Abstract
The molecular mechanisms of hepatitis C virus (HCV) persistence and pathogenesis are poorly understood. The design of an effective HCV vaccine is challenging despite a robust humoral immune response against closely related strains of HCV. This is primarily because of the huge genetic diversity of HCV and the molecular evolution of various virus escape mechanisms. These mechanisms are steered by the presence of a high mutational rate in HCV, structural plasticity of the immunodominant regions on the virion surface of diverse HCV genotypes, and constant amino acid substitutions on key structural components of HCV envelope glycoproteins. Here, we review the molecular basis of neutralizing antibody (nAb)-mediated immune response against diverse HCV variants, HCV-steered humoral immune evasion strategies and explore the essential structural elements to consider for designing a universal HCV vaccine. Structural perspectives on key escape pathways mediated by a point mutation within the epitope, allosteric modulation of the epitope by distant mutations and glycan shift on envelope glycoproteins will be highlighted (abstract graphic).
Keywords: Hepatitis C Virus, HCV, Neutralizing Antibodies, Antigenic Domains, Vaccine Design, Immune Evasion
Graphical Abstract
Introduction
Hepatitis C Virus (HCV), is a blood-borne pathogen identified more than 30 years ago causing acute (30%) and chronic hepatitis (70%) [1–3]. According to the WHO, an estimated 71 million individuals globally have chronic hepatitis C infection. HCV is a major cause of hepatocellular carcinoma leading to about half a million worldwide deaths annually. Acute HCV infections are increasing in the USA from 0.3/100,000 in 2009 to 1.2/100,000 in 2018 and the rate is increasing highest among young adults of 3./100,000 [4]. Illicit injection drug use is a major contributing factor and unfortunately during the COVID-19 pandemic, drug overdose is accelerating [5]. HCV is classified into seven genetically distinct genotypes (1 to 7) with substantial differences in geographical distribution [6,7]. Direct acting antivirals (DAAs) can cure more than 95% of chronic infections upon early detection and treatment availability [8]. However, reinfection will occur after successful treatment [9]. No preventive vaccines are available against HCV due in part to its high genetic variability and frequent mutations to escape the immune response [10–13].
HCV, a member of the family Flaviviridae and genus Hepacivirus, is an enveloped positive stranded RNA virus. The ~9.6 Kb genome encodes a single polyprotein of about 3011 amino acids, which is processed into ten proteins (3 structural) by viral and host proteases [14]. The N-terminal region of the HCV genome encodes three structural proteins: the core or capsid protein that protects the viral genome and the heterodimeric envelope glycoproteins (GPs), E1 and E2. The neutralization antibody (nAb) response against HCV, which is under constant immunological pressure in natural infections, is largely directed against the two surface accessible envelope GPs, E1 and E2. Although E1 is less immunogenic when compared to E2 in natural infections, a wide-range of genotype-specific and broadly neutralizing Abs against E1, E2 and E1E2 heterodimers have been identified and characterized, as reviewed in Keck, M et al., 2018 [15]. However, sequence and structural plasticity of highly variable regions on the extensively glycosylated E1 and E2 GPs facilitates viral escape mechanisms, thus demonstrating a major challenge for efficacious vaccine design with broader specificity. Limited knowledge about the three-dimensional (3D) structures of the E1E2 GPs and the structural components of the HCV is an added constraint to decipher the surface accessibility of epitopes.
Here, we review the molecular basis of nAb mediated immune response against diverse HCV variants, HCV-steered humoral immune evasion strategies and explore the essential elements to consider for designing a universal HCV vaccine.
The architecture of HCV and its glycoproteins
HCV exists as lipoviral particles (LVP) in patient serum and is associated with very low density lipoprotein particles (VLDL) or low density lipoprotein particles (LDL) [16]. The LVPs are heterogeneous in size, ranging in diameter from 40 to 100 nm, and are associated with apolipoproteins, cholesterol and triglycerides [17] [18]. Most of the cell-culture produced particles although heterogenous in size, appear to have spherical shape, suggesting a regular arrangement of the GPs, E1 and E2 on the virus surface. E1 and E2 form heterodimers and are embedded in a host-derived lipid bilayer membrane [19,20]. Oligomeric E1E2 heterodimers shield the nucleocapsid core as shown in a cartoon representation of the structure in Figure 1. A few studies suggest that E1 might form trimers on the virus surface [21]. As observed in other flaviviruses such as Zika virus and dengue virus, particle morphology, the oligomeric state and the conformation of the GPs on the virus surface are a few factors influencing neutralization by diverse Abs and evolution of virus escape mechanisms [22].
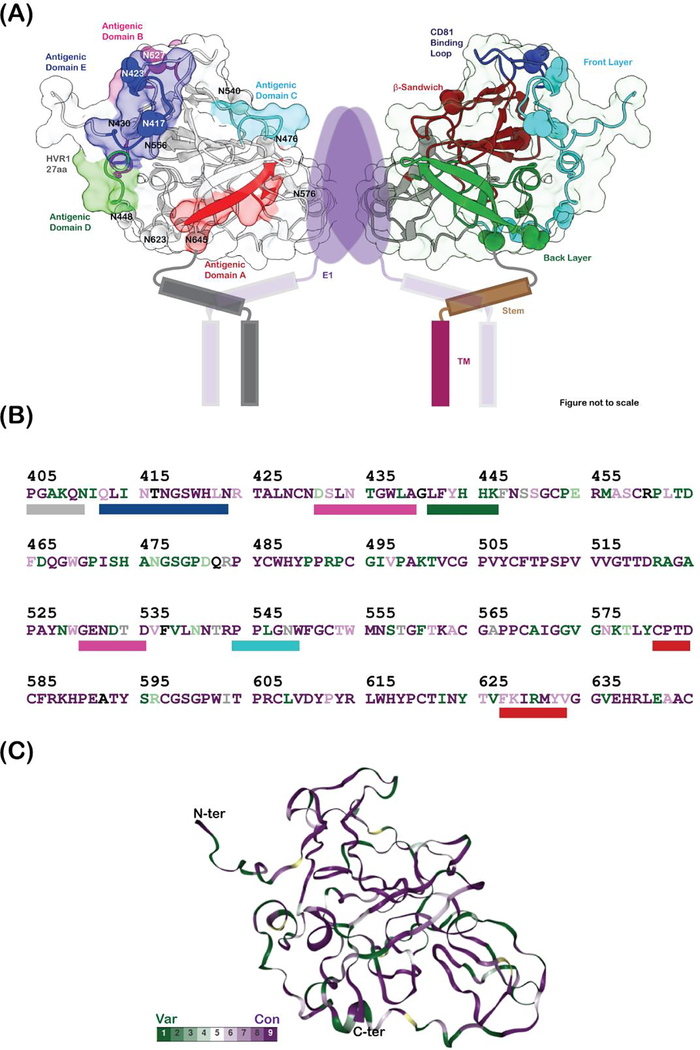
Figure 1. HCV envelope glycoproteins.
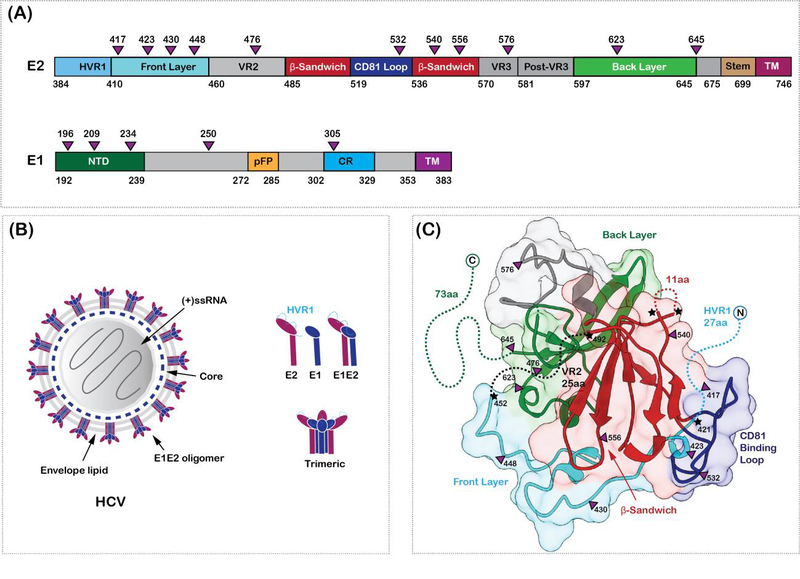
(A) Domain organization of HCV envelope glycoproteins, E1 and E2. The glycosylation sites are shown as purple triangles in (A) and (C). The hypervariable region 1 (HVR1), variable regions 2 and 3 (HVR2 and HVR3), the front layer, the back layer, the β-sandwich domains, stem and the transmembrane (TM) regions are labelled on E2. The N-terminal domain (NTD), putative fusion peptide (pFP), the conserved region (CR) and the transmembrane regions are labelled on E1. (B) Hepatitis C virion architecture. E1 (dark blue) and E2 (dark purple) form heterodimers which in turn might arrange into trimers on the virus surface. The components of the virion are labelled. (C) Crystal structure of E2 core domain (PDB ID: 4MWF). Missing regions in the structure are shown as dotted lines and highlighted with a star. The coloring scheme is similar to (A)
The 3D structure elucidation of native HCV particles has not been successful to date, due to the pleomorphic nature of the viruses and limitations in preparing large amounts of virus particles from cultured hepatic cells. However, in the past decade, numerous 3D structures of truncated HCV envelope GP E2, in complex with neutralizing antibodies (nAbs) were valuable in understanding the mechanisms of a broadly neutralizing immune response [19,23–36]. The GPs, E1 (residues 192–383) and E2 (residues 384–746) are type I transmembrane proteins with an N-terminal ectodomain and a C-terminal transmembrane domain [33] (Figure 1). The GP, E2 mediates cell entry by binding to the SR-B1 and CD81 co-receptors and other host factors [37,38]. The role of E1 is poorly understood but harbors a putative fusion loop. The ectodomains of the E1 and E2 proteins have five and eleven glycosylation sites, respectively [39]. The glycans are essential for the stability and antigenicity of the E1E2 proteins. The E1 and E2 ectodomains, also contain eight and eighteen cysteines, which can form inter- and intra-molecular disulfide bonds [40]. Both, the glycosylation and the disulfide bond formation play a role in the proper folding of the E1E2 proteins [41].
Significant but not adequate progress has been made in understanding the structure of the core domains of HCV E1 and E2 proteins [23,26,31,42]. The crystal structure of the N-terminal region of E1 shows a higher oligomeric, domain swapped and covalently linked complex with unexpected features [42]. Truncated E1 structure consists of a β-hairpin followed by an alpha-helix, sandwiched between a two and a three-stranded anti-parallel β-sheet structure. It was also shown that the E1 protein is similar to parts of phosphatidylcholine transfer protein, which is important for binding of lipid-like molecules. Whereas, the core domain of truncated forms of E2 adopts a central immunoglobulin-like fold (the β-sandwich domain) flanked by short alpha-helices and loops (the front layer and the back layer) as shown in Figure 1 [23,26]. The core domain of E2 consists of several conserved and variable residues for inducing a protective immune response. A large proportion of the structurally unresolved regions in E1 and E2 proteins essential for immunogenicity are predicted to be disordered. Attempts to resolve these disordered regions may be possible by examining the structure of E1E2 with bound nAbs that fix the variable residues in place.
Genetic diversity and immunogenic regions of E1 and E2
HCV is classified into seven distinct genotypes with a median intra-genotype and inter-genotype diversity at amino-acid level of 9.71% and 25.2%, respectively [7]. The highest overall diversity is displayed by the glycoprotein E2 (median diversity of 18.23%) [7]. The greatest variation is observed in the N-terminal region of E2, hypervariable region 1, HVR1 (384–410). Two other hypervariable regions of the E2 protein are the HVR2 (460–485) located proximal in 3D structure to the highly conserved CD81-binding region and the HVR3 or the intergenotype variable region (igVR; 570–580) located closer to the transmembrane domain of E2.
A majority of the nAbs are directed against the epitopes on the glycoprotein E2 followed by E1E2 heterodimer and the E1 glycoprotein (reviewed in [15]). The diversity of E2 might not influence the overall 3D structure of the protein but has a considerable impact on the surface accessible epitopes and immune response. The epitopes on E2 are either linear or conformational and can be further grouped into clusters of overlapping epitopes as antigenic domains (ADs): HVR1 and domains A-E or antigenic regions (ARs): AR1–3. Linear epitopes lie adjacent to each other or are separated by a few residues on the primary sequence of E2, while conformational epitopes are separated on the primary sequence and proximal in the 3D structure. Although, the epitope clusters, ADs and ARs are distinct in their properties, they also exhibit overlapping clusters. Several characteristics of the E2 antigenic domains and antigenic regions are listed in Table 1 and shown in Figure 2 along with examples of HCV nAbs.
Table 1.
Immunogenic regions of HCV E1 and E2 proteins and nAb examples.
| Antigenic Domains | Epitope type | Residues | Protein region | nAbs | Mechanism |
| HVR1 | Linear | 384–410 | N-terminus loop | H77.16; HEPC98; | Isolate-specific SR-B1 interaction |
| A | Conformational | 581–584 627–633 |
E2 back layer | CBH-4B; CBH-4D; CBH-20; CBH-21; A33; | Non-neutralizing |
| B | Conformational | 431–439 529–535 |
E2 surface layer | HC-1; HC-11; AR3A; AR3C; CBH-2; HEPC3; HEPC74; | Broadly neutralizing and CD81 binding |
| C | Conformational | 544–549 | E2 β-sandwich | CBH-7; CBH-23; AR1A; AR1B; HEPC50; | Weakly or non-neutralizing |
| D | Conformational | 441–446 616 |
E2 back layer | HC84.1; HC84.26; HC84.27; | Broadly neutralizing surface of E2 |
| E | Linear | 412–423 | N-terminus loop | HC33.1; HC33.4; HCV1; AP33; | Broadly neutralizing |
| Antigenic Region | Epitope type | Residues | Secondary structure | nAbs | Mechanism |
| AR1 | Conformational | 495, 519, 544, 545, 547–549, 632 | E2 apex | CBH-7; CBH-23; AR1A; AR1B; HEPC50; HEPC-167; | Weakly or non-neutralizing |
| AR2 | Conformational | 625, 628 | E2 back layer | Narrow neutralization | |
| AR3 | Conformational | 427–443 529–530 |
E2 neutralizing face | HC-1; HC-11; AR3A; AR3C; CBH-5; HEPC3; HEPC74; HC84.1; HC84.26; HC84.27; HEPC-122; HEPC-151–1; HEPC-153; HEPC-154; | Neutralizing and CD-81 binding |
| AR4 | E1E2 | 698 | E1E2 interface | AR4A; HEPC111; | Broadly neutralizing |
| AR5 | E1E2 | 639, 665 | E1E2 interface | AR5A; HEPC-130 | Broadly neutralizing |
| Others | Epitope type | Residues | Secondary structure | nAbs | Mechanism |
| AS108 | Conformational | 472,474, 543–549, 569; 585, 594; 597, 598, 635, | E2 VR2, β-sandwich, Post-VR3 and back layer | HEPC-108; HEPC-132; HEPC-158; | Broadly neutralizing |
| AS112 | Conformational | 321, 330, 517, 520, 529, 534, 535, 549 | E1 C-terminus, CD81 binding region and β-sandwich | HEPC-146; | Broadly neutralizing |
| AS146 | Conformational | 215,232, 233,246, 249, 252, 259, 260, 263,297, 299, 354, 361,378, 382 | E1 NTD and E1 stem region | HEPC-112; | Broadly neutralizing |
Figure 2. The immunogenic regions of HCV E2.
(A) Cartoon representation of heterodimeric E1E2 proteins. The dimer of E1E2 heterodimers representation was prepared to simultaneously depict the antigenic regions and the secondary structure definitions. This does not represent the functional oligomeric state of the E1E2 heterodimers on the virus surface. Figure prepared from a homology model of E2 (residues 405–645) calculated using the program modeler, where the missing loops of the crystal structure and part of the N-terminus are modeled. The antigenic domains A (581–584, 627–633), B (431–439, 529–535), C (544–549), D (441–446) and E (412–423) are colored red, magenta, cyan, green and blue, respectively, and shown as rectangular bars below the E2 sequence in (B). The hypervariable region 1 (HVR1), variable regions 2 and 3 (VR2 and VR3), the front layer, the back layer, the β-sandwich domains, stem, and the transmembrane (TM) regions are labelled on E2. The E1 protein, its TM domains, and the stem and TM helices of E2 are drawn as cartoon. The glycosylation sites are labelled and shown as spheres. (B) shows the sequence of E2 protein (subtype 1a, PDB ID: 6MEJ) colored according to conservation and mapped onto the E2 protein homology model in (C). Green depicts the least conserved and purple depicts the most conserved residues. The percent conservation was calculated using the ConSurf server. The N and C-termini are labelled in (C).
Immune response against HCV E1, E2 and E1E2
The antibody response against HCV can be classified into three categories: broadly neutralizing, strain-specific and non-neutralizing antibodies with structurally distinct epitopes. The glycosylation sites on each of the ADs or ARs of E2 regulate the surface accessibility of the epitopes. The core regions of E2 are highly conserved in contrast to the surface exposed loops and HVR regions similar to what has been observed in other viruses.
The immune response in acute and chronic infections mainly targets HVR regions on E2, which are under constant immune selection pressure. The large conformational space occupied by the HVRs and the glycosylation sites act as decoys shielding the more conserved regions on E2 and therefore assist in viral escape [43,44]. NAbs against HVR regions are usually strain-specific and linear [44–46]. The antigenic domain A (and AR1) on the E2 back layer primarily elicits a non-neutralization antibody response.
Despite the high sequence diversity several broadly neutralizing regions can be identified on the core structure of E2 (antigenic domains B-E) (Figure 2). Broadly neutralizing E2 Abs mostly target the highly conserved but flexible CD81 binding site and block cell entry [25]. This site consists of the AS412 region or antigenic domain E (residues 412–423), the front layer and the CD81 binding loop (residues 519–535). For example, the CD81 binding site bNAbs, HC33.1, HC33.4, HCV1 and AP33 recognize the domain E or AS412 region, whereas the antigenic domains B and D (AR3) Abs recognize the highly conserved E2 front layer (Table 1). The antigenic sites on E1 are not well-characterized because of the low immunogenicity of these epitopes. While a small number of NAbs have been identified to E1 and their epitopes are conserved [47,48], only one showed broad neutralization potential, IGH526 and crystal structural studies identified key contact residues at 314–324 [47–49]. The antigenic regions 4 and 5, formed by the E1E2 interface residues also mediate broad neutralization. Novel broadly neutralizing antigenic sites distinct from previously defined AR1–5 and E1 sites were recently identified: AS 108, AS112 and AS 146 [30]. However, it has also been observed that alternate conformation of HCV E2 neutralizating face alters the epitope presentation patterns on its surface uncovering novel antigenic sites [50].
Structural aspects of immune escape pathways in HCV
The underlying molecular mechanism(s) of HCV persistence and pathogenesis is yet to be elucidated. Viral escape in HCV is driven by three factors: a high mutational rate, its intrahost display as a large heterogenous population of closely related species with structural plasticity, and constant amino acid substitutions on key structural components of HCV glycoproteins. A few examples will be discussed in the next section elaborating some of the possible mechanisms.
Point mutation at a contact residue of the bNAb conformational epitope
CBH-2 is a neutralizing monoclonal antibody against a highly conserved conformational epitope on antigenic domain B. CBH-2 specifically inhibits binding of HCV to CD81. Alanine scanning mutagenesis identified two E2 regions, 430–435 and 520–540, and more specifically, D431, G523, G530 and D535 as essential residues for antibody-mediated binding and neutralization (Figure 3A). A point mutation atD431 results in loss of CBH-2-mediated neutralization [51]. Homology modelled E2 protein displays that the N-terminal long loop conformation of the front layer (421–460) can orient itself near to the CD81 binding loop and be part of nAb binding epitope. D431 is also proximal to a conserved glycosylation site (N430) in addition to a conserved cysteine (C429), which forms a disulfide bond with C503.
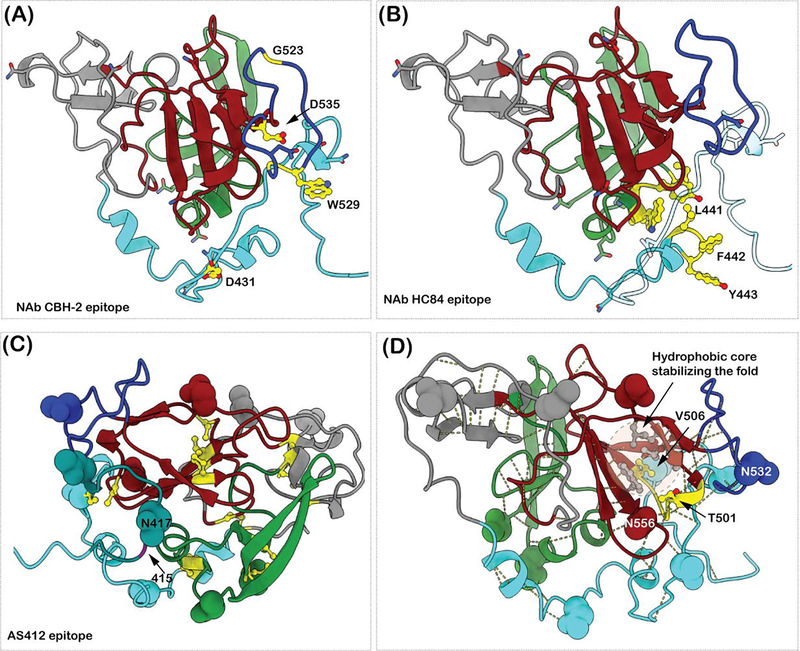
Figure 3. Examples of E2 epitope mutations leading to immune escape.
(A) The NAb CBH-2 crucial epitope residues are shown in yellow and ball and stick representation. G523, D535, W529 and G528 are farther from D431 in the primary sequence. (B) The epitope residues of NAb HC84 are shown in yellow and ball and stick representation. L441, F442 and Y443 adopt a helical conformation. In the homology model they are packed against the N-terminus of AS412 region. A single amino acid exchange of F442 to Ile or Leu decreases the HC84 binding affinity to E2 protein. (C) The AS412 epitope is shown in dark cyan. The positions of N417 and N415 are labeled. The cysteine residues forming disulfide bonds are in yellow ball and stick representation. The shifting of the glycan from N417 to N415 would cause steric hindrance for Ab binding. (D)The side chains (in ball and stick) of the residues 501 and 506 and the loop 500–506 are shown in yellow. The backbone structure is involved in various hydrogen bonds to keep the E2 core structure intact. Residue V506 is part of the hydrophobic core stabilizing the β-sandwich fold. The side chains of the residues involved in hydrophobic core stabilization (V497, V516, F537, L539, W554) are colored grey and rendered in ball and stick representation.
Escape from bNAbs that can be overcome by increasing antibody concentration
A group of bNAbs (HC84) to overlapping epitopes that were not associated with viral escape were identified [29,52]. These bNAbs bind to conformational epitopes overlapping the CD81 receptor binding site and segments 434–446 and 610–619 on E2 (Figure 3B). It was observed that after co-culturing infectious HCVcc at a critical antibody concentration, the majority of the infectious virions were neutralized. Structural studies confirmed that three residues are pivotal with these overlapping epitopes, 441, 442 and 443. Crystal structures of HC84–1 or HC84–27 in complex with a peptide mimicking 434–446 shows that the peptide adopts an alpha helical conformation similar to the homology model shown in Figure 3B. The C-terminal portion of the HVR1 region and the N-terminal portion of the E2 front layer shows interactions with this region in the homology model. The residue Y443 forms stackinig and hydrogen bonding interactions with the CDR loops of the HC-84 Ab, whereas the interactions between the residues 442–443 and the CDR loops are hydrophobic. The C-terminal HVR region in the homology model (shown as transparent loop in Figure 3B) shielding 441–443 is partly hydrophilic and can be easily displaced by a nAb with hydrophobic CDR loops. However, a mutation at the 442 position to either Ile or Leu leads to a decrease in binding of the Fab to E2. The mutation leads to a partial escape that can be overcome by increasing antibody concentration [52,53].
Multiple escape pathways associated with a region of mainly linear epitopes
The region downstream of HVR1, AD E/AS412 (412–423), is highly conserved and able to elicit protective antibodies against HCV isolates of different genotypes and subtypes. There are broadly nAbs, as represented by AP33 [54] or HCV1, that are associated with viral escape from a glycan shift at the 417 position to the 415 position (Figure 3C). N415 has been shown to be important for epitope recognition by AP33 and HCV1. Here, the region 412–423 adopts a beta-hairpin conformation, where the N415 appears to be buried in the Ab-peptide (peptide mimicking the 412–423 region) interface. Attachment of an N-glycan at N415 would sterically hinder the formation of E2-Ab complex and therefore cause HCV resistance to AP33 and HCV1.
However, when this occurs, escape is not associated with other broadly nAbs to this region, as represented by antibodies, HC33.1 and HC33.4. The co-crystal structure of these Abs with peptide mimicking the 412–423 region displayed a different conformation where the glycan on N415 is surface exposed. In fact, the glycan shift is associated with increased sensitivity to neutralization by the HC33 broadly nAbs. It was determined that the 415 position is associated with the AP33 epitope but not the HC33 epitopes [27,34,55]. Lastly, the binding of anti-HVRl Abs to the C-terminal region of HVR1 interferes with the binding of HC33.1 by steric hindrance. Conformational flexibility seems to be one of the main mechanisms used by HCV to evade immune response.
Escape from multiple bNAbs by distant mutations
Identification of mutations at E2 residues 501 and 506 positions leads to viral escape from many bNAbs to different regions in E2 that are reflected by decreased E2 binding to CD81. These residues flank the β-sandwich domain of the E2 core protein. These mutations must have an overall structural effect on E2. While viral fitness is compromised, viral persistence is observed [56]. In the homology modelled HCV E2, the residues 501 and 506 form several hydrogen bonds to maintain an intact 3D structure of the core domain (Figure 3D). The residue V506 is buried in the hydrophobic core stabilizing the β-sandwich fold. The hydrophobic core is formed by the residues: V497, V516, F537, L539 and W554. Any conformational change in the loop 500–506 or the β-sandwich core structure, in addition to altering the epitope accessibility, might also affect the conformation of the CD81 binding loop, which is stacked in front of the loop 500–506. When the mutations at 501 and 506 are converted back to an earlier isolate, N501S and A506V, wild-type infectivity is restored and sensitivity to bNAbs is re-established.
Escape from neutralization at sites distinct from the epitope are not uncommon in viruses. The complex folding and use of oligomeric assemblies create opportunities for dynamics that allow viral surface proteins increased freedom of movement. Thus, amino acid substitution at a flexible site may impose conformational influence at a distant site, such as an epitope. Identification of glycoprotein interactions and dynamics through structure-based approaches should help to further explain neutralizing antibody escape from mutations at a distance.
Conclusion
Several structures of nAbs in complex with truncated E2 core domains and E2-derived peptides were valuable in mapping epitopes and to understand neutralization mechanisms, a significant first step towards immunogen design. However, the structures of HVR1, HVR2 and the C-terminal regions of E2, the structure of E1E2 heterodimer and its oligomeric state on the HCV particle remain unknown. Furthermore, it has recently been shown that there is inherent structural flexibility in the E2 protein, which might be employed as an immune evasion strategy. The conformation of the unresolved regions of E2 and the structure of E1E2 heterodimers in the context of their interactions with nAbs and receptor molecules is a vital missing piece of the puzzle to understand immune evasion mechanisms and the design of vaccines based on structure. Utilization of structure-based design approaches similar to the introduction of proline mutations in SARS-CoV2, HIV, Influenza and RSV might assist in the development of a stable, conformation specific vaccine against HCV. The possibility of cooperativity within oligomeric association of E1E2 heterodimers on the virion surface adds additional complexity that is unclear at this time.
Acknowledgements
This work was supported in part by National Institute of Allergy and Infectious Diseases/NIH grants R21-AI126582 (SKHF), U19-AI123862 (SKHF) and R01-AI132213 (SKHF). It was also supported in part by the National Institute of Allergy and Infectious Disease/National Institutes of Health under contract HHSN272201700060 C to PIs Karla Satchell and RJK.
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M: Isolation of a Cdna Clone Derived from a Blood-Borne Non-a, Non B Viral-Hepatitis Genome. Science 1989, 244:359–362. [DOI] [PubMed] [Google Scholar]
- 2.Houghton M: Discovery of the hepatitis C virus. Liver International 2009, 29:82–88. [DOI] [PubMed] [Google Scholar]
- 3.Bukh J: The history of hepatitis C virus (HCV): Basic research reveals unique features in phylogeny, evolution and the viral life cycle with new perspectives for epidemic control. Journal of Hepatology 2016, 65:S2–S21. [DOI] [PubMed] [Google Scholar]
- 4.Ryerson AB, Schillie S, Barker LK, Kupronis BA, Wester C: Vital Signs: Newly Reported Acute and Chronic Hepatitis C Cases - United States, 2009–2018. Mmwr-Morbidity and Mortality Weekly Report 2020, 69:399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKnight-Eily LRO CA; Strine TW; Verlenden J; Hollis NT; Njai R; Mitchell EW; Board A; Puddy R; Thomas C;: Racial and Ethnic Disparities in the Prevalence of Stress and Worry, Mental Health Conditions, and Increased Substance Use Among Adults During the COVID-19 Pandemic — United States, April and May 2020. Morbidity and Mortality Weekly Report 2021, 70:162–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Messina JP, Humphreys I, Flaxman A, Brown A, Cooke GS, Pybus OG, Barnes E: Global Distribution and Prevalence of Hepatitis C Virus Genotypes. Hepatology 2015, 61:77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cuypers L, Li GD, Libin P, Piampongsant S, Vandamme AM, Theys K: Genetic Diversity and Selective Pressure in Hepatitis C Virus Genotypes 1–6: Significance for Direct-Acting Antiviral Treatment and Drug Resistance. Viruses-Basel 2015, 7:5018–5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pawlotsky JM: New Hepatitis C Therapies: The Toolbox, Strategies, and Challenges. Gastroenterology 2014, 146:1176–1192. [DOI] [PubMed] [Google Scholar]
- 9.Midgard H, Bjoro B, Maeland A, Konopski Z, Kileng H, Damas JK, Paulsen J, Heggelund L, Sandvei PK, Ringstad JO, et al. : Hepatitis C reinfection after sustained virological response. Journal of Hepatology 2016, 64:1020–1026. [DOI] [PubMed] [Google Scholar]
- 10.Forns X, Bukh J, Purcell RH: The challenge of developing a vaccine against hepatitis C virus. Journal of Hepatology 2002, 37:684–695. [DOI] [PubMed] [Google Scholar]
- 11.Houghton M, Abrignani S: Prospects for a vaccine against the hepatitis C virus. Nature 2005, 436:961–966. [DOI] [PubMed] [Google Scholar]
- 12.Houghton M: Prospects for prophylactic and therapeutic vaccines against the hepatitis C viruses. Immunological Reviews 2011, 239:99–108. [DOI] [PubMed] [Google Scholar]
- 13.Pierce BG, Keck ZY, Foung SKH: Viral evasion and challenges of hepatitis C virus vaccine development. Current Opinion in Virology 2016, 20:55–63.** - This article descirbes the challenges and efforts to develop a universal HCV vaccine.
- 14.Penin F, Dubuisson J, Rey FA, Moradpour D, Pawlotsky JM: Structural biology of hepatitis C virus. Hepatology 2004, 39:5–19. [DOI] [PubMed] [Google Scholar]
- 15.Keck ML, Wrensch F, Pierce BG, Baumert TF, Foung SKH: Mapping Determinants of Virus Neutralization and Viral Escape for Rational Design of a Hepatitis C Virus Vaccine. Frontiers in Immunology 2018, 9.** - The review outlines the organization of antigenic domains and epitopes within each domain of E2 that are associated with escape or arc more invariant and essential for viral fitness, receptor binding, and viral entry.
- 16.Wrensch F, Crouchet E, Ligat G, Zeisel MB, Keck ZY, Foung SKH, Schuster C, Baumert TF: Hepatitis C Virus (HCV)-Apolipoprotein Interactions and Immune Evasion and Their Impact on HCV Vaccine Design. Front Immunol 2018, 9:1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Catanese MT, Uryu K, Kopp M, Edwards TJ, Andrus L, Rice WJ, Silvestry M, Kuhn RJ, Rice CM: Ultrastructural analysis of hepatitis C virus particles. Proc Natl Acad Sci U S A 2013, 110:9505–9510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merz A, Long G, Hiet MS, Brugger B, Chlanda P, Andre P, Wieland F, Krijnse-Locker J, Bartenschlager R: Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J Biol Chem 2011, 286:3018–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freedman H, Logan MR, Law JL, Houghton M: Structure and Function of the Hepatitis C Virus Envelope Glycoproteins E1 and E2: Antiviral and Vaccine Targets. ACS Infect Dis 2016, 2:749–762. [DOI] [PubMed] [Google Scholar]
- 20.Dubuisson J, Hsu HH, Cheung RC, Greenberg HB, Russell DG, Rice CM: Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J Virol 1994, 68:6147–6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faison P, Bartosch B, Alsaleh K, Tews BA, Loquet A, Ciczora Y, Riva L, Montigny C, Montpellier C, Duverlie G, et al. : Hepatitis C Virus Envelope Glycoprotein E1 Forms Trimers at the Surface of the Virion. Journal of Virology 2015, 89:10333–10346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sevvana M, Kuhn RJ: Mapping the diverse structural landscape of the flavivirus antibody repertoire. Current Opinion in Virology 2020, 45:51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong L, Giang E, Nieusma T, Kadam RU, Cogburn KE, Hua Y, Dai X, Stanfield RL, Burton DR, Ward AB, et al. : Hepatitis C virus E2 envelope glycoprotein core structure. Science 2013, 342:1090–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pierce BG, Keck ZY, Lau P, Fauvelle C, Gowthaman R, Baumert TF, Fuerst TR, Mariuzza RA, Foung SKH: Global mapping of antibody recognition of the hepatitis C virus E2 glycoprotein: Implications for vaccine design. Proc Natl Acad Sci U S A 2016, 113:E6946–E6954.** - The article analyzes the E2 determinants critical for modulating modulating antibody recognition, local and global E2 stability, and viral escape.
- 25.Tzarum N, Wilson IA, Law M: The Neutralizing Face of Hepatitis C Virus E2 Envelope Glycoprotein. Front Immunol 2018, 9:1315.** - This review analyzes the structural aspects of E2 neutralizing sites in complex with broadly neutralizing antibodies, to understand the functional and preferred conformations for neutralization, and for viral escape.
- 26.Khan AG, Whidby J, Miller MT, Scarborough H, Zatorski AV, Cygan A, Price AA, Yost SA, Bohannon CD, Jacob J, et al. : Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature 2014, 509:381–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Pierce BG, Wang Q, Keck ZY, Fuerst TR, Foung SK, Mariuzza RA: Structural basis for penetration of the glycan shield of hepatitis C virus E2 glycoprotein by a broadly neutralizing human antibody. J Biol Chem 2015, 290:10117–10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pantua H, Diao J, Ultsch M, Hazen M, Mathieu M, McCutcheon K, Takeda K, Date S, Cheung TK, Phung Q, et al. : Glycan shifting on hepatitis C virus (HCV) E2 glycoprotein is a mechanism for escape from broadly neutralizing antibodies. J Mol Biol 2013, 425:1899–1914. [DOI] [PubMed] [Google Scholar]
- 29.Krey T, Meola A, Keck ZY, Damier-Piolle L, Foung SK, Rey FA: Structural basis of HCV neutralization by human monoclonal antibodies resistant to viral neutralization escape. PLoS Pathog 2013, 9:e1003364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colbert MD, Flyak AI, Ogega CO, Kinchen VJ, Massaccesi G, Hernandez M, Davidson E, Doranz BJ, Cox AL, Crowe JE Jr, et al. : Broadly Neutralizing Antibodies Targeting New Sites of Vulnerability in Hepatitis C Virus E1E2. J Virol 2019, 93.** - The articles reports novel broadly neutralizing antibody epitopes.
- 31.Cao L, Yu B, Kong D, Cong Q, Yu T, Chen Z, Hu Z, Chang H, Zhong J, Baker D, et al. : Functional expression and characterization of the envelope glycoprotein E1E2 heterodimer of hepatitis C virus. PLoS Pathog 2019, 15:e1007759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flyak AI, Ruiz S, Colbert MD, Luong T, Crowe JE Jr, Bailey JR, Bjorkman PJ: HCV Broadly Neutralizing Antibodies Use a CDRH3 Disulfide Motif to Recognize an E2 Glycoprotein Site that Can Be Targeted for Vaccine Design. Cell Host Microbe 2018, 24:703–716 e703.** - Here, the crystal structures of full-length E2 ectodomain complexes with HEPC3 and HEPC74, reveal lock-and-key antibody-antigen interactions, previously uncharacterized E2 core regions, and an antibody CDRH3 disulfide motif that interacts with a conserved epitope.
- 33.Nayak A, Pattabiraman N, Fadra N, Goldman R, Kosakovsky Pond SL, Mazumder R: Structure-function analysis of hepatitis C virus envelope glycoproteins E1 and E2. J Biomol Struct Dyn 2015, 33:1682–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meola A, Tarr AW, England P, Meredith LW, McClure CP, Foung SK, Mc Keating JA, Ball JK, Rey FA, Krey T: Structural flexibility of a conserved antigenic region in hepatitis C virus glycoprotein E2 recognized by broadly neutralizing antibodies. J Virol 2015, 89:2170–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flyak AI, Ruiz SE, Salas J, Rho S, Bailey JR, Bjorkman PJ: An ultralong CDRH2 in HCV neutralizing antibody demonstrates structural plasticity of antibodies against E2 glycoprotein. Elife 2020, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Potter JA, Owsianka AM, Jeffery N, Matthews DJ, Keck ZY, Lau P, Foung SK, Taylor GL, Patel AH: Toward a hepatitis C virus vaccine: the structural basis of hepatitis C virus neutralization by AP33, a broadly neutralizing antibody. J Virol 2012, 86:12923–12932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeisel MB, Fofana I, Fafi-Kremer S, Baumert TF: Hepatitis C virus entry into hepatocytes: molecular mechanisms and targets for antiviral therapies. J Hepatol 2011, 54:566–576. [DOI] [PubMed] [Google Scholar]
- 38.Bartosch B, Vitelli A, Granier C, Goujon C, Dubuisson J, Pascale S, Scarselli E, Cortese R, Nicosia A, Cosset FL: Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J Biol Chem 2003, 278:41624–41630. [DOI] [PubMed] [Google Scholar]
- 39.Goffard A, Dubuisson J: Glycosylation of hepatitis C virus envelope proteins. Biochimie 2003, 85:295–301. [DOI] [PubMed] [Google Scholar]
- 40.Castelli M, Clementi N, Pfaff J, Sautto GA, Diotti RA, Burioni R, Doranz BJ, Dal Peraro M, Clementi M, Mancini N: A Biologically-validated HCV E1E2 Heterodimer Structural Model. Sci Rep 2017, 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lavie M, Goffard A, Dubuisson J: Assembly of a functional HCV glycoprotein heterodimer. Curr Issues Mol Biol 2007, 9:71–86. [PubMed] [Google Scholar]
- 42.El Omari K, Iourin O, Kadlec J, Sutton G, Harlos K, Grimes JM, Stuart DI: Unexpected structure for the N-terminal domain of hepatitis C virus envelope glycoprotein E1. Nat Commun 2014, 5:4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prentoe J, Bukh J: Hypervariable Region 1 in Envelope Protein 2 of Hepatitis C Virus: A Linchpin in Neutralizing Antibody Evasion and Viral Entry. Front Immunol 2018, 9:2146.** - This review describes the functional aspects of HVR1 in modulating HCV entry via co-receptors and its role as a decoy epitope.
- 44.Lavie M, Hanoulle X, Dubuisson J: Glycan Shielding and Modulation of Hepatitis C Virus Neutralizing Antibodies. Frontiers in Immunology 2018, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prentoe J, Velazquez-Moctezuma R, Foung SKH, Law M, Bukh J: Hypervariable Region 1 Shielding of Hepatitis C Virus Is a Main Contributor to Genotypic Differences in Neutralization Sensitivity. Hepatology 2016, 64:1881–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Law JLM, Logan M, Wong J, Kundu J, Hockman D, Landi A, Chen C, Crawford K, Wininger M, Johnson J, et al. : Role of the E2 Hypervariable Region (HVR1) in the Immunogenicity of a Recombinant Hepatitis C Virus Vaccine. Journal of Virology 2018, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meunier JC, Russell RS, Goossens V, Priem S, Walter H, Depla E, Union A, Faulk KN, Bukh J, Emerson SU, et al. : Isolation and characterization of broadly neutralizing human monoclonal antibodies to the E1 glycoprotein of hepatitis C virus. Journal of Virology 2008, 82:966–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keck ZY, Sung VMH, Perkins S, Rowe J, Paul S, Liang T, Lai MMC, Foung SKH: Human monoclonal antibody to hepatitis C virus E1 glycoprotein that blocks virus attachment and viral infectivity. Journal of Virology 2004, 78:7257–7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kong L, Kadam RU, Giang E, Ruwona TB, Nieusma T, Culhane JC, Stanfield RL, Dawson PE, Wilson IA, Law M: Structure of Hepatitis C Virus Envelope Glycoprotein E1 Antigenic Site 314–324 in Complex with Antibody IGH526. Journal of Molecular Biology 2015, 427:2617–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tzarum N, Giang E, Kadam RU, Chen F, Nagy K, Augestad EH, Velazquez-Moctezuma R, Keck ZY, Hua YZ, Stanfield RL, et al. : An alternate conformation of HCV E2 neutralizing face as an additional vaccine target. Science Advances 2020, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keck ZY, Olson O, Gal-Tanamy M, Xia JM, Patel AH, Dreux M, Cosset FL, Lemon SM, Foung SKH: A point mutation leading to hepatitis C virus escape from neutralization by a monoclonal antibody to a conserved conformational epitope. Journal of Virology 2008, 82:6067–6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keck ZY, Xia J, Wang Y, Wang W, Krey T, Prentoe J, Carlsen T, Li AY, Patel AH, Lemon SM, et al. : Human monoclonal antibodies to a novel cluster of conformational epitopes on HCV E2 with resistance to neutralization escape in a genotype 2a isolate. PLoS Pathog 2012, 8:e1002653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keck ZY, Wang Y, Lau P, Lund G, Rangarajan S, Fauvelle C, Liao GC, Holtsberg FW, Warfield KL, Aman MJ, et al. : Affinity Maturation of a Broadly Neutralizing Human Monoclonal Antibody That Prevents Acute Hepatitis C Virus Infection in Mice. Hepatology 2016, 64:1922–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pantua H, Diao J, Ultsch M, Hazen M, Mathieu M, McCutcheon K, Takeda K, Date S, Cheung TK, Phung Q, et al. : Glycan Shifting on Hepatitis C Virus (HCV) E2 Glycoprotein Is a Mechanism for Escape from Broadly Neutralizing Antibodies. Journal of Molecular Biology 2013, 425:1899–1914. [DOI] [PubMed] [Google Scholar]
- 55.Keck ZY, Girard-Blanc C, Wang WY, Lau P, Zuiani A, Rey FA, Krey T, Diamond MS, Foung SKH: Antibody Response to Hypervariable Region 1 Interferes with Broadly Neutralizing Antibodies to Hepatitis C Virus. Journal of Virology 2016, 90:3112–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Keck ZY, Li SH, Xia J, von Hahn T, Balfe P, Mc Keating JA, Witteveldt J, Patel AH, Alter H, Rice CM, et al. : Mutations in Hepatitis C Virus E2 Located outside the CD81 Binding Sites Lead to Escape from Broadly Neutralizing Antibodies but Compromise Virus Infectivity. Journal of Virology 2009, 83:6149–6160. [DOI] [PMC free article] [PubMed] [Google Scholar]