Abstract
Forkhead box A2 (FOXA2) has emerged as a tumor inhibitor in several human malignancies. This work focused on the effect of FOXA2 on liver cancer (LC) cell invasion and migration and the involving molecules. FOXA2 expression in LC tissues and cell lines was determined. The potential target microRNA (miRNA) of FOXA2 was predicted via bioinformatic analysis and validated through a ChIP assay. The mRNA target of miRNA-103a-3p was predicted via bioinformatic analysis and confirmed via a luciferase assay. Altered expression of FOXA2, miR-103a-3p and GREM2 was introduced in cells to identify their roles in LC cell migration and invasion. Consequently, FOXA2 and GREM2 were poorly expressed while miR-103a-3p was highly expressed in LC samples. Overexpression of FOXA2 or GREM2 suppressed migration and invasion of LC cells, while up-regulation of miR-103a-3p led to inverse trends. FOXA2 transcriptionally suppressed miR-103a-3p to increase GREM2 expression. Silencing of GREM2 blocked the effects of FOXA2. GREM2 increased LATS2 activity and YAP phosphorylation and degradation. To conclude, this study demonstrated that FOXA2 suppressed miR-103a-3p transcription to induce GREM2 upregulation, which increased LATS2 activity and YAP phosphorylation to inhibit migration and invasion of LC cells.
Keywords: FOXA2, MiR-103a-3p, GREM2, LATS2, YAP, Liver cancer
Introduction
Although most of the risk factors such as obesity, excessive alcohol consumption, smoking, and hepatitis B and C virus (HBV and HCV) infection are potentially preventable, the incidence rate of liver cancer (LC) has increased most rapidly among human cancers by an annual 2 to 3% increase from 2007 to 2016 (Siegel et al. 2020). LC ranks the second highest cause of cancer death worldwide with hepatocellular carcinoma (HCC) representing the most common type (Momin et al. 2018). China has a particularly high mortality and morbidity rate in LC with estimated 466,100 new diagnoses and 422,100 deaths according to the cancer statistics in 2015, accounting for over half of all cases worldwide (Chen et al. 2016; De Mattia et al. 2019). In addition, due to the difficulty in early diagnosis, high malignancy with rapid progression, and the lack of effective targeted drugs, the survival rate of LC is seriously low (Fu and Wang 2018). Although surgical resection, liver transplantation and ablation are promising curative strategies for LC patients, these operations are only available for patients at early stages characterized by limited tumor in liver (Diaz-Gonzalez et al. 2016; Pinter and Peck-Radosavljevic 2018). Unfortunately, most patients are found at late stages with frequent malignant metastasis. Hence, developing novel ways in metastasis control may provide novel therapeutic options for LC treatment.
Forkhead box A2 (FOXA2) is a member of the forkhead class of DNA-binding proteins that are transcriptional activators for the regulation of cell differentiation and metabolism (Friedman and Kaestner 2006; Wang 2015). FOXA2 is a liver-specific transcription factor that regulates liver development, which plays a crucial role in glucose and lipid homeostasis maintenance and leads to the organismal energy balance (Shaalan et al. 2019). FOXAs may influence multiple cellular processes that are correlated with the initiation, metastasis, and development of cancers by different regulation manners with diverse networks (Bach et al. 2018). Interestingly, a previous study suggested the silencing of FOXA2 by microRNA (miR)-92a increased LC cell proliferation and invasion (Wang et al. 2017). However, the exact role of FOXA2 in LC cell migration and invasion and the mechanisms remain largely unstudied. Transcription factors are capable of binding to promoter regions of downstream target genes (Razaghi-Moghadam and Nikoloski 2020) including miRNAs. miRNAs are the mostly studied non-coding RNAs that are often involved in FOXA-mediated events (Peng et al. 2020). miRNAs comprise 17–25 nucleotides with a major role in gene degradation by binding to the target complementary mRNAs in a post-transcriptional level (Condrat et al. 2020). Aberrant expression of miRNAs is frequently observed in human malignancies, and they function as either oncogenes or tumor suppressors through the different post-transcriptional modifications in genes (Kai et al. 2018). Unsurprisingly, several miRNAs have been suggested to play key roles in LC pathogenesis by mediating cell malignancy and tumor metastasis, thus may serve as potential therapeutic approaches (Callegari et al. 2015). Here, our study suggested that FOXA2 bound to miR-103a-3p, and miR-103a-3p directly bound to target gremlin 2 (GREM2). miR-103a-3p has been suggested to work as a tumor promoter in oral squamous cell carcinoma (Zhang et al. 2020). On the other hand, it has been reported to suppress invasion and proliferation of prostate cancer cells (Ge et al. 2020). But its function in LC has not been investigated yet. As for GREM2, it was suggested to be lowly expressed in LC through the data on GEPIA (http://gepia.cancer-pku.cn/). Taken together, we hypothesized that FOXA2 could inhibit LC metastasis through miR-103a-3p suppression and the following GREM2 up-regulation, and cell migration and invasion experiments were performed to validate the hypothesis.
Materials and methods
Ethics statement
The study was approved by the Clinical Ethical Committee of the Second People’s Hospital of Liaocheng. This study adhered to the tenets of the Declaration of Helsinki. Signed informed consent was obtained from each eligible participant.
Clinical tissue sample collection
From January 2018 to February 2019, tumor tissues and the adjacent normal tissues (at least 3 cm away from the lesion sites) were collected from 50 LC patients treated in the Second People’s Hospital of Liaocheng. There were 39 male (78%) and 11 female (22%) patients with an average age of 51 ± 11.3 years. All enrolled patients were diagnosed by pathological examination without surgery history or other malignancies. The tissues were collected during surgery and instantly frozen at − 80 °C.
Cell culture and treatment
Immortalized human liver cell line THLE-2 and LC cell lines (HepG2 and SK-HEP-1) were purchased from American Type Culture Collection (Manassas, VA, USA). Cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen, Frederick, MD, USA) containing 10% fetal bovine serum (FBS, Gibco, Grand Island, NY, USA), streptomycin (0.1 mg/mL) and penicillin (100 U/mL) at 37 °C with 5% CO2 with constant humidity.
miR-103a-3p mimic and small interfering RNA (siRNA) targeting GREM2 (siRNA-GREM2) were designed and synthesized by GenePharma Co., Ltd. (Shanghai, China). The sequences of FOXA2 and GREM2 were synthesized and cloned to the pcDNA3.1 vector (pcDNA-FOXA2, pcDNA-GREM2, Invitrogen). The pcDNA3.1 empty vector and negative control (NC) for siRNA (siRNA-NC) and miRNA control (NC) were constructed as well. The vectors were transfected into cells using Lipofectamine 3000 Reagent (Invitrogen).
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNA in cells and tissues was extracted using the TRIzol Reagent (Invitrogen). The RNA was reverse-transcribed into cDNA utilizing a SuperScript RT kit (Invitrogen) according to the manufacturer’s instructions. Next, real-time qPCR was conducted using SYBR Green (Applied Biosystems, Foster City, CA, USA) on a CFX96 System (Bio-Rad, Hercules, CA, USA) in compliance with the manufacturer’s instructions. U6 was used as the internal reference for miR-103a-3p whereas GAPDH for other genes. Relative gene expression was determined using the 2−ΔΔCt method. Each procedure was performed three times. The primers are presented in Table 1.
Table 1.
Primer sequences in RT-qPCR
| Gene | Primer sequence (5′-3′) |
|---|---|
| FOXA2 | F: GAGGCGACAGCGTTAGCA |
| R: TACTCCATGGGACCCCTGTT | |
| miR-103a-3p | F: ACTGTAAAGAAGCCGAGGGC |
| R: CCCTATGTGTTTCTACTTTTTGGT | |
| GREM2 | F: TCCCTCCCCTATCTGTGTGG |
| R: ATGGAGGCTAGGGGTGGATT | |
| U6 | F: CTCGCTTCGGCAGCACA |
| R: AACGCTTCACGAATTTGCGT | |
| GAPDH | F: GCACCGTCAAGGCTGAGAAC |
| R: TGGTGAAGACGCCAGTGGA |
RT-qPCR reverse transcription quantitative polymerase chain reaction, FOXA2 forkhead box A2, GREM2 gremlin 2, GAPDH glyceraldehyde-3-phosphate dehydrogenase, F forward, R reverse
Immunohistochemical staining
The collected LC tissues were embedded in paraffin, cut into 5-μm sections, dewaxed and rehydrated. After antigen retrieval in 1 × ethylene diamine tetraacetic acid, the tissues were treated with 3% hydrogen peroxide to block the activity of endogenous peroxidase and treated with 5% bovine serum albumin (BSA) to block the non-specific binding. Next, the sections were incubated with the primary antibody against FOXA2 (1:500, ab108422, Abcam, Cambridge, UK) and GREM2 (1:100, ab228736, Abcam) at 4 °C overnight and then incubated with the secondary antibody goat-anti-rabbit immunoglobulin G (IgG) (HRP) (1:2000, ab205718, Abcam) at 23 °C for 30 min. Then, the sections were incubated with streptavidin–biotin complex for 30 min, stained with 3,3'-diaminobenzidine, counterstained with hematoxylin, and fixed for observation. The intensity of positive staining was evaluated by three independent monitors who had no idea of the grouping details. The positive staining intensity in cells was scored as follows: 0: no positive cells; 1: a 0–25% positive rate; 2: a 25–50% positive rate; 3: a 50–75% positive rate; 4: a 75–100% positive rate. The staining intensity was scored as follows: 1: weak; 2: moderate; 3: strong. Then, the integrated staining score was calculated as positive staining score × staining intensity score.
Transwell assay
Invasion and migration abilities of LC cells were determined using Transwell assays. For cell migration, 24 h after transfection, a total of 2 × 104 cells were sorted into the apical chambers supplemented with FBS-free RMPI-1640, and the basolateral chambers were loaded with 600 µL DMEM with 10% FBS. The Transwells were placed in an incubator at 37 °C for 24 h. The cells on the upper surface in the apical chambers were wiped out by cotton swabs. The cells migrated to the lower membrane were fixed in 4% paraformaldehyde (Beyotime Biotechnology Co., Ltd., Shanghai, China) for 20 min and then stained with 0.5% crystal violet (Beyotime) for 15 min. The invasion of cells was determined in a similar manner with additional pre-coating of Matrigel (BD-Biosciences, CA, USA) in apical chambers before cell seeding. The number of invading and migrating cells were observed and counted under an IX81 microscope (Olympus, Tokyo, Japan) with 5 fields randomly selected, and the average value was calculated.
Dual luciferase reporter gene assay
Luciferase reporter plasmids were constructed with the ligation of oligonucleotides containing the putative binding sequence (wide type, WT) between GREM2 3’UTR and miR-103a and mutant type (MT) binding sequence. The sequences were inserted into GV272 vectors (Genechem Co., Ltd., Shanghai, China) to construct luciferase vectors. The vectors were co-transfected with miR-103a mimic or mimic NC into HepG2 and SK-HEP-1 cells using a Lipofectamine 2000 kit (Invitrogen). After 48 h, the cells were lysed, and the luciferase activity was measured using a Dual Luciferase Reporter Assay Kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Three independent experiments were performed.
Chromatin immunoprecipitation (ChIP) assay
Cells were treated with 37% formaldehyde, collected and subjected to ultrasonication using a VCX750 (SONICS, USA) at a power of 25%. The cells were ultrasonicated for 4.5 s each time for a total of 14 times at a 9-s interval. Thereafter, the cells were centrifuged at 10,000×g at 4 °C for 10 min to discard the insolubles. Next, the antibody against FOXA2 (ab256493, Abcam) and the control IgG antibody (ab6721, Abcam) were added to combine with the target protein-DNA complexes. Next, Protein A was further administrated to precipitate the antibody-target protein-DNA complexes. The precipitate was successfully subjected to (1) low salt wash buffer-one wash; (2), high salt wash buffer-one wash; (3) LiCl wash buffer-one wash; (4) TE buffer-two washes. After that, the precipitates were eluted and de-crosslinked. The enriched DNA sequences were purified for qPCR. The primer sequence used were: Forward: 5′-TCACGGACATTCCAGG-3′; Reverse: 5′-TCGCAGTATTGCGACG-3′.
Western blot analysis
Cells were lysed in radio-immunoprecipitation assay cell lysis buffer on ice and then centrifuged at 14,000 rpm for 20 min with the lysis buffer discarded to collect total protein. The protein concentration was determined using a bicinchoninic acid kit (Pierce, Rockford, IL, USA). Next, an equal amount of protein (30–50 μg) was separated by 10% SDS-PAGE and transferred onto PVDF membranes (Merck Millipore, Billerica, MA, USA). The membranes were blocked in 5% non-fat milk for an hour and then incubated with the primary antibodies at 4 °C overnight and further with the secondary antibody at 20 °C for an hour. Then, the immunoreactive protein level was determined using an enhanced chemiluminescence kit (NEN, Dreieich, Germany). The antibodies used were against FOXA2 (1:1,000, ab108422,abcam), GREM2 (1:1,000, ab228736, Abcam), large tumor suppressor 2 (LATS2, 1:100, ab110780, Abcam), p-Yes‐associated protein (p-YAP, 1:1,000, Ser127, Cell Signaling Technology, MA, USA), YAP (1:500, 13,584–1-AP, Proteintech, Rosemont, USA), β-actin (1:2,500, AF5003, Beyotime) and horseradish peroxidase-labeled secondary antibody (1:3,000, ab6721, Abcam).
Statistical analysis
Measurement data were presented as mean ± standard deviation (SD) from at least three independent experiments. SPSS 21.0 (IBM Corp. Armonk, NY, USA) was applied for data analysis. Data were analyzed using the paired t test, Pearson’s Correlation Analysis, one-way or two-way analysis of variance (ANOVA) and Tukey’s multiple comparison test. The p value was obtained from two-tailed tests, and p < 0.05 was considered to show significant difference.
Results
FOXA2 is poorly expressed in LC and inhibits LC cell migration and invasion
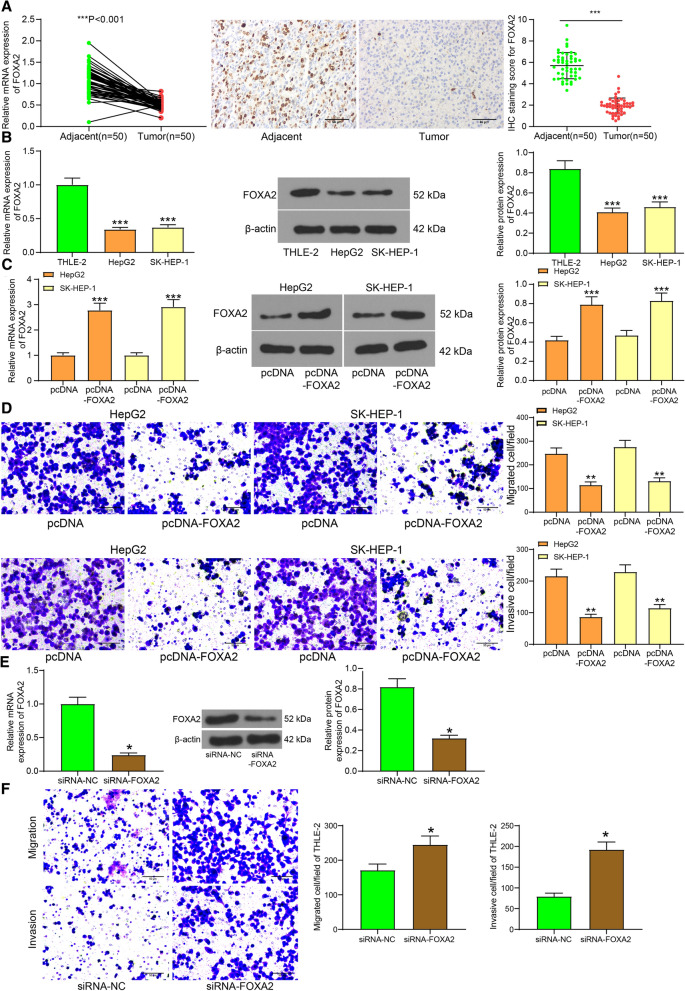
FOXA2 is a transcriptional activator for liver-specific genes. Importantly, FOXA2 has been suggested to be expressed at a lower level in LC tissues than that in normal liver (Convertini et al. 2019). Here, our study measured the FOXA2 expression in 50 pairs of LC tissues and the adjacent normal tissues using RT-qPCR, and the results suggested that the FOXA2 expression was much lower in tumor tissues than that in the paired normal ones (Fig. 1a). In cells, it was found that the LC cell lines HepG2 and SK-HEP-1 also presented lower levels of FOXA2 (Fig. 1b). Then, pcDNA-FOXA2 was administrated to artificially up-regulate FOXA2 expression in LC cells (Fig. 1c). Importantly, the invasion and migration abilities of cells were inhibited upon FOXA2 upregulation (Fig. 1d).
Fig. 1.
FOXA2 is poorly expressed in LC and inhibits LC cell migration and invasion. a FOXA2 expression in LC tumor tissues and in the paired adjacent normal tissues determined by RT-qPCR (n = 50, paired t test, ***, p < 0.001); b FOXA2 expression in THLE-2, HepG2, SK-HEP-1 cells determined by RT-qPCR and western blot analysis (one-way ANOVA, ***, p < 0.001); c transfection efficacy of pcDNA-FOXA2 determined by RT-qPCR and western blot analysis (one-way ANOVA, ***, p < 0.001); d migration and invasion abilities of cells determined by Transwell assays (one-way ANOVA, **, p < 0.01); e transfection efficacy of siRNA-FOXA2 in THLE-2 cells examined by RT-qPCR and western blot analysis (unpaired t test, *, p < 0.05); f migration and invasion of THLE-2 cells after siRNA-FOXA2 transfection examined by Transwell assays (unpaired t test, *, p < 0.05). Data were presented as the mean ± SD from three independent experiments
In addition, siRNA-FOXA2 and its control were transfected into THLE-2 cells to examine the function of FOXA2 in the behaviors of normal cells. The successful transfection was confirmed by RT-qPCR and western blot analysis (Fig. 1e). In this setting, the Transwell assays suggested that downregulation of FOXA2 significantly increased the migration and invasion abilities of THLE-2 cells (Fig. 1f). Namely, silencing of FOXA2 given tumor cell properties to normal cells.
FOXA2 transcriptionally suppresses miR-103a-3p
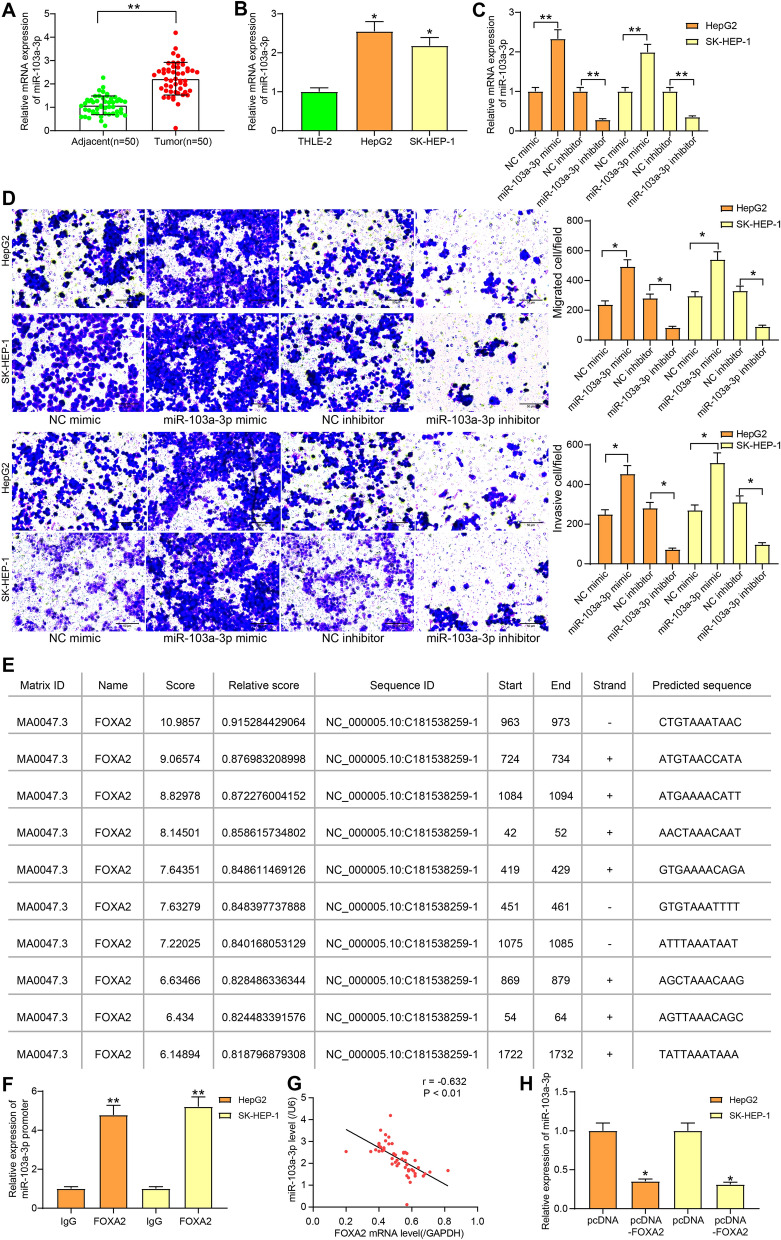
FOXA2 is a transcriptional factor that can specifically bind to the upstream sequences of genes at the 5’end to suppress gene expression. miR-103a-3p has been suggested to play oncogenic roles in human cancers and is linked to worse prognosis of cancer patients (Zhang et al. 2020). Here, we determined miR-103a-3p expression in LC and normal tissues. It was found that the LC tissues showed higher expression of miR-103a-3p (Fig. 2a). The similar trend was found in LC cell lines, where HepG2 and SK-HEP-1 cells had increased miR-103a-3p expression relative to THLE-2 cells (Fig. 2b). Then, miR-103a-3p mimic/inhibitor or the NC was transfected into cells, after which the miR-103a-3p expression in cells was correspondingly increased or decreased (Fig. 2c). Next, the Transwell assay results suggested that the migration and invasion abilities of cells were increased following miR-103a-3p up-regulation, while decreased after miR-103a03p down-regulation (Fig. 2d).
Fig. 2.
FOXA2 transcriptionally suppresses miR-103a-3p. a miR-103a-3p expression in LC tumor tissues and in the paired adjacent normal tissues determined by RT-qPCR (n = 50, paired t test, **, p < 0.01); b miR-103a-3p expression in THLE-2, HepG2, SK-HEP-1 cells determined by RT-qPCR (one-way ANOVA, *, p < 0.05); c miR-103a-3p expression in cells after miR-103a-3p mimic/inhibitor transfection determined by RT-qPCR; d migration and invasion abilities of cells after miR-103a-3p mimic/inhibitor transfection measured by Transwell assays; e putative binding sequence of miR-103a-3p and FOXA2 predicted on Jaspar (http://jaspar.genereg.net/); f enrichment of miR-103a-3p promoter sequence in the precipitation of FOXA2-DNA compound in the ChIP-qPCR assay (one-way ANOVA, **, p < 0.01); g correlation between miR-103a-3p expression and FOXA2 expression in 50 pairs of LC tissues and the adjacent tissues analyzed by Pearson’s Correlation Analysis (r = -0.632, * p < 0.01); h miR-103a-3p expression in cells detected after pcNDA-FOXA2 transfection (one-way ANOVA, * p < 0.05). Data were presented as the mean ± SD from three independent experiments
According to the prediction on the bioinformatic system Jaspar (http://jaspar.genereg.net/), FOXA2 was suggested to have binding sites with miR-103a-3p at the promoter region (Fig. 2e). The site with highest relative score was selected for ChIP assay, and the following RT-qPCR suggested that miR-103a-3p was enriched by anti-FOXA2 rather than anti-IgG, indicating that FOXA2 could bind with the promoter region of miR-103a-3p (Fig. 2f). In addition, the RT-qPCR results revealed an inverse correlation between miR-103a-3p and FOXA2 expression (Fig. 2g). Following pcDNA-FOXA2 administration, the miR-103a-3p expression was significantly inhibited (Fig. 2h). The above experiments suggest that miR-103a-3p promotes LC cell migration and invasion, whereas FOXA2 transcriptionally suppresses miR-103a-3p expression.
miR-103a-3p directly targets GREM2
To further explore the potential downstream molecules, the target genes of miR-103a-3p were predicted on StarBase (http://starbase.sysu.edu.cn/), miRDB (http://mirdb.org/), TargetScan (http://www.targetscan.org/vert_72/) and miRwalk (http://mirwalk.umm.uni-heidelberg.de/), and four genes were found to be intersected (Fig. 3a). Among them, GREM2 was the only one poorly expressed in LC according the data in the GEPIA system (Fig. 3b). GREM2 was found as a putative target of miR-103a-3p in StarBase (Fig. 3c). We then speculated that miR-103a-3p might exert functions through binding with GREM2. First, RT-qPCR and immunohistochemical staining results validated that GREM2 was poorly expressed in LC tissues relative to the normal ones (Fig. 3d). Similarly, the mRNA and protein expression of GREM2 was decreased in LC cell lines compared to that in THLE-2 cells (Fig. 3e). Then, pcDNA-GREM2 and siRNA-GREM2 were transfected into LC cell lines, and then the GREM2 expression was successfully up- or down-regulated, accordingly (Fig. 3f). The Transwell assay results suggested that pcDNA-GREM2 significantly inhibited the migration and invasion of LC cell lines. On the contrary, the migration and invasion abilities of cells were promoted when they were treated with siRNA-GREM2 (Fig. 3g).
Fig. 3.
miR-103a-3p directly targets GREM2. a the downstream target mRNAs of miR-103a-3p predicted on StarBase, TargetScan and miRDB; b GREM2 expression in LC predicted on GEPIA; c putative binding sites of miR-103a-3p and GREM2 predicted on Starbase; d GREAM2 expression in LC tumor tissues and in the paired adjacent normal tissues determined by RT-qPCR and immunohistochemical staining (n = 50, paired t test, **, p < 0.01); e mRNA and protein expression of GREM2 in THLE-2, HepG2, SK-HEP-1 cells determined by RT-qPCR and western blot analysis, respectively (one-way ANOVA, **, p < 0.01); f GREM2 expression in LC cell lines after pcDNA-GREM2 or siRNA-GREM2 transfection determined by RT-qPCR and western blot analysis, respectively (one-way ANOVA, **, p < 0.01); g number of migrated and invaded LC cells after pcDNA-GREM2 or siRNA-GREM2 transfection determined by Transwell assays (one-way ANOVA, *, p < 0.05); h correlation between GREM2 and miR-103a-3p expression in 50 pairs of LC tissues and the adjacent tissues analyzed by Pearson’s Correlation Analysis (r = -0.721, **, p < 0.01); i mRNA and protein expression of GREM2 in LC cell lines after miR-103a-3p transfection determined by RT-qPCR and western blot analysis (one-way ANOVA, *, p < 0.05); j binding relationship between miR-103a-3p and GREM2 validated through a dual luciferase reporter gene assay (two-way ANOVA, * p < 0.05); Data were presented as the mean ± SD from three independent experiments
In addition, GREM2 presented a negative correlation with miR-103a-3p expression in the collected tumor tissues (Fig. 3h). Further, after miR-103a-3p mimic transfection, it was found that the GREM2 expression was significantly decreased according to RT-qPCR and western blot analysis (Fig. 3i). To further validate the binding relationship between miR-103a-3p and GREM2, a dual luciferase reporter gene assay was performed, which suggested that the luciferase activity in cells co-transfected with miR-103a-3p mimic and GREM2-WT sequence was reduced, while the luciferase activity in cells subjected to other co-transfection showed little differences (Fig. 3j).
GREM2 activates LATS2 to promote YAP phosphorylation
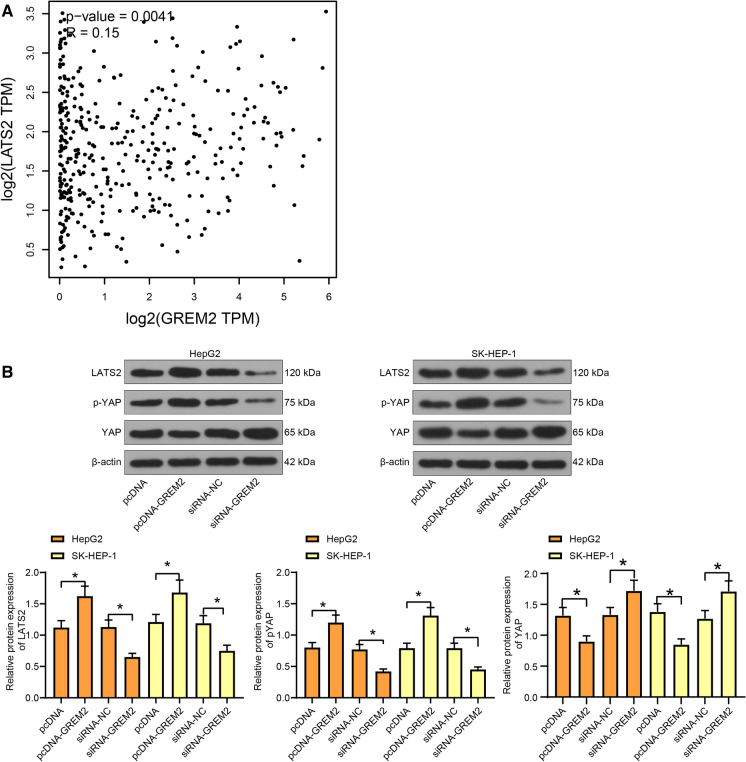
The downstream effector YAP of the Hippo signaling pathway is a publicly accepted oncogene. The nuclear translocated YAP can bind with the transcription factor TEAD to promote the transcription of proliferation and invasion-related genes (Zhang and Zhou 2019). According to the GEPIA database of The Cancer Genome Atlas (TCGA), LATS2, a core kinase of the Hippo signaling pathway, was suggested to show a positive correlation with the expression of GREM2 in LC (Fig. 4a). We therefore speculated that GREM2 possibly affects LATS2 expression to regulate the Hippo signaling pathway. To validate this, pcDNA-GREM2, siRNA-GREM2 or the corresponding NC vectors were transfected into LC cells. The LATS2 expression and YAP phosphorylation were increased but the total YAP expression was decreased when GREM2 was upregulated. Accordingly, siRNA-GREM2 led to a significant decline in LATS2 expression and YAP phosphorylation, but an increase in total YAP expression (Fig. 4b). The phosphorylation of YAP blocks its potential in nuclear translocation, leading to a degradation in cytoplasm. These results indicated that GREM2 increases the LATS2 kinase expression to trigger YAP phosphorylation, namely to activate the Hippo signaling pathway in LC cells.
Fig. 4.
GREM2 activates LATS2 to promote YAP phosphorylation. a correlation between LATS2 and GREM2 expression in LC in TCGA database; b expression of LATS2 and YAP and phosphorylation of YAP in LC cells after pcDNA-GREM2/siRNA-GREM-2 transfection determined by western blot analysis (one-way ANOVA, *, p < 0.05); Data were presented as the mean ± SD from three independent experiments
FOXA2 inhibits migration and invasion of LC cells through the miR-103a-3p/GREM2/LATS2/YAP axis
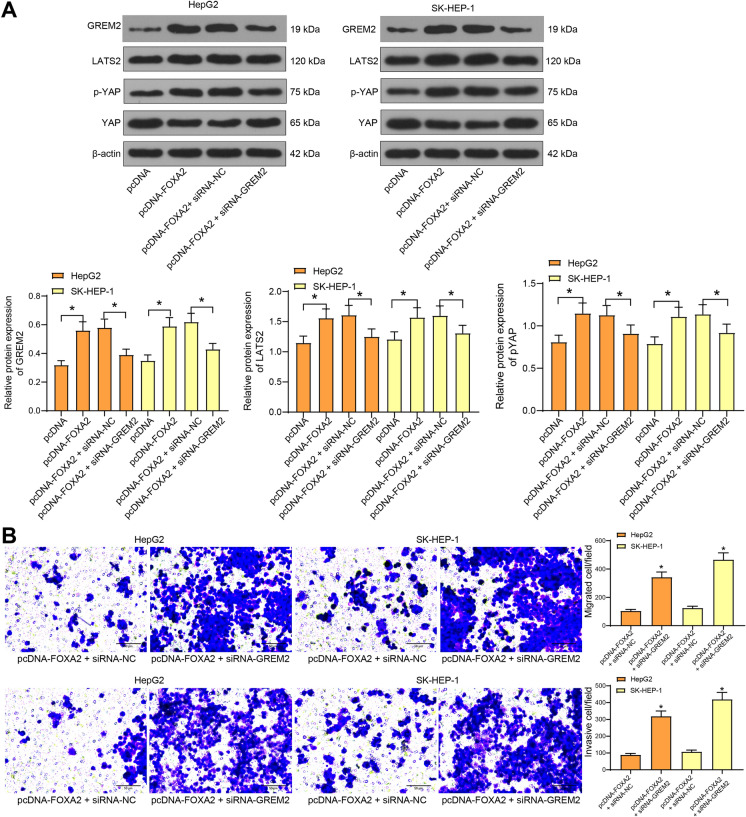
Then, pcDNA-FOXA2, pcDNA-FOXA2 + siRNA-GREM2 or the NC vectors were transfected into cells. Consequently, the GREM2 expression was increased by pcDNA-FOXA2 but suppressed by the further administration of siRNA-GREM2. In addition, pcDNA-FOXA2 led to an increase in LATS2 expression and YAP phosphorylation but a decrease in total YAP expression, but further administration of siRNA-GREM2 resulted in inverse trends in cells (Fig. 5a). These results indicated that downregulation of GREM2 blocked the regulatory functions of FOXA2 on the LATS2/YAP axis. In addition, in cells co-transfected with pcDNA-FOXA2 and siRNA-GREM2, the migration and invasion abilities of cells inhibited by pcDNA-FOXA2 were recovered after GREM2 downregulation (Fig. 5b). These findings infer that the GREM2 is at least partially implicated in the functions of FOXA2, which activates the LATS2/YAP axis to suppress the invasiveness of LC cells.
Fig. 5.
FOXA2 inhibits migration and invasion of LC cells through the miR-103a-3p/GREM2/LATS2 axis. a protein levels of LATS2 and YAP and phosphorylation of YAP in cells determined by western blot analysis (one-way ANOVA, *, p < 0.05); b migration and invasion abilities of cells transfected with pcDNA-FOXA2 and siRNA-GREM2 determined by Transwell assays (one-way ANOVA, *, p < 0.05). Data were presented as the mean ± SD from three independent experiments
Discussion
Owing to the always late diagnosis and lack of effective treatments, LC patients at advanced stages presented very unsatisfactory prognosis, leaving the development of new therapeutic options of great urgency. FOXA factors were initially known as hepatocyte nuclear factor 3 and as a family of pioneer transcription factors for liver-specific genes (Cho et al. 2012; Zhao and Li 2015). Studies have demonstrated the suppressing effects of FOXA2 in several human neoplastic diseases (Li et al. 2018; Bow et al. 2020). The present study identified that by suppressing miR-103a-3p transcription, FOXA2 upregulated the expression of GREM2, which induced LATS2 activation and YAP phosphorylation and suppressed migration and invasion of LC cells.
The FOXA subfamily participates in the differentiation and regulation of metabolic tissues including liver, pancreas and adipose tissues (Friedman and Kaestner 2006). The initial finding of the study was that FOXA2 was expressed at low levels in tissues from LC patients as well as in LC cell lines HepG2 and SK-HEP-1 relative to the paired normal liver tissues or cells. This was partly in line with a previous report where researchers found a decline in FOXA2 expression in LC cells and HepG2 cell line compared to normal liver cells (Convertini et al. 2019). A recent study by Chen et al. suggested that FOXA2 was transcriptionally suppressed by reduced linc00261, indicating that the poor expression of FOXA2 in LC cells might be induced by transcriptional inactivation by other molecules (Chen et al. 2021). Importantly, this selective regulation in cellular gene by the FOXA pioneer factors may offer the chance for the specific suppression in HBV gene expression and the following resolution of chronic HBV infections which cause approximately one million of deaths each year globally (Okumura et al. 2015; McFadden et al. 2017). In addition, our study confirmed that FOXA2 inhibited migration and invasion abilities of LC cell lines. The FOXA factors may differ from each other in terms of tumorigenesis in different cancer types (Yamashita et al. 2017; Bach et al. 2018; Thanan et al. 2020). In particular, FOXA2 has been suggested as a tumor suppressor in several cases. Unlike FOXA1 and FOXA3, FOXA2 was suggested to be significantly decreased in lung cancer cell lines and was positively linked to prognosis and better survival rate of lung cancer patients (Huang et al. 2019). More relevantly, FOXA2 has been documented to inhibit matrix metalloproteinase-9 expression to suppress HCC metastasis (Wang et al. 2014). FOXA2 was also suggested to fulfill key roles at enhancer regions of epithelial genes to maintain the enhancer structure and function, thus suppressing the epithelial-mesenchymal transition, a key process during invasion and metastasis (Song et al. 2010; Jagle et al. 2017). In addition, it was found that FOXA2 inhibition rendered migration and invasion abilities to THLE-2 cells and given the THLE-2 cells with sort of tumor cell properties. Although the THLE-2 cells belong to normal liver cells in the biological perspective, they may proliferate infinitely and acquire certain tumor cell properties (such as migration and invasion) upon immortalization.
The preliminary findings above triggered us to explore the possible downstream molecules. FOXA factors can recognize some specific patterns in DNA sequences and consequently bind to chromatin and promote the activities of other regulators (Bach et al. 2018). This study identified that FOXA2 bound to the promoter region of miR-103a-3p through online prediction and ChIP assays. miR-103a-3p expression was found to be highly expressed in LC tissues and cell lines, presenting an inverse correlation with FOXA2. Then, it was found that up-regulation of miR-103a-3p by miRNA mimic increased the migratory and invasive potentials of LC cell lines. miR-103a-3p has involved in cancer progression in several cancer types. For instance, it presented a high-expression profile in the tissues and plasma of patients with colorectal cancer (Zhang et al. 2019). An in vitro study suggested miR-103a-3p promoted proliferation of gastric cancer cells by targeting ATF7 (Hu et al. 2018). Likewise, inhibition of miR-103a-3p was found to inhibit the proliferation of oral squamous cell carcinoma (Zhang et al. 2020). Our finding here indicates the inhibiting potential of miR-103a-3p in LC. Moreover, our study further identified that GREM2 as a putative target of miR-103a-3p. GREM2 was suggested to be lowly expressed in LC according to the data on GEPIA bioinformation system. The role of GREM2 in cancers has been rarely investigated, though, it is a well-known antagonist of bone morphogenetic proteins (BMP) (Nolan et al. 2016), which have been revealed to promote the progression of several human cancers (Augeri et al. 2016; Di et al. 2019; Wang et al. 2019). Here, our study identified that GREM2 was decreased in LC tissues and cells, with its artificial up-regulation inhibited cell migration and invasion.
Intriguingly, our study found that GREM2 increased the activity of LATS2 and the further phosphorylation of YAP. LATS2 activation and the following YAP phosphorylation are typical signals of Hippo activation, whose dysfunction is involved in a large number of human diseases including cancer (Zheng and Pan 2019). By phosphorylation, YAP is prevented from nuclear accumulation and consequently degraded in cytoplasm, while the non-phosphorylated YAP is localized in nuclear and that promotes cell proliferation and organ growth, which was inhibited by LATS1/2 (Wennmann et al. 2014). Activation of the Hippo signaling has been found to inhibit the malignant behaviors including growth, migration and invasion of LC cell lines (Li et al. 2019; Ou et al. 2019). The experimental findings that pcDNA-FOXA2 increased LATS2 expression and YAP phosphorylation, and siRNA-LATS2 recovered the migration and invasion abilities inhibited by pcDNA-FOXA2 evidenced that the LATS2 was involved in the FOXA2-mediated events.
Conclusion
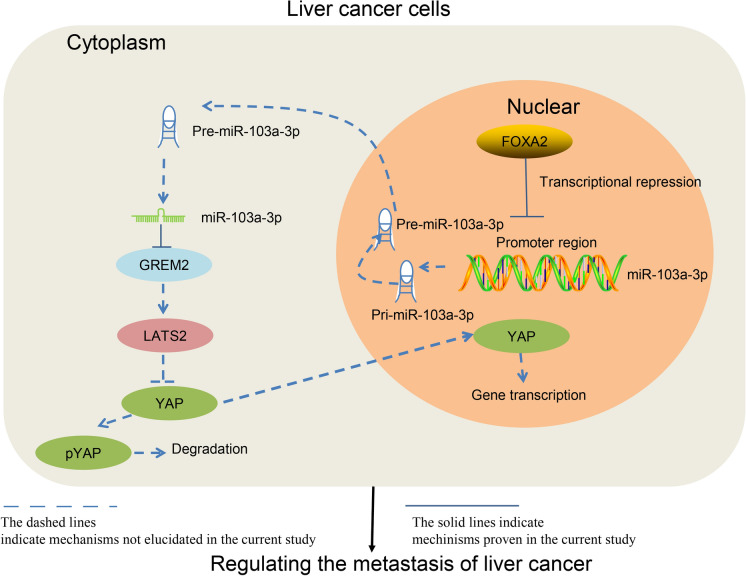
To sum up, our study demonstrated that FOXA2 fulfills tumor suppressing roles in LC by binding to the promoter region of miR-103a-3p to repress its transcription, after which GREM2 was up-regulated, which triggers LATS2 activation and YAP phosphorylation and degradation (Fig. 6). Although the paper was limited in exploring the migration and invasion in the cell perspective, we hope the current finding may offer new insights into LC control. In addition, due to the time and fund limits, the exact mechanism for the persistent downregulation of FOXA2 in LC and the regulatory mechanism between GREM2 and LATS2 were not involved in the current study. We would like to focus on these issues and to explore the role of FOXA2 in other cell behaviors such as proliferation and apoptosis in our future studies.
Fig. 6.
A diagram presentation of the molecular mechanism. In the nuclear of LC cells, FOXA2 binds to the promoter region of miR-103a-3p to repress miR-103a-3p transcription. This triggers the following GREM2 up-regulation and LATS2 activation, leading to further YAP phosphorylation and degradation in cytoplasm, resulting in a decline in cell migration and invasion
Declarations
Conflict of interest
The authors report no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Guangzhen Ma and Jirong Chen contributed equally to this work.
References
- Augeri DJ, Langenfeld E, Castle M, Gilleran JA, Langenfeld J. Inhibition of BMP and of TGFbeta receptors downregulates expression of XIAP and TAK1 leading to lung cancer cell death. Mol Cancer. 2016;15:27. doi: 10.1186/s12943-016-0511-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach DH, Long NP, Luu TT, Anh NH, Kwon SW, Lee SK (2018) The Dominant Role of Forkhead Box Proteins in Cancer. Int J Mol Sci 19 [DOI] [PMC free article] [PubMed]
- Bow YD, et al. Silencing of FOXA2 decreases E-cadherin expression and is associated with lymph node metastasis in oral cancer. Oral Dis. 2020;26:756–765. doi: 10.1111/odi.13282. [DOI] [PubMed] [Google Scholar]
- Callegari E, Gramantieri L, Domenicali M, D'Abundo L, Sabbioni S, Negrini M. MicroRNAs in liver cancer: a model for investigating pathogenesis and novel therapeutic approaches. Cell Death Differ. 2015;22:46–57. doi: 10.1038/cdd.2014.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, et al. Cancer statistics in China. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- Chen Z, et al. Epigenetically silenced linc00261 contributes to the metastasis of hepatocellular carcinoma via inducing the deficiency of FOXA2 transcription. Am J Cancer Res. 2021;11:277–296. [PMC free article] [PubMed] [Google Scholar]
- Cho JW, Lee CY, Ko Y. Therapeutic potential of mesenchymal stem cells overexpressing human forkhead box A2 gene in the regeneration of damaged liver tissues. J Gastroenterol Hepatol. 2012;27:1362–1370. doi: 10.1111/j.1440-1746.2012.07137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condrat CE et al. (2020) miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis Cells 9 [DOI] [PMC free article] [PubMed]
- Convertini P et al. (2019) Transcriptional regulation factors of the human mitochondrial aspartate/glutamate carrier gene, isoform 2 (SLC25A13): USF1 as Basal Factor and FOXA2 as activator in liver cells. Int J Mol Sci 20 [DOI] [PMC free article] [PubMed]
- De Mattia E, et al. Pharmacogenetics of the systemic treatment in advanced hepatocellular carcinoma. World J Gastroenterol. 2019;25:3870–3896. doi: 10.3748/wjg.v25.i29.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di L, et al. Discovery of a natural small-molecule compound that suppresses tumor EMT, stemness and metastasis by inhibiting TGFbeta/BMP signaling in triple-negative breast cancer. J Exp Clin Cancer Res. 2019;38:134. doi: 10.1186/s13046-019-1130-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Gonzalez A, Reig M, Bruix J. Treatment of Hepatocellular Carcinoma. Dig Dis. 2016;34:597–602. doi: 10.1159/000445275. [DOI] [PubMed] [Google Scholar]
- Friedman JR, Kaestner KH. The Foxa family of transcription factors in development and metabolism. Cell Mol Life Sci. 2006;63:2317–2328. doi: 10.1007/s00018-006-6095-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Wang H. Precision diagnosis and treatment of liver cancer in China. Cancer Lett. 2018;412:283–288. doi: 10.1016/j.canlet.2017.10.008. [DOI] [PubMed] [Google Scholar]
- Ge J et al. (2020) miR-103a-3p suppresses cell proliferation and invasion by targeting tumor protein D52 in prostate cancer. J Invest Surg:1–9 [DOI] [PubMed]
- Hu X, et al. miRNA-103a-3p promotes human gastric cancer cell proliferation by targeting and suppressing ATF7 in vitro. Mol Cells. 2018;41:390–400. doi: 10.14348/molcells.2018.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Liu J, Xiong B, Yonemura Y, Yang X. Expression and prognosis analyses of forkhead box A (FOXA) family in human lung cancer. Gene. 2019;685:202–210. doi: 10.1016/j.gene.2018.11.022. [DOI] [PubMed] [Google Scholar]
- Jagle S, Busch H, Freihen V, Beyes S, Schrempp M, Boerries M, Hecht A. SNAIL1-mediated downregulation of FOXA proteins facilitates the inactivation of transcriptional enhancer elements at key epithelial genes in colorectal cancer cells. PLoS Genet. 2017;13:e1007109. doi: 10.1371/journal.pgen.1007109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai K, Dittmar RL, Sen S. Secretory microRNAs as biomarkers of cancer. Semin Cell Dev Biol. 2018;78:22–36. doi: 10.1016/j.semcdb.2017.12.011. [DOI] [PubMed] [Google Scholar]
- Li JH, et al. microRNA-141–3p fosters the growth, invasion, and tumorigenesis of cervical cancer cells by targeting FOXA2. Arch Biochem Biophys. 2018;657:23–30. doi: 10.1016/j.abb.2018.09.008. [DOI] [PubMed] [Google Scholar]
- Li Y, et al. Artemisinin suppresses hepatocellular carcinoma cell growth, migration and invasion by targeting cellular bioenergetics and Hippo-YAP signaling. Arch Toxicol. 2019;93:3367–3383. doi: 10.1007/s00204-019-02579-3. [DOI] [PubMed] [Google Scholar]
- McFadden VC, et al. Hepatic deficiency of the pioneer transcription factor FoxA restricts hepatitis B virus biosynthesis by the developmental regulation of viral DNA methylation. PLoS Pathog. 2017;13:e1006239. doi: 10.1371/journal.ppat.1006239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momin B, Millman AJ, Nielsen DB, Revels M, Steele CB. Promising practices for the prevention of liver cancer: a review of the literature and cancer plan activities in the National Comprehensive Cancer Control Program. Cancer Causes Control. 2018;29:1265–1275. doi: 10.1007/s10552-018-1094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan K, Kattamuri C, Rankin SA, Read RJ, Zorn AM, Thompson TB. Structure of gremlin-2 in complex with GDF5 gives insight into DAN-family-mediated BMP antagonism. Cell Rep. 2016;16:2077–2086. doi: 10.1016/j.celrep.2016.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura N, Ikeda M, Satoh S, Dansako H, Sugiyama M, Mizokami M, Kato N. Negative regulation of hepatitis B virus replication by forkhead box protein A in human hepatoma cells. FEBS Lett. 2015;589:1112–1118. doi: 10.1016/j.febslet.2015.03.022. [DOI] [PubMed] [Google Scholar]
- Ou H, et al. Frizzled 2-induced epithelial-mesenchymal transition correlates with vasculogenic mimicry, stemness, and Hippo signaling in hepatocellular carcinoma. Cancer Sci. 2019;110:1169–1182. doi: 10.1111/cas.13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Q, et al. FOXA1 suppresses the growth, migration, and invasion of nasopharyngeal carcinoma cells through repressing miR-100–5p and miR-125b-5p. J Cancer. 2020;11:2485–2495. doi: 10.7150/jca.40709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter M, Peck-Radosavljevic M. Review article: systemic treatment of hepatocellular carcinoma. Aliment Pharmacol Ther. 2018;48:598–609. doi: 10.1111/apt.14913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razaghi-Moghadam Z, Nikoloski Z. Supervised learning of gene regulatory networks. Curr Protoc Plant Biol. 2020;5:e20106. doi: 10.1002/cppb.20106. [DOI] [PubMed] [Google Scholar]
- Shaalan UF, Ibrahim NL, Ehsan NA, Sultan MM, Naser GM, Abd El-Fatah MO. Reduced Immunohistochemical Expression of Hnf1beta and FoxA2 in Liver Tissue Can Discriminate Between Biliary Atresia and Other Causes of Neonatal Cholestasis. Appl Immunohistochem Mol Morphol. 2019;27:e32–e38. doi: 10.1097/PAI.0000000000000638. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- Song Y, Washington MK, Crawford HC. Loss of FOXA1/2 is essential for the epithelial-to-mesenchymal transition in pancreatic cancer. Cancer Res. 2010;70:2115–2125. doi: 10.1158/0008-5472.CAN-09-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanan R et al. (2020) Opposing roles of FoxA1 and FoxA3 in intrahepatic cholangiocarcinoma progression. Int J Mol Sci 21 [DOI] [PMC free article] [PubMed]
- Wang J, et al. FOXA2 suppresses the metastasis of hepatocellular carcinoma partially through matrix metalloproteinase-9 inhibition. Carcinogenesis. 2014;35:2576–2583. doi: 10.1093/carcin/bgu180. [DOI] [PubMed] [Google Scholar]
- Wang K. Molecular mechanisms of hepatic apoptosis regulated by nuclear factors. Cell Signal. 2015;27:729–738. doi: 10.1016/j.cellsig.2014.11.038. [DOI] [PubMed] [Google Scholar]
- Wang L, Wu J, Xie C. miR-92a promotes hepatocellular carcinoma cells proliferation and invasion by FOXA2 targeting. Iran J Basic Med Sci. 2017;20:783–790. doi: 10.22038/IJBMS.2017.9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Gu M, Jiang H, Zheng X (2019) BMP-2 upregulates the AKT/mTOR pathway in breast cancer with microcalcification and indicates a poor prognosis. Clin Transl Oncol [DOI] [PubMed]
- Wennmann DO, et al. Evolutionary and molecular facts link the WWC protein family to Hippo signaling. Mol Biol Evol. 2014;31:1710–1723. doi: 10.1093/molbev/msu115. [DOI] [PubMed] [Google Scholar]
- Yamashita H, et al. On a FOX hunt: functions of FOX transcriptional regulators in bladder cancer. Nat Rev Urol. 2017;14:98–106. doi: 10.1038/nrurol.2016.239. [DOI] [PubMed] [Google Scholar]
- Zhang G, Chen Z, Zhang Y, Li T, Bao Y, Zhang S (2020) Inhibition of miR-103a-3p suppresses the proliferation in oral squamous cell carcinoma cells via targeting RCAN1 Neoplasma [DOI] [PubMed]
- Zhang H, et al. A panel of seven-miRNA signature in plasma as potential biomarker for colorectal cancer diagnosis. Gene. 2019;687:246–254. doi: 10.1016/j.gene.2018.11.055. [DOI] [PubMed] [Google Scholar]
- Zhang S, Zhou D. Role of the transcriptional coactivators YAP/TAZ in liver cancer. Curr Opin Cell Biol. 2019;61:64–71. doi: 10.1016/j.ceb.2019.07.006. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Li Z. Interplay of estrogen receptors and FOXA factors in the liver cancer. Mol Cell Endocrinol. 2015;418:334–339. doi: 10.1016/j.mce.2015.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Pan D. The hippo signaling pathway in development and disease. Dev Cell. 2019;50:264–282. doi: 10.1016/j.devcel.2019.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]