Abstract
Emerging evidence has suggested a pivotal role of circular RNAs (circRNAs) in the progression of asthma. In this paper, we explored the mechanisms underlying the modulation of circRNA homeodomain interacting protein kinase 3 (circHIPK3, circ_0000284) in airway smooth muscle cell (AMSC) migration and proliferation induced by platelet-derived growth factor (PDGF). The stability of circHIPK3 was gauged by Ribonuclease R (RNase R) and Actinomycin D assays. Relative expression levels of circHIPK3, microRNA (miR)-375 and matrix metallopeptidase 16 (MMP-16) were measured by quantitative real-time polymerase chain reaction (qRT-PCR) and western blot. Cell proliferation, invasion, and apoptosis were evaluated by Cell Counting Kit-8 (CCK-8) assay, transwell assay, and flow cytometry, respectively. Cell migration was detected by wound-healing and transwell assays. Direct relationship between miR-375 and circHIPK3 or MMP-16 was verified by dual-luciferase reporter and RNA immunoprecipitation (RIP) assays. Our results indicated that PDGF induced the expression of circHIPK3 in human AMSCs (HAMSCs). CircHIPK3 silencing impeded proliferation, migration, invasion and promoted apoptosis of PDGF-treated HAMSCs. Mechanistically, circHIPK3 targeted miR-375 by directly binding to miR-375. MiR-375 was a downstream effector of circHIPK3 in controlling PDGF-induced proliferation, invasion and migration. MMP-16 was directly targeted and inhibited by miR-375, and circHIPK3 functioned as a post-transcriptional modulator of MMP-16 expression through miR-375. Moreover, miR-375-mediated inhibition of MMP-16 impacted HAMSC proliferation, invasion and migration induced by PDGF. Our findings identified the miR-375/MMP-16 axis as a novel mechanism for the modulation of circHIPK3 in PDGF-induced migration and proliferation in HASMCs.
Keywords: Asthma, circHIPK3, miR-375, MMP-16
Introduction
Asthma, one of the common respiratory diseases, remains a major cause of disease morbidity and a significant financial burden to society (Papi et al. 2018). Extending the knowledge of the pathophysiology of asthma is important to increase treatment options (Jones et al. 2018). Airway smooth muscle cells (ASMCs) play pivotal roles in the pathological process of asthma, and elevated migration and proliferation of ASMCs contribute to airway remodeling and thus aggravate asthma (James et al. 2018; Salter et al. 2017). This highlights that attenuating migration and proliferation of AMSCs may provide a potential therapeutic avenue for asthma.
Endogenously expressed circular RNAs (circRNAs) are naturally occurring RNA circles that exert importantly regulatory functions in eukaryotic cells by inhibiting the activity of microRNAs (miRNAs), which are well-known post-transcriptional regulators of gene expression (Hansen et al. 2013; Jeck and Sharpless 2014). Evidence is emerging that some circRNAs might regulate the development and management of various diseases, including asthma (Marques-Rocha et al. 2015; Bao et al. 2020). Huang et al. showed that circ_0002594, an overexpressed circRNA in CD4+ T cells of asthma, might have diagnostic and therapeutic potential for T helper 2-mediated asthma (Huang et al. 2021). They also demonstrated that circ_0005519 enhanced asthma progression through targeting let-7a-5p (Huang et al. 2019).
CircRNA homeodomain interacting protein kinase 3 (circHIPK3, circ_0000284), generated by exon 2 of HIPK3 mRNA, has been implicated in human diseases, such as cancer, coronary heart disease, and diabetics (Zhang et al. 2020; Sun et al. 2019; Wang et al. 2018). CircHIPK3 was also reported to control lung fibroblast-to-myofibroblast transition via reducing miR-338-3p activity (Zhang et al. 2019). Importantly, Lin et al. uncovered that circHIPK3 could regulate ASMC migration and proliferation induced by platelet-derived growth factor (PDGF) (Lin et al. 2020). Although this report also demonstrated the miR-326/stromal interaction molecule 1 (STIM1) axis as a downstream effector of circHIPK3 function (Lin et al. 2020), our understanding of its molecular basis is limited.
In this paper, we explored the mechanisms that circHIPK3 might be involved in the development of asthma. We showed that circHIPK3 directly targeted miR-375, a suppressor miRNA in PDGF-induced ASMC migration and proliferation (Ji et al. 2018). Furthermore, circHIPK3 targeted miR-375 to control the expression of matrix metallopeptidase 16 (MMP-16), a potential contributor for ASMC proliferation (Lou et al. 2020). Our study established a novel regulatory network that might be crucial in controlling the migration, proliferation and apoptosis of ASMCs.
Materials and methods
Isolation of cells
Bronchial biopsy specimens were obtained from the NO.2 Hospital of Baoding in accordance with procedures approved by the Ethics Committee of the NO.2 Hospital of Baoding. Written informed consent was provided by all 6 subjects without asthma and undergoing carcinoma resection. Human ASMCs (HASMCs) were isolated from the bronchial specimens as reported previously (Michaeloudes et al. 2011) and propagated in Dulbecco’s Modified Eagle Medium (Gibco, Tokyo, Japan) containing 10 % fetal bovine serum (Gibco) under standard conditions of 37 °C, 5 % CO2. HASMCs at passage 3–6 were used for all experiments.
Transient transfection of cells
CircHIPK3 overexpressing plasmid (circHIPK3) was created by inserting human circ_0000284 sequence synthesized by BGI (Shenzhen, China) into the pCD5-ciR vector (Geneseed, Guangzhou, China) opened with EcoR I and BamH I sites. MMP-16 overexpressing plasmid (pc-MMP-16) was made by cloning human MMP-16 (Accession: NM_005941.5) encoding sequence synthesized by BGI into the pcDNA3.1 vector (Invitrogen, Saint-Aubin, France) opened with Not I and Apa I sites. Negative control plasmids (vector and pc-NC) were generated with the scrambled control sequences in the same way. Knockdown of circHIPK3 and MMP-16 was accomplished by RNA interference using siRNA-circHIPK3 (si-circHIPK3, 5′-AUCUCGGUACUACAGGUAUGG-3′) and siRNA-MMP-16 (si-MMP-16, 5′-UCACUAUAUCUCCAAUAUCUG-3′), and a nontarget siRNA (si-NC, 5′-AAGACAUUGUGUGUCCGCCTT-3′) served as the control. Chemically modified miR-375 mimic (5′-UUUGUUCGUUCGGCUCGCGUGA-3′) and miRNA antisense oligonucleotide (miR-375-inhibitor, 5′-UCACGCGAGCCGAACGAACAAA-3′) were applied to alter miR-375 expression, and scrambled oligonucleotides (miR-NC mimic, 5′-UUCUCCGAACGUGUCACGUTT-3′ and miR-inhibitor, 5′-UGAGCUGCAUAGAGUAGUGAUUA-3′) served as their counterparts. All oligonucleotides were purchased from Ribobio (Guangzhou, China).
HASMCs were plated (100,000 cells/well) on 12-well culture dishes overnight and then transfected using Lipofectamine 3000 (Invitrogen) with 200 ng of plasmid, 100 nM of siRNA, or 50 nM of miRNA mimic or inhibitor, as per the accompanying guidance. After 8 h of transfection, media were replaced by the fresh growth medium, and cells were harvested 24 h later.
Actinomycin D treatment
Actinomycin D treatment was carried out to evaluate the stability of circHIPK3. Briefly, HASMCs were treated with 1 µM of Actinomycin D (Sigma-Aldrich, Toyko, Japan) for the indicated time points (0, 4, 8, 12 and 24 h) at 37 °C. Then, total RNA was isolated from the treated cells to gauge circHPIK3 and HIPK3 linear mRNA levels by quantitative real-time polymerase chain reaction (qRT-PCR) as below.
RNA preparation and Ribonuclease R (RNase R) assay
RNA preparation was done from cultured HASMCs using the RNeasy Mini Kit as per the manufacturing protocols (Qiagen, Valencia, CA, USA). RNA samples were quantified by a spectrophotometer and a total of 3 µg RNA from HASMCs was used for the assessment of the RNase R resistance of circHIPK3 by RNase R assay. Briefly, 3 µg RNA was incubated with 10 U of RNase R for 10 min at 37 °C as recommended by the manufacturers (Epicenter Technologies, Madison, WI, USA). Afterward, RNA was purified, and the levels of circHPIK3 and HIPK3 linear mRNA were gauged by qRT-PCR as below.
qRT-PCR
The levels of circHIPK3, mRNAs, and miR-375 were gauged by qRT-PCR under standard methods. For circHIPK3 and mRNAs analyses, cDNA was synthesized from 1 µg RNA samples using SuperScript III (Invitrogen) and random hexamer primers (Invitrogen); cDNA was subsequently analyzed in triplicate by qRT-PCR with SYBR Green (Invitrogen) and sequence-specific primers (synthesized by BGI) for circHIPK3 (5′-GGCAGCCTTACAGGGTTAAAG-3′-forward and 5′-GGGTAGACCAAGACTTGTGAGG-3′-reverse), HIPK3 (5′-GTCATAGCAGCTCAGGCACA-3′-forward and 5′-ACTCCTCACTCTTGCGCTTC-3′-reverse), MMP-16 (5′-TGAGCCATGGACACTAGGAAA-3′-forward and 5′-GGAGGAATAGAGCGGTGTGG-3′-reverse), or β-actin (5′-CTCGCCTTTGCCGATCC-3′-forward and 5′-GGGGTACTTCAGGGTGAGGA-3′-reverse). For miR-375 quantification, cDNA was prepared using TaqMan RT Kit (Applied Biosystems, Paisley, UK); cDNA was then analyzed in triplicate by qRT-PCR with TaqMan MiRNA Assay Kit (Applied Biosystems) and specific primers (BGI) for miR-375 (5′-GCCGAGTTTGTTCGTTCGGC-3′-forward and 5′-CTCAACTGGTGTCGTGGA-3′-reverse) or U6 (5′-CTCGCTTCGGCAGCACA-3′-forward and 5′-AACGCTTCACGAATTTGCGT-3′-reverse). The β-actin or U6 was included as a reference gene since they were constantly expressed in HASMCs. The comparative CT method (2−ΔΔCt) (Jaca et al. 2017) was used to determined relative expression.
Cell viability and proliferation assay
Cell viability and proliferation were examined by the Cell Counting Kit-8 (CCK-8) assay. For viability analysis, HASMCs (5000 cells/well) in 96-well culture dishes were exposed to various concentrations (0, 10, 20 and 30 ng/mL) of PDGF (Sigma-Aldrich) for 48 h. For proliferation analysis, transfected or untransfected HASMCs (2000 cells/well) were treated with 30 ng/mL of PDGF for the indicated time points (0, 24, 48 and 72 h) in triplicate. In both analyses, CCK-8 solution (10 µL, Beyotime, Shanghai, China) was added into each well and incubated at 37 °C for 4 h. The number of viable cells was proportional to the absorbance at 450 nm.
Flow cytometry
Cell apoptosis was evaluated by flow cytometry with the Apoptosis Assay Kit (BD Biosciences, Oxford, UK). In brief, transfected or untransfected HASMCs (50,000) were treated with 30 ng/mL of PDGF for 48 h. Subsequently, treated HASMCs were washed with ice phosphate buffered saline (Beyotime), suspended in 200 µL of binding buffer, and stained with 5 µL of fluorescein isothiocyanate (FITC)-labeled Annexin V and 10 µL of propidium iodide (PI) as per the instructions of the Apoptosis Assay Kit. A total of 10,000 cells per sample were analyzed using a FACScan flow cytometer with AccuriC6 software (BD Biosciences). Stained cells were designated as early (Annexin V+) and late (Annexin V+/PI+) apoptotic cells.
Wound-healing assay
A wound-healing assay was used to detect cell migration. In brief, transfected or untransfected HASMCs were exposed to 30 ng/mL of PDGF for 48 h. After that, treated cells were allowed to reach ~ 95 % confluence in 6-well culture dishes. An artificial wound was created with a 200 µL sterile tip in the cell monolayer. Images were captured at 0 and 24 h, and migrated distance was analyzed using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Transwell migration and invasion assays
Cell invasion and migration abilities were assessed by transwell assays. Transfected or untransfected HASMCs were stimulated with 30 ng/mL of PDGF for 48 h. For the migration experiments, treated cells in 200 µL serum-free medium were seeded in upper chambers of 24-Transwell inserts with 8 μm pores (BD Biosciences) at 50,000 cells per well. For the invasion experiments, treated cells were plated at 100,000 cells each well in upper chambers of 24-Transwell with Matrigel-coated inserts (BD Biosciences). Lower chamber was filled with complete growth medium. After 24 h, migratory or invasive cells located on the lower side of the chamber were scored using a microscope (Olympus, Shinjuku, Japan) at 100× magnification after being stained with crystal violet (0.5 %, Beyotime).
Western blot
Treated HASMCs were suspended in 1 % SDS Laemmli buffer (Sigma-Aldrich), sonicated 20 times at 30 s intervals, mixed with 1 × β-mercaptoethanol (Sigma-Aldrich), and then boiled for 5 min. Samples (30 µg per lane) were resolved on a 10 % SDS-polyacrylamide gel electrophoresis, and the resulting gel was electroblotted onto nitrocellulose membranes (Amersham, Buckinghamshire, UK). For immunoblotting, the membranes were probed with anti-proliferating cell nuclear antigen (anti-PCNA, ab92552; dilution 1:5,000), anti-MMP-3 (ab52915; dilution 1:5,000), anti-B-cell lymphoma 2 associated X (anti-Bax, ab182734; dilution 1:1,000), anti-MMP-16 (ab137322; dilution 1:2,000) and anti-GAPDH (ab181602; dilution 1:10,000) primary antibodies, and horseradish peroxidase-conjugated IgG secondary antibody (ab205718; dilution 1:10,000; All from Abcam, Cambridge, UK). Signals were developed using the enhanced chemiluminescent as described by the manufacturers (Amersham). Densitometric quantification of bands was carried out with Multi-Gauge software (Fujifilm, Tokyo, Japan).
Bioinformatics and dual-luciferase reporter assay
Bioinformatic analysis for the targeted miRNAs of circHIPK3 was done using the computer algorithm circInteractome (https://circinteractome.nia.nih.gov/). Analysis for miR-375 targets was conducted with starBase v.3 software (http://starbase.sysu.edu.cn/). The segments of circHIPK3 and MMP-16 3’UTR encompassing the predicted miR-375 complementary sites or mutated target sites were synthesized by BGI and cloned into the pMIR-REPORT vector (Invitrogen) downstream of the firefly luciferase coding region. HASMCs (100,000 cells) were co-transfected with 50 nM of miR-375 mimic or miR-NC mimic, 50 ng of pRL-TK control plasmid expressing Renilla luciferase (Promega, Beijing, China), and each reporter constructs at 200 ng. The cells were harvested after 36 h and assayed for luciferase activity using the Glomax 96 microplate luminometer (Promega).
RNA immunoprecipitation (RIP) assay
HASMCs were homogenized in RIPA lysis buffer as per the manufacturing guidance (Beyotime). Cell lysates were incubated with Argonaute 2 (Ago2, ab233727; dilution 1:50; Abcam) or isotype control IgG (ab172730; dilution 1:100; Abcam) antibody at 4 °C for 6 h prior to Protein A/G magnetic beads incubation for 3 h. Beads were collected and bound RNA was prepared to gauge the expression of circHIPK3, miR-375 and MMP-16 by qRT-PCR as above.
Statistical analysis
All experiments were done three independent biological replicates and data were expressed as mean ± standard deviation. A two-sided Student’s t-test was used for comparisons between two groups, and Tukey’s post hoc test was applied for analysis of three or more groups after analysis of variance. Differences at p-value under 0.05 were defined as statistical significance.
Results
PDGF induced circHIPK3 expression in HASMCs
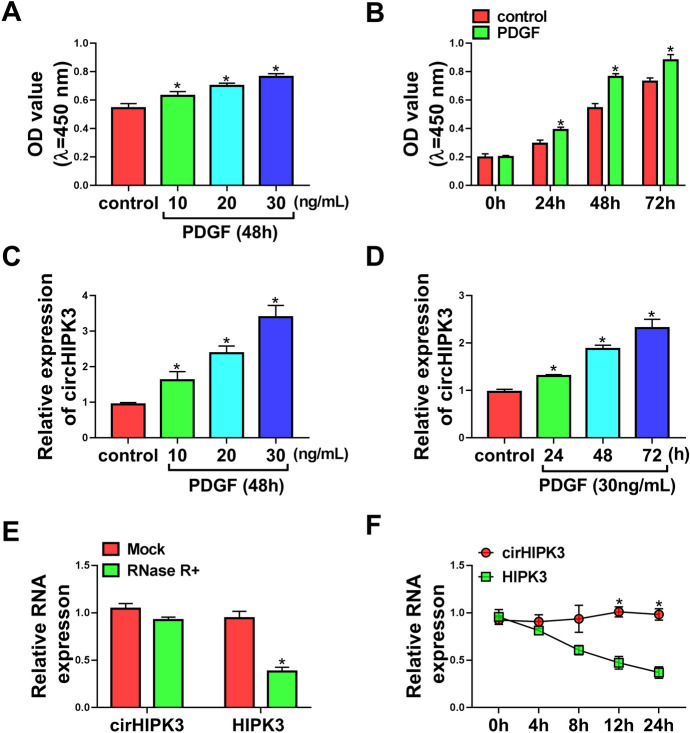
We firstly used CCK-8 assay to examine the influence of PDGF on cell viability and proliferation of HASMCs. In contrast to the control group, PDGF treatment led to a remarkable promotion in cell viability (Fig. 1a) and proliferation (Fig. 1b). We then evaluated whether PDGF could affect the expression of circHIPK3 in HASMCs by qRT-PCR assay. Notably, PDGF resulted in increased expression of circHIPK3 in dose- and time-dependent manners (Fig. 1c and 1d). To confirm the RNase R resistance of circHIPK3, we adopted RNase R assays by incubating total RNA with RNase R. As shown in Fig. 1e, RNase R treatment caused a reduction in the level of HIPK3 linear mRNA, and circHIPK3 was refractory to RNase R (Fig. 1e), indicating that circHIPK3 had a higher tolerance to exonucleases. To verify the stability of circHIPK3, we treated HASMCs with Actinomycin D and gauged circHIPK3 and HIPK3 linear mRNA levels by qRT-PCR. Incubation of HASMCs with Actinomycin D led to a striking reduction in the level of HIPK3 linear mRNA, and circHIPK3 expression did not reduce in the assayed time frame (Fig. 1f), demonstrating that circHIPK3 was unusually stable and its level was not affected by Actinomycin D. These results together indicated that circHIPK3 was an indeed circular transcript and its expression was increased in PDGF-treated HASMCs.
Fig. 1.
PDGF increased the expression of circHIPK3 in HASMCs. HASMCs were treated with various concentrations (0, 10, 20 and 30 ng/mL) of PDGF for 48 h or 30 ng/mL of PDGF for the indicated time points (0, 24, 48 and 72 h), followed by the assessment of cell proliferation by CCK-8 assay (a and b), circHIPK3 expression by qRT-PCR (c and d). e RNase R assays were done in HASMCs. f Actinomycin D assays were performed in HASMCs. *P < 0.05
Silencing of circHIPK3 counteracted PDGF-mediated pro-proliferation, pro-migration, pro-invasion, and anti-apoptosis effects in HASMCs
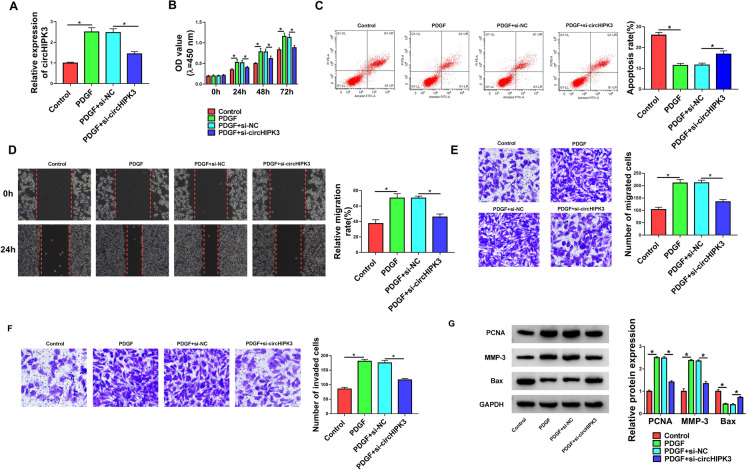
To gain insight into the biological role of circHIPK3 in PDGF-induced cell dysfunction, we carried out “phenocopy” silencing by siRNA targeting circHIPK3 (si-circHIPK3) in PDGF-treated HASMCs. Transient transfection of si-circHIPK3, but not the si-NC control sequence, remarkably decreased the expression of circHIPK3 in PDGF-treated HASMCs (Fig. 2a). CCK-8 assay showed that the reduced level of circHIPK3 significantly abolished PDGF-driven promotion of proliferation of HASMCs compared with the controls (Fig. 2b). We then used flow cytometry to assess the effect of circHIPK3 knockdown on cell apoptosis. By contrast, the down-regulation of circHIPK3 reversed PDGF-mediated HASMC apoptosis suppression (Fig. 2c). Using wound-healing and transwell assays, we found that the reduced expression of circHIPK3 counteracted PDGF-induced migration promotion (Fig. 2d and e) and invasion enhancement (Fig. 2f) in HASMCs. Moreover, the silencing of circHIPK3 abrogated the alteration of PDGF on PCNA, MMP-3 and pro-apoptotic protein Bax expression in HASMCs, which was confirmed by western blot analysis (Fig. 2g). All these findings suggested that the effects of PDGF might be in part due to the up-regulation of circHIPK3.
Fig. 2.
CircHIPK3 knockdown hampered PDGF-mediated pro-proliferation, pro-migration, pro-invasion, and anti-apoptosis effects in HASMCs. HASMCs were treated with control, PDGF (30 ng/mL), or transfected with si-NC or si-circHIPK3 before PDGF treatment. a Relative circHIPK3 expression was detected by qRT-PCR. b Cell proliferation was assessed by CCK-8 assay. c Cell apoptosis was determined by flow cytometry. d and e Cell migration was evaluated by wound-healing and transwell assays. f Cell invasion was measured by transwell assay. g The levels of PCNA, MMP-3, and Bax were gauged by western blot. *P < 0.05
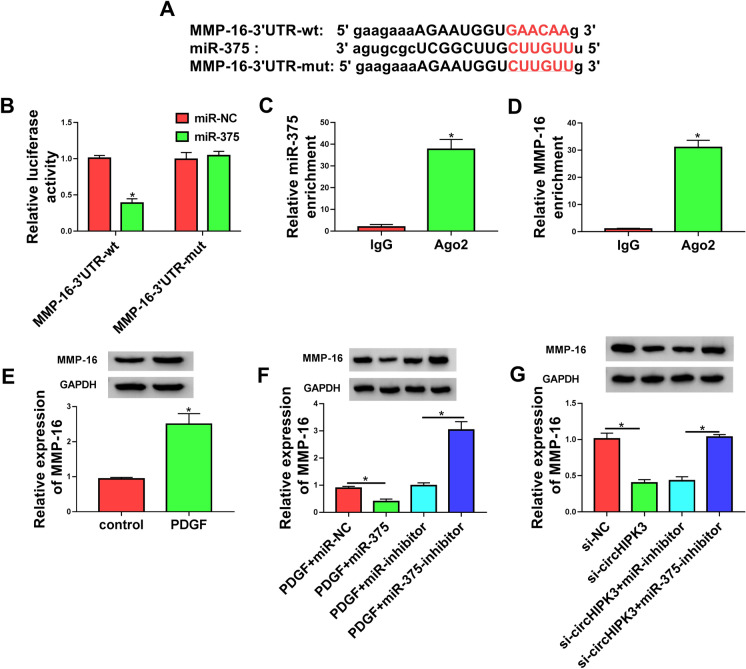
CircHIPK3 directly targeted miR-375
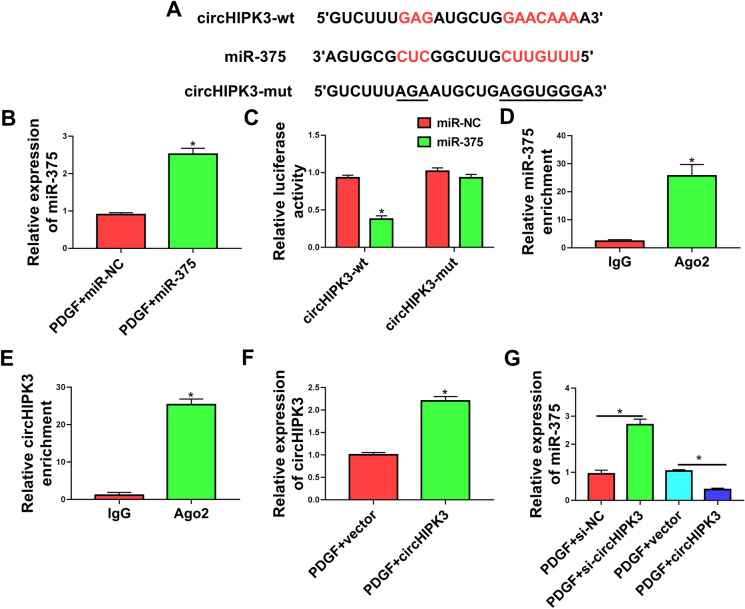
To further elucidate the mechanism by which circHIPK3 regulated PDGF-induced cell dysfunction, we performed a detailed analysis of its targeted miRNAs. Computer algorithm circInteractome predicted that circHIPK3 harbored a putative miR-375 pairing sequence (Fig. 3a). To validate this possibility, we adopted dual-luciferase reporter assays in HASMCs by co-transfecting circHIPK3 wild-type (circHIPK3-wt) or mutant-type (circHIPK3-mut) luciferase reporter construct and miR-375 mimic. The miR-375 overexpression efficacy of miR-375 mimic was validated by qRT-PCR (Fig. 3b). Remarkably, the overexpression of miR-375 inhibited the luciferase activity of circHIPK3-wt compare with the control group (Fig. 3c). However, mutation of the putative miR-375 binding sites in the reporter plasmid (circHIPK3-mut) abolished the inhibition of miR-375 on reporter gene expression (Fig. 3c). MiRNAs silence gene expression in RNA-induced silencing complexes (RISCs) that also contain Ago2 protein (Iwakawa and Tomari 2015). We thus performed RIP experiments by incubating cell lysates with the Ago2 antibody and magnetic beads. Interestingly, the enrichment levels of miR-375 and circHIPK3 were synchronously augmented by Ago2 antibody (Fig. 3d and e), suggesting a possibility for the relationship between circHIPK3 and miR-375 in HASMCs. To analyze whether circHIPK3 could influence the expression of miR-375 in PDGF-treated HASMCs, we manipulated circHIPK3 expression by using circHIPK3 overexpressing plasmid and si-circHIPK3. The effectiveness of circHIPK3 overexpressing plasmid in elevating circHIPK3 expression was verified by qRT-PCR (Fig. 3f). As expected, the reduced level of circHIPK3 by si-circHIPK3 transfection resulted in increased expression of endogenous miR-375, and the overexpression of circHIPK3 by circHIPK3 overexpressing plasmid transfection decreased miR-375 expression in PDGF-treated HASMCs (Fig. 3g). Taken together, these data demonstrated that circHIPK3 targeted miR-375 by binding to miR-375.
Fig. 3.
CircHIPK3 directly targeted miR-375. a Sequence of miR-375, the putative miR-375 binding sites in circHIPK3 and the mutation of the target region. b Relative miR-375 expression was assessed by qRT-PCR. c Dual-luciferase reporter assays were performed in HASMCs with circHIPK3 wild-type (circHIPK3-wt) or mutant-type (circHIPK3-mut) luciferase reporter plasmid. d and e RIP assays were done in HASMCs with Ago2 or IgG antibody. f Relative expression of circHIPK3 was gauged by qRT-PCR in PDGF-treated HASMCs transfected with vector or circHIPK3 overexpressing plasmid (circHIPK3). g The level of miR-375 was examined by qRT-PCR in PDGF-treated HASMCs transfected with si-circHIPK3, si-NC, vector or circHIPK3 overexpressing plasmid (circHIPK3). *P < 0.05
MiR-375 was a downstream mediator of circHIPK3 in modulating PDGF-induced cell dysfunction
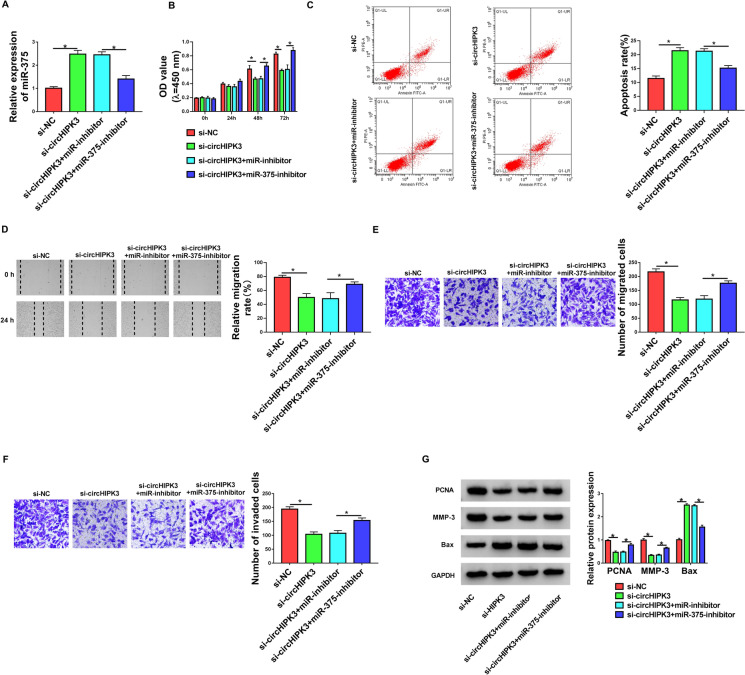
Having established that circHIPK3 depletion resulted in increased expression of miR-375, we then asked whether the effects of circHIPK3 silencing were due to the elevation of miR-375. To address this possibility, we reduced the expression of miR-375 with miR-375 inhibitor (miR-375-inhibitor) in the presence of si-circHIPK3 in PDGF-treated HASMCs (Fig. 4a). Indeed, the reduced expression of miR-375 abrogated circHIPK3 silencing-mediated anti-proliferation (Fig. 4b), pro-apoptosis (Fig. 4c), anti-migration (Fig. 4d and e), and anti-invasion (Fig. 4f) effects in PDGF-treated HASMCs. Additionally, the down-regulation of miR-375 reversed circHIPK3 silencing-driven the expression alteration of PCNA, MMP-3 and Bax in PDGF-treated HASMCs (Fig. 4g). These findings collectively supported our hypothesis that the effects of circHIPK3 depletion might be at least in part due to the up-regulation of miR-375.
Fig. 4.
CircHIPK3 silencing regulated cell proliferation, migration, invasion, and apoptosis by miR-375 in PDGF-treated HASMCs. HASMCs were transfected with si-circHIPK3, si-NC, si-circHIPK3 + miR-inhibitor or si-circHIPK3 + miR-375-inhibitor and then were treated with 30 ng/mL of PDGF for the indicated time points. a Relative expression of miR-375 in treated cells was detected by qRT-PCR. b Cell proliferation was gauged by CCK-8 assay. c Cell apoptosis was assessed by flow cytometry. d and e Cell migration was evaluated by wound-healing and transwell assays. f Cell invasion was determined by transwell assay. g Western blot was performed to measure PCNA, MMP-3 and Bax expression. *P < 0.05
CircHIPK3 controlled the expression of MMP-16 through miR-375
Next, we undertook to identify the downstream effectors of miR-375. Using starBase v.3 software, a predicted miR-375 binding sequence was found in the 3’UTR of MMP-16 (Fig. 5a). To confirm this finding, we generated MMP-16 3’UTR wild-type (MMP-16-3’UTR-wt) or mutant (MMP-163-’UTR-mut) luciferase reporter constructs and analyzed them by luciferase activity. Co-transfection of MMP-16-3’UTR-wt and miR-375 mimic into the HASMCs produced lower luciferase activity than cells cotransfected with miR-NC mimic but MMP-16-3’UTR-mut markedly abolished the repressive effect of miR-375 (Fig. 5b), demonstrating the viability of the binding sites for interaction. Furthermore, RIP experiments revealed that the enrichment levels of miR-375 and MMP-16 were simultaneously elevated by Ago2 antibody (Fig. 5c and 5d), implying the possibility that MMP-16 was a direct target of miR-375 in HASMCs. Additionally, in HASMCs, western blot showed that PDGF treatment remarkably increased the level of MMP-16 protein (Fig. 5e). To elucidate whether miR-375 operated as a post-transcriptional regulator of MMP-16, we manipulated miR-375 expression with miR-375 mimic or miR-375-inhibitor in PDGF-treated HASMCs. Notably, MMP-16 protein level was reduced by miR-375 overexpression and elevated as a result of miR-375 depletion in PDGF-treated HASMCs (Fig. 5f), reinforcing that MMP-16 was directly targeted and regulated by miR-375.
Fig. 5.
CircHIPK3 acted as a post-transcriptional modulator of MMP-16 expression through miR-375. a Sequence of miR-375, the putative miR-375 binding sites in the 3’UTR of MMP-16 and the mutation of the target region. b Dual-luciferase reporter assays were performed in HASMCs with MMP-16 3’UTR wild-type (MMP-163-’UTR-wt) or mutant (MMP-163-’UTR-mut) luciferase reporter constructs. c and d RIP assays were done in HASMCs with Ago2 or IgG antibody. e MMP-16 protein level was detected by western blot in PDGF-treated HASMCs transfected with miR-NC mimic, miR-375 mimic, miR-inhibitor or miR-375-inhibitor. f The level of MMP-16 protein was measured by western blot in HASMCs treated with PDGF. g Western blot was done to gauge the level of MMP-16 protein in PDGF-treated HASMCs transfected with si-NC, si-circHIPK3, si-circHIPK3 + miR-inhibitor or si-circHIPK3 + miR-375-inhibitor. *P < 0.05
Given our data that miR-375 directly bound to circHIPK3 and MMP-16, we decided to examine whether circHIPK3 could modulate MMP-16 expression. As wound be expected, the silencing of circHIPK3 led to a significant decrease in the level of MMP-16 protein in PDGF-treated HASMCs, and this effect was prominently abrogated by miR-375 down-regulation (Fig. 5g). Together, these observations strongly pointed to the role of circHIPK3 as a post-transcriptional modulator of MMP-16 expression through miR-375.
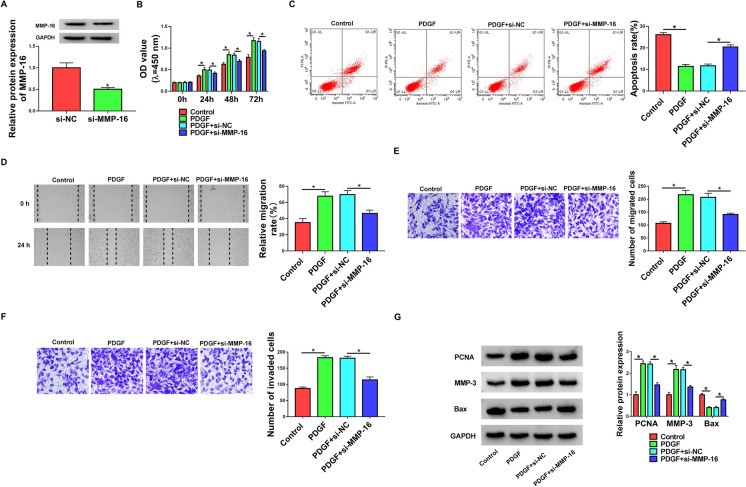
Silencing of MMP-16 reversed PDGF-mediated pro-proliferation, anti-apoptosis, pro-migration, and pro-invasion effects in HASMCs
Further, we decided to address the role of MMP-16 in controlling HASMC dysfunction induced by PDGF. The transfection of si-MMP-16 markedly inhibited the level of MMP-16 protein in PDGF-treated HASMCs (Fig. 6a), which expressed high level of MMP-16. In line with the effects of circHIPK3 silencing, the down-regulation of MMP-16 significantly abolished PDGF-mediated enhancement of proliferation (Fig. 6b), and repression of apoptosis (Fig. 6c), as well as promotion of migration (Fig. 6d and e) and invasion (Fig. 6 f) in HASMCs. Furthermore, the reduced level of MMP-16 reversed PDGF-mediated alteration of PCNA, MMP-3 and Bax levels in HASMCs (Fig. 6g). All these results demonstrated that MMP-16 knockdown abolished PDGF-induced HASMC dysfunction.
Fig. 6.
MMP-16 knockdown controlled proliferation, migration, invasion and apoptosis in PDGF-treated HASMCs. HASMCs were transfected with si-NC or si-MMP-16 and then treated with 30 ng/mL of PDGF for the indicated time points. a Relative level of MMP-16 protein in treated cells was detected by western blot. b Cell proliferation was measured by CCK-8 assay. c Cell apoptosis was gauged by flow cytometry. d and e Cell migration was determined by wound-healing and transwell assays. f Cell invasion was assessed by transwell assay. g Western blot was carried out to measure PCNA, MMP-3 and Bax expression. *P < 0.05
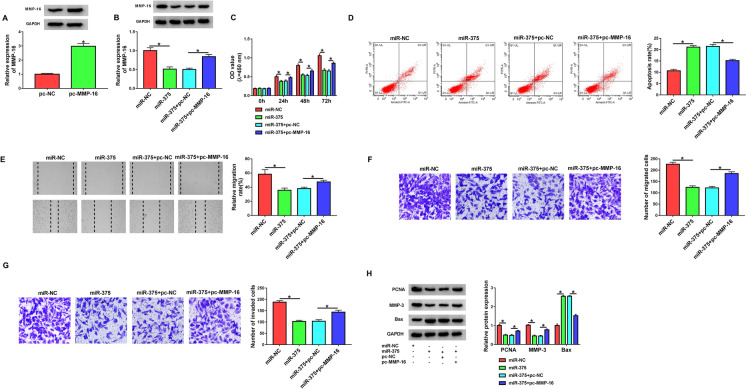
MiR-375-mediated inhibition of MMP-16 impacted cell dysfunction induced by PDGF
To directly elucidate whether inhibition of MMP-16 by miR-375 was responsible for the regulation of miR-375 in PDGF-induced cell dysfunction, we co-transfected MMP-16 overexpression plasmid in the presence of miR-375 mimic in HASMCs under PDGF treatment. The MMP-16 overexpression efficacy of MMP-16 overexpression plasmid was verified by western blot (Fig. 7a). By contrast, co-transfection of MMP-16 overexpression plasmid remarkably reversed miR-375 mimic-driven suppression of MMP-16 expression of PDGF-treated HASMCs (Fig. 7b). Remarkably, the enforced expression of miR-375 repressed cell proliferation (Fig. 7c), promoted apoptosis (Fig. 7d), as well as impeded migration (Fig. 7e, f) and invasion (Fig. 7g) in PDGF-treated HASMCs. Also, the overexpression of miR-375 elevated Bax expression and reduced the levels of PCNA and MMP-3 in HASMCs under PDGF treatment (Fig. 7h). Furthermore, these effects of miR-375 overexpression were significantly abrogated by the restoration of MMP-16 in PDGF-treated HASMCs (Fig. 7c–h). Collectively, these findings demonstrated that miR-375 overexpression regulated PDGF-induced HASMC dysfunction by down-regulating MMP-16.
Fig. 7.
MiR-375-mediated inhibition of MMP-16 suppressed proliferation, migration, invasion, and promoted apoptosis in PDGF-treated HASMCs. HASMCs were transfected with pc-NC, pc-MMP-16, miR-NC mimic, miR-375 mimic, miR-375 mimic + pc-NC or miR-375 mimic + pc-MMP-16 and then treated with 30 ng/mL of PDGF for the indicated time points. a and b Western blot was performed to gauge the level of MMP-16 protein in treated cells. c Cell proliferation was detected by CCK-8 assay. d Cell apoptosis was measured by flow cytometry. e and f Cell migration was evaluated by wound-healing and transwell assays. g Cell invasion was gauged by transwell assay. h The levels of PCNA, MMP-3 and Bax were determined by western blot. *P < 0.05
Discussion
Misrergulated circRNAs are being discovered to have relevance to human diseases, including asthma (Marques-Rocha et al. 2015; Bao et al. 2020). Understanding of the actions of circRNAs in regulating AMSC migration and proliferation would offer a potential therapeutic strategy to ameliorate asthma. In this paper, we treated HASMCs with PDGF to induce cell migration and proliferation, as had been reported previously (Tang and Luo 2018; Ji et al. 2018). In agreement with recent work (Lin et al. 2020), our data supported that the silencing of circHIPK3 impeded ASMC migration and proliferation induced by PDGF. Importantly, we provided a new molecular determinant for the modulation of circHIPK3 in PDGF-treated HASMCs. Such exploration was hampered at present by the lack of in vivo analysis using the animal models of asthma.
MiR-375 has been generally accepted as a tumor suppressor in various malignancies, such as head and neck squamous cell carcinoma, glioblastoma, and nasopharyngeal carcinoma (Jimenez et al. 2017; Li et al. 2019; Jia-Yuan et al. 2020). MiR-375 was also reported to be associated with the pathogenesis of acute myocardial infarction and type 2 diabetes (Ali Sheikh 2020; García-Jacobo et al. 2019). Moreover, recent studies uncovered the involvement of miR-375 in inflammation-related diseases, such as osteoarthritis and acute pancreatitis (Zou et al. 2019; Zhao et al. 2020b). More interestingly, the enforced expression of miR-375 hindered HASMC migration and proliferation induced by PDGF or cytokine (Ji et al. 2018; Zhao et al. 2020a). In this paper, we first showed that circHIPK3 directly targeted miR-375, and miR-375 was a downstream mediator of circHIPK3 function.
Using mRNA-target predicting starBase v.3 software, MMP-16 seemed to be a strong candidate for the miR-375 targets because MMP-16 was reported to enhance ASMC proliferation (Lou et al. 2020). Importantly, miR-192-5p attenuated inflammation and airway remodeling in asthma mice at least partially by targeting MMP-16 (Lou et al. 2020). We subsequently confirmed the targeted relationship between MMP-16 and miR-375. Moreover, miR-375-mediated inhibition of MMP-16 impeded PDGF-induced migration and proliferation of HASMCs, which was consistent with the effect of si-MMP-16 transfection. More importantly, we first pointed to the role of circHIPK3 as a post-transcriptional modulator of MMP-16 expression through miR-375, suggesting the miR-375/MMP-16 axis as an importantly downstream effector of circHIPK3 function. Recent work by Lin et al. had led to the identification of circHIPK3 that controlled ASMC migration and proliferation induced by PDGF via regulating STIM1 expression by targeting miR-326 (Lin et al. 2020). These findings suggest that there may be other miRNA/mRNA regulatory mechanisms that remain to be elucidated in the modulation of circHIPK3.
To conclude, the present work identified the miR-375/MMP-16 axis as a new mechanism for the regulation of circHIPK3 in PDGF-induced migration and proliferation in HASMCs. Knowing the mechanisms of action of asthma-related circRNAs will be of the essence in establishing the molecular targeted therapeutics against asthma.
Authors’ Contribution
XG designed the study. YJ conducted the experiments and drafted the manuscript. JQ collected and analyzed the data, edited the manuscript. All authors read and approved the final manuscript.
Funding
None.
Data Availability
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The research was authorized by the Ethics Committee of the NO. 2 Hospital of Baoding and was carried out according to the guidelines of Declaration of Helsinki. Each participant signed a written informed consent before conducting this study.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ali Sheikh MS. Overexpression of miR-375 Protects Cardiomyocyte Injury following Hypoxic-Reoxygenation Injury. Oxid Med Cell Longev. 2020;2020:7164069. doi: 10.1155/2020/7164069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao H, Zhou Q, Li Q, Niu M, Chen S, Yang P, et al. Differentially expressed circular RNAs in a murine asthma model. Mol Med Rep. 2020;22:5412–5422. doi: 10.3892/mmr.2020.11617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Jacobo RE, Uresti-Rivera EE, Portales-Pérez DP, González-Amaro R, Lara-Ramírez EE, Enciso-Moreno JA, et al. Circulating miR-146a, miR-34a and miR-375 in type 2 diabetes patients, pre-diabetic and normal-glycaemic individuals in relation to β-cell function, insulin resistance and metabolic parameters. Clin Exp Pharmacol Physiol. 2019;46:1092–1100. doi: 10.1111/1440-1681.13147. [DOI] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- Huang Z, Cao Y, Zhou M, Qi X, Fu B, Mou Y, et al. Hsa_circ_0005519 increases IL-13/IL-6 by regulating hsa-let-7a-5p in CD4(+) T cells to affect asthma. Clin Exp Allergy. 2019;49:1116–1127. doi: 10.1111/cea.13248. [DOI] [PubMed] [Google Scholar]
- Huang Z, Fu B, Qi X, Xu Y, Mou Y, Zhou M, et al. Diagnostic and Therapeutic Value of Hsa_circ_0002594 for T Helper 2-Mediated Allergic Asthma. Int Arch Allergy Immunol. 2021;182:388–398. doi: 10.1159/000511612. [DOI] [PubMed] [Google Scholar]
- Iwakawa HO, Tomari Y. The Functions of MicroRNAs: mRNA Decay and Translational Repression. Trends Cell Biol. 2015;25:651–665. doi: 10.1016/j.tcb.2015.07.011. [DOI] [PubMed] [Google Scholar]
- Jaca A, Govender P, Locketz M, Naidoo R. The role of miRNA-21 and epithelial mesenchymal transition (EMT) process in colorectal cancer. J Clin Pathol. 2017;70:331–356. doi: 10.1136/jclinpath-2016-204031. [DOI] [PubMed] [Google Scholar]
- James AL, Noble PB, Drew SA, Mauad T, Bai TR, Abramson MJ, et al. Airway smooth muscle proliferation and inflammation in asthma. J Appl Physiol. 2018;125:1090–1096. doi: 10.1152/japplphysiol.00342.2018. [DOI] [PubMed] [Google Scholar]
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Yang X, Su H. Overexpression of microRNA-375 impedes platelet-derived growth factor-induced proliferation and migration of human fetal airway smooth muscle cells by targeting Janus kinase 2. Biomed Pharmacother. 2018;98:69–75. doi: 10.1016/j.biopha.2017.12.012. [DOI] [PubMed] [Google Scholar]
- Jia-Yuan X, Wei S, Fang-Fang L, Zhi-Jian D, Long-He C, Sen L. miR-375 inhibits the proliferation and invasion of nasopharyngeal carcinoma cells by suppressing PDK1. Biomed Res Int. 2020;2020:9704245. doi: 10.1155/2020/3595402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez L, Lim J, Burd B, Harris TM, Ow TJ, Kawachi N, et al. miR-375 Regulates Invasion-Related Proteins Vimentin and L-Plastin. Am J Pathol. 2017;187:1523–1536. doi: 10.1016/j.ajpath.2017.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TL, Neville DM, Chauhan AJ. Diagnosis and treatment of severe asthma: a phenotype-based approach. Clin Med (Lond) 2018;18:s36–s40. doi: 10.7861/clinmedicine.18-2-s36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GF, Cheng YY, Li BJ, Zhang C, Zhang XX, Su J, et al. miR-375 inhibits the proliferation and invasion of glioblastoma by regulating Wnt5a. Neoplasma. 2019;66:350–356. doi: 10.4149/neo_2018_180714N484. [DOI] [PubMed] [Google Scholar]
- Lin J, Feng X, Zhang J. Circular RNA circHIPK3 modulates the proliferation of airway smooth muscle cells by miR-326/STIM1 axis. Life Sci. 2020;255:117835. doi: 10.1016/j.lfs.2020.117835. [DOI] [PubMed] [Google Scholar]
- Lou L, Tian M, Chang J, Li F, Zhang G. MiRNA-192-5p attenuates airway remodeling and autophagy in asthma by targeting MMP-16 and ATG7. Biomed Pharmacother. 2020;122:109692. doi: 10.1016/j.biopha.2019.109692. [DOI] [PubMed] [Google Scholar]
- Marques-Rocha JL, Samblas M, Milagro FI, Bressan J, Martínez JA, Marti A. Noncoding RNAs, cytokines, and inflammation-related diseases. FASEB J. 2015;29:3595–3611. doi: 10.1096/fj.14-260323. [DOI] [PubMed] [Google Scholar]
- Michaeloudes C, Sukkar MB, Khorasani NM, Bhavsar PK, Chung KF. TGF-β regulates Nox4, MnSOD and catalase expression, and IL-6 release in airway smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2011;300:L295–L304. doi: 10.1152/ajplung.00134.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papi A, Brightling C, Pedersen SE, Reddel HK. Asthma Lancet. 2018;391:783–800. doi: 10.1016/S0140-6736(17)33311-1. [DOI] [PubMed] [Google Scholar]
- Salter B, Pray C, Radford K, Martin JG, Nair P. Regulation of human airway smooth muscle cell migration and relevance to asthma. Respir Res. 2017;18:156. doi: 10.1186/s12931-017-0640-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Chen R, Lin S, Xie X, Ye H, Zheng F, et al. Association of circular RNAs and environmental risk factors with coronary heart disease. BMC Cardiovasc Disord. 2019;19:223. doi: 10.1186/s12872-019-1191-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Luo L. MicroRNA-20b-5p inhibits platelet-derived growth factor-induced proliferation of human fetal airway smooth muscle cells by targeting signal transducer and activator of transcription 3. Biomed Pharmacother. 2018;102:34–40. doi: 10.1016/j.biopha.2018.03.015. [DOI] [PubMed] [Google Scholar]
- Wang L, Luo T, Bao Z, Li Y, Bu W. Intrathecal circHIPK3 shRNA alleviates neuropathic pain in diabetic rats. Biochem Biophys Res Commun. 2018;505:644–650. doi: 10.1016/j.bbrc.2018.09.158. [DOI] [PubMed] [Google Scholar]
- Zhang JX, Lu J, Xie H, Wang DP, Ni HE, Zhu Y, et al. circHIPK3 regulates lung fibroblast-to-myofibroblast transition by functioning as a competing endogenous RNA. Cell Death Dis. 2019;10:182. doi: 10.1038/s41419-019-1430-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu Q, Liao Q. CircHIPK3: a promising cancer-related circular RNA. Am J Transl Res. 2020;12:6694–6704. [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Shi X, Wang N, Liu C, Wang J. YAP1, targeted by miR-375, enhanced the pro-angiogenesis of airway smooth muscle cells in asthma via STAT3 activation. Cell Cycle. 2020;19:1275–1284. doi: 10.1080/15384101.2020.1746874. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhao SP, Yu C, Xiang KM, Yang MS, Liu ZL, Yang BC. miR-375 Inhibits Autophagy and Further Promotes Inflammation and Apoptosis of Acinar Cells by Targeting ATG7. Pancreas. 2020;49:543–551. doi: 10.1097/MPA.0000000000001536. [DOI] [PubMed] [Google Scholar]
- Zou LX, Yu L, Zhao XM, Liu J, Lu HG, Liu GW, et al. MiR-375 Mediates Chondrocyte Metabolism and Oxidative Stress in Osteoarthritis Mouse Models through the JAK2/STAT3 Signaling Pathway. Cells Tissues Organs. 2019;208:13–24. doi: 10.1159/000504959. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.