Abstract
The main protease of SARS-CoV-2 is one of the key targets to develop and design antiviral drugs. There is no general agreement on the use of non-steroidal anti-inflammatory drugs (NSAIDs) in COVID-19. In this study, we investigated NSAIDs as potential inhibitors for chymotrypsin-like protease (3CLpro) and the main protease of the SARS-CoV-2 to find out the best candidates, which can act as potent inhibitors against the main protease. We also predicted the effect of NSAIDs on the arachidonic pathway and evaluated the hepatotoxicity of the compounds using systems biology techniques. Molecular docking was conducted via AutoDock Vina to estimate the interactions and binding affinities between selected NSAIDs and the main protease. Molecular docking results showed the presence of 10 NSAIDs based on lower binding energy (kcal/mol) toward the 3CLpro inhibition site compared to the co-crystal native ligand Inhibitor N3 (−6.6 kcal/mol). To validate the docking results, molecular dynamic (MD) simulations on the top inhibitor, Talniflumate, were performed. To obtain differentially-expressed genes under the 27 NSAIDs perturbations, we utilized the L1000 final Z-scores from the NCBI GEO repository (GSE92742). The obtained dataset included gene expression profiling signatures for 27 NSAIDs. The hepatotoxicity of NSAIDs was studied by systems biology modeling of Disturbed Metabolic Pathways. This study highlights the new application of NSAIDs as anti-viral drugs used against COVID-19. NSAIDs may also attenuate the cytokine storm through the downregulation of inflammatory mediators in the arachidonic acid pathway.
Keywords: COVID-19, NSAIDs, SARS-CoV2, Chymotrypsin-like protease, Arachidonic acid pathway, Molecular simulation, Molecular docking
1. Introduction
The incidence and mortality rate of COVID-19 caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have been dramatically on the rise. As of June 5th, 2021, more than 184.5 million cases have been reported worldwide, and the death toll has surpassed 3.993 million (https://www.worldometers.info/coronavirus/). Unfortunately, worldwide attempts have failed to introduce an ultimate therapy in fighting this global crisis, and COVID-19 still lacks specific treatment. Therefore, it is essential to identify new effective antiviral molecules for COVID-19 in a time-critical fashion. Drug repurposing can significantly contribute to the finding of new antiviral drugs and potent new targets. Besides the undeniable economic advantage, drug repurposing is very time-consuming since some clinical trial steps might not be required. Furthermore, drug repurposing possibly introduces the options to be used as combined therapy, leading to higher efficiency [1,2].
In recent decades, the molecular docking approach has been widely used as a computational tool capable of predicting and modeling the binding energies and interaction between macromolecules and receptors at the atomic level [3]. Essentially, the molecular docking technique aims to predict the ligand-receptor complex structure using computation methods. The docking process involves two basic steps: first, predicting the position and orientation of the ligand in the active site of the protein; then, ranking these conformations via a scoring function and assessment of the binding affinity [4]. Inexpensive and fast docking protocols are feasible before accurate but more costly molecular dynamics (MD) simulation to predict the protein-ligand complexes conveniently. In this combined protocol, for improving the drug-design process, docking method is used for the fast screening of large libraries, and MD simulations are then applied to explore conformations of the protein-ligand complexes, calculate accurate energies, and optimize the structures of the final complexes [5,6].
Non-steroidal anti-inflammatory drugs (NSAIDs) are widely used in managing fever and non-rheumatic pains [7]. The reports on the effect of NSAIDs on COVID-19 are limited and have been associated with conflicting results.
It has been claimed that ibuprofen can facilitate infection with COVID-19 in diabetic patients [8]. This possibly occurs through the upregulation of angiotensin-converting enzyme 2 (ACE2), the receptor by which the virus binds to its target cells [8,9]. Some recent papers have studied the antiviral activity of NSAIDs against SARS-CoV-2 protein. For example, they showed that the anti-viral activity of Ketotifen, Indomethacin, and Naproxen could reduce SARS-CoV-2 replication [[10], [11], [12]]. Furthermore, it was shown that indomethacin exerts anti-SARS-CoV activity in both in vivo and in vitro models [13]. This inhibitory effect is suggested to be mediated through inhibition of the viral replication process [13,14]. Several NSAIDs, including aspirin, ibuprofen, naproxen, acetaminophen, and lornoxicam, can also significantly inhibit other viral infections, such as Zika virus Env/HIV-1-pseudotyped viruses [15]. Interestingly, it has been shown that the combination of antiviral drug and ibuprofen decreases the clinical severity of the respiratory syncytial virus (RSV) in a bovine model when administered 3 and 5 days after infection [16].
Several structural and non-structural proteins are recognized in SARS-CoV-2, which can be used in COVID-19 targeted therapy [17]. The main protease of the virus is a key target in the fight against SARS-CoV-2, chymotrypsin-like protease (3CLpro, PDB code: 6LU7). This enzyme plays a crucial role in the viral replication and life cycle. Following entry, the release and un-coating of the incoming genomic RNA subject it to the immediate translation of two large open reading frames, ORF1a and ORF1b, resulting in the synthesis of polyproteins pp1a and pp1ab. A cysteine protease is automatically cleaved from these polyproteins, producing a mature protein. Subsequently, this enzyme cleaves the downstream nonstructural proteins (NSPs), leading to the release of sixteen NSPs from pp1a (nsp1–11) and pp1ab (nsp1–10, nsp12–16). The protease residing in nsp5 is commonly named 3CLpro, due to its similarities with the picornaviral 3C protease. In a different nomenclature, it is considered as the main protease (Mpro) due to its proteolytic processing of the majority of polyprotein cleavage sites [18,19]. 3CLpro is active in dimer form, and a catalytic dyad, CYS145–HIS41, is involved in its proteolytic mechanism [20]. Some residues, such as THR45, MET49, PHE140, ASN142, ASP187, ARG188, GLN189, MET165, HIS172, and GLU166, are believed to mediate substrate-enzyme interactions [20]. Several studies have attempted to find the compounds which specifically target this enzyme. China's National Health Commission has recommended the use of Lopinavir and Ritonavir as protease inhibitors used against HIV in the treatment of COVID-19 [21]. Given the critical role of the enzyme in the management of the viral life cycle and its uniqueness, because no similar enzyme is available in the host cells, 3CLpro is the most attractive target for the treatment of COVID-19.
The existing literature does not provide conclusive evidence for or against the use of NSAIDs in the treatment of COVID-19 patients [22]. Given the possible beneficial effects of NSAIDs on COVID-19 patients, the role of 3CLpro, and the lack of any kind of evidence on the effect of NSAIDs on the main protease of SARS-CoV-2, we attempted to examine the interaction of this group of drugs with the active site of the enzyme using molecular docking analysis and MD simulation. We also predicted the potential effects of NSAIDs on the expression of genes involved in the arachidonic pathway as well as the hepatotoxicity of NSAIDs using metabolic model and gene expression data.
2. Materials and methods
2.1. Molecular docking
Molecular docking was performed to evaluate the inhibitory function of NSAIDs with the SARS-CoV-2 main protease (3CLpro). The structure of 27 NSAIDs was obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). The 3D crystal structure of SARS-CoV-2 3CLpro/(PDB ID; 6LU7) was retrieved in PDB format from the protein data bank (PDB; (https://www.rcsb.org/). The native ligand of 6LU7 (N3) was re-docked as a potential noncovalent inhibitor of SARS-CoV 3CLpro [23] to validate the docking analysis. Furthermore, specific inhibitors against 3CLpro, including Lopinavir and nelfinavir, were used as positive controls [24]. The structure of 3CLpro included water molecules and co-crystal ligands. MGLTOOLS 1.5.6 [25] was applied for preparing protein, removing the native ligand and converting the structure into a PDBQT format, and adding gasteiger partial charges. Self-docking was used for the validation of the docking protocol. This procedure was performed with modified parameters in such a way to result in the root mean square deviations (RMSD) values less than 2 Å. The carbon 19 atom of the ligand with the coordinates of x = −11.993, y = 15.425, and z = 65.951 was considered as the center of the grid with dimensions of 40, 40, 40 in the active site of the enzyme. The exhaustiveness parameter was considered to be 8. The docking simulations were carried out using AutoDock Vina [26] on an 8-cores system. A cluster analysis was conducted on the result of docking poses to determine the ligands for which the molecules show fewer fluctuations in the binding cavity. For this purpose, the pairwise RMSD between 20 first top-ranked poses of each ligand was calculated through bash scripting. In the resulting matrix for each ligand, the average RMSD was calculated for each of the 20 calculated binding modes to afford a vector of the RMSD profile for each ligand across all binding modes. The binding profiles related to all ligands were merged to yield the final data frame before hierarchical cluster analysis. The output heatmap and the dendrogram were illustrated by the R software.
2.2. Molecular dynamics simulation
The molecular dynamics simulation of the best docking pose of Talniflumate in complex with 6LU7 was conducted using the Gromacs simulation package version 5.1, which was run on a Linux GPU server. Amber99SB force field was used to generate force field parameters and define the atom types. Coordinate files and the topology of the molecule were created using ACPYPE program. The complex was solvated by TIP3P water molecules [27] in a cubic box with a minimum distance of 1.0 nm from each wall and then neutralized by adding 0.15 mol/L sodium chloride. The steepest descent algorithm was run to minimize the system. Equilibration of the system was maintained using NVT and NPT with 100 ps in each step via a V-rescale Berendsen thermostat and Parrinello-Rahman, respectively. The temperature was controlled near 300 K, and the pressure of the system was stabilized to 1 atm under the NVT and NPT ensemble, respectively. The periodic boundary simulation based on the particle mesh Ewald (PME) method was applied, and the SHAKE algorithm was used to constrain the covalent bond lengths. The molecular dynamic simulation was performed for 150 ns simulation time using time steps of 2 fs in periodic boundary conditions and on the well-equilibrated system at 300 K and a pressure of 1 atm. Then, RMSD and residue root mean square fluctuation (RMSF) were analyzed to define the equilibrium time and investigate the stability of the protein backbone atoms of each snapshot during MD. The results containing trajectories were finally subjected to VMD to evaluate the binding mode of the ligand during simulation [28]. MM-PBSA (MM-Poisson–Boltzmann surface area) method was applied to calculate the van der Waals, electrostatic, and binding free energies [29].
2.3. Gene expression analysis
To obtain differentially expressed genes under the 27 NSAIDs perturbations, we utilized the L1000 final Z-scores from the NCBI GEO repository (GSE92742)..
2.4. Evaluation of the effects of NSAIDs on the arachidonic acid metabolism pathway
The genes of the arachidonic acid metabolism pathway were extracted from the KEGG (https://www.genome.jp/kegg) database and mapped to the gene expression dataset. There were 50 genes in the arachidonic acid pathway that corresponded to 23 genes in our gene expression dataset. Gene expression Z-score threshold was set to >2.0 and <-2.0 for up- and down-regulated genes, respectively.
2.5. Evaluation of hepatotoxicity of NSAIDs
To evaluate the hepatotoxicity of NSAIDs, we used the method developed by Carbonell et al. [30]. To put it shortly, a metabolic model of human metabolism global reconstruction (Recon 2), MODEL1109130000 [31], was downloaded from the Biomodels database [32]. The model contains reactions annotated with information such as gene-protein-reaction (GPR) associations as Boolean function [33]. By applying GPR rules to up-/downregulation of each gene involved in a reaction flux, the main perturbed reactions under each of the 27 NSAIDs for the liver hepatocyte cells were obtained. Finally, we used the COBRApy package [34] to perform a flux variability analysis (FVA) to determine the distribution of fluxes for each reaction [35]. To get the elasticity parameter for each compound, we integrated the reactions perturbation and reactions sensitivity.
3. Results
3.1. Molecular docking
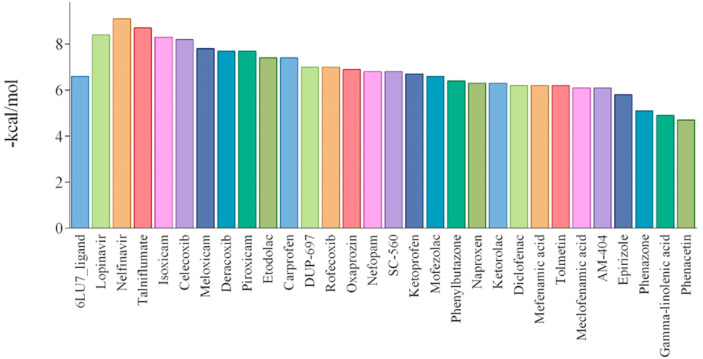
The molecular docking method has been widely used to predict the bioactive compounds or repurpose the drugs against different drug targetable proteins in infections and diseases [36]. This study applied the molecular docking simulation method for the selected NSAIDs to predict their potential inhibitory effect against 3CLpro. The docking results and the binding energy for 3CLpro-NSAIDs complexes are shown in Table 1 and Fig. 1 . All the docked drugs and N3 inhibitor showed significant docking confirmation with binding affinity energies > -6 kcal/mol in the active site of 3CLpro with respect to the N3 inhibitor. The binding energies related to two exhaustiveness 8 and 100 show reasonable convergence (r = 0.95).
Table 1.
Types of NSAID interactions with amino acids involved in enzyme catalysis (SARS-CoV-2 3CLpro).
| CID | Compound | EX-8 | Ex-100 | N-HP | N-HB | N-Cation-π | N-π - π |
|---|---|---|---|---|---|---|---|
| 156391 | Naproxen | −6.3 | −6.8 | 12 | 2 | 0 | 0 |
| 180540 | Ketoprofen | −6.7 | −6.9 | 16 | 5 | 0 | 0 |
| 181817 | Ketorolac | −6.3 | −7.2 | 8 | 2 | 0 | 0 |
| 184147 | Nefopam | −6.8 | −7.8 | 9 | 1 | 0 | 0 |
| 2206 | Phenazone | −5.1 | −6 | 6 | 1 | 0 | 0 |
| 25273599 | Talniflumate | −8.7 | −9.3 | 21 | 2 | 1 | 0 |
| 2662 | Celecoxib | −8.2 | −8.2 | 6 | 3 | 0 | 0 |
| 3033 | Diclofenac | −6.2 | −6.6 | 3 | 3 | 0 | 0 |
| 3058754 | Deracoxib | −7.7 | −8 | 8 | 4 | 0 | 0 |
| 3177 | DUP-697 | −7 | −7.4 | 8 | 1 | 0 | 0 |
| 3242 | Epirizole | −5.8 | −6.7 | 10 | 1 | 0 | 0 |
| 4037 | Meclofenamic acid | −6.1 | −6.9 | 12 | 2 | 0 | 0 |
| 4044 | Mefenamic acid | −6.2 | −7 | 13 | 2 | 0 | 0 |
| 4237 | Mofezolac | −6.6 | −6.9 | 16 | 2 | 0 | 0 |
| 4306515 | SC-560 | −6.8 | −7.1 | 10 | 2 | 0 | 0 |
| 4614 | Oxaprozin | −6.9 | −6.9 | 13 | 2 | 0 | 0 |
| 4754 | Phenacetin | −4.7 | −5.4 | 6 | 0 | 0 | 0 |
| 4781 | Phenylbutazone | −6.4 | −6.9 | 14 | 0 | 0 | 0 |
| 5090 | Rofecoxib | −7 | −7.1 | 14 | 1 | 0 | 0 |
| 5280933 | Gamma-linolenic acid | −4.9 | −5.3 | 16 | 2 | 0 | 0 |
| 54676228 | Piroxicam | −7.7 | −8.1 | 8 | 3 | 0 | 0 |
| 54677470 | Meloxicam | −7.8 | −8.1 | 8 | 2 | 0 | 0 |
| 54677972 | Isoxicam | −8.3 | −8.5 | 9 | 3 | 0 | 1 |
| 5509 | Tolmetin | −6.2 | −6.4 | 8 | 2 | 0 | 0 |
| 6604822 | AM-404 | −6.1 | −6.5 | 28 | 1 | 0 | 0 |
| 688461 | Etodolac | −7.4 | −7.6 | 12 | 2 | 0 | 0 |
| 6997572 | Carprofen | −7.4 | −7.8 | 11 | 4 | 0 | 0 |
CID= Compound ID number.
EX-8 = ΔG (kcal.mol-1 for exhaustiveness 8).
EX-100 = ΔG (kcal.mol-1 for exhaustiveness 100).
N-HP = number of hydrophobic contacts.
N-HB = number of hydrogen bonding.
N-Cation-π = number of cation-π contacts.
N- π - π = number of π - π contacts.
Correlation-coef (EX-8,EX-100) = 0.95.
Fig. 1.
The binding energy of 27 NSAIDs, Lopinavir, Nelfinavir, and the co-crystallized ligand of SARS-CoV-2 3CLpro.
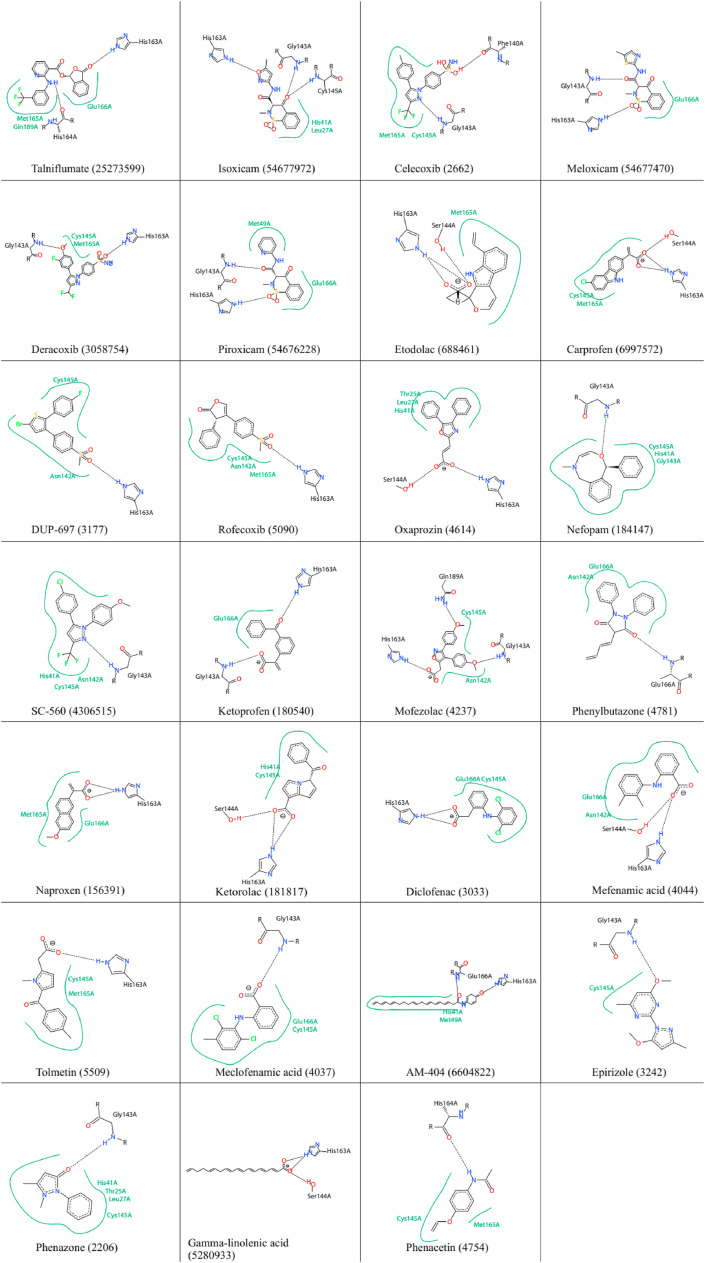
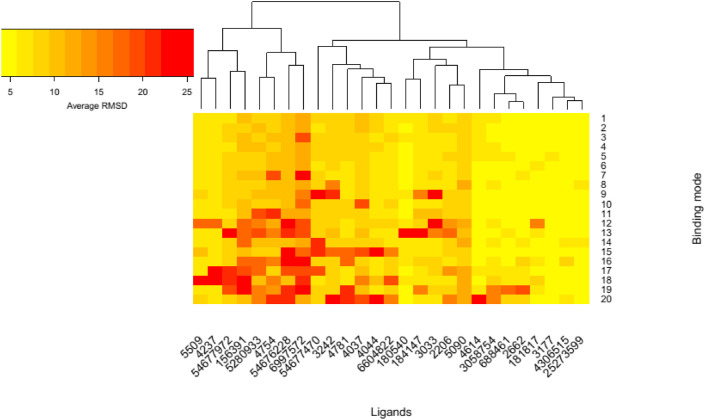
For example, the top 10 of these compounds, based on lower binding energy (kcal/mol), were Talniflumate (−8.7), Isoxicam (−8.3), Celecoxib (−8.2), Meloxicam (−7.8), Deracoxib (−7.7), Piroxicam (−7.7), Etodolac (−7.4), Carprofen (−7.4), DUP-697 (−7) and Rofecoxib (−7). The binding energy is related to different interactions like the number of H-bonds formed with the amino acids in the active site pocket of the 3CLpro [37]. The calculated binding energy values for the native ligand (N3), Nelfinavir, and Lopinavir were −6.6, −9.1, and −8.4, respectively. Docking analysis also revealed the possible H-bonds interactions between the active site amino acids and the drugs (Table 1). The native ligand (N3) formed H-bonds with the residues including HIS164, GLN189, MET165, ASN142, THR26, and ASN142 in the active site of 3CLpro. Nelfinavir formed H-bonds with catalytic residues, CYS145 and HIS41, as well as MET165, GLN189, and GLU166 in the active site of the enzyme. On the other hand, Talniflumate was involved in three H-bonds with the active site residues of 3CLpro (CYS145, ASP187, and MET165). The other control compound, Lopinavir, interacted with catalytic residue, GLU166, MET165, PHE140, and GLN189. Interestingly, the five studied compounds, including Isoxicam (54677972), Nefopam (184147), SC-560 (4306515), Ketorolac (181817), and Phenomazon (2206), formed H-bonds with the catalytic residue. There were also some NSAIDs, which interacted with one of these residues, including Celecoxib (3055754), Derecoxib (3055754), Carprofen (69897572), Oxaprozin (4614), Rofecoxib (5090), Duo-697 (3177), and Diclofenac (30330) (see Fig. 2 ). The output heatmap and the dendrogram of cluster analysis to compare the stability of ligands in the active site of SARS-CoV-2 Mpro are presented in Fig. 3 .
Fig. 2.
Interactions of NSAIDs with amino acids at the substrate site as well as the active site of the SARS-CoV-2 3CLpro enzyme.
Fig. 3.
Heatmap and the dendrogram for the cluster analysis of the ligands in the binding cavity.
3.2. Molecular dynamic simulation analysis
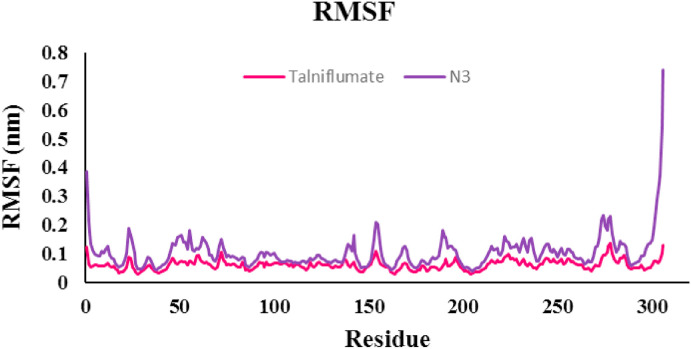
Molecular dynamics simulation was run to obtain more insights into the properties of the interaction of Talniflumate, the top docked ligand, complexed with 3CLpro with respect to co-crystal ligand N3 for 150 ns. From the MD simulation trajectory, the root mean square deviation (RMSD), root mean square fluctuation (RMSF), the number of hydrogen bonds, and clustering were analyzed to check the receptor-ligand conformational properties, such as flexibility and stability. Besides, the molecular mechanics Poisson-Boltzmann surface area (MM-PBSA) method was used to calculate the binding energy of receptor-ligand complexes throughout the whole MD simulation.
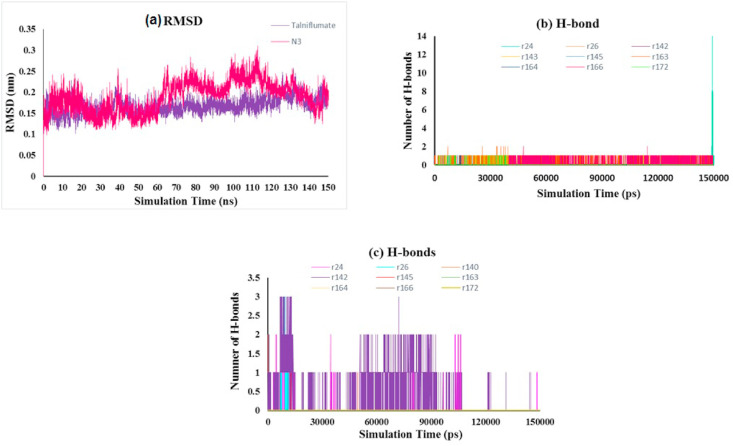
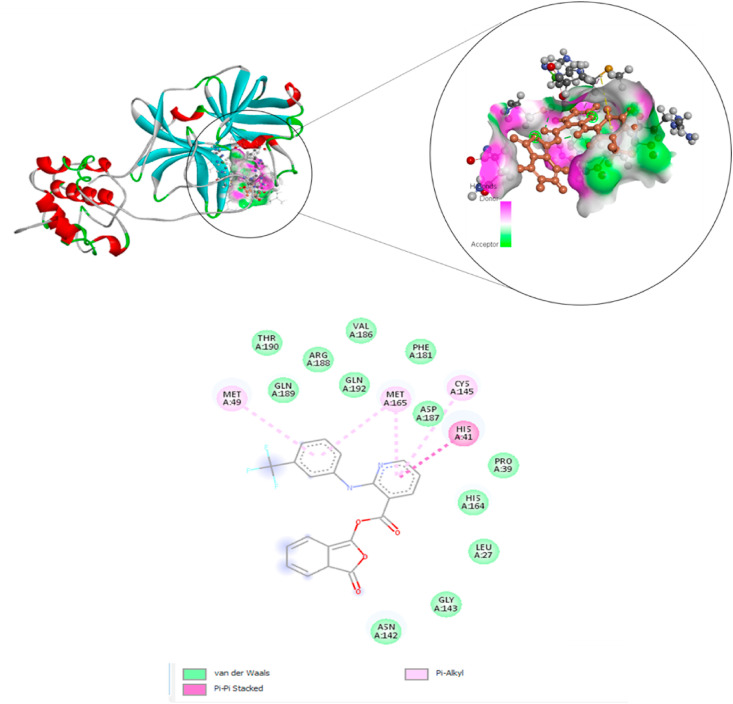
RMSD analysis of the protein backbone residues against the primary conformation was calculated to investigate the stability of the MD simulation system; Fig. 4 (a) displays the plot of RMSD for Talniflumate and N3 in complex with 3CLpro (pdb: 6LU7). Hydrogen bonds play an important role in the stability of protein-ligand complexes. The number of hydrogen bonds formed throughout the equilibrium time range of simulation between amino acid residues of the active site, ligand, and co-crystal ligand was calculated. The number of intermolecular hydrogen bonds in the ligand-protein complex is shown in Fig. 4b and c. The root means square fluctuation (RMSF) of the backbone residue of protein 6LU7 was calculated to investigate the flexibility and vibration of amino acid residues during the equilibrium simulation time. The overall average RMSF values of 3CLpro amino acid residues for Talniflumate and native ligand inhibitor N3 were 0.06 and 0.104 nm, respectively, as shown in Fig. 5 . The MM-PBSA method was carried out to evaluate the binding affinity of Talniflumate and native ligand inhibitor N3 against 3CLpro. Electrostatic (elec), van der Waals (vdW), and binding energies extracted from MM-PBSA calculation for the systems are reported in Table 2 . The obtained results were evaluated by averaging all three replicate simulations. A cluster analysis using the GROMOS method and a cut-off value of 0.15 was applied on trajectories throughout the equilibrium time range to identify the most representative frames. Fig. 6 shows the proposed binding mode and interactions of Talniflumate within the active site of 6LU7.
Fig. 4.
(a) RMSD plot of the protein backbone atoms of the complex, (b) number of hydrogen bonds between protein residues and ligand, (c) number of hydrogen bonds between protein residues and native ligand N3.
Fig. 5.
The root mean square fluctuation (RMSF) of backbone residues of protein (6LU7) in complex with Talniflumate.
Table 2.
Binding free energy, electrostatic, and van der Waals energy of ligand.
| Compound | Binding energy | Electrostatic energy | Van der waals energy | SASA energy | Polar solvation energy |
|---|---|---|---|---|---|
| Talniflumate | −123.382 ± 5.495 | −15.397 ± 0.655 | −202.147 ± 3.560 | −18.1407 ± 0.142 | 110.7627 ± 6.025 |
* Energy are in kilojoules per mole (kJ/mol).
Fig. 6.
The proposed binding mode and interactions of Talniflumate within the active site of 6LU7.
3.3. NSAIDs attenuate cytokine storm by downregulation of prostaglandin E synthase-1 microsomal
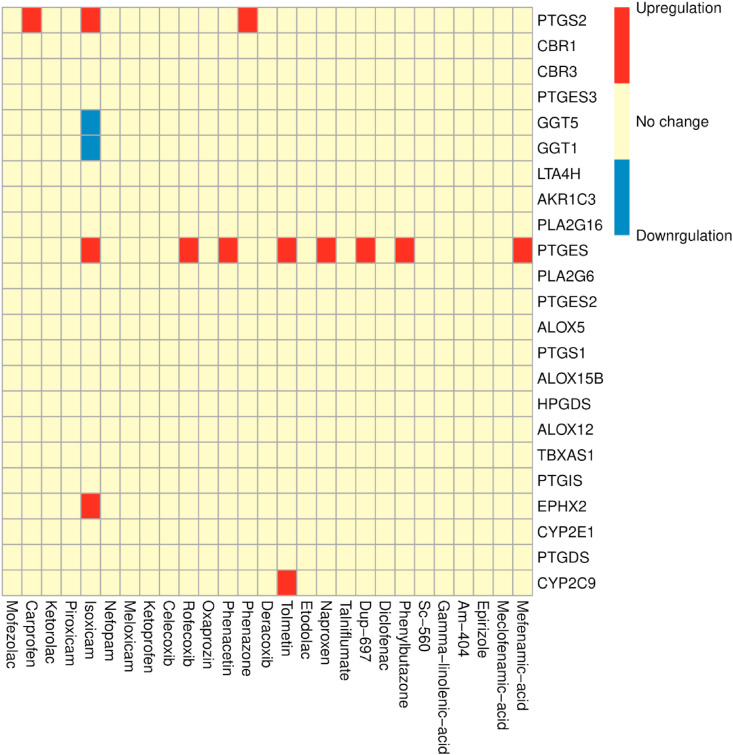
The obtained dataset included gene expression profiling signatures for 27 NSAIDs is provided in Table 3 . NSAIDs can change the expression of genes associated with different prostaglandins, which may lead to some inflammatory issues related to COVID-19. The PTGES gene, which encodes prostaglandin E synthase-1, underwent the most changes under the treatment of NSAIDs. Fig. 7 shows the up-/downregulated patterns of these genes under each of the 27 NSAIDs.
Table 3.
LINCS gene expression profiling signatures for 27 NSAIDs.
| Compound | LINCS gene expression profiling signatures | |
|---|---|---|
| 1 | Mofezolac | CPC012_PHH_24H:BRD-K49372556-001-01-7:10 |
| 2 | Etodolac | CPC014_PHH_24H:BRD-K99260425-001-01-2:10 |
| 3 | Carprofen | CPC015_PHH_24H:BRD-A17411484-001-05-1:10 |
| 4 | Ketorolac | CPC015_PHH_24H:BRD-A40639672-234-09-9:10 |
| 5 | Piroxicam | CPC015_PHH_24H:BRD-A57382968-001-18-3:10 |
| 6 | Isoxicam | CPC015_PHH_24H:BRD-A75552914-001-09-3:10 |
| 7 | Nefopam | CPC015_PHH_24H:BRD-A78877355-001-02-2:10 |
| 8 | Meloxicam | CPC015_PHH_24H:BRD-A84174393-236-03-0:10 |
| 9 | Ketoprofen | CPC015_PHH_24H:BRD-A97739905-001-15-8:10 |
| 10 | Celecoxib | CPC015_PHH_24H:BRD-K02637541-001-06-5:10 |
| 11 | Rofecoxib | CPC015_PHH_24H:BRD-K21733600-001-06-7:10 |
| 12 | Oxaprozin | CPC015_PHH_24H:BRD-K25394294-001-08-1:10 |
| 13 | Phenacetin | CPC015_PHH_24H:BRD-K38323065-001-09-0:10 |
| 14 | Phenazone | CPC015_PHH_24H:BRD-K46937689-001-08-5:10 |
| 15 | Deracoxib | CPC015_PHH_24H:BRD-K68558722-001-03-2:10 |
| 16 | Tolmetin | CPC015_PHH_24H:BRD-K82562631-236-03-8:10 |
| 17 | Naproxen | CPC017_PHH_24H:BRD-A87719232-001-02-4:10 |
| 18 | Talniflumate | CPC017_PHH_24H:BRD-A98378129-001-01-4:10 |
| 19 | Diclofenac | CPC017_PHH_24H:BRD-K08252256-236-17-1:10 |
| 20 | AM-404 | CPC017_PHH_24H:BRD-K21667562-001-01-4:10 |
| 21 | DUP-697 | CPC018_PHH_24H:BRD-K06221026-001-03-5:10 |
| 22 | Phenylbutazone | CPC018_PHH_24H:BRD-K10843433-001-12-8:10 |
| 23 | SC-560 | CPC018_PHH_24H:BRD-K14767410-001-01-5:10 |
| 24 | Gamma-linolenic-acid | CPC018_PHH_24H:BRD-K18059238-001-02-1:10 |
| 25 | Epirizole | CPC018_PHH_24H:BRD-K39339537-001-03-8:10 |
| 26 | Meclofenamic-acid | CPC018_PHH_24H:BRD-K50398167-236-12-8:10 |
| 27 | Mefenamic-acid | CPC018_PHH_24H:BRD-K92778217-001-17-8:10 |
Fig. 7.
Gene expression changes of 23 genes of the arachidonic acid metabolism pathway.
3.4. NSAIDs cause disorders in the metabolism of the hepatocytes
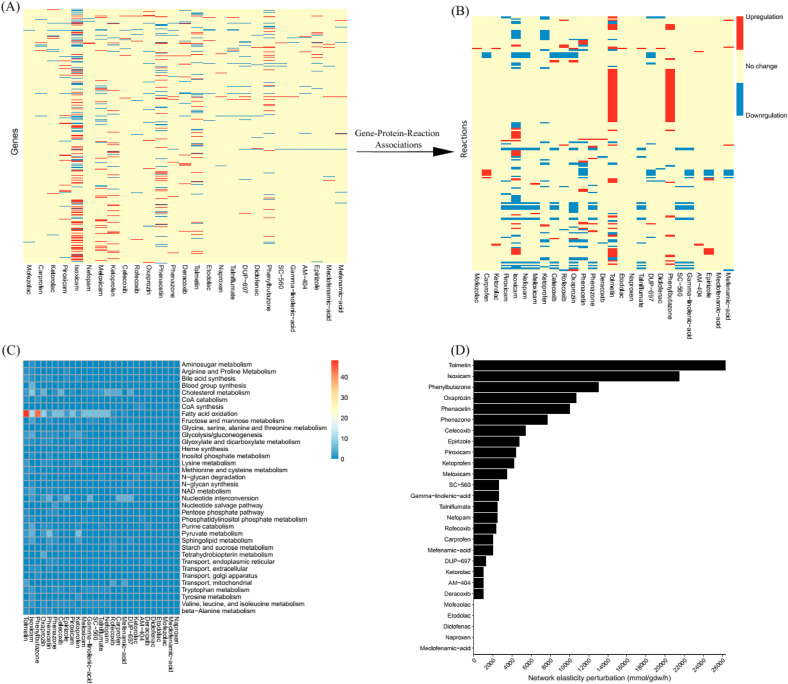
NSAIDs may interfere with the metabolism of the hepatocytes by disrupting some of the reactions in different pathways. Fig. 8 (A, B) shows the perturbed reactions that lead to disruption in different pathways (C). Pathway disruption also leads to perturbation in the metabolic network elasticity (Fig. 8 D).
Fig. 8.
A) Heatmap presents comparison of the up-regulated (red) and down-regulated genes (2936) in the presence of 27 NSAIDs. Mapping GPR rules to the gene regulation results in the second heatmap (B) for the reactions (171). C) Heatmap of altered pathways. Colormap indicates the number of reactions in each pathway. D) Network perturbation results for 27 NSAIDs.
4. Discussion
Due to the severity of the global health pandemic caused by COVID-19, discovering the drugs capable of inhibiting SARS-CoV-2 can be considered as a therapeutic target in the COVID treatment. In this regard, repurposing FDA-approved drugs, such as NSAIDs against COVID-19, can provide therapeutic alternatives which can be used as an effective, safe treatment for COVID-19 [12,38].
Here, using bioinformatics tools, we explored the potential antiviral properties of NSAIDs against the main protease (3CLpro) of the SARS-CoV-2 as well as the impact of NSAIDs on arachidonic acid metabolism pathways and potential hepatotoxicity.
Surprisingly, we showed that NSAIDs acted as inhibitors of the 3CLpro in a comparable potency with Lopinavir and Nelfinavir as the control compounds. To the best of our knowledge, this is the first study exploring the possible role of NSAIDs in inhibiting the viral life cycle. Gene expression analysis conducted on the effect of NSAIDs on arachidonic acid metabolism showed that NSAIDs down-regulated the expression of prostaglandin E synthase, and possibly due to the role of PGE2 in the viral life cycle, it may benefit COVID-19 patients. Finally, we selected some of the NSAIDs with the least hepatotoxic effects.
3CLpro plays a crucial role in the proteolytic maturation of the virus, and it has been considered as a potential key target to stop the viral life cycle [13]. Earlier efforts to target SARS-CoV resulted in the identification of several 3CLpro inhibitors targeting the catalytic dyad of the protein defined by His41 and Cys145 residues [39]. Given the promising outcomes of previous studies, availability of X‐ray crystal structure of the enzyme, and the crucial role in the viral life cycle, 3CLpro has been regarded as the most promising drug target in the fight against the SARS‐CoV‐2 [40]. Fortunately, specific inhibitors against the main protease, such as Lopinavir, Ritonavir, Remdesivir, and Nelfinavir, have created promising results in the fight against the HIV, Ebola virus, Marburg, MERS-CoV, SARS-CoV, respiratory syncytial virus, Nipah virus, and Hendra virus [24,41]. Our investigations on the inhibitory effect of NSAIDs on the main protease of the virus led to a surprising outcome. We showed that Talniflumate, Isoxicam, Celecoxib, Meloxicam, Deracoxib, Piroxicam, Etodolac, Carprofen, and DUP-697 interacted with the active site of the enzyme in comparable binding energy with control compounds, Lopinavir, and Nelfinavir. Interaction with both residues in the catalytic dyad, His41, and Cys145 was observed for Isoxicam, Nefopam, SC-560, Ketorolac, and Phenomazon. Furthermore, Celecoxib, Derecoxib, Carprofen, Oxaprozin, Rofecoxib, Duo-697, and Diclofenac formed H-bond with one of the residues in the active site. This clearly shows the considerable potency of NSAIDs in inhibiting the viral life cycle by blocking the activity of the main protease. In line with the results of our study, other researchers also presented shreds of evidence that NSAIDs can have an inhibitory effect on SARS-CoV-2 proteins. For example, Elmaaty and coworkers evaluated the inhibitory effect of a different set of FDA-approved NSAIDs against the main protease of SARS-CoV-2 using the molecular docking method. The results of the docking study showed that selected NSAID drugs (sulfinpyrazone, indomethacin, and auranofin) were proposed as potential antagonists of main COVID-19 protease with lower binding energy than the Native ligand (N3) with the main protease [42]. On the contrary, in the present study, by using a different set of NSAIDs, some compounds were found with better binding scores in comparison with the native ligand (N3). The top structures of this could be proposed as potential SARS-CoV-2 Mpro inhibitors. In another study, Ki Kwang Oh et al. investigated the inhibitory effect of NSAIDs on three proteins associated with the renin-angiotensin system (RAS). Among twenty NSAIDs, 6MNA, Rofecoxib, and Indomethacin revealed promising binding affinity with the highest docking score to these three identified target proteins. They concluded that 6MNA, Rofecoxib, and Indomethacin are the most potent NSAIDs against COVID-19 [43]. However, in the current study, another target (main protease) was used to evaluate the efficacy of NSAIDs in the SARS-COV2 treatment.
According to the dendrogram and heatmap of cluster analysis depicted in Fig. 3, the yellow color of NSAIDs, including Ketorolac (181817), Duo-697 (3177), SC-560 (4306515), and Talniflumate (25273599), demonstrated that these groups of NSAIDs have minimum fluctuation and more stability in the active site of SARS-CoV-2 Mpro. However, the red color attributed to Phenacetin (4754), Piroxicam (54676228), and Carprofen (6997572) indicates less stability and more fluctuations for these compounds in the active site of the protein.
As shown in Fig. 4 (a), there were acceptable fluctuations of the backbone residue of 3CLpro complexed with Talniflumate and N3 inhibitor until the end of the simulation. The RMSD of the Talniflumate-3CLpro complex reached a plateau form with a minimum fluctuation after nearly 40 ns from the beginning of the simulation. Thus, the ligand was stabilized in the active site of the protein. However, RMSD for the N3 inhibitor complex with 3CLpro showed variation and fluctuation between 60 and 110 ns throughout 150 ns MD simulation time. N3-3CLpro complex also presented a higher deviation of fluctuations due to the binding region of protein throughout the simulation trajectories.
The number of hydrogen bonds (H-bond) was computed to investigate the effect of H-bond in the active site of 3CLpro throughout the 150 ns MD simulations (Fig. 4b and c). The critical amino acids in the active site of 3CLpro were THR24, THR26, PHE140, ASN142, CYS145, IS163, HIS164, GLU166, and HIS172. It has been observed that the native co-crystal ligand N3 has formed H-bonds with critical residue in the active site of 3CLpro. The hydrogen bonds of the Talniflumate-3CLpro complex led to its conformational stability. Results indicated that Talniflumate formed several H-bonds with some critical residue in the active site compared to the native co-crystal ligand during the MD simulation. A maximum number of 14 hydrogen bonds were created between the ligand and THR24 of the active site. According to the results of the RMSF plot (Fig. 5), inconsiderable changes were observed for the involved residue in the active site, and there were no amino acid residues with RMSF value > 0.14 nm. The Talniflumate showed lower RMSF than that of the native co-crystal ligand. These results indicated that the binding of Talniflumate made the protein most flexible in all areas in contrast to the native co-crystal ligand during 150 ns simulation.
According to the results in Table 2, van der Waals energy played the most significant role as hydrophobic interactions in the binding positions of the ligand within the active site of the enzyme (Fig. 6). The first representative frame of the first cluster with the highest population showed the hydrogen bonds, pi-pi, and pi-alkyl interaction for Talniflumate within the active site of 6LU7. Accordingly, Talniflumate also established pi-alkyl interactions with CYS145, MET165, and MET49, and also, one pi-pi interaction with HIS41.
Some reports have addressed the potential mechanisms for the antiviral activity of NSAIDs. The effect of indomethacin as a well-known NSAID on the coronavirus family is studied [13,14]. For example, Indomethacin has been suggested as a potent antiviral drug against both SARS-CoV-2-infected Vero E6 cells in vitro and canine CoV-infected dogs in vivo [44]. A recent study also revealed that sustained-release formulations of Indomethacin could lead to a complete response for the treatment in patients infected by SARS-CoV2 [45,46]. It has shown that Indomethacin does not affect virus infectivity, binding, or entry into the target cells, but it acts early on the coronavirus replication cycle, selectively blocking viral RNA synthesis [14]. Furthermore, The antiviral effect of Indomethacin can be attributed to activation of protein kinase R (PKR) independently of interferons and double-stranded RNA [47], but it might also be attributed to the interaction with aldoketo-reductases, aldose reductases, PPAR-γ (Peroxisome Proliferator-Activated Receptor), and the cannabinoid CB2 receptor. PKR plays an important role in the antiviral defense mechanism acting as a sensor for replication of the virus, and upon activation, leads to eIF2α phosphorylation, and the protein synthesis is blocked in virally infected cells. The broad-spectrum antiviral activity of the Indomethacin depends on PKR and phosphorylation of eIF2α as critical targets [47].
Cytokine storm, overexpressed cytokines, such as interleukin 6 (IL-6), is one of the hallmarks in COVID-19 patients. Naturally, NSAIDs act in a way that they significantly reduce the level of these groups of immune system mediators [48,49]. It has also been shown that NSAIDs interfere with the metabolism of SARS-CoV-2 by preventing the activity of a nonstructural protein of the virus, Nsp7.
NSAIDs inhibit cyclooxygenase (COX) unselectively or selectively and affect the arachidonic metabolism. The anti-inflammatory effect of NSAIDs is not straightforward since prostaglandins like PGE2, PGD2, and prostacyclin (PGI2), which are inhibited by NSAIDs, can both promote and reduce inflammation [50]. On the other hand, the nucleocapsid (N) protein of SARS-CoV can bind directly to the COX-2 promoter and increase its expression, while PGE2 can inhibit replication of SARS-CoV, which is closely related to SARS-CoV-2 [14,51,52]. Gene expression analysis showed that NSAIDs changed the expression pattern of some genes in the arachidonic acid metabolism. Based on the results, the expression level of prostaglandin E synthase (PTGES) experiences a downregulation by Mefenamic acid, Phenylbutazone, DUP-697, Naproxen, Tolmetin, Phenacetin, Rofecoxib, and Isoxicam. PGE2 is hypothesized to be a crucial factor contributing to hyperinflammatory and immune responses of COVID-19 [53]. Interestingly, it has been shown that in different viral infections, PGE2 increases the viral pathogenicity by affecting the host immune system as well as the viral transcription, translation, and/or replication [[54], [55], [56]].
We also aimed to predict the hepatotoxicity of NSAIDs by systems biology modeling of disturbed metabolic pathways using gene expression data [57]. This is a protocol to predict organ-specific toxicity from gene regulation responses in the cells in silico, intending to increase the mechanistic understanding of the toxic effects of compounds. Notably, the highest number of altered reactions was observed in Tolmetin and Isoxicam. However, the lowest quantity of altered reactions was observed for Naproxen, Meclofenamic-acid, Diclofenac, Etodolac, Mofezolac, Deracoxib, AM-404, Ketorolac, DUP-697, Rofecoxib, Nefopam, Talniflumate, and SC-560. Interestingly, these NSAIDs with lower hepatotoxicity could interact with the catalytic dyad of the main protease of SARS-CoV2. This result shows that a group of NSAIDs can be used against SARS-CoV-2 infection with the lowest risk of damage to the patients’ liver.
5. Conclusion
Here, we have discussed the potential of repurposing NSAIDs to inhibit and bind to the active site of the SARS-CoV-2 main protease. The results of molecular docking studies revealed the stability and conformational flexibility of most of these drugs in the active site of the enzyme. Ten of the screened drugs (Talniflumate, Isoxicam, Celecoxib, Meloxicam, Deracoxib, Piroxicam, Etodolac, Carprofen, DUP-697, and Rofecoxib) showed the strongest binding affinities. It seems that NSAIDs can act as a potential therapeutic candidate to relieve COVID-19 by inhibiting the main protease of SARS-CoV-2 through down-regulation of PGE2. Furthermore, the interactions of molecular dynamics simulations of the top of the ten screened drugs-docking studies (Talniflumate) were confirmed by molecular docking analysis as a promising inhibitor of the main protease of SARS-CoV-2. Moreover, we suggest that NSAIDs may be considered by medicinal chemists as the lead compounds for the development of potent SARS-CoV-2 (Mpro) inhibitors. We suggest that these NSAIDs should be assessed further in a prospective clinical study as a treatment solution for COVID-19 patients.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
References
- 1.Rosa S.G.V., Santos W.C. Clinical trials on drug repositioning for COVID-19 treatment. Rev. Panam. Salud Públic. 2020;44 doi: 10.26633/RPSP.2020.40. e40-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mercorelli B., Palu G., Loregian A. Drug repurposing for viral infectious diseases: how far are we? Trends Microbiol. 2018;26(10):865–876. doi: 10.1016/j.tim.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lianingsih F., editor. Silico Analysis of Sponges Compound against Mpro COVID-19: A Review. Proceeding International Conference on Science and Engineering. 2021. [Google Scholar]
- 4.Meng X.-Y., Zhang H.-X., Mezei M., Cui M. Molecular docking: a powerful approach for structure-based drug discovery. Curr. Comput. Aided Drug Des. 2011;7(2):146–157. doi: 10.2174/157340911795677602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abdelsattar A.S., Dawoud A., Helal M.A. Interaction of nanoparticles with biological macromolecules: a review of molecular docking studies. Nanotoxicology. 2021;15(1):66–95. doi: 10.1080/17435390.2020.1842537. [DOI] [PubMed] [Google Scholar]
- 6.Alonso H., Bliznyuk A.A., Gready J.E. Combining docking and molecular dynamic simulations in drug design. Med. Res. Rev. 2006;26(5):531–568. doi: 10.1002/med.20067. [DOI] [PubMed] [Google Scholar]
- 7.Micallef J., Soeiro T., Jonville-Béra A.-P. Non-steroidal anti-inflammatory drugs, pharmacology, and COVID-19 infection. Therapie. 2020:355–362. doi: 10.1016/j.therap.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang L., Karakiulakis G., Roth M. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet Respir. Med. 2020;8(4) doi: 10.1016/S2213-2600(20)30116-8. e21-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;(7):94. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kiani P., Scholey A., Dahl T.A., McMann L., Iversen J.M., Verster J.C. Vitro assessment of the antiviral activity of ketotifen, indomethacin and naproxen, alone and in combination, against SARS-CoV-2. Viruses. 2021;13(4):558. doi: 10.3390/v13040558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Terrier O., Dilly S., Pizzorno A., Henri J., Berenbaum F., Lina B., et al. Broad-spectrum antiviral activity of naproxen: from Influenza A to SARS-CoV-2 Coronavirus. BioRxiv. 2020 doi: 10.1101/2020.04.30.069922. [DOI] [Google Scholar]
- 12.Terrier O., Dilly S., Pizzorno A., Chalupska D., Humpolickova J., Bouřa E., et al. Antiviral properties of the NSAID drug naproxen targeting the nucleoprotein of SARS-CoV-2 coronavirus. Molecules. 2021;26(9):2593. doi: 10.3390/molecules26092593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu T., Gao X., Wu Z., Selinger D.W., Zhou Z. Indomethacin has a potent antiviral activity against SARS CoV-2 in vitro and canine coronavirus in vivo. bioRxiv. 2020 doi: 10.1101/2020.04.01.017624. [DOI] [Google Scholar]
- 14.Amici C., Di Caro A., Ciucci A., Chiappa L., Castilletti C., Martella V., et al. Indomethacin has a potent antiviral activity against SARS coronavirus. Antivir. Ther. 2006;11(8):1021–1030. [PubMed] [Google Scholar]
- 15.Pan T., Peng Z., Tan L., Zou F., Zhou N., Liu B., et al. Nonsteroidal anti-inflammatory drugs potently inhibit the replication of Zika viruses by inducing the degradation of AXL. J. Virol. 2018;92(20) doi: 10.1128/JVI.01018-18. e01018–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walsh P., Lebedev M., McEligot H., Mutua V., Bang H., Gershwin L.J. A randomized controlled trial of a combination of antiviral and nonsteroidal anti-inflammatory treatment in a bovine model of respiratory syncytial virus infection. PloS One. 2020;(3):15. doi: 10.1371/journal.pone.0230245. e0230245-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., O’Meara M.J., et al. A SARS-CoV-2-human protein-protein interaction map reveals drug targets and potential drug-repurposing. nature. 2020;(583):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2020:1–16. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y., Tang L.V. Overview of targets and potential drugs of SARS-CoV-2 according to the viral replication. J. Proteome Res. 2021;20(1):49–59. doi: 10.1021/acs.jproteome.0c00526. [DOI] [PubMed] [Google Scholar]
- 20.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300(5626):1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 21.Chen Z.M., Fu J.F., Shu Q., Chen Y.H., Hua C.Z., Li F.B., et al. Diagnosis and treatment recommendations for pediatric respiratory infection caused by the 2019 novel coronavirus. World J. Pediatr. 2020;(16):240–246. doi: 10.1007/s12519-020-00345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Russell B., Moss C., Rigg A., Van Hemelrijck M. COVID-19 and treatment with NSAIDs and corticosteroids: should we be limiting their use in the clinical setting? Ecancermedicalscience. 2020;14:1023. doi: 10.3332/ecancer.2020.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khaerunnisa S., Kurniawan H., Awaluddin R., Suhartati S., Soetjipto S. 2020. Potential Inhibitor of COVID-19 Main Protease (Mpro) from Several Medicinal Plant Compounds by Molecular Docking Study; pp. 1–14. Prepr doi10 20944/preprints202003 0226 v1. [Google Scholar]
- 24.Lin S., Shen R., He J., Li X., Guo X. Molecular modeling evaluation of the binding effect of Ritonavir, Lopinavir and darunavir to severe acute respiratory syndrome coronavirus 2 proteases. bioRxiv. 2020 doi: 10.1101/2020.01.31.929695. [DOI] [Google Scholar]
- 25.Morris G., Huey R., Olson A. Current Protocols in Bioinformatics. John Wiley & Sons; Malden, MA: 2008. Using AutoDock for ligand-receptor docking; p. 14. [DOI] [PubMed] [Google Scholar]
- 26.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jorgensen W.L., Madura J.D., Swenson C.J. Optimized intermolecular potential functions for liquid hydrocarbons. J. Am. Chem. Soc. 1984;106(22):6638–6646. [Google Scholar]
- 28.Humphrey W., Dalke A., Schulten K.V.M.D. Visual molecular dynamics. J. Mol. Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 29.Kumari R., Kumar R., Consortium OSDD. Lynn A. g_mmpbsa- A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014;54(7):1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 30.Carbonell P., Lopez O., Amberg A., Pastor M., Sanz F. Hepatotoxicity prediction by systems biology modeling of disturbed metabolic pathways using gene expression data. ALTEX-Alternatives to animal experimentation. 2017;34(2):219–234. doi: 10.14573/altex.1602071. [DOI] [PubMed] [Google Scholar]
- 31.Thiele I., Swainston N., Fleming R.M., Hoppe A., Sahoo S., Aurich M.K., et al. A community-driven global reconstruction of human metabolism. Nat. Biotechnol. 2013;31(5):419. doi: 10.1038/nbt.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Juty N., Ali R., Glont M., Keating S., Rodriguez N., Swat M., et al. BioModels: content, features, functionality, and use. CPT Pharmacometrics Syst. Pharmacol. 2015;4(2):55–68. doi: 10.1002/psp4.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hucka M., Finney A., Sauro H.M., Bolouri H., Doyle J.C., Kitano H., et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19(4):524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 34.Ebrahim A., Lerman J.A., Palsson B.O., Hyduke D.R. COBRApy: COnstraints-based reconstruction and analysis for python. BMC Syst. Biol. 2013;7(1):74. doi: 10.1186/1752-0509-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orth J.D., Thiele I. Palsson BØ. What is flux balance analysis? Nat. Biotechnol. 2010;28(3):245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bharadwaj S., Lee K.E., Dwivedi V.D., Yadava U., Panwar A., Lucas S.J., et al. Discovery of Ganoderma lucidum triterpenoids as potential inhibitors against Dengue virus NS2B-NS3 protease. Sci. Rep. 2019;9(1):1–12. doi: 10.1038/s41598-019-55723-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Politi A., Durdagi S., Moutevelis-Minakakis P., Kokotos G., Mavromoustakos T. Development of accurate binding affinity predictions of novel renin inhibitors through molecular docking studies. J. Mol. Graph. Model. 2010;29(3):425–435. doi: 10.1016/j.jmgm.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 38.Abdelgawad M., Allam S., Shaheen M.A., Ali M., Hussein H.A.E., Gaber A.A., et al. 2021. An Overview of COVID-19 Treatment: Possible Candidates Based on Drug Repurposing and Molecular Docking. [Google Scholar]
- 39.Paasche A., Zipper A. Schäfer S, Ziebuhr J, Schirmeister T, Engels B. Evidence for substrate binding-induced zwitterion formation in the catalytic Cys-His dyad of the SARS-CoV main protease. Biochemistry. 2014;53(37):5930–5946. doi: 10.1021/bi400604t. [DOI] [PubMed] [Google Scholar]
- 40.Park J.Y., Ko J.A., Kim D.W., Kim Y.M., Kwon H.J., Jeong H.J., et al. Chalcones isolated from Angelica keiskei inhibit cysteine proteases of SARS-CoV. J. Enzym. Inhib. Med. Chem. 2016;31(1):23–30. doi: 10.3109/14756366.2014.1003215. [DOI] [PubMed] [Google Scholar]
- 41.Mothay D., Ramesh K.V. Binding site analysis of potential protease inhibitors of COVID-19 using AutoDock. VirusDisease. 2020;31(2):194–199. doi: 10.1007/s13337-020-00585-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elmaaty A., Hamed M., Ismail M., Elkaeed E., Abulkhair H., Khattab M., et al. Computational insights on the potential of some NSAIDs for treating COVID-19: priority set and lead optimization. Molecules. 2021;26:3772. doi: 10.3390/molecules26123772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oh K.K., Adnan M., Cho D.H. Network pharmacology approach to decipher signaling pathways associated with target proteins of NSAIDs against COVID-19. Sci. Rep. 2021;11(1):1–15. doi: 10.1038/s41598-021-88313-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu T., Gao X., Wu Z., Selinger D.W., Zhou Z. Indomethacin has a potent antiviral activity against SARS CoV-2 in vitro and canine coronavirus in vivo. BioRxiv. 2020 doi: 10.1101/2020.04.01.017624. [DOI] [Google Scholar]
- 45.Gomeni R., Xu T., Gao X., Bressolle-Gomeni F. Model based approach for estimating the dosage regimen of indomethacin a potential antiviral treatment of patients infected with SARS CoV-2. J. Pharmacokinet. Pharmacodyn. 2020;47:189–198. doi: 10.1007/s10928-020-09690-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koch G., Schönfeld N., Jost K., Atkinson A., Schulzke S.M., Pfister M., et al. Caffeine preserves quiet sleep in preterm neonates. Pharmacol. Res. Perspect. 2020;8(3) doi: 10.1002/prp2.596. e00596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amici C., La Frazia S., Brunelli C., Balsamo M., Angelini M., Santoro M.G. Inhibition of viral protein translation by indomethacin in vesicular stomatitis virus infection: role of e if 2α kinase pkr. Cell Microbiol. 2015;17(9):1391–1404. doi: 10.1111/cmi.12446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen L., Liu H.G., Liu W., Liu J., Liu K., Shang J., et al. [Analysis of clinical features of 29 patients with 2019 novel coronavirus pneumonia] Zhonghua Jiehe He Huxi Zazhi. 2020;43 doi: 10.3760/cma.j.issn.1001-0939.2020.0005. 0. E005. [DOI] [PubMed] [Google Scholar]
- 49.Yan Y., Guo T.M., Zhu C. Effects of nonsteroidal anti-inflammatory drugs on serum proinflammatory cytokines in the treatment of ankylosing spondylitis. Biochem. Cell. Biol. 2018;96(4):450–456. doi: 10.1139/bcb-2017-0267. [DOI] [PubMed] [Google Scholar]
- 50.Bergqvist F., Carr A., Wheway K., Watkins B., Oppermann U., Jakobsson P., et al. 2018. Prostacyclin: a Potential Novel Therapeutic Target to Treat Tendon Pain and Inflammation. [Google Scholar]
- 51.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yan X., Hao Q., Mu Y., Timani K.A., Ye L., Zhu Y., et al. Nucleocapsid protein of SARS-CoV activates the expression of cyclooxygenase-2 by binding directly to regulatory elements for nuclear factor-kappa B and CCAAT/enhancer binding protein. Int. J. Biochem. Cell Biol. 2006;38(8):1417–1428. doi: 10.1016/j.biocel.2006.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Smeitink J., Jiang X., Pecheritsyna S., Renkema H., van Maanen R., Beyrath J. preprints.org; 2020. Hypothesis: mPGES-1-Derived prostaglandin E2, a So far Missing Link in COVID-19 pathophysiology? [Google Scholar]
- 54.Kalinski P. Regulation of immune responses by prostaglandin E2. J. Immunol. 2012;188(1):21–28. doi: 10.4049/jimmunol.1101029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sander W.J., O'Neill H.G., Pohl C.H. Prostaglandin E2 as a modulator of viral infections. Front. Physiol. 2017;8:89. doi: 10.3389/fphys.2017.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mocarski E.S. Virus self-improvement through inflammation: no pain, no gain. Proc. Natl. Acad. Sci. Unit. States Am. 2002;99(6):3362–3364. doi: 10.1073/pnas.072075899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carbonell P., Lopez O., Amberg A., Pastor M., Sanz F. Hepatotoxicity prediction by systems biology modeling of disturbed metabolic pathways using gene expression data. ALTEX. 2017;34(2):219–234. doi: 10.14573/altex.1602071. [DOI] [PubMed] [Google Scholar]