Abstract
Background:
Acute rejection, which includes antibody-mediated rejection and acute cellular rejection, is a risk factor for lung allograft loss. Lung transplant patients often undergo surveillance transbronchial biopsies to detect and treat acute rejection before irreversible chronic rejection develops. Limitations of this approach include it’s invasiveness and high interobserver variability. We tested the performance of percent donor-derived cell-free DNA (%ddcfDNA), a non-invasive blood test, to detect acute rejection.
Methods:
This multicenter cohort study monitored 148 lung transplant subjects over a median of 19.6 months. We collected serial plasma samples contemporaneously with TBBx to measure %ddcfDNA. Clinical data was collected to adjudicate for acute rejection. The primary analysis consisted of computing the area-under-the-receiver-operating-characteristic-curve (AUROC) of %ddcfDNA to detect acute rejection. Secondary analysis determined %ddcfDNA rule-out thresholds for acute rejection.
Results:
ddcfDNA levels were high after transplant surgery and decayed logarithmically. With acute rejection, ddcfDNA levels rose six-fold higher than controls. ddcfDNA levels also correlated with severity of lung function decline and histological grading of rejection. %ddcfDNA AUROC for acute rejection, AMR, and ACR were 0.89, 0.93, and 0.83, respectively. ddcfDNA levels of <0.5% and < 1.0% showed a negative predictive value of 96% and 90% for acute rejection, respectively. Histopathology detected one-third of episodes with ddcfDNA levels ≥ 1.0%, even though > 90% of these events were coincident to clinical complications missed by histopathology.
Conclusions:
This study demonstrates that %ddcfDNA reliably detects acute rejection and other clinical complications potentially missed by histopathology, lending support to its use as a non-invasive marker of allograft injury.
Keywords: early diagnosis, rejection, cell-free DNA
Introduction
After transplantation, the lung allograft is subject to injury from acute rejection (including acute cellular rejection, (ACR)(1) and antibody-mediated rejection (AMR)(2, 3)), infections and other complications. Among these, acute rejection remains a major risk factor for chronic lung allograft dysfunction (CLAD) and early death.(4–7) Early detection and treatment of acute rejection may prevent allograft dysfunction. Therefore, lung transplant patients often undergo surveillance bronchoscopy and transbronchial biopsy (TBBx) to obtain allograft tissue for histopathology and aid in the diagnosis of acute rejection. Transbronchial biopsy uses blind tissue sampling that often results in an inadequate sample for histopathology, which itself has high interobserver variability.(8) This approach exposes patients to the risks inherent to anesthesia and invasive procedures (e.g., bleeding and pneumothoraces), as well as communicable diseases such as coronavirus disease 2019. These risks and limitations call for better diagnostic and monitoring approaches.
The high sensitivity of genomic sequencing provides an opportunity to overcome the limitations of histopathology, the current gold standard for detecting acute rejection. When cells die, short cell-free DNA (cfDNA) fragments are released into circulation. In transplantation, the allograft releases donor-derived cell-free DNA (ddcfDNA) into the recipient’s circulation. Owing to the vast numbers of single nucleotide polymorphisms (SNPs) between the donor and recipient genomes, ddcfDNA can be easily and reproducibly quantified in the recipient’s plasma.(9–13)
Prior studies indicate that levels of percent ddcfDNA (%ddcfDNA) increase in the setting of acute rejection. (14–17) We have since shown that not only is using shotgun-sequencing-mediated %ddcfDNA reliable and reproducible,(9) but it also detects AMR earlier than histopathology.(18) This study, which utilizes the Genomic Research Alliance for Transplantation (GRAfT)(19) cohort, evaluated the performance of %ddcfDNA to detect acute rejection after lung transplantation.
Methods
Study design, setting, and participants
This multicenter prospective cohort study (ClinicalTrials.gov identifier NCT02423070) was supported by the GRAfT consortium, which includes the National Heart, Lung, and Blood Institute (NHLBI), and five hospitals in the Washington, DC metropolitan area.(19) Between 2015 and the present, three of the five hospitals (Johns Hopkins, Inova, University of Maryland) enrolled lung transplant waitlist patients who were at least 18 years old. After transplantation, subjects underwent surveillance transbronchial biopsy (TBBx) with bronchoalveolar lavage (BAL) to monitor for acute rejection and the presence of pathogens at months 1,3, 6, 12, 18 and 24 post-transplant (Suppl. Table 1). Patients also underwent routine post-transplant testing including for donor specific antibody (DSA) and pulmonary function. The study collected serial plasma samples and clinical data in the early post-transplant period on Days 1, 3, 7, 14, 21, and at the time of all bronchoscopies and when patients presented with signs or symptoms of allograft dysfunction. The collection schedule, induction and maintenance immunosuppression regimen were reported previously.(18, 20) Plasma samples were used to measure the levels of ddcfDNA. A committee of lung transplant providers reviewed relevant clinical data to adjudicate for acute rejection, the composite primary endpoint of ACR and AMR, as well as other relevant clinical endpoints. Patients were excluded from this analysis if they were included in a prior analysis(18) or died within 30 days of transplantation. We used a calculation of the area under the receiver operator characteristic curve (AUROC) to assess the performance of %ddcfDNA to detect acute rejection. Only data within the first two years after transplantation were included, as participating centers limited use of surveillance bronchoscopy to this period. The study was approved by institutional review boards at each institution and is reported following accepted standards. (21)
Clinical data and study endpoints
The primary study endpoints were treatable categories of acute rejection at participating centers, including clinical AMR, ACR of grade 2 or higher, and ACR grade 1 accompanied by allograft dysfunction. These endpoints were pre-specified by the GRAfT Steering Committee and adjudicated by a committee blinded to %ddcfDNA data. To be consistent with usual care practices, endpoints were adjudicated using center data, rather than consensus data (Supplementary Table 2), following adjudication protocols(18, 20) and internationally-accepted definitions.(1,22, 23) To reduce heterogeneity inherent to standard definitions of AMR,(22) only categories associated with allograft dysfunction were included. Allograft dysfunction was defined based on spirometry alone(18, 20) due to inconsistencies in provider documentation of signs/symptoms. Allograft dysfunction was defined as ≥ 10% FEV1 (forced expiratory volume in the first second) decline, and categorized as “no” (< 10%), “mild” (≥ 10% to < 15%), or “moderate to severe” (≥ 15%) allograft dysfunction. Pathogens were defined as any organism identified on bronchoalveolar lavage and further categorized as being associated with abnormal histopathology or not or being associated to a decline in pulmonary function test (PFT) or not. Endpoints were paired with %ddcfDNA levels drawn on the day that the endpoint (biopsy, BAL, PFT) occurred. Time points with normal histopathology, spirometry, and microbiological tests were used as no-rejection controls. Time points with incomplete data were excluded in the primary analysis.
Measurement of %ddcfDNA
Automated shotgun sequencing(9) was used. First, we genotype both the donor and recipient to identify distinguishing SNPs. After transplantation, recipient’s plasma cfDNA was isolated for library construction and paired-end shotgun sequencing (Illumina HiSeq 3000, 2X 50 bp). We assigned sequence reads as donor or recipient using the distinguishing SNPs identified by genotyping. The %ddcfDNA was calculated as a quotient of the number of donor reads to the number of donor plus recipient reads. Values for single lung transplants were doubled to relative lung mass of double lung transplants.
Sample size
The sample size was computed based on equations (6.2) and (6.5) from Zhou, Obuchowski and McClish.(24) The number of subjects with acute rejection, N, was determined using N = (3.84 * V) / L2, where V is variance of the AUROC and 2L is the length of the 95% confidence interval. The reported %ddcfDNA AUROC for different grades of acute rejection vary from 0.76 to 0.9,(15) so ~12 acute rejection episodes will correspond to a 95% confidence interval of length 2L = 0.25 when the AUROC = 0.76 (assuming the lower limit of reported AUROC(15)). Considering that at least 20% of the lung transplant subjects will have an acute rejection episode, we concluded that 60 subjects would be a reasonable sample size.
Statistical Analyses
Computation of the AUROC was used to test the performance of %ddcfDNA to detect acute rejection. To do this, we first assessed the post-transplant %ddcfDNA decay kinetics assuming a logarithmic decay.(15) We calculated the AUROC at several benchmark times (days 14, 21, 28, 45, 60, 90, 120, 180), which correspond to routine patient follow-ups. For example, to determine the AUROC for day 45 (1.5 months), data before day 45 was eliminated for the AUROC analysis. The dataset with the highest AUROC was selected for downstream analyses. The %ddcfDNA data were log transformed [log2(x + 0.1)] to normalize for a skewed distribution. The normalized values are reported in the manuscript. We also compared the log-transformed %ddcfDNA data between groups using a generalized estimating equation approach which accounted for repeated measures of ddcfDNA levels from individual subjects.
We then established the performance of a low rule-out and a high rule-in %ddcfDNA using 1.0% as the high rule-in threshold based on a reported sensitivity of 100% to detect high grade ACR in a prior study.(15) We arbitrarily selected 0.5%, half of 1.0%, as the low rule-out threshold. Given the known limitations of histopathology, we also performed a head-to-head comparison of %ddcfDNA against histopathology using a level of ddcfDNA > 1.0% as a positive %ddcfDNA test.
Results
Study population
182 waitlisted transplant patients were eligible for inclusion in the study. Of these, 7 declined participation, 23 patients were not transplanted and 4 died within 1 month of transplant, leaving 148 subjects included in the final analysis. (Figure 1) The average age was 57 years (range = 18–74) and average lung allocation score was 47 (range = 32–89). Interstitial lung disease was the most common reason for transplantation, and a third of transplant recipients received single lung transplants. The average donor age was 33 years (range = 17–61) and about half (44%) of donors died from head trauma (Table 1).
Figure 1. Flowchart of study design.
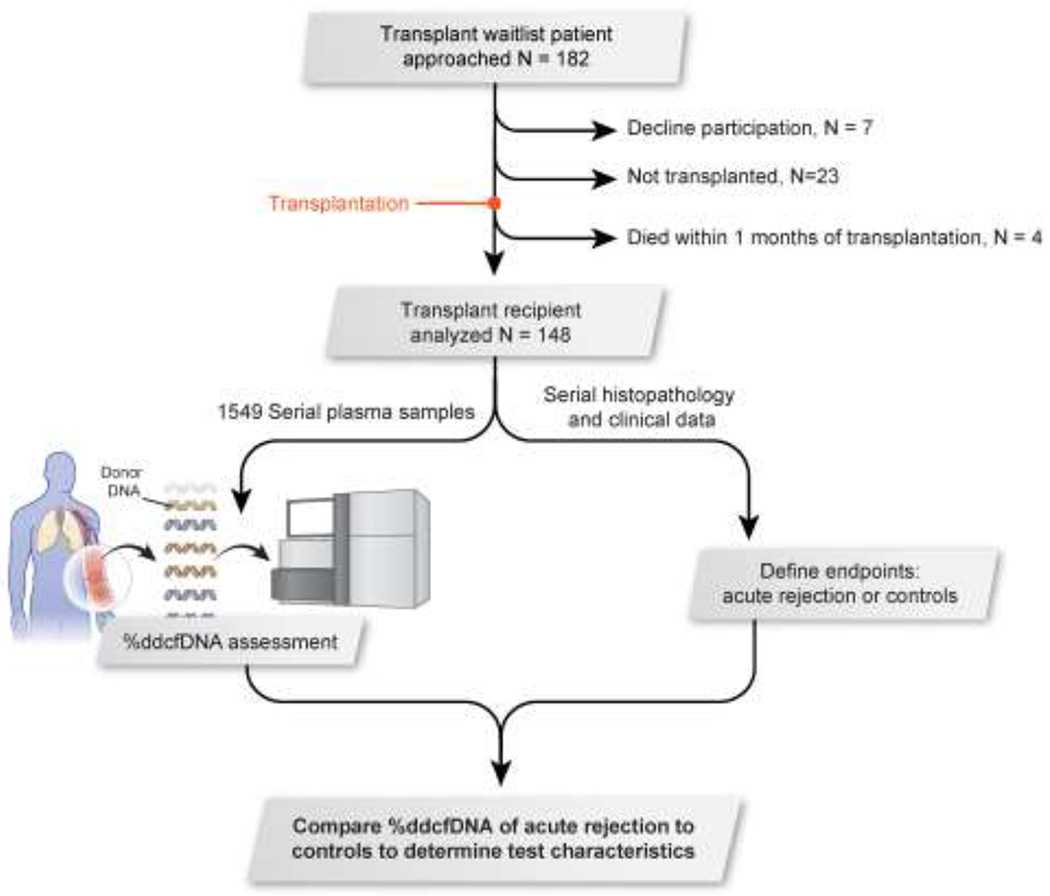
Prospective transplant patients were approached (n = 182). Subjects were excluded if they declined to participate (n = 7), did not undergo a transplantation (n = 23), or died within one month of transplantation (n = 4). Transplant patients who were analyzed (n = 148) underwent a %ddcfDNA assessment (via serial plasma samples) as well as had clinical data, including histopathology, collected to bin them into either the “acute rejection” or “control” endpoint. %ddcfDNA from “controls” and “acute rejection” subjects were compared to determine performance of %ddcfDNA, levels of donor-derived cell-free DNA.
Table 1.
Descriptive demographic variables.
| Recipient Factors (n = 148) | ||
| Demographics | ||
| Mean Age (Range) | 57 (18-74) | |
| Sex % (n) | ||
| Male | 47 (70) | |
| Female | 53 (78) | |
| Race % (n) | ||
| Black or African American | 18 (27) | |
| White | 79 (117) | |
| American Indian or Alaskan Native | 1 (1) | |
| Asian | 1 (1) | |
| Other | 1 (2) | |
| History of Smoking % (n) | ||
| Never | 60 (89) | |
| Past | 40 (59) | |
| BMI% (n) | ||
| BMI<30 | 74 (110) | |
| BMI >=30 | 26 (38) | |
| Transplantation Reason % (n) | ||
| COPD | 17 (25) | |
| CF | 11 (16) | |
| IPF | 34 (50) | |
| IPAH | 1 (1) | |
| Sarcoid | 6 (9) | |
| Other | 31 (46) | |
| Transplant LAS score | ||
| Mean (range) | 47 (32 – 89) | |
| Donor Factors | ||
| Demographics | ||
| Mean Age (Range) | 33 (17-61) | |
| Sex % (n) | ||
| Male | 69 (102) | |
| Female | 31 (46) | |
| Race % (n) | ||
| Black or African American | 24 (36) | |
| Caucasian/European American | 63 (93) | |
| Asian | 6 (9) | |
| Other | 7 (10) | |
| History of Smoking % (n) | ||
| Yes | 14 (24) | |
| No | 84 (124) | |
| History of Chest Trauma % (n) | ||
| Yes | 6 (9) | |
| No | 94 (139) | |
| Donor Cause of Death % (n) | ||
| Anoxia | 37 (55) | |
| CVA | 15 (22) | |
| Head Trauma | 44 (65) | |
| Other | 2(3) | |
| BMI % (n) | ||
| BMI<30 | 77 (114) | |
| BMI >=30 | 23 (34) | |
| Single/Double Lung % (n) | ||
| Single | 29 (43) | |
| Double | 71 (105) | |
Sampling and study endpoints
Over the median 19.6-months follow-up (IQR = 11.7–26.3), 654 biopsies were obtained for histopathology, including 651 transbronchial and 3 wedge biopsies. To account for %ddcfDNA decay, we eliminated histopathologies performed before day 45 or TBBx that lacked allograft tissue for histopathology, leaving 484 samples. Of these, 77.9% (n = 377) showed no abnormal findings, while 16.3% (n = 79) showed ACR, the most commonly detected abnormal histopathology finding. Of the ACR findings, 26.6% (n = 21) were mixed AMR/ACR. The remaining 58 ACR episodes without AMR were either of grade 1 (n = 35) or grade ≥ 2 (n = 23) Suppl. Figure 1. Pulmonary parenchyma ACR, termed “A-grade”, was more commonly detected (n = 38, 65.5%) than airway inflammation, termed “B-grade” (n = 14, 24.1%), or mixed A and B-subtypes (n = 6, 10.3%).
The primary endpoint of acute rejection was detected in 87 episodes, a composite endpoint that include the rejection categories treated at participating centers. This included ACR grade 1 accompanied by allograft dysfunction (n = 7), ACR grade ≥ 2 with and without allograft dysfunction (n = 23) and clinical AMR (n = 57). All AMR episodes showed positive DSA (HLA DSA = 55, non-HLA DSA = 2). Two-thirds of AMR episodes (67.2%) showed positive histopathology, of which ACR was the most common finding (n = 21). C4d, an immunohistochemical marker associated to AMR(25), was detected in only four of the AMR episodes (6.6%). “Possible” clinical AMR was twice as common as “probable/definite” AMR.
Half of respiratory samples sent for testing (n = 328 / 627) from 45 days and onwards had a pathogen detected. From these, 9.1% of the cases where a pathogen was detected were concurrent with abnormal histopathology findings, while 23.8% were concurrent with a decline in FEV1 ≥ 10%. Further details of study endpoints is provided in Supplementary Material.
Post-transplant trends of %ddcfDNA
%ddcfDNA was measured for 1,549 plasma samples (~10 per patient). Median sequencing depth was 12.7 million reads (Supplementary Table 3a). Sequencing data will be deposited publicly. Sequence reads showed a predominantly mononucleosomal pattern with a peak length of 158 base pairs (Supplementary Figure 2). The median level of ddcfDNA was 24.02% after transplant surgery (IQR = 18.05%-38.48%). Levels decayed logarithmically with a half-life of 0.9 days, at first, but then followed by a slower decay half-life of 20.2 days (Supplementary Table 3b). Median ddcfDNA levels reached 0.43% (IQR = 0.24–0.83) by 1.5 months after transplantation (Figure 2, Supplementary Table 3c), and reached the lowest baseline level of 0.21% by 4 months after transplantation. Median levels of ddcfDNA remained stable until 9 months, and then rose to reach 0.60%–0.68% between 18 and 24 months.
Figure 2. Median %ddcfDNA vs. time curve post-transplantation.
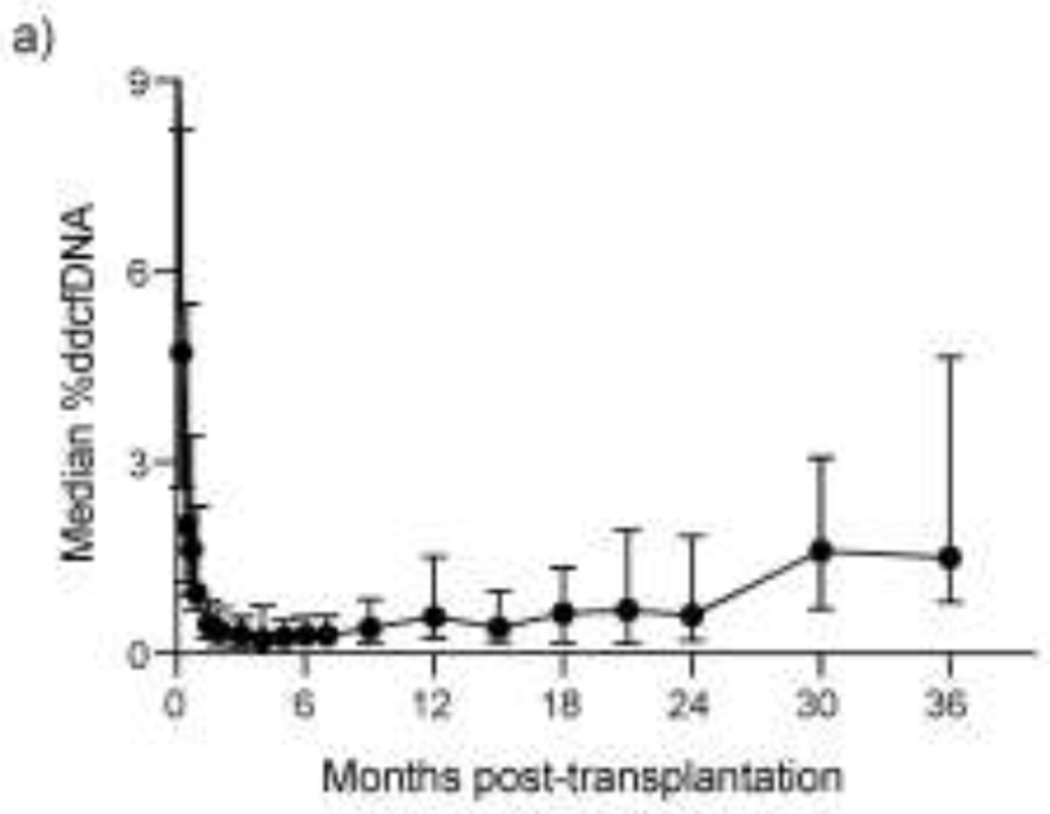
Median %ddcfDNA levels are shown over 24 months post-transplantation. %ddcfDNA, levels of donor-derived cell-free DNA.
%ddcfDNA correlates with measures of allograft injury and acute complications
Data before day 45 was excluded to account for post-transplant %ddcfDNA decay, leaving 484 histopathology events (88% of which had concurrent %ddcfDNA data). Levels of ddcfDNA correlated with histological grading of ACR defined by ISHLT criteria.(1) ACR grade 1 showed an approximate 1.5-fold higher level than ACR grade 0 (0.68% vs. 0.41%, p = 0.030), while ACR grade ≥ 2 showed an approximately two-fold higher level than ACR grade 1 (Figure 3a, Table 2a). Similarly, %ddcfDNA correlated with the severity of allograft dysfunction measured by the magnitude of FEV1 decline (p < 0.01, Figure 3b, Table 2a): levels of ddcfDNA were approximately three-fold higher in “mild” allograft dysfunction episodes compared to “no” allograft dysfunction, and approximately 1.7-fold higher for “moderate/severe” episodes compared to “mild” allograft dysfunction.
Figure 3. Correlation of %ddcfDNA with ACR and allograft dysfunction.
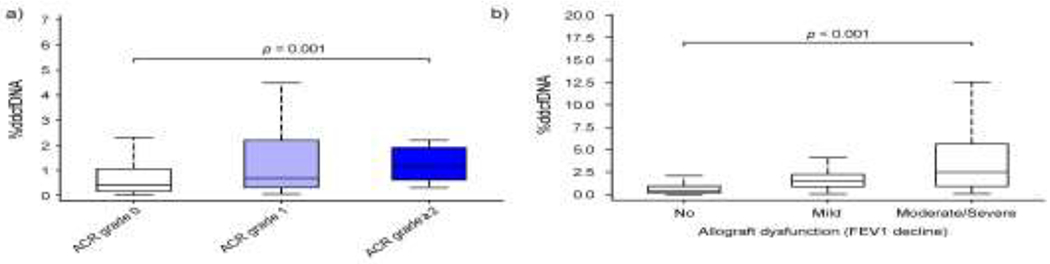
%ddcfDNA is measured by first excluding data before day 45. %ddcfDNA levels are shown for (a) grade 0, 1 and 2 ACR (per ISHLT criteria) and b) allograft dysfunction (as measured by FEV1 decline) categorized as “no”, “mild”, or “moderate/severe”. Number of subjects with each complication is shown in Table 2. %ddcfDNA, levels of donor-derived cell-free DNA.
Table 2.
(a) %ddcfDNA correlate with current measures of allograft dysfunction
| Clinical endpoint | Number of events | Number of subjects with events | Median %ddcfDNA | Interquartile range (%) | p-value compared to controls |
|
|---|---|---|---|---|---|---|
| Histological grading of ACR | Grade 0 | 426 | 148 | 0.41 | 0.15 – 1.06 | |
| Grade 1 | 35 | 27 | 0.68 | 0.29 – 3.10 | 0.03 | |
| Grade≥2 | 23 | 19 | 1.16 | 0.58 – 2.04 | <0.01 | |
| Clinical AMR | No AMR | 427 | 148 | 0.46 | 0.18 – 1.30 | |
| Possible | 43 | 27 | 2.32 | 1.58 – 7.05 | <0.01 | |
| Definite/Probable | 14 | 13 | 4.16 | 1.54 – 9.40 | <0.01 | |
| Allograft dysfunction | None | 1688 | 148 | 0.42 | 0.17 – 1.01 | |
| Mild | 109 | 65 | 1.48 | 0.85 – 2.26 | <0.01 | |
| Moderate to severe | 128 | 63 | 2.10 | 0.73 – 5.29 | <0.01 | |
| Pathogen | None | 309 | 148 | 0.47 | 0.17 – 0.92 | |
| Pathogens plus normal TBBx and PFTs | 243 | 62 | 0.53 | 0.22 – 1.31 | 0.17 | |
| Infection with abnormal TBBx | 30 | 24 | 1.55 | 0.72 – 2.51 | <0.01 | |
| Infection plus PFTs decline and normal TBBx | 53 | 36 | 1.61 | 0.48 – 4.18 | <0.01 | |
Some subjects showed more than one categories.
Subjects with concurrent ACR and AMR were excluded, p-value of determined by generalized estimating equation approach using log-transformed %ddcfDNA; however, normalized %ddcfDNA is reported
Performance of %ddcfDNA to detect acute rejection
Levels of ddcfDNA were approximately six-fold higher in acute rejection episodes compared to control episodes (defined by normal microbiological tests and spirometry) (p < 0.001, Table 2b, Figure 4a). The AUROC of %ddcfDNA increased in the early post-transplant period until day 45 (Supplementary Figure 3). At this point, the AUROC of using %ddcfDNA for detecting acute rejection (AR) was 0.89 (95% CI = 0.83 – 0.93: Figure 4b, Table 3a). At a threshold of 0.5% for AR, %ddcfDNA showed a sensitivity of 95%, specificity of 65%, a positive predictive value (PPV) of 51%, and a negative predictive value (NPV) of 96%. Sensitivity, specificity, PPV and NPV at the 1% %ddcfDNA threshold were 77%, 84%, 64% and 90%, respectively.
(b) %ddcfDNA levels for acute rejection and controls.
| Clinical endpoint |
Number of events |
Number of subjects |
Median %ddcfDNA |
Interquartile range (%) |
p-value compared to controls |
|
|---|---|---|---|---|---|---|
| AR | 87 | 51 | 1.95 | 1.14 – 5.04 | <0.01 | |
| AMR | 57 | 35 | 2.32 | 1.56 – 7.23 | <0.01 | |
| ACR | 30 | 22 | 1.23 | 0.65 - 2.03 | <0.01 | |
| Controls | - | 148 | 0.30 | 0.13 – 0.66 | - | |
Figure 4. %ddcfDNA trends between acute rejection phenotypes.
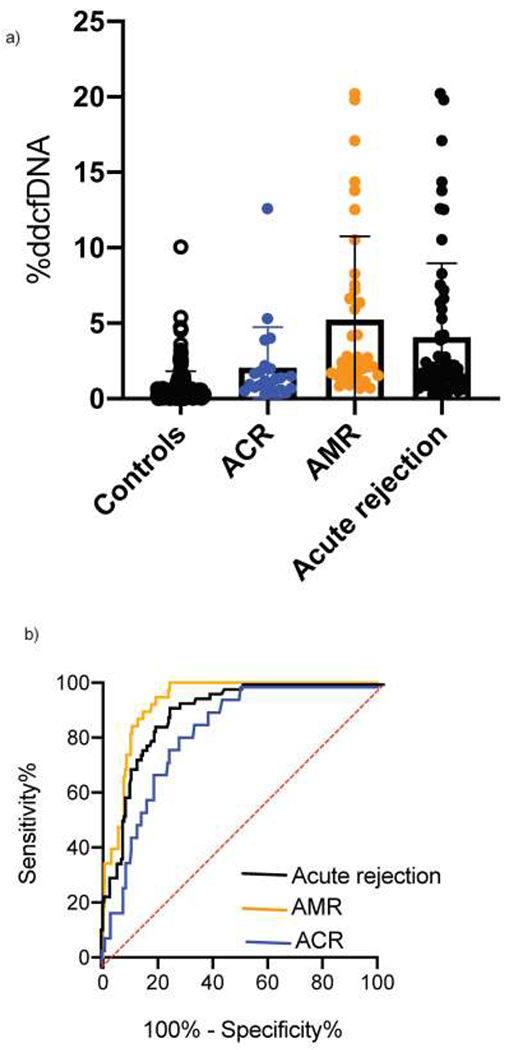
(a) %ddcfDNA levels are shown for controls, acute rejection, ACR and AMR. (b) Measuring the performance sensitivity and specificity of using %ddcfDNA to detect acute rejection, ACR and AMR. ACR, acute cellular rejection; AMR, antibody-mediated rejection; %ddcfDNA, levels of donor-derived cell-free DNA.
Table 3.
(a) %ddcfDNA test characteristics to detect AR defined by ISHLT criteria.
| Sensitivity (%) |
Specificity (%) |
Accuracy |
||||
|---|---|---|---|---|---|---|
| %ddcfDNA threshold | 0.5% | 1.0% | 0.5% | 1.0% | (95% CI) | |
| AR diagnosis | AR | 95 | 77 | 65 | 84 | 0.89 (0.83 – 0.93) |
| AMR | 100 | 89 | 65 | 84 | 0.93 (0.90 – 0.96) | |
| ACR | 85 | 55 | 65 | 84 | 0.83 (0.76 – 0.89) | |
Comparing %ddcfDNA to histopathology in the detection of clinical events
Given the potential limitations of histopathology alone to identify episodes of clinically relevant events such as ACR, AMR, infection or allograft dysfunction (>10% decline in FEV1), we compared histopathology to %ddcfDNA in the ability to detect these events using a threshold value of >1% as a positive test.(15)
After excluding data before day 45, there were 424 episodes with histopathology and %ddcfDNA data. Of these, 132 episodes showed %ddcfDNA > 1% (Table 3b). Within this group, histopathology showed an abnormal finding in only one-third (ACR, n = 22; bronchiolitis obliterans-organizing pneumonia, BOOP, n = 2; capillaritis, n = 2; non-specific inflammation, n = 14). However, 90.1% (n = 82) of the episodes with negative histopathology were concurrent to a clinical complication (AMR with negative biopsy, n = 13; pathogens, n = 36; PFT decline without identifiable cause, n = 23) or represented a rise in %ddcfDNA preceding a clinical event (n = 10).
(b) Comparing %ddcfDNA head-to-head with hispathology
| %ddcfDNA | |||
|---|---|---|---|
| ≥1.0 % (n=132) |
<1.0 % (n=292) |
||
| Histopathology | Any Abnormal finding | 31 | 15 |
| Normal | 69 | 85 | |
There were 292 episodes with ddcfDNA < 1%, 14.7% (n = 43) of which showed abnormal histopathology that were read as ACR grade ≥ 2 (n = 5), ACR grade 1 (n = 17) or non-specific inflammation (n = 20). Only 7 of the 292 episodes (2.4%) with negative %ddcfDNA tests showed abnormal histopathology findings that were treated (grade ≥ 2 ACR, n=5; non-specific findings with AMR diagnosis, n=2).
%ddcfDNA in relation to pathogens and DSA.
Pathogens with normal histopathology and spirometry showed similar levels of ddcfDNA to episodes with no pathogen (0.53, IQR 0.22–1.31 vs. 0.47, IQR 0.17–0.92, p = 0.17, Table 2a). Respiratory viruses, bacterial and fungi all showed similar %ddcfDNA levels (p=0.080). Pathogens associated with abnormal histopathology ora decline in PFTs showed three-fold higher levels of %ddcfDNA (1.55, IQR = 0.72–2.51, and 1.61, IQR = 0.76–4.18, respectively, Suppl. Figure 4). Similarly, detection of DSA alone showed similar %ddcfDNA levels as controls. However, detection of DSA in the context of AMR showed higher %ddcfDNA levels (Suppl. Figure 4).
Distinctive %ddcfDNA characteristics for AMR and ACR
Next, we assessed distinctive %ddcfDNA trends between the AMR and ACR phenotypes. AMR showed an approximately two-fold higher level of %ddcfDNA compared to ACR (p < 0.01, Table 2b, Figure 4a). This correlated with a greater decline in FEV1 (AMR median = −14%, IQR = −21% to −12% vs. ACR median = −5%, IQR = −8% to 4%; p < 0.01), and a higher AUROC compared to ACR (0.93 vs. 0.83). In addition, a rise in levels of %ddcfDNA preceding the diagnosis was encountered more often for AMR compared to ACR. For example, one month before diagnosis, 82% of AMR episodes showed a rise in levels of ddcfDNA (with levels ≥ 1%) compared to 18% of ACR episodes (Figure 5). The time between the rise in ddcfDNA levels and diagnosis was five-fold longer for AMR compared to ACR (AMR median = 2.6 months, IQR = 1.3–3.4 vs. ACR median = 0.5 months, IQR = 0–1.3 months).
Figure 5. Comparing the time course of elevated %ddcfDNA to time of diagnosis.
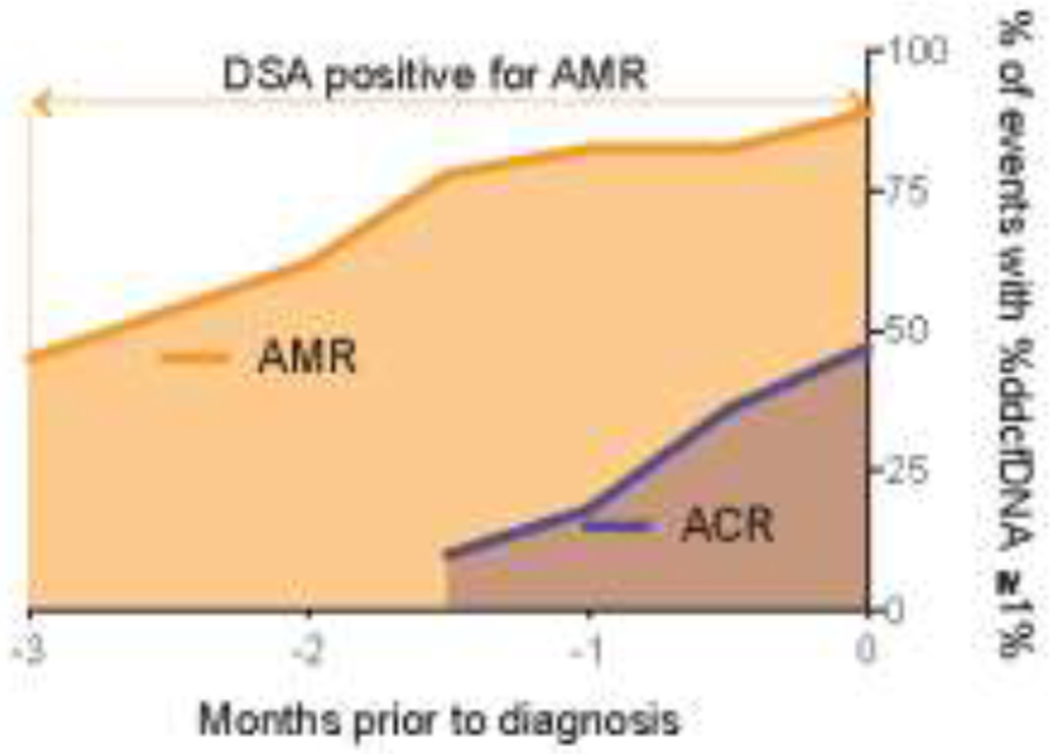
Shown are the number of episodes that showed a ≥ 1% rise in %ddcfDNA. ACR, acute cellular rejection; AMR, antibody-mediated rejection; %ddcfDNA, levels of donor-derived cell-free DNA. Donor-specific antibodies (DSA) was positive for AMR patients is shown.
Discussion
The study reports several important findings. First, %ddcfDNA correlates with current measures of allograft dysfunction, such as spirometry and histopathology. Second, %ddcfDNA reliably detects acute rejection, with a high negative predictive value. Third, a rise in %ddcfDNA levels often precedes a diagnosis of acute rejection using standard measures, particularly in the case of AMR, suggesting that %ddcfDNA may establish an earlier diagnosis. Lastly, a head-to-head comparison indicates that %ddcfDNA may detect clinically relevant events not identified by histopathology. These observations are consistent with prior experiences in other solid organ transplantation,(10–12, 26, 27) and suggest that %ddcfDNA can be safely used to monitor lung allograft health.
Conventional statistical methods that employ sensitivity and specificity to test the performance of new diagnostic tools against established standards are limited, particularly if the established standards exhibit poor performance.(28) In this study, histopathology detected only one-third of episodes where the levels of %ddcfDNA > 1%, a threshold that has been reported to correlate with 100% sensitivity in detecting high grade ACR.(15) While the remaining two-thirds of events with levels of %ddcfDNA > 1% were considered “false positives”, 90% of them were coincident to clinical complications, such as AMR, presence of pathogens or a decline in PFTs. These “false positives” also include events that represented a rise in %ddcfDNA levels preceding clinical AMR. The incorrect assignment of “false positives” may therefore underestimate the performance of %ddcfDNA.
Despite this underestimation in PPV, %ddcfDNA showed a high negative predictive value for acute rejection, indicating that a negative %ddcfDNA test would indicate a stable allograft. However, given the low specificity, high %ddcfDNA values may still require bronchoscopy plus other tests to identify the cause of allograft injury. As such, %ddcfDNA may serve a valuable role as a surveillance marker for underlying allograft injury in place of routine surveillance bronchoscopy. Under this scenario, patients with low levels of %ddcfDNA may defer undergoing surveillance bronchoscopy and avoid the unnecessary risks, time and cost of the procedure. A high %ddcfDNA level may serve as a trigger for bronchoscopy and other tests to identify the trigger of allograft injury. In our analysis, such an approach would avoid two-thirds of bronchoscopies and detect more clinically relevant events that would be missed by the conventional surveillance bronchoscopy monitoring approach. Four transplant centers recently adopted this monitoring approach as routine clinical care in place of surveillance bronchoscopy to reduce patient and staff exposure during the COVID-19 pandemic. Data on this practice is being collected retrospectively as part of the Analysis of Lung Allograft Remote Monitoring (ALARM) Study. Preliminary results will become available early this year.
Despite its limitations, bronchoscopy aids in the diagnosis of rejection and respiratory pathogens. Additional clinical, radiologic, histopathologic, and bronchoalveolar fluid cytology data is needed to distinguish pathogen colonization from infection.(23) This study does not examine infection, because the clinical data needed to define infection(23) was lacking or inconsistently reported in providers’ documentation. Nonetheless, our observations suggest that %ddcfDNA may distinguish infection from pathogens without infection: we observed similar ddcfDNA levels between pathogens and no pathogen, but three-fold higher ddcfDNA levels when pathogens were associated with either positive histopathology or a decline in PFTs (compared to pathogens without these signs of allograft dysfunction). A future study that prospectively captures clinical manifestation in real-time would be ideal to address this limitation and should test the performance of %ddcfDNA in relation to a composite endpoint of infection and acute rejection.
It is worth noting, that as a ratio, levels of %ddcfDNA are dependent on levels of underlying recipient cfDNA. Accordingly, absolute changes in recipient cfDNA, which is observed after transplantation, (29) may impact the value of %ddcfDNA and the test characteristics, despite relative changes in the amount of absolute ddcfDNA. Other limitations of this study include incomplete data at some time points and lack of a validation cohort. In addition, clinical practice and immunosuppression management vary among transplant providers and among the three GRAfT centers,(18, 20) which could affect the incidence of acute rejection and levels of %ddcfDNA. Nonetheless, this multicenter prospective study provides real-world evidence of the performance of %ddcfDNA to detect rejection.
In summary, our data demonstrates that %ddcfDNA may be superior to histopathology at monitoring for lung allograft injury. A %ddcfDNA monitoring approach may perform better than histopathology for detecting acute rejection and other complications. This work sets the stage for future randomized controlled clinical trials testing the utility of %ddcfDNA as a monitoring tool in place of surveillance bronchoscopy.
Supplementary Material
Acknowledgements
We thank the clinical coordinators at participating centers for recruiting and monitoring study subjects, organ procurement organizations (Living Legacy Foundation and Washington Regional Transplant Consortium) for providing donor blood samples for genotyping, the NHLBI Sequencing Core for shotgun sequencing, the National Cancer Institute sequencing core for genotyping, Kelly Byrne for editorial support, and Erina He for graphing support. We acknowledge Ulgen Fideli, Sasha Gorham, Yanqin Yang, Argit Marishta, Kenneth Bhatti for supporting the study enrollment and %ddcfDNA assays. The study was supported by intramural research funds of the National Heart, Lung, and Blood Institute and Cystic Fibrosis Grant AGBORE20Q10.
Disclosure statement
GRAfT and the cohort study are supported by intramural funding of the NHLBI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no competing interests to disclose.
References
- 1.Stewart S, Fishbein MC, Snell GI, Berry GJ, Boehler A, Burke MM, et al. Revision of the 1996 working formulation for the standardization of nomenclature in the diagnosis of lung rejection. J Heart Lung Transplant. 2007;26(12):1229–42. [DOI] [PubMed] [Google Scholar]
- 2.Roux A, Levine DJ, Zeevi A, Hachem R, Halloran K, Halloran PF, et al. Banff Lung Report: Current knowledge and future research perspectives for diagnosis and treatment of pulmonary antibody-mediated rejection (AMR). Am J Transplant. 2019;19(1):21–31. [DOI] [PubMed] [Google Scholar]
- 3.Roux A, Bendib Le Lan I, Holifanjaniaina S, Thomas KA, Hamid AM, Picard C, et al. Antibody-Mediated Rejection in Lung Transplantation: Clinical Outcomes and Donor-Specific Antibody Characteristics. Am J Transplant. 2016;16(4):1216–28. [DOI] [PubMed] [Google Scholar]
- 4.Lama VN, Murray S, Lonigro RJ, Toews GB, Chang A, Lau C, et al. Course of FEV(1) after onset of bronchiolitis obliterans syndrome in lung transplant recipients. Am J Respir Crit Care Med. 2007;175(11):1192–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sato M, Ohmori-Matsuda K, Saito T, Matsuda Y, Hwang DM, Waddell TK, et al. Time-dependent changes in the risk of death in pure bronchiolitis obliterans syndrome (BOS). J Heart Lung Transplant. 2013;32(5):484–91. [DOI] [PubMed] [Google Scholar]
- 6.Verleden GM, Raghu G, Meyer KC, Glanville AR, and Corris P. A new classification system for chronic lung allograft dysfunction. The Journal of Heart and Lung Transplantation. 2014;33(2):127–33. [DOI] [PubMed] [Google Scholar]
- 7.Witt CA, Gaut JP, Yusen RD, Byers DE, Iuppa JA, Bennett Bain K, et al. Acute antibody-mediated rejection after lung transplantation. J Heart Lung Transplant. 2013;32(10):1034–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhorade SM, Husain AN, Liao C, Li LC, Ahya VN, Baz MA, et al. Interobserver variability in grading transbronchial lung biopsy specimens after lung transplantation. Chest. 2013;143(6):1717–24. [DOI] [PubMed] [Google Scholar]
- 9.Agbor-Enoh S, Tunc I, De Vlaminck I, Fideli U, Davis A, Cuttin K, et al. Applying rigor and reproducibility standards to assay donor-derived cell-free DNA as a non-invasive method for detection of acute rejection and graft injury after heart transplantation. J Heart Lung Transplant. 2017;36(9):1004–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bloom RD, Bromberg JS, Poggio ED, Bunnapradist S, Langone AJ, Sood P, et al. Cell-Free DNA and Active Rejection in Kidney Allografts. J Am Soc Nephrol. 2017;28(7):2221–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.North PE, Ziegler E, Mahnke DK, Stamm KD, Thomm A, Daft P, et al. Cell-free DNA donor fraction analysis in pediatric and adult heart transplant patients by multiplexed allele-specific quantitative PCR: Validation of a rapid and highly sensitive clinical test for stratification of rejection probability. PLOS ONE. 2020;15(1):e0227385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schutz E, Fischer A, Beck J, Harden M, Koch M, Wuensch T, et al. Graft-derived cell-free DNA, a noninvasive early rejection and graft damage marker in liver transplantation: A prospective, observational, multicenter cohort study. PLoS Med. 2017;14(4):e1002286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharon E, Shi H, Kharbanda S, Koh W, Martin LR, Khush KK, et al. Quantification of transplant-derived circulating cell-free DNA in absence of a donor genotype. PLoS computational biology. 2017;13(8):e1005629–e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka S, Sugimoto S, Kurosaki T, Miyoshi K, Otani S, Suzawa K, et al. Donor-derived cell-free DNA is associated with acute rejection and decreased oxygenation in primary graft dysfunction after living donor-lobar lung transplantation. Sci Rep. 2018;8(1):15366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Vlaminck I, Martin L, Kertesz M, Patel K, Kowarsky M, Strehl C, et al. Noninvasive monitoring of infection and rejection after lung transplantation. Proc Natl Acad Sci U S A. 2015;112(43):13336–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knight SR, Thorne A, and Lo Faro ML. Donor-specific Cell-free DNA as a Biomarker in Solid Organ Transplantation. A Systematic Review. Transplantation. 2019;103(2):273–83. [DOI] [PubMed] [Google Scholar]
- 17.Zou J, Duffy B, Slade M, Young AL, Steward N, Hachem R, et al. Rapid detection of donor cell free DNA in lung transplant recipients with rejections using donor-recipient HLA mismatch. Hum Immunol. 2017;78(4):342–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agbor-Enoh S, Jackson AM, Tunc I, Berry GJ, Cochrane A, Grimm D, et al. Late manifestation of alloantibody-associated injury and clinical pulmonary antibody-mediated rejection: Evidence from cell-free DNA analysis. J Heart Lung Transplant. 2018;37(7):925–32. [DOI] [PubMed] [Google Scholar]
- 19.Agbor-Enoh S, Fideli U, Doveikis J, Zhu J, Tunc I, Shah P, et al. Genomic Research Alliance for Transplantation (GRAfT): A Model for Long Term Transplant Studies in Thoracic Organ Transplantation. The Journal of Heart and Lung Transplantation. 2016;35(4):S161. [Google Scholar]
- 20.Agbor-Enoh S, Wang Y, Tunc I, Jang MK, Davis A, De Vlaminck I, et al. Donor-derived cell-free DNA predicts allograft failure and mortality after lung transplantation. E Bio Medicine. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.von Elm E, Altman DG, Egger M, Pocock SJ, Gotzsche PC, Vandenbroucke JP, et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) Statement: Guidelines for Reporting Observational Studies. PLOS Medicine. 2007;4(10):e296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levine DJ, Glanville AR, Aboyoun C, Belperio J, Benden C, Berry GJ, et al. Antibody-mediated rejection of the lung: A consensus report of the International Society for Heart and Lung Transplantation. J Heart Lung Transplant. 2016;35(4):397–406. [DOI] [PubMed] [Google Scholar]
- 23.Husain S, Mooney ML, Danziger-Isakov L, Mattner F, Singh N, Avery R, et al. A 2010 working formulation for the standardization of definitions of infections in cardiothoracic transplant recipients. J Heart Lung Transplant. 2011;30(4):361–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou X ONaMD. Statistical Methods in Diagnostic Medicine, 2nd Edition. Wiley; 2011. [Google Scholar]
- 25.Ali HA, Pavlisko EN, Snyder LD, Frank M, and Palmer SM. Complement system in lung transplantation. Clin Transplant. 2018;32(4):e13208. [DOI] [PubMed] [Google Scholar]
- 26.De Vlaminck I, Valantine HA, Snyder TM, Strehl C, Cohen G, Luikart H, et al. Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci Transl Med. 2014;6(241):241ra77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khush KK, Patel J, Pinney S, Kao A, Alharethi R, DePasquale E, et al. Noninvasive detection of graft injury after heart transplant using donor-derived cell-free DNA: A prospective multicenter study. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2019;19(10):2889–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuhn TS. The structure of scientific revolutions. Chicago: University of Chicago Press; 1962. [Google Scholar]
- 29.Whitlam JB, Ling L, Skene A, Kanellis J, lerino FL, Slater HR, et al. Diagnostic application of kidney allograft-derived absolute cell-free DNA levels during transplant dysfunction. Am J Transplant 2019; 19(4): 1037–49. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


