Abstract
Introduction
Demodex and bacteria are both components of the ocular surface micro-ecology, constituting a complex interaction. This study aims to explore how ocular surface Demodex infestation (DI) affects ocular surface microbial communities and diversity.
Methods
We recruited 255 subjects, and examined the correlation between ocular surface mite infestation and clinical indicators such as age, blood glucose level, dry eye symptoms, and blood pressure. 16S rRNA sequencing was performed on the conjunctival swab samples of 14 patients with ocular DI (P group) and 17 healthy people (N group). For further analysis, the subjects were divided into four subgroups, i.e. N-NMGD (n = 11), N-MGD (n = 6), P-NMGD (n = 6), and P-MGD (n = 8), according to meibomian gland dysfunction (MGD) or no MGD (NMGD).
Results
There was no difference in the α-diversity of ocular surface microbial communities between the DI and healthy control groups. In linear discriminant analysis effect size (LEfSe), there were more Acinetobacter, Novosphingobium, and Anoxybacillus in the DI group and fewer Novosphingobium, Lactobacillus, and Candidatus Microthrix in the healthy control group. P-NMGD had more Thermaceae and fewer Pseudomonas than P-MGD. There were more Bacteroidetes in N-NMGD than in N-MGD. The α-diversity of P-NMGD was lower than that of N-NMGD (Shannon index, P = 0.027). At the same time, the α-diversity of N-MGD was lower than that of N-NMGD (Shannon, Simpson, and dominance index, P = 0.048). There was no significant difference in β-diversity or in the primary flora at the phylum and genus levels between groups and subgroups.
Conclusion
DI had no significant effect on the diversity of ocular surface microbial communities. DI primarily changed the dominant flora and relative abundance of ocular surface microbial communities. MGD may play an important role in this process.
Keywords: Demodex infestation, Meibomian gland dysfunction, Microbial communities, Ocular surface, 16S rRNA sequencing
Key Summary Points
| 1. We collected 255 subjects to analyze the relationship between ocular surface demodex infection and various clinical indicators. |
| 2. 16S rRNA sequencing of ocular surface microbial community was performed in 14 Demodex infected and 17 non infected individuals. We observed that Demodex infection has no significant effect on diversity of ocular surface microbial communities, but mainly changes the dominant flora and relative abundance of ocular surface microbial communities. |
| 3. After referring to whether the subjects have meibomian gland dysfunction, it seems that Demodex changes the bacterial ocular surface microbial community by affecting meibomian gland function. |
Digital Features
This article is published with digital features, including a summary slide, to facilitate understanding of the article. To view digital features for this article go to 10.6084/m9.figshare.14687535.
Introduction
Many Demodex species are found abundantly in nature, but only two species have been determined to inhabit the human body, namely, Demodex folliculorum and Demodex sebum [1, 2]. Demodex exist in the developed parts of sebaceous glands and hair follicles, such as those in the cheeks, forehead, chin, chest, back, and eyelids, and can cause acne, rosacea, blepharitis, pruritus of the external auditory canal, and other diseases [3–6]. Eye Demodex infestation (DI) is common in humans. The infestation rate of Demodex in the eyelashes of healthy people is generally between 17% and 72%, and it has been reported as high as 100% in people over 96 years of age [7]. However, according to the 18S rRNA sequencing results of one study, the detection rate of Demodex in dander samples scraped from the cheeks of adults over 18 years old was 100% [3]. Many factors may affect ocular surface mite infestation, such as age, diabetes mellitus (DM), dry eye disease (DED), and meibomian gland dysfunction (MGD) [8–11].
Ocular DI is also associated with MGD [5]. Approximately 20% of Europeans and 60% of Asians have MGD. A study from China showed that the positive rate of DI in MGD patients was 89.32%, which was significantly higher than that in the control group [4]. Our previous study also showed that the severity of DI was positively correlated with meibomian gland loss and could aggravate the symptoms of eye discomfort [12, 13].
Dry eye disease is a multifactorial ocular surface disease characterized by a steady-state imbalance of tear film accompanied by eye symptoms, in which the instability and hyperosmolarity of tear film, inflammation and damage to the ocular surface, and abnormal nerve sensation play important roles in the etiology of the disease [14]. Worldwide, between 5% and 34% of people suffer from dry eye disease, and the prevalence rate increases significantly with age. The subjective symptoms of dry eye disease usually include red eye, burning sensation, tingling, foreign body sensation, pruritus, and photophobia [15]. Additionally, Randon et al. showed that Demodex is associated with DED [16].
Many studies have suggested a relationship between Demodex and bacteria in the occurrence of diseases. Zhu et al. found that there was a correlation between Demodex and Propionibacterium acnes in blepharitis [17]. Demodex is an independent pathogenic factor of blepharitis and also a carrier of Bacillus oleronius. In most patients with severe blepharitis, Demodex infestation is often accompanied by B. oleronius infection [18]. In a study on rosacea, a relationship between the two microorganisms was also found [19].
To further study the effects of ocular surface mites on the ocular surface environment, we retrospectively analyzed 255 clinical patients and collected ocular surface bacterial samples from 31 patients for further 16S rRNA sequencing. Investigating the composition of the bacterial community, microbial diversity, and the factors that lead to these changes may help to elucidate the interaction between and possible mechanisms of Demodex and ocular surface microbial communities.
Methods
Participants
We conducted this cross-sectional study from June 2018 to October 2018 at the Zhujiang Hospital of the Southern Medical University in China. The study recruited 255 outpatients and inpatients from Zhujiang Hospital, Southern Medical University, Guangzhou, Guangdong, China. The exclusion criteria were as follows: recent acute complications and infestations such as diabetic ketoacidosis, history of eye surgery and trauma within 6 months, corneal contact lens wear history, elevated blood glucose caused by type 1 DM and other causes, dry eye caused by rheumatic immune disease and hyperthyroidism, pregnant or lactating women, and patients with mental illness. Ophthalmological interviews, questionnaires, and examinations were performed with all participants. Data from all participants included general information (age, sex, weight, height, and history of other diseases such as heart disease), routine blood tests, and hypertension. To facilitate the analysis of age, we divided age into several range groups: 0–9, 10–19, 20–29, 30–39, 40–49, 50–59, 60–69, 70–79, and 80–89. The outcome indices were the Ocular Surface Disease Index (OSDI) questionnaire, tear film break-up time, eyelid symptom score, total cholesterol, fasting blood glucose, fasting insulin, and HbA1c. Outcomes were cross-sectionally investigated with respect to factors associated with ocular mite infestation. Finally, 31 out of 255 subjects were selected for collection of samples of ocular surface flora and subsequent 16S rRNA sequencing. There were 14 patients with ocular surface Demodex infestation (group P, including 7 male and 7 female) and 17 healthy patients (group N, including 7 male and 10 female). The average age of group P was 52.43 ± 19.13 years, and the average age of group N was 42.18 ± 17.65 years. The average total OSDI score in group P was 15.51 ± 20.05, and that in group P was 10.41 ± 9.10. There were eight MGD subjects in group P and six in group N, accounting for 57.14% and 35.29%, respectively. At the same time, the subjects were divided into four subgroups according to the presence (N-MGD, P-MGD) or absence (N-NMGD, P-NMGD) of MGD.
Assessment of Ocular Surface Demodex Infestation
We removed two lashes from each eyelid from each subject with fine forceps under a slit-lamp microscope. The lashes from each subject’s eyelid were placed separately on a glass slide. Ocular surface mite detection and counting was completed by a professional technician who did not know the identity of the subjects. Saline or fluorescein-containing solution was dropped by a pipette to the edge of the coverslip before counting mites under a light microscope.
The positive diagnosis of ocular surface mite infestation is based on the 2018 expert consensus on the diagnosis and treatment of Demodex blepharitis in China diagnostic criteria: (1) Demodex mites in all phases are counted; (2) adult patients have a Demodex mite count of 3/3 eyelashes in any of the 4 eyelids; (3) less than the above criteria is suspected positive if combined with clinical manifestations; if necessary, other pathogenic microorganisms can be examined at the same time, such as bacteria and fungi. Therefore, the severity of ocular surface mite infestation is divided into 3 levels: negative, suspicious positive, and positive.
Assessment of Dry Eye Outcomes
Based on the American Academy of Ophthalmology and Dry Eye WorkShop (DEWS), subjects with at least one typical symptom plus one or more changes in the objective tests were diagnosed as having dry eye. Common symptoms of eye discomfort were evaluated, including gritty sensation, soreness, redness, itchiness, blurred vision (which improved with blinking), and excessive tearing. To analyze the severity of DED, we divided the OSDI scores into four levels: no obvious symptoms, OSDI score ≤ 12.0; mild, 12.00 < score ≤ 22.0; moderate, 22.0 < score ≤ 32.0; and severe, 32.0 < score ≤ 100. The main points of comparison were subjective complaints, conjunctival staining, objective findings on corneal staining with fluorescein, alterations in the meibomian glands, tear break-up time (BUT), results of the basal secretion test, and impression cytology of the conjunctival injection. Subjects also underwent objective clinical assessment for DED.
Sample Collection
For bacterial analysis, each participant underwent an ophthalmological examination at Zhujiang Hospital of the Southern Medical University, Guangzhou, Guangdong, China. Topical anesthesia was performed before sample collection. All operations were performed in a sterile room. We collected ocular specimens with a disposable aseptic dry cotton swab from the upper and lower palpebral, fornix conjunctiva, and caruncle of one randomly selected eye of each subject. At the same time, an aseptic dry cotton swab was instilled with topical anesthetic as a blank control. From June 2018 to September 2018, a total of 31 DNA samples were collected from participants (including 14 patients with DI and 17 healthy subjects). Prior to DNA extraction, the samples collected were stored in sterile tubes at −80 °C.
DNA Extraction
We extracted bacterial DNA from cotton swabs with a DNA extraction kit. The concentration and purity were measured with a NanoDrop One instrument (Thermo Fisher Scientific, MA, USA), following the manufacturer’s instructions. There was no bacterial DNA found in the swabs of blank controls. Twenty milliliters of elution buffer was added to each sample, after which they were immediately transferred to a freezer at −20 °C until polymerase chain reaction (PCR) analysis.
PCR Amplification and 16S rRNA Sequencing
Barcoded primers (12 bp) synthesized by Invitrogen (Carlsbad, CA, USA) were used for PCR analysis to amplify 16S rRNA V3-V4 fragments of bacteria. Processed DNA extracted from the conjunctiva of subjects was used as amplification template. PCRs contained 25 µl of CR reaction Taq (TaKaRa Bio, Dalian Co. Ltd., China), 3 µl of DNA (20 ng/µl) template, and 1 µl of each primer (10 mM) in a volume of 50 µl. The cycling steps of the PCR amplification were as follows: initialization at 94 °C for 30 s; followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 52 °C for 30 s, and extension at 72 °C for 30 s; finally, an extension at 72 °C for 10 min. The PCR products were electrophoresed in 1% agarose gel, and then sequenced with Illumina MiSeq (PE 300) in the MAGIGENE Genomics Institute. The PCR products were mixed at equal density according to GeneTools analysis software (version 4.03.05.0, Syngene).
Data Analyses
Sequence reads were preprocessed to remove the primers. Then we used QIIME (version 1.8.046.) for sequence reading. The following criteria were applied for QIIME quality trimming: (1) truncation of the sequence before three consecutive low-quality bases and re-evaluation for length, (2) no ambiguous base calls, and (3) a minimum sequence length of 100 bp after trimming. We performed rarefaction analysis of bacterial 16S rRNA sequences according to the operational taxonomic unit (OTU) table. The processing of results was based on R software (version 4.0.2). To show the microbial diversity of each sample, QIIME (V1.9.1) was used to analyze the sequence readings of samples. Alpha diversity reflects the species diversity in a single sample, including the richness and uniformity of the species composition [20]. The observed species, Simpson, dominance, Shannon, Chao1, and PD_whole_tree richness were performed to reflect the different degrees of richness, diversity, or uniformity of the ocular surface microbial community of the subjects. The results were processed in R software using the k-sample Fisher–Pitman permutation test. Beta diversity is the comparison of biodiversity between samples [20]. It can calculate the distance between two samples and obtain a distance matrix, which can be used for further diversity and visual statistical analysis. Weighted and unweighted UniFrac β-diversity indices and the Bray–Curtis distance matrix were calculated with QIIME to assess the differences in species complexity of all samples. The results were processed by QIIME and the ggplot2 package of R software, and principal coordinate analysis (PCoA) was performed to visualize multidimensional data. Linear discriminant analysis effect size (LEfSe) was used to analyze the significance of differences between the groups and to determine individual species in the groups. The linear discriminant analysis (LDA) score can be used to evaluate the dominant position of the species, with the threshold of LDA score > 3.
Statistical Analysis
The Statistical Package for the Social Sciences (SPSS) version 24.0 software (IBM Corp., Armonk, NY, USA) was used for data analysis. The descriptive statistics are presented as the mean ± SD for normally distributed continuous variables. Nonnumerical data are described as presence (yes) or absence (no). Jonckheere–Terpstra tests were used to compare the continuous variables of the groups; the chi-square test was used to compare the categorical variables of the groups. The Mann–Whitney U test was used to analyze the difference in relative abundance between groups. In LEfSe, the Kruskal–Wallis test was used to detect species with significant differences in relative abundance between different groups, and the Wilcoxon test was used to analyze differences between groups. The k-sample Fisher–Pitman permutation test was used to assess the differences in biodiversity between groups. P values < 0.05 were considered significant.
Ethics Compliance
The study was registered in the Chinese clinical trial registry (ChiCTR1800016357). The study protocol and one amendment to the protocol were reviewed and approved by the Zhujiang Hospital Human Experimental Committee. The study was performed in accordance with the Helsinki Declaration of 1964 and its later amendments, and written informed consent was provided by all participants before starting study treatment. All participants were informed that their information would be used for study and publication. All participants consented to participate in the research.
Results
Analysis of Related Factors Affecting Ocular Surface Mite Infestation
Basic Clinical Information of Subjects
After applying all entry criteria, 444 eyes from 255 participants (aged 18–84) were included in the analysis. As shown in Table 1, there was a significant correlation between age and mite infestation in subjects (P < 0.001). We also found that ocular surface DI was closely related to MGD (P = 0.002), DED (P = 0.013), and the OSDI score (P = 0.030). DM (P < 0.001) and duration of DM (P < 0.001) were also significantly correlated with mite infestation. In addition, the prevalence of mite infestation was higher in patients with hypertension (P = 0.016). However, no association was exhibited between sex and ocular surface mite infestation (P > 0.05) or between hyperlipidemia and ocular surface mite infestation (P > 0.05). Further analysis of hematological parameters showed that there was no association between ocular surface mite infestation and HbA1c, high-density lipoprotein (HDL), low-density lipoprotein (LDL), triglyceride (TG), or total cholesterol (TC).
Table 1.
Basic data of subjects and Demodex infection on ocular surface
| Characteristic | Negative | Suspicious positive | Positive | P value |
|---|---|---|---|---|
| Total n (%) | 192 (43.2%) | 148 (33.3%) | 104 (23.4%) | – |
| Age (years) | 49.84 ± 17.76 | 54.79 ± 4.13 | 62.12 ± 12.81 | < 0.001a |
| Sex (%) | ||||
| Male | 41.6% | 35.0% | 23.4% | 0.771b |
| Female | 44.5% | 32.0% | 23.5% | |
| MGD (%) | ||||
| Yes | 40.8% | 36.7% | 22.4% | 0.002b |
| No | 62.3% | 21.8% | 15.8% | |
| DM (%) | ||||
| Yes | 37.9% | 28.0% | 34.1% | < 0.001b |
| No | 49.1% | 37.5% | 13.4% | |
| DED (%) | ||||
| Yes | 35.5% | 38.1% | 26.3% | 0.013b |
| No | 50.2% | 29.7% | 20.1% | |
| Hypertension (%) | ||||
| Yes | 33.1% | 38.0% | 28.9% | 0.016b |
| No | 48.2% | 30.7% | 21.1% | |
| Hyperlipidemia (%) | ||||
| Yes | 41.9% | 32.4% | 25.7% | 0.803b |
| No | 44.4% | 32.9% | 22.7% | |
| DM duration (years) | 2.56 ± 5.51 | 3.10 ± 5.91 | 6.34 ± 6.68 | < 0.001a |
| HbA1C (%) | 8.22 ± 2.89 | 8.26 ± 2.33 | 8.34 ± 2.72 | 0.586a |
| OSDI score | 11.63 ± 11.23 | 14.88 ± 14.43 | 16.16 ± 16.65 | 0.030a |
| HDL (mg/dl) | 1.27 ± 0.43 | 1.18 ± 0.29 | 1.32 ± 0.35 | 0.338a |
| LDL(mg/dl) | 3.22 ± 0.96 | 2.86 ± 0.90 | 3.39 ± 1.24 | 0.681a |
| TG (mg/dl) | 1.89 ± 1.91 | 1.96 ± 1.21 | 1.66 ± 1.26 | 0.927a |
| TC (mg/dl) | 5.22 ± 2.27 | 4.90 ± 1.26 | 5.67 ± 2.59 | 0.173a |
aJonckheere–Terpstra test
bChi-square test
Difference in Relative Abundance of Ocular Surface Flora Between Patients with Ocular Surface Demodex infestation and Healthy Controls
We collected samples of the ocular surface flora from 31 subjects and performed 16S rRNA sequencing on the samples. Among the subjects, ocular surface infestation with Demodex was found in 14 patients (group P), while no infestation was found for 17 people, who were considered healthy (group N).
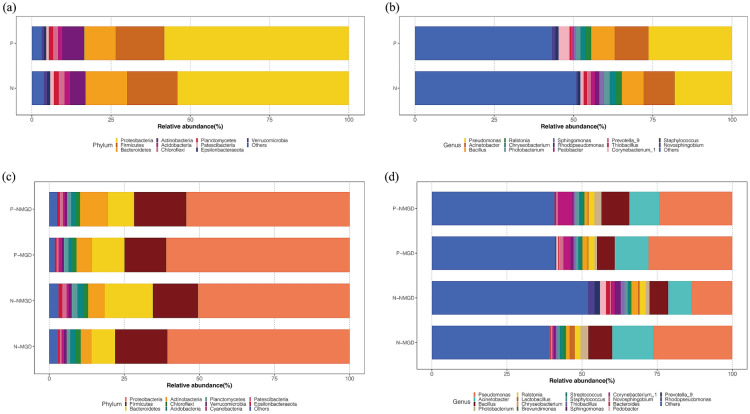
To explore the influence of DI on the composition of the ocular surface flora, we analyzed the relative abundance of the subjects’ ocular surface flora at the phylum and genus levels. Relative abundance refers to the ratio of the abundance of certain bacteria to the overall abundance of the flora, which is used to evaluate the richness of the bacteria in the flora. The composition of the ocular surface flora in the P and N groups was similar at the phylum level and was mainly composed of 10 phyla (Fig. 1a). The remaining phyla accounted for less than 4% of the total. Among the common phyla, Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria were the main bacteria in DI and healthy people, accounting for 90.4% and 87.9% of the total bacteria, respectively. At the genus level, the two groups were also similar. We mainly observed 14 genera on the ocular surface of the subjects (Fig. 1b). These genera accounted for 56.8% and 49.1% of the total bacteria, respectively. Pseudomonas, Acinetobacter, and Bacillus were the top three genera in the two groups, accounting for 44.4% and 34.8% of the total bacteria, respectively. A significant difference was found in the relative abundance of Novosphingobium (1.1% vs 0.4%) between patients with DI and healthy subjects (P < 0.012) (Table 2).
Fig. 1.
The relative abundance of the ocular surface flora between the groups was different at the phylum and genus level. The classification of bacteria is indicated with different colors. a Two groups at the phylum level. b Two groups at the genus level. c Four subgroups at the phylum level. d Four subgroups at the genus level. Patients with ocular surface Demodex infestation: group P, healthy controls: group N; MGD meibomian gland dysfunction, NMGD non-MGD
Table 2.
Relative abundance of ocular surface flora between patients with Demodex infestation on the ocular surface (group P) and healthy controls (group N)
| P group (%) | N group (%) | P value | |
|---|---|---|---|
| Phylum | |||
| Proteobacteria | 58.2 | 54.1 | 0.279 |
| Firmicutes | 15.4 | 15.9 | 0.681 |
| Bacteroidetes | 9.9 | 13.1 | 0.493 |
| Actinobacteria | 7.0 | 4.9 | 0.118 |
| Chloroflexi | 1.5 | 1.7 | 0.653 |
| Planctomycetes | 1.4 | 1.6 | 0.769 |
| Acidobacteria | 1.4 | 1.8 | 0.128 |
| Patescibacteria | 0.8 | 1.1 | 0.710 |
| Verrucomicrobia | 0.7 | 1.0 | 0.518 |
| Epsilonbacteraeota | 0.7 | 1.0 | 0.173 |
| Others | 3.1 | 3.8 | |
| Genus | |||
| Pseudomonas | 26.4 | 18.1 | 0.071 |
| Acinetobacter | 10.7 | 9.8 | 0.173 |
| Bacillus | 7.4 | 6.9 | 0.739 |
| Corynebacterium_1 | 3.5 | 1.0 | 0.084 |
| Ralstonia | 2.0 | 2.0 | 0.799 |
| Photobacterium | 1.5 | 1.7 | 0.739 |
| Chryseobacterium | 1.4 | 1.9 | 0.518 |
| Novosphingobium | 1.1 | 0.4 | 0.012* |
| Staphylococcus | 1.0 | 0.9 | 0.493 |
| Sphingomonas | 0.7 | 1.5 | 0.059 |
| Thiobacillus | 0.7 | 1.1 | 0.653 |
| Pedobacter | 0.4 | 1.3 | 0.493 |
| Prevotella_9 | 0.2 | 1.2 | 0.493 |
| Rhodopseudomonas | 0.01 | 1.4 | 0.953 |
| Others | 43.2 | 50.9 |
Statistics based on Mann–Whitney U test. Significant differences between the means of the different groups are indicated by (*) (P < 0.05)
We detected more Acinetobacter calcoaceticus, Novosphingobium, and Anoxybacillus (LDA score > 3) in the ocular surface samples of the P group (Fig. 2a). The eyes of group N contained more Sphingomonas, Lactobacillus, Candidatus Microthrix, Rhodobacterales, Rhodobacteraceae, Phycisphaerae, Spirochaetes, Spirochaetia, Spirochaetales, and Spirochaetaceae (LDA > 3) (Fig. 2a).
Fig. 2.
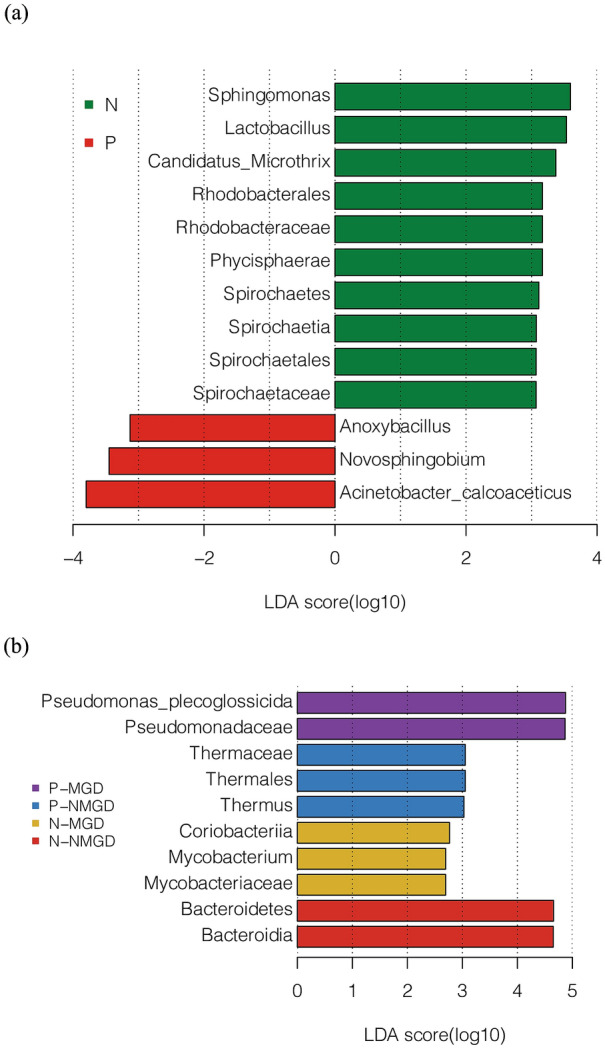
a Analysis of differences in the use of LEfSe between patients with ocular surface Demodex infestation (group P) and healthy controls (group N). b Analysis of differences in the use of LEfSe between patients with Demodex infestation on the ocular surface (group P) and healthy controls (group N, and between subjects with MGD and NMGD. The group and subgroup classifications are indicated with different colors
Difference in Relative Abundance of Ocular Surface Flora Between MGD and NMGD
To explore the influence of MGD on the composition of ocular surface flora, we conducted subgroup analysis of subjects in the ocular surface DI group (P group) and healthy control group (N group) based on whether they had MGD. The subjects were divided into four subgroups (N-MGD, N-NMGD, P-MGD, P-NMGD) according to the presence or absence of DI on the ocular surface and absence of MGD.
At the phylum level, the subjects’ ocular surface flora still consisted of four main phyla, Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria (Fig. 1c). In the healthy control group, Bacteroidetes (7.8% vs 16.0%) was lower in the N-MGD group than in the N-NMGD group (P < 0.05), reduced by more than 50% (Table 2). Among subjects without MGD, P-NMGD had higher Actinobacteria (9.3% vs 5.5%) than N-NMGD, while Bacteroidetes (8.7% vs 16.0%) had lower abundance (Table 2). At the genus level, Pseudomonas, Acinetobacter, and Bacillus were the main components found. Compared with N-MGD, Corynebacterium_1 (2.4% vs 5.4%) was lower in P-MGD, and the difference was statistically significant (P = 0.020).
As shown in Fig. 2b, the dominant phyla in the eyes of N-NMGD subjects were Bacteroidetes and Bacteroidia (LDA score > 4). The ocular surface of P-NMGD subjects contained more Thermaceae, Thermales, and Thermus (LDA score > 3) than the P-MGD group, while Pseudomonas plecoglossicida and Pseudomonadaceae were the dominant flora in the eyes of P-MGD subjects (LDA score > 4) (Fig. 2b).
Diversity Analysis of Ocular Surface Flora
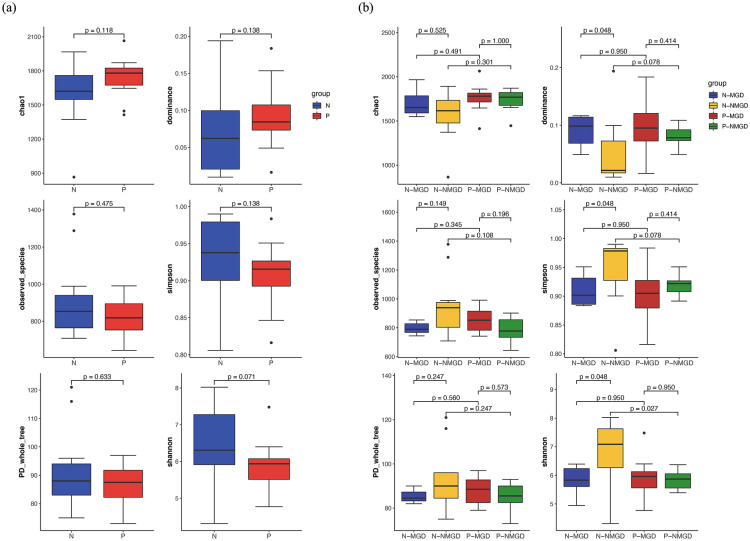
To compare the diversity of the ocular surface flora between the groups, we used six indicators (PD_whole_tree, Chao1, dominance, observed_species, Shannon, and Simpson) to analyze the α-diversity of the ocular surface flora. However, the difference in microbial α-diversity between the two groups was not statistically significant (P > 0.05) (Fig. 3a). The α-diversity was also compared between the four subgroups. The Shannon index of P-NMGD was lower than that of N-NMGD (5.84 vs 6.85), and the difference was statistically significant (P = 0.027) (Fig. 3b). There were significant differences in the dominance, Shannon, and Simpson indices between N-MGD and N-NMGD (P = 0.048), which indicated the lower α-diversity in eyes with MGD.
Fig. 3.
a The difference in the α-diversity index of ocular surface flora between patients with Demodex infestation on the ocular surface (group P) and healthy controls (group N). Groups are indicated by different colors. b The difference in the α-diversity index of ocular surface flora between patients with Demodex infestation on the ocular surface (group P) and healthy controls (group N) and between subjects with MGD and NMGD. Groups and subgroups ae indicated by different colors
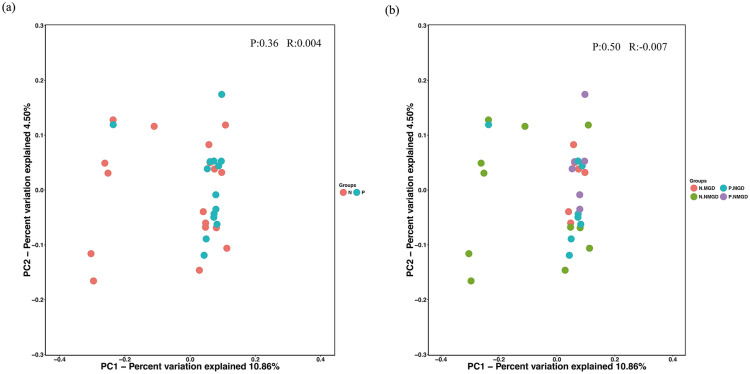
We also calculated the weighted and unweighted UniFrac β-diversity index using QIIME software. Principal coordinate analysis (PCoA) was performed to obtain the principal coordinates and visualize complex multidimensional data. There was no significant difference between the P group and the N group, indicating that DI did not have a significant effect on the diversity of the ocular surface flora (Fig. 4a). When examining β-diversity among subgroups, there was no significant difference among the four subgroups (Fig. 4b). However, except for N-NMGD, the three subgroups were relatively more concentrated in the PCoA (Fig. 4b), which suggests that MGD and DI may play a similar role in the process of changes in the diversity of the ocular surface flora.
Fig. 4.
a Weighted UniFrac was used to calculate β-diversity and PCoA in patients with ocular surface Demodex infestation (group P) and the healthy control group (group N). b Weighted UniFrac was used to calculate β-diversity and PCoA in MGD and NMGD subjects from patients with ocular surface Demodex infestation (group P) and the healthy control group (group N). Groups and subgroups are indicated with different colors
Discussion
Here we included 255 participants in our study. However, not all of the patients who participated in this study underwent all of the tests involved in the study. For this part of the lost data, we adopted an adjacent point interpolation method to account for it. Fortunately, we found strong associations between ocular surface mite infestation and age, MGD, DED, OSDI, DM, DM duration, and hypertension. However, no notable correlation was found between ocular surface mite infestation and sex or hyperlipidemia. Türk et al., on the other hand, reported that men had a higher rate of DI than women [21]. Keskin ’s study found an increase in the density of Demodex in patients with gestational DM [22]. Yamashita found that DI were more common in diabetic patients than in healthy volunteers [23]. Our research further confirms that DM is undoubtedly a risk factor for ocular surface mite infestation. DM has an adverse influence on the microvasculature in multiple organs, which aggravates ocular surface mite infestation through biochemical pathways involved in facilitating and abrogating microvascular injury and reducing local mucosal reaction, making it easier for ocular surface mite infestation to occur [24]. Through the analysis of 255 subjects, it was observed that DI was related to MGD. It has been shown that the greater the infestation frequency of Demodex, the more serious the damage to the microstructure of the meibomian gland [25]. DI is usually associated with MGD in symptomatic patients [16]. Ocular Demodex can destroy the lacrimal and meibomian glands, which causes a reduction in tears and surface oils, leading to dry eye disease. However, as tears have an immunomodulatory function on the ocular surface, DED results in reduced tear secretion and weakens the innate immunity of the ocular surface [26]. Overall, ocular surface DI and DED interact to jointly worsen ocular surface conditions.
We collected ocular surface bacterial samples from 31 subjects (including 14 patients with ocular surface DI and 17 healthy people) and sequenced 16S rRNA. However, there was no significant difference in α-diversity between the two groups (P > 0.05). PCoA showed that there was no significant difference between the patients with ocular surface DI and the healthy control group. This means that the ocular surface DI had no significant effect on the diversity of ocular surface flora.
Though the composition of ocular surface flora of both patients with DI and healthy controls was similar at the phylum and genus levels, there are still some differences. The results of LEfSe suggest that Acinetobacter calcoaceticus, Novosphingobium, and Anoxybacillus were dominant in eyes infested with Demodex, and could be classified as pathogens of DI. Novosphingobium, Lactobacillus, and Candidatus Microthrix can be regarded as the flora of a healthy eye.
According to Kugadas et al., coagulase-negative Staphylococcus spp. (CNS spp.), which include Staphylococcus epidermidis, are the most common bacteria on a healthy eye surface, while Propionibacterium spp. (P. acnes), Corynebacterium spp., S. aureus, Streptococcus spp., Micrococcus spp., Bacillus spp., and Lactobacillus spp. are less common [27]. Gram-negative bacteria were rarely detected on the surface of healthy eyes, while a large number of Pseudomonas and Acinetobacter were observed in healthy people and patients with ocular surface DI.
Research based on high-throughput sequencing has shown different results from the traditional culture methods. Graham et al. sequenced 57 healthy conjunctival samples based on 16S rRNA, and confirmed the presence of CNS spp., Bacillus spp., Rhodococcus spp., Corynebacterium spp., Propionibacterium spp., Klebsiella spp. and Ervinia spp. [28]. Zhou et al. reported that Corynebacterium, Streptococcus, Propionibacterium, Bacillus, Staphylococcus, and Ralsontia were the six most abundant genera in healthy conjunctiva [29]. The high relative abundance of Pseudomonas and Acinetobacter was not shown in the above studies. The high abundance of Bacillus in our study is consistent with the results reported in many other studies.
Mites can serve as carriers of bacteria to play a role in ocular lesions. Demodex can cause blepharitis by carrying bacteria (including Streptococcus and Staphylococcus) on its surface [30]. Staphylococcus epidermidis and Staphylococcus aureus are the main pathogens of conjunctivitis in children, and Demodex may be the carrier of these bacteria in the eyes [31]. In addition, some scholars postulate that Demodex plays a role in a balanced ocular ecology, acting as lash cleaners by grazing on bacteria and defending against other mite species, and as immune regulators and buffers [4, 32, 33]. Relevant studies have pointed out that after anti-mite treatment, eye symptoms become more serious, and that combination antibacterial treatment can ameliorate disease symptoms, which shows that mites and ocular surface bacteria can interact [1]. These reports, together with our study, indicate that there is a complex interaction between Demodex and bacteria in ocular surface diseases.
In addition to investigating the effect of DI on ocular surface flora, we also conducted subgroup analysis according to the presence or absence of MGD. There was no significant difference in α-diversity between group N and group P without considering the presence or absence of MGD. This may be because the microbial diversity of the ocular surface flora is more closely related to MGD than to DI. Even without DI, MGD can reduce the microbial diversity of ocular surface flora. Similarly, with NMGD, ocular infestation with Demodex can reduce the microbial diversity of the ocular surface. This indicates that both DI and MGD are associated with decreased microbial diversity of the ocular surface flora, and MGD may have a greater impact on microbial diversity than DI. Additionally, similar differences did not appear in the other two subgroups, which may mean that DI and MGD are not independent factors causing changes to the ocular surface flora.
Jiang et al. showed that the positive rate of bacterial culture in MGD patients was positively correlated with disease severity [34]. Zhang et al. found that the bacterial species in the conjunctival sac in MGD patients were more complex than those of healthy people [35]. However, according to results reported by Watters et al., the microbiota in the conjunctival sac was similar regardless of the presence or absence of MGD [36]. Another team used 16S rDNA sequencing to compare conjunctival sac bacteria in patients with and without MGD, and found that the distribution of ocular surface microorganisms in MGD patients was similar to that of healthy people, but at the phylum level there were more Proteus and Firmicutes and fewer Actinobacteria, which is consistent with our results. At the genus level, the relative abundance of Staphylococcus and Novosphingobium in MGD was higher than that in the healthy control group, while Corynebacterium was lower than that in the healthy control group [37]. Demodex can block the sebaceous duct, resulting in epithelial hyperplasia and hyperkeratosis [38]. The enzyme activity of the mite damages glands and epithelial cells in hair follicles, leading to inflammation [18]. Demodex folliculorum may be a carrier and common pathogenic factor of Bacillus oleronius in the occurrence of blepharitis [39].
The change in relative abundance and LEfSe results showed that MGD and DI led to an increase in Pseudomonas. Our previous studies also showed a correlation between MGD and Pseudomonas [40]. In addition, research showed that surfactant protein D (SP-D) played an important role in resisting Pseudomonas in a mouse model of dry eye disease [41]. Considering the close relationship between ocular surface DI and MGD and dry eye disease, this may provide a partial explanation for the increase in Pseudomonas in the eyes of subjects.
MGD may be an important link in the process of changes in ocular surface flora caused by DI of the ocular surface. Our retrospective study found a correlation between MGD and DI on the ocular surface. Demodex can cause microstructural changes in the meibomian glands, causing or aggravating MGD: the greater the DI, the more serious the structural damage [25]. Tear fluid has an immunoregulatory function on the ocular surface, and the tear film has a decontamination effect as the first line of defense against bacterial invasion. Meibomian gland function is important for maintaining the normal shape and composition of the tear film. Eye mites can damage the lacrimal and meibomian glands, weaken the ocular surface barrier, and increase the risk of bacterial infection in the eye [42]. In addition, Demodex may worsen the corneal epithelial barrier function by activating IL-17/MMP-9 signaling [11, 43].
This study has the following limitations. First, the sample was small, including only 14 patients with DI and 17 healthy subjects. Based on the size of the sample, we can only draw limited conclusions about the relationship between DI and the composition of ocular surface flora. Secondly, although there were differences in the relative abundance of several bacteria between eyes with and without DI, they were generally similar at the phylum and genus levels. The α-diversity between the two groups showed statistical differences in only a few indices, which may be related to our limited sample size.
Conclusion
Our research has shown that ocular surface mite infestation is significantly related to DED, MGD, age, hypertension and DM. Specifically, the infestation of Demodex on the ocular surface has no significant effect on the diversity of the ocular surface flora. It mainly changes the composition of the dominant flora, especially the Pseudomonas and Novosphingobium on the ocular surface. We suspect that DI leads to changes in the ocular surface flora, while MGD may play an important role in the process. However, whether DI and MGD are independent factors that change the ocular surface flora or play a role in different stages of the process of changing the ocular surface flora community still needs stronger evidence-based research.
Acknowledgements
The authors thank all participants for their contributions.
Funding
This study and the Rapid Service Fee was supported by National Natural Science Foundation of China (no: 81800804), Natural Science Foundation of Guangdong Province (no: 2017A030310108), Clinical Research Startup Program of Southern Medical University by High-level University Construction Funding of Guangdong Provincial Department of Education (no: LC2016YM017), and Innovation and Entrepreneurship for Undergraduates (no: 201812121171). The sponsor or funding organization had no role in the design or conduct of this research.
Authorship
Xiaotian Liang, Yingli Li, Ke Xiong, Shuze Chen, Zhenhao Li, Zhihan Zhang, Zhaoxia Xia, Guoguo Yi, Min Fu: All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Authorship Contributions
Min Fu, Xiaotian Liang and Ke Xiong contributed equally to this manuscript. Xiaotian Liang acquired and selected all the samples used in this study, compiled the database, wrote the manuscript, and provided critical revision. Ke Xiong and Zhenhao Li developed the study design, acquired and selected all the samples used in the study, and compiled the database. Shuze Chen compiled the database and performed the statistical analysis and data interpretation. Zhihan Zhang acquired and selected all the samples used in this study and wrote the manuscript. Yingli Yi conceived and developed the study design. Min Fu and Guoguo Yi conceived the study, developed the study design, acquired and selected all the samples used in this study, wrote the manuscript, and provided critical revision. Min Fu and Guoguo Yi agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Disclosures
All authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Compliance with Ethics Guidelines
The study was registered in the Chinese clinical trial registry (ChiCTR1800016357). The study protocol and one amendment to the protocol were reviewed and approved by the Zhujiang Hospital Human Experimental Committee. The study was performed in accordance with the Helsinki Declaration of 1964 and its later amendments, and written informed consent was provided by all participants before starting study treatment. All participants were informed that their information would be used for study and publication. All participants consented to participate in the research.
Data Availability
The raw data of sequencing results have been submitted to NCBI SRA (National Center for Biotechnology Information Sequence Read Archive). Raw data and project information are available via BioProject ID (PRJNA692647) and the following website: http://www.ncbi.nlm.nih.gov/bioproject/692647. The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.
Footnotes
Xiaotian Liang and Yingli Li contributed equally to the manuscript.
Contributor Information
Guoguo Yi, Email: yigg@mail.sysu.edu.cn.
Min Fu, Email: min_fu1212@163.com.
References
- 1.Nicholls SG, Oakley CL, Tan A, Vote BJ. Demodex species in human ocular disease: new clinicopathological aspects. Int Ophthalmol. 2017;37(1):303–312. doi: 10.1007/s10792-016-0249-9. [DOI] [PubMed] [Google Scholar]
- 2.Rufli T, Mumcuoglu Y. The hair follicle mites Demodex folliculorum and Demodex brevis: biology and medical importance. A review. Dermatologica. 1981;162(1):1–11. doi: 10.1159/000250228. [DOI] [PubMed] [Google Scholar]
- 3.Thoemmes MS, Fergus DJ, Urban J, Trautwein M, Dunn RR. Ubiquity and diversity of human-associated Demodex mites. PLoS One. 2014;9(8):e106265. doi: 10.1371/journal.pone.0106265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fromstein SR, Harthan JS, Patel J, Opitz DL. Demodex blepharitis: clinical perspectives. Clin Optom (Auckl) 2018;10:57–63. doi: 10.2147/OPTO.S142708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J, Sheha H, Tseng SC. Pathogenic role of Demodex mites in blepharitis. Curr Opin Allergy Clin Immunol. 2010;10(5):505–510. doi: 10.1097/ACI.0b013e32833df9f4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhandari V, Reddy JK. Blepharitis: always remember demodex. Middle East Afr J Ophthalmol. 2014;21(4):317–320. doi: 10.4103/0974-9233.142268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vargas-Arzola J, Reyes-Velasco L, Segura-Salvador A, Márquez-Navarro A, Díaz-Chiguer DL, Nogueda-Torres B. Prevalence of Demodex mites in eyelashes among people of Oaxaca, Mexico. Acta Microbiol Immunol Hung. 2012;59(2):257–262. doi: 10.1556/AMicr.59.2012.2.10. [DOI] [PubMed] [Google Scholar]
- 8.Gökçe C, Aycan-Kaya Ö, Yula E, et al. The effect of blood glucose regulation on the presence of opportunistic Demodex folliculorum mites in patients with type 2 diabetes mellitus. J Int Med Res. 2013;41(5):1752–1758. doi: 10.1177/0300060513494730. [DOI] [PubMed] [Google Scholar]
- 9.Biernat MM, Rusiecka-Ziółkowska J, Piątkowska E, Helemejko I, Biernat P, Gościniak G. Occurrence of Demodex species in patients with blepharitis and in healthy individuals: a 10-year observational study. Jpn J Ophthalmol. 2018;62(6):628–633. doi: 10.1007/s10384-018-0624-3. [DOI] [PubMed] [Google Scholar]
- 10.Mongi F, Laconte L, Casero RD. Ácaros del género Demodex: ¿parásitos colonizadores de personas sanas o asociados a patología ocular? [Demodex genus: colonizing parasites of healthy people or mites associated with ocular pathology?] Rev Argent Microbiol. 2018;50(4):369–373. doi: 10.1016/j.ram.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 11.Zhang XB, Ding YH, He W. The association between demodex infestation and ocular surface manifestations in meibomian gland dysfunction. Int J Ophthalmol. 2018;11(4):589–592. doi: 10.18240/ijo.2018.04.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Yi G, Ke X, Li S, Li Z, Chen X. Effect of Demodex on ocular surface function in patients with meibomian gland dysfunction. Int Eye Sci. 2019;19(07):1228–1231 (Chinese with English abstract).
- 13.Li Z, Gong Y, Chen S, Li S, Zhang Y, Zhong H, Wang Z, Chen Y, Deng Q, Jiang Y, Li L, Fu M, Yi G. Comparative portrayal of ocular surface microbe with and without dry eye. J Microbiol (Seoul, Korea) 2019;57(11):1025–1032. doi: 10.1007/s12275-019-9127-2. [DOI] [PubMed] [Google Scholar]
- 14.Nelson JD, Craig JP, Akpek EK, et al. TFOS DEWS II introduction. Ocul Surf. 2017;15(3):269–275. doi: 10.1016/j.jtos.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 15.Messmer EM. The pathophysiology, diagnosis, and treatment of dry eye disease. Dtsch Arztebl Int. 2015;112(5):71–82. 10.3238/arztebl.2015.0071. [DOI] [PMC free article] [PubMed]
- 16.Randon M, Liang H, El Hamdaoui M, et al. In vivo confocal microscopy as a novel and reliable tool for the diagnosis of Demodex eyelid infestation. Br J Ophthalmol. 2015;99(3):336–341. doi: 10.1136/bjophthalmol-2014-305671. [DOI] [PubMed] [Google Scholar]
- 17.Zhu M, Cheng C, Yi H, Lin L, Wu K. Quantitative analysis of the bacteria in blepharitis with Demodex infestation. Front Microbiol. 2018;9:1719. doi: 10.3389/fmicb.2018.01719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerber PA, Kukova G, Buhren BA, Homey B. Density of Demodex folliculorum in patients receiving epidermal growth factor receptor inhibitors. Dermatology. 2011;222(2):144–147. doi: 10.1159/000323001. [DOI] [PubMed] [Google Scholar]
- 19.Jarmuda S, O’Reilly N, Żaba R, Jakubowicz O, Szkaradkiewicz A, Kavanagh K. Potential role of Demodex mites and bacteria in the induction of rosacea. J Med Microbiol. 2012;61(Pt 11):1504–1510. doi: 10.1099/jmm.0.048090-0. [DOI] [PubMed] [Google Scholar]
- 20.Lozupone CA, Knight R. Species divergence and the measurement of microbial diversity. FEMS Microbiol Rev. 2008;32(4):557–578. doi: 10.1111/j.1574-6976.2008.00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Türk M, Oztürk I, Sener AG, Küçükbay S, Afşar I, Maden A. Comparison of incidence of Demodex folliculorum on the eyelash follicule in normal people and blepharitis patients. Turkiye Parazitol Derg. 2007;31(4):296–297. [PubMed] [Google Scholar]
- 22.Keskin Kurt R, Aycan Kaya O, Karateke A, et al. Increased density of Demodex folliculorum mites in pregnancies with gestational diabetes. Med Princ Pract. 2014;23(4):369–372. doi: 10.1159/000363244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamashita LS, Cariello AJ, Geha NM, Yu MC, Hofling-Lima AL. Demodex folliculorum on the eyelash follicle of diabetic patients. Arq Bras Oftalmol. 2011;74(6):422–424. doi: 10.1590/s0004-27492011000600008. [DOI] [PubMed] [Google Scholar]
- 24.Kirkman MS, Mahmud H, Korytkowski MT. Intensive blood glucose contd vascular outcomes in patients with type 2 diabetes mellitus. Endocrinol Metam Clin N Am. 2018;47:81–96. doi: 10.1016/j.ecl.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Cheng S, Zhang M, Chen H, Fan W, Huang Y. The correlation between the microstructure of meibomian glands and ocular Demodex infestation: a retrospective case-control study in a Chinese population. Medicine (Baltimore) 2019;98(19):e15595. doi: 10.1097/MD.0000000000015595Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jee K. Encyclopedia of ophthalmology. Berlin: Springer; 2018. [Google Scholar]
- 27.Kugadas A, Gadjeva M. Impact of microbiome on ocular health. Ocul Surf. 2016;14(3):342–349. doi: 10.1016/j.jtos.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graham JE, Moore JE, Jiru X, et al. Ocular pathogen or commensal: a PCR-based study of surface bacterial flora in normal and dry eyes. Invest Ophthalmol Vis Sci. 2007;48(12):5616–5623. doi: 10.1167/iovs.07-0588. [DOI] [PubMed] [Google Scholar]
- 29.Zhou Y, Holland MJ, Makalo P, et al. The conjunctival microbiome in health and trachomatous disease: a case control study. Genome Med. 2014;6(11):99. doi: 10.1186/s13073-014-0099-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolf R, Ophir J, Avigad J, Lengy J, Krakowski A. The hair follicle mites (Demodex spp.). Could they be vectors of pathogenic microorganisms?. Acta Derm Venereol. 1988;68(6):535–537. [PubMed]
- 31.Liang L, Safran S, Gao Y, Sheha H, Raju VK, Tseng SC. Ocular demodicosis as a potential cause of pediatric blepharoconjunctivitis. Cornea. 2010;29(12):1386–1391. doi: 10.1097/ICO.0b013e3181e2eac5. [DOI] [PubMed] [Google Scholar]
- 32.Knop E, Knop N, Millar T, Obata H, Sullivan DA. The international workshop on meibomian gland dysfunction: report of the subcommittee on anatomy, physiology, and pathophysiology of the meibomian gland. Invest Ophthalmol Vis Sci. 2011;52(4):1938–1978. doi: 10.1167/iovs.10-6997c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rynerson JM, Perry HD. DEBS—a unification theory for dry eye and blepharitis. Clin Ophthalmol. 2016;10:2455–2467. doi: 10.2147/OPTH.S114674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang X, Deng A, Yang J, et al. Pathogens in the Meibomian gland and conjunctival sac: microbiome of normal subjects and patients with Meibomian gland dysfunction. Infect Drug Resist. 2018;11:1729–1740. doi: 10.2147/IDR.S162135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang SD, He JN, Niu TT, et al. Bacteriological profile of ocular surface flora in meibomian gland dysfunction. Ocul Surf. 2017;15(2):242–247. doi: 10.1016/j.jtos.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 36.Watters GA, Turnbull PR, Swift S, Petty A, Craig JP. Ocular surface microbiome in meibomian gland dysfunction. Clin Exp Ophthalmol. 2017;45(2):105–111. doi: 10.1111/ceo.12810. [DOI] [PubMed] [Google Scholar]
- 37.Dong X, Wang Y, Wang W, Lin P, Huang Y. Composition and diversity of bacterial community on the ocular surface of patients with meibomian gland dysfunction. Invest Ophthalmol Vis Sci. 2019;60(14):4774–4783. doi: 10.1167/iovs.19-27719. [DOI] [PubMed] [Google Scholar]
- 38.Lacey N, Kavanagh K, Tseng SC. Under the lash: demodex mites in human diseases. Biochem (Lond) 2009;31(4):2–6. [PMC free article] [PubMed] [Google Scholar]
- 39.Szkaradkiewicz A, Chudzicka-Strugała I, Karpiński TM, et al. Bacillus oleronius and Demodex mite infestation in patients with chronic blepharitis. Clin Microbiol Infect. 2012;18(10):1020–1025. doi: 10.1111/j.1469-0691.2011.03704.x. [DOI] [PubMed] [Google Scholar]
- 40.Li Z, Gong Y, Chen S, et al. Comparative portrayal of ocular surface microbe with and without dry eye. J Microbiol. 2019;57(11):1025–1032. doi: 10.1007/s12275-019-9127-2. [DOI] [PubMed] [Google Scholar]
- 41.Heimer SR, Evans DJ, Mun JJ, Stern ME, Fleiszig SM. Surfactant protein D contributes to ocular defense against Pseudomonas aeruginosa in a murine model of dry eye disease. PLoS One. 2013;8(6):e65797. doi: 10.1371/journal.pone.0065797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Georgiev GA, Eftimov P, Yokoi N. Structure-function relationship of tear film lipid layer: a contemporary perspective. Exp Eye Res. 2017;163:17–28. doi: 10.1016/j.exer.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 43.Pflugfelder SC, Farley W, Luo L, et al. Matrix metalloproteinase-9 knockout confers resistance to corneal epithelial barrier disruption in experimental dry eye. Am J Pathol. 2005;166(1):61–71. doi: 10.1016/S0002-9440(10)62232-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data of sequencing results have been submitted to NCBI SRA (National Center for Biotechnology Information Sequence Read Archive). Raw data and project information are available via BioProject ID (PRJNA692647) and the following website: http://www.ncbi.nlm.nih.gov/bioproject/692647. The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.