Abstract
Background
COVID-19 diagnosis in symptomatic patients is an important factor for arranging the necessary lifesaving facilities like ICU care and ventilator support. For this purpose, we designed a computer-aided diagnosis and severity detection method by using transfer learning and a back propagation neural network.
Method
To increase the learning capability, we used data augmentation. Most of the previously done works in this area concentrate on private datasets, but we used two publicly available datasets. The first section diagnose COVID-19 from the input CT image using the transfer learning of the pre-trained network ResNet-50. We used ResNet-50 and DenseNet-201 pre-trained networks for feature extraction and trained a back propagation neural network to classify it into High, Medium, and Low severity.
Results
The proposed method for COVID-19 diagnosis gave an accuracy of 98.5% compared with the state-of-the-art methods. The experimental evaluation shows that combining the ResNet-50 and DenseNet-201 features gave more accurate results with the test data. The proposed system for COVID-19 severity detection gave better average classification accuracy of 97.84% compared with the state-of-the-art methods. This enables medical practitioners to identify the resources and treatment plans correctly.
Conclusions
This work is useful in the medical field as a first-line severity risk detection that is helpful for medical personnel to plan patient care and assess the need for ICU facilities and ventilator support. A computer-aided system that is helpful to make a care plan for the huge amount of patient inflow each day is sure to be an asset in these turbulent times.
Keywords: Computed tomography, COVID-19, DenseNet-201, ResNet-50, Transfer learning, Neural network
Introduction
Coronaviruses cause infections in the lungs and lead to more severe Middle East Respiratory Syndrome and Severe Acute Respiratory Syndrome cases. The World Health Organization (WHO) announced COVID-19 as a Public Health Emergency of International Concern (PHEIC) on January 31, 2020 [1]. On 23rd April, the COVID-19 update by the WHO announced that globally, there were 144,358,956 confirmed cases of COVID-19, including 3,066,113 deaths. Coronaviruses are zoonotic, which means it can be transmitted between animals and people, and the spreading rate is also high. It is essential to diagnose the disease so that the patient can be isolated as soon as possible.
Many methods have been used to confirm the suspected COVID-19 cases by clinicians, like real-time Reverse Transcription Polymerase Chain Reaction (RT-PCR), nonPCR tests such as isothermal nucleic acid amplification technology [2] non-contrast chest Computed Tomography (CT) and radiographs [3], etc. The RT-PCR test has been widely used to confirm COVID-19. The serial CT imaging is helpful to assess the severity of the lung involvement for COVID-19 patients. Number of works were reported for the detection of COVID-19 from X-ray image data set. The COVIDX-Net, a deep learning framework was developed by Hemdan et al. [4]. In this work, they analysed the performance of DenseNet-201,VGG19, InceptionV3, ResNetV2, Inception, Xception ResNetV2, and MobileNetV2. They used the data by [5], [6]. From the analysis, the results obtained from VGG19 and DenseNet-201 models performed well with an accuracy of 90%. A tailored deep learning-based framework, COVID-Net, was developed by Wang and Wong to detect COVID-19 from chest X-ray images [7]. The architecture consists of 1 × 1 convolutions, depth-wise and residual architecture. They proposed a multi-classification as normal, viral infection, bacterial infection, and COVID-19 infection. They used the data set provided by [5], [8] and got an accuracy of 83.5%. A study conducted by Aditya Borakati et al. analyzed the efficiency of common imaging modalities, chest X-ray (CXR) and CT, for diagnosis of COVID-19 [9]. In conclusion, the CT has more diagnostic performance than the CXR for COVID-19. CT should be strongly considered in the initial assessment for suspected COVID-19. There exist many methods for the detection of COVID-19 from the CT images. To diagnose the COVID-19, a self-supervised learning with transfer learning was proposed by He et al. [10]. Zhao et al. proposed a multi-tasking learning approach for the binary classification for COVID-19 [11]. To avoid the scarcity of the limited benchmark datasets for COVID-19, especially in chest CT images, Loey et al. proposed a method that uses data augmentation and CGAN to generate more CT images [12]. Liu et al. proposed a Lesion-Attention Deep Neural Network to perform the binary classification from the chest CT images [13]. The machine learning methods were explored by Barstugan et al. by making use of different feature extraction methods and support vector machine as a classifier [14]. Wang et al. proposed a method which utilizes a weakly supervised framework for COVID-19 classification [15]. They used the UNet architecture and got an accuracy of 90.10%.
Pathak et al. developed a Deep Transfer Learning method based on ResNet-50 and the 2D Convolutional Neural Network (CNN) [16]. An attention-based deep learning method was proposed by Han et al. [17]. Harmon et al. developed a lung segmentation algorithm to identify and localize whole lung regions for the prediction of COVID-19 disease [18].
COVID-19 can lead to pneumonia, Acute Respiratory Distress Syndrome (ARDS), and similar life-threatening situations. Hence, timely detection and assessment of severity and prognosis are important in the procurement of life-saving measures like ICU care and ventilator support. CT imaging is very helpful in diagnosing and assessing the severity of COVID-19 patients. The CO-RADS classification is a standardised reporting system for patients with suspected COVID-19 infection developed for a moderate to high prevalence setting. Based on the CT findings, the level of suspicion of COVID-19 infection is graded from very low or CO-RADS 1 up to very high or CO-RADS 5 and the severity and stage of the disease is determined with remarks on comorbidity and a differential diagnosis.
We are proposing a two-stage architecture for symptomatic COVID-19 patients; first, COVID-19 Detection – to detect whether a patient has COVID-19, second, COVID-19 Severity Detection – to detect the severity condition of the patient. This work advances state-of-the-art methods in COVID-19 detection and severity prediction. To improve the performance of deep learning architecture, we used data augmentation to increase the number of samples. The training and testing are performed on the publicly available data set. To get a proper dataset, combined the SARS-CoV-2 CT-scan dataset [19] and the COVID CT data set [11]. To avoid the gradient vanishing problem, we used the ResNet-50 and the DenseNet-201 architecture. The results obtained are totally in-line with the CO-RADS clinical finding, which provides emphasis to our proposal.
Materials and methods
Dataset
For the COVID-19 Severity detection, the metadata of the patient's CT images is also needed. So here we used two publicly available data set which was really proven.
SARS-CoV-2 CT-scan dataset
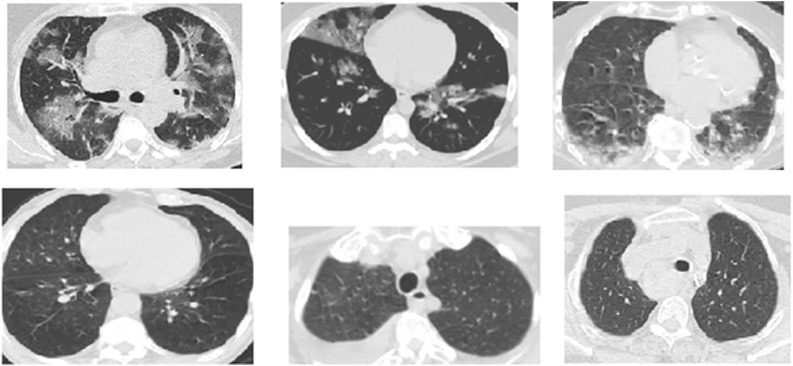
The SARS-CoV-2 CT-scan dataset [19] contains both positive and negative data; the data is collected from both males and females. The data set consists of 2482 CT scans, 1252 images are positive, and 1230 images are negative. The images have no standard size. Fig. 1 . shows a sample image from this data set.
Fig. 1.
Examples of CT COVID-19 images (positive cases top row) (negative cases bottom row) from the SARS-CoV-2 CT-scan dataset.
COVID-CT dataset
The authors [11] used PyMuPDF software to extract images of patients infected with COVID-19 from scientific articles, and images were collected from hospitals. Metadata were manually extracted and associated with each image: patient age, gender, location, medical history, scan time, the severity of COVID-19, and medical report. The data set consists of 349 COVID-19 images and 463 non-COVID-19 CT images. But, the non-COVID images consist of other lung infections.
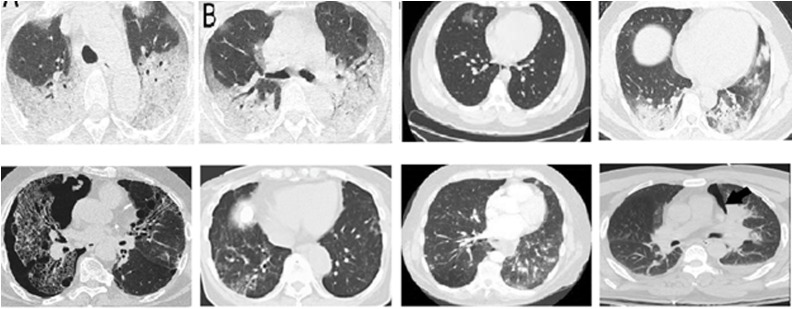
Comparing with the SARS-CoV-2 CT-scan dataset, the non-COVID CT images are more complex, and it is really a challenging task to distinguish as either COVID or non-COVID. Fig. 2 shows the sample COVID-19 and non-COVID-19 images.
Fig. 2.
Examples of CT COVID-19 images (positive cases top row) (negative cases bottom row) from the COVID-CT dataset.
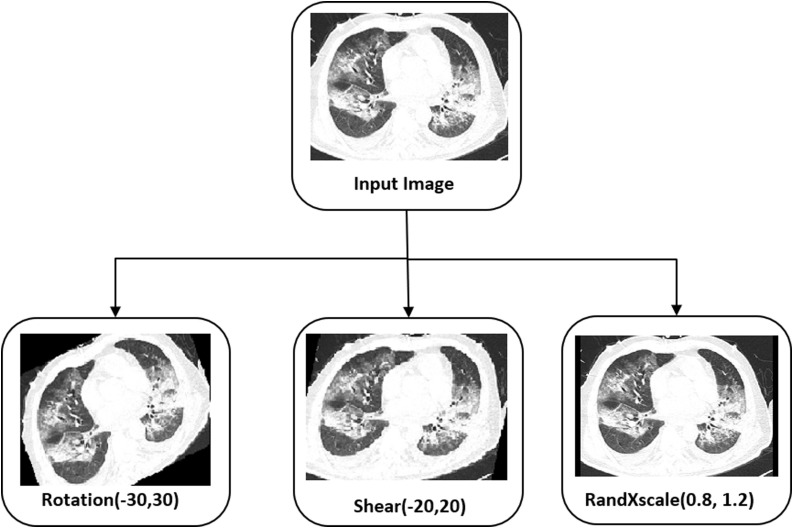
The proposed work aims to find whether a person is COVID positive and assess the patient’s severity condition so that the medical team can prepare the facilities needed in advance, such as a ventilator, availability of oxygen, etc. The [11] contains 349 COVID-19 images with the details to predict each patient's severity condition. To build a fully automatic system, we consider the data set [11], [19]. Since the metadata regarding the COVID-19 severity is available with [11], we took COVID-19 positive samples from [11]. 60% of positive data randomly chosen for testing including high, low, medium severity conditions. For the remaining data, augmentation is applied to get many samples. We used the data augmentation rotation in the range of −30 to 30, shear in the range of −20 to 20, and RandXscale in the range of 0.8 to 1.2. Since we have enough COVID-19 negative sample images, we do not apply any augmentation for non-COVID images and the images are taken from the dataset [19]. Fig. 3 shows the augmentation performed in the proposed method to improve the classification accuracy for COVID-19 severity detection. Table 1 shows the splitting up of data for the training, validation, and testing.
Fig. 3.
Augmentation used by the proposed method.
Table 1.
Dataset used for the proposed work with data augmentation.
| Category | Data | Training data | Validation data | Testing data |
|---|---|---|---|---|
| COVID-19 +ve images | 349 | 466 (augmented) | 82 (augmented) | 212 |
| COVID-19 −ve images | 736 | 466 | 82 | 188 |
Methodology
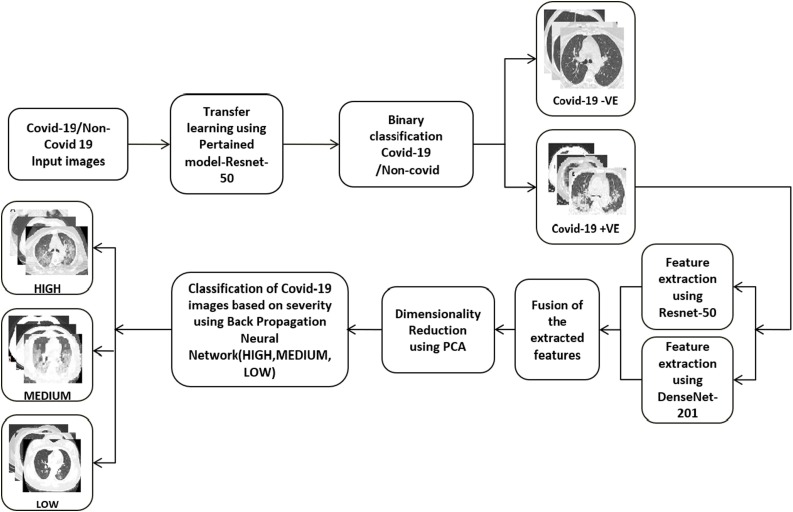
We developed a two-stage architecture for the severity detection of COVID-19 patients. If the input CT image is inputted to the architecture, The COVID-19 detection module detects whether that patient has COVID-19 or not. If the patient is diagnosed with COVID-19, the COVID-19 severity detection module finds out the severity of that patient. For the detection of severity, serial CT images of a single patient over two weeks are analyzed to create the severity prediction model. The overall architecture of the proposed system is shown in Fig. 4 .
Fig. 4.
Architecture of the proposed system.
In the COVID-19 detection, after preprocessing the input images, a ResNet-50 architecture is used to identify COVID-19. In the COVID-19 severity detection system, we utilize two pre-trained networks, Resnet-50, DenseNet-201, and a backpropagation network, to determine the severity of the COVID-19.
Preprocessing
The preprocessing is carried out to make the input images are of same size and same bit depth. The images are resized to the corresponding CNN's input size.
COVID-19 detection
Deep learning is a subfield of an artificial neural network consisting of algorithms inspired by the brain's function. In Deep Learning, feature extraction is done by itself with the help of several convolutional filters. Each layer of the deep learning architecture learns different features. By changing the filter size, it can generate more feature maps. These tensor of feature maps are utilized to solve the problem. Different deep learning architectures are designed by varying the number of convolutional layers, activation functions, pooling methodologies, etc. There exists several pre-trained deep learning architectures such as AlexNet, GoogLeNet, ResNet, VGG-16, VGG-19, etc. When we used a pre-trained system without its last layer, it acts as a good feature extractor for the input images. We analyzed the performance of various pre-trained networks such as Alexnet, VGG-16, VGG-19, ResNet-50, ResNet-101, and with DenseNet-201 for detecting the COVID-19 disease. The results are shown in Table 2 . From the experiments, it was observed that ResNet-50 provided the best results.
Table 2.
Evaluation of different pre-trained network.
| Pre-trained network | Accuracy (%) |
|---|---|
| AlexNet | 93.71 |
| VGG-16 | 94.62 |
| VGG-19 | 93.56 |
| ResNet-50 | 98.5 |
| ResNet-101 | 95.32 |
| DenseNet-201 | 97.95 |
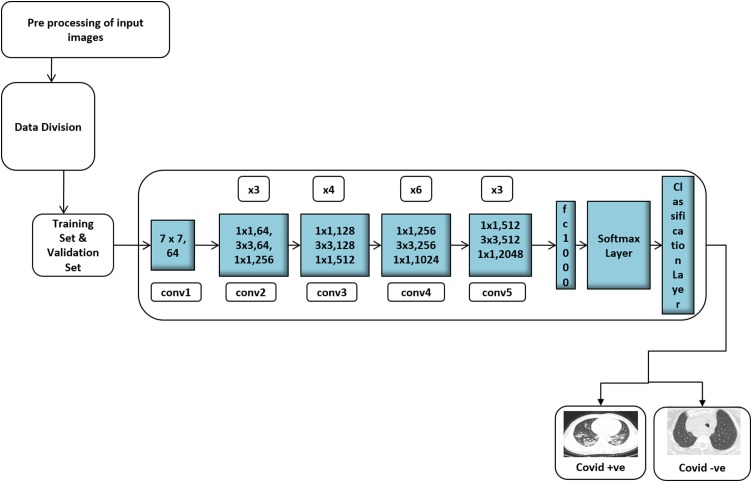
ResNet, can input an image of size 224 × 224 × 3 is the short name of Residual Network that supports Residual Learning. Fig. 5 shows the detailed architecture of the COVID-19 detection. In ResNet-50, at the first layer, a kernel size of 7 * 7 convolution and 64 different kernels, all with a stride of size two, is used. After that, max-pooling with a stride of size two is used. In the second convolution, there exists a kernel size of 1 × 1, 3 × 3, 1 × 1, 64, 64, and 256 kernels, respectively. These 3 layers are repeated three times, thus getting a 9-layer feature map. The third block consists of a kernel size of 1 × 1, 128 kernel, 3 × 3 128 kernels, and 1 × 1 512 kernels. It is repeated for four times, giving a feature map of 12 layers. The next block has a kernel size of 1 × 1, 3 × 3, 1 × 1, 256, 256, and 1024 kernels. This process is repeated six times, giving a total of 18 layers. Then next block contains 1 × 1,512 kernels with two more of 3 × 3, 512 and 1 × 1, 2048, which was repeated three times, giving us a total of 9 layers. Finally, the network has an average pool followed by a fully connected layer with 1000 nodes and a softmax function. The gradient vanishing problem caused while CNN addresses the complex problem can be solved using skip connections. By using skip connections, the layer activations of a particular layer are added directly to the activation of some other layer that is deeper in the network. There exist more skip connections and the Rectified Linear Unit (ReLU) activation function used in the ResNet-50. It is a convolutional neural network that has 50 layers deep.
Fig. 5.
Architecture for the COVID-19 diagnosis system.
COVID-19 severity detection
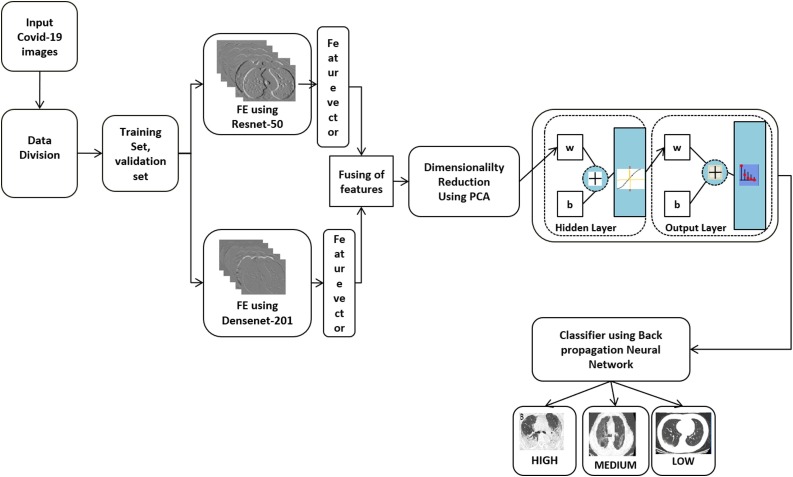
If the system diagnosed the input image as a COVID-19, the Severity Detection module processes the CT image. Fig. 6 shows the architecture of the severity detection method for COVID-19 patients. Here COVID-19 positive images of various severity levels are given as input. The metadata of 349 images in the dataset are augmented to get a good number of samples. While analyzing, it is observed that the ResNet-50 and DenseNet-201 are giving the most accurate results. So the training sets and the validation sets features are extracted using the well-known ResNet-50 and DenseNet-201. From the fully connected layer ’fc1000’ of ResNet-50 we got 1000 features.
Fig. 6.
Architecture for the COVID-19 severity detection system.
In DenseNet-201, network connects each layer to every succeeding layer in a feed-forward manner. So, it is referred to as Dense Convolutional Network (DenseNet-201). The feature maps of all the preceding layers are used as input for each succeeding layer. In this network, each layer receiving a ’collective knowledge’ from all the preceding layers. In the DenseNet-201, the connection is added from every previous layer. The advantages of the DenseNet are, the vanishing problem is reduced since it contains feature maps of all preceding layers. It strengthens feature propagations, and it also has less number of parameters.
For this work, the features are extracted from the fully connected layer ’fc1000’. That is, in total, we got 2000 image features. Since we have many images, and for each image, we extracted 2000 image features, the dimensionality is reduced using Principal Component Analysis (PCA). The reduced feature set is fed as the input to the feed-forward network and is used to classify the input images into High, Medium, and Low severity. The number of hidden layer neurons used is 10, and the activation function used is Sigmoid. Since weights are adjusted in the steepest descent direction, the backpropagation algorithm does not guarantee the fastest convergence. To avoid that, in this work, we used the scaled conjugate gradient backpropagation. Because of the searching along with the conjugate directions in this method produce faster convergence.
Results and discussions
The results of this study indicate that it was possible to construct an accurate model to find the detection and the severity condition of the patients. To measure the prediction performance in this study, we utilized common evaluation metrics such as Specificity, Sensitivity, Accuracy, and F1-Score.
Specificity: It is defined as the ratio of true negatives (the classifier returned) to the total negatives and can be calculated as:
| (1) |
Sensitivity: It is defined as the ratio of true positives to the total actual positive and can be calculated as:
| (2) |
Accuracy: The accuracy will give the recognition rate of the classifier. It is defined as the ratio of the correct predictions by the classifier to the total number of input data and can be calculated as:
| (3) |
F1-score: It is another metric to identity the usefulness of a classifier and it considers both the precision and recall. The higher the F1-score better the predictive power of the classification algorithm. It can be calculated as:
| (4) |
1. COVID-19 detection
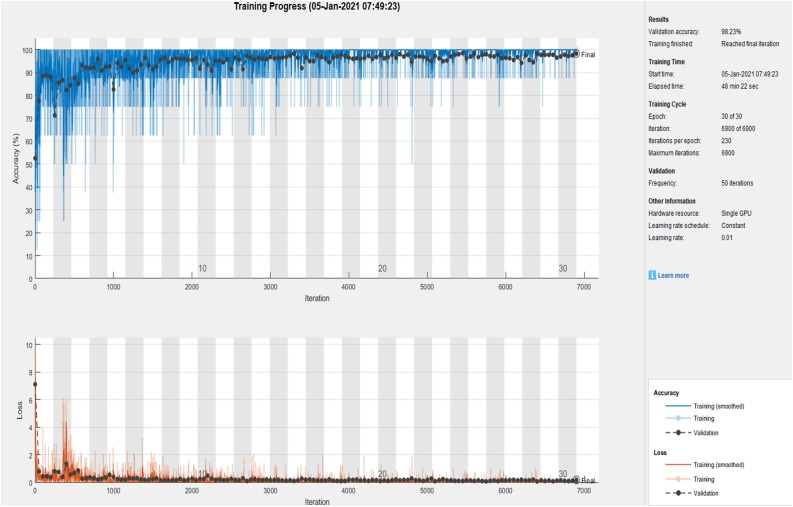
For COVID-19 detection, we trained the ResNet-50 with various initialization parameters. We tried with 10, 15, 25, and 30 epochs. The performance of COVID-19 detection is analyzed using Specificity, Sensitivity, Accuracy and F1-Score. Fig. 7 shows the Convergence graph of Accuracy and loss function using the ResNet-50 model for the 30th epoch.
Fig. 7.
Convergence graph of accuracy and loss function using ResNet-50.
Table 3shows the accuracy, while analyzed the network with the same hyper parameters with various epochs. From the experiments, we identified that the ResNet-50 with 30th iteration gives better performance.
Table 3.
Accuracy evaluation of the network with different epochs.
| Number of epochs | Validation (%) | Testing (%) |
|---|---|---|
| 15 | 93.27 | 92.80 |
| 25 | 96.34 | 97.10 |
| 30 | 98.23 | 98.5 |
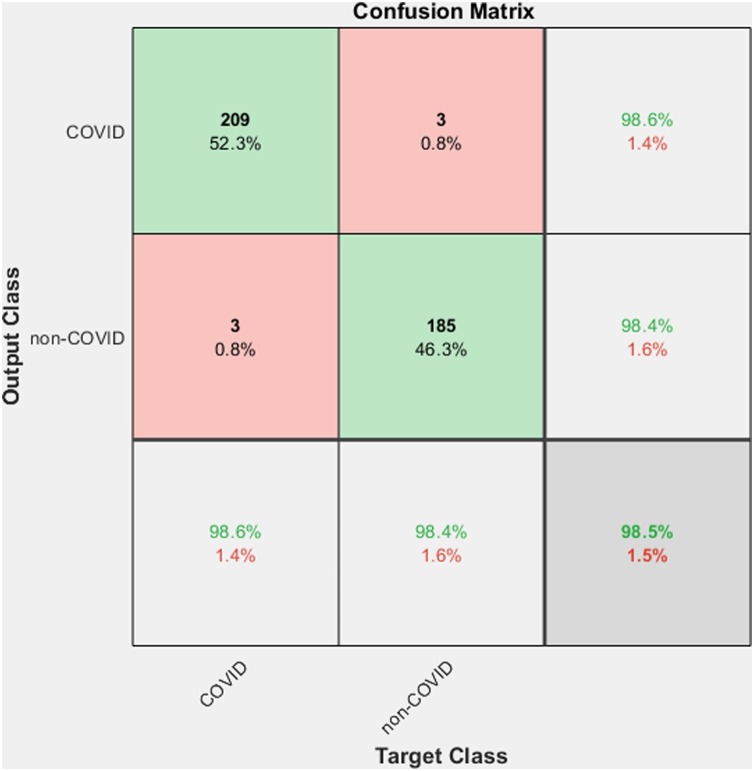
Table 4 shows the outputs we got from the first section of the proposed method. The method gives a better accuracy 98.5%. Fig. 8 shows the corresponding confusion matrix. Here out of 212 COVID-19 images, 209 images are classified into the corresponding class, and only 3 of them are misclassified in the non-COVID-19 class, thus getting a precision of 98.58%. And from a total of 188 non-COVID-19 images, only 3 of them are misclassified. The ROC curve corresponding to the output is shown in Fig. 9 . The value of Area Under the Curve (AUC) is 99.19%, which measures a classifier's performance.
Table 4.
Results obtained from the proposed COVID-19 detection method.
| Sensitivity (%) | Specificity (%) | Precision (%) | Accuracy (%) | F1-score (%) | AUC (%) |
|---|---|---|---|---|---|
| 98.58 | 98.40 | 98.58 | 98.5 | 98.58 | 99.19 |
Fig. 8.
Confusion matrix for the COVID-19 diagnosis system.
Fig. 9.
ROC for the COVID-19 diagnosis system.
Based on the literature and the survey conducted by Ozsahin et al. [20] several works are reported. Table 5 shows a comparison of the proposed method with the state-of-the-art methods. Various methods used a different number of images for training and testing.
Table 5.
Evaluation of COVID-19 detection system with different state-of-the-art methods.
| Number of subjects | Method | Sensitivity (%) | Specificity (%) | Accuracy (%) | F1-score (%) |
|---|---|---|---|---|---|
| 349 COVID-19397 non-COVID-19 | Self-supervised learning with transfer learning, DenseNet-201 [10] | NA | NA | 86 | 85 |
| 349 COVID-19 463 non-COVID-19 |
Multi-tasking learning approach [11] | NA | NA | 89 | 90 |
| 349 COVID-19 397 non-COVID-19 |
Different CNN models AlexNet, VGGNet16, VGGNet19, GoogleNet, ResNet50 [12] | NA | NA | 82.91 | NA |
| 564 COVID-19 660 non-COVID-19 |
VGG16 based lesion attention DNN [13] | 88.80 | NA | 88.60 | 87.9 |
| 313 COVID-19 229 non-COVID-19 |
UNet [15] | 90.70 | 91.1 | 90.10 | NA |
| 413 COVID-19 439 non-COVID-19 |
ResNet-50 + 2D CNN [16] | 91.46 | 94.78 | 93.02 | NA |
| 230 COVID-19 130 normal |
AD3D-MIL [17] | 97.90 | NA | 97.90 | 97.90 |
| 1029 COVID-19 1695 non-COVID-19 |
AH-Net DenseNet-201 [18] | 84.0 | 93.0 | 90.80 | NA |
| 496 COVID-19 1385 others |
CNN [21] | 94.06 | 95.47 | 94.98 | NA |
| 349 COVID-19 397 non-COVID-19 |
ResNet18 [22] | 100 | 98.60 | 99.40 | 99.5 |
| 760 COVID-19 by augmentation 736 non-COVID-19 |
ResNet-50 with data augmentation [proposed work] | 98.58 | 98.40 | 98.5 | 98.58 |
He et al. used transfer learning approach and the COVID-19 CT dataset, which contains 349 positive CT scans with clinical findings of COVID-19 and 397 negative images and gets an accuracy of 86% [10].
Zhao et al. created a COVID-19 data set and proposed a multitasking learning approach for the binary classification of COVID-19 [11]. They got an accuracy of 89%.
Loey et al. explored five different CNN models, namely, VGG16, AlexNet, VGG19, ResNet-50, and GoogleNet used 349 COVID CT scans and 397 non-COVID CT scans [12]. By analysis, they identified that ResNet-50 is the best performing model and achieved an accuracy of 82.91%. LA-DNN proposed by Liu et al. used 746 public chest positive CT images [13]. Here they performed a binary classification and a description of five lesions on the CT images associated with the positive cases. They got an accuracy of 88.6%.
The work proposed by Wang et al. used 3D CT volumes for COVID-19 classification [15]. Each patient's lung region is extracted using a trained UNet and inputted to a 3D deep neural network to predict the probability of COVID-19 infected portions. Here 499 CT volumes were used for training, and 131 CT volumes were used for testing, got an accuracy of 90.1%.
Pathak et al. used the ResNet-50 and a 2D CNN for the classification [16]. Additionally, a top-2 smooth loss function with cost-sensitive attributes is also utilized to handle noisy and imbalanced COVID-19 dataset kind of problems. They got an accuracy of 93.02%
In the work proposed by Han et al., each 3D chest CT was assigned a patient-level label [17]. 3D instances of all infection areas semantically generated by AD3D-MIL. The AD3D-MIL learned Bernoulli distributions of the bag level labels. They got an accuracy of 97.9%.
The work by Harmon et al. the lung segmentation model was trained using AH-Net architecture [18]. The network trained with LIDC dataset consisting of 1018 images and 95 in-house CT volumes. DenseNet-121 architecture is used for classification purposes. They got an accuracy of 90.8%. Jin et al. proposed a method based on CNN and they achieved an accuracy of 94.98% with a data set of 496 COVID-19 and 1385 non-COVID 19 images [21]. Ahuja et al. used data augmentation and used ResNet-18 for the classification [22]. They got an accuracy of 99.4% with a data set of 349 COVID-19 and 397 non-COVID-19 images. The proposed method used publicly available datasets and got an accuracy of 98.5% for the COVID-19 diagnosis.
2. Severity detection
Every hospital has a limited number of ICU facilities and ventilators and hence, the detection of the severity of lung involvement and subsequent need for advanced life support care is extremely important. With the staggering number of people being admitted into hospitals each day, a system which can assess patient condition and make a calculated risk analysis has become the need of the hour. Since this study's main objective is to classify the severity level of patients, various statistical measures like confusion matrix, specificity, sensitivity, F1-score, and accuracy is employed for this purpose.
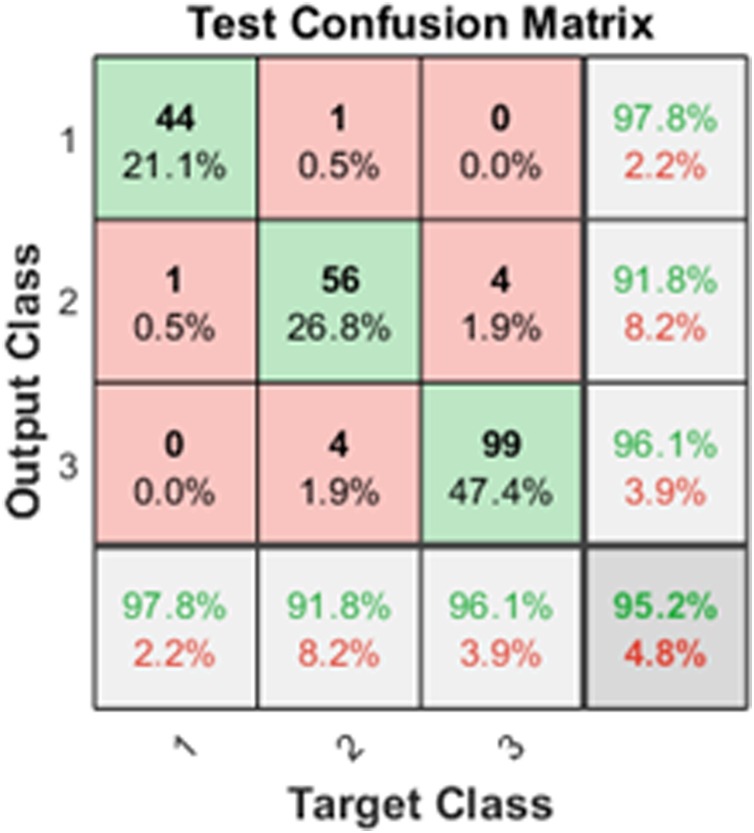
The results obtained for severity detection are shown in Table 6 . Since it is a multiclass problem, the performance measures are calculated for each of the classes [23]. Confusion matrix obtained from the Back propagation Neural Network shown in Fig. 10 .
Table 6.
Results obtained from the proposed COVID-19 severity detection method.
| Classes | Sensitivity (%) | Specificity (%) | Precision (%) | Accuracy (%) | F1-score (%) |
|---|---|---|---|---|---|
| High severity | 97.78 | 99.39 | 97.78 | 99.04 | 97.78 |
| Medium severity | 91.80 | 96.62 | 91.80 | 95.22 | 98.03 |
| Low severity | 96.12 | 96.23 | 96.12 | 96.17 | 96.12 |
Fig. 10.
Confusion matrix for the COVID-19 severity detection (High, Medium, Low).
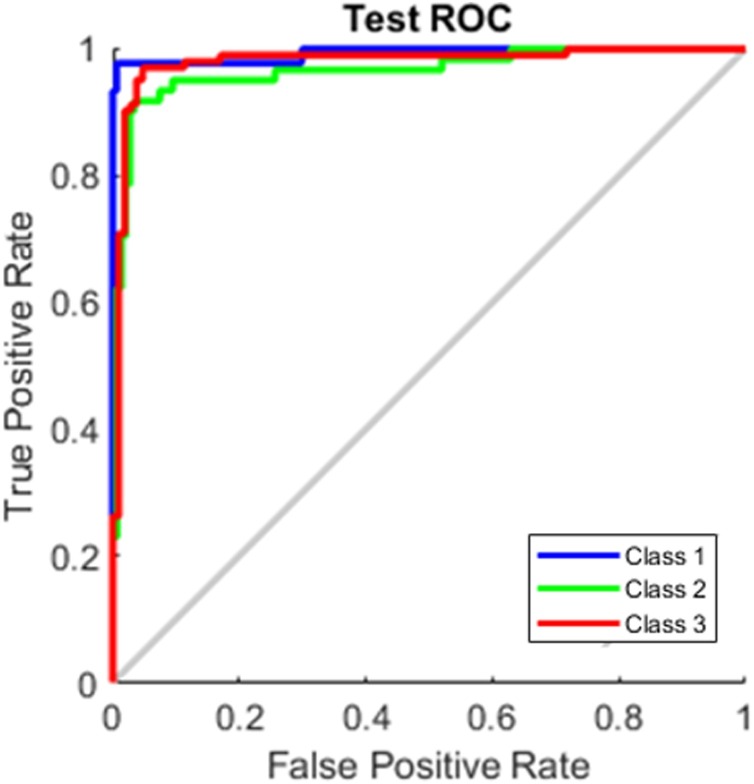
The method achieved a good result with an overall average accuracy of 97.84%. Here class 1 is High severity, class 2 is Medium severity, and class 3 is low severity. Out of 45 class1 (High) test data, 44 are classified correctly. Out of 61 samples from class 2 (Medium), one is classified in class 1 (High), and 4 is classified in class 3 (Low). For class 3, from a total of 103 images, four images are misclassified into class2 (Medium). The ROC curve corresponding to the output shown in Fig. 11 .
Fig. 11.
ROC for the COVID-19 severity detection (High, Medium, Low).
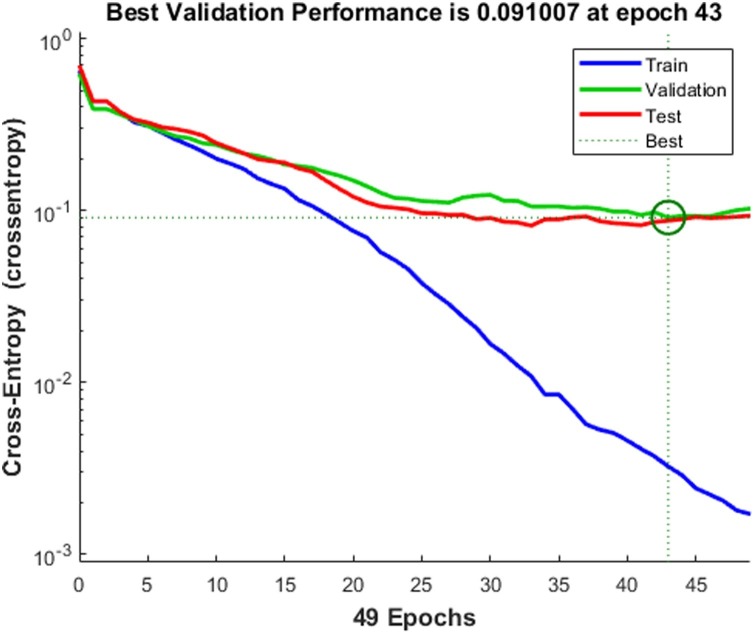
Fig. 12 shows the best validation performance at epoch 43. Several experiments with a different number of units in the hidden layer and the number of epochs are carried out. After analyzing, we fixed the number of epochs as 50. Fig. 12 shows the validation performance of the neural network.
Fig. 12.
Validation performance of the back propagation neural network.
Comparison with other methods
Most of the existing works in this area have not used standard datasets. Table 7 compares the proposed work with the state-of-the-art methods. To the best of our knowledge, this is the first work that evaluates the accuracy of the severity detection method in a standard data set and detects three types of severity (High, Low, Medium).
Table 7.
Evaluation of the proposed severity detection method with state-of-the-art methods.
| Author | Class | Subject | Method | Performance metric | Value |
|---|---|---|---|---|---|
| Changati et al. | Lung infection percentage | 9749 chest CT volume | Deep learning, deep reinforcement learning Ref. [2] | Pearson correlation coefficient | 0.92 for percentage of opacity (P < 0.001) |
| Shen et al. | Severe/non severe | CT images from 44 patients | Thresholding and adaptive region growing Ref. [26] | Pearson correlation coefficient | r ranged from 0.7679 to 0.837, P < 0.05 |
| Xiao et al. | Severe/non severe | 23,812 Covid-19 images | ResNet-34 Ref. [24] | Precision AUC | 81.3% 98.7% |
| Shan et al. | Segment and quantify infection regions | 549 CT volumes | VBNet Ref. [27] | Dice similarity coefficient | 91.6%10.0 |
| Pu et al. | COVID-19 severity and progression | 72 COVID-19 and 120 others volumes | UNetBER Algorithm Ref. [25] | Sensitivity Specificity |
95%84% |
| Tang et al. | Severe/non severe | Chest CT images of 176 patients | Random forest algorithm Ref. [28] | Accuracy | 87.5% |
| Proposed method | Covid 19 severity (High, Moderate, Low) | COVID-CT data set (public) (349 images with metadata) | ResNet-50 and Densenet-201 algorithm | Sensitivity SpecificityPrecision Accuracy |
95.23%97.41%95.3%97.84% |
To find out the severity, Changati et al. used a DenseUNet with anisotropic kernels for segmenting the infected regions [2]. The severity of lung and lobe involvement was measured in this study. For the COVID-19 cases, the Pearson Correlation Coefficient between prediction and ground truth is calculated. Percentage of Opacity (PO), Percentage of High Opacity (PHO), Lung Severity Score (LSS), and Lung High Opacity Score (LHOS) are calculated.
The method proposed by Shen et al. used thresholding and adaptive region growing for segmenting the lung volume [26]. Then the pulmonary vessels are segmented and subtracted from the lung region. After that the detection of pneumonia is performed and calculated Pearson correlation between lesion percentage determined by radiologists and the proposed method.
Xiao et al. proposed a binary classification method to categorize the image as either severe or non-severe with the help of ResNet-34 [24]. Five-fold cross-validation is performed and used 23,812 CT images. He achieved the prediction quality of detecting severity and nonseverity of 87.50% and 78.46%.
Shan et al. proposed VB-Net neural network for infection segmentation [27]. He used the Dice similarity coefficient as an evaluation metric and got a Dice similarity coefficient of 91.6% ± 10.0 between the ground truth and the proposed method.
Pu et al. used UNet architecture to first segment the lung boundary and the major vessels; then these two images are registered with the help of bidirectional elastic registration algorithm [25]. To detect the regions associated with pneumonia, a threshold is calculated by finding out the average density of the middle of the lungs. They used a private data set that consists of 72 COVID-19 and 120 other images. He achieved a sensitivity of 95% and specificity of 84%.
Tang et al. explore the machine learning algorithm, Random forest, for detecting the severity of COVID-19 patients as severe or non-severe, got an accuracy of 87.5% [28].
The proposed method classified the COVID-19 CT image data set into High, Medium, and Low severity and attained an excellent classification accuracy of 97.84%.
Conclusion
As we are aware, the world is now struggling to contain the spread of the new mutant strain of the COVID-19 virus, theoretically supposed to have a 70% more transmission rate. By this work, we are proposing a computer-aided system for rapid diagnosis and prognosis of a large number of CT samples in the shortest possible time. Here we have used the effectiveness of the pre-trained model ResNet-50 for COVID-19 detection and feature extraction by ResNet-50 and DenseNet-201 to estimate the severity of the patients. This is the first model of its kind that can, in a single architecture, both detect and assess the severity of the condition. Earlier works in this area have built models using a significantly less number of datasets. But we have taken a large number of datasets and also yielded an impactful result from the same. The proposed method gave an accuracy of 98.5% with a sensitivity of 98.58% of positivity detection. Similarly, we got an average accuracy of 97.84% for severity detection, which is a highly superlative result compared to earlier works in the field. The proposed work can have profound use in the medical field as a first-line severity risk detection method that can help medical personnel plan patient care, assess the need for ICU facilities and ventilator support. A computer-aided system that can help make a care plan for the huge amount of patient inflow each day is sure to be an asset in these turbulent times. A future scope that we see for this proposed work is a comparison study between the rate of transmission, the severity of lung involvement, and prognosis of patients affected with a new mutant strain of COVID-19 and patients affected with an older strain.
Limitations of the study
In this study, authors have used ResNet-50 architecture which has an image input size of 224 × 224 for the covid-19 detection. The combined features of ResNet-50 and DensNet-201 are used to detect the severity condition of the patient. DenseNet-201 also has an input processing capability of 224 × 224. By reducing the input images’ size to give as input to the pre-trained network, there is a chance of losing image information. Improved image warping packages has been used while manipulating the images which has helped to minimise such information mislays. Another issue while dealing with the severity detection was the number of samples with the ground truth this has been solved to a great extend by data augmentation. A possible extension or future enhancement of this work can be focusing on severity detection from 3D CT volumes.
Funding
We also confirm that no Funding/Research Grants has been obtained for this work.
In addition to this, non-financial interests like personal relationships or personal beliefs, position on editorial board, advisory board or board of directors or other type of management relationships, writing and/or consulting for educational purposes, expert witness, mentoring relations has not been utilised.
Authors have no financial or proprietary interests in any material discussed in this article.
This article does not contain any studies with human participants or animals performed by the author.
Conflicts of interest
Authors hereby confirm that there is no conflict of interest or competing interest for this work submitted in this journal.
Biographies

Aswathy A.L. received her Post graduation degree in Computer Science with the specialisation in digital image computing from the Department of Computer Science, University of Kerala, India. She is currently working as lecturer and a research scholar in the same department. Her prime research areas include medical image processing, machine learning algorithms and computer vision. She has came up with various deep learning architectures which helped in various medical diagnosis.

Anand Hareendran S. is currently working as Associate Professor at Muthoot Institute of Technology and Science, Kochi in Department of Computer Science & Engineering. He obtained his Ph.D. in Computer Science from University of Kerala and M. Tech from Anna University, Chennai. His current area of research includes machine learning algorithms, association rule mining and bioinspired methodologies. He has authored four books, a modest number of research journal publications and has five IPRs in machine learning algorithms. He has designed and implemented various rule mining algorithms, which finds application in medical field, route mapping and frequent item search. He is a reviewer in various international conferences and journals. He is also an active member of Machine Intelligence Research group.

Vinod Chandra S.S. is working as Professor in the Department of Computer Science, University of Kerala. Since 1999, he has taught in various Engineering Colleges in Kerala. He has obtained his Ph.D. from University of Kerala and M. Tech from CUSAT. He has discovered four microRNAs in the human cell. He has five IPRs and a patent in the machine learning field. He authored seven books and a modest number of research publications. He is a reviewer of many international journals and conferences. His research area includes machine intelligence algorithms and nature inspired computing. He is heading Machine Intelligence Research (MIR) group, a pinpoint research group in machine intelligence and nature inspired techniques. He is leading many e-Governance projects associated with Universities and Government. He holds many consultancy activities for Government organizations.
References
- 1.Li X., Geng M., Peng Y., Meng L., Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. J Pharm Anal. 2020 doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaganti S., Balachandran A., Chabin G., Cohen S., Flohr T., Georgescu B. 2020. Quantification of tomographic patterns associated with covid-19 from chest CT. arXiv preprint arXiv:2004.01279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee E.Y.P., Ng M.-Y., Khong P.-L. COVID-19 pneumonia: what has CT taught us? Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hemdan E.E.-D., Shouman M.A., Karar M.E. 2020. Covidx-net: a framework of deep learning classifiers to diagnose COVID-19 in X-ray images. [Google Scholar]
- 5.Cohen J.P., Morrison P., Dao L. 2020. COVID-19 image data collection. [Google Scholar]
- 6.Rosebrock A. 2020. Detecting COVID-19 in X-ray images with Keras, TensorFlow, and Deep Learning. [Google Scholar]
- 7.Wang L., Wong A. 2020. Covid-net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest radiography images. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaggle . 2020. Kaggle’s chest X-ray images (pneumonia) dataset. [Google Scholar]
- 9.Borakati A., Perera A., Johnson J., Sood T. Diagnostic accuracy of X-ray versus CT in COVID-19: a propensity-matched database study. BMJ Open. 2020;10 doi: 10.1136/bmjopen-2020-042946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He X., Yang X., Zhang S., Zhao J., Zhang Y., Xing E. Sample-efficient deep learning for COVID-19 diagnosis based on CT scans. MedRxiv. 2020 [Google Scholar]
- 11.Zhao J., Zhang Y., He X., Xie P. COVID-CT-Dataset: a CT scan dataset about COVID-19. 2020 [Google Scholar]
- 12.Loey M., Smarandache F., Khalifa N.E.M. 2020. A deep transfer learning model with classical data augmentation and CGAN to detect COVID-19 from chest CT radiography digital images. Preprints. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu B., Gao X., He M., Liu L., Yin G. A fast online COVID-19 diagnostic system with chest CT scans. Proceedings of KDD 2020, New York. 2020 [Google Scholar]
- 14.Barstugan M., Ozkaya U., Ozturk S. 2020. Coronavirus (COVID-19) classification using CT images by machine learning methods. arxiv:https://arXiv:2003.09424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X., Deng X., Fu Q. A weakly-supervised framework for COVID-19 classification and lesion localization from chest CT. IEEE Trans Med Imaging. 2020;39(8):2615–2625. doi: 10.1109/TMI.2020.2995965. [DOI] [PubMed] [Google Scholar]
- 16.Pathak Y., Shukla P.K., Tiwari A., Stalin S., Singh S., Shukla P.K. Deep transfer learning-based classification model for COVID-19 disease. IRBM. 2020 doi: 10.1016/j.irbm.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han Z., Wei B., Hong Y. Accurate screening of COVID19 using attention-based deep 3D multiple instance learning. IEEE Trans Med Imaging. 2020;39(8):2584–2594. doi: 10.1109/TMI.2020.2996256. [DOI] [PubMed] [Google Scholar]
- 18.Harmon S.A., Sanford T.H., Xu S. Artificial intelligence for the detection of COVID-19 pneumonia on chest CT using multinational datasets. Nat Commun. 2020;11(1):4080. doi: 10.1038/s41467-020-17971-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soares E., Angelov P., Bianco S., Froes M.H., Abe D.K. SARS-CoV-2 CT-scan dataset: a large dataset of real patients CT scans for SARS-CoV-2 identification. medRxiv. 2020 [Google Scholar]
- 20.Ozsahin I., Sekeroglu B., Musa M.S., Mustapha M.T., UzunOzsahin D. 2020. Review on Diagnosis of COVID-19 from chest CT images using artificial intelligence, computational and mathematical methods in medicine, Hindawi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin C., Chen W., Cao Y. Development and evaluation of an AI system for COVID-19 diagnosis. MedRxiv. 2020 doi: 10.1038/s41467-020-18685-1. https://medRxiv.org/abs/2020.03.20.20039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahuja S., Panigrahi B.K., Dey N., Gandhi T., Rajinikanth V. Deep transfer learning-based automated detection of COVID-19 from lung CT scan slices. Appl Intell. 2020 doi: 10.1007/s10489-020-01826-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vinod Chandra S.S. Smell detection agent based optimization algorithm. J Inst Eng Ser B. 2015;97(3):431–436. [Google Scholar]
- 24.Xiao L., Li P., Sun F. Development and validation of a deep learning-based model using computed tomography imaging for predicting disease severity of coronavirus disease 2019. Front Bioeng Biotechnol. 2020;8 doi: 10.3389/fbioe.2020.00898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pu J., Leader J.K., Bandos A. Automated quantification of COVID-19 severity and progression using chest CT images. Eur Radiol. 2020 doi: 10.1007/s00330-020-07156-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen C. Quantitative computed tomography analysis for stratifying the severity of Coronavirus Disease 2019. J Pharm Anal. 2020;10(2):123–129. doi: 10.1016/j.jpha.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shan F. 2020. Lung infection quantification of COVID-19 in CT images with deep learning. arXiv preprint arXiv:2003.04655. [Google Scholar]
- 28.Tang Z. 2020. Severity assessment of coronavirus disease 2019 (COVID-19) using quantitative features from chest CT images. arXiv preprint arXiv:2003.11988. [Google Scholar]