Abstract
In recent years, accumulating articles have revealed that long non-coding RNAs (lncRNAs) play crucial roles in ischemic stroke (IS). A previous study found that lncRNA zinc finger antisense 1 (ZFAS1) was down-regulated in IS patients compared with healthy controls. However, the precise function of ZFAS1 in IS and its associated mechanism remain unclear. Cell viability was assessed by cell counting kit-8 (CCK8) assay. Cell apoptosis was analyzed by flow cytometry. Western blot assay and quantitative real-time polymerase chain reaction (qRT-PCR) were conducted to measure protein and RNA expression. The interaction between microRNA-186-5p (miR-186-5p) and ZFAS1 or MCL1 apoptosis regulator, BCL2 family member (MCL1) was confirmed by dual-luciferase reporter assay, RNA-pull down assay and RNA immunoprecipitation (RIP) assay. IS cell model was established through exposing N2a cells to oxygen and glucose deprivation (OGD). OGD exposure restrained the viability and induced the apoptosis of N2a cells. OGD exposure down-regulated the expression of ZFAS1 and up-regulated the level of miR-186-5p in a time-dependent manner. ZFAS1 overexpression alleviated OGD-mediated injury in IS cell model. MiR-186-5p was identified as a direct target of ZFAS1, and OGD-induced injury in IS cell model was attenuated by the silence of miR-186-5p. MiR-186-5p interacted with the 3′ untranslated region (3′UTR) of MCL1 messenger RNA (mRNA). ZFAS1 positively regulated MCL1 mRNA expression by sequestering miR-186-5p in N2a cells. ZFAS1 overexpression-mediated protective effects in IS cell model were partly overturned by the overexpression of miR-186-5p. MCL1 silencing partly counteracted the protective effects mediated by miR-186-5p silencing in IS cell model. In conclusion, ZFAS1 overexpression exerted a protective role in IS cell model to attenuate OGD-induced injury through targeting miR-186-5p/MCL1 axis. ZFAS1/miR-186-5p/MCL1 signaling might be a novel diagnostic marker and promising treatment target for IS patients.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10616-021-00481-4.
Keywords: Ischemic stroke, OGD, ZFAS1, miR-186-5p, MCL1
Introduction
Ischemic stroke (IS) is the most common type of stroke (accounts for 80% of all stroke cases) that is featured by sudden interruption of blood flow in the brain, leading to neurological dysfunction (Hankey 2003; Allen and Bayraktutan 2009; Benjamin et al. 2018). Due to the high mortality and disability rate, IS remains one of the most resource-consuming diseases. Both the environmental and genetic factors affect the incidence of stroke (Hassan and Markus 2000). Understanding the underlying molecular mechanisms of IS might provide effective treatment strategies for IS.
Long non-coding RNAs (lncRNAs) are a class of non-coding RNAs with more than 200 nucleotides in length (Shi et al. 2013). LncRNAs are implicated in multiple important biological processes, including chromosome remodeling, transcriptional activation and post-transcriptional interference (Quinn and Chang 2016). LncRNAs play pivotal roles in brain development, and they are likely associated with brain damage (Barry 2014). Moreover, lncRNAs in central nervous system are involved in the pathology and physiology of neurons (Clark and Blackshaw 2014). Accumulating articles have pointed out the important regulatory roles of lncRNAs in cell apoptosis, inflammation and angiogenesis in IS (Bao et al. 2018). For instance, Bin et al. found that lncRNA ANRIL protected PC-12 cells against oxygen and glucose deprivation (OGD)-mediated damage via miR-127/MCL1 axis (Liu et al. 2019). Zhu et al. found that high expression of MIAT was associated with the stroke severity and infarct volume in IS patients (Zhu et al. 2018). Previous study found that the expression of ZFAS1 was decreased in IS patients compared with healthy subjects (Wang et al. 1993). We aimed to explore the biological function of zinc finger antisense 1 (ZFAS1) in IS using OGD-induced cell model in vitro.
LncRNAs can function as microRNA (miRNA) sponges in human diseases (Qi et al. 2015; Huang 2018; Chen et al. 2018). MiRNAs are a group of short non-coding RNAs with 19-24 nucleotides in length. MiRNAs are implicated in the development of neurons (Diaz et al. 2014). Furthermore, many miRNAs, including miR-134, miR-148b-3p, miR-151b and miR-27b-3p, have been identified as promising markers for acute IS (Zhou et al. 2018; Cheng et al. 2018). Through bioinformatic prediction, miR-186-5p was a possible target of ZFAS1. In this study, we explored the interaction between ZFAS1 and miR-186-5p and their functional association in IS cell model in vitro.
MiRNAs modulate gene expression through directly interacting with messenger RNAs (mRNAs), causing the translational repression or degradation of target mRNAs (Siddeek et al. 2014). Bioinformatic database revealed that MCL1 apoptosis regulator, BCL2 family member (MCL1) was a possible downstream target of miR-186-5p. MCL1 is an important pro-survival protein in many cell types (Mojsa et al. 2014). Moreover, MCL1 acted as the target of many miRNAs (miR-193a-3p, miR-125b, miR-320a) to regulate the progression of multiple cancers (Huang et al. 2018; Wu et al. 2015; Zhou et al.2018). Previous study reported that MCL1 acted as the target of ANRIL/miR-127 axis to protect PC-12 cells against OGD-induced injury (Liu et al. 2019). Here, the molecular mechanism of MCL1 in OGD-induced IS cell model was explored.
IS cell model was established through exposing N2a cells to OGD in vitro. We first analyzed the role of ZFAS1 in IS cell model. Subsequently, the downstream molecules of ZFAS1 were predicted by bioinformatic database, and the working mechanism was verified by rescue experiments. Our study might provide a novel insight to understand the molecular mechanism of IS.
Materials and methods
Cell lines
Mouse neuroblastoma cell lines (Neuro-2a (N2a) and 293T cell lines) were purchased from BeNa Culture Collection (Beijing, China). N2a and 293T cell lines were cultured with Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Carlsbad, CA, USA) added with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin/streptomycin at 37 °C with 5% CO2.
Oxygen and glucose deprivation (OGD) cell model
For OGD exposure, the complete culture medium was replaced with FBS-free and glucose-free DMEM medium, and N2a cells were grown in the anaerobic (5% CO2 and 95% N2) incubator for 24 h (Liu et al. 2019). The control group was cultured in normal DMEM plus 10% FBS under normoxic condition (37 °C, 5% CO2, 21% O2 and 74% N2).
Cell counting kit-8 (CCK8) assay
Transfection or OGD exposure was conducted after N2a cells attaching to the 96-well plates. After different treatment, CCK8 reagent (Dojindo, Tokyo, Japan) was added to the wells to incubate with the viable N2a cells for 2 h. The optical density (OD) value at 450 nm was measured to analyze the viability of N2a cells under different conditions.
Flow cytometry
The Annexin-V Apoptosis Detection Kit (BD Biosciences, San Jose, CA, USA) was utilized to analyze the apoptosis rate. N2a cells were collected and re-suspended in Annexin-V binding buffer. Annexin V-fluorescein isothiocyanate (Annexin V-FITC) and propidium iodide (PI) were simultaneously added to the binding buffer to mark N2a cells for 15 min in a dark room. The apoptosis rate (the percentages of FITC+/PI− N2a cells and FITC+/PI+ N2a cells) was analyzed on a FC-500 flow cytometer (Beckman Coulter, Pasadena, CA, USA).
Western blot assay
Protein samples were prepared using whole cell lysis buffer (Abcam, Cambridge, MA, USA). The concentrations of different protein samples were determined by BCA method using the BCA assay kit (Bio-Rad, Hercules, CA, USA). The proteins (30 μg) were separated by the 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel at 70 V for 120 min and blotted onto the polyvinylidene fluoride (PVDF) membrane (Bio-Rad). 5% skim milk was used to block the membrane for 1 h, and then the membrane was incubated with primary antibodies including anti-B cell leukemia/lymphoma 2 (anti-Bcl-2, ab185002, Abcam), anti-Bcl-2 associated X, apoptosis regulator (anti-Bax, ab32503, Abcam), anti-cleaved caspase-3 (ab49822, Abcam), anti-MCL1 (ab243136, Abcam), anti-cyclin D1 (ab16663, Abcam), anti-insulin-like growth factor 1 (anti-IGF-1, ab223567, Abcam) and anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH, ab181602, Abcam) overnight at 4 °C. The membrane was then washed for three times and incubated with secondary antibody labeled by horseradish peroxidase (HRP) for 2 h at room temperature. The protein bands were visualized using the enhanced chemiluminescent visualization (ECL) system kit (Pierce Biotechnology, Rockford, IL, USA). The quantification was performed by gray analysis using Image J software.
Quantitative real-time polymerase chain reaction (qRT-PCR)
RNA isolation was conducted with TRIzol reagent (Life Technologies, Carlsbad, CA, USA). PCR reaction was performed using SYBR Green PCR Master Mix Kit (Applied Biosystems, Foster City, CA, USA) on Bio-Rad CFX96 (Bio-Rad). GAPDH was used as the reference gene for ZFAS1 and MCL1, and U6 was used as the reference gene for miR-186-5p. The primer sequences were listed as below.
Mouse ZFAS1 forward primer (F; 5′-3′): CTACATTTCCCAGGACGCCA
Mouse ZFAS1 reverse primer (R; 5′-3′): ATCGGGAACAAAGCAAACGC
Mouse miR-186-5p F: GCCGAGCAAAGAATTCTCCT
Mouse miR-186-5p R: CAGTGCAGGGTCCGAGGTAT
Mouse MCL1 F: AACGGGACTGGCTTGTCAAA
Mouse MCL1 R: CTGATGCCGCCTTCTAGGTC
Mouse GAPDH F: CAGTGCCAGGTGAAAATCGC
Mouse GAPDH R: ATCCGTTCACACCGACCTTC
Mouse U6 F: CTCGCTTCGGCAGCACA
Mouse U6 R: AACGCTTCACGAATTTGCG
Cell transfection
ZFAS1 ectopic expression plasmid (ZFAS1) and empty vector (vector), the oligonucleotides of miR-186-5p mimics (miR-186-5p), miR-186-5p inhibitor (anti-miR-186-5p) and their negative controls (miR-NC and anti-NC) were obtained from Genepharma (Shanghai, China). Small interfering (si)RNA targeting MCL1 (si-MCL1), siRNA targeting IGF-1 (si-IGF-1) and si-NC were purchased from Sangon Biotech (Shanghai, China). N2a cells were plated into 6-well plates to settle down. When the confluence reached about 70–80%, Lipofectamine™ 3000 (Invitrogen, Carlsbad, CA, USA) was used to conduct transient transfection. After transfection for 6 h, the supernatant was replaced by fresh complete medium.
Dual-luciferase reporter assay
ZFAS1-miRNA interactions and miR-186-5p-mRNA interactions were predicted through starbase software. Dual-luciferase reporter assay was carried out to confirm the predicted interactions. The partial sequence in ZFAS1 or MCL1 3′untranslated region (3′UTR), containing wild-type putative miR-186-5p-binding sites or the mutant miR-186-5p-binding sites, was inserted to the downstream of the firefly luciferase gene of pGL3 reporter vector (Promega, Madison, WI, USA). 293T cells and N2a cells were co-transfected with miR-NC or miR-186-5p and the re-constructed reporter plasmids. The luciferase activities were measured using the Dual-luciferase reporter assay kit (Promega). The luciferase intensity of firefly was normalized to the luciferase intensity of Renilla.
RNA-pull down assay
All procedures were conducted under RNase-free condition. N2a cells were transfected with miR-186-5p. After transfection for 24 h, Biotin-labeled miR-186-5p (Bio-miR-186-5p) and Bio-NC were incubated with cell lysate. After incubation for 1 h, streptavidin-coupled Dynabeads (Invitrogen) were then incubated with the mixture, and the enrichment of ZFAS1 in the bound complex was analyzed using qRT-PCR.
RNA immunoprecipitation (RIP) assay
Sepharose beads (Bio-Rad, Hercules, CA, USA) were coated with antibody against Argonaute-2 (Ago2) or Immunoglobulin G (IgG), and pre-coated Sepharose beads were incubated with cell extracts. The Sepharose beads were washed for three times, and qRT-PCR was utilized to determine the enrichment of ZFAS1 and miR-186-5p.
Statistical analysis
All data were displayed as mean ± standard deviation. Data analysis was conducted using Student’s t test between two groups and one-way analysis of variance (ANOVA) with Tukey multiple comparison test in multiple groups. P < 0.05 was identified as statistically significant.
Results
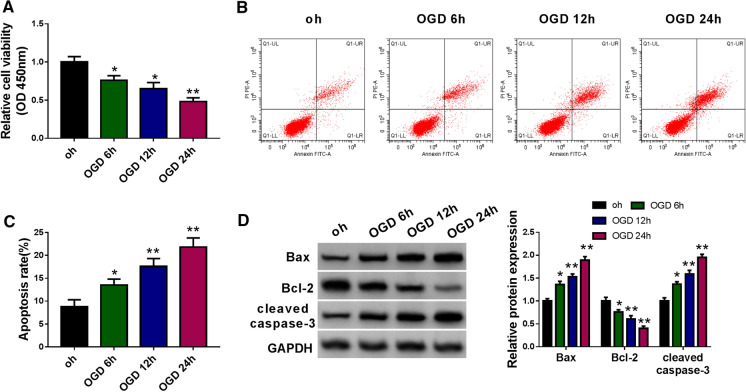
OGD induces the injury of N2a cells
To explore the potential mechanism of ischemic stroke (IS), we established IS cell model through exposing N2a cells to OGD for 0 h, 6 h, 12 h or 24 h. Cell viability and apoptosis were assessed by CCK8 assay and flow cytometry, respectively. OGD exposure reduced cell viability in a time-dependent manner (Fig. 1A). Also, OGD treatment time-dependently elevated the apoptosis rate of N2a cells (Fig. 1B and C). Western blot assay was conducted to detect the expression of apoptosis-related proteins to further verify the effect of OGD exposure on the apoptosis of N2a cells. After exposure to OGD, the expression of pro-apoptotic proteins (Bax and cleaved caspase-3) was up-regulated, especially in 24 h treatment group (Fig. 1D). The level of anti-apoptotic protein Bcl-2 revealed an opposite trend to Bax and cleaved caspase-3 (Fig. 1D), which further demonstrated that OGD exposure induced the apoptosis of N2a cells in a time-dependent manner. Overall, IS cell model was successfully established in vitro.
Fig. 1.
OGD induces the injury of N2a cells. A Cell viability of N2a cells following exposure to OGD for 0 h, 6 h, 12 h or 24 h was analyzed by CCK8 assay. B and C Flow cytometry was utilized to detect the apoptosis rate of N2a cells. The apoptosis rate indicates the percentages of apoptotic N2a cells in early and late stages. D Western blot assay was conducted to detect the protein expression of apoptosis-related proteins. *P < 0.05, **P < 0.01
ZFAS1 and miR-186-5p expression is abnormally regulated in OGD-induced N2a cells
Previous study showed that ZFAS1 expression was significantly down-regulated in IS patients compared with healthy volunteers (Wang et al. 1993). However, the exact role of ZFAS1 in IS remains to be disclosed. Here, we found that OGD exposure decreased the level of ZFAS1 in N2a cells in a time-dependent manner, and OGD treatment time-dependently up-regulated miR-186-5p level in N2a cells (Fig. 2A and B). The aberrant expression of ZFAS1 and miR-186-5p in IS cell model might imply their important roles.
Fig. 2.
ZFAS1 and miR-186-5p expression is abnormally regulated in OGD-induced N2a cells. A and B The expression of ZFAS1 and miR-186-5p was examined in N2a cells exposed to OGD for 0 h, 6 h, 12 h or 24 h by qRT-PCR. *P < 0.05, **P < 0.01
ZFAS1 overexpression partly offsets OGD-induced injury in N2a cells
To analyze the biological significance behind the abnormal down-regulation of ZFAS1 in IS cell model, we performed gain-of-function experiments. OGD treatment reduced the level of ZFAS1, and the addition of ZFAS1 overexpression plasmid rescued the expression of ZFAS1 in N2a cells (Fig. 3A). Furthermore, the addition of ZFAS1 plasmid largely recovered cell viability (Fig. 3B). OGD-induced apoptosis in N2a cells was also largely attenuated by the overexpression of ZFAS1 (Fig. 3C). The expression of Bax and cleaved caspase-3 was up-regulated by OGD exposure, and the addition of ZFAS1 overexpression plasmid reduced the expression of Bax and cleaved caspase-3 again (Fig. 3D). Bcl-2 level was down-regulated in OGD group, and its expression was largely recovered in OGD and ZFAS1 co-treated group (Fig. 3D), suggesting that OGD-induced apoptosis was largely diminished by the addition of ZFAS1 plasmid in N2a cells. Previous study found that ZFAS1 contributed to gastric cancer progression, and it elevated the expression of cyclin D1 (Xu et al. 2018). Additionally, cyclin D1 was an important stimulus for proliferation in N2a cells (Munoz et al. 2003). The expression of cyclin D1 was determined in N2a cells by Western blot assay. OGD stimulation down-regulated the expression of cyclin D1, and the level of cyclin D1 was largely rescued by the introduction of ZFAS1 plasmid (Supplementary Fig. S1A), suggesting the important role of cyclin D1 in ZFAS1-mediated protective role in OGD cell model. These findings demonstrated that OGD induced the injury of N2a cells partly through down-regulating ZFAS1.
Fig. 3.
ZFAS1 overexpression partly offsets OGD-induced injury in N2a cells. A–D Un-transfected N2a cells exposed to OGD for 24 h were termed as OGD group. N2a cells transfected with empty vector or ZFAS1 overexpression plasmid followed by exposure to OGD for 24 h were termed as OGD + vector or OGD + ZFAS1 group, respectively. Un-treated N2a cells were termed as control group. A The expression of ZFAS1 was analyzed by qRT-PCR. B CCK8 assay was used to assess the viability of N2a cells. C The apoptosis rate of N2a cells was evaluated using flow cytometry. D The expression of pro-apoptotic proteins (Bax and cleaved caspase-3) and anti-apoptotic protein (Bcl-2) was analyzed by Western blot assay. *P < 0.05, **P < 0.01
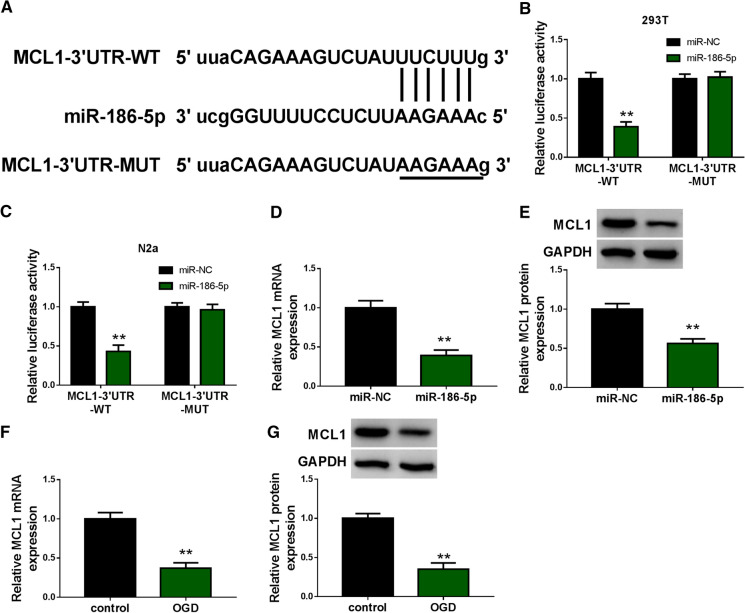
MiR-186-5p is a direct target of ZFAS1 in N2a cells
LncRNAs can serve as miRNA sponges to regulate cellular biological behaviors (Paraskevopoulou and Hatzigeorgiou 1402). We aimed to seek the downstream miRNAs of ZFAS1 to illustrate its working mechanism in IS cell model. The target relationship between miR-186-5p and ZFAS1 was predicted by starbase software (Fig. 4A). To test if the putative binding sites were required for the interaction between miR-186-5p and ZFAS1, we mutated these sites to “UAAGAAA” (Fig. 4A), and dual-luciferase reporter assay was conducted to verify the interaction between miR-186-5p and ZFAS1. 293T cells and N2a cells were co-transfected with luciferase reporter plasmids and miR-NC or miR-186-5p. MiR-186-5p overexpression significantly reduced the luciferase activity of wild-type reporter plasmid (ZFAS1-WT) compared with miR-NC and ZFAS1-WT co-transfected group (Fig. 4B and C), suggesting the binding relationship between ZFAS1 and miR-186-5p. The luciferase intensities of mutant reporter plasmid (ZFAS1-MUT) remained almost unchanged with the co-transfection of miR-NC or miR-186-5p (Fig. 4B and C), suggesting that the putative binding sites were required for the interaction between ZFAS1 and miR-186-5p. RNA-pull down assay revealed that ZFAS1 was pulled down when using Bio-miR-186-5p compared with Bio-NC group (Fig. 4D). The results of RIP assay showed that there was spatial interaction between ZFAS1 and miR-186-5p in N2a cells (Fig. 4E). The results of RNA-pull down assay and RIP assay further confirmed the interaction between ZFAS1 and miR-186-5p. We wondered whether the up-regulation of miR-186-5p was important for OGD-induced injury of N2a cells, and N2a cells were divided into four groups: control, OGD, OGD + anti-NC or OGD + anti-miR-186-5p. The knockdown efficiency of anti-miR-186-5p was high in N2a cells (Fig. 4F). OGD exposure-induced suppressive effect on cell viability and promoting effect on cell apoptosis were both partly reversed by the introduction of anti-miR-186-5p in N2a cells (Fig. 4G and H). Also, the results of Western blot assay revealed that OGD-triggered apoptosis was largely overturned by the silence of miR-186-5p in N2a cells (Fig. 4I). Overall, miR-186-5p was a direct target of ZFAS1, and OGD-induced injury of N2a cells was partly based on the up-regulation of miR-186-5p.
Fig. 4.
MiR-186-5p is a direct target of ZFAS1 in N2a cells. A The wild-type (WT) and mutant type (MUT) binding sites with miR-186-5p in ZFAS1 were shown. The target relationship between miR-186-5p and ZFAS1 was predicted by starbase software. B and C The direct interaction between miR-186-5p and ZFAS1 in 293T cells and N2a cells was assessed via dual-luciferase reporter assay. 293T cells and N2a cells were co-transfected with ZFAS1-WT or ZFAS1-MUT and miR-NC or miR-186-5p. D RNA-pull down assay was conducted to verify the interaction between miR-186-5p and ZFAS1 in N2a cells. E RIP assay was employed to validate if miR-186-5p was a target of ZFAS1 in N2a cells. F The knockdown efficiency of miR-186-5p inhibitor (anti-miR-186-5p) was evaluated in N2a cells by qRT-PCR. G–I N2a cells were treated with three groups: OGD, OGD + anti-NC or OGD + anti-miR-186-5p. Un-treated N2a cells were used as the control group. G Cell viability in four groups was analyzed by CCK8 assay. H The apoptosis rate in four groups was analyzed by flow cytometry. I The levels of Bax, Bcl-2 and cleaved caspase-3 in N2a cells were examined using Western blot assay. The quantification was carried out via gray analysis. *P < 0.05, **P < 0.01
MCL1 is a direct target of miR-186-5p in N2a cells
The putative binding sites between miR-186-5p and MCL1 were predicted by starbase database (Fig. 5A). The luciferase activities were notably decreased in miR-186-5p and MCL1-3′UTR-WT group compared with miR-NC and MCL1-3′UTR-WT group (Fig. 5B and C). However, the co-transfection of miR-NC or miR-186-5p had no effect on the luciferase activity of MCL1-3′UTR-MUT group. These findings suggested that MCL1 was a direct target of miR-186-5p. MCL1 mRNA and protein levels were down-regulated by the overexpression of miR-186-5p in N2a cells (Fig. 5D and E), suggesting the negative regulatory relationship between miR-186-5p and MCL1 in N2a cells. OGD exposure significantly reduced the mRNA and protein expression of MCL1 in N2a cells (Fig. 5F and G). Overall, miR-186-5p directly interacted with the 3′UTR of MCL1 in N2a cells.
Fig. 5.
MCL1 is a direct target of miR-186-5p in N2a cells. A The interaction between MCL1 and miR-186-5p was predicted by starbase database. The binding sites with miR-186-5p in MCL1 3′UTR were mutated to “AAGAAA”. B and C Dual-luciferase reporter assay was conducted to verify the binding relationship between miR-186-5p and MCL1. 293T cells and N2a cells were co-transfected with miR-NC or miR-186-5p and MCL1-3′UTR-WT or MCL1-3′UTR-MUT. The luciferase activities in the above-mentioned four transfected groups were detected. D and E The mRNA and protein expression of MCL1 was examined in N2a cells transfected with miR-186-5p mimics (miR-186-5p) or miR-NC by qRT-PCR and Western blot assay. F and G N2a cells were exposed to OGD for 24 h or not, and the mRNA and protein levels of MCL1 were detected by qRT-PCR and Western blot assay. **P < 0.01
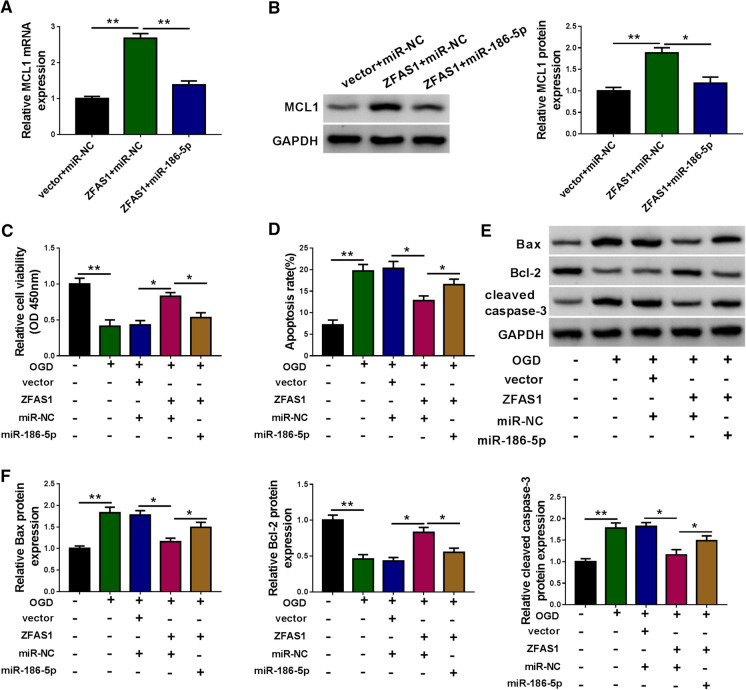
ZFAS1 attenuates OGD-induced injury of N2a cells through targeting miR-186-5p/MCL1 axis
To analyze the regulatory relationship among ZFAS1, miR-186-5p and MCL1 in N2a cells, MCL1 mRNA and protein expression was examined in N2a cells transfected with vector + miR-NC, ZFAS1 + miR-NC or ZFAS1 + miR-186-5p. As shown in Fig. 6A and B, ZFAS1 overexpression significantly elevated the mRNA and protein levels of MCL1, and the co-transfection of ZFAS1 and miR-186-5p decreased the mRNA and protein levels of MCL1 in N2a cells, suggesting that ZFAS1 up-regulated MCL1 mRNA and protein levels by sponging miR-186-5p in N2a cells. We also conducted rescue experiments in OGD-exposed N2a cells through transfecting ZFAS1 alone or together with miR-186-5p. ZFAS1 overexpression protected N2a cells against OGD-induced reduction in cell viability, and this protective effect was largely attenuated by the addition of miR-186-5p (Fig. 6C). Cell apoptosis was alleviated in ZFAS1 overexpression group in OGD-induced N2a cells, and the addition of miR-186-5p elevated the apoptosis rate again (Fig. 6D). The protein expression of Bax and cleaved caspase-3 was down-regulated in OGD-exposed N2a cells with the accumulation of ZFAS1, and the addition of miR-186-5p enhanced the expression of these two pro-apoptotic proteins (Fig. 6E and F). The expression trend of anti-apoptotic protein Bcl-2 revealed an opposite result to Bcl-2 and cleaved caspase-3 in N2a cells (Fig. 6E and F). Taken together, ZFAS1 protected N2a cells against OGD-induced injury through up-regulating MCL1 via sponging miR-186-5p. The addition of miR-186-5p mimics reduced the expression of cyclin D1 in ZFAS1-overexpressed N2a cells upon OGD exposure (Supplementary Fig. S1B), suggesting that cyclin D1 was regulated by ZFAS1/miR-186-5p axis.
Fig. 6.
ZFAS1 attenuates OGD-induced injury of N2a cells through targeting miR-186-5p/MCL1 axis. A and B N2a cells were transfected the following three groups: vector + miR-NC, ZFAS1 + miR-NC or ZFAS1 + miR-186-5p. The mRNA and protein expression of MCL1 was examined in N2a cells by qRT-PCR and Western blot assay. C–F N2a cells were divided into five groups: control, OGD, OGD + vector + miR-NC, OGD + ZFAS1 + miR-NC or OGD + ZFAS1 + miR-186-5p. C CCK8 assay was performed to detect the viability of N2a cells. D The apoptotic N2a cells in the early and late stages were identified by flow cytometry, and the apoptosis rate was analyzed. E and F Western blot assay was used to examine the protein expression of Bax, Bcl-2 and cleaved-caspase 3 in N2a cells. *P < 0.05, **P < 0.01
It has been reported that miR-186-5p accelerated the apoptosis in SH-SY5Y OGD model through targeting IGF-1 (Wang et al. 2018). Here, we aimed to analyze if miR-186-5p/IGF-1 axis worked in N2a OGD model and compare the effects between miR-186-5p/IGF-1 axis and miR-186-5p/MCL1 axis in N2a OGD model. As mentioned in Supplementary Fig. S2A and S2B, we found that miR-186-5p silencing up-regulated the mRNA and protein levels of MCL1 and IGF-1 in N2a cells, and the introduction of si-MCL1 or si-IGF-1 reduced the expression of MCL1 or IGF-1, respectively. Through performing rescue experiments, we found that MCL1 knockdown overturned miR-186-5p silencing-mediated effects in OGD-induced N2a cells better than the silence of IGF-1 (Supplementary Fig. S2C-F). Furthermore, we found that miR-186-5p silencing up-regulated the level of cyclin D1 in IS cell model, and the addition of si-MCL1 or si-IGF-1 decreased the protein level of cyclin D1 (Supplementary Fig. S2G), suggesting that cell cycle progression-associated protein cyclin D1 was involved the regulation of ZFAS1/miR-186-5p/MCL1 axis in IS cell model. These results demonstrated that miR-186-5p targeted MCL1 more specifically than IGF-1 in N2a OGD model.
Discussion
In the current study, we explored the role and working mechanism of ZFAS1 on the viability and apoptosis of OGD-induced IS cell model in vitro. ZFAS1 overexpression alleviated OGD-induced injury in N2a cells. Furthermore, the direct interaction between miR-186-5p and ZFAS1 or MCL1 was identified in this study for the first time. ZFAS1 protected N2a cells against OGD-mediated damage through up-regulating pro-survival protein MCL1 by sponging miR-186-5p. These findings will help us deeply understand the patho-mechanism of IS.
IS is characterized by the sudden interruption of cerebral blood, leading to high mortality and disability rate (Hankey 2003). Accumulating evidence found that non-coding RNAs, including lncRNAs and miRNAs, were implicated in the pathophysiologic mechanism of IS. For instance, Zhao et al. found that SNHG12 accelerated angiogenesis through modulating miR-150/VEGF axis following IS (Zhao et al. 2018). Zheng et al. demonstrated that miR-130a protected PC-12 cells against OGD-induced damage via PTEN/PI3K/AKT axis (Zheng et al. 1091). ZFAS1 is a newly identified lncRNA that located in chromosome 20q13.13. ZFAS1 was found to be dysregulated and it exerted crucial functions in many malignancies, including gastric cancer, bladder cancer, cervical cancer and thyroid cancer (Nie et al. 2017; Yin et al. 2018; Feng et al. 2019; Han et al. 2019). For instance, Wang et al. claimed that ZFAS1 was up-regulated in bladder cancer, and it accelerated the progression of bladder cancer by targeting miR-329 (Wang et al. 2018). As for IS, ZFAS1 level was found to be significantly reduced in IS patients compared with healthy controls, and it might be an underlying marker for stroke (Wang et al. 1993).
N2a cells were exposed to OGD to establish IS cell model in vitro. OGD exposure suppressed the viability and triggered the apoptosis of N2a cells, suggesting that the establishment of IS cell model was successful. OGD exposure time-dependently reduced the level of ZFAS1 in N2a cells, which was consistent with the clinical data described by the former article (Wang et al. 1993). Further study demonstrated that ZFAS1 exerted a neuroprotective effect to attenuate OGD-induced injury in N2a cells.
LncRNAs regulate cellular physiological and pathological processes through acting as miRNA sponges (Paraskevopoulou and Hatzigeorgiou 1402). For instance, lncRNA OC1 accelerated ovarian cancer progression through promoting cell proliferation and migration by targeting miR-34a and miR-34c (Tao et al. 2018). Liu et al. found that lncRNA HOTAIR promoted the proliferation and metastasis of gastric cancer cells through targeting miR-331-3p/HER2 axis (Liu et al. 2014). MiR-186-5p was identified as a direct molecular target of ZFAS1 in N2a cells. MiR-186-5p was reported to exert pro-tumor or anti-tumor role in diverse cancers. MiR-186-5p restrained the proliferation of non-small cell lung cancer cells through modulating proliferation-related cyclin D1, CDK2 and CDK6 (Cai et al. 2013). However, miR-186-5p acted as an oncogene in colon cancer through promoting the proliferation, colony formation and migration of colon cancer cells via down-regulating FAM134B (Islam et al. 2017). Furthermore, MiR-186-5p was reported to facilitate the apoptosis of OGD-treated SH-SY5Y cells via targeting IGF-1 (Wang et al. 2018). Here, we found that OGD exposure up-regulated miR-186-5p level in N2a cells in a time-dependent manner. Also, miR-186-5p silencing attenuated OGD-induced damage in N2a cells, suggesting that OGD mediated the injury of N2a cells partly by up-regulating miR-186-5p.
The interaction between miR-186-5p and MCL1 was confirmed in N2a cells. MCL1 is a member of Bcl-2 family, and it is generally known as a pro-survival protein in many types of cells. Zhou et al. claimed that SNHG12 elevated the doxorubicin resistance of osteosarcoma cells through up-regulating the expression of MCL1 via sponging miR-320a (Zhou et al. 2018). Liu et al. found that ANRIL exerted a neuroprotective effect in PC-12 cells through elevating MCL1 expression via sponging miR-127 (Liu et al. 2019). OGD exposure down-regulated the expression of MCL1 in N2a cells. Furthermore, rescue experiments revealed that ZFAS1 attenuated OGD-mediated injury of N2a cells through targeting miR-186-5p/MCL1 axis. Moreover, we found that cyclin D1 was involved in the regulation of ZFAS1/miR-186-5p/MCL1 axis in IS cell model. Previous study demonstrated that miR-186-5p promoted the apoptosis in SH-SY5Y OGD model via suppressing IGF-1 (2018). We found that miR-186-5p targeted MCL1 more specifically than IGF-1 in N2a OGD model.
Conclusion
In conclusion, we proposed a novel signal axis of ZFAS1/miR-186-5p/MCL1 that was involved in the viability and apoptosis regulation in IS cell model. More in vivo experiments need to be conducted to further verify the role of ZFAS1/miR-186-5p/MCL1 axis for IS treatment.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 ZFAS1 overexpression rescues the expression of cyclin D1 in OGD-induced N2a cells. (A) The protein expression of cyclin D1 was determined in N2a cells in the following four groups by Western blot assay: control, OGD, OGD + vector and OGD + ZFAS1. (B) The level of cyclin D1 was measured in N2a cells in the following five groups by Western blot assay: control, OGD, OGD + vector + miR-NC, OGD + ZFAS1 + miR-NC and OGD + ZFAS1 + miR-186-5p. *P < 0.05, **P < 0.01. (TIFF 452 kb)
Supplementary file2 The silence of MCL1 counteracts miR-186-5p knockdown-induced effects in N2a cells upon OGD stimulation better than the silence of IGF-1. (A and B) The mRNA and protein expression of IGF-1 was determined in N2a cells transfected with anti-NC + si-NC, anti-miR-186-5p + si-NC and anti-miR-186-5p + si-IGF-1. (C-G) N2a cells were divided into six groups: control group, OGD group, OGD + anti-NC + si-NC group, OGD + anti-miR-186-5p + si-NC group, OGD + anti-miR-186-5p + si-MCL1 group and OGD + anti-miR-186-5p + si-IGF-1 group. (C) CCK8 assay was applied to analyze cell viability. (D) Flow cytometry was employed to assess cell apoptosis rate. (E and F) Western blot assay was conducted to determine the expression of Bax, Bcl-2 and cleaved caspase-3 in N2a cells. (G) The level of cyclin D1 was determined in N2a cells by Western blot assay. *P < 0.05, **P < 0.01. (TIFF 1159 kb)
Abbreviations
- lncRNA
Long non-coding RNA
- miRNA
MicroRNA
- ZFAS1
Zinc finger antisense 1
- IS
Ischemic stroke
- MCL1
MCL1 apoptosis regulator, BCL2 family member
- OGD
Oxygen and glucose deprivation
Funding
This work was funded by Yancheng Development Project of Medical Science and Technology in 2020 (YK2020061)
Data availability
All data generated or analyzed during this study are included in this article.
Declarations
Conflicts of interest
The authors declare that they have no financial conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Allen CL, Bayraktutan U. Oxidative stress and its role in the pathogenesis of ischaemic stroke. Int J Stroke. 2009;4:461–470. doi: 10.1111/j.1747-4949.2009.00387.x. [DOI] [PubMed] [Google Scholar]
- Bao MH, Szeto V, Yang BB, Zhu SZ, Sun HS, Feng ZP. Long non-coding RNAs in ischemic stroke. Cell Death Dis. 2018;9:281. doi: 10.1038/s41419-018-0282-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry G. Integrating the roles of long and small non-coding RNA in brain function and disease. Mol Psychiatry. 2014;19:410–416. doi: 10.1038/mp.2013.196. [DOI] [PubMed] [Google Scholar]
- Benjamin EJ, Virani SS, Callaway CW, et al. Heart disease and stroke statistics-2018 update: a report from the American Heart Association. Circulation. 2018;137:e67–e492. doi: 10.1161/CIR.0000000000000558. [DOI] [PubMed] [Google Scholar]
- Cai J, Wu J, Zhang H, et al. miR-186 downregulation correlates with poor survival in lung adenocarcinoma, where it interferes with cell-cycle regulation. Cancer Res. 2013;73:756–766. doi: 10.1158/0008-5472.CAN-12-2651. [DOI] [PubMed] [Google Scholar]
- Chen L, Zhou Y, Li H. LncRNA, miRNA and lncRNA-miRNA interaction in viral infection. Virus Res. 2018;257:25–32. doi: 10.1016/j.virusres.2018.08.018. [DOI] [PubMed] [Google Scholar]
- Cheng X, Kan P, Ma Z, et al. Exploring the potential value of miR-148b-3p, miR-151b and miR-27b-3p as biomarkers in acute ischemic stroke. Biosci Rep. 2018;38:25. doi: 10.1042/BSR20181033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark BS, Blackshaw S. Long non-coding RNA-dependent transcriptional regulation in neuronal development and disease. Front Genet. 2014;5:164. doi: 10.3389/fgene.2014.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz NF, Cruz-Resendiz MS, Flores-Herrera H, Garcia-Lopez G, Molina-Hernandez A. MicroRNAs in central nervous system development. Rev Neurosci. 2014;25:675–686. doi: 10.1515/revneuro-2014-0014. [DOI] [PubMed] [Google Scholar]
- Feng LL, Shen FR, Zhou JH, Chen YG. Expression of the lncRNA ZFAS1 in cervical cancer and its correlation with prognosis and chemosensitivity. Gene. 2019;696:105–112. doi: 10.1016/j.gene.2019.01.025. [DOI] [PubMed] [Google Scholar]
- Han CG, Huang Y, Qin L. Long non-coding RNA ZFAS1 as a novel potential biomarker for predicting the prognosis of thyroid cancer. Med Sci Monit. 2019;25:2984–2992. doi: 10.12659/MSM.912921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankey GJ. Long-term outcome after ischaemic stroke/transient ischaemic attack. Cerebrovasc Dis. 2003;16:14–19. doi: 10.1159/000069936. [DOI] [PubMed] [Google Scholar]
- Hassan A, Markus HS. Genetics and ischaemic stroke. Brain. 2000;123:1784–1812. doi: 10.1093/brain/123.9.1784. [DOI] [PubMed] [Google Scholar]
- Huang Y. The novel regulatory role of lncRNA-miRNA-mRNA axis in cardiovascular diseases. J Cell Mol Med. 2018;22:5768–5775. doi: 10.1111/jcmm.13866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Luo H, Li F, et al. LINC00152 down-regulated miR-193a-3p to enhance MCL1 expression and promote gastric cancer cells proliferation. Biosci Rep. 2018;38:40. doi: 10.1042/BSR20171607. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Islam F, Gopalan V, Vider J, et al. MicroRNA-186-5p overexpression modulates colon cancer growth by repressing the expression of the FAM134B tumour inhibitor. Exp Cell Res. 2017;357:260–270. doi: 10.1016/j.yexcr.2017.05.021. [DOI] [PubMed] [Google Scholar]
- Liu XH, Sun M, Nie FQ, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Cao W, Xue J. LncRNA ANRIL protects against oxygen and glucose deprivation (OGD)-induced injury in PC-12 cells: potential role in ischaemic stroke. Artif Cells Nanomed Biotechnol. 2019;47:1384–1395. doi: 10.1080/21691401.2019.1596944. [DOI] [PubMed] [Google Scholar]
- Mojsa B, Lassot I, Desagher S. Mcl-1 ubiquitination: unique regulation of an essential survival protein. Cells. 2014;3:418–437. doi: 10.3390/cells3020418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz JP, Sanchez JR, Maccioni RB. Regulation of p27 in the process of neuroblastoma N2A differentiation. J Cell Biochem. 2003;89:539–549. doi: 10.1002/jcb.10525. [DOI] [PubMed] [Google Scholar]
- Nie F, Yu X, Huang M, et al. Long noncoding RNA ZFAS1 promotes gastric cancer cells proliferation by epigenetically repressing KLF2 and NKD2 expression. Oncotarget. 2017;8:38227–38238. doi: 10.18632/oncotarget.9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA-LncRNA interactions. Methods Mol Biol. 2016;1402:271–286. doi: 10.1007/978-1-4939-3378-5_21. [DOI] [PubMed] [Google Scholar]
- Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52:710–718. doi: 10.1136/jmedgenet-2015-103334. [DOI] [PubMed] [Google Scholar]
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- Siddeek B, Inoubli L, Lakhdari N, et al. MicroRNAs as potential biomarkers in diseases and toxicology. Mutat Res. 2014;764:46–57. doi: 10.1016/j.mrgentox.2014.01.010. [DOI] [PubMed] [Google Scholar]
- Tao F, Tian X, Lu M, Zhang Z. A novel lncRNA, Lnc-OC1, promotes ovarian cancer cell proliferation and migration by sponging miR-34a and miR-34c. J Genet Genom. 2018;45:137–145. doi: 10.1016/j.jgg.2018.03.001. [DOI] [PubMed] [Google Scholar]
- Wang R, Bao H, Zhang S, Li R, Chen L, Zhu Y. miR-186-5p promotes apoptosis by targeting IGF-1 in SH-SY5Y OGD/R Model. Int J Biol Sci. 2018;14:1791–1799. doi: 10.7150/ijbs.25352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JS, Liu QH, Cheng XH, Zhang WY, Jin YC. The long noncoding RNA ZFAS1 facilitates bladder cancer tumorigenesis by sponging miR-329. Biomed Pharmacother. 2018;103:174–181. doi: 10.1016/j.biopha.2018.04.031. [DOI] [PubMed] [Google Scholar]
- Wang J, Ruan J, Zhu M, et al. Predictive value of long noncoding RNA ZFAS1 in patients with ischemic stroke. Clin Exp Hypert. 2019;41:615–621. doi: 10.1080/10641963.2018.1529774. [DOI] [PubMed] [Google Scholar]
- Wu S, Liu F, Xie L, et al. miR-125b suppresses proliferation and invasion by targeting MCL1 in gastric cancer. Biomed Res Int. 2015;2015:365273. doi: 10.1155/2015/365273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, He L, Li Y, Tan Y, Zhang F, Xu H. Silencing of lncRNA ZFAS1 inhibits malignancies by blocking Wnt/beta-catenin signaling in gastric cancer cells. Biosci Biotechnol Biochem. 2018;82:456–465. doi: 10.1080/09168451.2018.1431518. [DOI] [PubMed] [Google Scholar]
- Yin S, Du W, Wang F, et al. MicroRNA-326 sensitizes human glioblastoma cells to curcumin via the SHH/GLI1 signaling pathway. Cancer Biol Ther. 2018;19:260–270. doi: 10.1080/15384047.2016.1250981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Wang J, Xi X, Tan N, Zhang L. SNHG12 promotes angiogenesis following ischemic stroke via regulating miR-150/VEGF pathway. Neuroscience. 2018;390:231–240. doi: 10.1016/j.neuroscience.2018.08.029. [DOI] [PubMed] [Google Scholar]
- Zheng T, Shi Y, Zhang J, et al. MiR-130a exerts neuroprotective effects against ischemic stroke through PTEN/PI3K/AKT pathway. Biomed Pharmacoth. 2019;117:109117. doi: 10.1016/j.biopha.2019.109117. [DOI] [PubMed] [Google Scholar]
- Zhou J, Chen L, Chen B, et al. Increased serum exosomal miR-134 expression in the acute ischemic stroke patients. BMC Neurol. 2018;18:198. doi: 10.1186/s12883-018-1196-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Li L, Li Y, Su H, Zeng C. Long noncoding RNA SNHG12 mediates doxorubicin resistance of osteosarcoma via miR-320a/MCL1 axis. Biomed Pharmacother. 2018;106:850–857. doi: 10.1016/j.biopha.2018.07.003. [DOI] [PubMed] [Google Scholar]
- Zhu M, Li N, Luo P, et al. Peripheral blood leukocyte expression of lncRNA MIAT and its diagnostic and prognostic value in ischemic stroke. J Stroke Cerebrovascul Dis. 2018;27:326–337. doi: 10.1016/j.jstrokecerebrovasdis.2017.09.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file1 ZFAS1 overexpression rescues the expression of cyclin D1 in OGD-induced N2a cells. (A) The protein expression of cyclin D1 was determined in N2a cells in the following four groups by Western blot assay: control, OGD, OGD + vector and OGD + ZFAS1. (B) The level of cyclin D1 was measured in N2a cells in the following five groups by Western blot assay: control, OGD, OGD + vector + miR-NC, OGD + ZFAS1 + miR-NC and OGD + ZFAS1 + miR-186-5p. *P < 0.05, **P < 0.01. (TIFF 452 kb)
Supplementary file2 The silence of MCL1 counteracts miR-186-5p knockdown-induced effects in N2a cells upon OGD stimulation better than the silence of IGF-1. (A and B) The mRNA and protein expression of IGF-1 was determined in N2a cells transfected with anti-NC + si-NC, anti-miR-186-5p + si-NC and anti-miR-186-5p + si-IGF-1. (C-G) N2a cells were divided into six groups: control group, OGD group, OGD + anti-NC + si-NC group, OGD + anti-miR-186-5p + si-NC group, OGD + anti-miR-186-5p + si-MCL1 group and OGD + anti-miR-186-5p + si-IGF-1 group. (C) CCK8 assay was applied to analyze cell viability. (D) Flow cytometry was employed to assess cell apoptosis rate. (E and F) Western blot assay was conducted to determine the expression of Bax, Bcl-2 and cleaved caspase-3 in N2a cells. (G) The level of cyclin D1 was determined in N2a cells by Western blot assay. *P < 0.05, **P < 0.01. (TIFF 1159 kb)
Data Availability Statement
All data generated or analyzed during this study are included in this article.