Abstract
The Neurofibromatosis type 2 gene encodes the Nf2/merlin tumor suppressor protein that is responsible for the regulation of cell proliferation. Once activated, Nf2/merlin modulates adhesive signaling pathways and thereby inhibits cell growth. Nf2/merlin controls oncogenic gene expression by modulating the Hippo pathway. By responding to several physical and biochemical stimuli, Hippo signaling determines contact inhibition of proliferation as well as organ size. The large tumor suppressor (LATS) serine/threonine-protein kinase is the key enzyme in the highly conserved kinase cascade that negatively regulates the activity and localization of the transcriptional coactivators Yes-associated protein (YAP) and its paralogue transcriptional coactivator with PDZ-binding motif (TAZ). Nf2/merlin belongs to the band 4.1, ezrin, radixin, moesin (FERM) gene family that links the actin cytoskeleton to adherens junctions, remodels adherens junctions during epithelial morphogenesis and maintains organized apical surfaces on the plasma cell membrane. Nf2/merlin and ERM proteins have a globular N-terminal cloverleaf head domain, the FERM domain, that binds to the plasma membrane, a central α-helical domain, and a tail domain that binds to its head domain. Here we present the high-resolution crystal structure of Nf2/merlin bound to LATS1 which shows that LATS1 binding to Nf2/merlin displaces the Nf2/merlin tail domain and causes an allosteric shift in the Nf2/merlin α-helix that extends from its FERM domain. This is consistent with the fact that full-length Nf2/merlin binds LATS1 ~10-fold weaker compared to LATS1 binding to the Nf2/merlin-PIP2 complex. Our data increase our understanding of Nf2/merlin biology by providing mechanistic insights into the Hippo pathway that are relevant to several diseases in particular oncogenic features that are associated with cancers.
Keywords: Actin, Cancer, Cell adhesion, Cell junction, Cell migration, Cell signaling, Inositol phospholipid, Neurofibromatosis type 2, Plasma membrane
Introduction
Neurofibromatosis type 2 (NF2) is a rare genetic, neoplastic disease that is characterized by several slow-growing tumors, especially ependymomas, meningiomas, and schwannomas [1], [2]. Unfortunately, conventional chemotherapy is ineffective as a possible treatment. The protein responsible for NF2 is the tumor suppressor Nf2/merlin, aka neurofibromin 2 or schwannomin. Inactivation of Nf2/merlin results in spontaneously forming malignant mesothelioma or schwannomas [3] and the targeted deletion of Nf2/merlin in mouse Schwann cells results in tumor formation [4], [5]. Nf2/merlin-null cells grow to high density due to their impaired contact inhibition of growth [6].
Nf2/merlin localizes to Schmidt-Lanterman clefts (myelin incisures) and paranodes in myelinating Schwann cells, specialized junctional structures that resemble epithelial adherens and tight junctions [7], [8]. Nf2/merlin is found in liquid-ordered, glycosphingolipid-, and cholesterol-rich membrane microdomains in cultured cells [7], [9], [10]. Such raft localization of Nf2/merlin is mediated by its binding to phosphatidylinositol 4,5-bisphosphate (PIP2) that is necessary for Nf2/merlin-mediated suppression of growth [11], [12]. Indeed, Nf2/merlin functions as a tumor suppressor mainly at the plasma membrane [13], [14], [15], [16], [17], [18].
Nf2/merlin-null cells have activated β-catenin, Rac, Ras, Src, and oncogenic signaling pathways [19], [20], [21], [22], [23], [24]. Nf2/merlin seems to promote contact inhibition by reducing the availability of the cell surface for the epidermal growth factor receptor and the ErbB2 and E-cadherin growth factor receptors [13], [25]. Nf2/merlin also plays a role in intracellular vesicular trafficking (growth factor endo- and exocytosis) [14], [15], [26], [27], [28], [29]. Furthermore, by its binding to angiomotin, Nf2/merlin inhibits Rac1 activity [30]. In the nucleus, Nf2/merlin mediates contact inhibition and suppresses tumorigenesis by inhibiting the cullin4A-RING (CRL4) E3 ubiquitin ligase DDB1 and cul4 associate factor 1 (DCAF1) [31], [32], [33], [34]. Nf2/merlin also activates the growth inhibitory kinase cascade, known as the Hippo pathway, that results in the phosphorylation of the yes-associated protein 1 (YAP1) growth-promoting transcription factor and its degradation [35], [36], [37], [38].
Nf2/merlin also recruits the large tumor suppressor (LATS) serine/threonine-protein kinases to the cell membrane where LATS is activated by the serine/threonine protein kinases called macrophage stimulating 1 (MST1) which drives phosphorylation and yes-associated protein 1 (YAP) and transcription coactivator with PDZ-binding motif (TAZ) [1], [39]. Nf2/merlin binding to LATS1 and LATS2 is facilitated by angiomotin which binds Nf2/merlin and thereby severs the Nf2/merlin auto-inhibitory head–tail interaction [40]. In cells that are inhibited in their contact, unphosphorylated/activated Nf2/merlin translocates to the nucleus where Nf2/merlin inhibits CRL4-DCAF1, which prevents this E3 ligase from ubiquitinating and inhibiting LATS1 and LATS2.
Nf2/merlin belongs to the band 4.1, ezrin, radixin, moesin (FERM) gene family [41], [42], [43], hence the acronym for moesin-ezrin-radixin-like protein, which plays essential roles in linking the actin cytoskeleton to adherens junctions, in remodeling adherens junctions during epithelial morphogenesis, and in maintaining organized apical surfaces on the plasma cell membrane [44]. ERM proteins have a ~300 residue globular N-terminal cloverleaf head domain comprised of 3 subdomains (F1, F2, and F3) [45], a central α-helical domain, and a tail domain that binds to filamentous actin (Fig. 1A). In contrast, Nf2/merlin seems to lack such a C-terminal F-actin binding site [46], [47] and instead seems to bind directly to actin via its FERM domain [44], [48], [49] or indirectly via heterotypic interactions with ERM proteins [42]. The FERM domains also bind to their tail domains as well as to the plasma membrane. The severing of the autoinhibitory head–tail interactions activates ERM proteins to bind to binding proteins with much higher affinity compared to their closed conformers [11], [40], [50], [51].
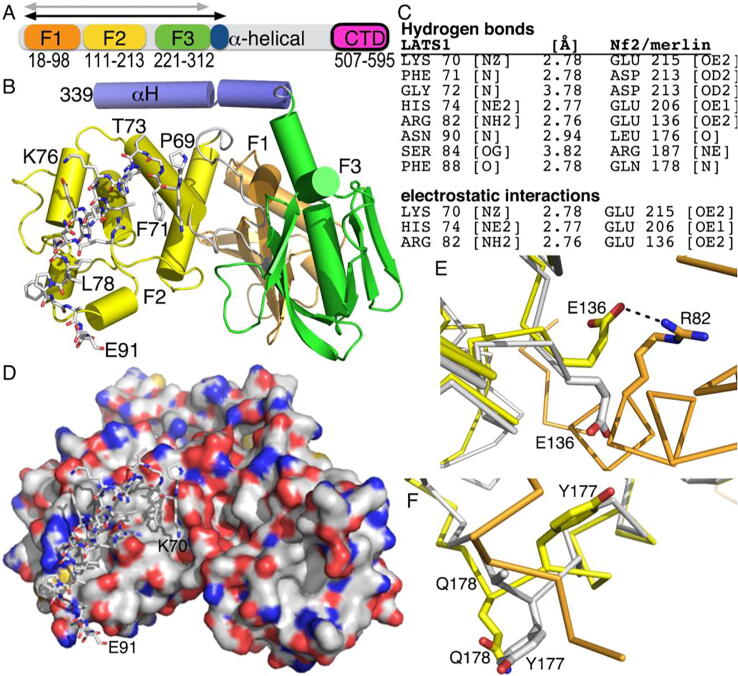
Fig. 1.
The crystal structure of Nf2/merlin bound to LATS1. A. Schematic of the Nf2/merlin domain structure. The N-terminus harbors the three FERM subdomains (F1, orange, residues 18–98; F2, yellow, residues 111–213; F3, green, residues 221–312) followed by the α-helical domain that harbors one α-helix that extends upon activation (blue, residues 315–338). The C-terminal domain (CTD, residues 507–595) of isoform 1 is shown in magenta (isoform 2 is truncated by five amino acids, Δ590-595, with residues 580- PQAQGRRPICI-590 unique to Nf2/merlin isoform 2) [78]. The gray double arrow indicates the truncated Nf2/merlin structures, and the black double arrow indicates the construct used here. B. Cartoon of the Nf2/merlin FERM domain (same color coding as in panel A). The Nf2/merlin first α-helix of the central helical domain (αH) (blue, as in panel A) in complex with LATS1 residues 69–91 are shown in stick representation. Some LATS1 residues and Nf2/merlin FERM subdomains are labeled as well as the last (339) Nf2/merlin residue. C. Residues contributing to the Nf2/merlin interface with LATS1. D. Surface representation of Nf2/merlin (red, oxygen; blue, nitrogen; white, carbon; yellow, sulfur) with LATS1 shown in stick representation. N- (K70) and C-terminal (E91) residues are indicated. E. Superposition of the apo (gray) and LATS1-bound (orange) Nf2/merlin (F2, yellow) structures shows flipping of Glu-136 upon LATS1 binding to engage in electrostatic interaction with LATS1 residue Arg-82. F. Superposition of the apo (gray) and LATS1-bound (orange) Nf2/merlin (F2, yellow) structures shows re-organization of the Nf2/merlin loop upon LATS1 binding to avoid steric clashes and widening of the grove to accommodate LATS1. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Nf2/merlin tail truncations in Drosophila resulted in a constitutively active protein [52]. This result suggested that, like ERM proteins, the open form of Nf2/merlin is active in vivo [53]. In mammals, alternative splicing of NF2 gives rise to an Nf2/merlin isoform with a unique C-terminus, known as isoform 2. The first 579 residues are identical for the two isoforms, and the last sixteen residues for isoform 1 (residues 580–595) are LTLQSAKSRVAFFEEL while the last eleven residues (580–590) for the shorter isoform 2 are PQAQGRRPICI. Isoform 2 exists in a constitutively open conformation [54], [55] and might therefore differ functionally from isoform 1. However, both isoforms are fully functional as tumor suppressors and compensate for the loss of the other isoform during development and in most adult organs [56].
Structural data on Nf2/merlin are sparse. This knowledge gap adds to the several contradictions in the literature about the molecular mechanisms of the tumor-suppressive functions of Nf2/merlin which is therefore poorly understood. The known Nf2/merlin structures center on its minimal FERM domain alone (Nf2/merlin residues 20–311, PDB entries 1h4r [57] and 3u8z [58]) or in complex with its binding partners, such as with its mutant (S518D, A585W) tail (Nf2/merlin residues 507–595; PDB entry 4zrj) [40], DCAF (PDB entry 4p7i) [reference is PMCID = PMC4031523], DCAF1/VprBP (residues 1478–1506; PDB entry 3wa0) [59], LATS1 (residues 69–89; PDB entry 4zrk) [40], and LATS2 (residues 71–89; PDB entry 4zri) [40]. The recent head–tail domain structure from the Drosophila melanogaster counterpart of Nf2/merlin (PDB entry 7edr) [60] revealed an additional head–tail interface compared to the mammalian mutant tail interaction with its FERM domain. The extended Nf2/merlin FERM domain structure, that includes the first α-helix of the central helical domain (αH) (residues 15–339; PDB entry 1isn) [61] or bound to lipids (phosphatidylinositol 4,5-bisphosphate; PDB entry 6cds) [62] showed that lipid binding causes α-helix αH to undergo a large movement to extend the last α-helix of the FERM domain, thereby severing the head–tail interaction. We resolved an almost two-decade-long debate by showing that conformational changes in Nf2/merlin upon binding to the plasma membrane are necessary for Nf2/merlin to inhibit cell proliferation and that the interaction of Nf2/merlin with the membrane is necessary for Nf2/merlin to exert its cell growth-inhibiting functions as well as to inhibit YAP activity [11].
Here we determined the 1.6 Å crystal structure of Nf2/merlin in complex with LATS1 which shows that LATS1 binding to Nf2/merlin displaces the Nf2/merlin C-terminus and causes an allosteric shift in the Nf2/merlin extended α-helix. This is consistent with the fact that the Nf2/merlin tail domain binds the Nf2/merlin head domain 2-fold weaker compared to LATS1 (~3 μM versus ~1.4 μM) [40] and that full-length Nf2/merlin binds LATS1 10-fold weaker compared to LATS1 binding to the Nf2/merlin-PIP2 complex [11]. Collectively, our data suggest a mechanism whereby PIP2 facilitates LATS1 binding. Such insights are directly relevant to our understanding of the Hippo pathway that impacts several diseases in particular cancer.
Results
LATS1 binding to Nf2/merlin
We used over 1000 commercially available crystallization screens, varied the concentrations of Nf2/merlin and LATS1 as well as the crystallization temperature, to attempt to grow LATS1-bound Nf2/merlin crystals but our efforts were unsuccessful. Thus, we resorted to the high-throughput crystallization screening facility at the Hauptman-Woodward Medical Research Institute [63]. Out of the 1536 crystallization conditions that were screened, crystals appeared from 500 mM imidazole (pH 6.5) and 15% (w/v) polyethylene glycol 3350 using the micro batch-under-oil method. We adapted this condition from the micro batch-under-oil method for hanging drop vapor diffusion by varying the imidazole and polyethylene glycol 3350 concentrations to grow crystals that diffracted X-ray beyond 1.6 Å Bragg spacings.
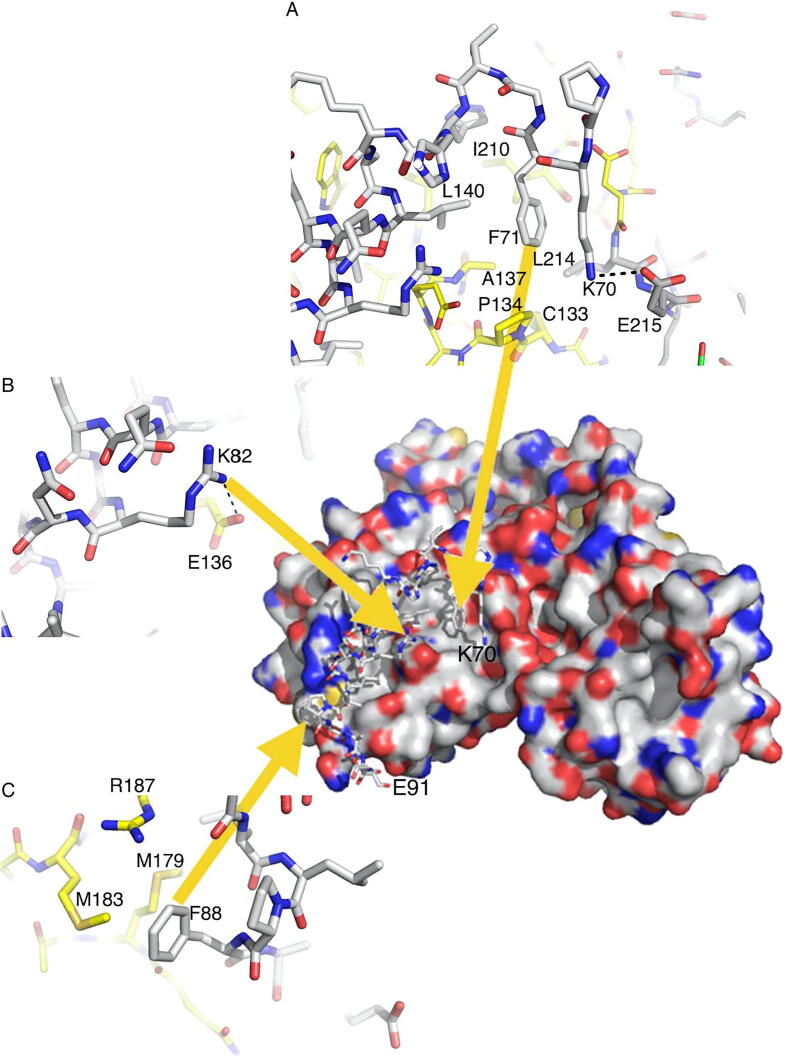
We determined the 1.6 Å resolution structure of the Nf2/merlin head domain, residues 1–339, bound to LATS1, residues 69–91 (Fig. 1A, 1B). LATS1 binding buries a surface area is ~840 Å2 and the shape complementarity [64] is 0.749. LATS1 has a central amphipathic α-helix that is extended by a randomly coiled structure at either terminus. LATS1 forms an extensive network of hydrophobic as well as hydrogen bonding interactions with the Nf2/merlin F2 FERM subdomain (Fig. 1C). LATS1 interacts with Nf2/merlin predominantly through core hydrophobic interactions mediated by LATS1 residues Ala-77, Leu-78, Ile-81, Leu-85, and Phe-88. In addition, LATS1 residues His-74 along with Phe-88 provides an end-capping interaction for LATS1 with Nf2/merlin in which the side chain of LATS1 residue Phe-88 provides a stable π-cation interaction on the C-terminus with the exposed hydrophobic surface formed by the helix-loop-helix region of Nf2/merlin F2 domain encompassing residues Nf2/merlin residues Tyr-177, Met-179, Met-183, and Arg-187 (Fig. 1D; Fig. 2). In contrast, LATS1 residue His-74 provides a dual-mode stable interaction at the N-terminus by electrostatic interactions with Nf2/merlin residue Glu-206 and simultaneous hydrophobic interactions with Leu-140 and Ile-210 of Nf2/merlin. Additionally, the central region of LATS1 is stabilized by another electrostatic interaction between LATS1 residue Arg-82 and Nf2/merlin residue Glu-136. The main hydrophobic interactions contributed by LATS1 are provided by Leu-85 that is stacked against Nf2/merlin residues Try-177, Met-179, Trp-184, and Arg-187, while LATS1 residue Ile-81 is surrounded by Nf2/merlin residues Glu-136, Val-139, and Trp-191, and LATS1 residue Leu-78 is probing the hydrophobic environment of Nf2/merlin residues Glu-136, Ala-137, and Leu-140. Ala-77 of LATS1 interacts with Nf2/merlin residue Trp-191. Interestingly, LATS1 residue Phe-71 situated in the random coil region extending from the LATS1 N-terminal α-helix, participates in hydrophobic stacking interactions with Ile-210, Asp-213, and Leu-214 of Nf2/merlin. Another electrostatic interaction originating from LATS1 residue Lys-70 and Nf2/merlin residue Glu-215 also resides in the same extended coil region. The latter two interactions contributed by Phe-71 and Lys-70 of LATS1 appear largely influenced by the crystal contacts emanating from the Nf2/merlin residues 311–314 of the adjacent symmetry molecule.
Fig. 2.
The interactions between Nf2/merlin and LATS1. Center, surface representation of Nf2/merlin (red, oxygen; blue, nitrogen; white, carbon; yellow, sulfur) with LATS1 shown in stick representation. Zoomed-in views of LATS1 (A) at the N-terminal region wherein Phe-71 engages the hydrophobic groove of Nf2/merlin (residues Ala-137, Pro-134, Leu-140, Ile-210, and Leu-214) through hydrophobic interactions, (B) residue Arg-82 engaging in electrostatic interactions with Nf2/merlin residue Glu-136, and (C) residue Phe-88 that stabilizes the exposed hydrophobic surface (Nf2/merlin residues Met-179, Met-183, and Arg-187) via hydrophobic interactions. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Superposition of the apo Nf2/merlin structure (PDB entry 1isn) [61] with our LATS1-bound structure shows that the LATS1 binding groove easily accommodates LATS1 which is blocked in apo structure by the electrostatic interaction between Glu-136 and Arg-187 of the Nf2/merlin FERM domain (Fig. 1E). Nf2/merlin residue Glu-136 is flipped upon binding to LATS1, thereby opening the groove to form electrostatic interactions with LATS1 residue Arg-82. LATS1 residue Arg-187 also undergoes positional rearrangement to stabilize the interaction (Fig. 1E). The side chains of Nf2/merlin residues Trp-184 and Trp-191 reposition to provide a favorable hydrophobic environment upon LATS1 binding. Additional rearrangement occurs to avoid a steric clash with LATS1 at the loop region where Nf2/merlin residues Tyr-177 and Gln-178 are also flipped to widen the groove (Fig. 1F). This loop region is further stabilized by polar interactions, locking the interface in the unique conformational state.
LATS1 and the Nf2/merlin tail domain bind to the same site on the Nf2/merlin FERM domain
Superposition of our LATS1-bound Nf2/merlin structure onto the Nf2/merlin head–tail structure (S518D, A585W mutant, PDB entry 4zrj) [40] highlights the overlapping binding site (Fig. 3; Supplementary Table S1). The FERM domains (residues 18–312) are very similar and superimpose with root means squares deviations of 0.64 Å for 1661 atoms (residues 21–312). The F1 and F2 subdomains of the FERM domain superimpose best, with root means squares deviations of 0.43 Å for 1208 atoms (residues 21–215).
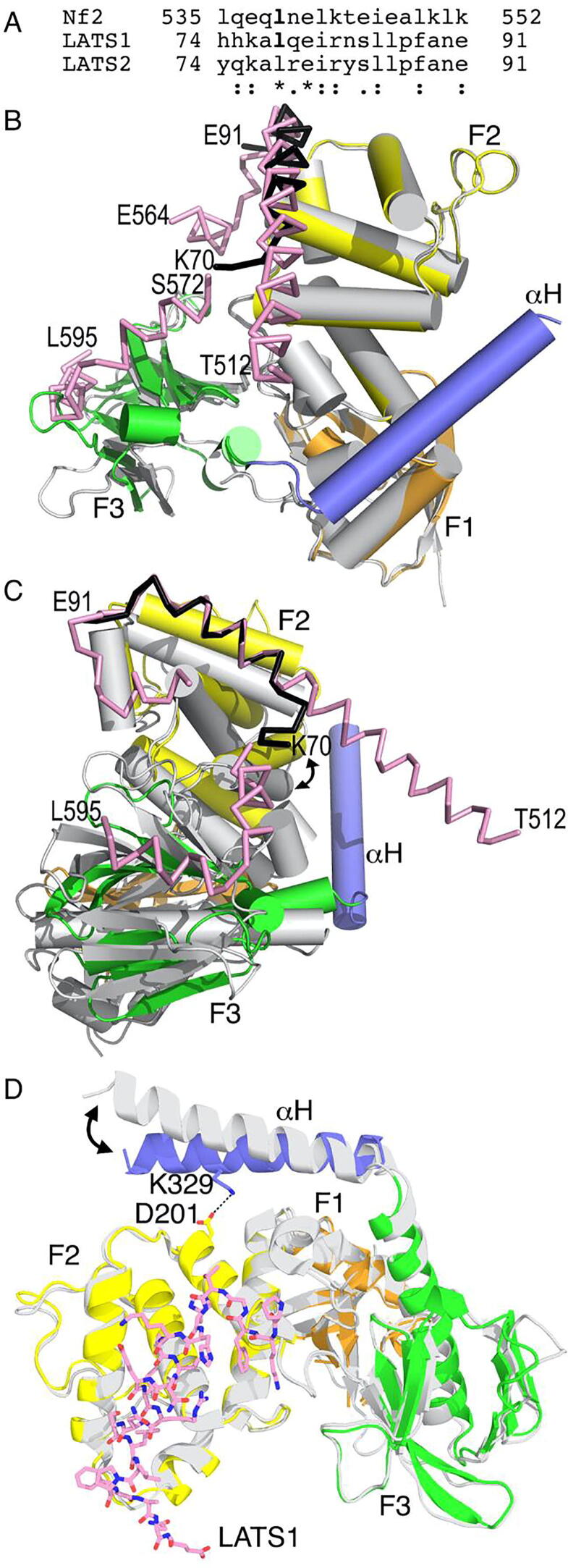
Fig. 3.
The LATS1 and Nf2/merlinC-terminus binding sites overlap on the Nf2/merlin FERM domain. A. Structure-based alignment of the Nf2/merlin tail and LATS. B. Superposition of our LATS1-bound Nf2/merlin structure (FERM domain: F1, orange; F2, yellow; F3, green; the first α-helix of the central helical domain αH, blue) with LATS1 shown as a black ribbon onto the Nf2/merlin FERM (gray) domain in complex with its mutant A585W tail (pink) domain (PDB entry 4zrj) [40]. LATS1 termini (K70, E91), Nf2/merlin tail domain termini (T512, E564, S572, and L595) and Nf2/merlin FERM subdomains are labeled. C. Superposition of our LATS1-bound Nf2/merlin structure onto the Nf2/merlin FERM domain in complex with its mutant A585W tail. The view is 90° rotated along the long vertical center of panel A. In panel A, the two FERM domains are superimposed. In this panel, LATS1 is superimposed onto the Nf2/merlin mutant S518D, A585W tail domain with the double arrow showing the relative Nf2/merlin FERM domain movements when comparing the LATS1-bound state with the Nf2/merlin tail-bound state. D. Superposition of our LATS1-bound Nf2/merlin structure onto the unbound Nf2/merlin FERM domain (PDB entry 1isn) [61] shown in gray with root means squares deviations of 0.923 Å for 1980 atoms. The double arrow shows the relative movements of the first α-helix of the central helical domain (αH) when comparing the LATS1-bound state with the unbound Nf2/merlin structure. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Sequence alignment of LATS1, LATS2, and the Nf2/merlin tail domain (Fig. 3A) shows that both LATS sequences have two (Leu-78 and Glu-80) out of 17 residues identical with Nf2/merlin (Leu-539 and Glu-541). The two LATS sequences are identical except for their distinct N-termini (residues 73–75) as well as residues 79 and 83. Given this high similarity, it was not surprising that our structures show that LATS1 and the Nf2/merlin tail domain have overlapping binding sites on F2 (Fig. 3B) whereby Nf2/merlin residues 536–551 can be superimposed onto LATS1 residues 75–90 with root means squares deviations of 0.21 Å for 52 atoms (Fig. 3C). Despite the low number of superimposed atoms, such structural comparison still shows the similarity of the binding pockets. The relative shifts of the F2 subdomain are less than 1 Å (as measured for example measured at F2 residues 177) of the tail-bound compared to the LATS1-bound Nf2/merlin FERM domains and up to ~8 Å when measured further away from the LATS1 and Nf2/merlin tail binding sites (as measured for example on F2 residues 112). The core hydrophobic residues of LATS1 and the Nf2/merlin tail domain engage in binding to the F2 domain similarly (Leu-78, Ile-81, Leu-85, and Phe-88 of LATS1 and Leu-539, Leu-542, Ile-546 and Leu-549 of the Nf2/merlin mutant A585W tail domain). Interestingly, four out of the six (or five out of eleven in the head–tail structure) hydrogen bonds between the Nf2/merlin FERM domain and LATS1 or the tail domain are provided by the backbone. One such interaction that does therefore stand out is Nf2/merlin residue Glu-215 that interacts with LATS1 residue Lys-70 as it does with Nf2/merlin Lys-573. Thus, the trace of LATS1 is in line with the Nf2/merlin mutant A585W tail domain in the Nf2/merlin head–tail complex structure (PDB entry 4zrj) [40], suggesting that Nf2/merlin adopts a flexible interaction with other proteins allowing multiple intermediate states (closed, open, or semi-open) that produce physiological relevant conformations.
Allosteric LATS1 binding to Nf2/merlin
Interestingly, the extended apo Nf2/merlin structure that also comprises the first α-helix of the central helical domain (αH) (residues 15–339; PDB entry 1isn) [61] has this αH α-helix closer to the LATS1 binding site compared to its position seen in our LATS1-bound structure (Fig. 3D). LATS1 binding to the F2 Nf2/merlin FERM subdomain causes about 10° movements of that α-helix αH by allostery without any contacts between this Nf2/merlin α-helix and LATS1. In the LATS1-bound structure, the Nf2/merlin α-helix αH is closer to the Nf2/merlin F2 subdomain, an interaction that is stabilized by electrostatic interaction between Nf2/merlin residues Asp-201 and Lys-201 that is not seen in the unbound Nf2/merlin structure. Thus, the Nf2/merlin α-helix αH is closer to the F2 domain in the LATS1-bound structure (Fig. 3D).
In our LATS1-bound structure, the extended C-terminal α-helix adopts a confirmation closer to the base of the F2 domain that is stabilized through numerous water-mediated interactions. Met-321 at the N-terminus of the extended Nf2/merlin C-terminal α-helix engages in hydrophobic interactions with Arg-57 and Glu-58 residing on the Nf2/merlin F1 domain and Val-110 from the F2 domain. Gln-324 from the same region exhibits hydrogen bonding interactions with Arg-57. These interactions are similar to the extended C-terminal helix interaction observed in the apo-structure previously reported (PDB entry 1isn) [61] with the notable exception of the hydrogen bonding interaction between Arg-57 and Asn-104 which is absent in our LATS1-bound Nf2/merlin structure. Overall, the extended C-terminus adopts a different conformation compared to the apo structure previously reported (PDB entry 1isn) [61] (Fig. 3D). Additional interactions that lead to the difference in this arrangement are the electrostatic interactions of Lys-322 with Glu-107 and Asp-314 which is present in the apo structure while only the Lys-322 with Asp-314 electrostatic interaction is retained in our LATS1-bound Nf2/merlin structure. This is in part due to the abrupt twist of the last turn of the C-terminal α-helix of the F3 domain in our LATS1-bound structure, because of the proximity of the LATS1 N-terminal random coil region to the adjacent symmetry molecule. In addition, the C-terminus of the extended α-helix region is stabilized by interactions with the F3 domain of different symmetry molecules.
Nf2/merlin attaches to the plasma membrane independently of LATS1
The association of Nf2/merlin with the plasma membrane is important for Nf2/merlin to regulate growth. While Nf2/merlin binds to several phosphorylated phosphoinositides, phosphatidylinositol 4,5-bisphosphate (PIP2) seems the physiological phosphorylated phosphoinositide that recruits and attaches Nf2/merlin to the plasma membrane. PIP2 is also the most abundant phospholipid in the membrane and in particular in lipid rafts, where Nf2/merlin localizes.
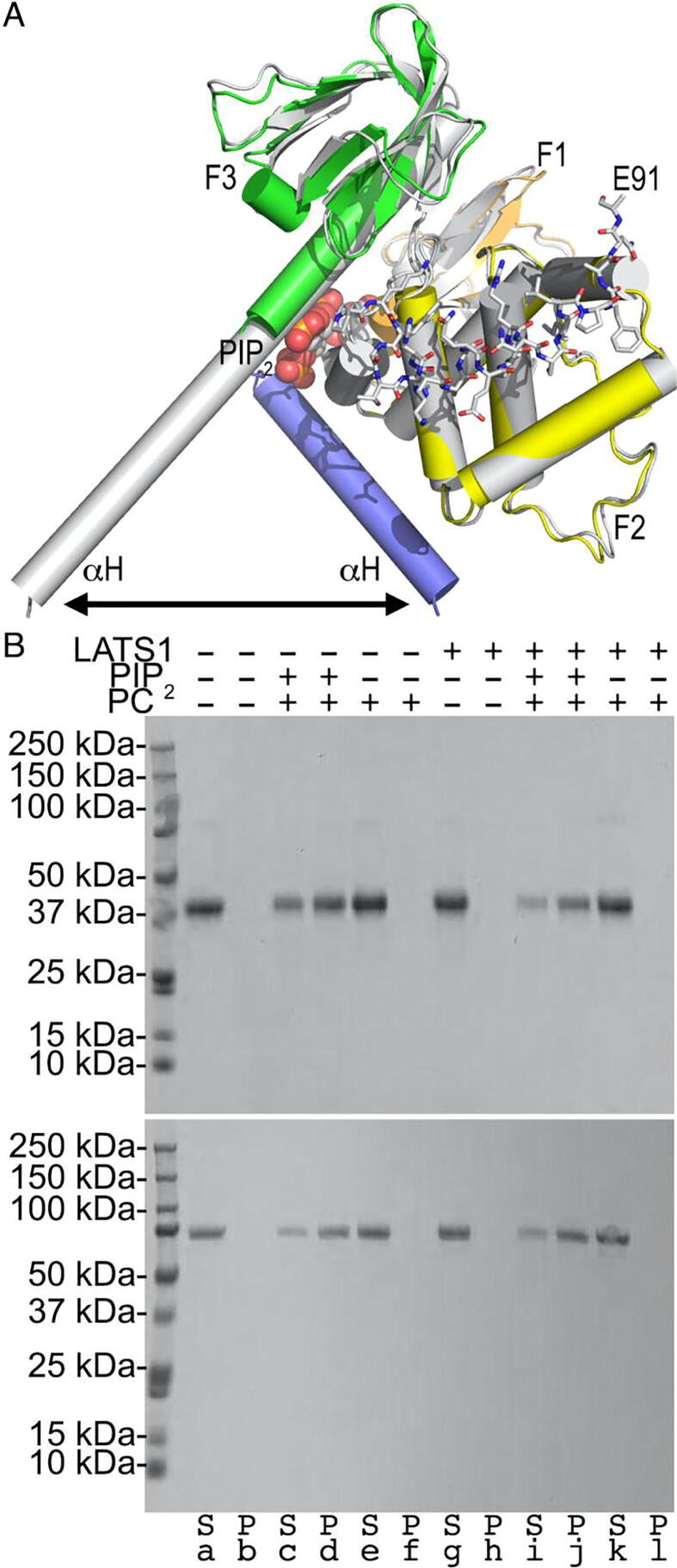
We recently determined that Nf2/merlin binding to the plasma membrane causes a conformational switch that activates the tumor-suppressive activity of Nf2/merlin [11]. In particular, the first α-helix (residues 315–340) of the central Nf2/merlin α-helical domain that follows the FERM domain, is extended in the lipid-bound Nf2/merlin structure thereby severing the Nf2/merlin FERM-tail interaction (Fig. 4A). However, given that the LATS- and the lipid-binding sites differ on the Nf2/merlin FERM domain, we wondered about their binding affecting each other. To test this mechanism, we performed co-sedimentation assays using liposomes comprised of 80% phosphatidylcholine and 20% PIP2 (Fig. 4B). The Nf2/merlin head domain (residues 1–339) only pelleted in the presence of the liposomes and the addition of LATS1 did not affect this interaction. The same results were obtained when using full-length Nf2/merlin. Thus, Nf2/merlin attaches to the membrane and this interaction is permissive for Nf2/merlin binding to LATS1.
Fig. 4.
LASTS1 binding is independent of Nf2/merlin attachment to the membrane. A. Superposition of our LATS1-bound Nf2/merlin structure (FERM domain: F1, orange; F2, yellow; F3, green; the first α-helix αH of the central helical domain, blue) in complex with LATS1 shown in stick representation onto the Nf2/merlin (gray) domain in complex with PIP2 (PDB entry 6cds) [62]. The LATS1 C-terminus, Nf2/merlin FERM subdomains, and PIP2 are labeled. The double arrow indicates the movement of the α-helix (αH) that extends upon activation. B. Lipid co-sedimentation assay of the Nf2/merlin head domain (top) and of full-length Nf2/merlin (bottom) as analyzed by sodium dodecyl sulfate–polyacrylamide electrophoresis gel. Nf2/merlin (top, head domain, residues 1–339; bottom full-length) does not pellet in the absence of liposomes (lanes a and b) or with phosphatidylcholine (PC) liposomes (lanes e and f) or when bound to LATS1 (lanes g and h) alone or in the presence of PC liposomes (lanes k and l). Both Nf2/merlin proteins (top and bottom) bind to phosphatidylinositol 4,5-bisphosphate (PIP2) containing liposomes in the absence (lanes d) or presence of LATS1 (lanes j). S, supernatant; P, pellet. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Discussion
The Hippo signaling pathway allows cells to sense and respond to local signals arising from changes in mechanical forces, the extracellular matrix, metabolites, as well as growth factors and to translate these signals into changes in gene expression. This pathway activates the large tumor suppressor (LATS) family of kinases, which then phosphorylate the transcriptional regulators yes-associated protein (YAP/tafazzin [TAZ]) [65]. Their dysregulation contributes to several diseases in particular oncogenic features that are associated with cancers.
Phosphatidic acid was identified as an inhibitor of LATS kinases whereby elevated phosphatidic acid levels potently induce nuclear YAP/TAZ activity as well as aberrant YAP/TAZ activation in cancer [66]. Phosphatidic acid is a key metabolite in membrane phospholipid biosynthesis and an important signaling molecule that increases cancer cell growth [67]. Phosphatidic acid binds to three regions on the C-terminal kinase domain of LATS1 [66] as well as to Nf2/merlin although earlier reports show the Nf2/merlin interaction with phosphatidic acid is weak [66]. While phosphatidic acid-binding to Nf2/merlin inhibits LATS1 recruitment to the cell membrane, an important event that couples the LATS kinases with upstream activating kinases [36], its binding do not affect Nf2/merlin recruitment to the membrane.
Nf2/merlin binds phospholipids in the lipid-ordered microdomains [10], [12], which is necessary for Hippo pathway activation. Specifically, Nf2/merlin is necessary for LATS (and YAP) phosphorylation [68]. We recently showed that the binding region of other phosphorylated phosphoinositides, such as phosphatidylinositol 4,5-bisphosphate (PIP2), reside within the Nf2/merlin FERM domain and that PIP2 binding induces Nf2/merlin activation via a conformational change allowing recruitment to the plasma membrane which increases LATS1 binding by ten-fold [11]. Our PIP2-binding deficient mutant (T59V, W60E, R309Q, R310Q) showed a decrease in LATS phosphorylation and activity [68]. Six further point mutations (K79N, K80N, E270N, K278N, K279N) prevented Nf2/merlin binding to several phospholipids, especially phosphatidylinositol (3,4)-bisphosphate, PIP2, and phosphatidylinositol (3,4,5)-triphosphate (PIP3), and completely abolished LATS phosphorylation [68]. Functionally relevant, the leading edge of migrating cells is enriched in PIP3 rather than the PIP2 signaling lipid [69], [70] and while PIP2 accounts for about 10% of the total phosphorylated phosphoinositides in the cell membrane, PIP2 is indeed concentrated in cholesterol- and glycosphingolipid-rich membrane microdomains [33], [71]. Further, binding to PIP2 recruits Nf2/merlin to lipid rafts and it is at these microdomains that Nf2/merlin recruits and activates LATS [36].
The Nf2/merlin tail binding site overlaps with the LATS1 binding site on the Nf2/merlin head domain. Thus, in its inactive and closed conformer, the LATS binding site on Nf2/merlin is occupied by the Nf2/merlin tail domain. PIP2 binding severs the Nf2/merlin head–tail binding. Consistently, LATS1 binding to the PIP2-bound Nf2/merlin increased ten-fold compared to unbound Nf2/merlin [11]. Further, our Nf2/merlin structure in complex with LATS1 and lipid co-sedimentation assays show that membrane attachment and simultaneous binding of LATS1 is possible. Thus, while the binding of phosphatidic acid to Nf2/merlin hinders LATS1 recruitment, the binding of PIP2 to Nf2/merlin is permissive for its binding to LATS1. Therefore, it seems that different phospholipids have specific mechanisms of Nf2/merlin regulation of the Hippo signaling pathway. It is possible that activation of phospholipase D that leads to phosphatidic acid formation might break down PIP2 and result in the phosphatidic acid-bound form of Nf2/merlin. Since phosphatidic acid-binding to Nf2/merlin inhibits LATS1 recruitment, while PIP2 binding to Nf2/merlin is permissive for binding to LATS1, the different lipids affect LATS kinase activation differently.
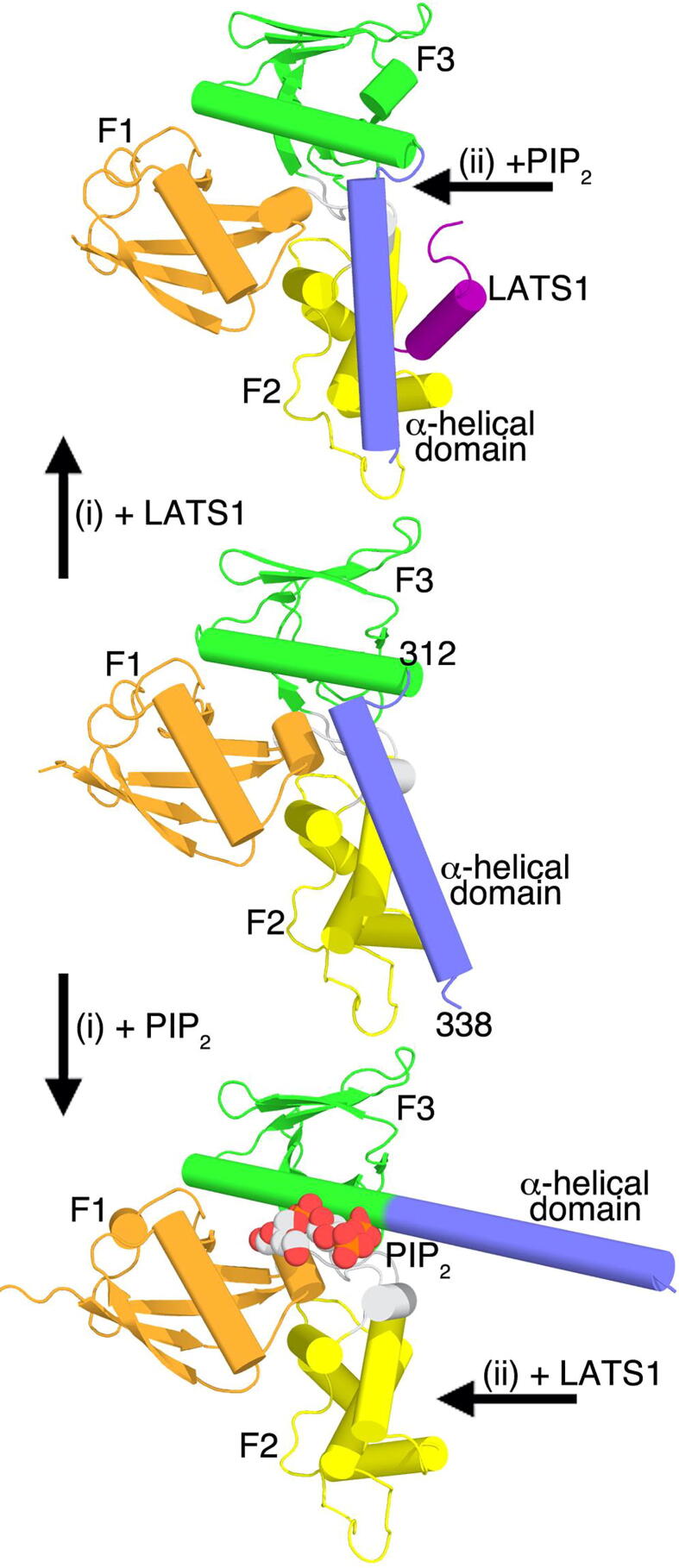
The significant novelty that came out of our LATS1-bound Nf2/merlin structure is the conformational changes in the functionally crucial α-helix that follows the Nf2/merlin FERM domain. This α-helix is the key to switching Nf2/merlin between its open and closed conformers which correlates to its tumor-suppressive functions [11]. The lower resolution (2.3 Å and 2.7 Å) truncated Nf2/merlin structures (residues 21–311) bound to LATS1 (PDB entry 4zrk) [40] and LATS2 (PDB entry 4zri) [40] that have this functionally key α-helix truncated and missing in their structures, additionally miss the allosteric LATS1 binding seen in our 1.6 Å structure (Fig. S2). Since the LATS1 binding site overlaps with the Nf2/merlin tail domain binding site on the Nf2/merlin FERM domain, LATS binding releases and replaces the Nf2/merlin C-terminal tail domain. However, LATS binding is not sufficient to release the central Nf2/merlin domain that seems necessary to activate Nf2/merlin (Fig. 5). These findings are consistent with the fact that angiomotin has been shown necessary to release the Nf2/merlin auto-inhibitory conformation [40] and that the affinity of LATS1 increases upon Nf2/merlin activation by its binding to the membrane [11]. Collectively, our study provides new mechanistic insights into the Hippo pathway that are relevant to several diseases in particular oncogenic features that are associated with cancers.
Fig. 5.
Nf2/merlin mechanistic events in Hippo signaling.Center, in its closed conformation, Nf2/merlin has its α-helix C-terminal of its FERM domain in contact with its F2 subdomain. (i) LATS1 (top) or the membrane (bottom) bind to the closed (center) Nf2/merlin conformation whereby only PIP2 causes dramatic structural activation rearrangements. LATS1 binding results in some allosteric movements of the α-helix from the α-helical domain. (ii) The membrane-bound (bottom) or LATS1-bound (top) Nf2/merlin are both permissive for ternary complex formation.
Methods
DNA constructs
We used our previously described Nf2/merlin construct, residues 1–339 [11], which was cloned into the pGEX-2T expression vector (GE Life Sciences) by using the full-length human Nf2/merlin plasmid (Addgene plasmid number #11629). The human Nf2/merlin full-length construct, residues 1–595, was cloned into a pET-32a expression vector (Novagen) with a hexahistidine tag followed by a thioredoxin tag. LATS1 (residues 69–91) was synthesized by GenScript USA Inc.
Protein preparation
Nf2/merlin (residues 1–339) was expressed in Escherichia coli strain BL21(DE3) Rosetta2 (Novagen) at 25 °C for 20 h. Cells were induced for protein expression with 0.2 mM isopropyl β-D-1-thiogalactopyranoside and harvested by centrifugation at 5,000 × g for 15 min. Nf2/merlin (residues 1-339) purification was performed as described previously [11] with minor modificationsas follows. We used a glutathione S-transferase column (GE Healthcare) equilibrated in 20 mM Tris, pH 7.5, 400 mM NaCl, and 0.1 mM ethylenediaminetetraacetic acid, and the Nf2/merlin protein was eluted with 10 mM glutathione. The glutathione S-transferase tag was removed by overnight cleavage with PreScission protease. After digestion, the protein and glutathione S-transferase were separated using the glutathione S-transferase column. The Nf2/merlin protein in elution buffer (20 mM Tris, pH 7.5, 400 mM NaCl, 0.1 mM ethylenediaminetetraacetic acid, and 10 mM glutathione) was concentrated to 5 ml and further purified by size exclusion chromatography (SEC) using a 26/60 Superdex 75 column (GE Healthcare) that was pre-equilibrated with 20 mM Tris (pH 7.5), 400 mM NaCl, 1 mM dithiothreitol, and 0.1 mM ethylenediaminetetraacetic acid.
Human full-length Nf2/merlin, residues 1–595, was expressed in Escherichia coli strain BL21-CodonPlus (DE3)-RIL (Novagen). Protein expression was induced by 0.2 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at 25 °C for 20 h. Cells were harvested by centrifugation at 5000 × g for 15 min at 4 °C. Obtained pellets were resuspended in lysis buffer (20 mM Tris, pH 7.5, 400 mM NaCl, and 0.1 mM ethylenediaminetetraacetic acid), lysed by sonication, and clarified by ultracentrifugation at 100,000 x g for 30 min. The supernatant was loaded onto a nickel affinity chromatography column (GE Healthcare), equilibrated in 20 mM Tris-HCl (pH 7.5) and 400 mM NaCl. The Nf2/merlin protein was eluted using a 0.5 M imidazole gradient. Histidine and thioredoxin tags were removed by overnight cleavage with PreScission protease at 4 °C and dialyzed in 20 mM Tris (pH 7.5), 400 mM NaCl, 2 mM thioredoxin, and 0.1 mM ethylenediaminetetraacetic acid. Nf2/merlin was further purified by SEC using a 16/60 Superdex 200 column (GE Healthcare) pre-equilibrated with 20 mM Tris (pH 7.5), 400 mM NaCl, 1 mM dithiothreitol, and 0.1 mM ethylenediaminetetraacetic acid. After purification, proteins were concentrated, and aliquots were frozen in liquid nitrogen and stored at −20 °C.
LATS1-bound Nf2/merlin crystallization
We obtained needle-shaped crystals within two days at room temperature by hanging drop vapor diffusion in 24 well VDX plates from Hampton Research (catalog number HR3-171) from a reservoir containing 600 mM imidazole (pH 6.5) and 15% (w/v) polyethylene glycol 3350. The protein to reservoir ratio was 1:1 (0.5 μl Nf2/merlin plus 0.5 μl LATS1). We used concentrations of 250 μM (10 mg/ml) for Nf2/merlin and 300 μM (795 μg/ml) for LATS1 (i.e. a 1:1.2 Nf2/merlin to LATS1 molar ratio). Crystals were transferred briefly to a drop containing the reservoir solution that included 15% glycerol as a cryoprotectant, mounted with no preferred orientation, and flash-frozen in liquid nitrogen.
LATS1-bound Nf2/merlin X-ray data collection and processing
LATS1 residues 69–91 in complex with Nf2/merlin residues 1–339 crystals diffracted X-rays at beamline ID22 at the Advanced Photon Source, Argonne National Laboratory beyond 1.6 Å Bragg spacings. The beamline was equipped with a Dectris Eiger X16M pixel array detector and a complete dataset of 1,800 frames was collected at a wavelength of 1 Å with a 0.2° oscillation range. X-ray data were indexed, integrated, and scaled using XDS and AIMLESS as implemented in autoPROC [72]. X-ray data reduction statistics are provided in Table 1.
Table 1.
X-ray data reduction statistics for our LATS1 (residues 69–91)-bound human Nf2/merlin (residues 1–339) structure.
| Space group | P 212121 |
|---|---|
| Unit cell dimensions: | |
| a, b, c | 52.24 Å, 84.24 Å, 95.86 Å |
| α = β = γ | 90° |
| Resolution (last shell) | 63.3 Å–1.6 Å (1.63 Å–1.60 Å) |
| Total measurements | 754,632 (37,434) |
| Number of unique reflections | 55,750 (2,710) |
| Wavelength | 1.0 Å |
| R-pim * | 0.024 (0.325) |
| Signal-to-noise I/σ (I) | 18.5 (2.3) |
| Completeness | 0.99 (0.97) |
| Multiplicity | 13.5 (13.8) |
| CC1/2 # | 0.999 (0.877) |
* Rp.i.m. is the precision indicating merging R-factor given as
# CC1/2 is a Pearson’s correlation coefficient calculated between the average intensities of each random half of measurements of unique reflections.
LATS1-bound Nf2/merlin crystal structure determination and crystallographic refinement
Phases were obtained by molecular replacement using the program PHASER [73] by using one Nf2/merlin polypeptide chain (residues 1–320) (PDB entry 4zrk) [40] as the search model. A unique solution was obtained and one molecule in the asymmetric unit was identified in space group P 212121 resulting in a calculated volume to mass ratio of 2.51 Å3/Da, which corresponds to a solvent content of 0.51. Initial crystallographic refinement without translation, libration, screw rotation (TLS) parameters was performed until convergence using autoBuster [74]. The model was manually built using Coot [75]. Nf2/merlin residues 321–339 that were missing from the search model as well as LATS1 residues 69–91 were unambiguously built with clear electron density after initial crystallographic refinement with BUSTER and were manually fit using Coot [75], [76]. Water molecules were included in further refinement runs using autoBuster and validated in Coot. Optimized imidazole and glycerol coordinates and restraints were obtained from the Grade web server (grade.globalphasing.org). The quality of the final model was assessed using MolProbity [77], which revealed no outliers, and the final crystallographic refinement statists are provided in Table 2.
Table 2.
Crystallographic refinement statistics for our LATS1 (residues 69–91)-bound human Nf2/merlin (residues 1–339) structure.
| Resolution (last shell) | 63.28 Å–1.61 Å (1.62 Å–1.61 Å) |
| No. of reflections, working set | 55,750 (1053) |
| No. of reflections, test set | 2748 (62) |
| R-factor * | 0.200 (0.233) |
| R-free, 5% of reflections omitted | 0.227 (0.308) |
| No. of non-hydrogen atoms: | |
| protein | 2944 |
| ligands | 21 |
| solvent | 426 |
| Average B-factor: | |
| protein | 30.35 Å2 |
| ligand | 40.38 Å2 |
| solvent | 42.66 Å2 |
| Root-mean-square deviation from ideal values: | |
| bond lengths | 0.008 Å |
| bond angles | 0.87° |
| Ramachandran plot, favoured | 99.41% |
| Ramachandran plot, allowed | 0.29% |
| Ramachandran plot, outliers | 0.30% |
* R-factor =.
Lipid co-sedimentation assays
Lipid binding to Nf2/merlin (residues 1–339) and full-length Nf2/merlin (residues 1–595) was assayed as described previously [62].
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We are indebted to the staff of the South Eastern Regional Access Team (Argonne National Laboratory) for synchrotron support. We thank Joe Kissil (The Scripps Research Institute) for insightful discussions. TI is supported by grants from the Department of Defense, The National Science Foundation, and by start-up funds provided to The Scripps Research Institute from the State of Florida. The coordinates and structure factors were deposited with the Protein Data Bank, accession code 7lwh. The plasmids used in this study were deposited at Addgene, with plasmid number #79308 for Nf2/merlin (residues 1-339), and plasmid number #107148 for Nf2/merlin full-length (residues 1-595). Our dataset was deposited at the SBGrid consortium: doi:10.15785/SBGRID/822. All other data are within the manuscript. This is publication #30072 from The Scripps Research Institute.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mbplus.2021.100074.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Rouleau G.A., Merel P., Lutchman M., Sanson M., Zucman J., Marineau C., Hoang-Xuan K., Demczuk S., Desmaze C., Plougastel B., Pulst S.M., Lenoir G., Bijlsma E., Fashold R., Dumanski J., Jong P.d., Parry D., Eldrige R., Aurias A., Delattre O., Thomas G. Alteration in a new gene encoding a putative membrane-organizing protein causes neuro-fibromatosis type 2. Nature. 1993;363:515–521. doi: 10.1038/363515a0. [DOI] [PubMed] [Google Scholar]
- 2.MacCollin M., Mohney T., Trofatter J., Wertelecki W., Ramesh V., Gusella J. DNA diagnosis of neurofibromatosis 2. Altered coding sequence of the merlin tumor suppressor in an extended pedigree. JAMA. 1993;270:2316–2320. doi: 10.1001/jama.270.19.2316. [DOI] [PubMed] [Google Scholar]
- 3.Petrilli A.M., Fernández-Valle C. Role of Merlin/NF2 inactivation in tumor biology. Oncogene. 2016;35:537–548. doi: 10.1038/onc.2015.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giovannini M., Robanus-Maandag E., van der Valk M., Niwa-Kawakita M., Abramowski V., Goutebroze L., Woodruff J.M., Berns A., Thomas G. Conditional biallelic Nf2 mutation in the mouse promotes manifestations of human neurofibromatosis type 2. Genes & Development. 2000;14:1617–1630. [PMC free article] [PubMed] [Google Scholar]
- 5.Gehlhausen J.R., Park S.J., Hickox A.E., Shew M., Staser K., Rhodes S.D., Menon K., Lajiness J.D., Mwanthi M., Yang X., Yuan J., Territo P., Hutchins G., Nalepa G., Yang F.C., Conway S.J., Heinz M.G., Stemmer-Rachamimov A., Yates C.W., Wade Clapp D. A murine model of neurofibromatosis type 2 that accurately phenocopies human schwannoma formation. Human Molecular Genetics. 2015;24:1–8. doi: 10.1093/hmg/ddu414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morrison H., Sherman L.S., Legg J., Banine F., Isacke C., Haipek C.A., Gutmann D.H., Ponta H., Herrlich P. The NF2 tumor suppressor gene product, merlin, mediates contact inhibition of growth through interactions with CD44. Genes & Development. 2001;15:968–980. doi: 10.1101/gad.189601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scherer S.S., Gutmann D.H. Expression of the neurofibromatosis 2 tumor suppressor gene product, merlin, in Schwann cells. Journal of Neuroscience Research. 1996;46:595–605. doi: 10.1002/(SICI)1097-4547(19961201)46:5<595::AID-JNR8>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 8.Denisenko N., Cifuentes-Diaz C., Irinopoulou T., Carnaud M., Benoit E., Niwa-Kawakita M., Chareyre F., Giovannini M., Girault J.-A., Goutebroze L. Tumor suppressor schwannomin/merlin is critical for the organization of Schwann cell contacts in peripheral nerves. Journal of Neuroscience. 2008;28:10472–10481. doi: 10.1523/JNEUROSCI.2537-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzalez-Agosti C., Xu L., Pinney D., Beauchamp R., Hobbs W., Gusella J., Ramesh V. The merlin tumor suppressor localizes preferentially in membrane ruffles. Oncogene. 1996;13:1239–1247. [PubMed] [Google Scholar]
- 10.Stickney J.T., Bacon W.C., Rojas M., Ratner N., Ip W. Activation of the tumor suppressor merlin modulates its interaction with lipid rafts. Cancer Research. 2004;64:2717–2724. doi: 10.1158/0008-5472.can-03-3798. [DOI] [PubMed] [Google Scholar]
- 11.Chinthalapudi K., Mandati V., Zheng J., Sharff A.J., Bricogne G., Griffin P.R., Kissil J., Izard T. Lipid binding promotes the open conformation and tumor-suppressive activity of neurofibromin 2. Nature Communications. 2018;9:1338. doi: 10.1038/s41467-018-03648-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mani T., Hennigan R.F., Foster L.A., Conrady D.G., Herr A.B., Ip W. FERM domain phosphoinositide binding targets merlin to the membrane and is essential for its growth-suppressive function. Molecular and Cellular Biology. 2011;31:1983–1996. doi: 10.1128/MCB.00609-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curto M., Cole B.K., Lallemand D., Liu C.H., McClatchey A.I. Contact-dependent inhibition of EGFR signaling by Nf2/Merlin. The Journal of Cell Biology. 2007;177:893–903. doi: 10.1083/jcb.200703010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cole B.K., Curto M., Chan A.W., McClatchey A.I. Localization to the cortical cytoskeleton is necessary for Nf2/merlin-dependent epidermal growth factor receptor silencing. Molecular and Cellular Biology. 2008;28:1274–1284. doi: 10.1128/MCB.01139-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lallemand D., Saint-Amaux A.L., Giovannini M. Tumor-suppression functions of merlin are independent of its role as an organizer of the actin cytoskeleton in Schwann cells. Journal of Cell Science. 2009;122:4141–4149. doi: 10.1242/jcs.045914. [DOI] [PubMed] [Google Scholar]
- 16.McClatchey A.I. Merlin and ERM proteins: unappreciated roles in cancer development? Nature Reviews Cancer. 2003;3:877–883. doi: 10.1038/nrc1213. [DOI] [PubMed] [Google Scholar]
- 17.McClatchey A.I., Giovannini M. Membrane organization and tumorigenesis - the NF2 tumor suppressor, Merlin. Genes & Development. 2005;19:2265–2277. doi: 10.1101/gad.1335605. [DOI] [PubMed] [Google Scholar]
- 18.Maitra S., Kulikauskas R.M., Gavilan H., Fehon R.G. The tumor suppressors Merlin and Expanded function cooperatively to modulate receptor endocytosis and signaling. Current Biology. 2006;16:702–709. doi: 10.1016/j.cub.2006.02.063. [DOI] [PubMed] [Google Scholar]
- 19.Shaw R.J., Paez J.G., Curto M., Yaktine A., Pruitt W.M., Saotome I., O'Bryan J.P., Gupta V., Ratner N., Der C.J., Jacks T., McClatchey A.I. The Nf2 tumor suppressor, merlin, functions in Rac-dependent signaling. Developmental Cell. 2001;1:63–72. doi: 10.1016/s1534-5807(01)00009-0. [DOI] [PubMed] [Google Scholar]
- 20.Xiao G.-H., Beeser A., Chernoff J., Testa J.R. p21-activated kinase links Rac/Cdc42 signaling to merlin. Journal of Biological Chemistry. 2002;277:883–886. doi: 10.1074/jbc.C100553200. [DOI] [PubMed] [Google Scholar]
- 21.Okada T., Lopez-Lago M., Giancotti F.G. Merlin/NF-2 mediates contact inhibition of growth by suppressing recruitment of Rac to the plasma membrane. Journal of Cell Biology. 2005;171:361–371. doi: 10.1083/jcb.200503165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Houshmandi S.S., Emnett R.J., Giovannini M., Gutmann D.H. The neurofibromatosis 2 protein, merlin, regulates glial cell growth in an ErbB2- and Src-dependent manner. Molecular and Cellular Biology. 2009;29:1472–1486. doi: 10.1128/MCB.01392-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou L., Ercolano E., Ammoun S., Schmid M.C., Barczyk M.A., Hanemann C.O. Merlin-deficient human tumors show loss of contact inhibition and activation of Wnt/beta-catenin signaling linked to the PDGFR/Src and Rac/PAK pathways. Neoplasia. 2011;13:1101–1112. doi: 10.1593/neo.111060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morrison H., Sperka T., Manent J., Giovannini M., Ponta H., Herrlich P. Merlin/neurofibromatosis type 2 suppresses growth by inhibiting the activation of Ras and Rac. Cancer Research. 2007;67:520–527. doi: 10.1158/0008-5472.CAN-06-1608. [DOI] [PubMed] [Google Scholar]
- 25.McClatchey A.I., Fehon R.G. Merlin and the ERM proteins–regulators of receptor distribution and signaling at the cell cortex. Trends in Cell Biology. 2009;19:198–206. doi: 10.1016/j.tcb.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hughes S.C., Fehon R.G. Phosphorylation and activity of the tumor suppressor Merlin and the ERM protein Moesin are coordinately regulated by the Slik kinase. The Journal of Cell Biology. 2006;175:305–313. doi: 10.1083/jcb.200608009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hennigan R.F., Moon C.A., Parysek L.M., Monk K.R., Morfini G., Berth S., Brady S., Ratner N. The NF2 tumor suppressor regulates microtubule-based vesicle trafficking via a novel Rac, MLK and p38(SAPK) pathway. Oncogene. 2013;32:1135–1143. doi: 10.1038/onc.2012.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muranen, T., Gronholm, M., Lampin, A., Lallemand, D., Zhao, F., Giovannini, M., and Carpen, O. (2007) The tumor suppressor merlin interacts with microtubules and modulates Schwann cell microtubule cytoskeleton. Human Molecular Genetics 16, 1742-1751. [DOI] [PubMed]
- 29.Bensenor L.B., Barlan K., Rice S.E., Fehon R.G., Gelfand V.I. Microtubule-mediated transport of the tumor-suppressor protein Merlin and its mutants. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:7311–7316. doi: 10.1073/pnas.0907389107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yi C., Troutman S., Fera D., Stemmer-Rachamimov A., Avila J., Christian N., Persson N., Shimono A., Speicher D., Marmorstein R., Holmgren L., Kissil J. A tight junction-associated Merlin-angiomotin complex mediates Merlin's regulation of mitogenic signaling and tumor suppressive functions. Cancer Cell. 2011;19:527–540. doi: 10.1016/j.ccr.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kressel M., Schmucker B. Nucleocytoplasmic transfer of the NF2 tumor suppressor protein merlin is regulated by exon 2 and a CRM1-dependent nuclear export signal in exon 15. Human Molecular Genetics. 2002;11:2269–2278. doi: 10.1093/hmg/11.19.2269. [DOI] [PubMed] [Google Scholar]
- 32.Muranen T., Grönholm M., Renkema G.H., Carpén O. Cell cycle-dependent nucleocytoplasmic shuttling of the neurofibromatosis 2 tumour suppressor merlin. Oncogene. 2005;24:1150–1158. doi: 10.1038/sj.onc.1208283. [DOI] [PubMed] [Google Scholar]
- 33.Li W., You L., Cooper J., Schiavon G., Pepe-Caprio A., Zhou L.u., Ishii R., Giovannini M., Hanemann C.O., Long S.B., Erdjument-Bromage H., Zhou P., Tempst P., Giancotti F.G. Merlin/NF2 suppresses tumorigenesis by inhibiting the E3 ubiquitin ligase CRL4(DCAF1) in the nucleus. Cell. 2010;140:477–490. doi: 10.1016/j.cell.2010.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cooper J., Li W., You L., Schiavon G., Pepe-Caprio A., Zhou L., Ishii R., Giovannini M., Hanemann C.O., Long S.B., Erdjument-Bromage H., Zhou P., Tempst P., Giancotti F.G. Merlin/NF2 functions upstream of the nuclear E3 ubiquitin ligase CRL4DCAF1 to suppress oncogenic gene expression. Science Signaling. 2011;4:pt6. doi: 10.1126/scisignal.2002314. [DOI] [PubMed] [Google Scholar]
- 35.Hamaratoglu F., Willecke M., Kango-Singh M., Nolo R., Hyun E., Tao C., Jafar-Nejad H., Halder G. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nature Cell Biology. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- 36.Yin F., Yu J., Zheng Y., Chen Q., Zhang N., Pan D. Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell. 2013;154:1342–1355. doi: 10.1016/j.cell.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moya I.M., Halder G. Discovering the Hippo pathway protein-protein interactome. Cell Research. 2014;24:137–138. doi: 10.1038/cr.2014.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwon Y., Vinayagam A., Sun X., Dephoure N., Gygi S.P., Hong P., Perrimon N. The Hippo signaling pathway interactome. Science. 2013;342:737–740. doi: 10.1126/science.1243971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cooper J., Giancotti F.G. Molecular insights into NF2/Merlin tumor suppressor function. FEBS Letters. 2014;588:2743–2752. doi: 10.1016/j.febslet.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y., Zhou H., Li F., Chan S.W., Lin Z., Wei Z., Yang Z., Guo F., Lim C.J., Xing W., Shen Y., Hong W., Long J., Zhang M. Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway. Cell Research. 2015;25:801–817. doi: 10.1038/cr.2015.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trofatter J.A., MacCollin M.M., Rutter J.L., Murrell J.R., Duyao M.P., Parry D.M., Eldridge R., Kley N., Menon A.G., Pulaski K., Haase V.H., Ambrose C.M., Munroe D., Bove C., Haines J.L., Martuza R.L., MacDonald M.E., Seizinger B.R., Short M.P., Buckler A.J., Gusella J.F. A novel moesin-, ezrin-, radixin-like gene is a candidate for the neurofibromatosis 2 tumor suppressor. Cell. 1993;75:826. doi: 10.1016/0092-8674(93)90501-g. [DOI] [PubMed] [Google Scholar]
- 42.Bretscher A., Edwards K., Fehon R.G. ERM proteins and merlin: integrators at the cell cortex. Nature Reviews Molecular Cell Biology. 2002;3:586–599. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- 43.Li Q., Nance M.R., Kulikauskas R., Nyberg K., Fehon R., Karplus P.A., Bretscher A., Tesmer J.J. Self-masking in an intact ERM-merlin protein: an active role for the central alpha-helical domain. Journal of Molecular Biology. 2007;365:1446–1459. doi: 10.1016/j.jmb.2006.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brault E., Gautreau A., Lamarine M., Callebaut I., Thomas G., Goutebroze L. Normal membrane localization and actin association of the NF2 tumor suppressor protein are dependent on folding of its N-terminal domain. Journal of Cell Science. 2001;114:1901–1912. doi: 10.1242/jcs.114.10.1901. [DOI] [PubMed] [Google Scholar]
- 45.Pearson M.A., Reczek D., Bretscher A., Karplus P.A. Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain. Cell. 2000;101:259–270. doi: 10.1016/s0092-8674(00)80836-3. [DOI] [PubMed] [Google Scholar]
- 46.Huang L., Ichimaru E., Pestonjamasp K., Cui X., Nakamura H., Lo G.Y.H., Lin F.I.K., Luna E.J., Furthmayr H. Merlin differs from moesin in binding to F-actin and in its intra- and intermolecular interactions. Biochemical and Biophysical Research Communications. 1998;248:548–553. doi: 10.1006/bbrc.1998.9009. [DOI] [PubMed] [Google Scholar]
- 47.Turunen O., Wahlstrom T., Vaheri A. Ezrin has a COOH-terminal actin-binding site that is conserved in the ezrin protein family. Journal of Cell Biology. 1994;126:1445–1453. doi: 10.1083/jcb.126.6.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.James M.F., Manchanda N., Gonzalez-Agosti C., Hartwig J.H., Ramesh V. The neurofibromatosis 2 protein product merlin selectively binds F-actin but not G-actin, and stabilizes the filaments through a lateral association. The Biochemical Journal. 2001;356:377–386. doi: 10.1042/0264-6021:3560377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu H.-M., Gutmann D.H. Merlin differentially associates with the microtubule and actin cytoskeleton. Journal of Neuroscience Research. 1998;51:403–415. doi: 10.1002/(SICI)1097-4547(19980201)51:3<403::AID-JNR13>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 50.Nguyen R., Reczek D., Bretscher A. Hierarchy of merlin and ezrin N- and C-terminal domain interactions in homo- and heterotypic associations and their relationship to binding of scaffolding proteins EBP50 and E3KARP. Journal of Biological Chemistry. 2001;276:7621–7629. doi: 10.1074/jbc.M006708200. [DOI] [PubMed] [Google Scholar]
- 51.Hennigan R.F., Foster L.A., Chaiken M.F., Mani T., Gomes M.M., Herr A.B., Ip W. Fluorescence resonance energy transfer analysis of merlin conformational changes. Molecular and Cellular Biology. 2010;30:54–67. doi: 10.1128/MCB.00248-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.LaJeunesse D.R., McCartney B.M., Fehon R.G. Structural analysis of Drosophila merlin reveals functional domains important for growth control and subcellular localization. Journal of Cell Biology. 1998;141:1589–1599. doi: 10.1083/jcb.141.7.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sher I., Hanemann C.O., Karplus P.A., Bretscher A. The tumor suppressor merlin controls growth in its open state, and phosphorylation converts it to a less-active more-closed state. Developmental Cell. 2012;22:703–705. doi: 10.1016/j.devcel.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meng J.-J., Lowrie D.J., Sun H., Dorsey E., Pelton P.D., Bashour A.-M., Groden J., Ratner N., Ip W. Interaction between two isoforms of the NF2 tumor suppressor protein, merlin, and between merlin and ezrin, suggests modulation of ERM proteins by merlin. Journal of Neuroscience Research. 2000;62:491–502. doi: 10.1002/1097-4547(20001115)62:4<491::AID-JNR3>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 55.Gonzalez-Agosti C., Wiederhold T., Herndon M.E., Gusella J., Ramesh V. Interdomain interaction of merlin isoforms and its influence on intermolecular binding to NHE-RF. Journal of Biological Chemistry. 1999;274:34438–34442. doi: 10.1074/jbc.274.48.34438. [DOI] [PubMed] [Google Scholar]
- 56.Zoch A., Mayerl S., Schulz A., Greither T., Frappart L., Rübsam J., Heuer H., Giovannini M., Morrison H., Schlatt S. Merlin isoforms 1 and 2 both act as tumour suppressors and are required for optimal sperm maturation. PLoS ONE. 2015;10:e0129151. doi: 10.1371/journal.pone.0129151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kang B.S., Cooper D.R., Devedjiev Y., Derewenda U., Derewenda Z.S. The structure of the FERM domain of merlin, the neurofibromatosis type 2 gene product. Acta Crystallographica. Section D, Biological Crystallography. 2002;58:381–391. doi: 10.1107/s0907444901021175. [DOI] [PubMed] [Google Scholar]
- 58.Yogesha S.D., Sharff A.J., Giovannini M., Bricogne G., Izard T. Unfurling of the band 4.1, ezrin, radixin, moesin (FERM) domain of the merlin tumor suppressor. Protein Science. 2011;20:2113–2120. doi: 10.1002/pro.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.T. Mori, S. Gotoh, M. Shirakawa, T. Hakoshima (2014) Structural basis of DDB1-and-Cullin 4-associated Factor 1 (DCAF1) recognition by merlin/NF2 and its implication in tumorigenesis by CD44-mediated inhibition of merlin suppression of DCAF1 function. Genes to cells: devoted to molecular & cellular mechanisms. [DOI] [PubMed]
- 60.F. Zhang, B. Liu, Y. Gao, J. Long, H. Zhou (2021) The crystal structure of the FERM and C-terminal domain complex of Drosophila Merlin. Biochemical and Biophysical Research Communications 553, 92-98. [DOI] [PubMed]
- 61.Shimizu T., Seto A., Maita N., Hamada K., Tsukita S., Tsukita S., Hakoshima T. Structural basis for neurofibromatosis type 2. Crystal structure of the merlin FERM domain. Journal of Biological Chemistry. 2002;277:10332–10336. doi: 10.1074/jbc.M109979200. [DOI] [PubMed] [Google Scholar]
- 62.Chinthalapudi K., Rangarajan E.S., Brown D.T., Izard T. Differential lipid binding of vinculin isoforms promotes quasi-equivalent dimerization. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:9539–9544. doi: 10.1073/pnas.1600702113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luft J.R., Collins R.J., Fehrman N.A., Lauricella A.M., Veatch C.K., DeTitta G.T. A deliberate approach to screening for initial crystallization conditions of biological macromolecules. Journal of Structural Biology. 2003;142:170–179. doi: 10.1016/s1047-8477(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 64.Lawrence M.C., Colman P.M. Shape complementarity at protein/protein interfaces. Journal of Molecular Biology. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 65.Yu F.-X., Zhao B., Guan K.-L. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Han H., Qi R., Zhou J.J., Ta A.P., Yang B., Nakaoka H.J., Seo G., Guan K.-L., Luo R., Wang W. Regulation of the hippo pathway by phosphatidic acid-mediated lipid-protein interaction. Molecular Cell. 2018;72:328–340.e8. doi: 10.1016/j.molcel.2018.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Park J.B., Lee C.S., Jang J.-H., Ghim J., Kim Y.-J., You S., Hwang D., Suh P.-G., Ryu S.H. Phospholipase signalling networks in cancer. Nature Reviews Cancer. 2012;12:782–792. doi: 10.1038/nrc3379. [DOI] [PubMed] [Google Scholar]
- 68.Hong A.W., Meng Z., Plouffe S.W., Lin Z., Zhang M., Guan K.-L. Critical roles of phosphoinositides and NF2 in Hippo pathway regulation. Genes & Development. 2020;34:511–525. doi: 10.1101/gad.333435.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Postma M., Roelofs J., Goedhart J., Loovers H.M., Visser A.J., Van Haastert P.J. Sensitization of Dictyostelium chemotaxis by phosphoinositide-3-kinase-mediated self-organizing signalling patches. Journal of Cell Science. 2004;117:2925–2935. doi: 10.1242/jcs.01143. [DOI] [PubMed] [Google Scholar]
- 70.Cai H., Devreotes P.N. Moving in the right direction: how eukaryotic cells migrate along chemical gradients. Seminars in Cell & Developmental Biology. 2011;22:834–841. doi: 10.1016/j.semcdb.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Phair R.D., Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 72.Vonrhein C., Bricogne G. AutoPROC - a framework for automated data processing. Acta Crystallographica Section F: Structural Biology and Crystallization Communications. 2008;64:C78. [Google Scholar]
- 73.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.G. Bricogne, E. Blanc, M. Brandl, C. Flensburg, P. Keller, P. Paciorek, P. Roversi, A. Sharff, O.S. Smart, C. Vonrhein, T.O. Womack (2011) BUSTER version 2.9. Cambridge, United Kingdom: Global Phasing Ltd.
- 75.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica. Section D, Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 76.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta crystallographica Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Davis I.W., Murray L.W., Richardson J.S., Richardson D.C. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Research. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gutmann D.H., Wright D.E., Geist R.T., Snider W.D. Expression of the neurofibromatosis 2 (NF2) gene isoforms during rat embryonic development. Human Molecular Genetics. 1995;4:471–478. doi: 10.1093/hmg/4.3.471. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.