Abstract
Soil microbial communities regulate global biogeochemical cycles and respond rapidly to changing environmental conditions. However, understanding how soil microbial communities respond to climate change, and how this influences biogeochemical cycles, remains a major challenge. This is especially pertinent in alpine regions where climate change is taking place at double the rate of the global average, with large reductions in snow cover and earlier spring snowmelt expected as a consequence. Here, we show that spring snowmelt triggers an abrupt transition in the composition of soil microbial communities of alpine grassland that is closely linked to shifts in soil microbial functioning and biogeochemical pools and fluxes. Further, by experimentally manipulating snow cover we show that this abrupt seasonal transition in wide-ranging microbial and biogeochemical soil properties is advanced by earlier snowmelt. Preceding winter conditions did not change the processes that take place during snowmelt. Our findings emphasise the importance of seasonal dynamics for soil microbial communities and the biogeochemical cycles that they regulate. Moreover, our findings suggest that earlier spring snowmelt due to climate change will have far reaching consequences for microbial communities and nutrient cycling in these globally widespread alpine ecosystems.
Subject terms: Metagenomics, Climate-change ecology, Microbial ecology, Biogeochemistry, Soil microbiology
Introduction
By regulating global biogeochemical cycles, soil microbial communities control the availability of key plant nutrients, soil carbon (C) storage, and a host of other ecosystem functions [1]. However, mechanistic understanding of the linkages between soil microbial community composition, functioning and biogeochemical cycling is limited [2], and the effects of climate change on these linkages remains poorly understood [3]. This hinders the predictive power of global biogeochemical models, which currently tend to treat the soil microbial community as a ‘black box’ [4]. If future global biogeochemical cycles are to be predicted accurately, an improved mechanistic understanding of the links between microbial community composition and functioning, and biogeochemical cycling, in the context of a changing climate is urgently needed.
Uncertainty regarding climate change impacts on ecosystem functioning is particularly problematic in alpine ecosystems. Alpine regions occur at almost all latitudes, they store significant amounts of C, harbour much biodiversity, and provide many ecosystem services [5, 6]. However, they are disproportionately vulnerable to climate change. Winter snow cover is predicted to decrease by around 50% at 2500 m elevation by the end of the century under a high emissions scenario [7], with spring snowmelt occurring 50–60 days earlier [8]. Changes of this magnitude will likely have severe ecological impacts due to the importance of winter snow for alpine ecosystem functioning. Snow insulates soil and thereby enables microbial communities to remain active during winter [9, 10]. Reductions in winter snow cover will diminish this insulating effect, resulting in more frequent and severe freeze-thaw cycles [11], with uncertain consequences for microbial-mediated biogeochemical cycles.
Reductions in snow cover are associated with earlier spring snowmelt [8, 12], which is likely to have significant ecological impacts because alpine soil microbial communities and biogeochemical cycling show strong seasonality [13–15]. Past studies have shown that ‘winter’ microbial communities with high microbial biomass and abundance of fungi relative to bacteria, immobilise large amounts of C, nitrogen (N) and phosphorus (P), whereas taxonomically distinct ‘summer’ microbial communities have lower biomass, are more bacterially dominated, and immobilise less C, N and P [16–18]. Spring snowmelt triggers the winter to summer transition by causing sudden changes in osmotic potential due to thawing, and an end to relatively stable soil microclimatic conditions beneath the snow [19–21]. This is associated with microbial cell lysis, leading to large pulses in soil nutrient availability [20, 22]. However, it remains unknown how tightly coupled seasonal transitions in alpine soil microbial community composition, including the relative abundances of specific bacterial and fungal taxa, are to broader shifts in microbial functioning and biogeochemical cycles during spring snowmelt. It also remains unclear how climate change-induced reductions in winter snow cover and earlier spring snowmelt might influence seasonal transitions in soil microbial communities and the biogeochemical cycles they regulate. Indeed, an increased frequency and severity of freeze-thaw cycles in soil due to reduced snow cover during the winter months could impact microbial and biogeochemical cycles during snowmelt, regardless of its timing. Any or all of these winter and spring climate change impacts could potentially lead to cross-season legacy effects [23, 24]. Earlier spring snowmelt, in particular, could disrupt the temporal dynamics of plant and soil microbial resource demands [16, 25], with potential consequences for ecosystem C and N retention in these widespread and vulnerable mountain ecosystems [26].
Here, we address three research questions. First, are seasonal transitions in soil microbial community composition during spring snowmelt closely linked to shifts in soil microbial functioning and biogeochemical cycling in high alpine grassland? Second, do changes in snowmelt timing alter the timing of these transitions? Third, do preceding winter conditions affect soil microbial and biogeochemical processes occurring during the phenological phase of snowmelt, irrespective of changes in the timing of snowmelt?
Materials and methods
Experimental design
Our experiment was established on high alpine grassland at Hohe Mut (2650 m) near Obergurgl, Austria (lat. 46.84862, long. 11.02957). We used 15 5 m × 5 m plots that were randomly allocated to one of the three snow manipulation treatments (n = 5 replicates per treatment): snow removal, snow addition and an untreated control treatment, with treatments arranged in a completely randomised design across the site. Snow was manipulated four times between March and the end of April 2017. During each snow manipulation, snow was removed from the removal plots to a depth of <10 cm using a ‘snow blower’ machine or by hand with shovels, and was evenly added to addition plots. Snow in control plots was left undisturbed. We recorded the exact melt-out date, i.e. snowmelt timing, of each plot. Snow manipulations altered snowmelt timing (Supplementary Fig. S1) and had a clear impact on soil temperature and moisture, which were monitored throughout the experiment using HOBO data loggers (Onset, MA, USA). Plots were located directly adjacent to each other over a topographically homogenous area of ca. 375 m2. The soil type, a shallow podzol [27], and the vegetation, dominated by the graminoid Carex curvula All., were also homogenous across the site. Plots were fenced to prevent human and animal trampling. Mean annual air temperature recorded at the closest location (Innsbruck University weather station on Hohe Mut, ca. 1.1 km from our site and at 50 m lower elevation), was −0.3 °C in 2016 and 2017. Total annual precipitation recorded at the closest location (Central Institute for Meteorology and Geodynamics [ZAMG] Obergurgl weather station, ca. 2.2 km from our site and ca. 700 m lower elevation) was 814 mm in 2016 and 906 mm in 2017.
Soil sampling
All sampling was conducted at least 1 m from the edge of the plots to avoid edge effects. Soil was sampled at six time points throughout the experiment, which spanned the seasonal transition from late winter to early summer in 2017. Sampling took place in late winter (28th March; ‘winter 28/3’ [day/month format]), during snowmelt (1st and 8th of June; ‘snowmelt 1/6’ and “snowmelt 8/6”, respectively), spring (12th and 18th of June; ‘spring 12/8’ and “spring 18/6”, respectively), and early summer (8th July; ‘summer 8/7’). Soil cores were taken (Ø = 2 cm, depth = 7 cm, where possible, although some cores were shallower when soil was frozen) using a steel corer from five randomly chosen locations in each plot. Soil cores from the same plot were pooled and homogenised, any vegetation or litter was separated and discarded. Five sub-samples (~200 mg) were taken for molecular analyses, lysed in the field and then stored at −80 °C (see SI methods for details). Soil samples for enzyme and biogeochemical analyses were sieved (4 mm), stored at 4 °C for up to 2 weeks, and shipped to Manchester (UK) for further analysis.
PLFA analyses
Phospholipid fatty acid (PLFA) analyses were used to assess microbial biomass in total as well as to characterise the soil microbial community across different kingdoms, based on the methods of Bligh and Dyer [28] as described by Bardgett et al. [29] (see SI methods for details).
DNA extraction and profiling of bacterial and fungal communities
DNA was extracted using ZR soil microbe DNA kit (Zymo research, CA, USA) under manufacturer’s recommendations with a few amendments to account for sample preparation (see SI methods for details). Bacterial and fungal community structure was assessed using rarefied sequence abundance of the genetic regions encoding for 16S Small subunit ribosomal RNA (16S rRNA) and the internal transcribed spacer region 2 (ITS2)—targeting bacteria and fungi respectively. Amplicons were generated under a 2-step amplification approach (see SI methods), using Illumina TruSeq tagged primers based upon the primers 784F [30], V6R2 [31] and flTS73 [32], ITS4R4 [33], with the addition of a unique custom barcode combination corresponding to each sample [34]. PCR Products were normalised using Sequalprep normalisation plates (Invitrogen, CA, USA). Pooled amplicon libraries were vacuum concentrated and gel purified. Resultant libraries were quantified using a Qubit dsDNA HS Assay kit (Invitrogen) and the pool sequenced at a concentration of 5.4 pM with a 0.6 pM addition of Illumina generated PhiX control library. Sequencing was performed on an Illumina MiSeq platform using V3 chemistry (Illumina Inc., CA, USA).
Sequences were trimmed, quality filtered, de-replicated, and amplicon sequence variant (ASV) tables were generated using the DADA2 [35] pipeline in R V.3.0.17 [36]. Sequences were processed in R using DADA2 to quality filter, merge, denoise and assign taxonomies. 16S rRNA amplicon reads were trimmed to 250 and 220 bases, forward and reverse respectively. ITS2 amplicons reads were trimmed to 270 and 220 bases, forward and reverse respectively. Filtering settings were maximum number of Ns (maxN) = 0, maximum number of expected errors (maxEE) = (1, 1). The primer sequences were removed using trimLeft = c(20, 20). Sequences were dereplicated and the DADA2 core sequence variant inference algorithm applied. mergePairs was used to merge sequences and actual sequence variant (ASV) tables were constructed. Chimeric sequences were removed using removeBimeraDenovo default settings. ASVs were subject to taxonomic assignment using assignTaxonomy at default settings; training databases were GreenGenes v13.8 [37] and Unite v7.2 [38] for 16S rRNA and ITS, respectively.
After quality filtering a total of 4,857,571 bacterial (16S rRNA) and 2,398,955 fungal (ITS2) sequences were used in the analysis. To account for the effect of sequencing bias the resultant ASV tables were rarefied to an even depth of 10,083 (16S rRNA) and 5,349 (ITS2), within the ‘phyloseq’ R package [39].
Shotgun metagenomic sequencing
Major functional traits of the soil microbiome were investigated at three key sampling time points: (i) winter 28/3, (ii) snowmelt 1/6 and (iii) summer 8/7 on a random subset of three samples of each treatment (n = 27 samples total). Approx. 280 ng of extracted DNA per sample was sheared for 55 s using an E220 Focused-ultrasonicator (Covaris Inc., MA, USA) with the following settings: 175 W incident power, 5% duty factor and 200 cycles per burst. Shotgun metagenomic libraries were prepared using the NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs Inc., MA, USA), with the following modifications on the manufacturer’s protocol. NEBNext Adaptor for Illumina was diluted (1:10) and the Adaptor-ligated DNA samples were purified with size selection for 400‒500 bp insert size using AMPure XP Beads (Beckman Coulter Inc., CA, USA). The purified samples were amplified using 3.5 μl of i7 and i5 primers of the NEBNext Multiplex Oligos for Illumina (Dual Index Primers Set 1) kit (New England Biolabs) and 8 Denaturation/Annealing PCR cycles. The final products were purified by the Cleanup of PCR reaction protocol twice with different rations of template DNA:AMPure XP Beads (0.6 and 0.8, respectively) to completely remove the PCR primers. The size distribution and concentrations of DNA libraries were evaluated on a Fragment Analyzer Automated CE System (Advanced Analytical Technologies Inc., CA, USA) using the DNF-473 Standard Sensitivity NGS Fragment Analysis Kit (1–6000 bp). The libraries were diluted to 2 nM and sequenced on a HiSeq 2500 Sequencing System (2 × 250 bp Rapid Run mode, Illumina Inc.) using the HiSeq Rapid SBS v2 (500 cycles) and HiSeq PE Rapid Cluster v2 kits after equimolar pooling.
Sequenced metagenomic datasets were demultiplexed using the bcl2fastq v.2.20.0.422 software [40] with the no-lane-splitting option. Demultiplexed datasets were processed following the suggestions of Johan Sáenz et al. [41]. Forward and reverse reads with minimum alignment length of 30 bases were merged, adaptor sequences and low quality bases (<15) were trimmed from the ends of the reads and short reads (<100 bases) were discarded using AdapterRemoval v. 2.1.7 [42]. Reads identified as PhiX (>90% alignment coverage threshold) were also discarded using DeconSeq v.0.4.3 [43]. Fastq files were converted to fasta format using the read_fastq and write_fasta commands of the Biopieces framework (http://www.biopieces.org). After quality filtering a total of 7–20 million sequences were used in the analysis. All the remaining reads were taxonomically assigned against the non-redundant NCBI database (January 2017) using Kaiju v.1.4.4 [44]. The reads were additionally annotated to the genes related to the nitrogen cycle against the NCyc database (100% identity) [45] using Diamond v0.9.24.125, and genes of carbohydrate-active enzymes against the dbCAN-HMM V7 database using the hmmscan command of HMMER 3.1b2 (http://hmmer.org/). Several steps were run in parallel using the GNU Parallel programme [46]. Results were normalised by calculating relative (%) abundances.
Extracellular enzyme assays
To interrogate soil microbial community functioning, the potential activity of eight soil extracellular enzymes was measured (µmol prod. g−1 dry soil h−1). These included the lignocellulose degrading enzymes β-glucosidase (GLC), cellobiohydrolase (CBH), β-xylosidase (XYL), phenol oxidase (POX) and peroxidase (PER), alongside N-acetylglucosaminidase (NAG), phosphatase (PHO) and urease (URE). GLC, CBH, XYL, NAG, PHO, POX and PER were measured photometrically [47, 48], and URE activity was evaluated by ammonia production [49], see SI methods for full details. To calculate potential enzyme activity relative to microbial biomass, i.e. microbial biomass-specific enzyme activity (nmol prod. nmol−1 PLFA h−1), we divided potential enzyme activities by total PLFA.
Biogeochemical cycling and other soil properties
To measure plant available NH4+-N and NO3−-N, and dissolved organic nitrogen (DON), 5 g (fresh weight) of soil was extracted with 25 ml of 1 M KCl for NH4+-N and NO3--N, or 35 ml ultrapure (Milli-Q®) H2O for DON, shaken for 1 h (or 10 min for DON), and then extracts were analysed on a Seal AA3 Segmented Flow Multi-chemistry analyser (Mequon, WI, USA). To calculate DON we measured total dissolved N and total dissolved inorganic N (NH4+-N and NO3−-N) simultaneously in each extract, and then subtracted total dissolved inorganic N from total dissolved N [50]. Net NH4+-N and NO3−-N mineralisation rate was measured as the release of NH4+-N and NO3−-N after incubation of soil samples (5 g) for 14 days at 25 °C [51]. Dissolved (water-extractable) organic carbon (DOC) was determined by extracting 5 g (fresh weight) of soil in 35 ml ultrapure (Milli-Q®) water and analysed using a 5000A TOC analyser (Shimadzu, Japan). All extracts were filtered through Whatman no. 42 filter papers. Soil pH (1:2.5, soil:water) was determined using a pH metre (Mettler Toledo, UK), and soil water content was determined gravimetrically. Soil total C and N content was measured on oven-dried soil (105 °C for 48 h) using an Elementar Vario EL elemental analyser (Hanau, Germany). Soil respiration was measured in the field using a soil respiration chamber with built-in fan (SRC; PP Systems, MA, USA) attached to an infra-red gas analyser (IRGA; EGM4, PP Systems, MA, USA). Soil respiration was only measured during the snow-free period, see SI methods for details.
Statistical analysis
6All statistical analyses were performed in R v.3.6.0 [36]. Linear mixed effects models were used to test our three research questions, with sampling time point, treatment, and their interaction as fixed effects, and plot as a random effect due to repeated measures being taken on the same plots, using the R package ‘nlme’ (see SI methods for details). To further investigate the effect of snowmelt timing on seasonal transitions, we also used linear regression to assess how soil microbial and biogeochemical parameters changed during the snowmelt (1/6) sampling time point, which corresponded with the majority of snowmelt across all plots and maximum treatment effects (see SI methods for details).
Results
Seasonal dynamics of soil microbial communities and biogeochemical cycling
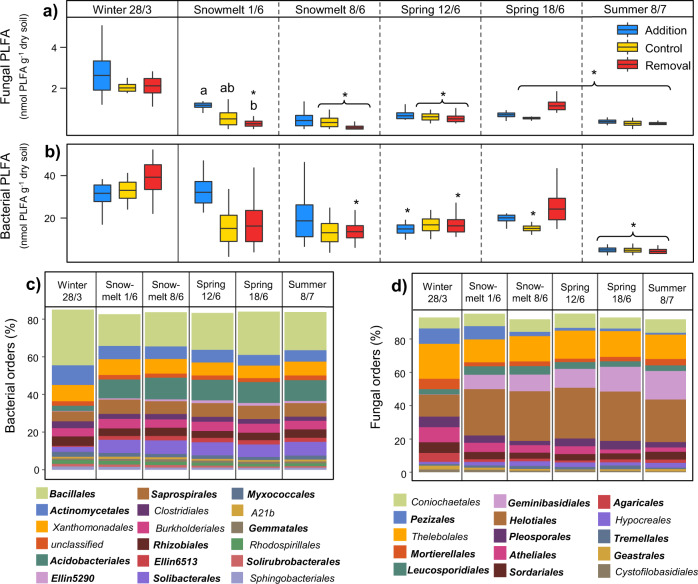
Our data revealed a marked winter to summer transition in the abundance and composition of soil bacterial and fungal communities, as well as the relative abundances of genes coding for enzymes catalysing key processes in C and N cycling and potential enzyme activities. Further, these changes in soil microbial community composition and functioning were accompanied by extensive changes in biogeochemical pools and fluxes. The biomass of the active soil microbial community, measured using total PLFA abundance, decreased by 88% from late winter to summer, with fungi and bacteria both showing similar declines (Fig. 1a and b, Supplementary Fig. S2 and Supplementary Table S1). Bacteria were more abundant than fungi throughout the sampling period, especially during snowmelt (ratio of fungal: bacterial PLFA markers; fig. S2), and Gram negative bacteria dominated over Gram positive bacteria, particularly in winter (ratio of Gram+: Gram- PLFA markers; Supplementary Fig. S2). The composition of soil microbiomes, assessed using molecular barcoding approaches, indicated marked differences between seasons for bacterial and fungal orders (Fig. 1c, d, Supplementary Fig. S2 and Supplementary Table S1). Notably, Acidobacteriales increased in relative abundance by 301% from winter to summer and became the second most abundant order. In contrast, Actinomycetales, the second most abundant order in winter, decreased in relative abundance by 42% in summer. The relative abundances of the fungal orders Pleosporales, Atheliales and Pezizales decreased by 50%, 71% and 88%, respectively from winter to summer, whereas Geminibasidiales increased by >8000%. The largely root-associated fungal order Helotiales increased in relative abundance by 93% from late winter to summer, and became the dominant fungal order.
Fig. 1. Seasonal transitions and the effect of snow manipulation treatments on soil microbial community composition.
PLFA marker abundance for a fungi and b bacteria; only relevant significant differences (p < 0.05) are shown; which demonstrate (1) how timing of the seasonal transition differs between treatments (i.e., significant differences between winter treatment means and means of the same treatment in other sampling time points) indicated with *; and (2) any significant differences between treatment means within individual time points (lowercase letters). Mean relative abundance of c bacterial and d fungal orders, with orders that differed significantly across sampling time points in bold. Boxplots show mean, ±SE and range.
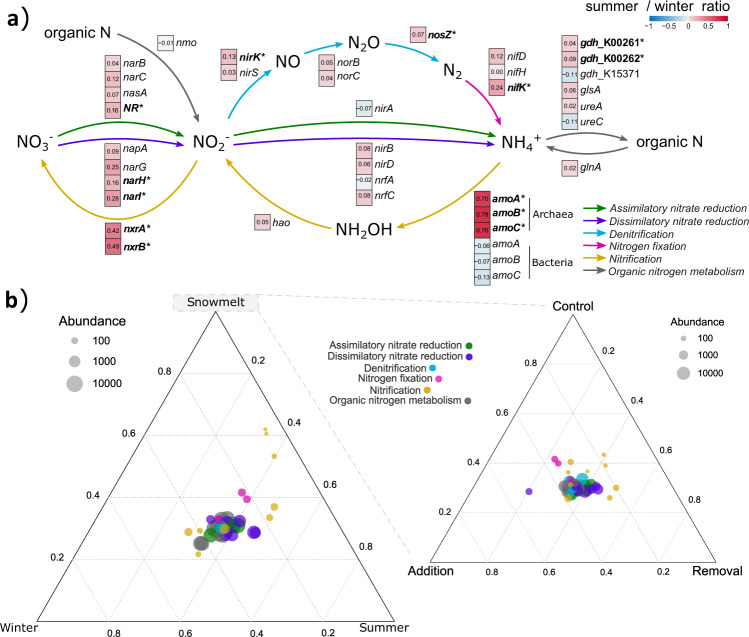
The seasonal transition in soil microbial community composition was accompanied by a marked shift in potential microbial functioning. Relative abundances of various genes coding for enzymes involved in key N-cycling pathways increased significantly in summer (Fig. 2a). Large increases were detected for nitrification genes, including the ammonia-oxidising archaeal (AOA) genes amoA, amoB and amoC, and in the nitrite-oxidising genes nxrA and nxrB (Fig. 2a). We also detected smaller, but significant increases in the relative abundance of nitrogen fixation (nifK), denitrification (nirK and nosZ), nitrate reduction (NR, narH and narI) and organic N synthesis (gdh_K00261 and gdh_k00262) genes. Similarly, the relative abundance of nine subfamilies of modules of cellobiohydrolases (CBH), β-glycosidases (GLC), and β-xylosidases (XYL), which are important for C cycling, increased in summer (Supplementary Fig. S3), whereas the relative abundances of seven GH43 subfamilies harboured only by XYL decreased in summer (Supplementary Fig. S3).
Fig. 2. Reconstruction of the complete nitrogen cycle based on gene relative abundances.
a Relative potential for each pathway presented as the log-ratio of summer (8/7): winter (28/3), with zero showing no change (white boxes), positive numbers (red boxes) showing higher potential in summer and negative numbers (blue boxes) showing higher potential in winter. Asterisks and bold names indicate statistically significant changes in gene relative abundances. b Distribution and abundance of the nitrogen cycle genes during different seasons (left) and in treatments during snowmelt (1/6) (right). Different pathways are depicted with distinct colours and circle size indicates relative abundances of genes.
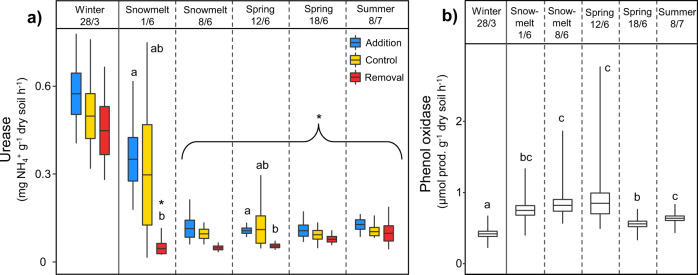
Microbial biomass-specific enzyme activity increased by 118–274% in summer for most measured enzymatic activities involved in C-cycling, along with phosphatase (PHO) activity (Supplementary Table S2). However, their potential activity decreased in summer relative to winter (Supplementary Fig. S4). Similar decreases were detected in potential urease (URE) and N-acetylglucosaminidase (NAG) activity (Fig. 3a, Supplementary Fig. S4). In contrast, potential activities of phenol oxidase (POX) and peroxidase (PER) increased in summer relative to winter by over 50% (Fig. 3b, Supplementary Table S2), while their microbial biomass-specific activity increased by >1000% (Supplementary Table S2). Soil respiration, an integrated measure of soil activity in situ, was highest directly after snowmelt, but decreased in summer (Supplementary Fig. S4).
Fig. 3. Seasonal transitions and the effect of snow manipulation treatments on soil extracellular enzyme activities.
Potential enzymatic activity of a urease and b phenol oxidase. For a only relevant significant differences (p < 0.05) are shown; which demonstrate (1) how timing of the seasonal transition differs between treatments (i.e., differences between winter treatment means and means of the same treatment in other sampling time points), indicated with *; and (2) any significant differences between treatment means within individual sampling time points (lowercase letters). For b significant differences (p < 0.05) are indicated with lowercase letters. Boxplots show mean, ±SE and range.
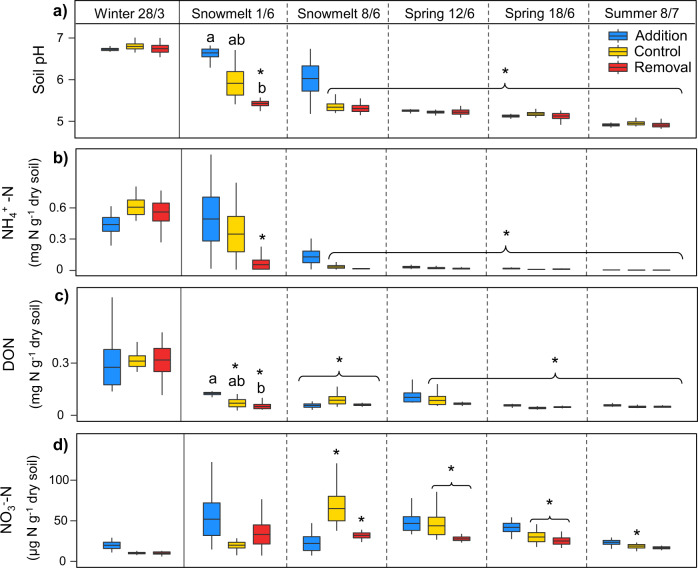
Marked transitions in abiotic soil properties accompanied observed shifts in soil microbial community composition and functioning. We detected large declines in soil pH (from 6.8 to 4.9), available NH4+ (−99%), dissolved organic nitrogen (DON, −83%), dissolved total organic carbon (DOC, −92%) and gravimetric soil water content (−44%) between late winter and summer (Fig. 4a–c, Supplementary Fig. S5, Supplementary Table S3). In contrast, available NO3- and the ratio of DON to available inorganic N increased by 46% and 329%, respectively in summer compared to late winter (Fig. 4d, Supplementary Fig. S5, Supplementary Table S3). Furthermore, during lab incubation of soil samples at 25 °C for 14 days, rates of ammonification and nitrification changed from −22 (±4; mean ± SE) and 60 (±7) µg N g−1 dry soil day−1, respectively in winter, to 1.7 (±0.2) and −0.4 (±0.1) µg N g−1 dry soil day−1 in summer (Supplementary Table S3).
Fig. 4. Seasonal transitions and the effect of snow manipulation treatments on soil biogeochemistry and pH.
a Soil pH, b NH4+-N concentration, c dissolved organic nitrogen (DON), d NO3--N concentration. Only relevant significant differences (p < 0.05) are shown; which demonstrate (1) how timing of the seasonal transition differs between treatments (i.e., differences between winter treatment means and means of the same treatment in other sampling time points) indicated with *; and (2) any significant differences between treatment means within individual time points, indicated with lowercase letters. Boxplots show mean, ±SE and range.
Effects of snowmelt timing and preceding winter conditions
The snow manipulation treatments significantly affected the timing of transitions in soil microbial community composition, functioning and biogeochemical cycling. The snow removal treatment brought snowmelt timing forward by 3 and 10 days on average relative to the control and snow addition treatments, respectively (Supplementary Fig. S1). Compared to the control treatment, snow removal led to earlier declines in microbial biomass (Fig. 1a and b, Supplementary Table S4) and shifts in community composition (Supplementary Fig. S2 and Supplementary Table S4), which were associated with earlier decreases in potential activities of URE, NAG, GLC, CBH and XYL (Fig. 3a, b, Supplementary Fig. S4, and Supplementary Table S4), soil pH and available NH4+ (Fig. 4a and b, Supplementary Table S4). Snow removal also consistently decreased microbial biomass-specific urease activity, and consistently increased DON: available inorganic N ratios, compared to the snow addition treatment (Supplementary Fig. S5, Supplementary Table S4). The relative abundance of the analysed genes was not as strongly affected by the snow manipulation treatments, although some responses to snow manipulation, albeit not significant, were detected. Genes involved in C and N cycling that showed significantly higher relative abundances in summer (e.g., amoA, amoB and amoC; Fig. 2a), also tended to show higher abundance in the snow removal treatment during snowmelt (1/6) (Fig. 2b), whereas genes that showed higher relative abundances in winter (e.g., subfamilies harboured only by β-xylosidases) tended to show higher abundances in the snow addition treatment during snowmelt (1/6) (Supplementary Fig. S3).
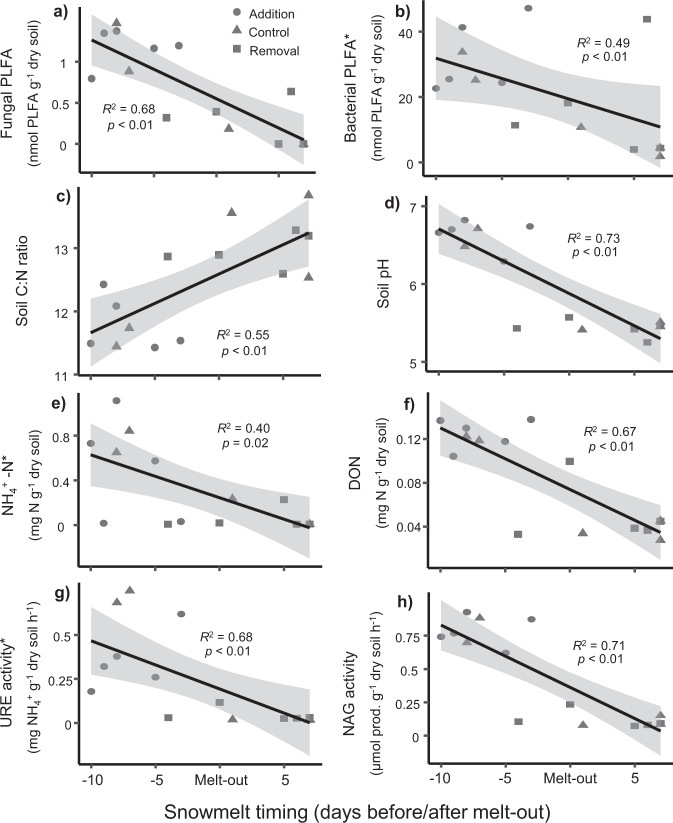
During the second sampling time point (snowmelt 1/6), which corresponded with the majority of snowmelt across our site and the maximum differences between treatments, snowmelt timing significantly (p < 0.05) explained 40–85% of the variation (based on R2 values) in 22 measured soil parameters. These included: total, fungal and bacterial biomass; the abundance of fungi relative to bacteria (i.e., F:B ratio); the relative abundances of the bacterial orders Ellin6513 and Ellin5290 and fungal orders Pezizales and Geminibasidiales; the potential activity of six enzymes (URE, NAG, PHO, CBH, XYL and GLC); the microbial biomass-specific activity of POX and PER; and a range of soil abiotic properties, including pH, DOC, available NH4+, DON, CN ratio and gravimetric water content (Fig. 5 and Supplementary Table S5). By this time point (snowmelt 1/6), 80% of the snow removal plots had melted completely, whereas none of the snow addition plots had.
Fig. 5. The effect of snowmelt timing on seasonal transition in soil microbial community composition, functioning and biogeochemical cycling during snowmelt (1/6).
PLFA marker abundance for a fungi and b bacteria, c soil C:N ratio, d pH, e NH4+-N concentration, f dissolved organic nitrogen (DON), g potential urease (URE) activity, and h potential N-acetylglucosaminidase (NAG) activity. Snowmelt timing refers to the time (days) when each plot was sampled in relation to when it melted-out (i.e., day zero). *log10 transformed to meet model assumptions (untransformed data shown). See Supplementary Table S5 for full list of variables. Shaded area represents 95% confidence interval.
Preceding winter conditions did not affect soil microbial or biogeochemical processes during the phenological phase of snowmelt, irrespective of snowmelt timing. There were no significant differences (p < 0.05) between the removal treatment on 01/06, the control treatment on 08/06, and the addition treatment on the 12/06, for any of the parameters that we measured (Figs. 1–4 and Supplementary Figs. S1–S4).
Discussion
Our data show that spring snowmelt in high alpine grassland triggers an abrupt transition in soil microbial community composition, which is accompanied by simultaneous shifts in a wide array of soil microbial functions, and biogeochemical pools and fluxes. Our findings also demonstrate that reduced snow cover, which is expected under climate change in alpine regions [7], advances this seasonal transition. The relatively modest (max. 10 day) advance in snowmelt timing observed in our study was ecologically significant, yet more extreme advances in snowmelt timing are forecast for the end of the century. Our findings suggest that these future advances in snowmelt timing will have far-reaching implications for annual C and N fluxes, plant community composition, and ecosystem productivity, in globally widespread alpine grasslands.
Our study advances current understanding in microbial ecology and biogeochemistry by demonstrating that temporal transitions in soil microbial community composition, under field conditions, are explicitly linked to shifts in soil microbial functioning and biogeochemical cycling. High microbial biomass during winter was associated with high potential soil enzyme activities. This shows that key microbial-mediated biogeochemical processes continue to operate under snow in high alpine grasslands, as in other ecosystems [9, 17]. In particular, our findings indicate that winter soil microbial communities contribute to breaking down organic compounds, such as xylan, cellulose and chitin [52, 53], which constitute much of the dead plant, invertebrate and fungal residues in winter soils [17]. Our data also reveal that the microbial taxa that dominate soil microbial communities in winter are associated with these microbial functions. Specifically, the fungal order Thelebolales, and bacterial orders Actinomycetales and Bacilliales, are all closely associated with cellulose degradation [54–56]. Cellulose fibres are hard to access, but the hyphal growth of fungi and cellulolytic actinomycetes can penetrate pores in the plant cell wall, and thereby bring the cellulases into close contact with the cellulose polymers [55]. While potential enzyme activities were high in winter, microbial biomass-specific enzyme activities increased in summer. This indicates that high microbial biomass likely explains high potential enzyme activities in winter, but that microbes typically invested more in enzyme production, especially the lignin-degrading enzymes POX and PER, in summer. The pH of our buffer solution (5.0) corresponded closely with in-situ summer soil pH (mean = 4.9), but was lower than winter soil pH (mean = 6.8). Given that soil pH influences potential enzyme activities [57], the potential activities we measured in winter, and in the addition treatment during snowmelt, may not have been as accurate as those measured in summer, or in the removal treatment during snowmelt (Fig. 4a). Nevertheless, the stable soil temperature and moisture (fig. S1), high substrate availability, and high microbial biomass typically found under the snow in winter [13, 17], suggest that while the rate of in-situ soil enzyme activity is likely to be lower than that measured in the lab, total enzyme activity over the winter will still be high [17]. This would also explain the high concentrations of dissolved organic C and N, along with available NH4+, in winter soil.
The composition of soil microbial communities in summer was closely related to the key functions occurring in summer, and were typical for the more acidic (pH = 4.9), nutrient-poor summer soil conditions [58]. Specifically, Acidobacteriales and Solibacterales, which are both strongly associated with acidic soils and lignin degradation [56], became abundant members of bacterial communities. This likely explains the higher potential activity and microbial investment in lignin-degrading enzymes POX and PER in summer. Lignin degradation may indicate that microbes are resorting to mining N from more recalcitrant organic matter pools due to severely N-limited conditions [53, 59], which corresponds with the dramatic decreases in DON (−83%) and available NH4+ (−99%). We also detected higher relative abundances of genes involved in nitrification, particularly amoA, amoB and amoC of AOA. The increasing importance of AOA likely reflects the combination of lower soil pH in summer and a legacy of high rates of nitrification following snowmelt, fuelled by large amounts of NH4+ that accumulated during winter [60, 61]. High nitrification rates during snowmelt likely contributed to the substantial reductions in soil pH observed during snowmelt, given that H+ ions are produced during nitrification. The much lower microbial biomass and soil N availability observed in summer was largely due to rapid reductions in bacterial and fungal abundances, DON and available NH4+ that occurred during snowmelt. The N contained in these pools was likely either lost via nitrification and subsequent nitrate leaching in meltwater [62], as suggested by the high relative abundance of nitrifying AOA genes and peak in soil NO3- concentrations in spring, or taken up by alpine plants [63]. Alongside reductions in microbial biomass and soil N availability, snowmelt also triggered abrupt shifts in microbial composition (e.g., relative abundance of Geminibasidiales), potential enzyme activities involved in C and N cycling (e.g., URE, NAG, CBH, XYL and GLC), and soil pH. Our findings demonstrate the central role of spring snowmelt in controlling the temporal dynamics of soil microbial community composition, which are closely connected to changes in soil N cycling under field conditions in alpine grassland.
Our findings show that changes in the timing of spring snowmelt alter the timing of seasonal transitions in soil microbial community composition, functioning and biogeochemical cycling in alpine grassland. However, we found no evidence that preceding winter conditions, including increased frequency and severity of freeze-thaw cycles, influenced the microbial and biogeochemical processes occurring during snowmelt. Rather, the main mechanism that underpinned the ecological impact of reduced winter snow cover was the earlier transition in soil microbial communities and biogeochemical cycling due to earlier snowmelt. This advances our understanding of soil microbial responses to climate change because it demonstrates that seasonal transitions in soil microbial community composition, functioning and biogeochemical cycling will likely occur earlier with reduced snow cover, which is expected in alpine regions due to climate change [7]. As a consequence, the important ecosystem functions performed by winter soil microbial communities, such as the breakdown of complex litter residues [16, 17] and associated build-up of high soil DON and available NH4+, will likely be reduced in alpine regions under future climates. This could result in a potential mismatch between seasonal soil N fluxes and the onset of plant growth during spring [25], with implications for annual ecosystem C and N fluxes. The peak in available NO3− following snowmelt is important because alpine plant species often prefer NO3− as their N source [64], and the N made available to alpine plants during snowmelt likely fulfils much of their annual N demand [65]. However, the onset of plant growth in spring may not keep pace with earlier snowmelt [25]. Snow removal also led to a consistent increase in the DON: inorganic N ratio compared with snow addition. Reduced snow cover may therefore shift soil N availability towards organic N, possibly as a result of inorganic N losses during freeze-thaw cycles or nitrate leaching during snowmelt [62], which could affect plant community composition [66, 67].
Together, our findings demonstrate that abrupt transitions in soil microbial community composition during snowmelt are closely linked to shifts in a wide array of microbial functions and biogeochemical cycling. Further, we show through observation and experimental snow manipulation that this abrupt seasonal transition is advanced by earlier snowmelt, although the processes that take place during snowmelt were not altered by preceding winter conditions. Given that climate change is expected to lead to reduced snow cover and earlier snowmelt, our results suggest that the transition in soil microbial communities from winter to summer will occur earlier in the year. This could potentially lead to altered annual C and N fluxes in seasonally snow-covered ecosystems. While the differences in snowmelt timing due to our experimental treatments were ecologically significant, they were also modest (max. 10 days) compared to those predicted for the end of this century, with snowmelt expected to occur 50–130 days earlier in the European Alps [8]. Changes in snowmelt timing of this magnitude will likely have far reaching consequences for biogeochemical cycling, and plant growth, in these vulnerable, globally widespread ecosystems.
Data and materials availability
The raw shotgun metagenomic dataset has been deposited in the National Center for Biotechnology Information’s Sequence Read Archive under the accession number PRJNA634461. Molecular taxon raw sequencing reads were deposited at the European Nucleotide Archive, under accession number PRJEB38678. All other data, including processed OTU and ASV tables for molecular taxa, are deposited in the NERC Environmental Information Data Centre public repository 10.5285/255320dd-38ae-4a07-93bb-4f90439309f8.
Supplementary information
Acknowledgements
The research was funded by a grant from the UK Natural Environment Research Council (NERC) (NE/N009452/1) initiated and led by RDB. IC was supported by a Ramon Areces Foundation Research Fellowship and Biotechnology and Biological Sciences Research Council Discovery Fellowship (BB/S010661/1). This study was supported by the Alpine Research Center Obergurgl of the University of Innsbruck (AFO, Alpine Forschungsstätten). We would like to thank Veronika Brune, Daniel Bahn, Julia Wagner, Julian Maindok and Yannick Pütz for invaluable assistance in the field, along with the Obergurgl–Hochgurgl Ski resort and the agricultural community Rotmoos–Kippele–Alpe for land access and ski lift support. We also thank Susanne Kublik for sequencing the prepared metagenomic libraries. Three anonymous reviewers provided constructive feedback on the manuscript, which was much appreciated.
Author contributions
RDB initiated and gained funding for the study, with significant input from MB, RIG and MS. The experiment was designed by RDB, MB, HSKS, NS, RK, RIG and MS. AADB, HSKS, AM, WJP, LN, IC, TG, NS, RK and RDB collected and/or analysed resulting data, and all authors contributed to data interpretation. AADB conducted statistical analysis. AADB wrote the manuscript in close consultation with RDB, and all authors contributed substantially to revising the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Arthur A.D. Broadbent, Helen S.K. Snell
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-021-00922-0.
References
- 1.Bardgett RD, Van Der Putten WH. Belowground biodiversity and ecosystem functioning. Nature. 2014;515:505–11. doi: 10.1038/nature13855. [DOI] [PubMed] [Google Scholar]
- 2.Fierer N. Embracing the unknown: disentangling the complexities of the soil microbiome. Nat Rev Microbiol. 2017;15:579–90. doi: 10.1038/nrmicro.2017.87. [DOI] [PubMed] [Google Scholar]
- 3.De Vries FT, Shade A. Controls on soil microbial community stability under climate change. Front Microbiol. 2013;4:1–16. doi: 10.3389/fmicb.2013.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allison SD, Martiny JBH. Resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci. 2008;105:11512–9. doi: 10.1073/pnas.0801925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leifeld J, Zimmermann M, Fuhrer J, Conen F. Storage and turnover of carbon in grassland soils along an elevation gradient in the Swiss Alps. Glob Chang Biol. 2009;15:668–79. doi: 10.1111/j.1365-2486.2008.01782.x. [DOI] [Google Scholar]
- 6.Schirpke U, Leitinger G, Tasser E, Schermer M, Steinbacher M, Tappeiner U. Multiple ecosystem services of a changing Alpine landscape: past, present and future. Int J Biodivers Sci Ecosyst Serv Manag. 2013;9:123–35. doi: 10.1080/21513732.2012.751936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beniston M. Is snow in the Alps receding or disappearing? Wiley Interdiscip Rev Clim Chang. 2012;3:349–58. doi: 10.1002/wcc.179. [DOI] [Google Scholar]
- 8.Beniston M, Keller F, Koffi B, Goyette S. Estimates of snow accumulation and volume in the Swiss Alps under changing climatic conditions. Theor Appl Climatol. 2003;76:125–40. doi: 10.1007/s00704-003-0016-5. [DOI] [Google Scholar]
- 9.Monson RK, Burns SP, Williams MW, Delany AC, Weintraub M, Lipson DA. The contribution of beneath-snow soil respiration to total ecosystem respiration in a high-elevation, subalpine forest. Glob Biogeochem Cycles. 2006;20:1–13. doi: 10.1029/2005GB002684. [DOI] [Google Scholar]
- 10.Zhang Y, Wang S, Barr AG, Black TA. Impact of snow cover on soil temperature and its simulation in a boreal aspen forest. Cold Reg Sci Technol. 2008;52:355–70. doi: 10.1016/j.coldregions.2007.07.001. [DOI] [Google Scholar]
- 11.Campbell JL, Ollinger SV, Flerchinger GN, Wicklein H, Hayhoe K, Bailey AS. Past and projected future changes in snowpack and soil frost at the Hubbard Brook Experimental Forest, New Hampshire, USA. Hydrol Process. 2010;24:2465–80. [Google Scholar]
- 12.Pederson GT, Gray ST, Woodhouse CA, Betancourt JL, Fagre DB, Littell JS, et al. The unusual nature of recent snowpack declines in the North American Cordillera. Science. 2011;333:332–5. doi: 10.1126/science.1201570. [DOI] [PubMed] [Google Scholar]
- 13.Gavazov K, Ingrisch J, Hasibeder R, Mills RTE, Buttler A, Gleixner G, et al. Winter ecology of a subalpine grassland: effects of snow removal on soil respiration, microbial structure and function. Sci Total Environ. 2017;590–591:316–324. doi: 10.1016/j.scitotenv.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 14.Buckeridge KM, Banerjee S, Siciliano SD, Grogan P. The seasonal pattern of soil microbial community structure in mesic low arctic tundra. Soil Biol Biochem. 2013;65:338–47. doi: 10.1016/j.soilbio.2013.06.012. [DOI] [Google Scholar]
- 15.Puissant J, Cécillon L, Mills RTE, Robroek BJM, Gavazov K, De Danieli S, et al. Seasonal influence of climate manipulation on microbial community structure and function in mountain soils. Soil Biol Biochem. 2015;80:296–305. doi: 10.1016/j.soilbio.2014.10.013. [DOI] [Google Scholar]
- 16.Bardgett RD, Bowman WD, Kaufmann R, Schmidt SK. A temporal approach to linking aboveground and belowground ecology. Trends Ecol Evol. 2005;20:634–41. doi: 10.1016/j.tree.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt SK, Costello EK, Nemergut DR, Cleveland CC, Reed SC, Weintraub MN, et al. Biogeochemical consequences of rapid microbial turnover and seasonal succession in soil. Ecology. 2007;88:1379–85. doi: 10.1890/06-0164. [DOI] [PubMed] [Google Scholar]
- 18.Schadt CW, Martin AP, Lipson DA, Schmidt SK. Seasonal dynamics of previously unknown fungal lineages in Tundra soils. Science. 2003;301:1359–61. doi: 10.1126/science.1086940. [DOI] [PubMed] [Google Scholar]
- 19.Jefferies RL, Walker NA, Edwards KA, Dainty J. Is the decline of soil microbial biomass in late winter coupled to changes in the physical state of cold soils? Soil Biol Biochem. 2010;42:129–35. doi: 10.1016/j.soilbio.2009.10.008. [DOI] [Google Scholar]
- 20.Buckeridge KM, Grogan P. Deepened snow increases late thaw biogeochemical pulses in mesic low arctic tundra. Biogeochemistry. 2010;101:105–21. doi: 10.1007/s10533-010-9426-5. [DOI] [Google Scholar]
- 21.Schimel J, Balser TC, Wallenstein M. M. Ecology. 2007;88:1386–94. doi: 10.1890/06-0219. [DOI] [PubMed] [Google Scholar]
- 22.Buckeridge KM, Grogan P. Deepened snow alters soil microbial nutrient limitations in arctic birch hummock tundra. Appl Soil Ecol. 2008;39:210–22. doi: 10.1016/j.apsoil.2007.12.010. [DOI] [Google Scholar]
- 23.Väisänen M, Gavazov K, Krab EJ, Dorrepaal E. The legacy effects of winter climate on microbial functioning after snowmelt in a subarctic Tundra. Micro Ecol. 2019;77:186–90. doi: 10.1007/s00248-018-1213-1. [DOI] [PubMed] [Google Scholar]
- 24.Darrouzet-Nardi A, Steltzer H, Sullivan PF, Segal A, Koltz AM, Livensperger C, et al. Limited effects of early snowmelt on plants, decomposers, and soil nutrients in Arctic Tundra soils. Ecol Evol. 2019;9:1820–44. doi: 10.1002/ece3.4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ernakovich JG, Hopping KA, Berdanier AB, Simpson RT, Kachergis EJ, Steltzer H, et al. Predicted responses of arctic and alpine ecosystems to altered seasonality under climate change. Glob Chang Biol. 2014;20:3256–69. doi: 10.1111/gcb.12568. [DOI] [PubMed] [Google Scholar]
- 26.Li W, Wu J, Bai E, Jin C, Wang A, Yuan F, et al. Response of terrestrial carbon dynamics to snow cover change: a meta-analysis of experimental manipulation (II) Soil Biol Biochem. 2016;103:388–93. doi: 10.1016/j.soilbio.2016.09.017. [DOI] [Google Scholar]
- 27.Neuwinger I Bodenökologische. Untersuchungen im Gebiet Obergurgler Zirbenwald—Hohe Mut. In: Patzelt G (Hrsg.. (ed). MaB-Projekt Obergurgl. 1987. Universitätsverlag Wagner, Innsbruck, Austria, pp 173-90.
- 28.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/y59-099. [DOI] [PubMed] [Google Scholar]
- 29.Bardgett RD, Hobbs PJ, Frostegard A. Changes in soil fungal:bacterial biomass ratios following reductions in the intensity of management of an upland grassland. Biol Fertil Soils. 1996;22:261–4. doi: 10.1007/BF00382522. [DOI] [Google Scholar]
- 30.Andersson AF, Lindberg M, Jakobsson H, Bäckhed F, Nyrén P, Engstrand L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS ONE. 2008;3:e2836. doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arenz BE, Schlatter DC, Bradeen JM, Kinkel LL. Blocking primers reduce co-amplification of plant DNA when studying bacterial endophyte communities. J Microbiol Methods. 2015;117:1–3. doi: 10.1016/j.mimet.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 32.Ihrmark K, Bödeker ITM, Cruz-Martinez K, Friberg H, Kubartova A, Schenck J, et al. New primers to amplify the fungal ITS2 region—evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol Ecol. 2012;82:666–77. doi: 10.1111/j.1574-6941.2012.01437.x. [DOI] [PubMed] [Google Scholar]
- 33.White TJ, Bruns T, Lee S, Taylor J. PCR protocols. 1990. Academic Press.
- 34.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–20. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.R Core Team. R: a language and environment for statistical computing. 2019. R Foundation for Statistical Computing.
- 37.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–72. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kõljalg U, Larsson KH, Abarenkov K, Nilsson RH, Alexander IJ, Eberhardt U, et al. UNITE: A database providing web-based methods for the molecular identification of ectomycorrhizal fungi. N. Phytol. 2005;166:1063–8. doi: 10.1111/j.1469-8137.2005.01376.x. [DOI] [PubMed] [Google Scholar]
- 39.McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Illumina. bcl2fastq and bcl2fastq2 Conversion software. 2020. https://support.illumina.com/sequencing/sequencing
- 41.Sáenz JS, Marques TV, Barone RSC, Cyrino JEP, Kublik S, Nesme J, et al. Oral administration of antibiotics increased the potential mobility of bacterial resistance genes in the gut of the fish Piaractus mesopotamicus. Microbiome. 2019;7:1–14. doi: 10.1186/s40168-019-0632-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schubert M, Lindgreen S, Orlando L. AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res Notes. 2016;9:1–7. doi: 10.1186/s13104-016-1900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmieder R, Edwards R. Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PLoS ONE. 2011;6:e17288. doi: 10.1371/journal.pone.0017288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Menzel P, Ng KL, Krogh A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun. 2016;7:11257. doi: 10.1038/ncomms11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tu Q, Lin L, Cheng L, Deng Y, He Z. NCycDB: a curated integrative database for fast and accurate metagenomic profiling of nitrogen cycling genes. Bioinformatics. 2019;35:1040–8. doi: 10.1093/bioinformatics/bty741. [DOI] [PubMed] [Google Scholar]
- 46.First Y, Job P. GNU parallel: the command-line power tool | USENIX. 3: 42–47.
- 47.Jackson CR, Tyler HL, Millar JJ. Determination of microbial extracellular enzyme activity in waters, soils, and sediments using high throughput microplate assays. J Vis Exp. 2013;80:e50399. doi: 10.3791/50399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Long JR, Semchenko M, Pritchard WJ, Cordero I, Fry EL, Jackson BG, et al. Drought soil legacy overrides maternal effects on plant growth. Funct Ecol. 2019;33:1400–10. doi: 10.1111/1365-2435.13341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kandeler E, Gerber H. Short-term assay of soil urease activity using colorimetric determination of ammonium article in biology and fertility of soils. Biol Fertil Soils. 1988;6:68–72. doi: 10.1007/BF00257924. [DOI] [Google Scholar]
- 50.Jones DL, Willett VB. Experimental evaluation of methods to quantify dissolved organic nitrogen (DON) and dissolved organic carbon (DOC) in soil. Soil Biol Biochem. 2006;38:991–9. doi: 10.1016/j.soilbio.2005.08.012. [DOI] [Google Scholar]
- 51.Ross DJ. Influence of sieve mesh size on estimates of microbial carbon and nitrogen by fumigation-extraction procedures in soils under pasture. Soil Biol Biochem. 1992;24:343–50. doi: 10.1016/0038-0717(92)90194-3. [DOI] [Google Scholar]
- 52.De Boer W, Folman LB, Summerbell RC, Boddy L. Living in a fungal world: Impact of fungi on soil bacterial niche development. FEMS Microbiol Rev. 2005;29:795–811. doi: 10.1016/j.femsre.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 53.Moorhead DDL, Sinsabaugh RRL. A theoretical model of litter decay and microbial interaction. Ecol Monogr. 2006;76:151–74. doi: 10.1890/0012-9615(2006)076[0151:ATMOLD]2.0.CO;2. [DOI] [Google Scholar]
- 54.Zhou Y, Pope PB, Li S, Wen B, Tan F, Cheng S, et al. Omics-based interpretation of synergism in a soil-derived cellulose-degrading microbial community. Sci Rep. 2014;4:1–6. doi: 10.1038/srep05288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev. 2002;66:506–77. doi: 10.1128/MMBR.66.3.506-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhatnagar JM, Peay KG, Treseder KK. Litter chemistry influences decomposition through activity of specific microbial functional guilds. Ecol Monogr. 2018;88:429–44. doi: 10.1002/ecm.1303. [DOI] [Google Scholar]
- 57.Sinsabaugh RL, Lauber CL, Weintraub MN, Ahmed B, Allison SD, Crenshaw C, et al. Stoichiometry of soil enzyme activity at global scale. Ecol Lett. 2008;11:1252–64. doi: 10.1111/j.1461-0248.2008.01245.x. [DOI] [PubMed] [Google Scholar]
- 58.Fierer N, Lauber CL, Ramirez KS, Zaneveld J, Bradford MA, Knight R. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 2012;6:1007–17. doi: 10.1038/ismej.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Broadbent AAD, Orwin KH, Peltzer DA, Dickie IA, Mason NWH, Ostle NJ, et al. Invasive N-fixer impacts on litter decomposition driven by changes to soil properties not litter quality. Ecosystems. 2017;20:1–13. doi: 10.1007/s10021-016-0099-3. [DOI] [Google Scholar]
- 60.Prosser JI, Nicol GW. Archaeal and bacterial ammonia-oxidisers in soil: the quest for niche specialisation and differentiation. Trends Microbiol. 2012;20:523–31. doi: 10.1016/j.tim.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 61.Verhamme DT, Prosser JI, Nicol GW. Ammonia concentration determines differential growth of ammonia-oxidising archaea and bacteria in soil microcosms. ISME J. 2011;5:1067–71. doi: 10.1038/ismej.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brooks PD, Williams MW, Schmidt SK. Inorganic nitrogen and microbial biomass dynamics before and during spring snowmelt. Biogeochemistry. 1998;43:1–15. doi: 10.1023/A:1005947511910. [DOI] [Google Scholar]
- 63.Jaeger CH, Monson RK, Fisk MC, Schmidt SK. Seasonal partitioning of nitrogen by plants and soil microorganisms in an alpine ecosystem. Ecology. 1999;80:1883–91. doi: 10.1890/0012-9658(1999)080[1883:SPONBP]2.0.CO;2. [DOI] [Google Scholar]
- 64.Ashton IW, Miller AE, Bowman WD, Suding KN. Niche complementarity due to plasticity in resource use: plant partitioning of chemical N forms. Ecology. 2010;91:3252–60. doi: 10.1890/09-1849.1. [DOI] [PubMed] [Google Scholar]
- 65.Bilbrough CJ, Welker JM, Bowman WD. Early spring nitrogen uptake by snow-covered plants: a comparison of Arctic and alpine plant function under the snowpack. Arct, Antarct Alp Res. 2000;32:404–11. doi: 10.1080/15230430.2000.12003384. [DOI] [Google Scholar]
- 66.Michelsen A, Schmidt IK, Jonasson S, Quarmby C, Sleep D. Leaf 15N abundance of subarctic plants provides field evidence that ericoid, ectomycorrhizal and non-and arbuscular mycorrhizal species access different sources of soil nitrogen. Oecologia. 1996;105:53–63. doi: 10.1007/BF00328791. [DOI] [PubMed] [Google Scholar]
- 67.Wookey PA, Aerts R, Bardgett RD, Baptist F, Bråthen K, Cornelissen JHC, et al. Ecosystem feedbacks and cascade processes: understanding their role in the responses of Arctic and alpine ecosystems to environmental change. Glob Chang Biol. 2009;15:1153–72. doi: 10.1111/j.1365-2486.2008.01801.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.