Abstract
With a growing number of parasites and pathogens experiencing large-scale range expansions, monitoring diversity in immune genes of host populations has never been so important because it can inform on the adaptive potential to resist the invaders. Population surveys of immune genes are becoming common in many organisms, yet they are missing in the honey bee (Apis mellifera L.), a key managed pollinator species that has been severely affected by biological invasions. To fill the gap, here we identified single nucleotide polymorphisms (SNPs) in a wide range of honey bee immune genes and developed a medium-density assay targeting a subset of these genes. Using a discovery panel of 123 whole-genomes, representing seven A. mellifera subspecies and three evolutionary lineages, 180 immune genes were scanned for SNPs in exons, introns (< 4 bp from exons), 3’ and 5´UTR, and < 1 kb upstream of the transcription start site. After application of multiple filtering criteria and validation, the final medium-density assay combines 91 quality-proved functional SNPs marking 89 innate immune genes and these can be readily typed using the high-sample-throughput iPLEX MassARRAY system. This medium-density-SNP assay was applied to 156 samples from four countries and the admixture analysis clustered the samples according to their lineage and subspecies, suggesting that honey bee ancestry can be delineated from functional variation. In addition to allowing analysis of immunogenetic variation, this newly-developed SNP assay can be used for inferring genetic structure and admixture in the honey bee.
Subject terms: Biological techniques, Genetics
Introduction
The worldwide movements of humans and goods, coupled with climate change, have led to the introduction, and often successful spread, of many pathogens and parasites into novel environments, and this phenomenon is occurring at unprecedented temporal and spatial scales1. The range and host shifts resulting from these introductions are threatening many organisms across the globe, from mammals2,3, birds4 amphibians5, to fishes6. Within insects, the honey bee Apis mellifera L. has been particularly impacted by introduced parasites and pathogens, most notably the mite Varroa destructor and the microsporidian Nosema ceranae7–10. These are both native to Asia and have rapidly spread worldwide, after a host shift from Apis cerana to Apis mellifera7,10. V. destructor suppresses the bee immunity, but most importantly it acts as a reservoir, incubator and transmission route for several viruses11, including one of the prime honey bee pathogens: the Deformed Wing Virus (DWV). N. ceranae is an intracellular parasite, which decreases colony longevity by inducing oxidative stress and by causing changes in metabolism and immune response8,9,12.
With emerging diseases becoming major selective pressures, colony survival will ultimately depend on how honey bees are able to successfully activate immune mechanisms to protect themselves against foreign pathogens, at both colony (social immunity) and individual (individual immunity) levels. Social immunity arises from behavioural cooperation and includes removal of adult corpses (necrophoric behaviour), removal of diseased or parasitized larvae (hygienic behaviour), and over-production of heat (thermoregulatory behaviour; reviewed by Evans and Spivak13 and DeGrandi-Hoffman and Chen14). Individual immunity entails different lines of defence, ranging from physical barriers (e.g. exoskeleton cuticle and peritrophic membranes lining the digestive tract) to cellular (e.g. phagocytosis, nodulation, encapsulation mediated by hemocytes, and melanization of hemolymph catalysed by phenoloxidase) and humoral responses (synthesis and secretion of antimicrobial peptides, AMPs, such as abaecin, hymenoptaecin, apidaecin, and defensin14–16. In addition to cellular and humoral responses, RNA interference (RNAi) has been described as a major mechanism of antiviral defence in honey bees14,17–19.
The honey bee immune system is triggered when structural motifs on the surface of pathogens (pathogens-associated molecular patterns—PAMPs) bind to its receptors (pattern recognition receptors—PRRs) activating different pathways, depending on the pathogen. These pathways include Toll, IMD (immune deficiency), Jak-STAT (Janus kinase and signal transducer and activator of transcription), and c-Jun N-terminal kinase (JNK). The honey bee has fewer immune genes than Drosophila melanogaster, Aedes aegypti, or Anopheles gambiae17,20. Nonetheless, all the main components of the major pathways, as well as the AMPs, identified in these Dipteran model species are shared by the honey bee17.
Although immune genes are highly conserved across species, some of them also display substantial intra-specific genetic variation21–24. Different studies show that this level of variation affects disease resistance in a wide range of taxa, from mammals25,26 to insects22,24,27. In fact, there is evidence that a small number of large-effect genetic variants has a substantial contribution to differential susceptibility to many diseases in humans28 as well as in arthropods27,29. In honey bees, studies on immune response to infection have been conducted at the gene-expression level for important pathogens and parasites (e.g.11,30–33). However, the link of these findings to inter-individual variability at the DNA level is lacking.
Single nucleotide polymorphisms (SNPs) have been widely used to screen genetic diversity in immune genes (e.g.21,23,33–36) and to identify genes associated with susceptibility to pathogens and parasites in many organisms2,23,36,37 but not in honey bees. As a first step towards filling this gap, here we developed a medium-density-SNP assay for genotyping immune genes in the honey bee. This assay contains 91 quality-proved SNPs located in functional regions of 89 immune genes and these can be readily genotyped with the high-sample-throughput iPLEX MassARRAY system. To the best of our knowledge, this is the first medium-density assay tailored for genotyping SNPs involved in immune response in honey bees.
Results
Assay design, quality control and genotyping accuracy
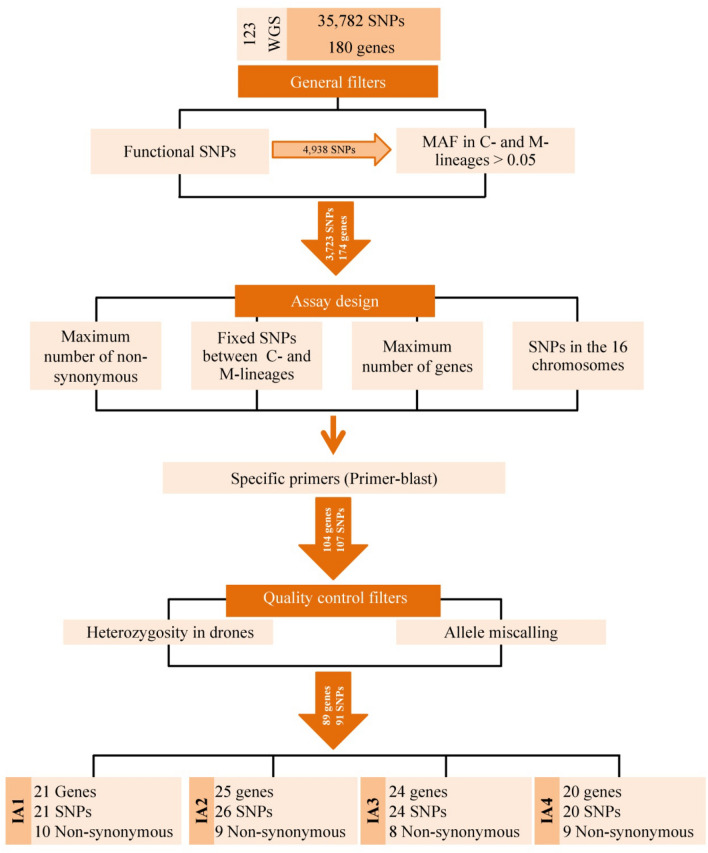
A total of 209 immune genes were compiled from the literature review, but 29 could not be found in the reference genome Amel_4.5. The remaining 180 genes were scanned for polymorphisms in the 123 WGs (Fig. 1; Supplementary Table S1).
Figure 1.
Geographic locations of the 123 whole-genome samples (represented by dots) used for constructing the SNP assay and of the 156 independent samples (represented by stars) used for testing (map generated by QGIS 3.12.2- București- https://qgis.org/es/site/forusers/download.html).
These genes are marked by 35,782 SNPs of which 20,308 are intronic and 11,602 intergenic (Supplementary Table S2). The genes containing the highest number of SNPs are GB46759 (3486) and GB49918 (3032), both related with antiviral defence17. Of the initial 35,782 SNPs, only 4938 could be assigned to putative functional regions, including 2876 (2307 synonymous; 569 non-synonymous) in protein coding, 36 in intronic < 4 bp from exons, 1030 in intergenic < 1 Kb upstream of the transcription start site, 183 in 5’ UTR, and 813 in 3’ UTR. These functional SNPs cover 177 genes, with GB50482 (serine protease) and GB55483 (immunoglobulin-like and fibronectin type III domain) genes harbouring the highest numbers with 233 and 399 SNPs, respectively (Supplementary Table S2).
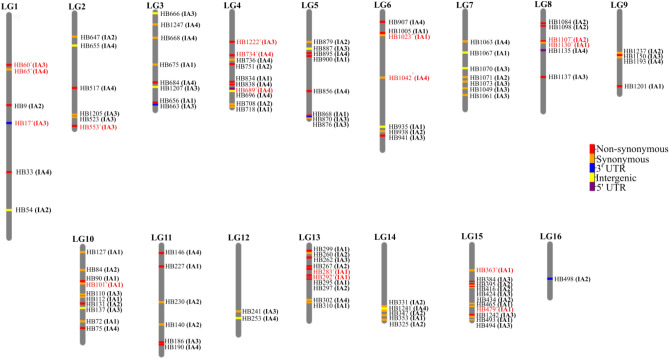
Among the 4938 functional SNPs only those that revealed to be polymorphic (MAF > 0.05) within the M- and C- European lineages were retained, allowing further downsizing to 3723 SNPs. In the subsequent filtering step, which was linked to the MassARRAY MALDI-TOF genotyping system, the flanking regions of the 3723 SNPs were used as input in the AssayDesign software. Despite the high number of possible SNP combinations, none of the final designed multiplexes reached the desired 30-plex size, due to predicted hairpin and dimmer formation artefacts, with IA1 and IA3 containing 28 SNPs, IA2 27, and IA4 only 24 (Fig. 2). Altogether, the four assays allow genotyping 107 SNPs, of which 44 (41%), 61 (57%) and 72 (67%) were polymorphic for the C- (A. m. carnica and A. m. ligustica), M- (A. m. mellifera and A. m. iberiensis) and A-lineages (A. m. intermissa and A. m. sahariensis), respectively, and 46 (43%) were fixed (FST = 1) between C- and M- lineages (Supplementary Table S3). These SNPs cover 104 immune genes distributed across the 16 honey bee chromosomes (Fig. 3).Genomic information for the 107 SNPs as well as their flanking sequences and the PCR and iPLEX reaction primers, for genotyping in the MassARRAY MALDI-TOF platform, are shown in Supplementary Table S4.
Figure 2.
Diagram depicting the different criteria and steps involved in the development of the four SNP assays (IA1, IA2, IA3, and IA4) using as a baseline whole‐genome sequence data from 123 individuals representing seven honey bee subspecies.
Figure 3.
Chromosome map showing the SNP positions in the four assays (IA1–IA4). Each functional state is represented by a different colour. SNPs coloured in red were discarded by at least one filtering criterion.
The SNP calling quality and genotyping accuracy obtained for the four assays with the iPLEX MassARRAY system was assessed in 18 haploid individuals that have been sequenced for the whole-genome in the HiSeq 2500 platform41,42. This endeavour was greatly facilitated by screening haploid drones because we could reliably detect false heterozygous loci. Of the total 1926 called genotypes (107 SNPs × 18 individuals), 131 (6.8%) revealed to be inaccurate (Table 1). The most common source of mismatch between the two platforms was due to heterozygous positions erroneously called in > 10% of the haploid individuals by the MassARRAY system (75 genotypes). However, most mismatches (96%) could be assigned to only 14 SNP loci. In addition to this problem, three SNPs (HB1107, HB60, HB734) exhibited inconsistent allele calls between the two platforms in > 10% of the samples. One of them, locus HB734, was also prone to be misidentified as heterozygous in the haploid individuals (Supplementary Table S5). After removing the 16 problematic loci, we ended up with 91 quality-proved SNPs distributed as follows: 21 in assay IA1, 26 in IA2, 24 in IA3, and 20 in IA4 (Fig. 2; Supplementary Table S4).
Table 1.
SNP calling quality assessed for the 107 SNPs genotyped in 18 haploid individuals (honey bee males) using the iPLEX MassARRAY system. Calls were compared with those obtained with the Illumina HiSeq 2500 platform for the same individuals.
| SNP calling | Number of genotypes |
|---|---|
| Miscalling—inconsistent alleles | 22 |
| Miscalling—heterozygosity | 75 |
| Missing data | 34 |
| Accurate calls | 1795 |
| Total | 1926 |
The 91 SNPs cover 89 immune genes that belong to a wide array of pathways and groups, being serine proteases the most representative (Fig. 4; Supplementary Table S4). Important immune-related pathways are marked by more than one SNP, including IMD, RNAi, JAK/STAT, and Toll, with 2, 3, 4, and 11 SNPs, respectively. Twenty three of the 89 genes have been found to be differentially expressed in honey bees infected with different viruses and Nosema spp. (Table 2; Supplementary Table S4). The 91 SNPs are putatively functional, with the great majority (75) laying in protein coding regions (39 synonymous and 36 non-synonymous). While sixteen SNPs are non-coding, they are likely of functional relevance as they are located in 5’ UTR (1), 3’ UTR (3) and in < 1 Kb upstream of the transcription start site regions (12), where the promoter is expected to be located21.
Figure 4.
Distribution of the 91 SNPs by functional state and pathway/group.
Table 2.
SNPs mapped to genes differentially expressed in honey bees infected by different pathogens.
| SNP | Gene | Pathway | Pathogens | References |
|---|---|---|---|---|
| HB663 | GB41227 | Undescribed | SINV | 17,43 |
| HB656 | GB41241 | Undescribed | SINV | 17,43 |
| HB1241 | GB41669 | Apoptosis | Nosema spp. | 33 |
| HB1237 | GB42706 | Apoptosis | Nosema apis | 33 |
| HB1067 | GB43303 | Ribosomal biosynthesis | IAPV | 17,44 |
| HB1061 | GB44311 | Ribosomal biosynthesis | IAPV | 17,44 |
| HB186 | GB45248 | Undescribed | IAPV | 17,39 |
| HB868 | GB45735 | Endocytosis | IAPV | 17,31 |
| HB498 | GB47575 | MAPK | IAPV | 17,31 |
| HB302 | GB47804 | PGRP | DWV; IAPV | 17,39,40 |
| HB310 | GB47805 | PGRP | DWV; IAPV | 17,39,40 |
| HB1207 | GB49154 | Apoptosis | Nosema spp. | 33 |
| HB1063 | GB49244 | Endocytosis | IAPV | 17,31 |
| HB465 | GB50020 | Jak/STAT | IAPV | 17,31 |
| HB1242 | GB50261 | Cell cycle | Nosema ceranae | 33 |
| HB494 | GB50290 | Undescribed | IAPV | 17,39 |
| HB736 | GB50418 | Toll/TLR | IAPV | 17,39 |
| HB395 | GB50508 | Undescribed | SINV | 17,43 |
| HB72 | GB50955 | RNAi | IAPV; SINV | 17,39,43 |
| HB75 | GB51047 | Undescribed | Nosema ceranae | 45 |
| HB253 | GB52015 | MAPK | IAPV | 17,31 |
| HB137 | GB52625 | MAPK | IAPV | 17,31 |
| HB1205 | GB55605 | Apoptosis | Nosema apis | 33 |
Applying the SNP assay
The 91 SNPs were genotyped on pools of workers collected from 156 colonies from France (Avignon and the island of Ouessant), Israel (Beit Dagan), Portugal (Bragança and the island of São Miguel) and Spain (Marchamalo, Fig. 1). The quality control of the MassARRAY-generated genotypes indicates a high genotyping success rate of the 156 samples. Missing data per locus ranged from 0% (36 SNPs) to 21.8%, although only two SNPs (HB1049 and HB895) exceeded 15% (Supplementary Table S5). Missing data per individual was also low, varying between 0 and 12.1%, with most individuals (83.3%) below 5% (Supplementary Table S6).
The MAF distribution for each SNP and population is shown in Supplementary Fig. S1. Locus HB938 is the least polymorphic (MAF = 0.05) whereas HB310 is the most polymorphic, with both alleles displaying equal frequencies (MAF = 0.5). For 75.8% of the SNPs, the greatest contrast in allelic distribution is exhibited between Beit Dagan plus Avignon and the remaining populations (Ouessant, Bragança, São Miguel, Marchamalo). This contrasting pattern is further reflected by membership proportions (Q-values) inferred by ADMIXTURE for one to seven K genetic groups. According to the cross-validation error, the most likely K is three (Supplementary Fig. S2), although it is only at K = 4 that the two M-lineage subspecies (A. m. mellifera and A. m. iberiensis) split (Fig. 5). At K = 4, populations from Avignon (average Q-value = 0.575, Supplementary Table S6) and Beit Dagan (average Q-value = 0.774, Supplementary Table S6) share an important genetic component with the eastern European A. m. carnica and A. m. ligustica reference subspecies (orange cluster). This suggests that both populations are mostly of C-lineage commercial ancestry, although they also have a background assigned to the clusters of the reference subspecies A. m. mellifera (green cluster) and the north African A. m. intermissa and A. m. sahariensis subspecies (purple cluster). On the other hand, the remaining populations show negligible levels of C-lineage ancestry and they are assigned to clusters that match the historical distributional ranges of the subspecies, i.e. samples from Portugal and Spain are assigned mostly to the A. m. iberiensis cluster (blue) and samples from Ouessant (France) are assigned to the A. m. mellifera cluster (green).
Figure 5.
Admixture plots showing the membership partitioning (Q-value) from two (K = 2) to four clusters (K = 4). Each individual is represented by a vertical bar. The black vertical lines separate individuals from different populations (Bragança and São Miguel-Portugal; Marchamalo-Spain; Avignon and Ouessant- France; Beit Dagan- Israel).
Discussion
In the last decades, international trade in honey bees and their products has facilitated global dissemination of exotic parasites and pathogens46,47, many of which became major causes of colony losses7,10,48. In addition to be a vehicle for disseminating parasites and pathogens at the global scale, trade can also help exotic gene flow within the A. mellifera natural range, leading to introgressive hybridization or even displacement of native subspecies49–51. In previous studies, we developed SNP-based tools for estimating introgression of C-derived commercial honey bee stocks into M-lineage subspecies from Western Europe42,52,53. In this study, we expanded the existing SNP-tool box by developing a medium-density assay for screening polymorphisms in immune genes. Of note is that, in addition to allowing surveys of putatively functional variation in 89 immune genes14,17,30,33,38,45,54–56, this assay can discriminate subspecies belonging to different evolutionary lineages and detect C-derived introgression in colonies with diverse genetic background.
Development of the medium-density assay begun with detection of SNPs across the honey bee immunome. The entire coding regions of 180 genes (< 1 kb upstream of the transcription start site, exons, introns < 4 bp from exons, and UTRs) were scanned on 123 whole genomes, representing seven subspecies and three evolutionary lineages (A, M, and C). By sampling such an important fraction of the A. mellifera diversity, this discovery panel enabled detection of SNPs with low frequency of the minor allele (average MAF = 0.19), therefore minimizing ascertainment bias57. With the exception of the recently published study of Momeni, et al.58, this is the largest and broadest panel used for SNP discovery in the honey bee. Previous works screened narrower discovery panels, suited to the goals being addressed by the constructed assays. While Momeni, et al.58 screened 14 subspecies to construct a ~ 4 K SNP assay for diagnosing European subspecies, others used discovery panels with less than four subspecies to construct high- to medium-density SNP assays tailored for addressing multiple objectives, including analysis of variation linked to defensive behaviour against V. destructor59,60 genomic selection and breeding60, identification of Africanised honey bees61,62, and inference of C-lineage introgression into M-lineage subspecies42,52,63.
Selection of SNPs for the medium-density assay was done in multiple steps using several criteria, with emphasis put on exonic SNPs while at the same time attempting a good representation of the immunome. Due to potential hairpin and dimmer artefacts, the AssayDesign software only allowed multiplexing 107 SNPs, covering 58% (104 in 180) of the polymorphic immune genes. After validation, the number of usable variation was downsized to 91 SNPs, representing only 89 immune genes (49%) but yet covering a wide array of important pathways, including IMD, RNAi, JAK/STAT. Besides, 23 of these genes might be of adaptive relevance because they are differentially expressed in bees infected with different viruses and Nosema spp.17,31,33,39,40,43–45.
The 107 selected SNPs were validated on 18 independent drones (haploid males) by comparing the SNP calls generated by the iPLEX MassARRAY system with those obtained in the Illumina HiSeq 2500 platform. This endeavour was greatly facilitated by the haplodiploid sex determination system in A. mellifera because, by screening drones instead of the traditionally used workers (diploid non-fertile females), we were able to detect allelic drop-in artefacts in the erroneously typed heterozygous loci. Fourteen SNPs showed heterozygous calls in > 10% of the drones, which was the main source of mismatch (14 out of 16 problematic SNPs) between the two platforms. This 13% (14 out of 107 typed SNPs) allele drop-in error was considerably higher than that obtained (2%, 3 out of 127 typed SNPs) with a SNP assay designed in a previous study for detecting introgression in A. m. mellifera, which was also typed with the iPLEX MassARRAY system in an independent dataset52. Allele drop-in could not be due to contamination because the 14 problematic SNPs were spread across the four assays and the remaining loci were correctly typed as homozygous and matched the calls of the Illumina HiSeq 2500 platform. Instead, allele drop-in could be explained by gene homology52, although this is an unlikely source of error because the primers designed by the AssayDesign software were all verified in silico for potential binding to multiple targets. Nonetheless, this hypothesis cannot be completely ruled out because the honey bee genome is not fully curated. Given that the genotyping with the two different assays was outsourced to different laboratories52, which have distinct MassARRAY models, the difference in the error rates could be explained by a technical artefact. After removing the problematic SNPs, the remaining 91 were accurately called in the 18 drone samples and successfully genotyped in the 156 independent samples, as reflected by the low rate of missing data (3.5%). Among the 91 quality-proved SNPs, of particular interest are 36 that result in non-synonymous changes, as these are likely to have a functional effect. However, the remaining 55 are also promising as they may affect gene regulation or expression and thus also have a functional role64.
Evidence from many organisms indicates that sequence variation in immune genes affects host susceptibility to infections22,24–27. In honey bees, the interaction between the immune system and pathogens has been addressed only through employment of gene expression assays11,30–33. These assays showed differential expression in immune genes when honey bees are infected with SINV43, IAPV31,43, DWV40 and Nosema spp.33,45. But more importantly, these assays showed differential expression between tolerant and sensitive bee strains to infections of N. ceranae and V. destructor30,38,65. Despite the shallow coverage of the honey bee immunome, the SNP assay developed here can be used as a very first step towards unravelling the genetic basis of honey bee susceptibility to infections through case–control association tests, similar to studies on other organisms37,66–69. Our assay is promising for this type of analyses for different reasons. First, although the assay only partially samples the immunome (average one SNP per gene and 89 of the 180 annotated genes), it includes SNPs located in the main pathways, such as IMD, JAK-STAT, Toll and RNAi, and there is evidence that a small number of large-effect genes have a substantial contribution to the genetic variation of host susceptibility27–29, increasing the probability of finding associations. Second, 82% of the SNPs lay in protein-coding regions and the non-synonymous SNPs may directly change protein function. Third, while the remaining SNPs fall outside of protein-coding regions, they are not only mapped to other relevant regions, potentially affecting transcription regulation (5’UTR), messenger RNA stability (3’UTR), and binding of the transcription initiation complex to the promoter (< 1 Kb upstream of the transcription start site21,64), but also in linkage with the coding region. Finally, some of the SNPs are located in genes that showed to be differentially expressed in honey bees infected with SINV43, IAPV31,43, DWV40 and Nosema spp.33,45.
The medium-density immune assay developed here, as well as other dedicated assays42,52,59–62, is not recommended for standard population genetics analyses relying on allele frequency spectrum in different genomic regions or neutrality (e.g. nucleotide diversity, population size, migration, population structure). This is because genetic inference can be distorted due to ascertainment bias70,71 as well as to biases introduced from choosing high-graded SNPs segregating among target populations72, or from targeting only functional regions73, which might not be neutral. Particularly, SNPs in immune genes are expected to be under selective pressure by the spread of new parasites and pathogens or by evolution of the existing ones28,74,75, which can act to increase the frequency of beneficial alleles or to maintain polymorphisms. Therefore, caution is warranted when using this biased subset of the honey bee genome to infer broader patterns of differentiation because, as shown for other organisms, disease-related genes may have excessive SNPs with lower or higher levels of population differentiation due to purifying76, balancing77 or directional selection76. Notwithstanding these biases, inclusion of non-neutral or highly informative markers in dedicated assays may be advantageous when attempting to (i) assign individuals to source populations, (ii) estimate genetic admixture or even (iii) infer structure on ecological (rather than evolutionary) time-scale (reviewed by Helyar, et al.78). In this study, the ADMIXTURE analysis clustered the reference samples according to their lineage and subspecies, suggesting that our medium-density assay can be used to infer population structure, consistent with findings reported for other organisms with immune SNPs73,79. Furthermore, the admixture patterns inferred for the 156 samples are consistent with the conservation status and the genetic background that has been documented for the populations from where they were collected. Samples originating from the A. m. mellifera population of Ouessant and from A. m. iberiensis of mainland Portugal and Spain show a high genetic integrity as well as a shared ancestry, as expected for these sister subspecies41,80,81. In contrast, samples from the Azorean island of São Miguel and, more prominently, samples from mainland France and Israel are severely introgressed, as a result of beekeeper-mediated processes linked to recurrent use of commercial stock typically of C-lineage ancestry50,51,82–84.
Here, we detailed the steps involved in developing a medium-density assay for genotyping immune-related SNPs with the iPLEX MassARRAY system. We provided the genomic information along with the PCR and iPLEX primers for the 91 quality-proved SNPs, enabling immediate application of the immune assay and analysis of the output does not require powerful computational resources or advanced bioinformatics skills. The 91 SNPs are mapped to coding regions (and some even involve amino acid changes) or in their close vicinity. Whether they are directly associated to immune response to pathogens or in linkage with causative SNPs not included in our assay requires functional or expression studies85. The assay was tailored for assessing diversity in immune genes and allows for high-throughput screening of samples in a time- and cost-effective manner. In addition to potential application in studies searching for associations between molecular markers and response to pathogens, the assay could be useful for delineating ancestry in honey bees.
Methods
Samples and whole-genome re-sequencing
The immune SNPs were identified from scanning whole genomes (WGs) previously re-sequenced for 123 individuals originating from a wide geographical area41,42,52 (Fig. 1; Supplementary Table S7). This WG dataset represents seven subspecies from three evolutionary lineages, with sample sizes varying from 95 for lineage M (87 A. m. iberiensis and 8 A. m. mellifera), 19 for lineage A (12 A. m. intermissa and 7 A. m. sahariensis), to 9 for lineage C (4 A. m. ligustica, 3 A. m. carnica, and 2 A. m. siciliana; Fig. 1). The 123 samples were run on an Illumina HiSeq 2500 platform with sequencing libraries generated using Illumina TruSeq Sample Preparation Kit (see41 for further details). Mapping and variant calling were performed following Henriques, et al.41. A total of 2,525,621 SNPs with a minor allele frequency (MAF) ≥ 0.05 were identified after the quality control step. The functional state (non-synonymous, synonymous, intronic, UTR, or intergenic) of each SNP was annotated using the reference genome Amel_4.5, the Official Gene Set 3.2 (BEEBASE) and the Entrez Gene of NCBI.
SNP assay design
A list of 209 immune genes putatively involved in honey bee defence against viruses14,17,54,55 and Nosema spp. infections30,33,38,45,56 was compiled from the literature (Supplementary Table S1). These were scanned for polymorphisms in the 123 WGs, leading to detection of 35,782 SNPs located in only 180 of the 209 genes. Following application of several filters and criteria (Fig. 2), the 35,782 SNPs were narrowed down to fit the optimal plex size of 30 SNPs per assay, as recommended by Agena BioScience for the iPLEX chemistry and the MassARRAY MALDI-TOF platform (herein abbreviated to iPLEX MassARRAY). Of the three chip formats (24, 96, and 384) available for genotyping with the iPLEX MassARRAY system, we chose the 384-format because it offers the best trade-off between number of loci and number of samples. Using a medium-density SNP panel (4 assays × 30 SNPs = 120 SNPs maximum) it is possible to screen 96 individuals in one chip.
To downsize the 35,782 SNPs, while assuring that the final medium-density assay contained SNPs with functional relevance, we discarded putatively non-functional SNPs and only retained those located in (i) protein coding (synonymous and non-synonymous), (ii) intergenic < 1 Kb upstream of the transcription start site, where the promoter is expected to be located, (iii) 3’UTR, (iv) 5’UTR, and (v) intronic < 4 bp from exons, where alternative splicing occurs86. To further narrow down the SNP number, while assuring that the final panel contained informative SNPs for subspecies important to beekeeping in Europe (A. m. iberiensis and A. m. mellifera in lineage M; A. m. ligustica and A. m. carnica in lineage C), we discarded those with MAF ≤ 0.05 identified in a subset comprising only C- and M-lineage subspecies (Fig. 2).
The ensuing step was linked to the iPLEX MassARRAY system (Fig. 2). The 501-bp flanking regions (250 bp on either side) of all retained SNPs (3723) served as input for delineating the four assays (IA1, IA2, IA3, and IA4) with the AssayDesign Suite software (http://agenabio.com/assay-design-suite-20-software), using the default settings. The 3723 SNPS were prioritized to assure that the final panel contained the highest possible number of non-synonymous SNPs, functional SNPs with FST = 1 between M- and C- lineages (calculated in PLINK 1.987), immune genes, and chromosomes. The AssayDesign Suite searched for optimal areas within the 501-bp flanking region of the 3723 SNPs for designing the PCR primers, while constructing all possible multiplex combinations. The optimal 30-SNP multiplexing capacity was attempted by the software whilst preventing hairpin and dimmer formation. In addition to the PCR primers, the software designed the iPLEX extension primers placed immediately adjacent to each SNP. The quality of each designed primer was assessed using the Primer-BLAST tool available in NCBI. This additional step in the SNP selection process allowed identification of primers matching multiple targets in the honey bee genome, and subsequent deletion of potentially problematic SNPs. Lastly, the genomic positions of the SNPs incorporated in the final panel were upgraded in the latest honey bee genome assembly Amel_HAv3.1 by using the BLASTn tool in NCBI (Supplementary Table S4).
IPLEX MassARRAY
The four newly-designed SNP assays were genotyped using the iPLEX MassARRAY MALDI-TOF system. Briefly, the PCR primers were pooled to a final concentration of 500 nM. The PCR primer pool was used to amplify 10 ng of DNA in a 5 µl volume reaction with 1 U of FastStart Taq Polymerase (Roche, Indianapolis, IN) and 4 mM of Mg2Cl. PCRs were performed in a 5 µl volume using a standard 384-well plate format according to the specifications provided by Agena BioScience. The plates were cycled 45 times with an annealing temperature of 56 °C. After PCR, shrimp alkaline phosphatase (SAP) was used to dephosphorylate any remaining dNTPs to render them unusable for ensuing polymerase reactions. A total of 0.5 U of SAP and buffer (Agena Bioscience) was added to the PCR tubes and incubated for 40 min at 37 °C, followed by 5 min at 85 °C. Single base extension reactions were performed on the PCR reactions with the iPLEX Gold Kit (Agena Bioscience) and 0.8 µl of the custom UEP (unextended primer) pool. The kit contains mass-modified terminator nucleotides that increase the mass difference between extended UEPs, allowing for greater accuracy in genotyping. The mass difference with unmodified terminator nucleotides ranges from 9 to 40 kDa, depending on the two nucleotides compared. With the mass-modified terminator nucleotides the mass difference increases to 16–80 kDa. The single base extension reactions were cycled with a nested PCR protocol that used five cycles of annealing and extension nested with a denaturation step in a cycle that was repeated 40 times for a total of 200 annealing and extension steps. The goal was to extend nearly all of the UEPs. Following single base extension, the reactions were diluted with 16 µl of deionized water and with 6 ng of resin. After a 20-min. deionizing step, the reactions were dispensed onto SpectroChipArrays with a Nanodispenser (Agena Bioscience). The speed of dispensation was optimized to deliver an average of 20 nL of each reaction to a matrix pad on the SpectroChip. An Agena Bioscience Compact MassArray Spectrometer was used to perform MALDI-TOF mass spectrometry according to the iPLEX Gold Application Guide88. The Typer 4 software package (Agena Bioscience) was used to analyse the resulting spectra and the composition of the target bases was determined from the mass of each extended oligo.
SNP assay assessment
Eighteen drones (4 A. m. iberiensis, 3 A. m. mellifera, 4 A. m. ligustica, 3 A. m. carnica, 4 A. m. intermissa) were chosen from the WG set for assessing SNP calling quality and genotyping accuracy of the newly developed IA1, IA2, IA3, and IA4 assays (Fig. 2). The DNAs of the 18 drones (haploid males) were previously isolated with a phenol-clorophorm protocol89 and these were genotyped herein using the four assays and the iPLEX MassARRAY system, as described above. The missing genotype data were calculated using PLINK. The SNP calls generated by the iPLEX MassARRAY system were compared with those obtained previously with the Illumina HiSeq 2500 platform41,42,52 for the WGS and the number of inconsistent genotypes was recorded.
SNP assay application
The four assays were applied to 156 samples collected between 2018 and 2019 from colonies deployed in Spain (N = 34), France (N = 36), Israel (N = 40), and Portugal (N = 46; Fig. 1; Supplementary Table S6). Each of the 156 samples represented a single colony and consisted of a pool of 10 workers (diploid non-fertile females). Total DNA was extracted from each pool (20 front legs) using the Ron’s Tissue DNA Mini Kit (Bioron), following manufacturer instructions with slight modifications. The 156 samples were genotyped using the iPLEX MassARRAY system, as described above.
The allele frequencies were plotted for each SNP using the R script described in Jenkins, et al.90 with slights modifications. The membership proportions (Q-values) were inferred for the 156 pooled samples with ADMIXTURE91 for a number of K genetic groups ranging from one to seven, using 10,000 iterations in 20 independent runs. The convergence between iterations was monitored by comparing log-likelihood scores (LLS) using the default termination criterion set to stop when LLS increases by < 0.0001 between iterations. CLUMPAK92 was used to summarize and visualize the Q-plots. A subset of the whole-genome sequenced individuals (15 A. m. iberiensis, 12 A. m. intermissa, 4 A. m. ligustica, 3 A. m. carnica, 8 A. m. mellifera, 7 A. m. sahariensis and 2 A. m. siciliana) was used as a reference in the ADMIXTURE analysis. The most probable K was estimated for this dataset using the 5- fold cross-validation (CV) error.
Supplementary Information
Acknowledgements
We are deeply indebted to Frank Aguiar, Luís Silva, Edgardo Melo, João Martins, João Melo, Manuel Moura, Manuel Viveiros, and Ricardo Sousa from "Direção Regional da Agricultura e Desenvolvimento Rural dos Açores" (Portugal), and to Laura Garreau, Laurent Maugis, Pascale Sauvage and Jacques Kermagoret, from “Association Conservatoire de l’Abeille Noir Bretonne” (France), for sampling the apiaries in São Miguel, Santa Maria, and Ouessant islands. Genotyping was outsourced to the Epigenetics and Genotyping laboratory, Central Unit for Research in Medicine (UCIM), University of Valencia, Spain. Data analyses were performed using computational resources at the Research Centre in Digitalization and Intelligent Robotics (CeDRI), Instituto Politécnico de Bragança. Ana Rita Lopes is supported by a PhD scholarship (SFRH/BD/143627/2019) from the Foundation for Science and Technology (FCT), Portugal. FCT provided financial support by national funds (FCT/MCTES) to CIMO (UIDB/00690/2020).This research was funded through the projects BEEHAPPY (POCI-01-0145-FEDER-029871, FCT and COMPETE/QREN/EU) and BEEHEAL. BEEHEAL was funded by the ARIMNet2 2016 Call by the following agencies: INIA (Spain), MOARD (Israel), ANR (France) and FCT (Portugal). ARIMNet2 (ERA-NET) received funding from the European Union’s Seventh Framework Programme for research, technological development and demonstration under grant agreement no. 618127.
Author contributions
M.A.P. and D.H. conceived the ideas and designed the experiment. The analyses were performed by D.H. with assistance of A.R.L. D.H., M.A.P. and A.R.L. wrote the manuscript. M.A.P., A.R.L., N.C., V.S., Y.L.C., A.D., M.R.C., M.H., R.M.H. contributed with honey bee samples. All the authors critically reviewed the manuscript for important intellectual content.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-94833-x.
References
- 1.Prenter J, MacNeil C, Dick JT, Dunn AM. Roles of parasites in animal invasions. Trends Ecol. Evol. 2004;19:385–390. doi: 10.1016/j.tree.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 2.Donaldson ME, et al. Profiling the immunome of little brown myotis provides a yardstick for measuring the genetic response to white-nose syndrome. Evol. Appl. 2017;10:1076–1090. doi: 10.1111/eva.12514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McInnes C, et al. First cases of squirrelpox in red squirrels (Sciurus vulgaris) in Scotland. Vet. Rec. Case Rep. 2013;1:e528. doi: 10.1136/vetreccr.164.17.528rep. [DOI] [PubMed] [Google Scholar]
- 4.Van Riper C, Van Riper SG, Goff ML, Laird M. The epizootiology and ecological significance of malaria in Hawaiian land birds. Ecol. Monogr. 1986;56:327–344. doi: 10.2307/1942550. [DOI] [Google Scholar]
- 5.Lips KR, et al. Emerging infectious disease and the loss of biodiversity in a Neotropical amphibian community. Proc. Natl. Acad. Sci. USA. 2006;103:3165–3170. doi: 10.1073/pnas.0506889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauer O, Pugachev O, Voronin V. Study of parasites and diseases of sturgeons in Russia: a review. J. Appl. Ichthyol. 2002;18:420–429. doi: 10.1046/j.1439-0426.2002.00422.x. [DOI] [Google Scholar]
- 7.Traynor KS, et al. Varroa destructor: A complex parasite, crippling honey bees worldwide. Trends Parasitol. 2020;36:592–606. doi: 10.1016/j.pt.2020.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Antúnez K, et al. Immune suppression in the honey bee (Apis mellifera) following infection by Nosema ceranae (Microsporidia) Environ. Microbiol. 2009;11:2284–2290. doi: 10.1111/j.1462-2920.2009.01953.x. [DOI] [PubMed] [Google Scholar]
- 9.Dussaubat C, et al. Gut pathology and responses to the microsporidium Nosema ceranae in the honey bee Apis mellifera. PLoS ONE. 2012;7:e37017. doi: 10.1371/journal.pone.0037017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martín-Hernández R, et al. Nosema ceranae in Apis mellifera: A 12 years postdetection perspective. Environ. Microbiol. 2018;20:1302–1329. doi: 10.1111/1462-2920.14103. [DOI] [PubMed] [Google Scholar]
- 11.Yang X, Cox-Foster DL. Impact of an ectoparasite on the immunity and pathology of an invertebrate: Evidence for host immunosuppression and viral amplification. Proc. Natl. Acad. Sci. USA. 2005;102:7470–7475. doi: 10.1073/pnas.0501860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Botías C, Martín-Hernández R, Barrios L, Meana A, Higes M. Nosema spp infection and its negative effects on honey bees (Apis mellifera iberiensis) at the colony level. Vet. Res. 2013;44:25. doi: 10.1186/1297-9716-44-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans JD, Spivak M. Socialized medicine: individual and communal disease barriers in honey bees. J. Invertebr. Pathol. 2010;103:S62–S72. doi: 10.1016/j.jip.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 14.DeGrandi-Hoffman G, Chen Y. Nutrition, immunity and viral infections in honey bees. Curr. Opin. Insect Sci. 2015;10:170–176. doi: 10.1016/j.cois.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 15.Evans J, et al. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 2006;15:645–656. doi: 10.1111/j.1365-2583.2006.00682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosales C, Vonnie S. Insect Physiology and Ecology. InTeach Publication, CCBY; 2017. Cellular and molecular mechanisms of insect immunity; pp. 179–212. [Google Scholar]
- 17.Brutscher LM, Daughenbaugh KF, Flenniken ML. Antiviral defense mechanisms in honey bees. Curr. Opin. Insect Sci. 2015;10:71–82. doi: 10.1016/j.cois.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kingsolver MB, Huang Z, Hardy RW. Insect antiviral innate immunity: Pathways, effectors, and connections. J. Mol. Biol. 2013;425:4921–4936. doi: 10.1016/j.jmb.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larsen A, Reynaldi FJ, Guzmán-Novoa E. Fundaments of the honey bee (Apis mellifera) immune system. Review. Rev Mex Cienc Pecu. 2019;10:705–728. doi: 10.22319/rmcp.v10i3.4785. [DOI] [Google Scholar]
- 20.Weinstock GM, et al. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lazarus R, et al. Single nucleotide polymorphisms in innate immunity genes: Abundant variation and potential role in complex human disease. Immunol. Rev. 2002;190:9–25. doi: 10.1034/j.1600-065X.2002.19002.x. [DOI] [PubMed] [Google Scholar]
- 22.Tinsley M, Blanford S, Jiggins F. Genetic variation in Drosophila melanogaster pathogen susceptibility. Parasitology. 2006;132:767. doi: 10.1017/S0031182006009929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elbers JP, Brown MB, Taylor SS. Identifying genome-wide immune gene variation underlying infectious disease in wildlife populations: A next generation sequencing approach in the gopher tortoise. BMC Genom. 2018;19:64. doi: 10.1186/s12864-018-4452-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lazzaro BP, Sackton TB, Clark AG. Genetic variation in Drosophila melanogaster resistance to infection: A comparison across bacteria. Genetics. 2006;174:1539–1554. doi: 10.1534/genetics.105.054593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bishop S, Morris C. Genetics of disease resistance in sheep and goats. Small Rumin. Res. 2007;70:48–59. doi: 10.1016/j.smallrumres.2007.01.006. [DOI] [Google Scholar]
- 26.Turner AK, Begon M, Jackson JA, Bradley JE, Paterson S. Genetic diversity in cytokines associated with immune variation and resistance to multiple pathogens in a natural rodent population. PLoS Genet. 2011;7:e1002343. doi: 10.1371/journal.pgen.1002343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Magwire MM, et al. Genome-wide association studies reveal a simple genetic basis of resistance to naturally coevolving viruses in Drosophila melanogaster. PLoS Genet. 2012;8:e1003057. doi: 10.1371/journal.pgen.1003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hill AV. Evolution, revolution and heresy in the genetics of infectious disease susceptibility. Philos. Trans. R. Soc. B. 2012;367:840–849. doi: 10.1098/rstb.2011.0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luijckx P, Fienberg H, Duneau D, Ebert D. Resistance to a bacterial parasite in the crustacean Daphnia magna shows Mendelian segregation with dominance. Heredity. 2012;108:547–551. doi: 10.1038/hdy.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Q, Kryger P, Le Conte Y, Moritz RF. Survival and immune response of drones of a Nosemosis tolerant honey bee strain towards N. ceranae infections. J. Invertebr. Pathol. 2012;109:297–302. doi: 10.1016/j.jip.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 31.Chen YP, et al. Israeli acute paralysis virus: epidemiology, pathogenesis and implications for honey bee health. PLoS Pathog. 2014;10:e1004261. doi: 10.1371/journal.ppat.1004261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nazzi F, et al. Synergistic parasite-pathogen interactions mediated by host immunity can drive the collapse of honeybee colonies. PLoS Pathog. 2012;8:e1002735. doi: 10.1371/journal.ppat.1002735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martín-Hernández R, et al. Microsporidia infection impacts the host cell's cycle and reduces host cell apoptosis. PLoS ONE. 2017;12:e0170183. doi: 10.1371/journal.pone.0170183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morris KM, Wright B, Grueber CE, Hogg C, Belov K. Lack of genetic diversity across diverse immune genes in an endangered mammal, the Tasmanian devil (S arcophilus harrisii) Mol. Ecol. 2015;24:3860–3872. doi: 10.1111/mec.13291. [DOI] [PubMed] [Google Scholar]
- 35.Kongchum P, Palti Y, Hallerman EM, Hulata G, David L. SNP discovery and development of genetic markers for mapping innate immune response genes in common carp (Cyprinus carpio) Fish Shellfish Immunol. 2010;29:356–361. doi: 10.1016/j.fsi.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 36.Kosch T, et al. Genetic potential for disease resistance in critically endangered amphibians decimated by chytridiomycosis. Anim. Conserv. 2019;22:238–250. doi: 10.1111/acv.12459. [DOI] [Google Scholar]
- 37.Sackton TB, Lazzaro BP, Clark AG. Genotype and gene expression associations with immune function in Drosophila. PLoS Genet. 2010;6:e1000797. doi: 10.1371/journal.pgen.1000797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurze C, et al. Nosema tolerant honeybees (Apis mellifera) escape parasitic manipulation of apoptosis. PLoS ONE. 2015;10:e104174. doi: 10.1371/journal.pone.0140174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Galbraith DA, Yang X, Nino EL, Yi S, Grozinger C. Parallel epigenomic and transcriptomic responses to viral infection in honey bees (Apis mellifera) PLoS Pathog. 2015;11:e1004713. doi: 10.1371/journal.ppat.1004713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuster RD, Boncristiani HF, Rueppell O. Immunogene and viral transcript dynamics during parasitic Varroa destructor mite infection of developing honey bee (Apis mellifera) pupae. J. Exp. Biol. 2014;217:1710–1718. doi: 10.1242/jeb.097766. [DOI] [PubMed] [Google Scholar]
- 41.Henriques D, et al. Whole genome SNP-associated signatures of local adaptation in honeybees of the Iberian Peninsula. Sci. Rep. 2018;8:1–14. doi: 10.1038/s41598-018-29469-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henriques D, et al. Developing reduced SNP assays from whole-genome sequence data to estimate introgression in an organism with complex genetic patterns, the Iberian honeybee (Apis mellifera iberiensis) Evol. Appl. 2018;11:1270–1282. doi: 10.1111/eva.12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flenniken ML, Andino R. Non-specific dsRNA-mediated antiviral response in the honey bee. PLoS ONE. 2013;8:e77263. doi: 10.1371/journal.pone.0077263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boncristiani HF, et al. In vitro infection of pupae with Israeli acute paralysis virus suggests disturbance of transcriptional homeostasis in honey bees (Apis mellifera) PLoS ONE. 2013;8:e73429. doi: 10.1371/journal.pone.0073429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang Q, et al. Four quantitative trait loci associated with low Nosema ceranae (Microsporidia) spore load in the honeybee Apis mellifera. Apidologie. 2014;45:248–256. doi: 10.1007/s13592-013-0243-4. [DOI] [Google Scholar]
- 46.Mutinelli F. The spread of pathogens through trade in honey bees and their products (including queen bees and semen): Overview and recent developments. Rev. Sci. Tech. 2011;30:257. doi: 10.20506/rst.30.1.2033. [DOI] [PubMed] [Google Scholar]
- 47.Owen R. Role of human action in the spread of honey bee (Hymenoptera: Apidae) pathogens. J. Econ. Entomol. 2017;110:797–801. doi: 10.1093/jee/tox075. [DOI] [PubMed] [Google Scholar]
- 48.Potts SG, et al. Global pollinator declines: Trends, impacts and drivers. Trends Ecol. Evol. 2010;25:345–353. doi: 10.1016/j.tree.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 49.Soland-Reckeweg G, Heckel G, Neumann P, Fluri P, Excoffier L. Gene flow in admixed populations and implications for the conservation of the Western honeybee, Apis mellifera. J. Insect Conserv. 2009;13:317–328. doi: 10.1007/s10841-008-9175-0. [DOI] [Google Scholar]
- 50.Pinto MA, et al. Genetic integrity of the Dark European honey bee (Apis mellifera mellifera) from protected populations: A genome-wide assessment using SNPs and mtDNA sequence data. J. Apic. Res. 2014;53:269–278. doi: 10.3896/IBRA.1.53.2.08. [DOI] [Google Scholar]
- 51.Jensen AB, Palmer KA, Boomsma JJ, Pedersen BV. Varying degrees of Apis mellifera ligustica introgression in protected populations of the black honeybee, Apis mellifera mellifera, in northwest Europe. Mol. Ecol. 2005;14:93–106. doi: 10.1111/j.1365-294X.2004.02399.x. [DOI] [PubMed] [Google Scholar]
- 52.Henriques D, et al. High sample throughput genotyping for estimating C-lineage introgression in the dark honeybee: An accurate and cost-effective SNP-based tool. Sci. Rep. 2018;8:1–14. doi: 10.1038/s41598-018-26932-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muñoz I, et al. Reduced SNP panels for genetic identification and introgression analysis in the dark honey bee (Apis mellifera mellifera) PLoS ONE. 2015;10:e0124365. doi: 10.1371/journal.pone.0124365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Evans JD. Beepath: An ordered quantitative-PCR array for exploring honey bee immunity and disease. J. Invertebr. Pathol. 2006;93:135–139. doi: 10.1016/j.jip.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 55.McMenamin AJ, Daughenbaugh KF, Parekh F, Pizzorno MC, Flenniken ML. Honey bee and bumble bee antiviral defense. Viruses. 2018;10:395. doi: 10.3390/v10080395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Branchiccela B, et al. Characterization of Nosema ceranae genetic variants from different geographic origins. Microb. Ecol. 2017;73:978–987. doi: 10.1007/s00248-016-0880-z. [DOI] [PubMed] [Google Scholar]
- 57.Kim JM, et al. A high-density SNP chip for genotyping great tit (Parus major) populations and its application to studying the genetic architecture of exploration behaviour. Mol. Ecol. Resour. 2018;18:877–891. doi: 10.1111/1755-0998.12778. [DOI] [PubMed] [Google Scholar]
- 58.Momeni J, et al. Authoritative subspecies diagnosis tool for European honey bees based on ancestry informative SNPs. BMC Genom. 2021;22:1–12. doi: 10.1186/s12864-021-07379-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spötter A, Gupta P, Nürnberg G, Reinsch N, Bienefeld K. Development of a 44K SNP assay focussing on the analysis of a varroa-specific defence behaviour in honey bees (Apis mellifera carnica) Mol. Ecol. Resour. 2012;12:323–332. doi: 10.1111/j.1755-0998.2011.03106.x. [DOI] [PubMed] [Google Scholar]
- 60.Jones JC, et al. Tool for genomic selection and breeding to evolutionary adaptation: Development of a 100K single nucleotide polymorphism array for the honey bee. Ecol. Evol. 2020;10:6246–6256. doi: 10.1002/ece3.6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chapman NC, et al. An abbreviated SNP panel for ancestry assignment of honeybees (Apis mellifera) Apidologie. 2017;48:776–783. doi: 10.1007/s13592-017-0522-6. [DOI] [Google Scholar]
- 62.Chapman NC, et al. A SNP test to identify Africanized honeybees via proportion of ‘African’ancestry. Mol. Ecol. Resour. 2015;15:1346–1355. doi: 10.1111/1755-0998.12411. [DOI] [PubMed] [Google Scholar]
- 63.Parejo M, et al. Using whole-genome sequence information to foster conservation efforts for the European Dark Honey Bee, Apis mellifera mellifera. Front. Ecol. Evol. 2016;4:140. [Google Scholar]
- 64.Li MJ, Yan B, Sham PC, Wang J. Exploring the function of genetic variants in the non-coding genomic regions: Approaches for identifying human regulatory variants affecting gene expression. Brief. Bioinform. 2015;16:393–412. doi: 10.1093/bib/bbu018. [DOI] [PubMed] [Google Scholar]
- 65.Navajas M, et al. Differential gene expression of the honey bee Apis mellifera associated with Varroa destructor infection. BMC Genom. 2008;9:301. doi: 10.1186/1471-2164-9-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kumar S, et al. Association of Bovine CLEC7A gene polymorphism with host susceptibility to paratuberculosis disease in Indian cattle. Res. Vet. Sci. 2019;123:216–222. doi: 10.1016/j.rvsc.2019.01.016. [DOI] [PubMed] [Google Scholar]
- 67.Cao Z, et al. Genetic polymorphism of MHC class IIα alleles and its association with resistance/susceptibility to viral nervous necrosis virus (VNNV) in golden pompano (Trachinotus ovatus) Aquaculture. 2019;501:144–152. doi: 10.1016/j.aquaculture.2018.11.025. [DOI] [Google Scholar]
- 68.Ziakas PD, Prodromou ML, El Khoury J, Zintzaras E, Mylonakis E. The role of TLR4 896 A> G and 1196 C> T in susceptibility to infections: A review and meta-analysis of genetic association studies. PLoS ONE. 2013;8:e81047. doi: 10.1371/journal.pone.0081047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rossi UA, et al. A haplotype at intron 8 of PTPRT gene is associated with resistance to Brucella infection in Argentinian creole goats. Vet. Microbiol. 2017;207:133–137. doi: 10.1016/j.vetmic.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 70.Lachance J, Tishkoff SA. SNP ascertainment bias in population genetic analyses: Why it is important, and how to correct it. BioEssays. 2013;35:780–786. doi: 10.1002/bies.201300014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Albrechtsen A, Nielsen FC, Nielsen R. Ascertainment biases in SNP chips affect measures of population divergence. Mol. Biol. Evol. 2010;27:2534–2547. doi: 10.1093/molbev/msq148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Waples RS. High-grading bias: Subtle problems with assessing power of selected subsets of loci for population assignment. Mol. Ecol. 2010;19:2599–2601. doi: 10.1111/j.1365-294X.2010.04675.x. [DOI] [PubMed] [Google Scholar]
- 73.Elbers JP, Clostio RW, Taylor SS. Population genetic inferences using immune gene SNPs mirror patterns inferred by microsatellites. Mol. Ecol. Resour. 2017;17:481–491. doi: 10.1111/1755-0998.12591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Woolhouse ME, Webster JP, Domingo E, Charlesworth B, Levin BR. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat. Genet. 2002;32:569–577. doi: 10.1038/ng1202-569. [DOI] [PubMed] [Google Scholar]
- 75.Ferrer-Admetlla A, et al. Balancing selection is the main force shaping the evolution of innate immunity genes. J. Immunol. 2008;181:1315–1322. doi: 10.4049/jimmunol.181.2.1315. [DOI] [PubMed] [Google Scholar]
- 76.Wu D-D, Zhang Y-P. Different level of population differentiation among human genes. BMC Evol. Biol. 2011;11:1–7. doi: 10.1186/1471-2148-11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tschirren B, Andersson M, Scherman K, Westerdahl H, Råberg L. Contrasting patterns of diversity and population differentiation at the innate immunity gene toll-like receptor 2 (TLR2) in two sympatric rodent species. Evolution. 2012;66:720–731. doi: 10.1111/j.1558-5646.2011.01473.x. [DOI] [PubMed] [Google Scholar]
- 78.Helyar SJ, et al. Application of SNPs for population genetics of nonmodel organisms: New opportunities and challenges. Mol. Ecol. Resour. 2011;11:123–136. doi: 10.1111/j.1755-0998.2010.02943.x. [DOI] [PubMed] [Google Scholar]
- 79.Cherednichenko A, et al. Prevalence of gene polymorphisms associated with immune disorders in populations of Northern Eurasia. Mol. Biol. 2015;49:881–889. doi: 10.1134/S0026893315060047. [DOI] [PubMed] [Google Scholar]
- 80.Chávez-Galarza J, et al. Revisiting the I berian honey bee (Apis mellifera iberiensis) contact zone: Maternal and genome-wide nuclear variations provide support for secondary contact from historical refugia. Mol. Ecol. 2015;24:2973–2992. doi: 10.1111/mec.13223. [DOI] [PubMed] [Google Scholar]
- 81.Wragg D, et al. Whole-genome resequencing of honeybee drones to detect genomic selection in a population managed for royal jelly. Sci. Rep. 2016;6:1–13. doi: 10.1038/srep27168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Muñoz I, Pinto MA, De la Rúa P. Effects of queen importation on the genetic diversity of Macaronesian island honey bee populations (Apis mellifera Linneaus 1758) J. Apic. Res. 2014;53:296–302. doi: 10.3896/IBRA.1.53.2.11. [DOI] [Google Scholar]
- 83.Ferreira H, et al. Historical and contemporaneous human-mediated processes left a strong genetic signature on honey bee populations from the Macaronesian archipelago of the Azores. Apidologie. 2019;51:316–328. doi: 10.1007/s13592-019-00720-w. [DOI] [Google Scholar]
- 84.Soroker V, Yossi S, Chejanovsky N. Asian Beekeeping in the 21st Century. Springer; 2018. pp. 95–109. [Google Scholar]
- 85.Riveron JM, et al. Directionally selected cytochrome P450 alleles are driving the spread of pyrethroid resistance in the major malaria vector Anopheles funestus. Proc. Natl. Acad. Sci. USA. 2013;110:252–257. doi: 10.1073/pnas.1216705110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ast G. How did alternative splicing evolve? Nat. Rev. Genet. 2004;5:773–782. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- 87.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr. Protoc. Hum. Genet. 2009;60:2.12.11–12.12.18. doi: 10.1002/0471142905.hg0212s60. [DOI] [PubMed] [Google Scholar]
- 89.Chávez-Galarza J, et al. Signatures of selection in the Iberian honey bee (Apis mellifera iberiensis) revealed by a genome scan analysis of single nucleotide polymorphisms. Mol. Ecol. 2013;22:5890–5907. doi: 10.1111/mec.12537. [DOI] [PubMed] [Google Scholar]
- 90.R Script: Plot Allele Frequencies (Dryad, 2019).
- 91.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015;15:1179–1191. doi: 10.1111/1755-0998.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.