Abstract
Background
Type I interferon signature is one of the most important features of systemic lupus erythematosus (SLE), which indicates an active immune response to antigen invasion. Characteristics of type I interferon-stimulated genes (ISGs) in SLE patients have not been well described thus far.
Methods
We analyzed 35,842 cells of PBMC single-cell RNA sequencing data of five SLE patients and three healthy controls. Thereafter, 178 type I ISGs among DEGs of all cell clusters were screened based on the Interferome Database and AUCell package was used for ISGs activity calculation. To determine whether common ISG features exist in PBMCs and kidneys of patients with SLE, we analyzed kidney transcriptomic data from patients with lupus nephritis (LN) from the GEO database. MRL/lpr mice model were used to verify our findings.
Findings
We found that monocytes, B cells, dendritic cells, and granulocytes were significantly increased in SLE patients, while subsets of T cells were significantly decreased. Neutrophils and low-density granulocytes (LDGs) exhibited the highest ISG activity. GO and pathway enrichment analyses showed that DEGs focused on leukocyte activation, cell secretion, and pathogen infection. Thirty-one common ISGs were found expressed in both PBMCs and kidneys; these ISGs were also most active in neutrophils and LDGs. Transcription factors including PLSCR1, TCF4, IRF9 and STAT1 were found to be associated to ISGs expression. Consistently, we found granulocyte infiltration in the kidneys of MRL/lpr mice. Granulocyte inhibitor Avacopan reduced granulocyte infiltration and reversed renal conditions in MRL/lpr mice.
Interpretation
This study shows for the first time, the use of the AUCell method to describe ISG activity of granulocytes in SLE patients. Moreover, Avacopan may serve as a granulocyte inhibitor for treatment of lupus patients in the future.
Keywords: Single-cell RNA sequencing, Interferon, Granulocytes, Systemic lupus erythematosus, Lupus nephritis
Research in context.
Evidence before this study
The etiology of systemic lupus erythematosus (SLE) is related to comprehensive factors, including internal and external causes. Ethnicity, gene susceptibility, and environmental predisposing factors are highly relevant to the incidence of SLE. Pathogens or autoantigens can induce an aberrant immune response in susceptible SLE patients. Moreover, type I interferon response is a key contributor to effective antiviral responses, which have been found to be highly active in SLE patients. Interferon-stimulated genes (ISGs) are a gene set whose expression is regulated by interferon. However, the characteristics of type I ISGs in SLE and LN have not been clearly described.
Added value of this study
Our study described granulocyte ISG activity in SLE patients and found that transcription factors like PLSCR1, TCF4, IRF9 and STAT1 were associated to ISGs expression. Furthermore, Avacopan may serve as a granulocyte inhibitor for treatment of lupus patients in future.
Implications of all the available evidence
Granulocytes display highly active ISG expression in both peripheral blood mononuclear cells and in the kidneys of lupus patients. Drugs that target granulocytes, such as Avacopan, may reverse lupus-associated kidney damage. Regulatory pathways involving granulocytes or ISGs may be considered as potential targets for lupus treatment.
Alt-text: Unlabelled box
1. Introduction
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease that can affect many organs, including the kidney, which often indicates poor outcome of disease progression [1,2]. Lupus nephritis (LN) is a major risk factor for morbidity and mortality in SLE, and 10%–20% of patients will progress to end-stage renal disease (ESRD) within 5 years of diagnosis [3,4]. The etiology of SLE is related to comprehensive factors, including internal and external causes. Ethnicities, gene susceptibility, and environmental predisposing factors are highly relevant to SLE incidence [5]. Pathogens are the most common inducing factor in the environment, which could induce an aberrant immune response in SLE susceptible patients, and type I interferon response is a key contributor to effective antiviral responses, which have been found to be highly active in SLE patients [[6], [7], [8]].
Type I interferons can be produced by multiple cell types, such as monocytes, neutrophils, plasmacytoid dendritic cells, macrophages, and epithelial cells, when pathogens invade the human body [9]. Interferon-stimulated genes (ISGs) are a gene set whose expression is regulated by interferon. ISGs can be divided based on the activating class of interferon, namely type I, type II, or type III [10,11]. Specifically, type I interferons are antiviral cytokines that trigger ISGs that combat viral infections and are also involved in bacterial infections [12]. However, the characteristics of type I ISGs in SLE and LN have not been clearly described.
In this study, single-cell RNA sequencing data of peripheral blood mononuclear cells (PBMCs) from patients with SLE and microarray transcriptomics profile data from the kidneys of LN patients were used to investigate the expression characteristics of type I ISGs and the potential regulatory mechanism of LN. Furthermore, MRL/lpr and C57BL/6 mice were used for validation of the results.
2. Methods
2.1. Ethics
This study was approved by the Ethics Committee of the Chinese People's Liberation Army General Hospital (no.S2019-095-01). Informed consent was obtained from all participants. For animal experiments, the experimental protocol was carried out in accordance with the approved guidelines of the Institutional Animal Care and Use Committee at the Chinese PLA General Hospital.
2.2. Blood preparation for scRNA-seq
Fresh peripheral blood was obtained from healthy volunteers and patients who were newly diagnosed with SLE, prior to using any immunosuppressive drugs. The Human Leucocyte Separation Kit (P8670, Solarbio, Beijing) was used for leucocyte separation. After obtaining cell pellets, red blood cells were removed using red blood cell lysis buffer (R1010, Solarbio, Beijing). A dual fluorescence cell counter (LUNA-FLTM, Logos Biosystems, South Korea) was used for cell count and viability test. Single-cell transcriptomic amplification and library preparation were performed using single Cell 3′ v3 chemistry (10X Genomics) according to the manufacturer's instructions. Approximately 8 000 cells were captured for each sample following the standard 10X capture, and then sequenced using a NovaSeq 6 000 sequencing system (PE150, Illumina).
2.3. Single-cell raw data quality control
The sequencing data from 10 x Genomics were aligned and quantified using the CellRanger software package (version 3.1) against the human reference genome (hg19). R (version 4.0.2) and the Seurat R package (version 3.2.1) were applied for further analyses [13]. The MergeSeurat function was used to merge multiple datasets. According to the median number of genes and the percentage of mitochondrial genes in the kidney samples, cells with <200 and >2,500 genes, and a mitochondrial gene percentage of >10% were filtered (Supplemental Fig. S1A). Gene expression matrices were normalized to the total cellular unique molecular identifiers (UMI) count. The normalized expression was scaled by regressing the total cellular UMI counts. After data normalization, all highly variable genes in single cells were identified after controlling for the relationship between average expression and dispersion. Subsequently, we used principal component analysis (PCA) with variable genes as the input and identified significant principal components (PCs) based on the jackStraw function. Twenty PCs were selected as the input for uniform manifold approximation and projection (UMAP).
2.4. Removing multiplets
During single cell capture, two or more cells could be captured within the same microfluidic droplet and tagged with the same barcode. Such artifactual multiplets can confound downstream analyses. We applied the DoubletFinder R package to remove these multiplets [14]. After multiplet removal, 35,842 cells were retained for further analyses (Supplemental Fig. S1B).
2.5. Batch effect correction and cell type identification
Since these data were obtained from different samples, R package “Harmony” was used for batch correction (version 1.0) [15] (Supplemental Fig. S1C) in order to avoid the batch effect disrupting downstream analysis. At a resolution of 0.4, cells were clustered by the FindClusters function and classified into 20 different cell types. Next, we used the FindAllMarkers function to find differentially expressed genes of each cell type. Cluster identification was performed based on the top differentially expressed genes (DEGs) of each cluster, and each gene was manually checked on the CellMarker database to match cell types.
2.6. ISGs score
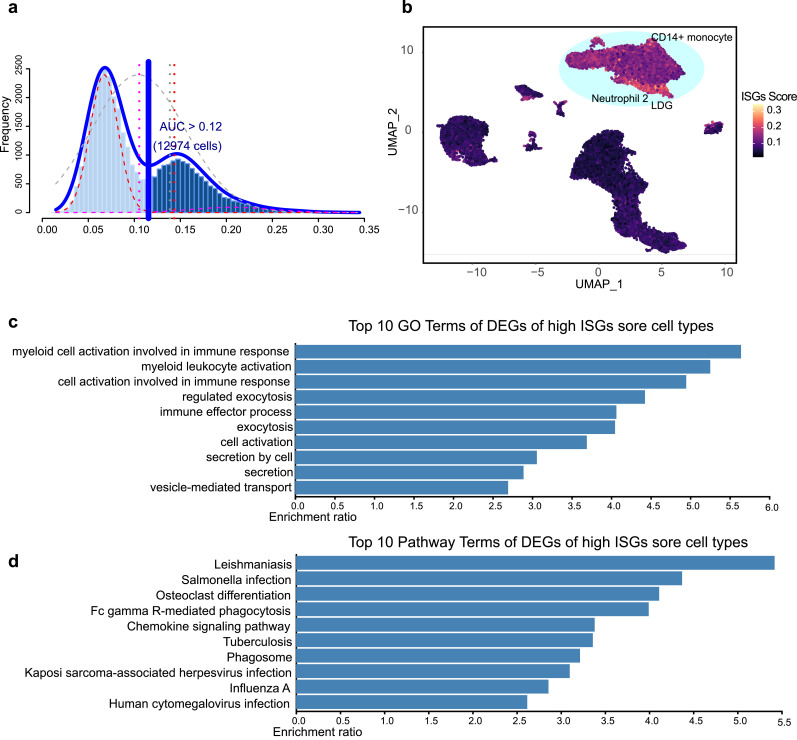
Differentially expressed genes (logFC > 0.25) of each cluster were set as input to generate type I interferon-related ISGs based on the Interferome database, and 178 ISGs gene sets were obtained for ISGs scoring using the AUCell R package (Version 1.12.0) [16]. These 178 ISG sets were set as input data for area under the curve (AUC) value calculation. According to the AUC value, gene-expression rankings were built for each cell. The AUC estimates the proportion of genes in the gene set that are highly expressed in each cell. Cells expressing many genes from the gene set will have higher AUC values than cells expressing fewer genes. Function “AUCell_exploreThresholds” was used to calculate the threshold that could be used to consider the current gene-set active (Fig. 2a). Then, cell clustering UMAP embedding was colored based on the AUC score of each cell to show which cell clusters were active in the ISG gene set (Fig. 2b).
Fig. 2.
ISG score of SLE PBMC cell clusters. (a) Score of 178 screened ISG sets. The threshold was chosen as 0.12 and the ISG score of 12 974 cells exceeded the threshold value. (b) UMAP plots based on the ISG score of each cell. High ISG score cell clusters are highlighted. (c and d) GO and pathway enrichment analysis of DEGs in high ISG score cell clusters.
2.7. Transcriptomic data obtained from the GEO database
Raw data from the expression profiles of GSE113342, GSE104954/104948, GSE99325/99340, and GSE32592/32591 were obtained from the GEO database. Thereafter, differentially expressed genes (DEGs) were calculated using the Biobase, GEOquery, and limma R package. The DEGs of the dataset with an absolute log2 fold change (FC) > 1 and an adjusted P value of <0.05 were considered for subsequent analyses.
2.8. Animals
Female C57BL/6 mice were purchased from Vital River Laboratory (Beijing, China) and MRL/lpr mice from the Jackson Laboratory (Bar Harbor, ME, USA). Animals were housed in cages in a room with constant temperature on a 12 h day/night cycle with access to drinking water and food ad libitum.
2.9. Administration of Avacopan to mice
The Avacopan solution (HY-17627, MedCheExpress, Monmouth Junction, NJ, USA) was dissolved in a vehicle solution consisting of PEG-400/solutol-HS-15 (42966, Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer's instructions. C57BL/6 mice were divided into normal and vehicle control groups. MRL/lpr mice were divided into the disease group and Avacopan group. A minimum of 3 mice were allocated to each group, and 40 mice in total were used in this study. At the beginning of observation, all mice were 12 weeks old. After fasting for one night, Avacopan (30 mg/kg) or vehicle (PEG-400/solutol-HS-15) were administered to mice every morning by gavage for two weeks. Mice were sacrificed at 14, 15, and 16 weeks of age, and the kidneys, blood, and urine of mice were collected for further study.
2.10. Measurement of mouse urinary protein and creatinine
Urine albumin was determined using a commercial enzyme-linked immunosorbent assay kit (C035-2-1, Jiancheng, Nanjing, China). Urine creatinine levels were measured in the same samples using the Creatinine Colorimetric Assay Kit (C011-2-1, Jiancheng, Nanjing, China) according to the manufacturer's instructions.
2.11. Kidney histology
Renal tissues fixed in 4% paraformaldehyde were embedded in paraffin, and 2 -µm-thick sections were cut for morphological analysis. These sections were stained with hematoxylin and eosin (H&E) and periodic acid-Schiff (PAS) stains. Images were captured using an Olympus BX53 microscope. Cell numbers around microvessels (3 fields per mouse (n=3) were observed) were counted using ImageJ (NIH, Bethesda, MD, USA).
2.12. Immunofluorescence staining
Immunofluorescence staining was conducted on paraffin kidney sections using standard procedures. Briefly, kidney sections were incubated with primary antibodies at 4°C overnight. The primary antibodies used were anti-CD16b (Thermo Fisher Scientific Cat# MA1-7633, RRID:AB_2103889). After washing with PBS, sections were incubated with fluorescein (FITC) or cyanine dye 3 (Cy3)-labeled secondary antibodies at room temperature for 1 h. Slides were mounted with 6-diamidino-2-phenylindole (ab104139, Abcam). Images were acquired using a confocal microscope (Leica-SP8). Positive cell numbers per field (>3 fields were observed) were counted using ImageJ (NIH, Bethesda, MD, USA).
2.13. Western blotting and co-immunoprecipitation
Western blotting was performed on kidney tissue lysates to detect IFNα1, IFNAR2, PLSCR1, and TCF4. Proteins were separated using sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS-PAGE), transferred onto a polyvinylidene fluoride (PVDF) membrane, and subjected to immunoblot analysis. Blotting was performed using antibodies against IFNα1 (LS-C330899, LifeSpan BioSciences, Seattle, WA, USA), IFNAR2 (Abcam Cat# ab56070, RRID:AB_880736), PLSCR1 (ab180518, Abcam), and TCF4 (Abcam Cat# ab185736, RRID:AB_2801300). After rinsing with Tris-buffered saline supplemented with 0.1% Tween 20 (TBST), the membranes were incubated with a horseradish peroxidase (HRP)-conjugated anti-rabbit antibody for 60 min at room temperature. For co-immunoprecipitation, equal amounts of protein from groups were immunoprecipitated with saturating amounts of STAT1 (Abcam Cat# ab2071, RRID:AB_2302805) overnight at 4°C, followed by a 1 h incubation with 50 μL protein A-sepharose beads at 4 °C. The beads were then washed, boiled, and the supernatants were immunoblotted with antibodies against IRF9 (LS-C666494, LifeSpan BioSciences). Specific signals were detected using enhanced chemiluminescence (ECL, Amersham Biosciences, Little Chalfont, UK) on the ChemiDoc™ Touch Imaging System (Bio-Rad, Hercules, CA, USA). Quantification was performed by measuring the intensity of the gels using ImageJ.
2.14. Transmission electron microscopy
Kidneys were cut into tissue blocks (1 mm3) and fixed in 2.5% glutaraldehyde in 0.01 mol/L phosphate buffer at 4°C, followed by 2% osmium tetroxide. They were then dehydrated in a series of graded ethanol solutions. Ethanol was then substituted with propylene oxide, and the tissue was embedded in epoxy resin. Ultrathin sections were double-stained with uranyl acetate and lead and examined under a JEM1200EX transmission electron microscope (JOEL) at 80 kV.
2.15. Statistical analyses
Data are expressed as mean ± standard deviation (SD). The statistical significance of comparisons between two groups was analyzed using the Student's t test. Comparisons between multiple groups were performed using one-way or two-way ANOVA followed by Bonferroni's post hoc test. The Shapiro-Wilk test was used to test normality and Levene's test was used to assess the equality of variances for all measurement data. A minimum of three biological replicates were included for all mice experiments, including for histopathology, immunofluorescence, and western blotting, all of which were performed once; representative experiments are shown. Values of P < 0.05 were considered to indicate statistical significance.
2.16. Sample size estimation
To cover rare cell types in the PBMCs of lupus patients a larger number of cells need to be sequenced. An online web tool, https://satijalab.org/howmanycells/, was used to calculate how many cells we would need to sequence to cover rare cell types in the PBMCs of lupus patients. We set the number of cell type as 20, minimum fraction of rarest cell type as 0.02 and minimum cells per type as 5, and it showed that at least 734 cells in total would have to be sampled to obtain a minimum of 5 cells from each of those cell types (99% confidence level). For animal experiments, we used software G*Power for sample size calculation[17]. We used serum creatinine values of 16-week-old mice from a previous published article to calculate the sample size[18]. Mean serum creatinine of the MRL/lpr group was input as 180 and of the normal group, vehicle group and Avacopan group was input as 70. The results showed that 3 mice of each group would obtain a power of 0.99.
2.17. Data deposition
Data generated during this study have been deposited in the Gene Expression Omnibus (GEO) with the accession code GSE162577.
3. Results
3.1. Single-cell transcriptomic data revealed the complexity of SLE PBMC
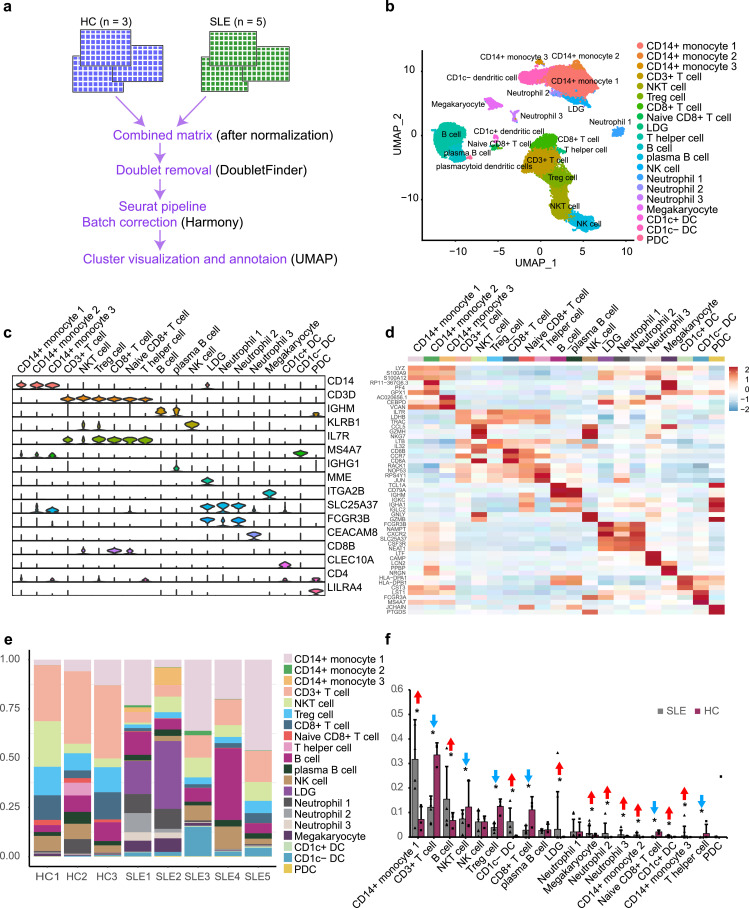
We analyzed the transcriptomic data of 38 748 PBMC cells from three healthy controls (one of which was obtained from our own data, and the other two datasets were obtained from the 10 × Genomics official website) and five SLE patients (two of which were obtained from our own data, and the other three datasets were obtained from the Gene Expression Omnibus (GEO) database (GSE142016)) using 10 × Genomics sequencing (Fig. 1a). After stringent raw data processing and filtration, 35 842 cells were retained for further analyses (Supplementary Fig. 3). After normalization of gene expression and principal component analyses (PCA), we used graph-based clustering to partition the cells into 20 clusters (Fig. 1b). These clusters could be assigned to known cell lineages through marker genes or top 3 differentially expressed genes (Fig. 1c, d). The proportion of each cell lineage varied greatly among different individuals (Fig. 1e, Supplemental Table S1). Clusters, such as monocytes, B cells, dendritic cells, and granulocytes, were significantly increased in SLE patients and subsets of T cells were significantly decreased in SLE patients, as confirmed by individual quantification of the cell composition (Fig. 1f).
Fig. 1.
Single-cell RNA sequencing revealed the complexity of SLE PBMC. (a) Pipeline of single cell RNA sequencing data processing. (b) A UMAP plot representing the 20 clusters across 258 868 PBMCs from eight individuals (five SLE patients and three healthy controls (HC)). (c) Violin plots showing expression of marker genes for 20 distinct cell types. (d) Heatmap showing expression of the top 3 DEGs in each cell type. (e) Bar plots showing the proportion of cell types in each sample. (f) Ration comparison of each cell type in SLE and HC groups. The red arrow represents significantly elevated cell types in the SLE group, while the blue arrow indicates the opposite. Data were analyzed using the Chi-Squared Test. * P < 0.01 (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
3.2. ISG scores of SLE PBMC cell clusters
To investigate the type I ISG expression characteristics of PBMCs from SLE patients, differentially expressed genes (logFC > 0.25) of each cluster were set as the input to generate type I interferon-related ISGs based on the Interferome database, and 178 ISG sets were obtained for further analyses (Supplemental Table S2). In order to calculate ISG activity for each cell, R package AUCell was used for ISG set scoring, which could estimate the proportion of genes in the gene set that are highly expressed in each cell. Cells expressing many genes from the gene set will exhibit higher AUC values than cells expressing fewer genes. We found two peaks in the AUC values of all cells, while 12 974 cells showed relatively higher AUC values when the AUC value threshold was set to 0.12 (Fig. 2a). These cells were mainly in CD14+ monocyte 1, CD14+ monocyte 2, CD14+ monocyte 3, CD1c− dendritic cell, Neutrophil 2, and LDG (Fig. 2b). Gene Ontology and pathway enrichment analyses were carried out to investigate functional characteristics of these cell subsets. We found that the expression features were mainly related to leukocyte activation, cell secretion, and pathogen infection (Fig. 2c, d).
3.3. Differentially expressed genes (DEGs) of LN kidney screening from GEO transcriptomic datasets
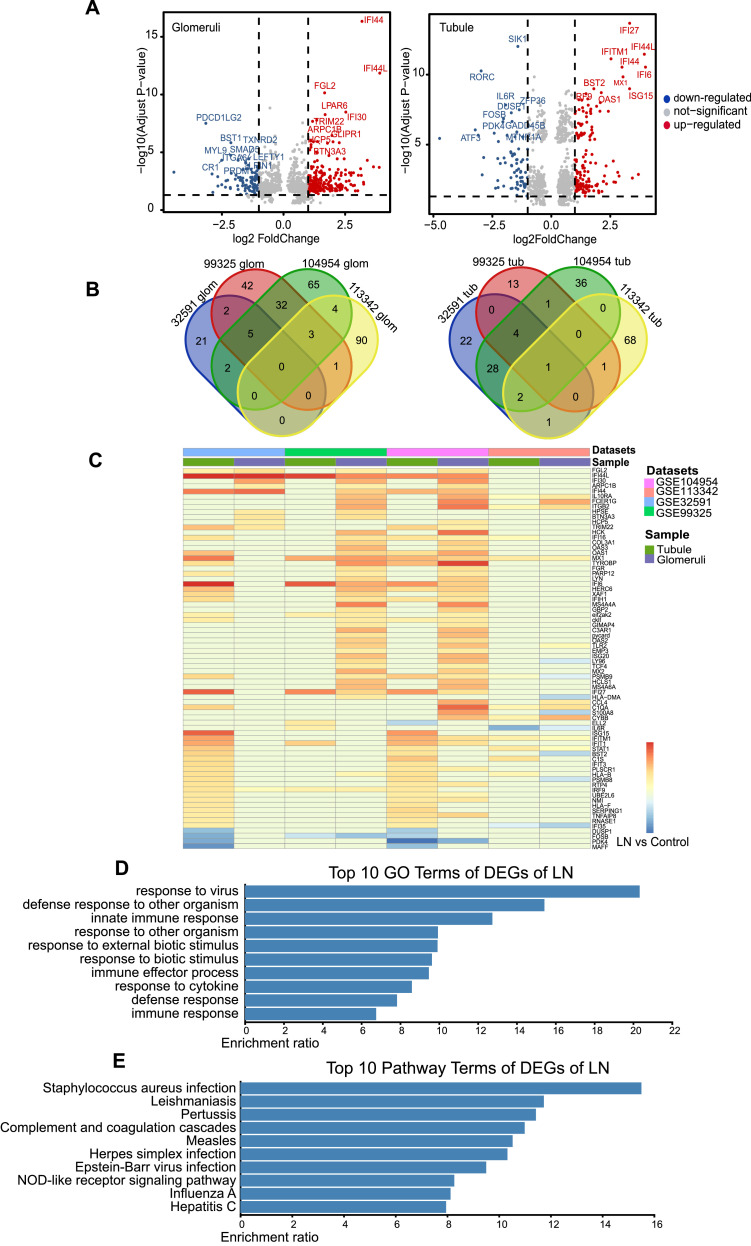
To investigate expression features of kidney tissue in LN, we searched transcriptomics datasets in the GEO database with term “systemic lupus erythematosus” and filter setting “Homo sapiens”, and more than 270 SLE-related datasets were shown. The datasets that met the following criteria were included: 1. There are articles related to the current dataset being published; 2. Data were available for download; 3. Kidneys were used as samples for transcriptome profiling. As a result, four GEO microarray datasets (GSE11342, GSE104954/104948, GSE99325/99340, and GSE32592/32591), in which kidney tissue was isolated from glomeruli and tubules for transcriptome profiling, were finally screened (Supplemental Fig. S2, Supplemental Table S3). DEGs present in at least two GSE datasets with |logFC| > 1 and adjusted P value < 0.05 were considered for further analyses. Thereafter, 49 glomeruli DEGs and 38 tubule DEGs were retained (Fig. 3a, b). Their expression in different GSE datasets is shown in Fig. 3c. Gene Ontology and pathway enrichment analyses were carried out to elucidate the functional characteristics of these DEGs. Consistent with the expression features of PBMCs in SLE, we found that most of these genes were focused on immune response and pathogen infection, such as Staphylococcus aureus, Leishmania parasites, Bordetella pertussis, measles virus, herpes simplex virus, Epstein-Barr virus, influenza virus, and hepatitis virus (Fig. 3d, e).
Fig. 3.
DEGs of LN kidney screening from GEO transcriptomic datasets. (a) Volcano plot showing expression of DEGs (|logFC|>1 and adjusted P value<0.05) in glomeruli (left panel) and tubule (right panel) of LN patient kidneys. (b) DEGs of glomeruli (left panel) and tubule (right panel) expressed in more than two GSE datasets were selected for further analysis. (c) Heatmap showing expression of selected DEGS of glomeruli and tubule in each GSE dataset. (d and e) GO and pathway enrichment analysis of DEGs.
3.4. Common expression characteristics of ISGs and relevant regulatory transcription factors (TFs)
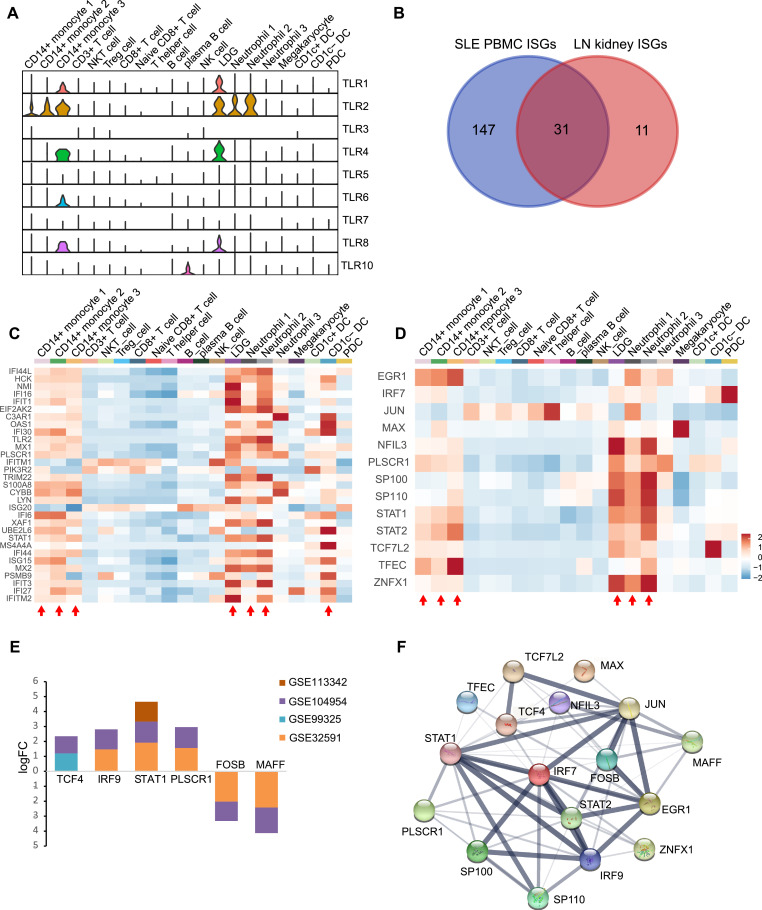
As Gene Ontology and pathway enrichment analyses of SLE PBMC and LN kidney mainly focused on pathogen infection, expression of the Toll-like receptor family, which could recognize molecules that are broadly shared by pathogens, were shown to explore which were involved. In general, TLR1, TLR2, TLR4, and TLR8 showed higher expression mainly in monocytes and granulocytes, and LDG and CD14+ monocyte 3 were the most active cell subsets (Fig. 4a). To explore the common expression characteristics of ISGs between SLE PBMC and LN kidney, a list of ISGs that were expressed in both groups were obtained based on the Interferon database, and 31 ISGs were retained (Fig. 4b, Supplemental Table S1). Expression of these genes was mainly active in CD14+ monocyte 1, CD1C- dendritic cell, LDG, neutrophil 1, neutrophil 2, CD14+ monocyte 2, and CD14+ monocyte 3 (Fig. 4c). To investigate the transcriptionally regulated activity of ISGs, a list of TFs was obtained based on the database AnimalTFDB 3.0; 13 TFs in SLE PBMC and six TFs in LN kidney were retained. These genes were mainly active in CD14+ monocyte 1, LDG, neutrophil 1, neutrophil 2, CD14+ monocyte 2, and CD14+ monocyte 3 (Fig. 4d, e). Furthermore, STAT1 and PLSCR1 were expressed in both SLE PBMC and LN kidney. Key transcription factors and genes may be regulated by them in the protein-protein interaction (PPI) network (Fig. 4f and Supplemental Fig. S3).
Fig. 4.
Combined analysis of ISGs in PBMC and kidney of SLE and LN patients. (a) Violin plots showing TLR family expression in each cell type. (b) Thirty-one common ISGs expressed in PBMC and kidney were selected for further analysis. (c) Heatmap showing expression of 31 common ISGs in each cell type. The red arrow indicates which cell types are active regarding the 31 ISGs. (d) Heatmap showing expression of transcription factors related to ISG regulation in PBMCs from SLE patients. The red arrow indicates which cell types are active regarding these transcription factors. (e) Expression of transcription factors related to ISG regulation in kidneys of SLE patients. (f) Protein-Protein Interaction (PPI) network of transcription factors in PBMCs and kidneys of SLE patients (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
3.5. Granulocytes infiltrated the kidney of MRL/lpr mice, while Avacopan may reduce granulocyte infiltration and delay kidney exacerbation
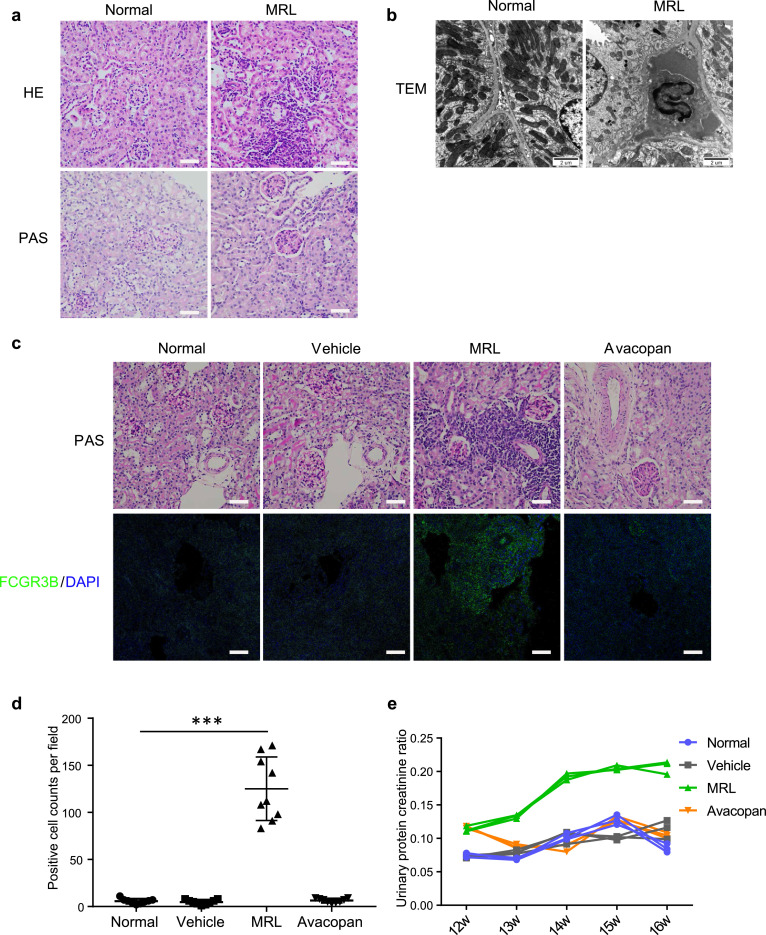
The above results suggest that granulocytes, like LDG and neutrophils, may infiltrate the kidney during lupus nephritis and contribute to exacerbation of the kidney condition. MRL/lpr mice, which are a murine model of spontaneous lupus nephritis, were used for validation. MRL/lpr mice, at 16 weeks of age, were used for observation. Consistently, inflammatory cell infiltration was observed in the kidney, and mesangial cell proliferation was also observed via histopathological staining (Fig. 5a). Further clear nuclear morphometry of granulocytes in the renal interstitium of MRL/lpr mice was observed, compared to the back-to-back tubule structure of normal mice using transmission electron microscopy (Fig. 5b).
Fig. 5.
Avacopan inhibited granulocyte infiltration and alleviated kidney exacerbation in MRL/lpr mice. (a) Representative histopathological staining and TEM images of kidney sections from MRL/lpr mice and C57BL/6 mice at 16 weeks of age. Bar = 50 μm. (b) Representative TEM images of kidney sections from MRL/lpr mice and C57BL/6 mice at 16 weeks of age. Bar = 2 μm. (c and d) Representative histopathological staining and FCGR3B immunofluorescent staining images of kidney sections from MRL/lpr mice and C57BL/6 mice at 16 weeks of age after administration of Avacopan to MRL/lpr mice for two weeks. Cells positively stained with FCGR3B were counted at three fields of different sections per mouse (n=3). Bar = 50 μm. * * * P < 0.001. (e) Urinary protein creatinine ratio was tested and calculated since Avacopan was administered to MRL/lpr mice at 12 weeks of age (n=3).
C5a is a potent granulocyte chemoattractant. Avacopan is a potent selective inhibitor of the C5a receptor, which could block C5a-mediated chemotaxis and neutrophil margination. Here, Avacopan was administered to MRL/lpr mice by gavage for two weeks to observe whether kidney condition could be improved when granulocyte function was inhibited. Granulocyte infiltration decreased significantly in the Avacopan group compared to the MRL group (Fig. 5c, d). Urine protein and creatinine were tested to evaluate kidney function. Kidney function was observed to return to normal upon Avacopan administration, while kidney function increased obviously in the MRL group as mice aged (Fig. 5e).
3.6. ISG regulation related transcription factors expressed in the kidney of MRL/lpr mice
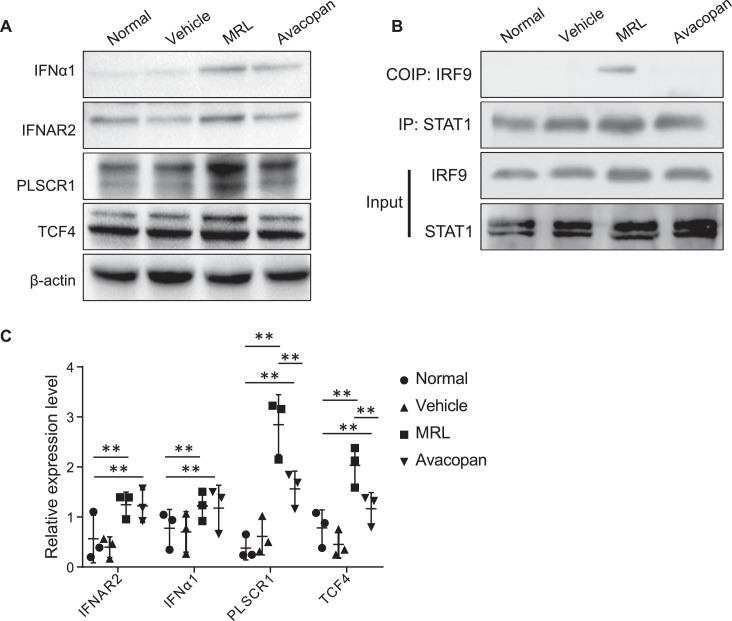
In transcriptomic data obtained from the kidney of LN patients, we found that transcription factors IRF9, STAT1, PLSCR1, and TCF4 were highly expressed and may play an important role in the regulation of ISGs. Thus, we investigated whether these TFs were expressed in MRL/lpr mice. We found that IFNα1 and its receptor IFNAR2 were highly expressed in MRL/lpr mice, and Avacopan significantly inhibited their expression (Fig. 6a, c). Consistent with LN patients, transcription factors IRF9, STAT1, PLSCR1, and TCF4 were also highly expressed in MRL/lpr mice (Fig. 6a–c). Furthermore, IRF9 was found to combine with STAT1 to regulate downstream pathways by co-immunoprecipitation (Fig. 6b).
Fig. 6.
ISG regulation-related transcription factors expressed in kidney of MRL/lpr mice. (a) Western blot results for IFNα1, IFNAR2, PLSCR1, and TCF4 in the kidney at 14 weeks of age after administration of Avacopan for two weeks. (b) Co-immunoprecipitation of IRF9 and STAT1 in the kidney at 14 weeks of age after administration of Avacopan for two weeks. (c) Semi-quantification of IFNα1, IFNAR2, PLSCR1, and TCF4. * * P < 0.01.
4. Discussion
ISG features driven by interferon are one of the most important transcription signatures of patients with SLE. Here, we used single-cell RNA sequencing to resolve ISG features of leucocyte clusters in patients with SLE. Main leucocyte types were obtained, and subpopulations of granulocytes and monocytes were found in SLE patients. Subpopulations of nonspecific immune cell types in SLE may indicate an active immune response to external or internal antigens. Our results agree with a study by Djamel Nehar-Belaid et al., as they also found altered PBMC composition in pediatric patients with SLE by performing single-cell RNA sequencing using PBMCs [19]. After quality control of cells, 35 842 cells were used for analysis and the number of neutrophils was 2 305 cells in total representing only 6.43% of all PBMC cells (Fig. 1e). We considered that this could be caused by two reasons. One is a large consumption of neutrophils in lupus patients induced by infection or autoantigen exposure. The other reason may be related to the technical procedure, as neutrophil loss is a possibility during sample preparation.
Interferons have been found to play a key role in coping with antigens that often appear during the early phase of the immune process to induce amounts of ISGs [10,20]. In particular, type I interferon has been found to be highly active in SLE. Thus, we calculated the ISG score of each cell to evaluate the ISG activity of the cell types (Fig. 2a and b). We found that subpopulations of granulocytes and monocytes, especially neutrophils 2 and LDG, were most active in ISG activity. Consistently, GO and pathway enrichment analyses of DEGs in these cell types mainly focused on leukocyte activation, cell secretion, and pathogen infection. Studies have found that neutrophil IFN signature is associated with a more aggressive course of SLE and the occurrence of lupus nephritis by analyzing the transcriptome of PBMCs from SLE patients [21,22]. Furthermore, LDG, which has been found to be a pathogenic neutrophil subset, is elevated in SLE patients [[23], [24], [25]]. These findings suggest that the IFN immune response of neutrophils and LDG are highly correlated with the severity of SLE disease. Our findings also support the highly active status of neutrophils and LDG in SLE patients. Furthermore, our analyses indicate that external pathogens may be the main factors inducing SLE onset, and neutrophils may protect against pathogens by producing high levels of ISGs.
Further, we observed the expression of ISGs by analyzing transcription profiles of kidneys obtained from LN patients. Compared with the list of ISGs between PBMCs and kidneys, we obtained 31 ISGs that were expressed in both PBMCs and kidney samples (Fig. 4b). Unsurprisingly, these 31 ISGs were highly expressed in granulocyte-like neutrophil 2 and LDG (Fig. 4c). These findings may indicate that granulocytes infiltrate the kidney and initiate a resident immune response. Several studies have found that neutrophils can infiltrate the kidney of active lupus nephritis and cause resident inflammatory responses, primarily through the formation of neutrophil extracellular traps (NETS) [[26], [27], [28]]. Applying single-cell RNA sequencing to the kidneys of patients with LN, Dong et al. found that T and B cells infiltrated the kidney of LN patients exhibiting high ISG activity, but did not mention granulocytes [29]. This may be due to granulocyte loss during sample preparation. We also found that classical interferon regulatory transcription factors, such as IRF7, IRF9, EGR1, STAT1, and STAT2, were expressed. In particular, PLSCR1 is expressed in both PBMCs and kidneys, and few studies have reported its expression in lupus nephritis, but it has been found to positively regulate the expression of ISGs [30,31].
We validated the above-mentioned results in the MRL/lpr mouse model. Consistently, we found neutrophil infiltration in the kidneys of MRL/lpr mice. C5aR has been found to be expressed primarily in granulocytes, and Avacopan, as its antagonist, has been used for therapy of ANCA-associated vasculitis, which can reduce granulocyte numbers in blood circulation [32]. Indeed, we found that few granulocytes infiltrated the kidneys of MRL/lpr mice, and renal function was also obviously reversed after administration of Avacopan, compared to the disease control group (Fig. 5). Currently, drugs targeting neutrophils are used clinically or in clinical trials, and these drugs treat diseases, such as rheumatoid arthritis, asthma, and bronchiolitis obliterans syndrome. Transcription factors TCF4, PLSCR1, STAT1, and IRF9, which we found being expressed in kidneys of LN patients, were also expressed in MRL/lpr mice at the protein level. This may imply a potential regulatory mechanism of the interferon-related pathway, which deserves further exploration.
Since SLE is a complex and heterogeneous disease, one limitation of our study is the small sample size of lupus patients. The low number of samples may not cover all possible disease changes caused by individual factors such as genetic or environmental influences. A study with a larger sample size will be carried out in our lab to further validate our findings.
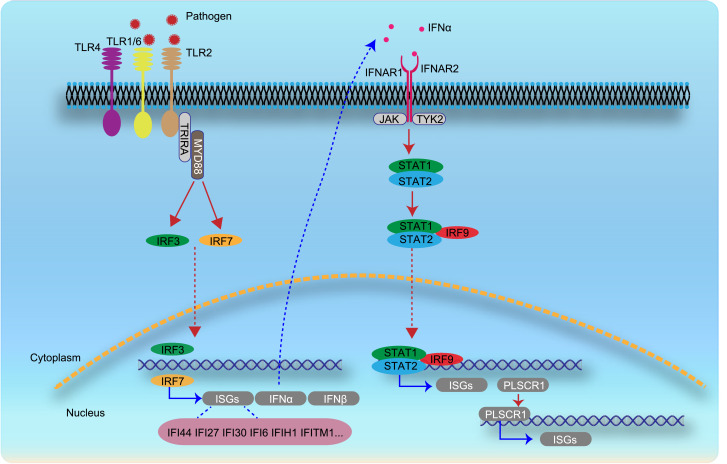
In conclusion, we described ISG expression features of leucocyte clusters and delineated possible ISG regulation pathways in SLE and LN patients. Transcription factors PLSCR1, TCF4, IRF9 and STAT1 may especially play an important role for ISGs expression regulation (Fig. 7). Granulocytes, as the most active cell type in ISGs, may be targeted as early as possible to prevent overwhelming activation of downstream pathways. Moreover, Avacopan, a C5aR antagonist, may serve as a potential clinical treatment option for SLE or lupus nephritis.
Fig. 7.
Hypothesis of possible ISG regulation pathway based on our analyses and findings in SLE and LN patients. Various pathogens could infect human cells through the TLR family, such as TLR1, TLR2, TLR4, and TLR6, and activate interferon regulatory factors, such as IRF3 and IRF7, which are expressed in PBMCs of SLE patients. IRF3 and IRF7 may positively regulate expression of interferon and ISGs. Production of interferon α may combine with interferon receptors and further promote the expression of ISGs and other positive ISG regulation transcription factors, like PLSCR1.
Data sharing statement
The data that support the findings of this study are available from the corresponding author upon request.
Contributors
YD and XC designed the study, developed the methodology, analyzed the data and prepared the manuscript. Y.D. constructed the experiment with assistance from YZ, DL, QH, MZ, and YD performed bioinformatics. YD and XC prepared the manuscript. All of the authors (YD, YZ, DL, QH, MZ, BF, LW, XW, WS, YZ, JC, KS, XL, SS, GC and XC) discussed the results and approved the manuscript.
Declaration of Competing Interest
None.
Acknowledgments
This work was supported by National Natural Science Foundation of China [Grant Nos. 81830019 and 8203000957].
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103477.
Appendix. Supplementary materials
References
- 1.Costenbader K.H., Desai A., Alarcon G.S. Trends in the incidence, demographics, and outcomes of end-stage renal disease due to lupus nephritis in the US from 1995 to 2006. Arthritis Rheum. 2011;63(6):1681–1688. doi: 10.1002/art.30293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dörner T., Furie R. Novel paradigms in systemic lupus erythematosus. Lancet. 2019;393(10188):2344–2358. doi: 10.1016/S0140-6736(19)30546-X. [DOI] [PubMed] [Google Scholar]
- 3.Almaani S., Meara A., Rovin BH. Update on lupus nephritis. Clin J Am Soc Nephrol. 2017;12(5):825–835. doi: 10.2215/CJN.05780616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hahn B.H., McMahon M.A., Wilkinson A. American College of Rheumatology guidelines for screening, treatment, and management of lupus nephritis. Arthritis Care Res. 2012;64(6):797–808. doi: 10.1002/acr.21664. (Hoboken) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaul A., Gordon C., Crow M.K. Systemic lupus erythematosus. Nat Rev Dis Prim. 2016;2:16039. doi: 10.1038/nrdp.2016.39. [DOI] [PubMed] [Google Scholar]
- 6.Rönnblom L., Alm GV. An etiopathogenic role for the type I IFN system in SLE. Trends Immunol. 2001;22(8):427–431. doi: 10.1016/s1471-4906(01)01955-x. [DOI] [PubMed] [Google Scholar]
- 7.Ronnblom L., Alm G.V., Eloranta ML. The type I interferon system in the development of lupus. Semin Immunol. 2011;23(2):113–121. doi: 10.1016/j.smim.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 8.Illescas-Montes R., Corona-Castro C.C., Melguizo-Rodriguez L., Ruiz C., Costela-Ruiz V.J. Infectious processes and systemic lupus erythematosus. Immunology. 2019;158(3):153–160. doi: 10.1111/imm.13103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McNab F., Mayer-Barber K., Sher A., Wack A., O'Garra A. Type I interferons in infectious disease. Nat Rev Immunol. 2015;15(2):87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schneider W.M., Chevillotte M.D., Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schoggins J.W. Interferon-stimulated genes: what do they all do? Annu Rev Virol. 2019;6(1):567–584. doi: 10.1146/annurev-virology-092818-015756. [DOI] [PubMed] [Google Scholar]
- 12.Schoggins J.W. Recent advances in antiviral interferon-stimulated gene biology. F1000Res. 2018;7:309. doi: 10.12688/f1000research.12450.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stuart T., Butler A., Hoffman P. Comprehensive integration of single-cell data. Cell. 2019;177(7) doi: 10.1016/j.cell.2019.05.031. 1888-902 e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGinnis C.S., Murrow L.M., Gartner Z.J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019;8(4) doi: 10.1016/j.cels.2019.03.003. 329-37 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korsunsky I., Millard N., Fan J. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16(12):1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aibar S., Gonzalez-Blas C.B., Moerman T. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14(11):1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nehar-Belaid D., Hong S., Marches R. Mapping systemic lupus erythematosus heterogeneity at the single-cell level. Nat Immunol. 2020;21(9):1094–1106. doi: 10.1038/s41590-020-0743-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Talemi S.R., Hofer T. Antiviral interferon response at single-cell resolution. Immunol Rev. 2018;285(1):72–80. doi: 10.1111/imr.12699. [DOI] [PubMed] [Google Scholar]
- 19.Banchereau R., Hong S., Cantarel B. Personalized immunomonitoring uncovers molecular networks that stratify lupus patients. Cell. 2016;165(6):1548–1550. doi: 10.1016/j.cell.2016.05.057. [DOI] [PubMed] [Google Scholar]
- 20.Jourde-Chiche N., Whalen E., Gondouin B. Modular transcriptional repertoire analyses identify a blood neutrophil signature as a candidate biomarker for lupus nephritis. Rheumatology. 2016;56(3):477–487. doi: 10.1093/rheumatology/kew439. [DOI] [PubMed] [Google Scholar]
- 21.Mistry P., Nakabo S., O’Neil L. Transcriptomic, epigenetic, and functional analyses implicate neutrophil diversity in the pathogenesis of systemic lupus erythematosus. Proc Natl Acad Sci U S A. 2019;116(50):25222–25228. doi: 10.1073/pnas.1908576116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denny M.F., Yalavarthi S., Zhao W. A distinct subset of proinflammatory neutrophils isolated from patients with systemic lupus erythematosus induces vascular damage and synthesizes type I IFNs. J Immunol. 2010;184(6):3284–3297. doi: 10.4049/jimmunol.0902199. (Baltimore, Md: 1950) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rahman S., Sagar D., Hanna R.N. Low-density granulocytes activate T cells and demonstrate a non-suppressive role in systemic lupus erythematosus. Ann Rheum Dis. 2019;78(7):957–966. doi: 10.1136/annrheumdis-2018-214620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Villanueva E., Yalavarthi S., Berthier C.C. Netting neutrophils induce endothelial damage, infiltrate tissues, and expose immunostimulatory molecules in systemic lupus erythematosus. J Immunol. 2011;187(1):538–552. doi: 10.4049/jimmunol.1100450. (Baltimore, Md: 1950) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hakkim A., Fürnrohr B.G., Amann K. Impairment of neutrophil extracellular trap degradation is associated with lupus nephritis. Proc Natl Acad Sci U S A. 2010;107(21):9813–9818. doi: 10.1073/pnas.0909927107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta S., Kaplan M.J. The role of neutrophils and NETosis in autoimmune and renal diseases. Nat Rev Nephrol. 2016;12(7):402–413. doi: 10.1038/nrneph.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arazi A., Rao D.A., Berthier C.C. The immune cell landscape in kidneys of patients with lupus nephritis. Nat Immunol. 2019;20(7):902–914. doi: 10.1038/s41590-019-0398-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metz P., Dazert E., Ruggieri A. Identification of type I and type II interferon-induced effectors controlling hepatitis C virus replication. Hepatology. 2012;56(6):2082–2093. doi: 10.1002/hep.25908. [DOI] [PubMed] [Google Scholar]
- 29.Dong B., Zhou Q., Zhao J. Phospholipid scramblase 1 potentiates the antiviral activity of interferon. J Virol. 2004;78(17):8983–8993. doi: 10.1128/JVI.78.17.8983-8993.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jayne D.R.W., Bruchfeld A.N., Harper L. Randomized trial of C5a receptor inhibitor avacopan in ANCA-associated vasculitis. J Am Soc Nephrol. 2017;28(9):2756–2767. doi: 10.1681/ASN.2016111179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bekker P., Dairaghi D., Seitz L. Characterization of pharmacologic and pharmacokinetic properties of CCX168, a potent and selective orally administered complement 5a receptor inhibitor, based on preclinical evaluation and randomized phase 1 clinical study. PLoS One. 2016;11(10) doi: 10.1371/journal.pone.0164646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nemeth T., Sperandio M., Mocsai A. Neutrophils as emerging therapeutic targets. Nat Rev Drug Discov. 2020;19(4):253–275. doi: 10.1038/s41573-019-0054-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.