionally, during Aspergillus fumigatus pneumonia, B cell deficiency (Ighm−/−) or impaired LXA4 (Alox15−/−) production results in increased neutrophil recruitment. Therefore, we have uncovered a regulatory system requiring recirculating peripheral B cells that attenuates excessive neutrophil recruitment and activation in the lung in vivo. We have additionally developed a companion online B cell atlas tool to allow further interrogation of lung B cell states (http://www.biernaskielab.ca/b_cell or http://www.biernaskielab.com/b_cell). Together, this knowledge contributes to new understandings of lung inflammation and pneumonia that could drive novel therapeutic options for treating diseases such as ARDS that result from neutrophil-mediated acute lung injury and infection.
Results
The lung capillaries contain marginated intravascular B cells
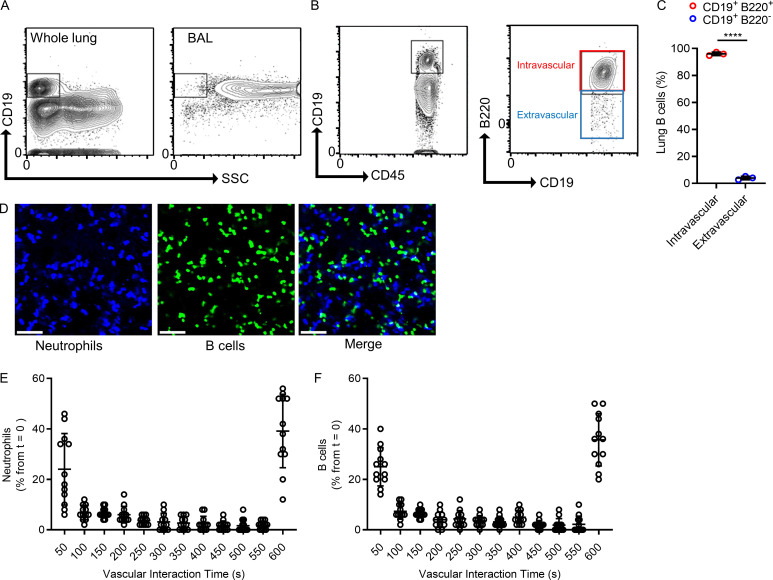
B cells are a major cellular component of the circulation, and the lungs contain a large volume of blood within their vast vascular network; yet, the study of peripheral B cells within the lung vasculature remains limited. We used flow cytometry to identify B cells (CD19+) in homogenized whole lungs (Fig. 1 A). B cells were localized to the lung tissue but not the airways, because few CD19+ cells were found in bronchoalveolar lavage (BAL) samples (Fig. 1 A). To anatomically localize the lung B cells within the tissue, we distinguished intravascular from extravascular B cells using a dual antibody staining technique (Barletta et al., 2012). Intravenous fluorochrome-conjugated anti-B220 mAb was administered i.v. 5 min before lungs were harvested, followed by a fluorochrome-conjugated anti-CD19 mAb added to the lung single-cell suspension for total B cells (Fig. 1 B). This revealed that most lung B cells were intravascular (Fig. 1 C). To investigate the positioning and localization of these intravascular B cells, we used lung intravital confocal microscopy. To visualize B cells, we crossed mice with homozygous expression of Cre recombinase from Cd19 [B6.129P2(C)-Cd19tm1(cre)Cgn/J] with a mouse homozygous for the expression of a fluorescent protein ZsGreen1 from the Rosa26 locus [B6.Cg-Gt(ROSA)26Sortm6(CAG-ZsGreen1)Hze/J] to generate a B cell reporter mouse termed “Cd19ZsGreen1” (Fig. 1 D and Video 1). Surprisingly, B cells sequestered to the endothelium for >30 s, a time point used in leukocyte recruitment studies to signify firm adhesion (Yipp et al., 2017). Moreover, 35.5% of lung B cells visualized at time point 0 adhered for the entire 10 min of imaging, whereas 24.9% transited through the capillary microcirculation within seconds. Lung neutrophils can firmly adhere to the endothelium in a process termed “margination” (Yipp et al., 2017; Lien et al., 1987); however, this behavior is not reported in B cells. Like capillary marginated neutrophils, B cells firmly adhered to the endothelium like neutrophils did (Fig. 1, E and F; and Video 1). Therefore, the lung microcirculation harbors peripheral B cells that localize within the vasculature via firm adhesion consistent with the phenomenon known as margination.
Figure 1.
The lung capillaries contain marginated intravascular B cells. (A) Flow cytometry compared B cells (fluorescently conjugated anti-CD19 mAb) within a whole-lung homogenate with BAL fluid. SSC, side scatter. (B and C) Representative flow cytometry and quantification of intravascular (CD19+/B220+) versus extravascular (CD19+/B220−) B cells using a two-antibody staining technique (A–C, n = 3 independent experiments with six mice total). (D) Lung intravital microscopy visualized both intravascular neutrophils (fluorescently conjugated anti-Ly6G mAb) and B cells (Cd19ZsGreen1). Scale bar, 70 µm. (E and F) The transit times of neutrophils and B cells were quantified using intravital microscopy. Marginated cells were observed in the FOV for up to 600 s. Cells at time point 0 were followed for their duration in the lung vasculature and expressed as a percentage of the initial total B cell numbers in each FOV (D–F, n = 4 independent experiments using four mice total; each dot represents one FOV). Pooled data are presented as mean ± SD. Exact P values were determined using Student’s t test. ****, P < 0.0001.
Video 1.
B cells marginate in the lung capillaries. Related to Fig. 1. Intravital lung imaging was used to assess CD19+ B cell (Cd19ZsGreen1; green) and Ly6G+ neutrophil (fluorochrome-conjugated anti-Ly6G antibody; blue) marginated behavior in the lung capillaries. The red arrows point to examples of marginated B cell behavior. Transit time was quantified from the B cells and neutrophils present at time 0. Experiments were repeated three times (n = 3). Playback speed is 12 frames/s, 20 min actual imaging time.
The lung intravascular space contains eight subsets of genetically distinct B cells
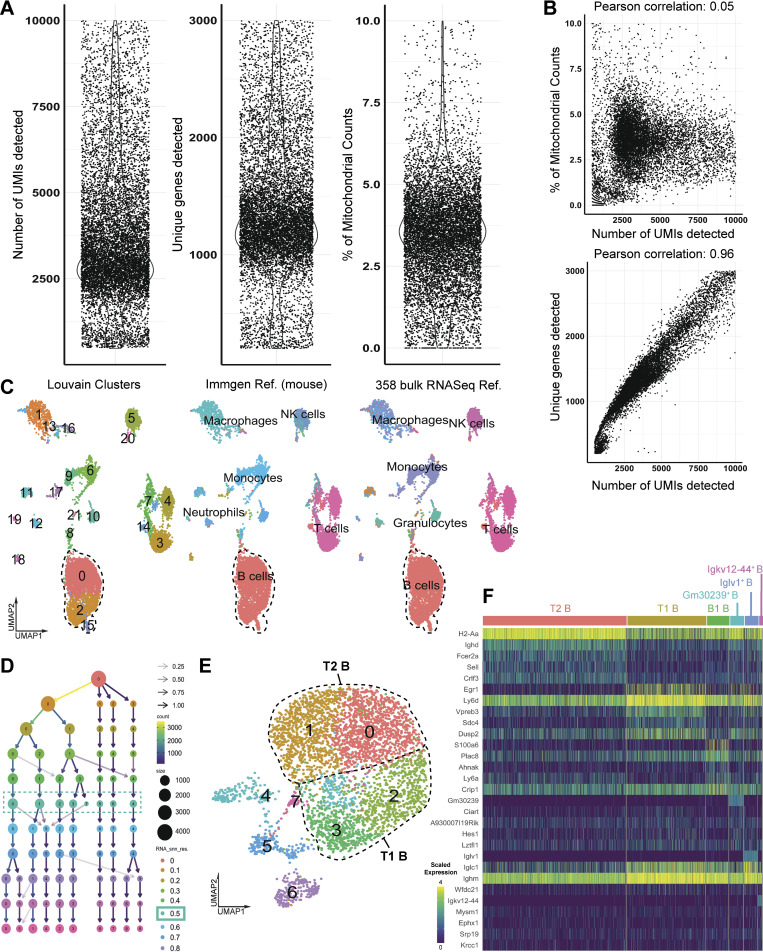
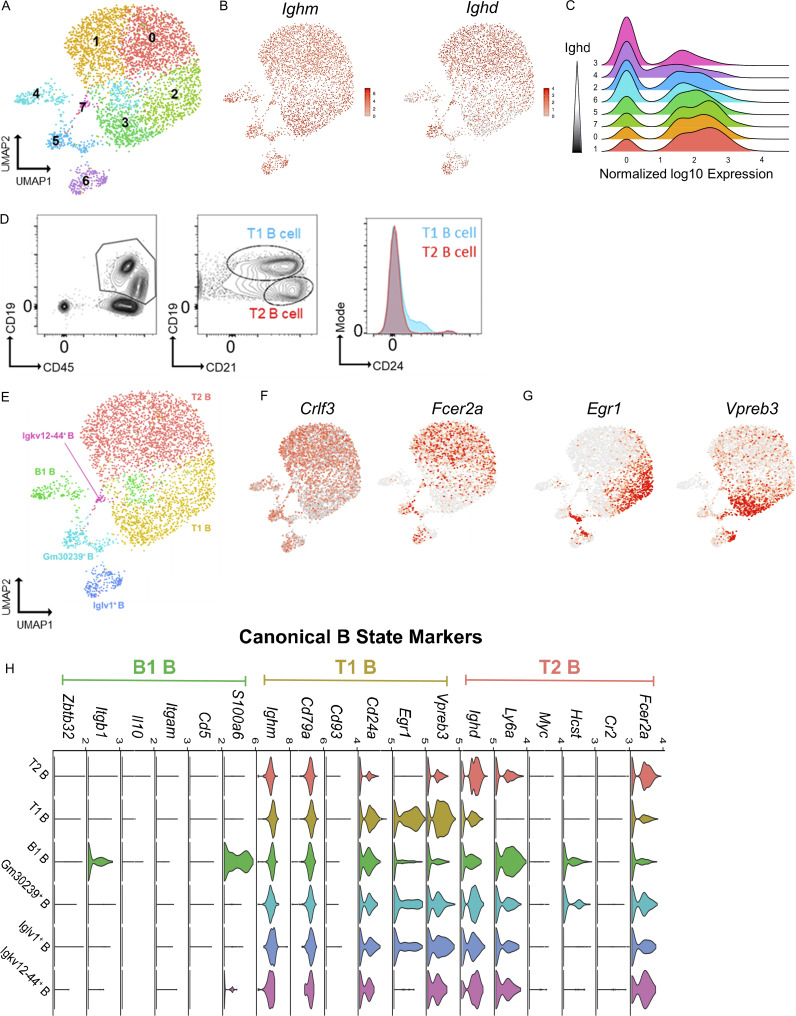
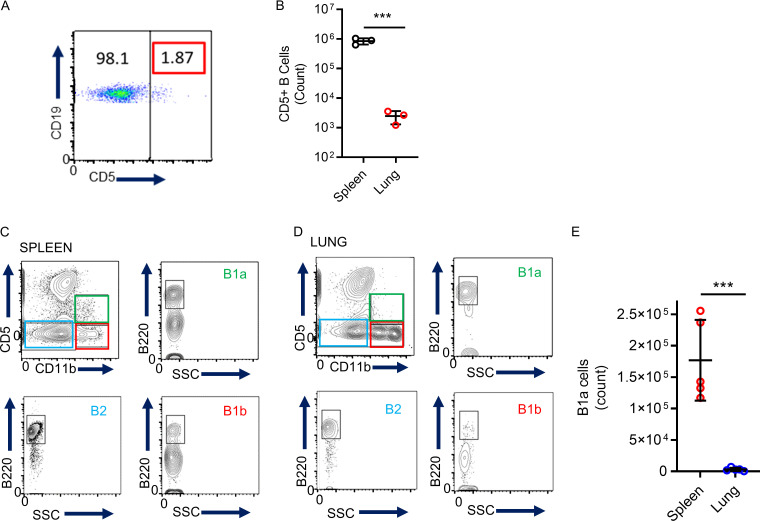
No precedent data exist characterizing lung marginating B cells; therefore, we performed scRNAseq of lung B cells to establish their identities. Lungs from seven female C57BL/6 mice were homogenized and pooled, and live leukocytes (CD45+) were isolated by cell sorting. 12,492 cells met quality control criteria (Fig. S1, A–C) and were included in subsequent analyses. The hierarchical tree algorithm using Seurat’s Louvain algorithm revealed eight distinct B cell subsets (Figs. 2 A and S1 D). The B lymphocyte marker Ighm was expressed throughout all lung B cells, whereas Ighd was enriched in four of eight subclusters (Fig. 2, B and C). Coexpression of IgM and IgD is related to B cell maturity, with less mature naive B cells having lower levels of IgD (Hobeika et al., 2016). RNA transcripts of Ighd revealed comparable frequency and expression levels between clusters 0 and 1 and between clusters 2 and 3 (Fig. 2 C), suggesting shared states of maturity. Further analysis of cell surface expression of CD21and CD24 using flow cytometry demonstrated that the two largest groups of lung B cells were T1B cells (CD45+ CD19+ CD21+ CD24+) and T2B cells (CD45+ CD19+ CD21+ CD24lo; Teague et al., 2007; Loder et al., 1999; Evans et al., 2007; Benitez et al., 2014; Fig. 2 D). Therefore, because most lung B cells could be divided by cell surface markers into T1B and T2B cells with compatible Ighd levels (Su and Rawlings, 2002; Su et al., 2004), we grouped clusters 2 and 3 as T1B and clusters 0 and 1 as T2B (Fig. 2 E). Analysis of the T1B cell cluster RNAseq revealed increased levels of Ighm, Cd24a, Vpreb3, and Egr1 compared with T2B, which had higher levels of Crlf3 and Fcer2a (CD23; Fig. 2, F–H). B cells were also identified with high RNA transcripts of Ighm, S100a6, and Itgb1, which have been reported in innate B1 B cells (Francés et al., 2006; Mabbott and Gray, 2014); yet, they had low levels of transcripts of other B1 markers, such as Cd5, Itgam, and Il10 (Fig. 2 H). Using flow cytometry, we analyzed lung B cells for established markers of B1a and B1b innate B cells (Haas et al., 2005), which showed low numbers of B1 cells defined by cell surface expression of CD5 in the lung compared with the spleen (Fig. S2). Importantly, we could not detect CSF2 mRNA in any of our B cell subsets, indicating that these cells do not belong to the recently identified innate response activator B cells, which can have regulatory properties (Rauch et al., 2012).
Figure S1.
scRNAseq quality control metrics and B cell annotations.(A) Unique molecular identifier (UMI), unique gene, and mitochondrial counts for each individual cell. (B) Pearson correlation of UMI counts with percentage of mitochondrial genes and number of unique genes detected for each individual cell. (C) Automated annotations of all Cd45+ immune cells using established SingleR databases (mouse RNAseq and ImmGen) confirmed the B cell identity of outlined clusters. NK, natural killer. (D) Subclustering of B cells was performed over a range of resolutions from 0 to 0.8 in steps of 0.1 to determine an optimal clustering resolution. (E) UMAP plot revealing eight distinct B cell clusters (at resolution 0.5). (F) Heatmap of the top five marker genes (determined by nonparametric Wilcoxon rank-sum test) for each of the six B cell states described in Fig. 2.
Figure 2.
scRNAseq reveals distinct B cell states within the lung. (A) B cells were analyzed using scRNAseq, and unsupervised clustering was achieved using a hierarchical tree algorithm using Seurat’s Louvain algorithm and visualized by UMAP. (B) Comparison of Ighm and Ighd transcripts across all B cells. (C) Ridge plots of Ighd transcripts between clusters 0 and 7. (D) Flow cytometry of lungs demonstrates two main B cell subtypes, T1B and T2B, using fluorescently conjugated mAbs to detect cell surface expression of CD45, CD19, CD21, and CD24. Data represent n = 3 independent experiments using three mice total. (E) Reclustering of B cells to map T1B and T2B cell states. (F and G) Relative RNA expression of representative genes associated with B cell states, including Crlf3, Fcer2a (T2), and Egr1 and Vpreb3 (T1). (H) Violin plots of RNA transcripts grouped by B cell states. For scRNAseq data in A–C and E–H, 4,044 B cells were from seven pooled mice, and sequencing was performed once.
Figure S2.
Characterizing B cell subsets in the lung. (A and B) Flow cytometry was used to analyze lung CD5+ B cells (A) and to compare the quantitative counts (B) between spleen and lung (unpaired Student’s t test, n = 3 independent experiments using three mice in total). (C–E) Flow cytometry was used to identify B1a, B1b, and B2 B cell subsets in the spleen (C) and lung (D) and to compare the two organs (E). Unpaired Student’s t test; n = 5 independent experiments using five mice in total. Data is mean ± SD. SSC, side scatter. ***, P < 0.001.
Transcription factors, such as PAX5 (Cobaleda et al., 2007) and IKAROS (Kwon et al., 2008; Schwickert et al., 2014), and gene regulatory networks (GRNs) are critical determinants of cellular fates and behaviors for both early B cell development and late terminal differentiation into memory and plasma cells (Laidlaw et al., 2020; Laidlaw and Cyster, 2021). However, transcriptional regulation pathways are less clear in directing and controlling transitional B cell states. To understand the transcriptional regulation of lung B cell subsets, we performed single-cell regulatory network inference and clustering (SCENIC) analysis (Fig. 3). Regulon specificity scores revealed unique regulons associated with T1B (Mafk, Ddit3), T2B (Chd2, Sp1), and B1 B cells (Tcf4, Nap1l1; Fig. 3, A–C). We further interrogated known B cell developmental transcription factors, such as EBF1 (Kwon et al., 2008), SpiB (Willis et al., 2017), and TCF3 (also known as E2A), which were dominant regulons of T1B but were not active in T2B (Fig. 3 D). Interestingly, EBF1 mRNA (yellow) was abundant throughout all lung B cell clusters; yet, the active GRN (blue) was confined to the T1 clusters, suggesting additional levels of cell state regulation (Fig. 3 D). The driving regulons to establish a T2B fate were found to be Sp1 and Chd2, which had high GRN activity in T2B cells but not in T1B cells, despite this early state having mRNA present (Fig. 3 E). Together, these data reveal important molecularly defined alterations between the T1 and T2 states and provide new pathways that determine B cell fates. Finally, we discovered novel transcriptional regulons for B1 B cells that require TCF4 (also known as E2-2) and Nap1l1 (Fig. 3 F). Interestingly, Bhlhe41, a known factor for development of innate B1a cells (Kreslavsky et al., 2017), was higher in mRNA for our B1 defined subset (Fig. 3 F), but the Bhlhe41 regulon was active throughout all the subsets, suggesting a more complex role of this transcription factor in the development of innate B1 cells. Together, our genetic analyses reveal distinct IgM B cells defined by discrete RNA profiles and GRNs that are found in the lung, where most B cells are intravascular, and these subsets are dominated by T1B and T2B cells.
Figure 3.
T1B, T2B, and B1 cells occupy distinct regulatory states. (A) GRN-based clustering color coded by B cell state and projected on a t-distributed stochastic neighbor embedding (t-SNE) map reveals that T1B, T2B, and B1 have distinct regulatory network structures. (B) GRN density projected onto SCENIC t-SNE map. (C) Rank-ordered regulon specificity scores for transcriptionally defined B cell states. (D–F) t-SNE map plotting gene expression (yellow), binarized regulon activity (active, blue; inactive, gray), and kernel density line area under the curve (AUC) histogram plotting inferred transcription factor (TF) activation. Examples of differentially active GRNs in T1B (D), T2B (E), and B1 (F) states.
T2B cells localize to the lung vasculature via CD49e-mediated adhesion
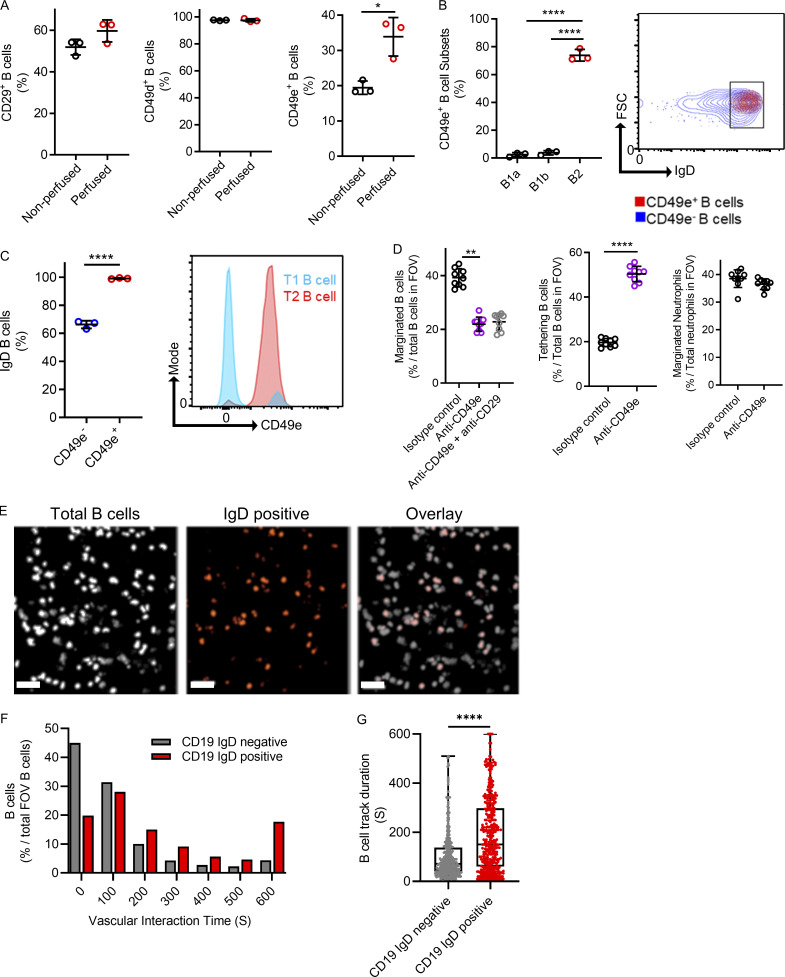
To ascertain the relationship between intravascular firm adhesion observed during intravital microscopy and the IgM subsets of lung B cells determined by scRNAseq, we used flow cytometry to compare cell surface molecule expression between nonperfused (containing circulating and marginated B cells) and perfused lungs devoid of circulating blood (to enrich adhered B cells). We assessed cell surface integrins because these are known to be important in B cell retention in lymphoid tissues (Lu and Cyster, 2002), and our scRNAseq revealed that the B1 cluster expressed Itgb1 (CD29, β1 integrin). However, after flushing out nonadhered B cells, no difference was observed in the population of B cells expressing CD29, suggesting that the adhered B cells were not B1 cells (Fig. 4 A). Similarly, CD49d is a reported adhesion molecule that was also not enriched after removing nonadhered B cells (Fig. 4 A). The adhesion molecule CD49e has been reported on early B cells (Jensen et al., 2016); therefore, we repeated our perfusion assay and found a significant increase in the percentage of B cells expressing CD49e, suggesting enrichment after the removal of circulating nonadhered B cells (Fig. 4 A). We reexamined the scRNAseq data for CD49e expression, but we could not identify the RNA. This, however, does not exclude protein expression. We next used flow cytometry to assess the cell surface protein expression of CD49e in defined B cell subsets that showed CD49e expression on IgD-expressing cells with T2B markers compared with cells with either T1B or B1 cell markers (Fig. 4, B and C). Using lung intravital microscopy, we assessed B cell firm adhesion after the i.v. administration of either a CD49e blocking mAb alone or in combination with a CD29 blocking antibody or an appropriate isotype control antibody. B cells had significantly less margination, defined by firm adhesion of ≥30 s when CD49e is blocked compared with control mice, and this resulted in a significant increase in transient B cell–endothelial interactions, termed “tethering” (Fig. 4 D and Video 2). However, the addition of an anti-CD29 inhibitory antibody did not further reduce B cell margination, suggesting a limited role of the β1 integrin component of VLA5 (CD49e/CD29). Neither the control nor the anti-CD49e antibody impacted neutrophil margination (Fig. 4 D). Therefore, B cells marginate within the lung microvasculature via CD49e-mediated firm adhesion.
Figure 4.
Marginated B cells are marked by CD49e, which mediates endothelial interactions. (A) Flow cytometry compared cell surface molecule expression on B cells obtained from nonperfused lungs (circulating blood remaining) with perfused lungs (circulating blood removed). (B) Flow cytometry expression of CD49e using a fluorescently conjugated anti-CD49e mAb. B cells were pregated for prototypical B1a, B1b, and B2 markers, and the percentage of CD49e-positive B cells is shown per group. FSC, forward scatter. (C) The expression of cell surface IgD was assessed by flow cytometry and compared between CD49e-positive B cells versus CD49e-negative B cells (A–C, n = 3 independent experiments using a total of six mice). (D) Intravital microscopy was used to visualize vascular B cells in mice treated i.v. with an inhibitory anti-CD49e mAb with or without an inhibitory anti-CD29 antibody compared with an appropriate isotype mAb control (pooled FOV replicates shown for n = 3 independent experiments using nine mice in total). (E) Intravital microscopy was performed with Cd19ZsGreen1 and costained for IgD-positive B cells. Scale bar, 50 µm. (F and G) Vascular interaction times and track duration are shown as frequency distributions and pooled together to compare IgD-negative and IgD-positive B cells (n = 3 independent experiments with three mice in total). Exact P values were determined using Student’s t test or ANOVA. *, P < 0.05; **, P < 0.01; ****, P < 0.0001. Pooled data are presented as mean ± SD or box-and-whisker plots showing median and interquartile range.
Video 2.
B cells require CD49e for margination in the lung. Related to Fig. 4. Intravital lung imaging was used to evaluate the role of CD49e on CD19+ B cell (fluorochrome-conjugated anti-CD19 antibody; green) marginated behavior in the lung capillaries. The mice were treated with either an i.v. injection of the isotype control antibody (left) or anti-CD49e blocking antibody (right). Videos were recorded 30 min after antibody injection. Experiments were repeated three times (n = 3). Playback speed is 12 frames/s, 10 min actual imaging time.
To more specifically investigate if the B cells that marginated were transitional B cells, we used a fluorescently conjugated anti-IgD antibody to mark transitional B cells in our total B cell reporter mouse (Cd19ZsGreen1; Fig. 4 E and Video 3). IgD+ CD19+ B cells represented an increased population of cells that stayed attached to the lung vasculature for at least 10 min compared with IgD− CD19+ B cells. Quantifying and comparing the vascular interaction times of all imaged B cells showed significantly increased durations of IgD+ B cells in the lung capillaries (Fig. 4, F and G).
Video 3.
IgD+ B cells marginate in the lung vasculature. Related to Fig. 4. Intravital lung microscopy was used to investigate if transitional B cells marginate in the lung vasculature. Cd19ZsGreen1 mice (white in video) were costained with anti-IgD fluorescently conjugated antibody (red). Experiments were repeated three times (n = 3). Playback speed is 24 frames/s, 10 min actual imaging time per segment.
The CXCR5–CXCL13 axis modulates the level of B cell margination in the lung
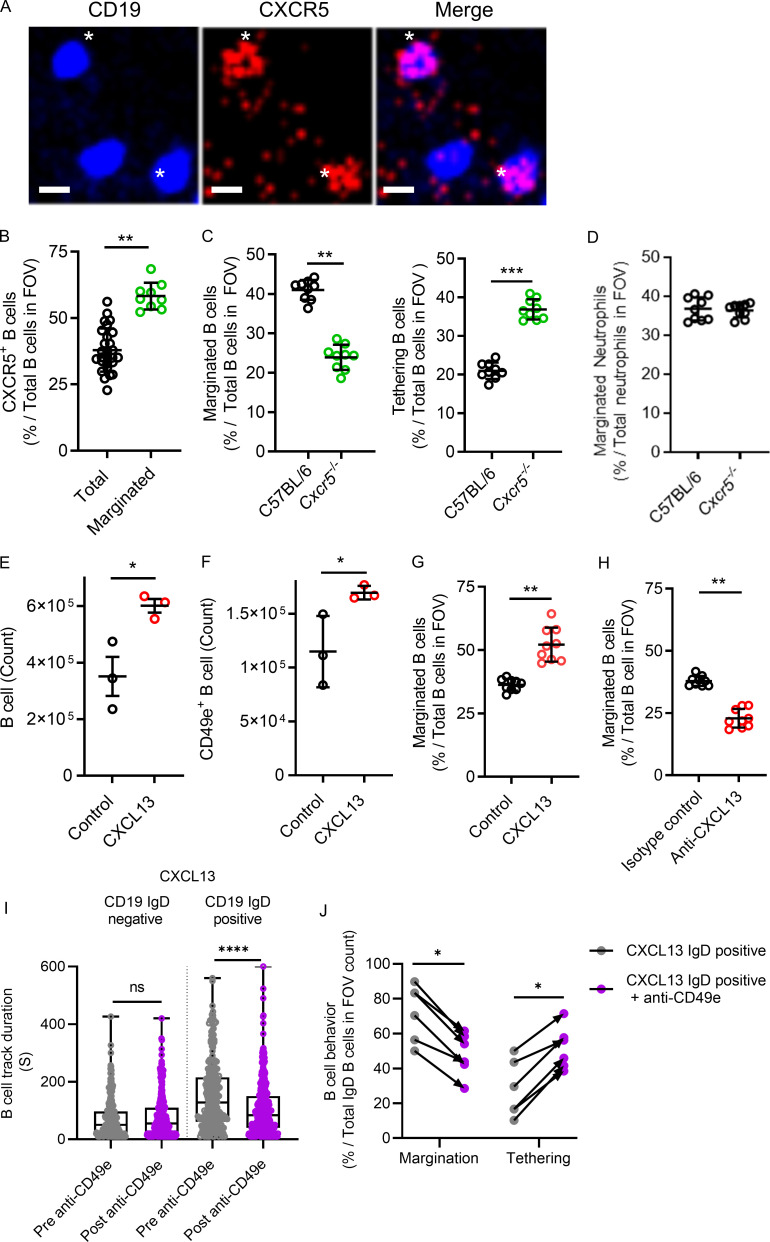
Chemokines and their receptors are important for localizing leukocyte populations (Ansel et al., 2002; Ansel et al., 2000). CXCR5, a known chemokine receptor on B cells essential for homing and localization (Denton et al., 2019; Ansel et al., 2000), was detected at low levels broadly in our lung B cells by scRNAseq (see the online genetic atlas at http://www.biernaskielab.ca/b_cell or http://www.biernaskielab.com/b_cell). We used lung intravital microscopy to assess if marginated B cells in the microvasculature expressed CXCR5 and found that, in vivo, a subset of intravascular B cells stained positive for CXCR5 using a fluorescently conjugated anti-CXCR5 mAb (Fig. 5 A). Moreover, a larger percentage of marginated B cells stained positive for CXCR5 than total B cells in the lung vasculature (Fig. 5 B). To determine if CXCR5 is required for B cell margination within the lung microvasculature, we investigated Cxcr5−/− mice using lung intravital microscopy and found fewer marginated but more tethering B cells during homeostasis than in C57BL/6 mice (Fig. 5 C and Video 4). Cxcr5−/− did not impact neutrophil margination (Fig. 5 D). CXCL13 (Ansel et al., 2002; Cinamon et al., 2004) is the only known cognate chemokine for CXCR5, so we tested if exogenous CXCL13 could induce B cell margination in the lung. Indeed, intratracheal (i.t.) CXCL13 significantly increased total B cells and CD49e+ B cells as quantified by flow cytometry (Fig. 5, E and F). Marginated B cells were also increased in CXCL13-treated mice compared with control animals as quantified by intravital lung imaging (Fig. 5 G and Video 5). Importantly, systemic administration of a neutralizing anti-CXCL13 mAb resulted in a significant decrease in marginated B cells compared with control isotype antibody-treated mice analyzed by intravital microscopy, demonstrating that endogenous CXCL13 is required for margination (Fig. 5 H and Video 6). To further confirm the role of transitional B cells in margination, we treated Cd19Zsgreen mice with CXCL13 and imaged the IgD+ versus the IgD− B cells before and after CD49e inhibition because T2B cells had high levels of CD49e expression by flow cytometry. CD19+ IgD− duration in the lung vasculature was not altered by the addition of inhibitory anit-CD49e antibodies; however, CD19+ IgD+ B cells had a significant decrease in the duration of time spent adhered to the vasculature (Fig. 5 I). Moreover, the number of marginated IgD+ B cells induced by CXCL13 was reduced, and transient tethering increased after i.v. inhibitory anti-CD49e antibody administration (Fig. 5 J). Therefore, transitional B cell margination within the lung microvasculature requires the CXCL13–CXCR5 chemokine axis and CD49e adhesion.
Figure 5.
The CXCR5–CXCL13 axis modulates the level of B cell margination in the lung. (A) Intravital lung microscopy revealed B cells (Cd19ZsGreen1) that were visible for CXCR5 using i.v. administered, fluorescently conjugated anti-CXCR5 mAb and that were marginated. For clarity, B cells were falsely colored blue, and CXCR5 is red. Asterisks show two examples of CXCR5-positive B cells (representative images from n = 3 independent experiments using three mice; scale bars, 7 µm). (B) CXCR5+ B cells were compared with total B cells, as was the proportion of CXCR5+ B cells that marginated compared with total B cells observed in the FOV. (C and D) Intravital lung microscopy compared B cell margination and tethering in C57BL/6 versus Cxcr5−/− mice (C), while D shows marginated neutrophils. (E and F) Cd19Zsgreen1 mice were pretreated with exogenous CXCL13 i.t., and flow cytometry quantified total and CD49e+ lung B cells. (G and H) Intravital lung microscopy quantified B cell margination in purified CXCL13-pretreated Cd19Zsgreen1 mice or in Cd19Zsgreen1 mice that received systemic pretreatment with an isotype control antibody versus a neutralizing anti-CXCL13 mAb. (I) CXCL13 i.t. treated Cd19ZsGreen1 mice were costained for IgD, and B cells were quantified by track duration before and after administration of inhibitory anti-CD49e antibodies. (J) B cells were phenotyped for margination and tethering before and after anti-CD49e antibody administration. For imaging experiments, n = 3 individual experiments were performed. Pooled FOV replicates are shown for B–D, G, and H. I and J represent n = 3 independent experiments using six mice in total. Exact P values were determined using Student’s t test, except where groups were compared using the Mann-Whitney test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Pooled data are presented as the mean of n values ± SD or as box-and-whisker plots showing median and interquartile range. Statistical testing was performed using the n values.
Video 4.
CXCR5 deficiency reduces B cell margination. Related to Fig. 5. Intravital lung microscopy was used to investigate the role of CXCR5 in B cell margination. B cells were stained with anti-CD19 antibody. C57BL/6 mice are compared with Cxcr5−/− animals. Playback speed is 10 frames/s, 10 min actual imaging time.
Video 5.
CXCL13 enhances B cell margination in the lung. Related to Fig. 5. Intravital lung microscopy was used to investigate the role of exogenously provided CXCL13 on CD19+ B cell (Cd19ZsGreen1; green) margination in the lung capillaries. 12 h after i.t. CXCL13 injection, B cell marginated behavior was assessed. Ly6G+ neutrophils (fluorochrome-conjugated anti-Ly6G antibody; blue) were also labeled. Experiments were repeated three times (n = 3). Playback speed is 12 frames/s, 10 min actual imaging time.
Video 6.
Endogenous CXCL13 maintains B cell margination. Related to Fig. 5. Intravital lung microscopy was used to investigate the role of endogenous CXCL13 on CD19+ B cell (Cd19ZsGreen1; green) margination in the lung capillaries. B cell margination was assessed 12 h after i.p. injection of either the isotype control antibody (left) or anti-CXCL13 neutralizing antibody (right). Experiments were repeated three times (n = 3). Playback speed is 12 frames/s, 10 min actual imaging time.
Marginated B cells dampen neutrophil intravascular inflammation during lung injury
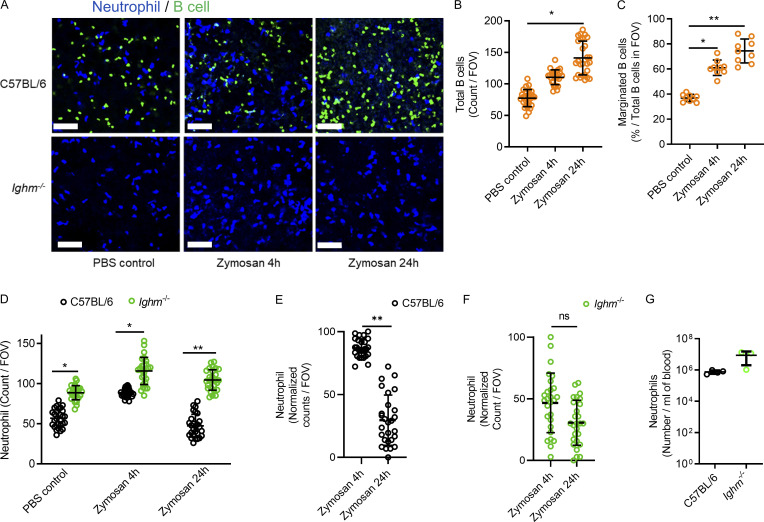
Prepositioning or homing of leukocytes to various tissues is generally purposeful; however, there are no reports establishing a purpose for B cells to marginate within the lung microcirculation. T2B cells found in the spleen have established regulatory properties in the context of joint inflammation (Evans et al., 2007; Khoder et al., 2014; Rosser et al., 2014); therefore, we hypothesized that T2B cells marginating in the lung microcirculation may perform regulatory functions. Zymosan, a potent fungal inflammatory stimulus causing airway inflammation and lung injury (Han et al., 2013; Cone et al., 2003), was used to investigate the role of marginated B cells during lung inflammation. Zymosan-coated beads were injected i.t. into C57BL/6 mice, which were compared with mice genetically deficient in B cells (Ighm−/−). Lung intravital microscopy revealed that intravascular B cells were recruited in C57BL/6 mice during zymosan-induced lung injury after 4 h and 24 h of stimulation compared with PBS treatments, but no B cells were visualized in Ighm−/− mice (Fig. 6, A and B; and Video 7). Additionally, the percentage of B cells marginating in the microvasculature of C57BL/6 mice significantly increased 4 h and 24 h after lung injury (Fig. 6 C). Neutrophil recruitment was quantified using lung intravital microscopy, and significantly more intravascular neutrophils were observed in Ighm−/− mice treated with PBS and with 4 h or 24 h zymosan stimulation compared with C57BL/6 mice (Fig. 6 D and Video 7). Resolution of neutrophil recruitment was observed between 4 h and 24 h of zymosan-induced lung injury in C57BL/6 mice, but his did not occur in the Ighm−/− B cell–deficient mice (Fig. 6, E and F). The margination of neutrophils in B cell–deficient mice was not due to significantly increased circulating neutrophil numbers (Fig. 6 G). Thus, airway inflammation during lung injury leads to increases in marginated lung B cells that restrain excessive neutrophil inflammation and participate in inflammatory resolution in the lung.
Figure 6.
Marginated B cells increase during direct zymosan-induced lung injury and restrain excessive neutrophil recruitment.(A) Lung intravital microscopy was performed on either C57BL/6 or Ighm−/− mice that were treated with PBS i.t. as a control or with 106 zymosan-coated beads i.t. Neutrophils and B cells were visualized using i.v. fluorescently conjugated anti-Ly6G mAb and fluorescently conjugated anti-CD19 mAb. Scale bar, 70 µm. (B and C) Total visualized B cells and marginated B cells were quantified in C57BL/6 mice, and no B cells were visualized in Ighm−/− mice. (D–F) Neutrophils were quantified and compared between mice that received PBS control or 4 h or 24 h of zymosan i.t. (G) Peripheral blood neutrophil counts were compared. For A–G, n = 3 independent experiments using nine mice in total. All individual imaging FOV replicates are shown for B–F. B and C were analyzed using one-way ANOVA and Tukey’s post hoc test. D–G were analyzed using Student’s t test based on n. Pooled data are presented as mean of n ± SD. *, P < 0.05; **, P < 0.01.
Video 7.
B cells resolve focal neutrophil inflammation in vivo. Related to Fig. 6. C57BL/6 or Ighm−/− mice were i.t. treated with zymosan-coated beads (106) or sterile PBS. Lung intravital microscopy was performed at 4 h and 24 h after stimulation, and neutrophil recruitment and B cell margination were assessed. Playback speed is 30 frames/s, 10 min actual imaging time per segment.
Marginated B cells restrain excessive neutrophilic inflammation via LXA4
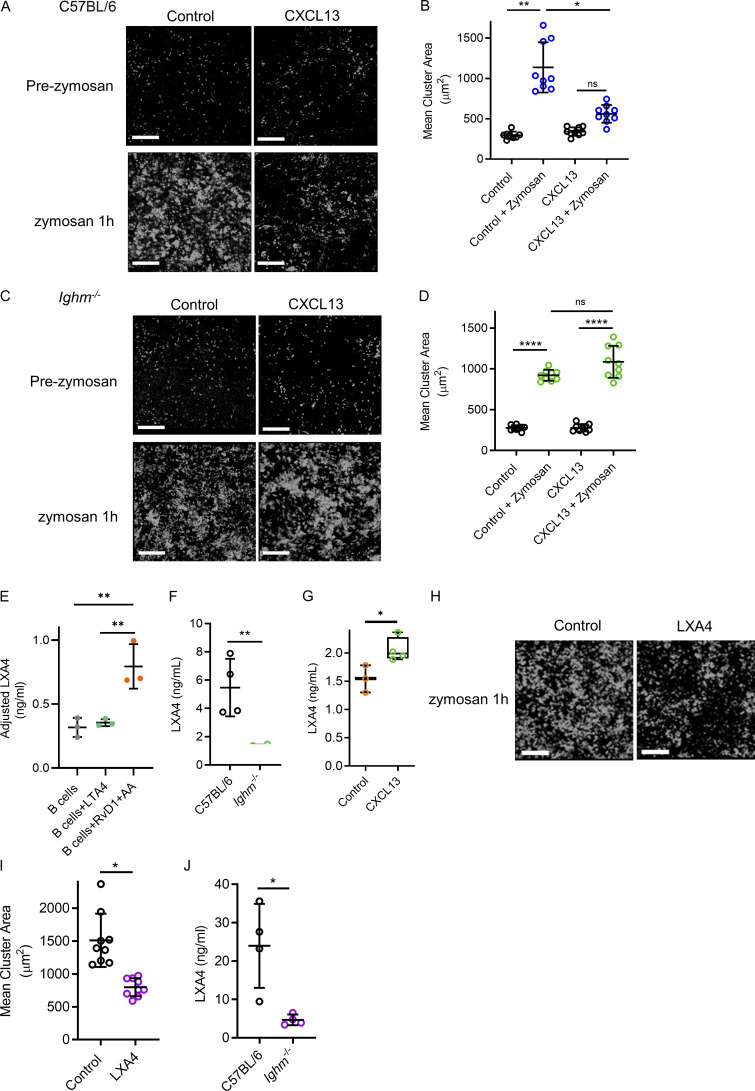
Lung injury can also occur secondary to systemic stimulation as opposed to direct airway stimulation. Therefore, we used a model of a pathogenic hyperinflammatory state with intense intravascular neutrophilic recruitment, swarming, and cluster formation resulting from systemic i.v. zymosan administration (Lee et al., 2018). Using lung intravital microscopy, we observed a significant increase in neutrophil recruitment, swarming, and cluster formation after administration of i.v. zymosan beads in C57BL/6 mice (Fig. 7, A and B; and Video 8). However, C57BL/6 mice pretreated with i.t. CXCL13, which increases marginated B cells, attenuated intravascular neutrophil clustering (Fig. 7, A and B; and Video 8). Ighm−/− mice were then assessed for intravascular neutrophil responses to i.v. zymosan-coated beads. Ighm−/− animals also generated neutrophil intravascular clusters, similar to tissue swarming and clusters (Lämmermann et al., 2013); however, unlike in C57BL/6 mice, CXCL13 did not attenuate clustering in B cell–deficient mice (Fig. 7, C and D). Neutrophil clustering requires the proinflammatory lipid mediator leukotriene B4 (Lämmermann et al., 2013; Lee et al., 2018), and this process can be reversed in vitro by specialized proresolving mediators (SPMs) such as LXA4 (Lee et al., 2018; Lämmermann et al., 2013; Reátegui et al., 2017). In addition, B cell–enriched tissues such as the bone marrow and tonsils have high levels of LXA4 (Ramon et al., 2014). We tested if murine B cells contributed to LXA4 generation in vitro. Isolated splenic B cells maintained in culture wells had low levels of LXA4 detected by ELISA (Fig. 7 E). Leukotriene A4 (LTA4) or resolvin D1 (RvD1) plus arachidonic acid (AA) can drive LXA4 generation (Samuelsson et al., 1987; Fredman et al., 2014). In vitro, RvD1 + AA, but not LTA4 alone, significantly increased the levels of soluble LXA4 in isolated B cells (Fig. 7 E). To establish that lung B cells contributed to LXA4 production, we tested the in vivo levels of pulmonary LXA4 in untreated mouse lung homogenates from C57BL/6 mice and compared them with Ighm−/− mice using ELISA. Indeed, among unstimulated mice, B cell–deficient mice had significantly less lung homogenate LXA4 than C57BL/6 mice (Fig. 7 F). Furthermore, we quantified the lung levels of LXA4 in mice treated with CXCL13 and found a significant increase compared with control (Fig. 7 G). We next tested if exogenous LXA4 could dampen excessive neutrophilic inflammation in vivo. We observed that LXA4 administration attenuated neutrophil intravascular clustering in vivo during systemic zymosan stimulation (Fig. 7, H and I). Finally, we treated C57BL/6 and Ighm−/− mice with i.v. zymosan-coated beads and compared the amount of LXA4 generated in whole-lung homogenates using ELISA. Congruent with our findings that B cells produce LXA4 in vitro, we found a significant increase in LXA4 only in mice with B cells but limited LXA4 generation in the B cell–deficient mice (Fig. 7 J). Therefore, lung B cells contribute to generating LXA4, which dampens excessive neutrophilic pulmonary inflammation during systemic stimulation.
Figure 7.
Manipulating the effects of marginated B cells restrains excessive neutrophilic inflammation in a model of systemic inflammation induced lung injury. (A and B) Lung intravital microscopy quantified neutrophil inflammation and cluster formation in control versus CXCL13 i.t. treated C57BL/6 mice before and after i.v. administration of zymosan-coated beads. Neutrophils (i.v. fluorescently conjugated anti-Ly6G mAb) and neutrophil clusters have been falsely colored white. Scale bars, 200 µm. (C and D) Neutrophil cluster formation and surface area were assessed using lung intravital microscopy in B cell–deficient mice (Ighm−/−) treated with or without exogenous CXCL13 i.t. before and after zymosan-coated bead administration. For A–D, n = 3 independent experiments using 12 mice in total; pooled FOV replicates are shown. Exact P values were calculated using one-way ANOVA with Tukey’s post hoc test. Scale bars, 200 µm. (E) Isolated B cells were cultured, and supernatants were tested for LXA4 levels using ELISA. Adjusted levels were determined by subtracting the control wells without B cells added. n = 3 independent experiments using nine mice total. Exact P values were determined using Student’s t test. (F and G) ELISA quantified LXA4 levels in lung homogenates for C57BL/6 and Ighm−/− mice (F) and mice treated with CXCL13 (G). n = 4 independent experiments using eight mice total. Exact P values were determined using Student’s t test. (H and I) Intravital lung microscopy quantified cluster formation in mice receiving zymosan-coated beads i.v. alone or with i.v. LXA4. Neutrophil clusters have been falsely colored white. Scale bar, 200 µm. Pooled FOV replicates are shown for n = 3 independent experiments using six mice total. Exact P values were determined using Student’s t test. (J) ELISA quantified LXA4 levels in lung homogenates for C57BL/6 and Ighm−/− mice 1 h after administration of i.v. zymosan-coated beads. n = 4 independent experiments using eight mice total. Exact P values were determined using Student’s t test. Pooled data are presented as mean ± SD. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
Video 8.
B cells and LXA4 act to diminish intense neutrophil inflammation. Related to Fig. 7. Intravital lung imaging was used to assess the role of B cell margination and LXA4 on zymosan bead (white/yellow)-induced neutrophil (fluorochrome-conjugated anti-Ly6G antibody; blue) clustering. The PBS + zymosan beads, CXCL13 pretreatment + zymosan beads, and zymosan beads + LXA4 images of dynamic neutrophil clustering support the differences in neutrophil clustering highlighted in Fig. 7. Playback speed is 12 frames/s, 10 min actual imaging time per segment.
B cells attenuate pulmonary neutrophil inflammation during A. fumigatus pneumonia
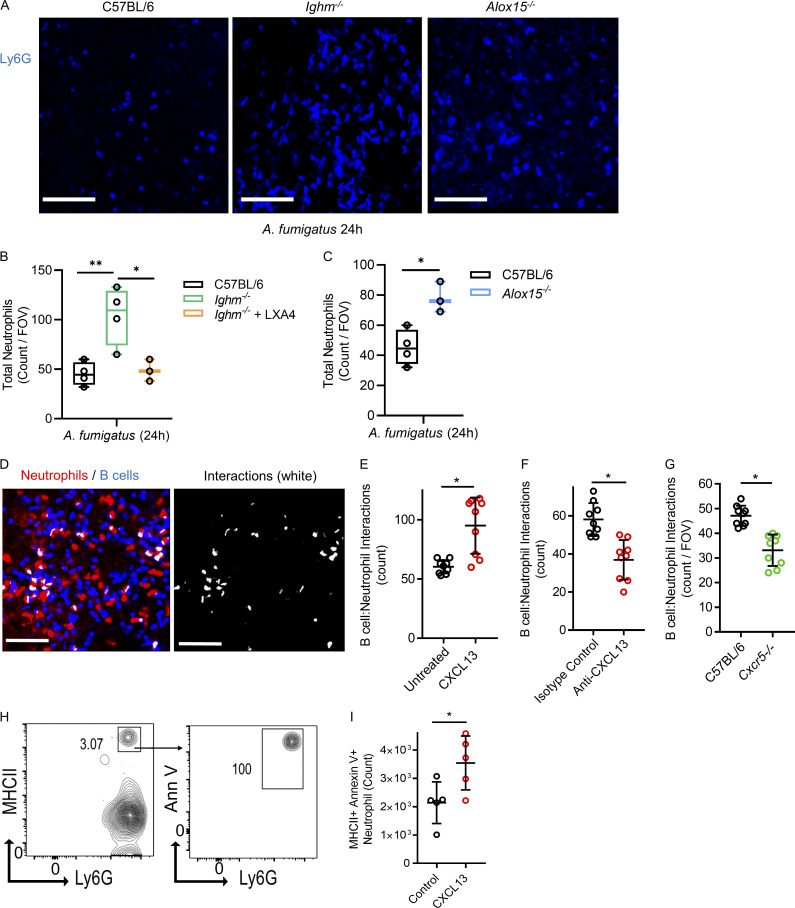
Neutrophils are essential for host defense, but overexuberant lung responses can be detrimental, so tight regulation is important. We investigated if B cells and LXA4 attenuated neutrophil inflammation during A. fumigatus pneumonia in vivo (Snarr et al., 2020). Compared with C57BL/6 mice, Ighm−/− mice had significantly elevated lung vascular neutrophils during fungal pneumonia, which were attenuated with exogenous LXA4 (Fig. 8, A and B). Furthermore, mice deficient in LXA4 production (Alox15−/−) showed significantly elevated lung vascular neutrophils during A. fumigatus pneumonia (Fig. 8, A and C). Finally, the induction of neutrophil apoptosis is a mechanism of inflammatory regulation during pneumonia; therefore, we tested if altering B cell margination impacted neutrophil apoptosis. We previously described that aged neutrophils were induced by physical contact with B cells in the lung to undergo programmed cell death (Kim et al., 2018). CXCL13 administration increased B cell–neutrophil interactions, which were diminished with the anti-CXCL13 inhibitory antibody and in the Cxcr5−/− mouse (Fig. 8, D–G). Lung neutrophils can be MHCII positive after physical interaction with B cells (Kim et al., 2018), consistent with our current finding that MHCII neutrophils in the lung were apoptotic and that apoptotic neutrophils increased in mice treated with CXCL13, owing to increased B cell–neutrophil interactions (Fig. 8, H and I). Therefore, B cells and LXA4 attenuate lung neutrophil inflammation during pneumonia, and B cells induce neutrophils to undergo apoptosis, an important mechanism to resolve inflammation.
Figure 8.
B cell regulation of lung neutrophils during fungal pneumonia. (A–C) Lung intravital microscopy was performed in mice infected with A. fumigatus (24 h), and neutrophils were imaged and quantified. Scale bars, 100 µm. (B) C57BL/6 mice were compared with Ighm−/− mice with and without LXA4 administration (5 µg i.v. at the time of infection). (C) C57BL/6 mice were compared with Alox15−/− mice. A–C represent 3–4 independent experiments using 14 total mice. (D) Neutrophil (i.v. fluorescently conjugated anti-Ly6G mAb, red) and B cell (Cd19Zsgreen1, blue) physical interactions were observed using lung intravital microscopy and highlighted as colocalization (white in the right panel). Scale bars, 70 µm. (E) Untreated Cd19Zsgreen1 mice were compared with CXCL13 i.t. pretreated mice. (F and G) B cell neutrophil interactions were quantified and compared between Cd19Zsgreen1 mice treated with i.v. isotype control antibody versus a neutralizing anti-CXCL13 mAb (F) or between C57BL/6 versus Cxcr5−/− mice (G). (H) The acquisition of MHCII from B cells to neutrophils and the initiation of neutrophil apoptosis using annexin V staining was evaluated using flow cytometry. (I) MHCII+ annexin V+ lung neutrophils were quantified by flow cytometry and compared between control and exogenous CXCL13 i.t. treated mice. For imaging experiments, D–G pooled FOV replicates are shown, and n = 3 independent experiments were performed per condition using a total of 18 mice. In H and I, n = 5 independent experiments using 10 mice in total. Exact P values were determined using Student’s t test to compare two groups or ANOVA to compare three groups. Pooled data are presented as mean ± SD. *, P < 0.05; **, P < 0.01.
Discussion
Pulmonary neutrophilic inflammation is both protective and pathological, depending on the disease; however, the regulation of lung neutrophilic inflammation remains underappreciated. Here, we discovered that eight unique populations of IgM+ B cells can be found in the lung, largely represented by T1B and T2B cells. T2B cells were directly observed to marginate, which is an unexpected finding, considering that marginating lung neutrophils have been investigated for decades without investigators reporting a similar phenomenon in B cells (Martin et al., 1987; Doerschuk et al., 1987). Indeed, limited evidence exists to document the life and behavior of IgM+ peripheral B cell populations such as T2B cells. Our data demonstrate that T2B cells specifically home to and populate the lung microcirculation via CD49e and CXCR5 and can dampen neutrophilic inflammation by producing resolving molecules such as LXA4.
The findings that B cells localizing in the lung capillaries participate in dampening excessive inflammation and aid in resolving acute inflammation via lipoxins were unexpected. Indeed, in previous reports, the in vivo cellular source of proresolving molecules involved transcellular communication pathways between granulocytes and platelets and could be generated by inflammatory exudates, apoptotic neutrophils, and M2 macrophages (Papayianni et al., 1995; Fiore and Serhan, 1990; Chiang et al., 2005; Dalli and Serhan, 2012); yet, the cellular sources are less clear in B cell–rich tissues such as tonsils, marrow, and spleen (Duffney et al., 2018). B cells were not known to be producers of lipoxins, but they can respond to lipoxins via the LXA4 receptor, also known as N-formyl peptide receptor 2 (Ramon et al., 2014; Ramon et al., 2012). Here, researchers found that LXA4 increased antibody production in activated splenic B cells (Ramon et al., 2012), but LXA4 treatment of memory B cells significantly decreased both IgM and IgG production (Ramon et al., 2014). The relevance of these observations was not clear, although it was suggested that this could keep B cells in check and potentially govern immune responses in organs affected by autoimmune diseases. Additionally, in allergic asthma, LXA4 diminishes IgE-producing pathological B cells, highlighting the potential for SPM therapeutically (Kim et al., 2016). A limitation in our study is that, despite our total B cell–deficient mouse having diminished lung LXA4 and our in vitro isolated B cells producing LXA4, we do not have direct experiments showing that T2B cells are the specific source of lung LXA4. In addition, it remains possible that T2B cells also participate in a transcellular LXA4 mechanism involving other cells within the lung. Additional future attention needs to be directed at determining if LXA4 administration can alter clinically relevant endpoints during pneumonia. Further studies will illuminate the contribution of B cell margination and immunoresolving activity to infectious diseases, vaccine development, inflammatory lung diseases such as asthma and chronic obstructive pulmonary disease, autoimmunity, and B cell cancers.
Excessive pulmonary neutrophilic inflammation has been observed in B cell–deficient models (Kozakiewicz et al., 2013; Maglione et al., 2007). Additionally, B cells regulate hepatic neutrophil inflammation during leishmaniasis (Smelt et al., 2000), and B cells can limit neutrophil migratory ability (Kondratieva et al., 2010). Interestingly, a clinically relevant biomarker for several lung diseases is the neutrophil/lymphocyte ratio. Patients have worse outcomes in both conventional ARDS and COVID-19–related ARDS if they have a high neutrophil/lymphocyte ratio specifically driven by lymphopenia that includes reduced peripheral circulating B cells (Qun et al., 2020; Qin et al., 2020; Li et al., 2019). The pathological mechanistic relationship between low peripheral lymphocytes and neutrophil-mediated lung injury has not been elucidated in human disease, but it remains plausible that it could be related to a decrease in lymphocytes with regulatory capacities, including T2B cells.
Our scRNAseq analysis revealed eight distinct clusters of B cells. Although we categorized clusters 0 and 1 as T2B cells and 2 and 3 as T1B cells, it is possible that four transitional B cell states exist or that these are intermediate states between T1 and T2. The other large subset of B cells identified in the lung had genetic signatures compatible with B1 cells. However, it was more challenging to find B1 cells by classical cell surface receptor markers or by the prototypical cytokine, IL-10. A temporal disconnect between RNA levels and protein is possible, but this remains untested in our models. Furthermore, the functional characteristics of these cells as true B1 cells has not been determined. The remaining B cell subsets are also not yet definitively characterized, and further exploration of these cells is warranted. We have created an online lung B cell genetic atlas that will assist researchers in further elucidating these interesting B cell subsets (http://www.biernaskielab.ca/b_cell or http://www.biernaskielab.com/b_cell).
In summary, the lung microvasculature is a specialized niche for neutrophils that must tightly balance host defense and excessive inflammation to protect against lung injury (Kim et al., 2018; Wu et al., 2016; Woodfin et al., 2011; Colom et al., 2015). Here, we have uncovered a regulatory system where peripheral IgM+ transitional B cells serve to dampen excessive neutrophil activity in the lung. To date, no specific therapy is clinically useful to treat neutrophil-mediated lung injury or ARDS. Our findings define new potential targets that can be manipulated pharmacologically to alter and attenuate diseases with excessive neutrophil lung inflammation, such as infectious pneumonia, that result in severe lung injury.
Materials and methods
Mice
Male and female 7–12-wk-old C57BL/6, B6.129P2(C)-Cd19tm1(cre)Cgn/J, B6.Cg-Gt(ROSA)26Sortm6(CAG-ZsGreen1)Hze/J, and B6.129S2-Ighmtm1Cgn/J mice were obtained from The Jackson Laboratory and were provided access to food and water ad libitum. The Cd19ZsGreen1 mouse model was generated by crossing homozygous B6.129P2(C)-Cd19tm1(cre)Cgn/J mice with homozygous B6.Cg-Gt(ROSA)26Sortm6(CAG-ZsGreen1)Hze/J mice, resulting in fluorescent B cell reporter Cd19ZsGreen1 mice. LXA4-deficient mice, termed “Alox15−/−” (B6.129S2-Alox15tm1Fun/J) were purchased from The Jackson Laboratory. All mice were housed in the environmentally controlled specific-pathogen-free facility at the University of Calgary. Sibling mice were housed together by sex and used between 6 and 12 wk of age. Male and female mice were used equally among experiments. All animal protocols were approved by the University of Calgary Animal Care Committee (protocol no. AC18-0071).
Lung intravital microscopy
Mice were anesthetized by i.p. injection of ketamine (100 mg/kg) and xylazine (10 mg/kg). Anesthetized mice were cannulated at the internal jugular veins to allow i.v. injection of antibodies, reagents, and additional anesthetics before and during imaging. Mice were placed on mechanical ventilation via a tracheal catheter. The left lung was exposed by removing two or three ribs. A vacuum chamber with a glass slide fitted on top was used to gently stabilize a portion of the lung for imaging (Looney et al., 2011). Anti-Ly6G (clone 1A8; BioLegend), anti-CD19 (clone 6D5; BioLegend), and in some cases anti-CD31 (clone MEC13.3; BioLegend) antibodies conjugated with either Alexa Fluor 647 or 594 fluorochrome were injected i.v. (7 µl per mouse) to visualize polymorphonuclear neutrophils and the vasculature, respectively. 1 µg of anti-IgD antibody conjugated with Alexa Fluor 647 (clone 11-26c.2a) was administered to image transitional B cells. A resonant scanner confocal microscope (Leica SP8) was used for all pulmonary imaging. A 25×/0.95 water objective lens was used for imaging for all videos and still images. A tunable multiline white light laser was used to simultaneously excite the required fluorochromes.
In specific experiments, 50 µg purified anti-CD29 (clone HMβ1-1) or anti-CD49e (clone HMα5-1) mAb or Armenian hamster IgG isotype control antibody was added i.v. after 10 min of intravital imaging from which information about B cell and neutrophil populations in the untreated lung would be collected. In specific experiments, 30 µg purified anti-CXCL13 or rat IgG2a isotype control was added via i.p. injection 12 h before intravital imaging to allow time for physiological changes to occur in response to the neutralizing effect of antibody on the chemokine. Videos were acquired and recorded ∼10 min after the administration of fluorochrome-conjugated antibodies specific to the cells of interest. Videos (each 10 min in length) from three different fields of view (FOVs) were recorded at identical time points, thereby representing intraexperimental replicates, using the resonant scanner confocal microscope (Leica SP8). Videos were processed using either Leica LAS AF or Volocity 6.30. Analysis of cell amount, behavior, and interactions was performed using Volocity 6.30 or Imaris 9.7 software.
Processing of single-cell suspensions and flow cytometry
Single-cell suspensions of the sample tissues were processed in the following manner. For collecting BAL samples, mice were sacrificed, and P90 tubing was inserted into the trachea attached to a 20-gauge needle and 1-ml syringe. The airway was washed with 5 ml PBS total in 1-ml increments. For processing lung tissue into a single-cell suspension, the lungs were first flushed by running 10 ml saline through the right ventricle to exsanguinate the pulmonary circulation. The lung tissue was then harvested and minced with surgical scissors in 5 ml PBS on ice and poured into a gentleMACS tissue dissociator tube. The minced lung was then processed with the gentleMACS tissue dissociator. The lung homogenate was passed through a 100-µm filter into a 50-ml Falcon tube with 10 ml PBS, centrifuged at 2,000 rpm at 4°C for 5 min, and resuspended in 10 ml PBS. The spleen was surgically extracted from the abdominal cavity. The spleen was then passed through a 100-µm filter into a 50-ml Falcon tube with 10 ml PBS, centrifuged at 2,000 rpm at 4°C for 5 min, and resuspended in 10 ml PBS. The samples were treated with 1× RBC Lysis Buffer (BioLegend) for 3 min. After the samples were washed three times with PBS, the tissue samples were incubated with anti-CD16/32 (Fc Block; Bio X Cell) for 30 min on ice. After the samples were transferred to a 96-well plate, the samples were stained with Zombie Aqua (1:1,000 dilution in PBS) for 20 min at room temperature for live/dead cell differentiation. After the samples were washed twice with PBS, antibody cocktails were added to the corresponding wells and stained on ice for 30 min. All antibodies were used at 1:200 dilution. After the samples were washed three times with FACS wash, the cells were resuspended in 200 µl FACS wash and processed using a BD FACSCanto flow cytometer. If needed, after staining, the samples were washed with 1× annexin V binding buffer (BioLegend) and were resuspended with 100 µl of the 1× annexin V binding buffer. 5 µl Alexa Fluor 647–conjugated annexin V (concentration varies by lot number) was added to the cell suspension and incubated in the dark at room temperature for 15 min. 400 µl of the 1× annexin V binding buffer was added to the cell suspension and processed using a BD FACSCanto flow cytometer. Analysis was performed using FlowJo software.
Cell isolation and FACS sorting
Cells were maintained at room temperature throughout our single-cell sample preparation. Intact lungs were dissected from seven females and placed in a FACS tube with 4 ml of 1× PBS. A gentleMACS Dissociator 2.01 was used to mechanically liberate lung cells, and the cell suspension was filtered through a 70-µm cell strainer. The cell strainer was rinsed with an additional 6 ml of 1× PBS to recover remaining cells. Tissue samples were centrifuged at 2,000 rpm for 5 min, and supernatant was discarded. To eliminate erythrocyte contamination in the single-cell suspension, 2 ml RBC lysis buffer was added for 3 min. RBC lysis buffer was diluted by adding 10 ml PBS. The resulting cell suspension was centrifuged at 2,000 rpm for 5 min, and supernatant containing the medium plus enzyme was discarded. Cells were resuspended in 500 µl PBS, and 12.5 µl Fc Block was added for 15 min to block Fc receptors before being exposed to anti–CD45-PE–cyanine 7 (Cy7) antibody. After another round of centrifugation at 2,000 rpm for 5 min, the cell pellet was first stained with 7-aminoactinomycin D viability dye (5 µl per 200 µl) for 7 min in the dark and then with 1 µl anti–CD45-PE-Cy7 in 200 µl of final volume for 20 min. Unbound antibodies were rinsed off by centrifuging cells at 2,000 rpm for 5 min and discarding the supernatant. Cells were resuspended in 300 µl PBS + 2% BSA, then filtered through a 100-µm cell strainer. 7-Aminoactinomycin D− CD45+ viable immune cells were sorted in PBS + 2% BSA. The percentage of CD45+ cells was determined by flow cytometry and varied between 92 and 97%. After centrifugation at 2,000 rpm for 10 min, 40,000 CD45+ single cells were resuspended in 50 µl PBS + 2% BSA + 1 µl RNase inhibitor solution.
Single-cell transcriptomics of healthy rodent lungs
To isolate immune cells comprising healthy lung tissue, 40,000 single, viable CD45+ cells were barcoded using 10X Genomics Chromium Single Cell 3′ Reagent Guidelines version 3 Chemistry as per the manufacturer’s protocol. Quality control and cDNA quantification were performed using DNA Tape Station D1000. Sequencing was performed over two rounds on an Illumina NovaSeq 125 Cycle S2 dual lane flow cell run. We recovered 12,492 cells sequenced to 91,931 reads/cell (89% sequencing saturation) with an estimated doublet rate of ∼8%. Read alignment to the prebuilt mm10 reference genome was performed using cellranger count (10X Genomics; Zheng et al., 2017). The resulting gene–barcode matrix was imported into Seurat version 3 (Stuart et al., 2019) for quality control, dimensionality reduction, cell clustering, and differential expression analysis (Fig. S1, A and B). 3 Ms4a1/Cd20+ B cell clusters identified during the first round of clustering were systematically subclustered at 10 clustering resolutions (0–1 at 0.1 intervals). Manual B cell annotations were also corroborated by automated SingleR version 1.0.6 mapping using two separate murine reference sets (Fig. S1 C). Optimal clustering resolution of 0.5, determined using clustree v.0.4.2 (Zappia and Oshlack, 2018), generated eight B cell subclusters (Fig. S1, E and F). T1B and T2B states (each comprising two distinct subclusters) were identified by querying canonical state-specific markers Vpreb3, Egr1, and Cd24a for T1 and Fcer2a and Ighd for T2. The B1 subcluster was identified by querying S100a6 and Itgb1 (Fig. S1 F). The remaining three subclusters were annotated by their highest expressed signature determined using the FindMarkers function in Seurat. To enable intuitive exploration of B cell profiles, a web portal interface (http://www.biernaskielab.ca/b_cell or http://www.biernaskielab.com/b_cell) was created using the RShiny (version 1.1.0), shinyLP (version 1.1.2), and shinythemes (version 1.1.2) packages.
GRN reconstruction
SCENIC analysis (Aibar et al., 2017) was used to reconstruct transcription factor networks active in distinct B cell states. Analysis was performed using recommended parameters described in SCENIC’s vignette (https://github.com/aertslab/SCENIC) with the mm9 RcisTarget database. To determine essential regulators of B cell states, the regulon specificity score was calculated (by reimplementing pySCENIC’s regulon_specificity_scores function) and plotted as a rank-ordered scatterplot using ggplot2. Kernel density line histograms showing area under the curve score distribution across different B cell states were plotted with ggplot2 version 3.1.1 using the regulon activity matrix (an output of the SCENIC workflow). GRNs of all transcription factors identified can be queried at http://www.biernaskielab.ca/b_cell or http://www.biernaskielab.com/b_cell. scRNAseq data are available in the Gene Expression Omnibus (accession no. GSE156311), which automatically makes a Sequence Read Archive deposit.
Flow cytometry investigation of extravascular and intravascular pulmonary B cells
To label vascular B cells with a specific marker to differentiate vascular versus tissue B cells, mice were first anesthetized, and a right internal jugular i.v. catheter was inserted to administer 2 µg anti-B220 allophycocyanin-Cy7 fluorescent antibodies. The antibody was allowed to circulate for 5 min before euthanasia (Barletta et al., 2012). Lung tissue was then harvested, homogenized, and stained with anti-CD19 fluorochrome-conjugated antibodies.
B cell isolation
Negative selection using a magnet (STEMCELL Technologies) was used to purify B cells from the spleen. The commercially available negative selection kit (EasySep Mouse Pan-B Cell Isolation Kit) includes a cocktail designed for isolating all B cells that contains biotinylated antibodies against non–pan B cells (CD4, CD8, CD11c, CD49b, CD90.2, Gr-1, TER119). After allowing the antibodies to bind and label the target cells, the cell sample was incubated with streptavidin-coated magnetic particles (RapidSpheres). The labeled cells were extracted from the solution by attaching to the magnet while the desired B cells were poured off into a new tube. RBC lysis was performed to remove RBCs from the B cell isolations.
Tissue handling for LXA4 ELISA
Splenic B cells were isolated as described above. Isolated B cells were resuspended in PBS and added to a 96-well plate (5 × 105 cells per well) and incubated at 37°C for 1 h. If required, the B cells were incubated with literature-reported precursors for the synthesis of LXA4 (10 µM LTA4 for 40 min or 1 nM RvD1 for 15 min + 10 µM AA for 40 min; Cayman Chemical). After incubation, the samples were cooled to room temperature because the LXA4 ELISA kit is temperature sensitive. After cooling, the supernatants from the samples were collected and analyzed for the presence of LXA4 using an LXA4 ELISA kit (Neogen). The lungs were processed into single-cell suspensions as described above. However, when passing the lung tissue through the 100-µm filter, instead of adding 10 ml PBS, the lung samples were adjusted to a final concentration of 100 µg/200 µl with PBS. The supernatants of the tissue homogenate samples were collected and analyzed for the presence of LXA4 using an LXA4 ELISA kit.
Zymosan beads and neutrophil clustering
i.v. injections of zymosan-coated fluorescently conjugated beads (Alexa Fluor 488) were used to investigate differences in neutrophil clustering in the lung capillaries. After anesthetizing the mouse with ketamine/xylazine as described above, the jugular vein was cannulated with a catheter, and the mouse was either immediately injected with zymosan-coated fluorescently conjugated beads (107 beads) for the LXA4 ELISA or was injected with zymosan beads after 10 min of intravital lung imaging where the dynamics of unstimulated neutrophil populations were captured. For the LXA4 ELISA, the lung samples were collected and processed 1 h after zymosan bead injection. For intravital lung imaging, neutrophil clustering in the lung capillaries was assessed before zymosan bead injection or 1 h after zymosan bead injection.
Assessing the impact of LXA4 on neutrophil clustering
To assess the impact of LXA4 on neutrophil clustering, 1 µg LXA4 (Cayman Chemical) was injected i.v. in conjunction with the zymosan beads. As previously described, neutrophil clustering was assessed before zymosan bead injection or 1 h after zymosan bead injection.
CXCL13 injections
To assess the impact of exogenous CXCL13 on B cell margination and associated outcomes, the mice received i.t. injections of CXCL13 (BioLegend). After the mouse was anesthetized and placed in a supine position, 10 ng/g CXCL13 was injected i.t. in 50-µl resuspensions of PBS. After 12 or 48 h, the murine lungs were assessed using intravital lung microscopy or flow cytometry.
Fungal strain and pneumonia infection
A fluorescently transgenic A. fumigatus strain, Af293, was previously generated (Campoli et al., 2013). Af293 conidia were grown on yeast-peptone-dextrose agar plates for 7 d at 37°C, at which point the conidia were harvested by gentle washing with PBS + 0.05% (vol/vol) Tween-20. Conidia were counted using a hemocytometer, and CFU count was verified by performing serial dilutions and plating on yeast-peptone-dextrose agar for 24 h. For infections, mice were anesthetized with isoflurane and i.t. infected with 5 × 107 Af293 conidia in 50 µl PBS + 0.05% (vol/vol) Tween-20 (Snarr et al., 2020). LXA4 was injected i.v. (5 µg) at the time of infection.
Quantification and statistical analysis
In vivo image analysis
The following video processing using Leica software was performed before analysis with Volocity. Blur function was used to smooth images on individual frames of the videos, and brightness/contrast was adjusted in a linear fashion. B cell–neutrophil interactions were determined by using the colocalization analysis function in Volocity. The count of interactions was then recorded manually. The B cell and neutrophil transit time and the crawling, tethering, and adhesion behavioral characteristics were manually analyzed. “Crawling” was defined as continuous interaction between a polarized neutrophil or B cell and the vascular wall and does not remain stationary. “Tethering” was defined by discrete instances when B cells or neutrophils arrest their circulatory movement for <30 s. “Adhesion” was defined as a cell that remained static for at least 30 s. “Transit time” was defined as the duration that the cells of interest left the standardized FOV of the pulmonary capillaries, beginning at set time points (time 0 s) and stopping at set endpoints (600 s).
Statistics
The data were represented as mean ± SD values or as box-and-whisker plots showing median and interquartile range. Each dataset was first tested for normality using the Shapiro-Wilks test. Normal data were analyzed using parametric tests, and nonnormal data were analyzed using nonparametric tests. For computing the statistical differences between two groups of normal data, a two-tailed Student’s t test was performed, and the Mann-Whitney test was used for nonparametric data. One-way ANOVA and Tukey’s multiple comparisons post hoc test were used when comparing the statistical differences between more than two groups. Paired analysis was used when assessing changes in B cell and neutrophil behavior over time in the same mouse after administering a blocking antibody (anti-CD49e). The data were analyzed using GraphPad Prism 7.03 for Windows. For all experiments, n is used to distinguish independent experiments. For some imaging experiments, the imaging replicates are shown as pooled data; however, all replicates were averaged in each independent experiment, and statistical testing was conducted according to independent experimental n values and not according to the replicate’s numbers.
Online supplemental material
Fig. S1 shows the quality control metrics and B cell annotations of the scRNAseq analysis. Fig. S2 shows how the B cell subsets in the lung were characterized using flow cytometry to distinguish B1a, B1b, and B2 B cell subsets. Video 1 shows, using intravital confocal lung imaging, B cells marginating in the lung capillaries. Video 2 shows, using intravital confocal lung imaging, that B cells require CD49e for margination in the lung. Video 3 shows, through intravital confocal lung imaging, that IgD+ B cells marginate in the lung vasculature. Video 4 shows, using intravital confocal lung microscopy, that CXCR5 deficiency reduces B cell margination. Video 5 shows, using lung intravital confocal microscopy, that exogenous CXCL13 enhances B cell margination in the lung. Video 6 shows, using intravital confocal lung microscopy, that endogenous CXCL13 maintains B cell margination. Video 7 shows, using intravital confocal lung microscopy, that B cells resolve focal neutrophil inflammation in vivo. Video 8 shows, using intravital confocal lung microscopy, that B cells and LXA4 act to diminish intense neutrophil inflammation.
Acknowledgments
We acknowledge the use of the Live Cell Imaging Facility, funded by the Calvin, Phoebe and Joan Snyder Institute for Chronic Diseases, and we acknowledge K. Poon at the Nicole Perkins Microbial Communities Core Labs flow cytometry core facility.
This work was supported with infrastructure funding provided by the Canada Foundation for Innovation John R. Evans Leaders Fund with matching support from the Alberta Enterprise and Advanced Education Research Capacity Program, an operating grant from the Canadian Institutes of Health Research (RS-342013), the Calgary Firefighters Burn Treatment Society (to J. Biernaskie), an Alberta Children’s Hospital Research Institute fellowship (to E. Labit), a Canadian Institutes of Health Research Vanier scholarship (to S. Sinha), a Killam doctoral scholarship (to S. Sinha), a Canadian Institutes of Health Research Frederick Banting and Charles Best Canada graduate scholarship (to J. Podstawka), and a tier II Canada Research Chair in Pulmonary Immunology, Inflammation and Host Defence (to B.G. Yipp).
Author contributions: Conceptualization: J. Podstawka, J.H. Kim, G. Andonegui, S. Sinha, C.H. Hiroki, N. Sarden, E. Granton, E. Labit, N.L. Rosin, J. Biernaskie, and B.G. Yipp; methodology: J. Podstawka, J.H. Kim, G. Andonegui, S. Sinha, C.H. Hiroki, N. Sarden, E. Granton, E. Labit, N.L. Rosin, Y. Lou, B.D. Snarr, D.C. Sheppard, J. Biernaskie, and B.G. Yipp; investigation: J. Podstawka, J.H. Kim, G. Andonegui, S. Sinha, C.H. Hiroki, N. Sarden, E. Granton, E. Labit, N.L. Rosin, J. Biernaskie, and B.G. Yipp; writing – original draft: J. Podstawka and B.G. Yipp; writing – review and editing: J. Podstawka, J.H. Kim, G. Andonegui, S. Sinha, C.H. Hiroki, N. Sarden, E. Granton, E. Labit, N.L. Rosin, J. Biernaskie, and B.G. Yipp; funding acquisition: B.G. Yipp; resources: J. Biernaskie and B.G. Yipp; supervision: J. Biernaskie and B.G. Yipp.
Data availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by Bryan G. Yipp (bgyipp@ucalgary.ca).
References
- Aibar, S., González-Blas C.B., Moerman T., Huynh-Thu V.A., Imrichova H., Hulselmans G., Rambow F., Marine J.C., Geurts P., Aerts J., et al. 2017. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods. 14:1083–1086. 10.1038/nmeth.4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansel, K.M., Ngo V.N., Hyman P.L., Luther S.A., Förster R., Sedgwick J.D., Browning J.L., Lipp M., and Cyster J.G.. 2000. A chemokine-driven positive feedback loop organizes lymphoid follicles. Nature. 406:309–314. 10.1038/35018581 [DOI] [PubMed] [Google Scholar]
- Ansel, K.M., Harris R.B.S., and Cyster J.G.. 2002. CXCL13 is required for B1 cell homing, natural antibody production, and body cavity immunity. Immunity. 16:67–76. 10.1016/S1074-7613(01)00257-6 [DOI] [PubMed] [Google Scholar]
- Barletta, K.E., Cagnina R.E., Wallace K.L., Ramos S.I., Mehrad B., and Linden J.. 2012. Leukocyte compartments in the mouse lung: distinguishing between marginated, interstitial, and alveolar cells in response to injury. J. Immunol. Methods. 375:100–110. 10.1016/j.jim.2011.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes, B.J., Adrover J.M., Baxter-Stoltzfus A., Borczuk A., Cools-Lartigue J., Crawford J.M., Daßler-Plenker J., Guerci P., Huynh C., Knight J.S., et al. 2020. Targeting potential drivers of COVID-19: Neutrophil extracellular traps. J. Exp. Med. 217:e20200652. 10.1084/jem.20200652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, T.C., Gomes A.C., Cyster J.G., and Pereira J.P.. 2014. CXCR4 and a cell-extrinsic mechanism control immature B lymphocyte egress from bone marrow. J. Exp. Med. 211:2567–2581. 10.1084/jem.20140457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez, A., Weldon A.J., Tatosyan L., Velkuru V., Lee S., Milford T.-A., Francis O.L., Hsu S., Nazeri K., Casiano C.M., et al. 2014. Differences in mouse and human nonmemory B cell pools. J. Immunol. 192:4610–4619. 10.4049/jimmunol.1300692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boothby, M.R., Hodges E., and Thomas J.W.. 2019. Molecular regulation of peripheral B cells and their progeny in immunity. Genes Dev. 33:26–48. 10.1101/gad.320192.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campoli, P., Perlin D.S., Kristof A.S., White T.C., Filler S.G., and Sheppard D.C.. 2013. Pharmacokinetics of posaconazole within epithelial cells and fungi: insights into potential mechanisms of action during treatment and prophylaxis. J. Infect. Dis. 208:1717–1728. 10.1093/infdis/jit358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candando, K.M., Lykken J.M., and Tedder T.F.. 2014. B10 cell regulation of health and disease. Immunol. Rev. 259:259–272. 10.1111/imr.12176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvelli, J., Demaria O., Vély F., Batista L., Chouaki Benmansour N., Fares J., Carpentier S., Thibult M.L., Morel A., Remark R., et al. Explore COVID-19 Marseille Immunopole group . 2020. Association of COVID-19 inflammation with activation of the C5a-C5aR1 axis. Nature. 588:146–150. 10.1038/s41586-020-2600-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang, N., Arita M., and Serhan C.N.. 2005. Anti-inflammatory circuitry: lipoxin, aspirin-triggered lipoxins and their receptor ALX. Prostaglandins Leukot. Essent. Fatty Acids. 73:163–177. 10.1016/j.plefa.2005.05.003 [DOI] [PubMed] [Google Scholar]
- Cinamon, G., Matloubian M., Lesneski M.J., Xu Y., Low C., Lu T., Proia R.L., and Cyster J.G.. 2004. Sphingosine 1-phosphate receptor 1 promotes B cell localization in the splenic marginal zone. Nat. Immunol. 5:713–720. 10.1038/ni1083 [DOI] [PubMed] [Google Scholar]
- Cobaleda, C., Schebesta A., Delogu A., and Busslinger M.. 2007. Pax5: the guardian of B cell identity and function. Nat. Immunol. 8:463–470. 10.1038/ni1454 [DOI] [PubMed] [Google Scholar]
- Colom, B., Bodkin J.V., Beyrau M., Woodfin A., Ody C., Rourke C., Chavakis T., Brohi K., Imhof B.A., and Nourshargh S.. 2015. Leukotriene B4-neutrophil elastase axis drives neutrophil reverse transendothelial cell migration in vivo. Immunity. 42:1075–1086. 10.1016/j.immuni.2015.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone, J.B., Ferrer T.J., Wallace B.H., Wang J., and Hauer-Jensen M.. 2003. Alterations in endothelial thrombomodulin expression in zymosan-induced lung injury. J. Trauma. 54:731–736. 10.1097/01.TA.0000054652.38788.5A [DOI] [PubMed] [Google Scholar]
- Cyster, J.G., and Allen C.D.C.. 2019. B cell responses: cell interaction dynamics and decisions. Cell. 177:524–540. 10.1016/j.cell.2019.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalli, J., and Serhan C.N.. 2012. Specific lipid mediator signatures of human phagocytes: microparticles stimulate macrophage efferocytosis and pro-resolving mediators. Blood. 120:e60–e72. 10.1182/blood-2012-04-423525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton, A.E., Innocentin S., Carr E.J., Bradford B.M., Lafouresse F., Mabbott N.A., Mörbe U., Ludewig B., Groom J.R., Good-Jacobson K.L., and Linterman M.A.. 2019. Type I interferon induces CXCL13 to support ectopic germinal center formation. J. Exp. Med. 216:621–637. 10.1084/jem.20181216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerschuk, C.M., Allard M.F., Martin B.A., MacKenzie A., Autor A.P., and Hogg J.C.. 1987. Marginated pool of neutrophils in rabbit lungs. J. Appl. Physiol. (1985). 63:1806–1815. 10.1152/jappl.1987.63.5.1806 [DOI] [PubMed] [Google Scholar]
- Duffney, P.F., Falsetta M.L., Rackow A.R., Thatcher T.H., Phipps R.P., and Sime P.J.. 2018. Key roles for lipid mediators in the adaptive immune response. J. Clin. Invest. 128:2724–2731. 10.1172/JCI97951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, J.G., Chavez-Rueda K.A., Eddaoudi A., Meyer-Bahlburg A., Rawlings D.J., Ehrenstein M.R., and Mauri C.. 2007. Novel suppressive function of transitional 2 B cells in experimental arthritis. J. Immunol. 178:7868–7878. 10.4049/jimmunol.178.12.7868 [DOI] [PubMed] [Google Scholar]
- Fiore, S., and Serhan C.N.. 1990. Formation of lipoxins and leukotrienes during receptor-mediated interactions of human platelets and recombinant human granulocyte/macrophage colony-stimulating factor-primed neutrophils. J. Exp. Med. 172:1451–1457. 10.1084/jem.172.5.1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francés, R., Tumang J.R., and Rothstein T.L.. 2006. Extreme skewing of annexin II and S100A6 expression identified by proteomic analysis of peritoneal B-1 cells. Int. Immunol. 19:59–65. 10.1093/intimm/dxl122 [DOI] [PubMed] [Google Scholar]
- Fredman, G., Ozcan L., Spolitu S., Hellmann J., Spite M., Backs J., and Tabas I.. 2014. Resolvin D1 limits 5-lipoxygenase nuclear localization and leukotriene B4 synthesis by inhibiting a calcium-activated kinase pathway. Proc. Natl. Acad. Sci. USA. 111:14530–14535. 10.1073/pnas.1410851111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, K.M., Poe J.C., Steeber D.A., and Tedder T.F.. 2005. B-1a and B-1b cells exhibit distinct developmental requirements and have unique functional roles in innate and adaptive immunity to S. pneumoniae. Immunity. 23:7–18. 10.1016/j.immuni.2005.04.011 [DOI] [PubMed] [Google Scholar]
- Han, H., Gong G., Bai X., Lin Y.C., Sun J., Wang W., Zhao Y., Yang L., Wang X., Zhang Z., et al. 2013. Inhibition of notch signaling protects mouse lung against zymosan-induced injury. Shock. 40:312–319. 10.1097/SHK.0b013e3182a102e5 [DOI] [PubMed] [Google Scholar]
- Hobeika, E., Maity P.C., and Jumaa H.. 2016. Control of B cell responsiveness by isotype and structural elements of the antigen receptor. Trends Immunol. 37:310–320. 10.1016/j.it.2016.03.004 [DOI] [PubMed] [Google Scholar]
- Jensen, C.T., Lang S., Somasundaram R., Soneji S., and Sigvardsson M.. 2016. Identification of stage-specific surface markers in early B cell development provides novel tools for identification of progenitor populations. J. Immunol. 197:1937–1944. 10.4049/jimmunol.1600297 [DOI] [PubMed] [Google Scholar]
- Khoder, A., Sarvaria A., Alsuliman A., Chew C., Sekine T., Cooper N., Mielke S., de Lavallade H., Muftuoglu M., Fernandez Curbelo I., et al. 2014. Regulatory B cells are enriched within the IgM memory and transitional subsets in healthy donors but are deficient in chronic GVHD. Blood. 124:2034–2045. 10.1182/blood-2014-04-571125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, N., Ramon S., Thatcher T.H., Woeller C.F., Sime P.J., and Phipps R.P.. 2016. Specialized proresolving mediators (SPMs) inhibit human B-cell IgE production. Eur. J. Immunol. 46:81–91. 10.1002/eji.201545673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.H., Podstawka J., Lou Y., Li L., Lee E.K.S., Divangahi M., Petri B., Jirik F.R., Kelly M.M., and Yipp B.G.. 2018. Aged polymorphonuclear leukocytes cause fibrotic interstitial lung disease in the absence of regulation by B cells. Nat. Immunol. 19:192–201. 10.1038/s41590-017-0030-x [DOI] [PubMed] [Google Scholar]
- Kondratieva, T.K., Rubakova E.I., Linge I.A., Evstifeev V.V., Majorov K.B., and Apt A.S.. 2010. B cells delay neutrophil migration toward the site of stimulus: tardiness critical for effective bacillus Calmette-Guérin vaccination against tuberculosis infection in mice. J. Immunol. 184:1227–1234. 10.4049/jimmunol.0902011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozakiewicz, L., Chen Y., Xu J., Wang Y., Dunussi-Joannopoulos K., Ou Q., Flynn J.L., Porcelli S.A., Jacobs W.R. Jr., and Chan J.. 2013. B cells regulate neutrophilia during Mycobacterium tuberculosis infection and BCG vaccination by modulating the interleukin-17 response. PLoS Pathog. 9:e1003472. 10.1371/journal.ppat.1003472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreslavsky, T., Vilagos B., Tagoh H., Poliakova D.K., Schwickert T.A., Wöhner M., Jaritz M., Weiss S., Taneja R., Rossner M.J., and Busslinger M.. 2017. Essential role for the transcription factor Bhlhe41 in regulating the development, self-renewal and BCR repertoire of B-1a cells. Nat. Immunol. 18:442–455. 10.1038/ni.3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, K., Hutter C., Sun Q., Bilic I., Cobaleda C., Malin S., and Busslinger M.. 2008. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity. 28:751–762. 10.1016/j.immuni.2008.04.014 [DOI] [PubMed] [Google Scholar]
- Laidlaw, B.J., and Cyster J.G.. 2021. Transcriptional regulation of memory B cell differentiation. Nat. Rev. Immunol. 21:209–220. 10.1038/s41577-020-00446-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laidlaw, B.J., Duan L., Xu Y., Vazquez S.E., and Cyster J.G.. 2020. The transcription factor Hhex cooperates with the corepressor Tle3 to promote memory B cell development. Nat. Immunol. 21:1082–1093. 10.1038/s41590-020-0713-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lämmermann, T., Afonso P.V., Angermann B.R., Wang J.M., Kastenmüller W., Parent C.A., and Germain R.N.. 2013. Neutrophil swarms require LTB4 and integrins at sites of cell death in vivo. Nature. 498:371–375. 10.1038/nature12175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBien, T.W., and Tedder T.F.. 2008. B lymphocytes: how they develop and function. Blood. 112:1570–1580. 10.1182/blood-2008-02-078071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, E.K.S., Gillrie M.R., Li L., Arnason J.W., Kim J.H., Babes L., Lou Y., Sanati-Nezhad A., Kyei S.K., Kelly M.M., et al. 2018. Leukotriene B4-mediated neutrophil recruitment causes pulmonary capillaritis during lethal fungal sepsis. Cell Host Microbe. 23:121–133.e4. 10.1016/j.chom.2017.11.009 [DOI] [PubMed] [Google Scholar]
- Li, W., Ai X., Ni Y., Ye Z., and Liang Z.. 2019. The association between the neutrophil-to-lymphocyte ratio and mortality in patients with acute respiratory distress syndrome: a retrospective cohort study. Shock. 51:161–167. 10.1097/SHK.0000000000001136 [DOI] [PubMed] [Google Scholar]
- Lien, D.C., Wagner W.W. Jr., Capen R.L., Haslett C., Hanson W.L., Hofmeister S.E., Henson P.M., and Worthen G.S.. 1987. Physiological neutrophil sequestration in the lung: visual evidence for localization in capillaries. J Appl Physiol (1985). 62:1236–1243. 10.1152/jappl.1987.62.3.1236 [DOI] [PubMed] [Google Scholar]
- Lloyd, C.M., and Marsland B.J.. 2017. Lung homeostasis: influence of age, microbes, and the immune system. Immunity. 46:549–561. 10.1016/j.immuni.2017.04.005 [DOI] [PubMed] [Google Scholar]
- Loder, F., Mutschler B., Ray R.J., Paige C.J., Sideras P., Torres R., Lamers M.C., and Carsetti R.. 1999. B cell development in the spleen takes place in discrete steps and is determined by the quality of B cell receptor-derived signals. J. Exp. Med. 190:75–89. 10.1084/jem.190.1.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looney, M.R., Thornton E.E., Sen D., Lamm W.J., Glenny R.W., and Krummel M.F.. 2011. Stabilized imaging of immune surveillance in the mouse lung. Nat. Methods. 8:91–96. 10.1038/nmeth.1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, T.T., and Cyster J.G.. 2002. Integrin-mediated long-term B cell retention in the splenic marginal zone. Science. 297:409–412. 10.1126/science.1071632 [DOI] [PubMed] [Google Scholar]
- Mabbott, N.A., and Gray D.. 2014. Identification of co-expressed gene signatures in mouse B1, marginal zone and B2 B-cell populations. Immunology. 141:79–95. 10.1111/imm.12171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglione, P.J., Xu J., and Chan J.. 2007. B cells moderate inflammatory progression and enhance bacterial containment upon pulmonary challenge with Mycobacterium tuberculosis. J. Immunol. 178:7222–7234. 10.4049/jimmunol.178.11.7222 [DOI] [PubMed] [Google Scholar]
- Martin, B.A., Wiggs B.R., Lee S., and Hogg J.C.. 1987. Regional differences in neutrophil margination in dog lungs. J. Appl. Physiol. (1985). 63:1253–1261. 10.1152/jappl.1987.63.3.1253 [DOI] [PubMed] [Google Scholar]
- Matthay, M.A., Zemans R.L., Zimmerman G.A., Arabi Y.M., Beitler J.R., Mercat A., Herridge M., Randolph A.G., and Calfee C.S.. 2019. Acute respiratory distress syndrome. Nat. Rev. Dis. Primers. 5:18. 10.1038/s41572-019-0069-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauri, C., and Bosma A.. 2012. Immune regulatory function of B cells. Annu. Rev. Immunol. 30:221–241. 10.1146/annurev-immunol-020711-074934 [DOI] [PubMed] [Google Scholar]
- Nehar-Belaid, D., Hong S., Marches R., Chen G., Bolisetty M., Baisch J., Walters L., Punaro M., Rossi R.J., Chung C.H., et al. 2020. Mapping systemic lupus erythematosus heterogeneity at the single-cell level. Nat. Immunol. 21:1094–1106. 10.1038/s41590-020-0743-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papalexi, E., and Satija R.. 2018. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol. 18:35–45. 10.1038/nri.2017.76 [DOI] [PubMed] [Google Scholar]
- Papayianni, A., Serhan C.N., Phillips M.L., Rennke H.G., and Brady H.R.. 1995. Transcellular biosynthesis of lipoxin A4 during adhesion of platelets and neutrophils in experimental immune complex glomerulonephritis. Kidney Int. 47:1295–1302. 10.1038/ki.1995.184 [DOI] [PubMed] [Google Scholar]
- Qin, C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., and Tian D.S.. 2020. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 71:762–768. 10.1093/cid/ciaa248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qun, S., Wang Y., Chen J., Huang X., Guo H., Lu Z., Wang J., Zheng C., Ma Y., Zhu Y., et al. 2020. Neutrophil-to-lymphocyte ratios are closely associated with the severity and course of non-mild COVID-19. Front. Immunol. 11:2160. 10.3389/fimmu.2020.02160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radermecker, C., Detrembleur N., Guiot J., Cavalier E., Henket M., d’Emal C., Vanwinge C., Cataldo D., Oury C., Delvenne P., and Marichal T.. 2020. Neutrophil extracellular traps infiltrate the lung airway, interstitial, and vascular compartments in severe COVID-19. J. Exp. Med. 217:e20201012. 10.1084/jem.20201012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon, S., Gao F., Serhan C.N., and Phipps R.P.. 2012. Specialized proresolving mediators enhance human B cell differentiation to antibody-secreting cells. J. Immunol. 189:1036–1042. 10.4049/jimmunol.1103483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon, S., Bancos S., Serhan C.N., and Phipps R.P.. 2014. Lipoxin A4 modulates adaptive immunity by decreasing memory B-cell responses via an ALX/FPR2-dependent mechanism. Eur. J. Immunol. 44:357–369. 10.1002/eji.201343316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch, P.J., Chudnovskiy A., Robbins C.S., Weber G.F., Etzrodt M., Hilgendorf I., Tiglao E., Figueiredo J.L., Iwamoto Y., Theurl I., et al. 2012. Innate response activator B cells protect against microbial sepsis. Science. 335:597–601. 10.1126/science.1215173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reátegui, E., Jalali F., Khankhel A.H., Wong E., Cho H., Lee J., Serhan C.N., Dalli J., Elliott H., and Irimia D.. 2017. Microscale arrays for the profiling of start and stop signals coordinating human-neutrophil swarming. Nat. Biomed. Eng. 1:0094. 10.1038/s41551-017-0094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosser, E.C., and Mauri C.. 2015. Regulatory B cells: origin, phenotype, and function. Immunity. 42:607–612. 10.1016/j.immuni.2015.04.005 [DOI] [PubMed] [Google Scholar]
- Rosser, E.C., Oleinika K., Tonon S., Doyle R., Bosma A., Carter N.A., Harris K.A., Jones S.A., Klein N., and Mauri C.. 2014. Regulatory B cells are induced by gut microbiota-driven interleukin-1β and interleukin-6 production. Nat. Med. 20:1334–1339. 10.1038/nm.3680 [DOI] [PubMed] [Google Scholar]
- Samuelsson, B., Dahlén S.E., Lindgren J.A., Rouzer C.A., and Serhan C.N.. 1987. Leukotrienes and lipoxins: structures, biosynthesis, and biological effects. Science. 237:1171–1176. 10.1126/science.2820055 [DOI] [PubMed] [Google Scholar]
- Scharer, C.D., Patterson D.G., Mi T., Price M.J., Hicks S.L., and Boss J.M.. 2020. Antibody-secreting cell destiny emerges during the initial stages of B-cell activation. Nat. Commun. 11:3989. 10.1038/s41467-020-17798-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwickert, T.A., Tagoh H., Gültekin S., Dakic A., Axelsson E., Minnich M., Ebert A., Werner B., Roth M., Cimmino L., et al. 2014. Stage-specific control of early B cell development by the transcription factor Ikaros. Nat. Immunol. 15:283–293. 10.1038/ni.2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smelt, S.C., Cotterell S.E., Engwerda C.R., and Kaye P.M.. 2000. B cell-deficient mice are highly resistant to Leishmania donovani infection, but develop neutrophil-mediated tissue pathology. J. Immunol. 164:3681–3688. 10.4049/jimmunol.164.7.3681 [DOI] [PubMed] [Google Scholar]
- Snarr, B.D., St-Pierre G., Ralph B., Lehoux M., Sato Y., Rancourt A., Takazono T., Baistrocchi S.R., Corsini R., Cheng M.P., et al. 2020. Galectin-3 enhances neutrophil motility and extravasation into the airways during Aspergillus fumigatus infection. PLoS Pathog. 16:e1008741. 10.1371/journal.ppat.1008741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M. III, Hao Y., Stoeckius M., Smibert P., and Satija R.. 2019. Comprehensive integration of single-cell data. Cell. 177:1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, T.T., and Rawlings D.J.. 2002. Transitional B lymphocyte subsets operate as distinct checkpoints in murine splenic B cell development. J. Immunol. 168:2101–2110. 10.4049/jimmunol.168.5.2101 [DOI] [PubMed] [Google Scholar]
- Su, T.T., Guo B., Wei B., Braun J., and Rawlings D.J.. 2004. Signaling in transitional type 2 B cells is critical for peripheral B-cell development. Immunol. Rev. 197:161–178. 10.1111/j.0105-2896.2004.0102.x [DOI] [PubMed] [Google Scholar]
- Tas, J.M., Mesin L., Pasqual G., Targ S., Jacobsen J.T., Mano Y.M., Chen C.S., Weill J.C., Reynaud C.A., Browne E.P., et al. 2016. Visualizing antibody affinity maturation in germinal centers. Science. 351:1048–1054. 10.1126/science.aad3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teague, B.N., Pan Y., Mudd P.A., Nakken B., Zhang Q., Szodoray P., Kim-Howard X., Wilson P.C., and Farris A.D.. 2007. Cutting edge: Transitional T3 B cells do not give rise to mature B cells, have undergone selection, and are reduced in murine lupus. J. Immunol. 178:7511–7515. 10.4049/jimmunol.178.12.7511 [DOI] [PubMed] [Google Scholar]
- Uhl, B., Vadlau Y., Zuchtriegel G., Nekolla K., Sharaf K., Gaertner F., Massberg S., Krombach F., and Reichel C.A.. 2016. Aged neutrophils contribute to the first line of defense in the acute inflammatory response. Blood. 128:2327–2337. 10.1182/blood-2016-05-718999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora, G.D., and Nussenzweig M.C.. 2012. Germinal centers. Annu. Rev. Immunol. 30:429–457. 10.1146/annurev-immunol-020711-075032 [DOI] [PubMed] [Google Scholar]
- Williams, A.E., and Chambers R.C.. 2014. The mercurial nature of neutrophils: still an enigma in ARDS? Am. J. Physiol. Lung Cell. Mol. Physiol. 306:L217–L230. 10.1152/ajplung.00311.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis, S.N., Tellier J., Liao Y., Trezise S., Light A., O’Donnell K., Garrett-Sinha L.A., Shi W., Tarlinton D.M., and Nutt S.L.. 2017. Environmental sensing by mature B cells is controlled by the transcription factors PU.1 and SpiB. Nat. Commun. 8:1426. 10.1038/s41467-017-01605-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodfin, A., Voisin M.-B., Beyrau M., Colom B., Caille D., Diapouli F.-M., Nash G.B., Chavakis T., Albelda S.M., Rainger G.E., et al. 2011. The junctional adhesion molecule JAM-C regulates polarized transendothelial migration of neutrophils in vivo. Nat. Immunol. 12:761–769. 10.1038/ni.2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, D., Zeng Y., Fan Y., Wu J., Mulatibieke T., Ni J., Yu G., Wan R., Wang X., and Hu G.. 2016. Reverse-migrated neutrophils regulated by JAM-C are involved in acute pancreatitis-associated lung injury. Sci. Rep. 6:20545. 10.1038/srep20545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yipp, B.G., Kim J.H., Lima R., Zbytnuik L.D., Petri B., Swanlund N., Ho M., Szeto V.G., Tak T., Koenderman L., et al. 2017. The lung is a host defense niche for immediate neutrophil-mediated vascular protection. Sci. Immunol. 2:eaam8929. 10.1126/sciimmunol.aam8929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappia, L., and Oshlack A.. 2018. Clustering trees: a visualization for evaluating clusterings at multiple resolutions. Gigascience. 7:giy083. 10.1093/gigascience/giy083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D., Chen G., Manwani D., Mortha A., Xu C., Faith J.J., Burk R.D., Kunisaki Y., Jang J.E., Scheiermann C., et al. 2015. Neutrophil ageing is regulated by the microbiome. Nature. 525:528–532. 10.1038/nature15367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, G.X.Y., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J., et al. 2017. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8:14049. 10.1038/ncomms14049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, Y., Yalavarthi S., Shi H., Gockman K., Zuo M., Madison J.A., Blair C., Weber A., Barnes B.J., Egeblad M., et al. 2020. Neutrophil extracellular traps in COVID-19. JCI Insight. 5:e138999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Further information and requests for resources and reagents should be directed to and will be fulfilled by Bryan G. Yipp (bgyipp@ucalgary.ca).