Abstract
Regulation of Kiss1 transcription is crucial to the development and function of the reproductive axis. The homeodomain transcription factor, ventral anterior homeobox 1 (VAX1), has been implicated as a potential regulator of Kiss1 transcription. However, it is unknown whether VAX1 directly mediates transcription within kisspeptin neurons or works indirectly by acting upstream of kisspeptin neuron populations. This study tested the hypothesis that VAX1 within kisspeptin neurons regulates Kiss1 gene expression. We found that VAX1 acts as a repressor of Kiss1 in vitro and within the male arcuate nucleus in vivo. In female mice, we found that the loss of VAX1 caused a reduction in Kiss1 expression and Kiss1-containing neurons in the anteroventral periventricular nucleus at the time of the preovulatory luteinizing hormone surge, but was compensated by an increase in Kiss1-cFos colocalization. Despite changes in Kiss1 transcription, gonadotropin levels were unaffected and there were no impairments to fertility.
Keywords: Kiss1, Kisspeptin, VAX1, gene regulation, hypothalamus
1. Introduction
Kisspeptin, encoded by the Kiss1 gene, is a critical peptide hormone in the reproductive neuroendocrine axis. Kisspeptin, released from neurons in the anteroventral periventricular nucleus (AVPV) and arcuate nucleus (ARC), directly stimulates gonadotropin-releasing hormone (GnRH) release by binding to its receptor (Kiss1R) located on GnRH neurons (Gottsch et al., 2004; Han et al., 2005; Irwig et al., 2004; Matsuda et al., 2019; Messager et al., 2005). GnRH, in turn, stimulates the gonadotrope cells of the pituitary to release luteinizing hormone (LH) and follicle-stimulating hormone (FSH) to regulate ovulation, folliculogenesis, and the synthesis of gonadal sex steroids (Kaprara and Huhtaniemi, 2018; Plant, 2015; Stamatiades and Kaiser, 2017). Disruptions to kisspeptin signaling by mutations in the Kiss1 and Kiss1R genes, in both mice and humans, lead to impairments in sexual maturation, gonadotropin and sex steroid secretion, and fertility (Gottsch et al., 2006; Kauffman, 2010; Topaloglu, 2017; Topaloglu et al., 2012).
Kiss1 expression in the AVPV and ARC is differentially regulated by sex steroid feedback, leading to divergent roles in reproductive function. Many ARC kisspeptin neurons coexpress the stimulatory peptide, neurokinin B, and the inhibitory peptide, dynorphin, and are thought to be responsible for stimulating basal pulsatile GnRH secretion (Clarkson et al., 2017; Goodman et al., 2007; Matsuda et al., 2019; Moore et al., 2018; Navarro et al., 2009; Wakabayashi et al., 2010). Sex steroids, such as androgens, estradiol (E2), and progesterone, are detected by sex steroid receptors on kisspeptin neurons in the ARC and produce a negative feedback response resulting in decreased Kiss1 expression and decreased LH pulse frequency (McQuillan et al., 2019; Smith et al., 2005a; Smith et al., 2005b). Conversely, AVPV kisspeptin neurons, which are sexually dimorphic and primarily found in females, increase Kiss1 expression in response to E2 (Smith et al., 2005a). This positive feedback, along with increased neuronal activity, is necessary for increased GnRH secretion, leading to the induction of the LH surge that prompts ovulation (Dror et al., 2013; Dungan et al., 2007).
Recently, several homeodomain transcription factors have emerged as critical regulators of the reproductive axis, yet little is known of their influence on kisspeptin signaling (Hoffmann et al., 2021; Hoffmann et al., 2019; Hoffmann et al., 2014; Pandolfi et al., 2020; Pandolfi et al., 2018; Pandolfi et al., 2019). Ventral anterior homeobox 1 (VAX1) is a highly conserved homeodomain transcription factor, with the amino acid sequence of the homeodomain binding domain identical between mouse and human, and was first recognized for its importance in the development of the rostral and ventral forebrain (Hallonet et al., 1998). In mice, Vax1 is expressed in the olfactory placode, hypothalamus, pituitary, and the testis, but not the ovary (Hoffmann et al., 2014). VAX1 has recently been implicated as a crucial regulator of reproduction. The loss of a single Vax1 allele in the Vax1 heterozygous animal reduces the number of GnRH-expressing neurons by half, and results in reproductive deficiencies, such as a reduction in total sperm and sperm motility in males and increased estrous cycle length in females (Hoffmann et al., 2014; Hoffmann et al., 2016). Complete loss of Vax1 results in no detectable GnRH-expressing cells, demonstrating a gene-dose dependent role of VAX1 (Hoffmann et al., 2016). Selective deletion of VAX1 from GnRH neurons abolishes GnRH expression, resulting in hypogonadism and complete infertility in both male and female mice (Hoffmann et al., 2016; Pandolfi et al., 2020), while VAX1 in the suprachiasmatic nucleus (SCN) regulates the LH surge (Hoffmann et al., 2021). However, these additional studies, do not fully explain the reproductive phenotypes observed in Vax1 heterozygous females. While the importance of Vax1 expression within GnRH neurons and the SCN has been demonstrated (Hoffmann et al., 2021; Hoffmann and Mellon, 2016; Pandolfi et al., 2020), the role of Vax1 in other regions and cell types within the hypothalamus has largely been unexplored.
Haploinsufficiency of Vax1 in heterozygous females leads to a robust increase in Kiss1 mRNA levels in the AVPV and a corresponding increase in circulating LH and E2 (Hoffmann et al., 2014), which was not observed in mice with selective deletion of Vax1 in GnRH neurons (Hoffmann et al., 2016). Several regions of both mouse and human Kiss1/KISS1 promoters contain the VAX1 consensus sequence (ATTA), yet it is unknown whether the increased Kiss1 transcription is due to a direct effect of VAX1 at the level of the kisspeptin neurons, or an indirect effect through altered steroid feedback to kisspeptin neurons. Additionally, it is unknown whether a homozygous loss of Vax1 within ARC kisspeptin neurons would lead to a change in Kiss1 gene expression. Our current study aims to test the hypothesis that VAX1 contributes to the regulation of Kiss1 transcription in ARC and AVPV kisspeptin populations and consequently regulates gonadotropin secretion. To address our hypothesis, we used immortalized kisspeptin cell lines, as well as transgenic mouse models with VAX1 deleted from Kiss1-expressing cells. We found that VAX1 regulates the human KISS1 promoter in vitro and Kiss1 transcription in vivo, but the loss of VAX1 from Kiss1-expressing cells in vivo does not alter fertility.
2. Materials and Methods
2.1. Animals
All animal procedures were performed in accordance with the University of California, San Diego Institutional Animal Care and Use Committee regulations. To produce mice lacking VAX1 in kisspeptin cells, we crossed Vax1Flox/Flox mice (RRID:MGI:6500817) (Hoffmann et al., 2016) with Kiss1Cre mice (RRID:IMSR_JAX:023426) (Cravo et al., 2011). Offspring were backcrossed to generate Vax1Flox/Flox:Kiss1Cre conditional knockouts (Vax1KissCre) or Vax1Flox/Flox:Kiss1WT (Vax1WT). Kiss1Cre reporter mice were generated by crossing Kiss1Cre mice with Ai9 Rosa-tdTomato mice (RRID:MGI:104735) (Madisen et al., 2010) to create mice in which Kiss1Cre expressing cells were identifiable by tdTomato expression. All mice were on a C57BL/6 background, group housed, and maintained on a 12-hour light, 12-hour dark cycle, with ad libitum chow and water. Animals were randomly assigned to experimental groups. Mice were sacrificed with CO2 or isoflurane overdose followed by rapid decapitation. All animals were sacrificed between zeitgeber time (ZT) 4-7, unless otherwise stated. Female mice were sacrificed during diestrus unless otherwise stated.
For genotyping, genomic DNA was extracted from hypothalamus, pituitary, testis, ovary, and tail tip using DNeasy Kit (Qiagen) according to manufacturer’s instructions. To differentiate between a Vax1 flox allele and wildtype allele we used the following primers: VaxFlox-Forward: 5’-GCCGGAACCGAAGTTCCTA-3’, VaxWT-Forward: 5’-CCAGTAAGAGCCCCTTTGGG-3’, and Vax-Reverse: 5’-CGGATAGAC CCCTTGGCATC-3’. To detect Vax1 recombination, the following primers were used: VaxRec-Forward: 5’-GCAGTGGCCTAGAGAGATCG-3’ and VaxRec-Reverse: 5-GCACTGTGTAGTGCTCCTAT-3’. CRE genotyping was performed using CRE-Forward: 5’-GCATTACCGGTCGTAGCAACGAGTG-3’ and CRE-Reverse: 5’-GAACGCTAGAGCCTGTTTTGCACGTTC-3’. tdTomato mice were genotyped using tdtF 1: 5’-GGCATTAAAGCAGCGTATCC-3’, tdtR1: 5;- CTGTTCCTGTACGGCATGG-3, tdtF2: 5’-CCGAAAATCTGTGGGAAGTC-3’, tdtR2: 5’- AAGGGAGCTGCAGTGGAGTA-3’. Mice that were positive for germline recombination of VAX1 were excluded from the study.
2.2. Immunohistochemistry
Kiss1Cre-tdTomato+/- mice were euthanized at ZT6. Brains were quickly dissected and submerged in 4% PFA overnight. The following day, brains were transferred to 30% sucrose until they sank. 40 pm sections containing the AVPV were sectioned using a cryostat and sections transferred to PBS. On the day of staining, sections underwent heated antigen retrieval in 1X Citra buffer (Biogenex). Sections were washed and processed with a Mouse-on-Mouse ABC kit (Vector Labs) according to manufacturer’s instruction. VAX1 antibody (RRID:AB_2723772) (Clone OTI4E5, Origene) was used at 1:100. After ABC processing for 30 minutes, sections were washed and incubated in 1:250 biotinylated tyramide (Akoya Biosciences) with 0.003% H2O2 for ten minutes. Sections were washed again and incubated in 1:200 Streptavidin DyLight 488 (Invitrogen) for 30 minutes. Sections were washed, mounted, and coverslipped with ProLong with DAPI (Invitrogen).
2.3. RNA Isolation and qPCR
Brains were collected on dry ice and immediately stored at −80°C. Two mm AVPV and ARC micropunches were collected on a cryostat from 0.3 mm thick sections as previously described (Di Giorgio et al., 2013; Tonsfeldt et al., 2019). RNA from micropunches was isolated with RNeasy-Micro Kit (Qiagen) according to manufacturer’s instructions. Purified RNA was converted to cDNA using iScript cDNA Synthesis Kit (Bio-Rad Laboratories). cDNA products were detected on a Bio-Rad CFX Connect quantitative real-time PCR system (Bio-Rad laboratories) using SYBR Green Supermix (Bio-Rad Laboratories). Data were analyzed by the 2-∆∆Ct method (Livak and Schmittgen, 2001), by normalizing the gene of interest to Gapdh, and represented as mean fold change compared to Vax1WT±SEM. The following primers were used: Kiss1-Forward: 5’-TGCTGCTTCTCCTCTGT-3’, Kiss1-Reverse: 5’-ACCGCGATTCCTTTTGC-3’ (Gottsch et al., 2011), Pdyn-Forward: 5’-GTGTGCAGTGAGGATTCAGG-3’, Pdyn-Reverse: 5’-AGTCATCCTTGCCACGGAGC-3’ (Schoeller et al., 2016), Tac2-Forward: 5’-CTGCTTCGGAGACT CT ACG-3’, Tac2-Reverse: 5’-GGTTGGCTGTTCCTCTTGC-3’ (Schoeller et al., 2016), Gapdh-Forward: 5’-TGCACCACCAACTGCTTAG-3’, Gapdh-Reverse: 5’-GGATGCAGGGATGATGTTC-3’.
2.4. Cell Culture
KTaR-1 (RRID:CVCL_VS93) and KTaV-3 (RRID:CVCL_VS94) cell lines (kindly provided by Patrick E. Chappell, Oregon State University) (Jacobs et al., 2016) were maintained in complete medium consisting of DM EM (Corning) containing 10% fetal bovine serum (FBS) (Omega Scientific) and 1% penicillin/streptomycin (HyClone) and incubated at 37°C with 5% CO2.
2.5. Hormones
17β-estradiol (E2) was obtained from Sigma-Aldrich (St. Louis, MO). For cell culture, E2 was dissolved in absolute ethanol and diluted to a 1 nM stock. Immediately prior to hormone treatment, 1 nM E2 stock was diluted 1:1000 in charcoal-stripped DMEM to a final concentration of 1 pM E2. For E2 pellets, E2 was dissolved in sesame oil to a concentration of 25 μg/ml. Methyltrienolone (R1881) was obtained from NEN Life Sciences (Boston, MA) and diluted to a 10 mM R1881 stock in absolute ethanol. Immediately prior to hormone treatments, 10 mM stock was diluted 1:1000 in charcoal-stripped DMEM to a final concentration of 10 μM R1881.
2.6. Site-Directed Mutagenesis
Site-directed mutagenesis of three regions of the −1313/+27 human-Kiss-Luc in a pGL2 backbone (hKiss-Luc) (Mueller et al., 2011), kindly provided by Alejandro Lomniczi and Sergio Ojeda, (Oregon National Primate Research Center), was performed using the Q5 Site-Directed Mutagenesis Kit (New England Biolabs) according to manufacturer’s instructions. The μ-1211-1193 sequence was mutated at −1211 to −1193 base pairs (bp) upstream of the transcriptional start site (TSS) from TAATGGGTGTGATAATAAT to CGGCGGGTGTGACGGCGGC. The μ-1111-1108 sequence was mutated at −1111 to −1108 bp upstream of the TSS from ATTA to CGGC. The μ-362-359 sequence was mutated at −362 to −359 bp upstream TSS from ATTA to CGGC.
2.7. Transient Transfections and Luciferase Assays
One day prior to transfections, KTAR-1 and KTAV-3 cells were seeded at 3 × 104 cells per well in 12-well plates with DMEM containing 10% FBS. Transient transfections were performed using Polyjet In Vitro DNA Transfection Reagent (SignaGen Laboratories), following manufacturer’s instructions. For all experiments, each well was transfected with 500 ng of reporter plasmid, −1313/+27 human-Kiss-Luc in a pGL2 backbone (hKiss-Luc) (Mueller et al., 2011) or reporter backbone, pGL2, and co-transfected with VAX1-CMV6 (VAX1) (Hoffmann et al., 2016) or CMV6-empty vector (EV) (TrueClone, Origene). 100 ng of a reporter plasmid containing β-galactosidase driven by the herpes virus thymidine kinase promoter (TK-βgal) was co-transfected to serve as an internal control for transfection efficiency. To determine optimal VAX1 concentration, 0 to 50 ng VAX1 or EV was co-transfected with reporter plasmids. To determine if VAX1 regulated the KISS1 promoter through regions containing ATTA sites, we co-transfected hKiss-Luc, μ-1211-1193, μ-1111-1108, and μ-362-359 or pGL2, with 50 ng VAX1 or EV. Polyjet/DNA complex-containing medium was removed after 24 hours and replaced with complete medium. Cells were harvested 48 hours from start of transfection.
For transfections involving hormone treatments, cells were seeded in charcoal-stripped FBS. 50 ng VAX1 or EV was transfected along with 100 ng ratAR-pSG5 (Ikonen et al., 1998) or 50 ng ERα-pcDNA3.1 to ensure adequate expression of steroid receptors. Polyjet/DNA complex-containing medium was replaced after 24 hours with charcoal-stripped medium containing 10 μM R1881 (synthetic androgen), 1 pM E2, or vehicle (100% ethanol). Cells were harvested 48 hours following hormone treatment. To harvest cells, medium was aspirated, cells were washed with 1X PBS and then lysed with 0.1 M K-phosphate buffer, pH 7.8, containing 0.2% Triton X-100. Luciferase and β-galactosidase assays were performed as previously described (Givens et al., 2005). Transfections were performed in triplicate. Within each well, luciferase values were normalized to TK-βgal values. Luciferase/TK-βgal triplicate values were averaged and then hKiss-Luc, μ-1211-1193, μ-1111-1108, and μ-362-359 values were normalized to pGL2 values. A minimum of three independent replicates per experiment were performed.
2.8. Hormone Analysis
Blood samples were collected at time of euthanasia, allowed to clot at room temperature for 1 hour, centrifuged at 2000 ×g for 15 minutes, and then serum was collected and stored at −20°C until assayed. Serum LH, FSH, estradiol, and testosterone were measured by The University of Virginia Center for Research in Reproduction Ligand Assay and Analysis Core. LH was measured using LH RIA with a reportable range between 0.02 – 75.0 ng/mL (Intra-assay CV=5.5%, Inter-assay CV=8.4%). FSH was measured using FSH RIA with a reportable range between 3.0 – 75.0 ng/mL (Intra-assay CV=6.7%, Inter-assay CV=8.7%). Estradiol was measured using an ELISA with reportable range between 3.0 – 300.0 pg/mL (Intra-assay CV=7.5%, Inter-assay CV=10.1%). Testosterone was measured using an ELISA with reportable range between 10.0 – 1600.0 ng/dL (Intra-assay CV=6.0%, Inter-assay CV=9.3%).
2.9. LH Surge
Between ZT 2 - 5, female mice (10-16 weeks old, weighing between 18-28 grams) were ovariectomized and a pellet containing 0.75 μg of 17-β estradiol dissolved in sesame oil subcutaneously implanted to mimic proestrus levels of E2 (Dror et al., 2013). Two days after surgery, mice were sacrificed either in the morning (AM), between ZT 4-5, or at the time of lights off (PM), between ZT 12-13. Blood and brains were collected at sacrifice. An LH surge was conservatively defined as LH values that were 3 standard deviations above the AM average (Dungan et al., 2007).
2.10. Fluorescent In Situ Hybridization (ISH)
20 μm serial coronal sections were collected from fresh frozen brains, spanning the length of the AVPV or ARC (Di Giorgio et al., 2013; Tonsfeldt et al., 2019). Sections were fixed in chilled 4% PFA, washed two times with 1X PBS, and dehydrated through a series of ethanol washes ranging from 50%-100% ethanol. RNAscope Multiplex Fluorescent v2 Assay (Advanced Cell Diagnostic, 323100) was performed according to manufacturer’s instructions with the following probes: Mm-Kiss1 (500141) and Mm-Fos-C2 (316921-C2). Sections were counterstained with DAPI and coverslipped with ProLong Gold (Invitrogen).
2.11. Microscopy and Image Analysis
Fluorescent microscopy was performed at the Nikon Imaging Core (UCSD) using a Nikon Eclipse Ti2-E microscope with Plan Apo objectives. Samples were excited by the Lumencor SpectraX and acquired with a DS-Qi2 CMOS camera using NIS-Elements software. Lighting was determined by using the minimum LED intensity and exposure time for each channel using positive and negative control slides, and then used for all subsequent acquisition. Images were imported into FIJI (NIH ImageJ) (Schindelin et al., 2012). To determine the number of Kiss1-positive cells in the AVPV, a defined 0.7 mm2 region encompassing both sides of the third ventricle was set. Kiss1-positive and cFos-positive cells were counted manually, using FIJI Cell Counter tool. Counts were performed by two independent experimenters who were both blinded to genotype.
2.12. Puberty, First Estrus, Estrous Cyclicity, and Fertility Assessment
Beginning at weaning (21 days of age), mice were checked daily for pubertal onset. Date of pubertal onset was marked by the occurrence of vaginal opening in females and by preputial separation in males. Following female pubertal onset, vaginal lavages were taken daily until the first occurrence of estrus was observed. Beginning at 12 weeks of age, vaginal smears were taken for 16 consecutive days to assess estrous cyclicity. The slides used for vaginal lavages were stained with 0.1% methylene blue stain and stage of cycle was determined by the composition of cell types present (Byers et al., 2012). To assess fertility, 12-15-week-old virgin Vax1KissCre or Vax1WT mice were paired with a Vax1WT breeder for 90 days. The number of offspring, latency to first litter, and total number of litters were recorded.
2.13. Sperm Motility and Total Sperm Count
Cauda epididymides were dissected and placed in M2 media (Sigma-Aldrich) at room temperature. The right epididymis was cut in half and forceps were used to manually expel sperm into M2 media and left undisturbed for 15 minutes. Motile sperm were counted on a hemocytometer and then placed on a 55°C heat block for 5 minutes to immobilize all sperm. Following immobilization, all intact sperm cells on the hemocytometer were counted. Motile sperm counts were divided by the total counts and multiplied by 100 to determine the percent motility. The left epididymis was minced, filtered through a 70 μM filter (Falcon), and diluted in water. Using a hemocytometer, all sperm heads were counted to determine total sperm count.
2.14. Statistical Analysis
Students t-test, Welch’s t-test, one-way ANOVA, and two-way ANOVA were used to determine differences between groups as indicated in the figure legends. For one-way and two-way ANOVA, significant effects were followed by Tukey’s Honest Significant Difference test. Graphpad PRISM 9 was used for statistical analysis, with p<0.05 indicating significance.
3. Results
3.1. Generation of VAX1Flox/Flox:Kiss1Cre mice
The loss of a single allele in Vax1 heterozygous mice has been shown to increase Kiss1 mRNA in diestrus females (Hoffmann et al., 2014). To test the hypothesis that the increase of Kiss1 is mediated by a direct effect of VAX1 within kisspeptin neurons, we first established whether Kiss1 and VAX1 were colocalized. Using a newly-available VAX1 monoclonal antibody, we confirmed VAX1 is expressed in the SCN (Fig. 1A), an area of high Vax1 mRNA expression in adulthood according to the Allen Brain Atlas (http://mouse.brain-map.org)(Allen Institute for Brain Science, 2011) (Fig. 1B). We also detected VAX1 protein in tdTomato-containing neurons from KissCre-tdTomato mice (Fig. 1C), establishing the presence of VAX1 in Kiss1-expressing neurons in adulthood. Then, we selectively deleted Vax1 from Kiss1-expressing cells in mice by generating Vax1Flox/Flox:Kiss1Cre (Vax1KissCre) and Vax1Flox/Flox:Kiss1WT (Vax1WT) mice (Fig. 1D). Exons 2 and 3 of the Vax1 gene, containing the homeodomain coding region, are flanked by LoxP sites, allowing for the functional protein to be present in Vax1WT mice and excised in Vax1KissCre mice. We collected genomic DNA and verified that recombination of the Vax1Flox allele occurred in tissues known to express Kiss1, such as the hypothalamus, pituitary, testis, and ovary (Ikeda et al., 2017; Merhi et al., 2016; Salehi et al., 2015; Smith et al., 2005a; Smith et al., 2005b), while recombination was absent from male pituitary and tail (Fig. 1E). There was no recombination detected in any tissues from Vax1WT mice (Fig. 1E), signifying intact Vax1 expression. These findings indicate the successful recombination of the Vax1Flox allele in areas known to express Kiss1.
Fig. 1. VAX1 is expressed in Kiss1 neurons and recombined in Kiss1-Cre-expressing tissues.
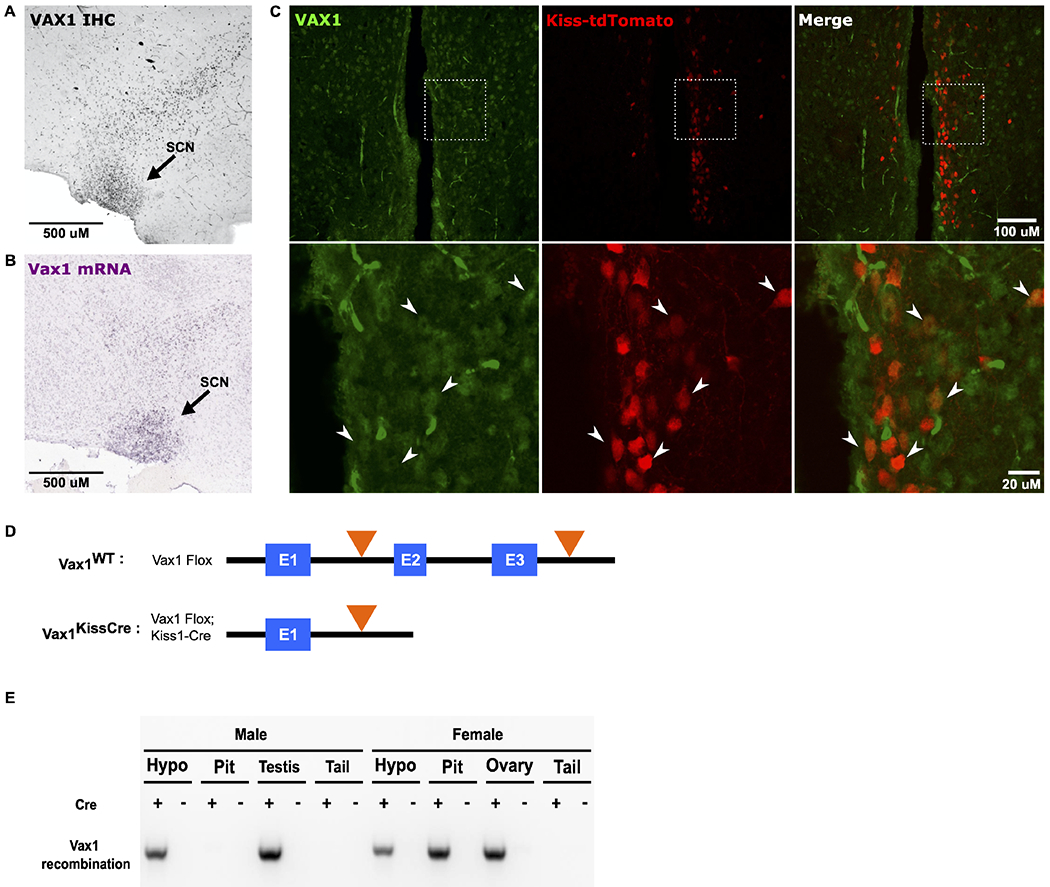
(A) Representative sagittal image of VAX1 immunoreactivity in the suprachiasmatic nucleus (SCN) and surrounding area in an adult WT mouse brain. (B) Vax1 in situ hybridization from adult male mouse, Allen Brain Atlas, image 18, Vax1 - RP_090303_04_G09 – sagittal, https://mouse.brain-map.org/gene/show/22083. (C) Fluorescent imaging showing VAX1 (green) and tdTomato (red) in Kiss1Cre-tdTomato mice. Arrows show colocalization of tdTomato-positive neurons and VAX1. (D) Schematic of the Vax1 gene in Vax1WT and Vax1KissCre mice. Black line depicts intronic sequence, blue boxes represent exons, and orange triangles depict LoxP sites. (E) Representative PCR to show recombination of the Vax1Flox allele from genomic DNA, collected from tissues known to either express Kiss1 (hypothalamus, pituitary, testis, ovary) or not express Kiss1 (tail) in Vax1KissCre (Cre+) and Vax1WT (Cre−) mice.
3.2. VAX1 regulates Kiss1 and Tac2 mRNA in the ARC of male mice
Using Vax1KissCre mice, we tested whether VAX1 plays a role in regulating Kiss1 gene expression in ARC kisspeptin neurons in vivo. We took micropunches of the ARC from Vax1WT and Vax1KissCre intact male and intact diestrus female mice. We found that Kiss1 was increased 2-fold in Vax1KissCre male mice compared to Vax1WT, while levels in diestrus female mice were unchanged (Fig. 2A, B). We also measured levels of Tac2 and Pdyn, which are coexpressed with Kiss1 in approximately 90% of ARC kisspeptin neurons (Navarro et al., 2009). We found that Tac2 mRNA was increased 1.4-fold in Vax1KissCre males, but unchanged in females (Fig. 2C, D). We found no change in Pdyn in mutants of either sex (Fig. 2E, F). We did not identify a sex difference in Vax1 mRNA expression in the ARC of intact male and intact diestrus-staged or OVX + E2-treated females. However, we did observe a significant reduction of Vax1 levels in OVX + E2-treated females compared to diestrus-staged females (Fig. 2G). These findings show that Kiss1 is differentially regulated by VAX1 in males and females in vivo.
Fig. 2. VAX1 is required for Kiss1 and Tac2 expression in male ARC.
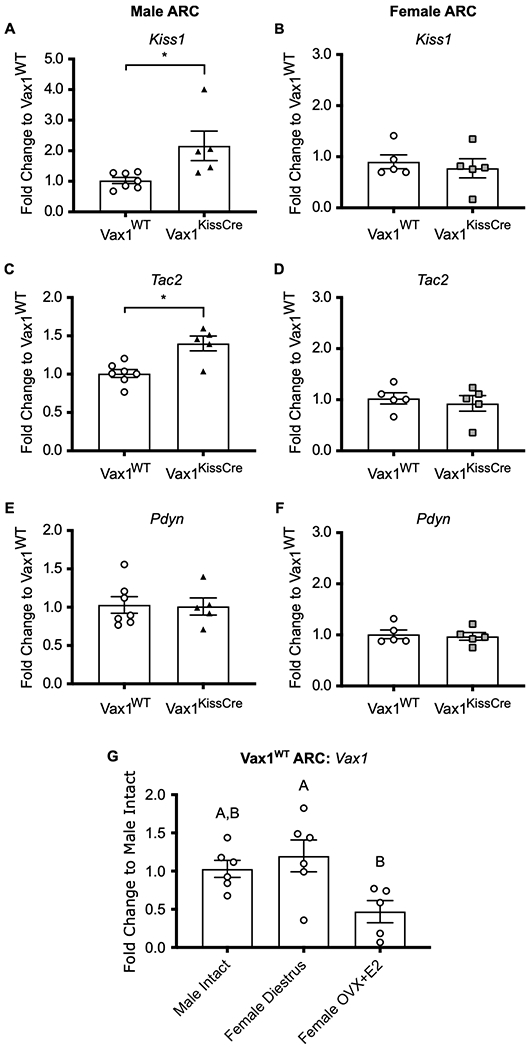
RT-qPCR profiles of (A,B) Kiss1, (C,D) Tac2, and (E,F) Pdyn in the ARC of intact males and intact diestrus females. Data were analyzed by the 2^-delta delta Ct method and represented as fold change compared to Vax1WT ± SEM. (G) RT-qPCR of Vax1 in ARC of intact Vax1WT males, diestrus-staged Vax1WT females, and OVX+E2 Vax1WT females. Data were analyzed by the 2^-delta delta Ct method and represented as fold change compared to Male Intact ± SEM. All samples were collected between ZT 4-7. All data were analyzed using (A-F) Student’s t-test or (G) One-way ANOVA. * or different letters indicate significance of p<0.05. N=5-7.
3.3. VAX1 regulates the KISS1 promoter in vitro
We next used a human −1313/+27 KISS1-Luciferase reporter (hKiss-Luc) (Mueller et al., 2011), which contains three ATTA sites located at −1211 to −1193, one ATTA site located at −1111 to −1108, and one ATTA site located at −362 to −359 bp upstream of the KISS1 transcriptional start site (TSS) (Fig. 3A), to examined whether VAX1 can regulate the kisspeptin promoter in vitro. We co-transfected hKiss-Luc with various concentrations of a VAX1 expression vector or an empty vector (EV) control into the immortalized mouse ARC (KTaR-1) kisspeptin cell line (Jacobs et al., 2016). We found that overexpression of VAX1 represses hKiss-Luc transcription in KTaR-1 cells in a dose-dependent manner (Fig. 3B). To determine if VAX1 mediated KISS1 repression through any of the three regions on the promoter that contain ATTA sites, we used site-directed mutagenesis to create cis-mutations at −1211 to −1193 bp (μ-1211-1193), −1111 to −1108 bp (μ-1111-1108), and −362 to −359 bp (μ-362-359) on the hKiss-Luc promoter (3A). We found that VAX1 was not able to significantly repress transcription on μ-1211-1193 and μ-362-359 as compared to the control (hKiss-Luc), while μ-1111-1108 was able to repress transcription to a similar degree as hKiss-Luc (Fig. 3C). These findings indicate that, within immortalized kisspeptin neurons derived from the female ARC, VAX1 acts upon two regions of the KISS1 promoter to mediate repression and are consistent with the effects seen in the male ARC in vivo.
Fig. 3. VAX1 regulates transcription from the KISS1 promoter in KTaR-3 cells.
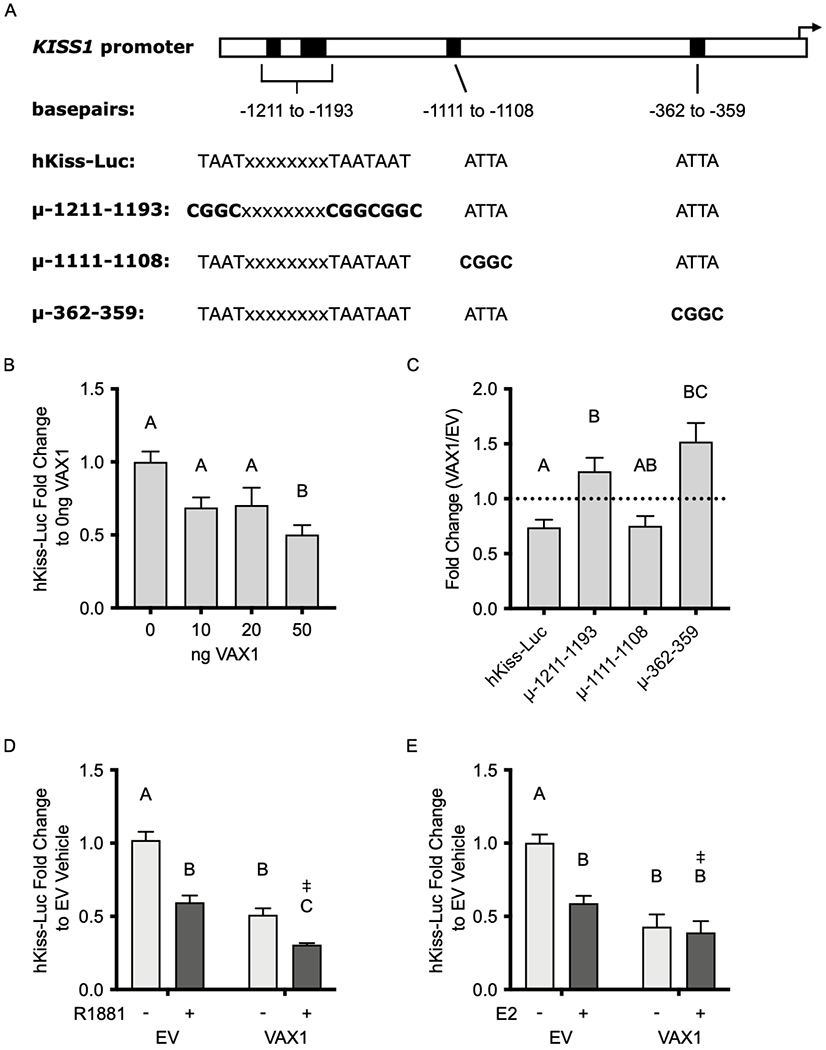
(A) Schematic of ATTA binding sites (black rectangles) on the KISS1 promoter and the sequence of these sites on the hKiss-Luc reporter or hKiss-Luc constructs containing cis-mutations at the indicated positions upstream of the transcriptional start site (black arrow). Bolded sequences indicate base pairs that were changed via site-directed mutagenesis. (B) hKiss-Luc was co-transfected with increasing concentrations of VAX1 expression vector into KTaR-1 cells. Data are represented as fold change as compared to 0 ng VAX1. (C) hKiss-Luc, μ-1211-1193, μ-1111-1108, and μ-362-359 were co-transfected into KTaR-1 cells with 50 ng VAX1 or EV. Data is represented as fold change of VAX1/EV. Dotted line indicates EV/EV. (D) KTaR-1 cells were transfected with hKiss-Luc or pGL2, 50 ng VAX1 or empty vector (EV), and 100 ng AR plasmids, and then treated with 10 μM R1881 (+) or vehicle (−). Data are represented as fold change as compared to EV vehicle. (E) KTaR-1 cells were transfected with hKiss-Luc or pGL2, 50 ng VAX1 or EV, and 50 ng ERα plasmids, and then treated with 1 pM E2 (+) or vehicle (−). Data are represented as fold change to EV vehicle. For all experiments, values represent means ± SEM. Data were analyzed using (B, C) One- or (D, E) Two-way ANOVA. Different letters denote significance, p<0.05. ‡ denotes a significant two-way factor interaction, p<0.05. N=3-4.
To better understand the differential regulation of Kiss1 expression observed between males and females in vivo, we determined whether sex steroids influence the effects of VAX1 on KISS1 promoter activity. We co-transfected KTaR-1 cells with 50 ng of VAX1 and 100 ng of androgen receptor (AR) in the presence or absence of 10 μM R1881, a synthetic androgen. We found that the combination of VAX1 overexpression and androgen-containing medium caused a significant reduction in hKiss-Luc transcription compared to VAX1 or R1881 alone, and that there was a significant interaction between VAX1 and R1881 (p<0.05) (Fig. 3D). To approximate the female hormonal milieu in vitro, we transfected KTaR-1 cells with 50 ng of VAX1 and 50 ng of estrogen receptor alpha (ERα), in the presence or absence of 1 pM E2. We show that E2, along with ERα can repress hKiss-Luc transcription (Fig. 3E). We found that in the presence of E2, the effects of VAX1 on the KISS1 promoter were masked and that the effects of the combination of E2 and VAX1 were not different than the effects of E2 or VAX1 alone. However we did observe a significant interaction between VAX1 and E2 (p<0.05) (Fig. 3E). These findings support our in vivo results, in which we saw an upregulation of Kiss1 following the loss of VAX1 in males, but not in females.
3.4. Basal serum gonadotropin levels are not altered by loss of Vax1 from Kiss1 cells
Because we observed that overexpression of VAX1 represses Kiss1 transcription in vitro and the loss of VAX1 in vivo led to increased Kiss1 transcription in males, we next wanted to determine whether the loss of VAX1 in vivo led to altered levels of circulating LH and FSH. We found that LH levels in Vax1WT and Vax1KissCre males were not significantly different (Fig. 4A). However, they had significantly different variances (F test, p = 0.005). We found that circulating levels of FSH (Fig. 4B) between Vax1WT and Vax1KissCre males were not different. We also detected no change in testosterone levels (Vax1WT: T = 318.3 ± 133.1 ng/dL, Vax1KissCre: T = 135.3 ± 67.5 ng/dL, N=10-13, p > 0.05), but found a significant difference in variance (F test, p = 0.021). We found no difference among LH and FSH levels (Fig. 4C, D) or E2 (Vax1WT: E2 = 3.2 ± 0.2 pg/mL, Vax1KissCre: E2 = 3.3 ± 0.2 pg/mL, N=7-8, p > 0.05, level of detection = 3.0 pg/mL, all values below level of detection were set to 3.0 pg/mL) between female Vax1KissCre and Vax1WT mice.
Fig. 4. Circulating hormone levels are comparable between intact male and female Vax1WT and Vax1KissCre mice.
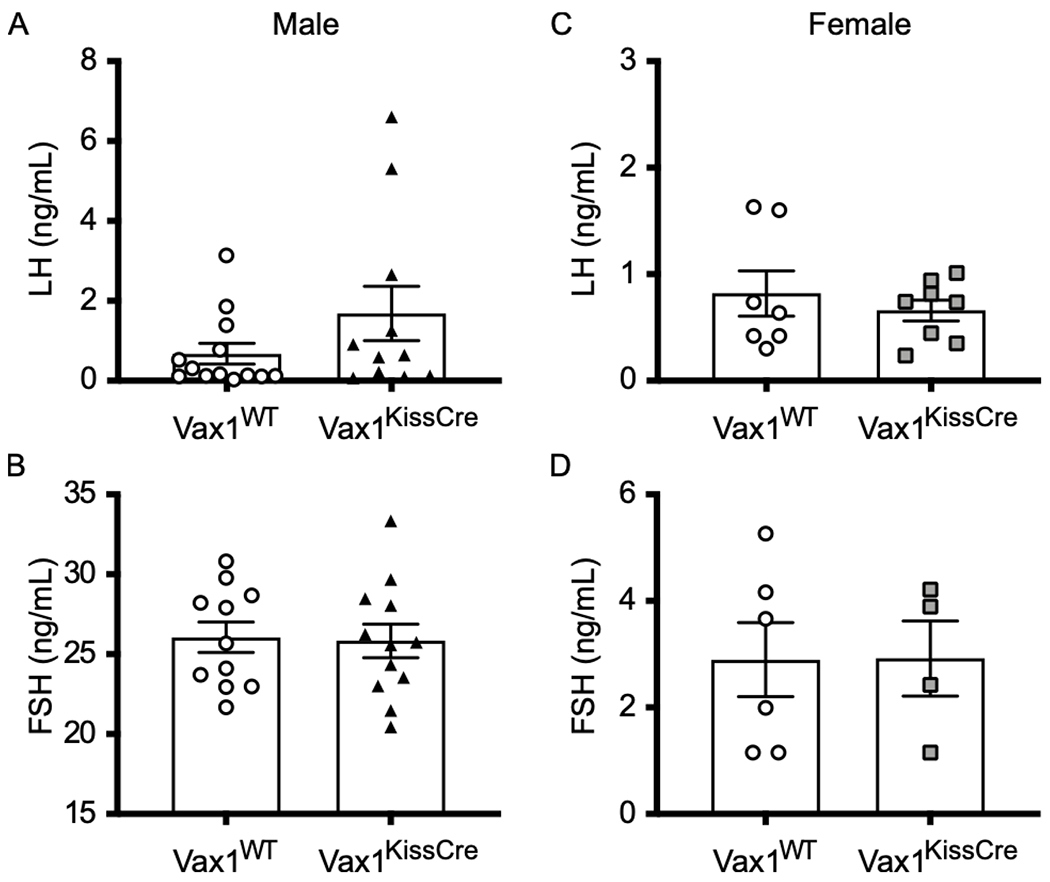
(A) Intact male LH levels. (B) Intact male FSH levels. (C) Diestrus-staged female LH levels. (D) Diestrus-staged female FSH levels. Each symbol represents a biological replicate. Bars represent mean ± SEM. Data were analyzed using (A) Welch’s t-test or (B-D) Student’s t-test. N=4-7.
3.5. VAX1 regulates Kiss1 in the AVPV of females during induced proestrus
After our data suggested that the effect of VAX1 on Kiss1 was steroid-dependent in the ARC, we next determined whether VAX1 has a role in the regulation of Kiss1 in the AVPV of female mice. We transfected immortalized mouse AVPV kisspeptin cells (KTaV-3) (Jacobs et al., 2016) with VAX1 and found that overexpression of VAX1 repressed the hKiss-Luc promotor, similar to what was observed in KTaR-1 cells (Fig. 5A). We then measured Kiss1 mRNA in micropunches of the AVPV from intact diestrus female mice, when E2 and AVPV Kiss1 levels are low (Marraudino et al., 2017; Wang et al., 2016). We found no change in AVPV Kiss1 levels between Vax1WT and Vax1KissCre females, differing from our in vitro findings (Fig. 5B).
Fig. 5. Kiss1 expression is reduced in the AVPV of E2-treated Vax1KissCre females.
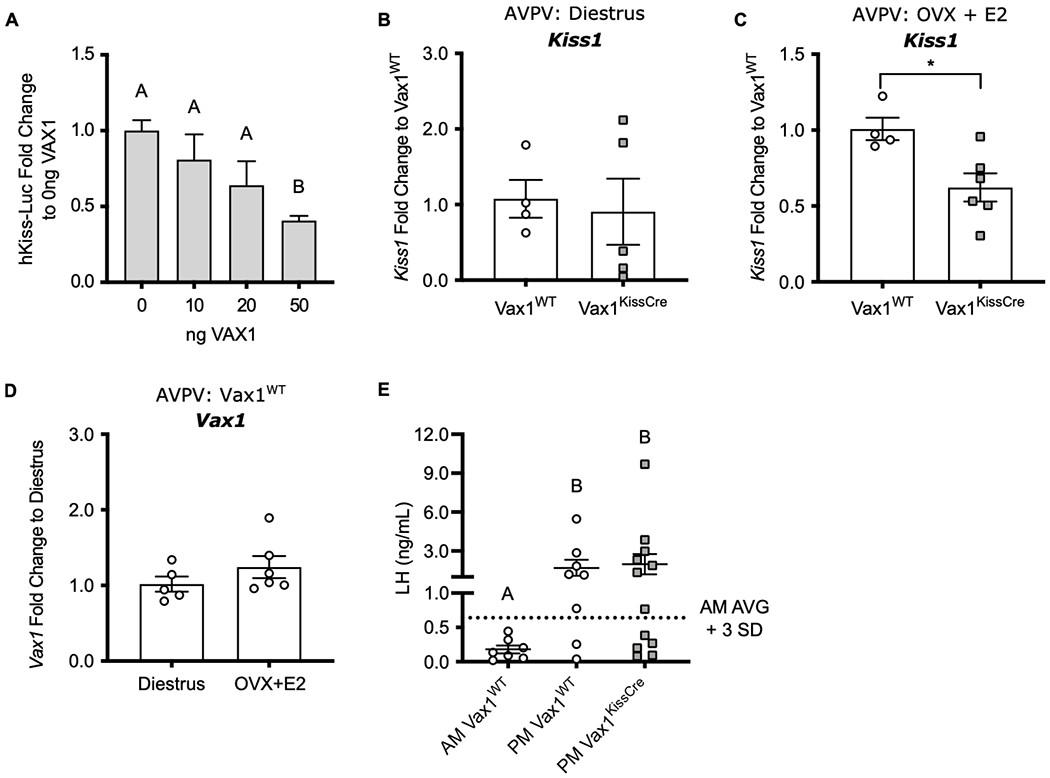
(A) hKiss-Luc or pGL2 was co-transfected with increasing concentrations of VAX1 expression vector into KTaV-3 cells. hKiss-Luc values were normalized to pGL2 values and represented as fold change compared to 0 ng VAX1. Values represent means ± SEM. N=4. RT-qPCR of Kiss1 in the AVPV of (B) diestrus-staged female mice collected at ZT 4-7 and (C) OVX + E2 treated females collected at ZT 12-13. (D) RT-qPCR of Vax1 from the AVPV of intact diestrus-staged Vax1WT collected at ZT 4-7 or OVX + E2-treated Vax1WT females collected at ZT 4-5. qPCR data were analyzed by the 2^-delta delta Ct method and represented as fold change compared to (B, C) Vax1WT or (D) Diestrus ± SEM. (E) Serum LH levels from OVX+E2 treated females, collected at ZT 4-5 (AM) or at ZT 12-13 (PM). Dotted line indicates surge threshold (AM average + 3 SD = 0.64 ng/mL), N=7-12. Data were analyzed using (A, E) One-way ANOVA or (B-D) Student’s t-test. Different letters or * indicate significance of p<0.05.
Next, we examined the role of VAX1 in the AVPV during high E2 conditions. We hypothesized that the loss of a repressive element would lead to higher Kiss1 expression. We induced an LH surge in Vax1WT and Vax1KissCre mice via ovariectomy (OVX) and implantation of an E2 pellet to mimic proestrus levels of circulating E2. In this model, Kiss1 mRNA and LH are low in the morning, and high in the evening, at the time of the expected LH surge. We performed qRT-PCR on punches from OVX+E2 treated females harvested in the evening of the induced proestrus. Unexpectedly, we found that AVPV Kiss1 mRNA was significantly reduced in Vax1KissCre mice compared to Vax1WT mice (Fig. 5C). Because we only observed a difference in Kiss1 gene regulation in OVX + E2-treated females, we wanted to determine if Vax1 was regulated by different E2 states. We found that AVPV Vax1 levels were comparable between diestrus-staged and OVX + E2-treated Vax1WT females, when collected between ZT 4-7. We then assessed circulating LH levels to determine if the changes in Kiss1 mRNA in the AVPV caused a physiological change in the ability of Vax1KissCre females to induce an LH surge. As expected, AM Vax1WT females maintained low LH levels, while PM Vax1WT females had significantly Increased levels of LH (Fig. 5E). Despite having reduced AVPV Kiss1 levels, the LH levels in PM Vax1KissCre females were not significantly different from PM Vax1WT mice (Fig. 5D). The proportion of Vax1WT (6 out of 8) and Vax1KissCre (7 out of 12) mice meeting the surge criteria was independent of genotype (X2= 0.586, df =1).
To understand the mechanisms enabling Vax1KissCre mice to show an LH surge despite reduced Kiss1 expression, we performed fluorescent in situ hybridization to assess Kiss1 and cFos expression (Fig. 6A). Only mice that met the surge criteria were assayed. We measured the overall number of Kiss1-containing neurons, as well as the number of Kiss1 neurons which colocalized with the immediate early gene cFos to assess neural activity. We found fewer Kiss1-positive neurons in the AVPV at the time of the induced surge in Vax1KissCre mice compared to Vax1WT mice (Fig. 6B). Despite a reduction in the number of overall AVPV Kiss1 cells, there was an increase in the percent of kisspeptin neurons that colocalized with cFos in Vax1KissCre mice compared to Vax1WT (Fig. 6C). These findings suggest that, despite having lower Kiss1 mRNA and fewer overall Kiss1-positive cells, Vax1KissCre mice have a larger proportion of activated AVPV kisspeptin neurons during the LH surge, which may compensate and result in surge-level LH concentrations.
Fig. 6. Vax1KissCre females have fewer Kiss1 neurons and increased cFos colocalization during an E2-induced LH surge.
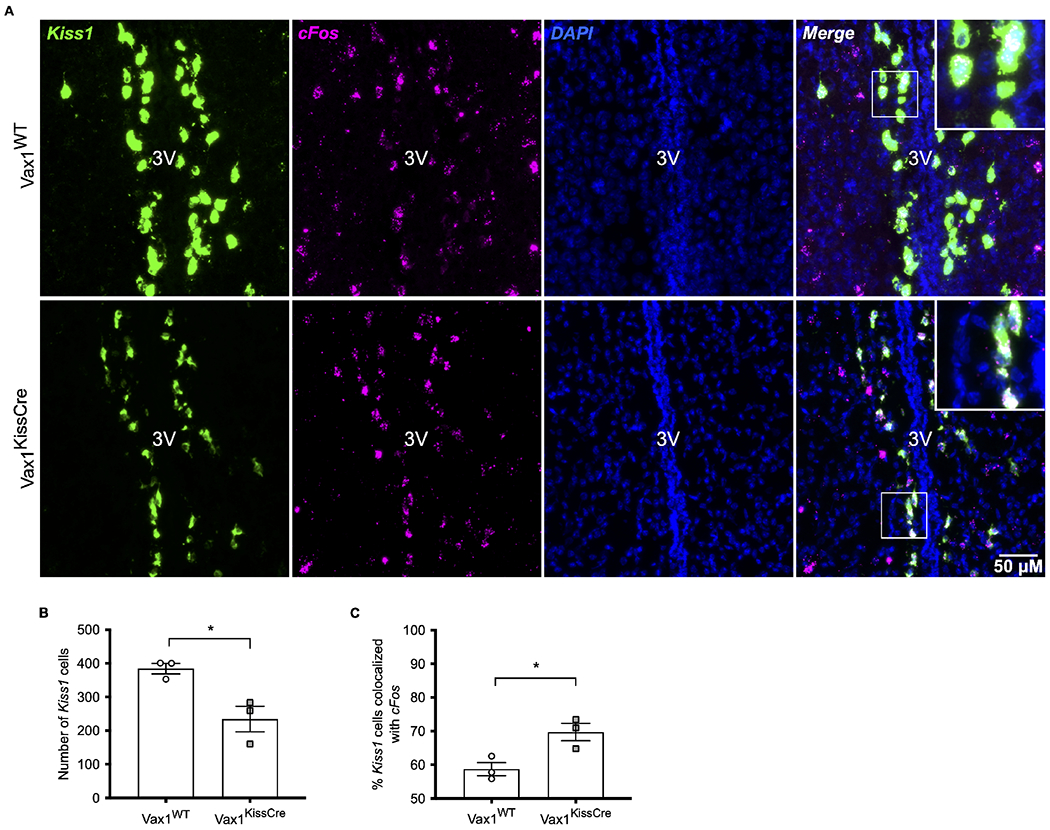
(A) Representative images of fluorescent in situ hybridization to detect Kiss1 (green) and cFos (magenta) in the AVPV of OVX+E2 treated females collected at ZT 12-13. Colocalization of Kiss1 and cFos are visualized in white. Sections were counterstained with DAPI (blue) for visualization of nuclei. (B) Quantification of the number of Kiss1-positive cells, N=3 per group. (C) Percentage of Kiss1 cells colocalized with cFos, N=3 per group. (B,C) Data were analyzed using Student’s t-test. *, p<0.05.
3.6. VAX1 in Kiss1-expressing cells is not required for reproductive function
To investigate whether deletion of Vax1 within kisspeptin neurons would impact gross reproductive function, we measured several parameters, including pubertal onset, estrous cyclicity, spermatogenesis, and fecundity. We found that female and male Vax1KissCre mice have normal timing of pubertal onset (Fig. 7A, B) and females also have normal timing of first estrus (Vax1WT: 41.6 ±1.4 days, Vax1KissCre: 42.5 ± 1.3 days, N=11-14, p > 0.05), suggesting that Kiss1-specific VAX1 does not alter sexual maturation in mice. Weights at pubertal onset were comparable between Vax1WT and Vax1KissCre mice for both females (Vax1WT: 12.24 ± 0.23 g, Vax1KissCre: 12.39 ± 0.26 g) and males (Vax1WT: 14.21 ± 0.28 g, Vax1KissCre: 14.77 ± 0. 35g). We observed no disruptions to estrous cyclicity in Vax1KissCre females as measured by cycle length (Fig. 7C) and time spent in each stage of estrous (Fig. 7D). However, male Vax1KissCre mice have significantly reduced total sperm counts compared to Vax1WT (Fig. 7E), but no difference in sperm motility (Fig. 7F). To measure fecundity, we performed a 120-day fertility assessment, in which we paired Vax1KissCre and Vax1WT mice with a male or female wildtype mouse. We found that both female and male Vax1KissCre mice had comparable number of litters to Vax1WT mice (Fig. 7G) and the time to produce the first litter was not different between groups (Fig. 7H). There were no significant differences in the number of pups born in each litter (Fig. 7I). Given these assessments, we found that the loss of VAX1 from kisspeptin cells does not impact gross fertility measures in either sex.
Fig. 7. Deletion of VAX1 from kisspeptin cells does not alter reproductive function.
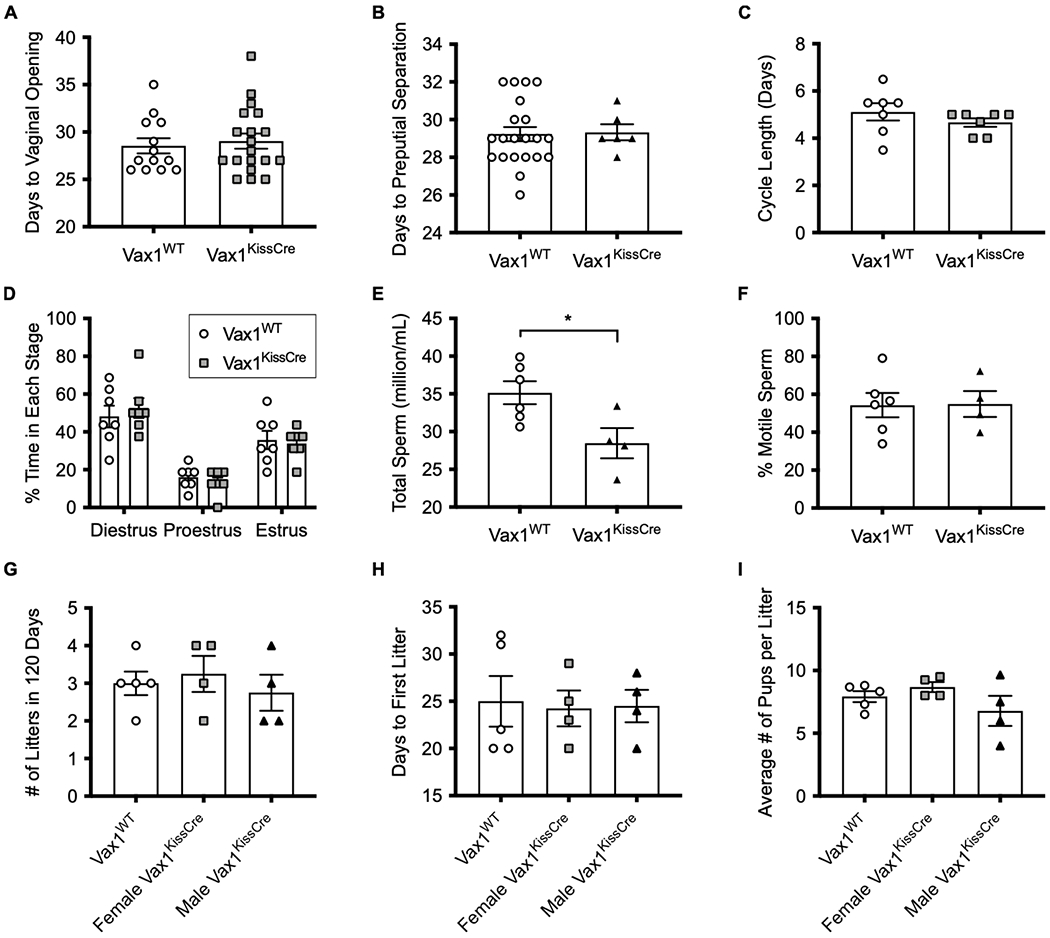
(A) Time to pubertal onset in females determined by the number of days to reach vaginal opening. N=13-18. (B) Time to pubertal onset in males determined by the number of days to preputial separation. N=6-21. (C) Average length in days to complete one estrous cycle in female mice. N=7. (D) Time spent in each stage of the estrous cycle within a 16-day period in female mice. N=7. (E) Total sperm concentration per epididymis. N=4-6. (F) Percentage of motile sperm. N=4-6. (G) Number of litters produced in 120 days. N=4. (H) Number of days until first litter was born. N=4. (I) Average number of pups per litter. N=4. For all experiments, values represent means ± SEM. Data were analyzed using One-way ANOVA or Student’s t-test. *, p<0.05.
4. Discussion
While the importance of kisspeptin signaling within the reproductive axis is unquestioned, the dynamics of Kiss1 gene regulation are not well established. We previously demonstrated that the heterozygous loss of the homeodomain transcription factor VAX1 leads to increased Kiss1 expression in the AVPV of female mice (Hoffmann et al., 2014). Using our Vax1KissCre mouse model and immortalized kisspeptin cell lines, we have established that VAX1 is a regulator of Kiss1 in vitro and in vivo and this regulation is dependent on brain region and hormone conditions. We found that VAX1 regulates Kiss1 expression in the male, but not female ARC, and in the female AVPV at the time of the LH surge, but these changes were not sufficient to significantly affect fertility.
KissCre-tdTomato mice were utilized to show that adult kisspeptin neurons express VAX1. We then selectively deleted Vax1 from Kiss1-expressing cells by creating Vax1KissCre mice and verified that the Vax1 allele was recombined in areas that are known to express Kiss1. Although Vax1 was recombined in the female pituitary of the Vax1KissCre mice, it is not expressed in αGSU-expressing cells, such as the gonadotrope cells (Hoffmann and Mellon, 2016). Correspondence with authors from Ho, et al., 2020, informed us that Vax1 is mainly constrained to melanotroph cells and was not detected in 8-week-old mouse gonadotropes, according to their single-cell RNA-seq analysis (Ho et al., 2020). Thus, the recombination detected in the pituitary should not impact gonadotrope function.
The loss of VAX1 alters expression of Kiss1 and Tac2, the gene that encodes for the stimulatory neuropeptide neurokinin B, in the male arcuate. Future studies would be needed to determine if the change in Tac2 is VAX1-dependent, or a compensatory response to the increase in Kiss1. Although we observed increased gene expression of Kiss1, we found no significant change in LH and FSH secretion in male Vax1KissCre mice. It is possible that the increase in Kiss1 was not sufficient to cause a physiologic change in LH secretion. However, while the difference in LH levels was not significantly different in male Vax1KissCre versus Vax1WT mice, we did find a significant difference in the variation between these groups. It is important to note that these LH values were taken from a single time point, and did not evaluate LH pulses. One possibility is that increased Kiss1 could increase LH pulse amplitude or pulse frequency, which would lead to a higher probability of detecting a pulse in a one-time measurement. However, profiling LH pulses in intact male mice is difficult due to pulses occurring only once every two to three hours (Steyn et al., 2013). Further exploration is needed to establish whether or not LH profiles are altered; however, our findings suggest any changes are insufficient to affect overall fertility.
We also found that Vax1KissCre males had a reduction in total sperm production, which is consistent with a phenotype observed in Vax1 heterozygous mice (Hoffmann et al., 2014). We observed a less severe sperm reduction in Vax1KissCre compared to that reported in the Vax1 heterozygotes, and we reported no impairments in reproductive capacity in Vax1KissCre males. Several studies have reported that male mice are able to maintain fertility and fecundity despite large reductions in total sperm. For example, Arl4-null mice had a 60% reduction and FSH-deficient mice had a 75% reduction in total sperm counts without impairments to litter size or frequency (Kumar et al., 1997; Schurmann et al., 2002). We cannot conclude whether the sperm reduction in our Vax1KissCre mice is due to a loss of Vax1 in kisspeptin neurons or if it is caused within the testis, as Vax1 and Kiss1 both are expressed therein (Hoffmann et al., 2014). Within the mouse testis, Kiss1 mRNA and kisspeptin protein have been reported in Leydig cells and elongated spermatids, and may play roles in sperm capacitation and spermatogenesis (Anjum et al., 2012; Hsu et al., 2014; Salehi et al., 2015; Sharma et al., 2020; Toolee et al., 2019). However, it remains to be determined which cell types in the testis express Vax1 as no Vax1 enrichment was detected in a microarray analysis from an HA-pulldown from either Leydig cell-specific or Sertoli cell-specific RiboTag mice (Sanz et al., 2013). Sperm production could also be affected by changes to upstream hormone secretion. LH binds to receptors on Leydig cells to promote the synthesis of testosterone, driving spermatogenesis (Ge et al., 2009; Zirkin and Papadopoulos, 2018). Correct levels of circulating LH are important for maintaining these processes as, in humans, increased LH levels are associated with decreased sperm counts (Andersson et al., 2004; Liu et al., 2017). While this association has not been investigated in mice, it leads us to postulate that an increase in LH levels or pulse frequency, driven by increased Kiss1, could lead to a reduction in total sperm production.
Using immortalized kisspeptin cell lines, we demonstrate that VAX1 acts as a repressor of the KISS1 promoter in vitro. VAX1 can regulate transcription through binding at ATTA consensus sites (Hoffmann et al., 2021; Hoffmann et al., 2016; Pandolfi et al., 2019; Slavotinek et al., 2012). The human KISS1 promoter contains 5 ATTA sites within the 1313 bp upstream of exon 1 (http://genome.ucsc.edu/) (Kent et al., 2002), presenting potential sites for VAX1 to act as a direct regulator of KISS1 transcription. Indeed, we found that two regions containing ATTA sites on the KISS1 promoter regulated VAX1-mediated repression of the promoter. The most distal −1211 to −1193 bp region, containing three ATTA sites, and the most proximal −362 to −359 bp region were both necessary for repression by VAX1. Although homology between the mouse and human Kiss1/KISS1 promoters is not high (~55% homology according to Needleman-Wunsch alignment between two sequences) (Altschul et al., 1997), the mouse promoter contains 8 ATTA sites within the 1313 bp upstream of the first exon, with multiple regions containing overlapping ATTA sites (http://genome.ucsc.edu/) (Kent et al., 2002), which could potentially facilitate a similar function.
Intriguingly, we observed that Kiss1 was differentially regulated by VAX1 in female mice compared to males in vivo. We postulate that this sex difference could be due to the interaction of VAX1 with androgens or estrogens. Both androgens and E2 inhibit ARC Kiss1 mRNA in vivo (Smith et al., 2005a; Smith et al., 2005b) and E2 represses Kiss1 mRNA in the ARC derived cell line (KTaR-1) in vitro (Jacobs et al., 2016). We showed, for the first time, that the synthetic androgen, R1881, and E2 can regulate transcription by repressing KISS1 promoter activity in KTaR-1 cells. Additionally, we observed that the combination of VAX1 and R1881 was able to repress the KISS1 promoter to a greater degree than either of the two independently, and that there was a significant interaction. The combined effects of VAX1 and E2, on the other hand, could not be differentiated. It is likely that R1881 and E2 are mediating their actions through their steroid receptors, AR and ERα, respectively. It is possible that binding of AR or ERα to the KISS1 promoter alters VAX1 binding efficiency, or conversely, that binding of VAX1 alters AR or ERα binding. Another possibility is that R1881 or E2 treatments could alter the expression of other genes that are endogenously expressed in the KTaR-1 cell line that could potentially modulate the effects of VAX1. One limitation of the study is that KTaR-1 cells were derived from a female mouse and the effects of androgens on kisspeptin neurons derived from male ARC could produce a different response (Jacobs et al., 2016). Further studies will be required to determine more specifically how steroid hormones acting via nuclear receptors might be interacting with VAX1 on the KISS1 promoter.
To understand the influence of VAX1 on female AVPV Kiss1 regulation, we assessed gene expression during diestrus, when we expect AVPV kisspeptin activity to remain low (Marraudino et al., 2017), as well as during the preovulatory LH surge, when there is active Kiss1 mRNA transcription (Robertson et al., 2009). Previously, it was observed that diestrus-staged Vax1 heterozygous females had elevated AVPV Kiss1, serum LH, and E2 levels compared to WT mice. In our study, we found that diestrus Vax1KissCre mice had normal AVPV Kiss1, E2, and LH levels. The differences in our data lead us to postulate that the increased AVPV Kiss1 seen in Vax1 heterozygotes could be due to positive feedback from the high E2 levels. The source of elevated E2 levels observed in the Vax1 heterozygous mice is unknown, however, as we did not observe a difference in E2 levels in our Vax1KissCre females, the increase in Kiss1 in Vax1 heterozygous mice is likely driven by the loss of Vax1 from cell populations outside of kisspeptin neurons. Vax1 heterozygous mice have a copy of Vax1 deleted from all Vax1-expressing cells in the body, including all of those in the hypothalamus, while the homozygous deletion of Vax1 in Vax1KissCre mice is confined to kisspeptin neurons. The differences in our data could also be due to a developmental effect, resulting from the timing of Kiss1Cre onset of expression. While the Vax1 haploinsufficiency is present throughout development (Bertuzzi et al., 1999), the Kiss1Cre is not expressed in AVPV kisspeptin neurons until Kiss1 expression begins postnatally (P10-16) (Semaan et al., 2010), potentially allowing Vax1KissCre females to overcome developmental modulation of the kisspeptin neurons that occurs prior to Kiss1 expression and thus, VAX1 deletion.
To examine Kiss1 levels during heightened AVPV kisspeptin activity, we used a validated E2-induced LH surge paradigm (Christian et al., 2005). The presence of both E2 and a circadian signal is required for the induction of an appropriately timed LH surge. We found that Kiss1 mRNA levels in Vax1KissCre mice were significantly lower than Vax1WT mice when collected at the time of lights off. These findings deviate from our in vitro findings, and suggest a complex relationship between VAX1, E2, and circadian regulation. Despite changes in Kiss1 gene expression, Vax1KissCre mice were still capable of inducing an LH surge. Upon closer examination of the molecular changes occurring at the time of the surge, we found that Vax1KissCre females had fewer Kiss1-positive cells in the AVPV than Vax1WT mice. We were unable to determine if this reduction was a true reduction in the number of kisspeptin cells, or a reflection of decreased Kiss1 mRNA that led to fewer cells being detected. Future experiments using lineage tracing will need to be performed to determine if VAX1 is necessary for the development or survival of kisspeptin neurons or for the maintenance of Kiss1 gene expression in adulthood. Although there was an overall reduction in the number of Kiss1-positive cells, there were approximately 10% more Kiss1-positive cells that colocalized with cFos in Vax1KissCre mice than in Vax1WT mice. This indicates that a higher proportion of Kiss1 neurons are active (cFos is expressed in activated neurons), potentially compensating for the overall reduction in Kiss1 neurons. These findings are consistent with the ability of Vax1KissCre females to produce an LH surge and maintain fertility.
This study shows for the first time that the homeodomain transcription factor VAX1 is a transcriptional repressor of Kiss1 transcription. We show that VAX1 differentially regulates ARC Kiss1 expression in males and females in vivo and VAX1 differentially regulates the KISS1 promoter in the presence of androgens and estrogens in vitro. We confirmed a previous association between loss of Vax1 and reduced sperm production, although it is insufficient to alter fecundity. In addition, we found that Kiss1 mRNA levels and the number of Kiss1-expressing neurons in the AVPV were reduced at the time of an E2-induced LH surge. The changes in Kiss1 expression did not significantly impact fertility, hormone levels, or the ability to mount an LH surge. Overall, these findings increase our understanding of the impacts of homeodomain transcription factors on Kiss1 gene regulation.
Highlights.
VAX1 differentially regulates Kiss1 gene expression in the ARC of male and female mice.
VAX1 represses the human KISS1 promoter under basal conditions and in the presence of androgens in vitro.
The loss of VAX1 reduces the number of Kiss1-expressing cells in the AVPV at the time of an induced-LH surge in female mice.
Acknowledgements
We thank Peng Hu for his correspondence regarding their single cell RNA-seq analysis of 8-week old mouse pituitaries. We thank Alejandro Lomniczi and Sergio Ojeda for providing the hKiss-Luc plasmid and Patrick E. Chappell for providing the KTaR-1 and KTaV-3 cell lines. We also thank Jessica S. Lee for her help setting up preliminary experiments and Ichiko Saotome and Austin Chin for technical assistance.
Funding Support:
This work was supported by NIH grants R01 HD082567, R01 HD100580, and R01 HD072754 (to P.L.M.) and by P50 HD012303 as part of the National Centers for Translational Research in Reproduction and Infertility (P.L.M.). P.L.M. was partially supported by P30 DK063491, P42 ES010337, and P30 CA023100, all of which also supported the UCSD Transgenic Mouse and Embryonic Stem Cell Core. S.N.L. was partially supported by P42 ES010337, T32 GM008666, and a supplement to P50 HD012303. T.C. was partially supported by the Ledell Family Research Undergraduate Research Scholarship Award for Science and Engineering. J.H. was partially supported by The Endocrine Society Summer Research Fellowship Award and the McNair Research Program. N.C.P.N. was partially supported by the McNair Research Program. K.J.T. was partially supported by T32 HD007203, F32 HD090837, P42 ES010337. H.M.H. was partially supported by K99/R00 HD084759. The University of Virginia Ligand Assay Core was supported by NIH P50 HD028934.
Footnotes
Publisher's Disclaimer: This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of record. This version will undergo additional copyediting, typesetting and review before it is published in its final form, but we are providing this version to give early visibility of the article. Please note that, during the production process, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure statement: The authors have nothing to disclose.
References
- Allen Institute for Brain Science (2011) Allen Mouse Brain Atlas. https://mouse.brain-map.org/gene/show/22083. February 2021. [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W and Lipman DJ, 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson AM, Jorgensen N, Frydelund-Larsen L, Rajpert-De Meyts E and Skakkebaek NE, 2004. Impaired Leydig cell function in infertile men: a study of 357 idiopathic infertile men and 318 proven fertile controls. J Clin Endocrinol Metab 89, 3161–3167. [DOI] [PubMed] [Google Scholar]
- Anjum S, Krishna A, Sridaran R and Tsutsui K, 2012. Localization of gonadotropin-releasing hormone (GnRH), gonadotropin-inhibitory hormone (GnIH), kisspeptin and GnRH receptor and their possible roles in testicular activities from birth to senescence in mice. J Exp Zool A Ecol Genet Physiol 317, 630–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertuzzi S, Hindges R, Mui SH, O’eary DD and Lemke G, 1999. The homeodomain protein vax1 is required for axon guidance and major tract formation in the developing forebrain. Genes Dev 13, 3092–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers SL, Wiles MV, Dunn SL and Taft RA, 2012. Mouse estrous cycle identification tool and images. PLoS One 7, e35538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian CA, Mobley JL and Moenter SM, 2005. Diurnal and estradiol-dependent changes in gonadotropin-releasing hormone neuron firing activity. Proc Natl Acad Sci USA 102, 15682–15687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson J, Han SY, Piet R, McLennan T, Kane GM, Ng J, Porteous RW, Kim JS, Colledge WH, Iremonger KJ and Herbison AE, 2017. Definition of the hypothalamic GnRH pulse generator in mice. Proc Natl Acad Sci U S A 114, E10216–E10223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cravo RM, Margatho LO, Osborne-Lawrence S, Donato J Jr., Atkin S, Bookout AL, Rovinsky S, Frazao R, Lee CE, Gautron L, Zigman JM and Elias CF, 2011. Characterization of Kiss1 neurons using transgenic mouse models. Neuroscience 173, 37–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giorgio NP, Catalano PN, Lopez PV, Gonzalez B, Semaan SJ, Lopez GC, Kauffman AS, Rulli SB, Somoza GM, Bettler B, Libertun C and Lux-Lantos VA, 2013. Lack of functional GABAB receptors alters Kiss1 , Gnrh1 and Gad1 mRNA expression in the medial basal hypothalamus at postnatal day 4. Neuroendocrinology 98, 212–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror T, Franks J and Kauffman AS, 2013. Analysis of multiple positive feedback paradigms demonstrates a complete absence of LH surges and GnRH activation in mice lacking Kisspeptin signaling. Biol Reprod 88, 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dungan HM, Gottsch ML, Zeng H, Gragerov A, Bergmann JE, Vassilatis DK, Clifton DK and Steiner RA, 2007. The role of kisspeptin-GPR54 signaling in the tonic regulation and surge release of gonadotropin-releasing hormone/luteinizing hormone. J. Neurosci. 27, 12088–12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge R, Chen G and Hardy MP, 2009. The Role of the Leydig Cell in Spermatogenic Function. In: Molecular Mechanisms in Spermatogenesis. (C.Y. C, ed.), Advances in Experimental Medicine and Biology, Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- Givens ML, Rave-Harel N, Goonewardena VD, Kurotani R, Berdy SE, Swan CH, Rubenstein JL, Robert B and Mellon PL, 2005. Developmental regulation of gonadotropin-releasing hormone gene expression by the MSX and DLX homeodomain protein families. J Biol Chem 280, 19156–19165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman RL, Lehman MN, Smith JT, Coolen LM, de Oliveira CV, Jafarzadehshirazi MR, Pereira A, Iqbal J, Caraty A, Ciofi P and Clarke IJ, 2007. Kisspeptin neurons in the arcuate nucleus of the ewe express both dynorphin A and neurokinin B. Endocrinology 148, 5752–5760. [DOI] [PubMed] [Google Scholar]
- Gottsch ML, Clifton DK and Steiner RA, 2006. Kisspepeptin-GPR54 signaling in the neuroendocrine reproductive axis. Mol Cell Endocrinol 254-255, 91–96. [DOI] [PubMed] [Google Scholar]
- Gottsch ML, Cunningham MJ, Smith JT, Popa SM, Acohido BV, Crowley WF, Seminara S, Clifton DK and Steiner RA, 2004. A role for kisspeptins in the regulation of gonadotropin secretion in the mouse. Endocrinology 145, 4073–4077. [DOI] [PubMed] [Google Scholar]
- Gottsch ML, Popa SM, Lawhorn JK, Qiu J, Tonsfeldt KJ, Bosch MA, Kelly MJ, Ronnekleiv OK, Sanz E, McKnight GS, Clifton DK, Palmiter RD and Steiner RA, 2011. Molecular properties of Kiss1 neurons in the arcuate nucleus of the mouse. Endocrinology 152, 4298–4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallonet M, Hollemann T, Wehr R, Jenkins NA, Copeland NG, Pieler T and Gruss P, 1998. Vax1 is a novel homeobox-containing gene expressed in the developing anterior ventral forebrain. Development 125, 2599–2610. [DOI] [PubMed] [Google Scholar]
- Han SK, Gottsch ML, Lee KJ, Popa SM, Smith JT, Jakawich SK, Clifton DK, Steiner RA and Herbison AE, 2005. Activation of gonadotropin-releasing hormone neurons by kisspeptin as a neuroendocrine switch for the onset of puberty. J Neurosci 25, 11349–11356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y, Hu P, Peel MT, Chen S, Camara PG, Epstein DJ, Wu H and Liebhaber SA, 2020. Single-cell transcriptomic analysis of adult mouse pituitary reveals sexual dimorphism and physiologic demand-induced cellular plasticity. Protein Cell 11, 565–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann H, Meadows J, Breuer J, Yaw A, Nguyen D, Tonsfeldt K, Chin A, Devries B, Trang C, Oosterhouse H, Lee J, Doser J, Gorman M, Wesh D and Mellon P, 2021. The transcription factors SIX3 and VAX1 are required for suprachiasmatic nucleus circadian output and fertility in female mice. Journal of Neuroscience Research, In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann HM and Mellon PL, 2016. A small population of hypothalamic neurons govern fertility: the critical role of VAX1 in GnRH neuron development and fertility maintenance. Neurosci Commun (Houst) 2, e1373. [PMC free article] [PubMed] [Google Scholar]
- Hoffmann HM, Pandolfi EC, Larder R and Mellon PL, 2019. Haploinsufficiency of homeodomain proteins Six3, Vax1, and Otx2 causes subfertility in mice via distinct mechanisms. Neuroendocrinology 109, 200–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann HM, Tamrazian A, Xie H, Perez-Millan MI, Kauffman AS and Mellon PL, 2014. Heterozygous deletion of ventral anterior homeobox (vax1) causes subfertility in mice. Endocrinology 155, 4043–4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann HM, Trang C, Gong P, Kimura I, Pandolfi EC and Mellon PL, 2016. Deletion of Vax1 from gonadotropin-releasing hormone (GnRH) neurons abolishes GnRH rxpression and leads to hypogonadism and infertility. J Neurosci 36, 3506–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu MC, Wang JY, Lee YJ, Jong DS, Tsui KH and Chiu CH, 2014. Kisspeptin modulates fertilization capacity of mouse spermatozoa. Reproduction 147, 835–845. [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Tagami A, Komada M and Takahashi M, 2017. Expression of Kisspeptin in Gonadotrope Precursors in the Mouse Pituitary during Embryonic and Postnatal Development and in Adulthood. Neuroendocrinology 105, 357–371. [DOI] [PubMed] [Google Scholar]
- Ikonen T, Palvimo JJ and Janne OA, 1998. Heterodimerization is mainly responsible for the dominant negative activity of amino-terminally truncated rat androgen receptor forms. FEBS Lett. 430, 393–396. [DOI] [PubMed] [Google Scholar]
- Irwig MS, Fraley GS, Smith JT, Acohido BV, Popa SM, Cunningham MJ, Gottsch ML, Clifton DK and Steiner RA, 2004. Kisspeptin activation of gonadotropin releasing hormone neurons and regulation of KiSS-1 mRNA in the male rat. Neuroendocrinology 80, 264–272. [DOI] [PubMed] [Google Scholar]
- Jacobs DC, Veitch RE and Chappell PE, 2016. Evaluation of Immortalized AVPV- and Arcuate-Specific Neuronal Kisspeptin Cell Lines to Elucidate Potential Mechanisms of Estrogen Responsiveness and Temporal Gene Expression in Females. Endocrinology 157, 3410–3419. [DOI] [PubMed] [Google Scholar]
- Kaprara A and Huhtaniemi IT, 2018. The hypothalamus-pituitary-gonad axis: Tales of mice and men. Metabolism 86, 3–17. [DOI] [PubMed] [Google Scholar]
- Kauffman AS, 2010. Coming of age in the kisspeptin era: sex differences, development, and puberty. Mol. Cell. Endocrinol 324, 51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM and Haussler D, 2002. The human genome browser at UCSC. Genome Res. 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar TR, Wang Y, Lu N and Matzuk MM, 1997. Follicle stimulating hormone is required for ovarian follicle maturation but not male fertility. Nat Genet 15, 201–204. [DOI] [PubMed] [Google Scholar]
- Liu Z, Shi X, Wang L, Yang Y, Fu Q and Tao M, 2017. Associations between male reproductive characteristics and the outcome of assisted reproductive technology (ART). Biosci Rep 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ and Schmittgen TD, 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES and Zeng H, 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraudino M, Miceli D, Farinetti A, Ponti G, Panzica G and Gotti S, 2017. Kisspeptin innervation of the hypothalamic paraventricular nucleus: sexual dimorphism and effect of estrous cycle in female mice. J Anat 230, 775–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda F, Ohkura S, Magata F, Munetomo A, Chen J, Sato M, Inoue N, Uenoyama Y and Tsukamura H, 2019. Role of kisspeptin neurons as a GnRH surge generator: Comparative aspects in rodents and non-rodent mammals. J Obstet Gynaecol Res 45, 2318–2329. [DOI] [PubMed] [Google Scholar]
- McQuillan HJ, Han SY, Cheong I and Herbison AE, 2019. GnRH Pulse Generator Activity Across the Estrous Cycle of Female Mice. Endocrinology 160, 1480–1491. [DOI] [PubMed] [Google Scholar]
- Merhi Z, Thornton K, Bonney E, Cipolla MJ, Charron MJ and Buyuk E, 2016. Ovarian kisspeptin expression is related to age and to monocyte chemoattractant protein-1. J Assist Reprod Genet 33, 535–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messager S, Chatzidaki EE, Ma D, Hendrick AG, Zahn D, Dixon J, Thresher RR, Malinge I, Lomet D, Carlton MB, Colledge WH, Caraty A and Aparicio SA, 2005. Kisspeptin directly stimulates gonadotropin-releasing hormone release via G protein-coupled receptor 54. Proc. Natl. Acad. Sci. USA 102, 1761–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AM, Lucas KA, Goodman RL, Coolen LM and Lehman MN, 2018. Three-dimensional imaging of KNDy neurons in the mammalian brain using optical tissue clearing and multiple-label immunocytochemistry. Sci Rep 8, 2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller JK, Dietzel A, Lomniczi A, Loche A, Tefs K, Kiess W, Danne T, Ojeda SR and Heger S, 2011. Transcriptional regulation of the human KiSS1 gene. Mol Cell Endocrinol 342, 8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro VM, Gottsch ML, Chavkin C, Okamura H, Clifton DK and Steiner RA, 2009. Regulation of gonadotropin-releasing hormone secretion by kisspeptin/dynorphin/neurokinin B neurons in the arcuate nucleus of the mouse. J. Neurosci. 29, 11859–11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandolfi EC, Breuer JA, Nguyen Huu VA, Talluri T, Nguyen D, Lee JS, Hu R, Bharti K, Skowronska-Krawczyk D, Gorman MR, Mellon PL and Hoffmann HM, 2020. The homeodomain transcription factors Vax1 and Six6 are required for SCN development and function. Mol Neurobiol 57, 1217–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandolfi EC, Hoffmann HM, Schoeller EL, Gorman MR and Mellon PL, 2018. Haploinsufficiency of SIX3 abolishes male reproductive behavior rhrough disrupted olfactory development, and impairs female fertility through disrupted GnRH neuron migration. Mol Neurobiol 55, 8709–8727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandolfi EC, Tonsfeldt KJ, Hoffmann HM and Mellon PL, 2019. Deletion of the homeodomain protein Six6 from GnRH neurons decreases GnRH gene expression, resulting in infertility. Endocrinology 160, 2151–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant TM, 2015. 60 YEARS OF NEUROENDOCRINOLOGY: The hypothalamo-pituitary-gonadal axis. J Endocrinol 226, T41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson JL, Clifton DK, de la Iglesia HO, Steiner RA and Kauffman AS, 2009. Circadian regulation of Kiss1 neurons: implications for timing the preovulatory gonadotropin-releasing hormone/luteinizing hormone surge. Endocrinology 150, 3664–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi S, Adeshina I, Chen H, Zirkin BR, Hussain MA, Wondisford F, Wolfe A and Radovick S, 2015. Developmental and endocrine regulation of kisspeptin expression in mouse Leydig cells. Endocrinology 156, 1514–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E, Evanoff R, Quintana A, Evans E, Miller JA, Ko C, Amieux PS, Griswold MD and McKnight GS, 2013. RiboTag Analysis of Actively Translated mRNAs in Sertoli and Leydig Cells In Vivo. PLoS One 8, e66179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P and Cardona A, 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeller EL, Clark DD, Dey S, Cao NV, Semaan SJ, Chao LW, Kauffman AS, Stowers L and Mellon PL, 2016. Bmal1 Is Required for Normal Reproductive Behaviors in Male Mice. Endocrinology 157, 4914–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurmann A, Koling S, Jacobs S, Saftig P, Krauss S, Wennemuth G, Kluge R and Joost HG, 2002. Reduced sperm count and normal fertility in male mice with targeted disruption of the ADP-ribosylation factor-like 4 (Arl4) gene. Mol Cell Biol 22, 2761–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semaan SJ, Murray EK, Poling MC, Dhamija S, Forger NG and Kauffman AS, 2010. BAX-dependent and BAX-independent regulation of Kiss1 neuron development in mice. Endocrinology 151, 5807–5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Thaventhiran T, Minhas S, Dhillo WS and Jayasena CN, 2020. Kisspeptin and Testicular Function-Is it Necessary? Int J Mol Sci 21. [Google Scholar]
- Slavotinek AM, Chao R, Vacik T, Yahyavi M, Abouzeid H, Bardakjian T, Schneider A, Shaw G, Sherr EH, Lemke G, Youssef M and Schorderet DF, 2012. VAX1 mutation associated with microphthalmia, corpus callosum agenesis, and orofacial clefting: the first description of a VAX1 phenotype in humans. Hum Mutat 33, 364–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JT, Cunningham MJ, Rissman EF, Clifton DK and Steiner RA, 2005a. Regulation of Kiss1 gene expression in the brain of the female mouse. Endocrinology 146, 3686–3692. [DOI] [PubMed] [Google Scholar]
- Smith JT, Dungan HM, Stoll EA, Gottsch ML, Braun RE, Eacker SM, Clifton DK and Steiner RA, 2005b. Differential regulation of KiSS-1 mRNA expression by sex steroids in the brain of the male mouse. Endocrinology 146, 2976–2984. [DOI] [PubMed] [Google Scholar]
- Stamatiades GA and Kaiser UB, 2017. Gonadotropin regulation by pulsatile GnRH: Signaling and gene expression. Mol Cell Endocrinol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyn FJ, Wan Y, Clarkson J, Veldhuis JD, Herbison AE and Chen C, 2013. Development of a methodology for and assessment of pulsatile luteinizing hormone secretion in juvenile and adult male mice. Endocrinology 154, 4939–4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonsfeldt KJ, Schoeller EL, Brusman LE, Cui LJ, Lee J and Mellon PL, 2019. The contribution of the dircadian gene Bmal1 to female fertility and the generation of the preovulatory Luteinizing Hormone surge. J Endocr Soc 3, 716–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toolee H, Rastegar T, Solhjoo S, Mortezaee K, Mohammadipour M, Kashani IR and Akbari M, 2019. Roles for Kisspeptin in proliferation and differentiation of spermatogonial cells isolated from mice offspring when the cells are cocultured with somatic cells. J Cell Biochem 120, 5042–5054. [DOI] [PubMed] [Google Scholar]
- Topaloglu AK, 2017. Update on the Genetics of Idiopathic Hypogonadotropic Hypogonadism. J Clin Res Pediatr Endocrinol 9, 113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topaloglu AK, Tello JA, Kotan LD, Ozbek MN, Yilmaz MB, Erdogan S, Gurbuz F, Temiz F, Millar RP and Yuksel B, 2012. Inactivating KISS1 mutation and hypogonadotropic hypogonadism. N Engl J Med 366, 629–635. [DOI] [PubMed] [Google Scholar]
- Wakabayashi Y, Nakada T, Murata K, Ohkura S, Mogi K, Navarro VM, Clifton DK, Mori Y, Tsukamura H, Maeda K, Steiner RA and Okamura H, 2010. Neurokinin B and dynorphin A in kisspeptin neurons of the arcuate nucleus participate in generation of periodic oscillation of neural activity driving pulsatile gonadotropin-releasing hormone secretion in the goat. J. Neurosci. 30, 3124–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, DeFazio RA and Moenter SM, 2016. Excitability and Burst Generation of AVPV Kisspeptin Neurons Are Regulated by the Estrous Cycle Via Multiple Conductances Modulated by Estradiol Action. eNeuro 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirkin BR and Papadopoulos V, 2018. Leydig cells: formation, function, and regulation. Biol Reprod 99, 101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]


