Abstract
Mycoplasma capricolum subsp.subsp. capripneumonia (Mccp) and Mycoplasma mycoides subsp.sbusp. capri (Mmc) cause caprine pleuropneumonia (CCPP) and mycoplasmal pneumonia in goats and sheep (MPGS), respectively. These diseases cannot be identified on clinical symptoms alone and it is laborious to distinguish them using biochemical methods. It is therefore important to establish a simple, rapid identification method for Mccp and Mmc. Here, we report a high-resolution melting (HRM) curve analysis using specific primers based on the Mmc 95010 strain MLC_0560 and Mccp F38 strain MCCPF38_00984 gene sequences. The method was highly specific with intra- and inter-batch coefficients of variation < 1%. The lower limit of detection for Mccp and Mmc was 55 copies/μL and 58 copies/μL, respectively. HRM and fluorescence qPCR results were compared using 106 nasal swabs and 47 lung tissue samples from goats (HRM-qPCR coincidence rate 94.8%; 145/153). Mycoplasma isolation and identification was performed on 30 lung tissue samples and 16 nasal swabs (HRM-culturing coincidence rate 87.0%; 40/46). HRM analysis was more sensitive than fluorescence qPCR and Mycoplasma isolation, indicating the practicality of HRM for accurate and rapid identification of Mccp and Mmc, and diagnosis and epidemiology of CCPP and MPGS.
Subject terms: Microbiology, Infectious-disease diagnostics
Introduction
Mycoplasma capricolumsubsp subsp. capripneumonia (Mccp) is the causative pathogen of contagious caprine pleuropneumonia (CCPP). Clinically, the disease is characterized by hyperpyrexia, rhinorrhea, cough, cellulose pleuropneumonia, abortion, and progressive emaciation of some ewes1. The incidence can reach 80–100%, and the mortality rate can reach 60–80%2,3. CCPP is one of the statutory reportable animal infectious diseases listed by the World Organization for Animal Health4. Mycoplasma mycoides subsp. capri (Mmc) is one of the causative pathogens of mycoplasmal pneumonia of goats and sheep (MPGS)5. The incidence of MPGS caused by Mmc can reach 50%, and the mortality rate can reach 40%6,7. In addition to causing pathological lung changes similar to Mccp in goats, Mmc can also cause mastitis, arthritis, and keratitis, among other conditions8, causing significant economic losses to the goat farming industry. Clinical symptoms and necropsy lesions of CCPP and MPGS are similar, and distinguishing them clinically is difficult. Mccp and Mmc are both members of the M. mycoides cluster and show high genetic and antigen similarity9. In fact, in China they have been traditionally referred to as CCPP collectively. However, only Mccp has been shown to conform to Koch’s postulate, and it is the only causative pathogen of CCPP4. Laboratory identification is typically conducted by mycoplasma pathogen isolation, culture, and identification combined with serological testing, but this method can only be carried out in a laboratory, is difficult to standardize, and is time-consuming, labor-intensive, and lacks proper biological safeguards. Therefore, it is important to establish a rapid identification method for Mccp and Mmc.
Established molecular detection methods for Mccp and Mmc mainly include PCR detection10–12, multiplex PCR13–15, nested PCR for Mmc specifically16, and real-time fluorescence quantitative PCR (qPCR) detection17–19. A high-resolution melting (HRM) curve is useful for detecting genetic mutations. The technique is based on the use of a novel saturated fluorescent dye first proposed by the Wittwer Laboratory of the University of Utah20. This method involves genotyping by monitoring the binding of double-stranded DNA with the fluorescent dye and PCR amplification products in real-time during the heating process according to the differences in fragment length, G + C content, and nucleic acid distribution21. It shows strong specificity and high sensitivity and is suitable for establishing Mccp and Mmc identification methods.
No HRM-based methods have been developed to identify Mccp and Mmc. Therefore, in this study, bioinformatics software was used to analyze the genome sequences of Mccp and Mmc to screen for conserved fragments with adequate differences to design primers that can distinguish the two microbes. The HRM detection method described here was established using the small-fragment amplification method in order to rapidly detect and identify Mccp and Mmc.
Materials and methods
Mycoplasmas, virus strains, bacterial strains, and clinical samples
Referring to Lin et al.22, the strains used in this study were: Mccp strain F38, Mccp strain California Kid, Mmc strain PG3, were provided by Prof. Yuefeng Chu at Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences (LVRI, CAAS). The M. leachii PG50 strain was a gift from Prof. Jiuqing Xin at Harbin Veterinary Research Institute, CAAS (HVRI, CAAS). Mmc strain Y-goat and M. agalactiae strain PG2 were gifts from Prof. Cheng Tang at the College of Life and Technology, Southwest University for Nationalities. E. coli strain ATCC25922 (Guangdong Huankai Microbial), Pm strain CVCC44801 (China Institute of Veterinary Drug Control), and Sa strain 261111 (National Institutes for Food and Drug Control) were gifts from Prof. Long-Fei Cheng, Institute of Animal Science and Veterinary Medicine, Fujian Academy of Agricultural Sciences. Mannheimia haemolytica, Mycoplasma arginini, Acholeplasma laidlawii, M. bovis, Mycoplasma ovipneumoniae (Movi), Mmc strain FJ-GT, Mmc strain FJ-CL, and Orf virus (ORFV) were all isolated, identified, and preserved in our laboratory. 153 clinical samples (106 nasal swabs and 47 lung tissue samples) of goats with clinically suspected MPGS were collected from the cities of Fuzhou, Ningde, Quanzhou, Putian, Sanming, and Nanping of Fujian Province between September 2014, and December 2018. All sample manipulations were performed in BSL-2 laboratories.
Reagents
MiniBEST Bacteria Genomic DNA Extraction Kit Ver. 3.0, MiniBEST Viral RNA/DNA Extraction Kit Ver. 5.0, MiniBEST Plasmid Purification Kit Ver. 4.0, EXTAQ, MiniBEST Agarose Gel DNA Extraction Kit Ver. 4.0, and pMD TM 18-T Vector Cloning Kit were purchased from TAKARA Biomedical Technology (Beijing, China). Syto9 was purchased from ThermoFisher Scientific (Waltham, MA, USA).
Primer design
The sequences of the conserved gene MLC_0560 of Mmc 95010 strain (FQ377874.1) and MCCP F38_00984 (LN515398.1) from Mccp F38 were retrieved from GenBank. Using BioEdit Sequence Alignment Editor software for multiple sequences alignment (Fig. 1), a pair of specific primers was designed using Primer Premier 6.0 software. The primer sequences selected were: MF-107: 5′-CAATACATCCATTAGAACTCTTGA-3′; and MR-107: 5′-GAATACATTCAGGTTGATTATTAGGA-3′. Primers were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China) targeted sequence length 107 bp. Melting temperature (Tm) of the sequence was calculated using uMELT online resource (https://www.dna.utah.edu/umelt/umelt.html).
Figure 1.
The design of primers for HRM. Conservation of the corresponding sequence of each primer was analysed using BioEdit sequence alignment editor v7.1.7 aligned, The corresponding sequence of each primer in all selected Mmc and Mccp isolates shown in bule colour, The different bases in all selected Mmc and Mccp isolates shown in yellow colour.
DNA extraction
All genomic DNAs were extracted using the MiniBEST Bacteria Genomic DNA Extraction Kit Ver. 3.0 according to manufacturer’s instructions, Viral DNAs was extracted with the MiniBEST Viral RNA/DNA Extraction Kit Ver. 5.0 according to manufacturer’s instructions. After DNA extraction, the concentration of each sample was measured with a UV–Vis spectrophotometer NanoDrop 2000 (Thermo Fisher, Wilmington, USA).
Construction of a positive plasmid standard
Mmc and Mccp genomic DNA were used as the templates and the primers MF-107 and MR-107 were used for PCR amplification of the target fragment from those templates. The amplified products were recovered and purified after agar gel electrophoresis, and the purified target gene was ligated into the pMD18-T cloning vector. DH5α competent cells were transformed with the vector, after which the MiniBEST Plasmid Purification Kit Ver. 4.0 was used to extract the plasmid for PCR identification and restriction enzyme digestion identification. Agarose gel electrophoresis revealed a band of approximately 107 bp, consistent with the expected fragment size. Sequencing results from transformed bacteria were compared and analyzed using BLAST. The results showed that the homology of the Mmc PCR amplification products with Mmc PG3 strains was 100% and the homology of Mccp PCR amplification products with Mccp F38 strains was also 100%. This plasmid was then used as the positive control for subsequent experiments. The concentration was determined with a NanoDrop 2000 and converted into a copy number (number of copies = (amount × 6.022 × 1023)/(length × 1 × 109 × 650)). The plasmids were stored at − 70 ℃ until use.
Preliminary establishment of an HRM detection method
The reaction mix consisted of the following: the primers MF-107 and MR-107 (0.2 μmol/L), 2 μL template DNA, 12.5 μL premix Taq, 1.25 µmol/L Syto9, and ddH2O to a final volume of 25 μL. The thermocycler sequence used was as follows: pre-denaturation at 95 °C for 5 min followed by 40 cycles of denaturation at 95 °C for 30 s, annealing at 60 °C for 30 s, and extension at 72 °C for 20 s. HRM analysis was performed using a Rotor-Gene Q Fluorescence qPCR instrument. The detection conditions were as follows: denaturation at 95 °C for 5 min, renaturation at 45 °C for 1 min, and thermal equilibration at 50 °C for 10 s. From 50 °C, the melting curve was collected at a heating rate of 0.3 °C/s to 95 °C.
Optimization of the HRM detection method
Annealing temperature optimization was performed by running scans at the following annealing temperatures based on the estimated Tm: 50 °C, 52 °C, 54 °C, 55 °C, 57 °C, 59 °C. 53 °C was chosen for subsequent experiments. After annealing temperature optimization, we performed primer concentration optimization. Reactions were performed with primer concentrations of 0.2 µmol/L, 0.4 µmol/L, 0.6 µmol/L, and 0.8 µmol/L. Based on these results, the optimum concentration was used for subsequent experiments.
Specificity
Mmc, Mccp, Movi, ORFV, E. coli, Salmonella, Pasteurella, and M. arginine genomic DNA were extracted using the MiniBEST Bacteria Genomic DNA Extraction Kit Ver. 3.0 and used as templates to determine the specificity of the HRM method, the protocol according to “Optimization of the HRM detection method” section. If no specific amplification curve was generated and no expected Tm values, the samples were considered as negative cases.
Sensitivity
The positive plasmid standard and Mmc and Mccp genomic DNA were diluted in tenfold serial dilutions with ddH2O, and PCR-HRM analysis was conducted under the optimal reaction conditions described in “Optimization of the HRM detection method” section to establish the minimum detection limit of the HRM method. Each dilution series, along with a blank control, was repeated three times. A standard curve diagram was drawn according to the Ct values and concentration. Ct values and melting curves were used to define the limit of detection (LOD). Data analysis and standard curve was performed with Rotor-Gene Q Series Software. For comparison with the conventional PCR assay, primers and reaction conditions used were the same as for the PCR step of the HRM method.
Reproducibility
The positive control plasmid was diluted to three gradients to determine the reproducibility of the HRM method. All measurements were repeated three times for each sample at each concentration in the batch; the inter-batch test was carried out three times at an interval of 3 days. The HRM reaction system and cycle parameters are as described in “Optimization of the HRM detection method” section. The intra- and inter-batch coefficients of variation were calculated according to the Tm values of the two pathogens. Statistical analysis was performed with SPSS Statistics 25. Tm values between the two groups were compared with the independent t-test. We considered two-sided P values of 0.05 to be significant.
DNA sequencing
To benchmark Mmc and Mccp detection with HRM analysis, DNA sequencing was performed. After the HRM reaction was completed, the HRM products were subjected to agarose gel electrophoresis, recovered, and purified. The purified target fragment was ligated into pMD 18-T and used to transform DH5α competent cells as described in “Construction of a positive plasmid standard” section. MF-107 and MR-107 were used for PCR to determine the positive clones, which sequenced at Takara Biotechnology (Dalian, China).
Mycoplasma isolation and identification
Lung tissues were homogenized with modified Hayfick's culture medium. Ten-fold serial dilutions were prepared by combining 0.3 mL of the homogenized supernatant and subsequent dilutions with 2.7 mL of modified Hayfick's culture medium resulting in a dilution series from 10–3 to 10–1. All dilutions were incubated at 37 °C in 5% CO2, and three blind passages were performed every 7 days. Afterwards, 0.1 mL of the cultures that turned yellow was diluted tenfold to 10–5 to 10–3, and 0.2 mL of each dilution were streaked onto FRIIS Agar plates and cultured at 37 °C in 5% CO2 for 5–10 days. A single characteristic colony was inoculated in liquid culture in Modified Hayflick’s Medium, Mccp and Mmc field strains were identified using PCR methods22.
Clinical sample testing
DNA was extracted from 153 clinical samples using the MiniBEST Bacteria Genomic DNA Extraction Kit Ver. 3.0. Samples were analyzed using the optimized HRM method in parallel with fluorescence qPCR19. Concurrently, detection using Mycoplasma isolation and identification described in “Mycoplasma isolation and identification” section was performed on 46 clinic samples (30 lung tissue samples and 16 nasal swabs). To evaluate the accuracy of the HRM method, samples with different test results between the three methods were sequenced.
Ethics statement
The research reported here was approved by the Animal Care and Use Committee of the Institute of Animal Husbandry and Veterinary Medicine, Fujian Academy of Agricultural Sciences. For all clinical samples, we obtained informed consent of the goat’s owner. All methods were performed in accordance with the relevant guidelines and regulations.
Results
Optimization of HRM reaction conditions
Optimized HRM assay conditions were optimized as follows. The optimized primer concentrations were 0.4 μmol/L and the overall reaction mix contained 1× premix Taq, 1.25 μmol/L Syto9, 1 μL template, and ddH2O to a final volume of 25 μL. The optimal annealing temperature was determined to be 53 °C; therefore, the reaction conditions were: pre-denaturation at 95 °C for 3 min followed by 40 cycles of 95 °C for 15 s, 53 °C for 15 s, and 72 °C for 10 s. The HRM analysis procedure was as follows: 95 °C for 1 min, 45 °C for 1 min, 50 °C for 10 s, and heating to 90 °C at 0.3 °C/s.
HRM specificity
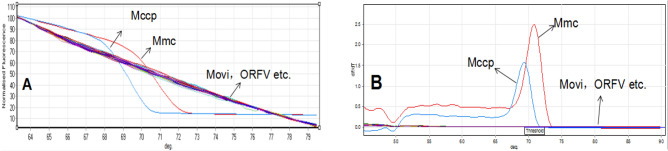
We next used the HRM method to detect Mmc PG3 strains and Mccp F38 strains. The results (Fig. 2) showed that only two specific melting peak curves of Mmc and Mccp were formed. The difference in Tm value of two dissolution curves was approximately 1.3, indicating that the two strains could easily be distinguished based on the melting curves, while no specific melting curves were generated from common goat pathogens as Movi and ORFV, indicating that this HRM method was specific for Mmc and Mccp.
Figure 2.
Specificity test of the HRM method for Mccp and Mmc. The pictures are generated by Rotor Gene Q. (A) Normalized characteristic curves of sample; (B) Melting curve of sample.
HRM reproducibility
As shown in Table 1, the intra- and inter-batch coefficients of variation for melting peak Tm1 were both within 0.08–0.11%, while for Tm2, they were 0.08–0.14% and 0.11–0.14%, respectively. As these values were all < 1%, the reproducibility of this method was verified. Tm values between the two groups were verified as statistically significant by the independent t-test (P = 0.000; P <0.01) (Table 1).
Table 1.
Reproducibility assay of the HRM method.
| No. | Intra-assay reproducibility | Inter-assay reproducibility | ||||||
|---|---|---|---|---|---|---|---|---|
| Tm1 mean ± SD |
CV/% | Tm2 mean ± SD | CV/% | Tm1 mean ± SD |
CV/% | Tm2 mean ± SD |
CV/% | |
| 1 × 10–2 | 69.53 ± 0.06B | 0.08 | 71.04 ± 0.09A | 0.14 | 69.47 ± 0.06B | 0.08 | 71.08 ± 0.08A | 0.11 |
| 1 × 10–3 | 69.46 ± 0.06B | 0.08 | 70.95 ± 0.09A | 0.12 | 69.48 ± 0.08B | 0.11 | 70.98 ± 0.08A | 0.11 |
| 1 × 10–4 | 69.41 ± 0.08B | 0.11 | 70.93 ± 0.06A | 0.08 | 69.53 ± 0.06B | 0.08 | 71.01 ± 0.1A | 0.14 |
Different uppercase letters in the same row mean extremely significant difference (P = 0.000 < 0.01).
HRM sensitivity
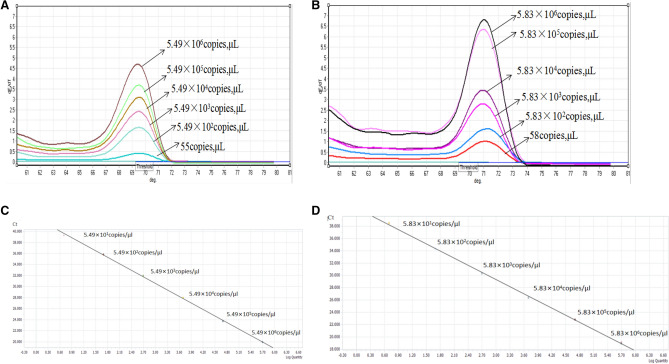
The positive control plasmids were diluted tenfold and detected using the HRM method described here as well as conventional PCR. The results (Fig. 3) indicated that specific melting curves and Tm values could be determined at Mccp and Mmc plasmid concentrations as low as 55 copies/μL and 58 copies/μL, respectively. The Ct values of Mccp and Mmc were both less than 40, there was a good correlation between copy number (105 to 101 copies/μL) and the Ct values, no reaction was detected at concentrations of 100copies/μL, Taken together, the minimum detectable limits of the HRM method for Mccp and Mmc were 55 and 58 copies/μL, respectively. The formula between Ct values and copy number (105 to 101 copies/μL) for Mccp and Mmc is y = − 3.5765logx + 44.83 (R2 = 0.9972) and y = − 3.6640logx + 44.41 (R2 = 0.9974) respectively(Fig. 3). The minimum detectable Ct values between the two groups were compared with independent t-test, the statistical analyses show the average Ct differences between the two pathogens were significant (P = 0.012, P < 0.05) (Table 2). The results of agarose gel electrophoresis showed that the corresponding minimum detection limits of conventional PCR were 5.49 × 104 copies/μL for Mccp and 5.83 × 104 copies/μL for Mmc, indicating that the HRM method was more sensitive than conventional PCR detection. This analysis was conducted using both the control plasmids and genomic DNA as the template. After the genomic DNA was diluted 10 times, the minimum detectable limit for Mmc and Mccp was determined as 300 fg and 400 fg, respectively.
Figure 3.
Sensitivity tests of the HRM method of Mccp and Mmc positive samples. The pictures are generated by Rotor Gene Q. (A) Sensitivity tests of the HRM method for Mccp; (B) Sensitivity tests of the HRM method for Mmc. (C) Standard curve of Mccp performed in a linear graph with R2 = 0.9972. (D) Standard curve of Mmc performed in a linear graph with R2 = 0.9974.
Table 2.
Comparison of LOD in the detection of Mmc and Mccp.
| No. | Mccp | Mmc | ||
|---|---|---|---|---|
| Ct mean ± SD |
CV/% | Ct mean ± SD |
CV/% | |
| 10–9 | 38.81 ± 0.03a | 0.09 | 38.36 ± 0.18b | 0.4 |
Different lowercase letters in the same row mean significant difference (0.01 < P < 0.05). The average Ct values differences between the two pathogens were significant (P = 0.012 < 0.05).
Clinical sample testing results
A total of 153 clinical samples from goats suspected of having mycoplasma infection were detected using HRM, real-time PCR, and pathogen isolation to compare the accuracy of the HRM method. The results (Table 3) showed that 31 Mmc positive samples were detected by HRM, but no Mccp were detected. A total of 23 Mmc HRM-positive samples were also detected with fluorescence qPCR while qPCR did not detect any Mccp in the samples. Using pathogen isolation methods, two Mmc positive samples were identified, while Mccp was not detected. Two samples positive for Mmc using mycoplasma isolation and identification were also positive for Mmc using the HRM and qPCR methods. Of the 31 HRM-positive samples for Mmc, only 23 were also determined positive with qPCR. The 8 samples that tested positive for Mmc by HRM but negative for Mmc by qPCR were sequenced, and the results were subjected to a BLAST search, which showed that the homology with Mmc was 100%. Thus, the coincidence rate of HRM and real-time PCR was 94.8% (145/153) and for HRM and pathogen isolation was 87.0% (40/46) (Table 3). Thus, the HRM method established in this study was more sensitive than fluorescence qPCR and pathogen isolation.
Table 3.
Results of clinical samples with real-time PCR and HRM methods.
| Detecting method | HRM | Total | ||
|---|---|---|---|---|
| Mmc positive number | Mmc negative number | |||
| Real-time PCR | Mmc positive number | 23 | 0 | 23 |
| Mmc negative number | 8 | 122 | 130 | |
| Sum | 31 | 122 | 153 | |
| Pathogen isolation and identification | Mmc positive number | 2 | 0 | 2 |
| Mmc negative number | 6 | 38 | 44 | |
| Sum | 8 | 38 | 46 | |
Discussion
Contagious caprine pleuropneumonia and mycoplasmal pneumonia in sheep and goats have been common in China for a long period of time. However, before 2007, mycoplasmal pneumonia in sheep and goats caused by Mmc was also regarded as contagious caprine pleuropneumonia in China23–25. This was mainly because Mccp and Mmc both belong to the M. mycoides cluster and have high homology and cause similar clinical symptoms. They show cross-reactivity in serology analysis making distinguishing these mycoplasmas with immunological methods difficult. Therefore, it is clinically important to establish a rapid and simple method for differentiating Mccp and Mmc.
Amores et al. have demonstrated that PCR is a faster and more sensitive method for detection of Mmc than mycoplasma isolation and identification26. Fluorescence qPCR is more rapid with greater sensitivity and specificity than conventional PCR, and has been widely used in the clinical detection of various pathogens27–29. Woubit30 established in 2007 a real-time PCR method for detecting the M. mycoides cluster, which can detect and identify Mmc and Mccp, but it requires two real-time PCR reactions.
HRM has been widely used for mutation scanning, methylation detection, mononucleotide polymorphism analysis, genotyping, sequence matching, and other applications because of its high sensitivity, good specificity, low cost, high-throughput detection. Douarre et al.31 described in 2012 a HRM-PCR assay for clearly differentiating the two main types of Mycobacterium avium subsp. paratuberculosis (cattle and sheep) and Gurtler et al.32 developed in the same year a HRM-PCR method for identification and van genotyping different Enterococcus species. In 2018, Liu et al.33 developed a highly specific multiplex high resolution melt-curve real-time PCR assay for the reliable detection of Salmonella serotypes. This approach is also suitable for establishing a method for rapidly identifying Mccp and Mmc.
In this study, the Mccp and Mmc sequences were compared to those published in NCBI to identify conserved gene fragments. A primer pair was designed using the Primer Premier 6.0 software to establish a HRM detection method for Mccp and Mmc. This method demonstrated specificity towards Mccp and Mmc without amplifying corresponding sequences from other common goat pathogens. Additionally, five positive samples detected using the HRM method were randomly selected for sequencing. The results showed that the sequence homology between the determined sequence and corresponding Mmc fragment was 100%, which demonstrated the high specificity of the method. The method showed strong reproducibility as melting curves of Mccp and Mmc were essentially the same as those of the positive control samples, and the intra- and inter-batch coefficients of variation for the two melting peaks were < 1%. Results showed that the minimum detection limits of Mccp- and Mmc-positive plasmids using this method were 55 copies/µL, and 58 copies/µL, respectively, whereas the minimum detection limit using SYBR GreenI qRT-PCR was 226 copies/µL and the detection limit with conventional PCR was 2.26 × 104 copies/L19. Furthermore, the minimum detection limits of the multiplex PCR method were 4.03 × 105 copies/µL and 6.78 × 105 copies/µL for Mmc and Mccp, respectively15, indicating that the HRM method was more sensitive than the conventional and fluorescence qPCR methods for Mmc and Mccp detection.
To verify the practicability of this method in clinical testing of Mmc and Mccp, a total of 153 clinical samples were detected using the HRM method established in this study and compared to the results of fluorescence qPCR and Mycoplasma isolation and identification. The results show that the detection rate of Mmc using HRM was 20.3%, whereas that of SYBR GreenI qRT-PCR Mmc analysis and Mycoplasma isolation was 15.0% and 4.3%, respectively. Additionally, eight samples that were detected to be positive for Mmc with HRM method but negative by SYBR Green I qRT-PCR method were sequenced. The sequencing results confirmed the consistency with the corresponding fragment sequence of Mmc to be 100%, supporting the accuracy of Mmc detection using the HRM method, and indicating that the sensitivity of the HRM method was also higher than that of fluorescence qPCR and Mycoplasma isolation in detecting clinical samples. Furthermore, Thiaucourt9 established a real-time PCR method for detecting the M. mycoides cluster, which can detect and differentiate Mmc and Mccp, but it requires two separate fluorescence qPCR methods. The HRM method established in this study can differentiate Mmc and Mccp in only one reaction, which is faster and less labor-intensive. Because Mccp has not been found in Fujian Province, it was not detected in any of the clinical samples, which is consistent with the results of Jiang et al.34.
In summary, detection of Mmc and Mccp based on the HRM analysis established in this study can quickly and accurately distinguish the two pathogens with high specificity, sensitivity, and reproducibility. Compared to conventional and fluorescence qPCR methods, this method showed higher sensitivity and convenient operation. It can be used for rapid detection and identification of Mmc and Mccp in clinical nasal swab samples, rapid diagnosis, and epidemiological investigation of goat mycoplasmal pneumonia and contagious caprine pleuropneumonia.
Acknowledgements
We thank Prof. Yuefeng Chu from the Lanzhou Veterinary Research Institute for providing the Mccp F38 and M1601, Mcc California Kid, Mmc PG3, strains. We would also like to thank Prof. Jiuqing Xin from the Harbin Veterinary Research Institute for providing M. leachii PG50. We thank Prof. Cheng Tang from the Collage of Life and Technology, Southwest University for Nationalities for providing the M. agalactiae strain PG2. We also thank Prof. Long-Fei Cheng from the Institute of Animal Science and Veterinary Medicine, Fujian Academy of Agricultural Sciences for providing the E. coli strain ATCC25922 (Guangdong Huankai Microbial), Pm strain CVCC44801 (China Institute of Veterinary Drug Control), and Sa strain 261111 (National Institutes for Food and Drug Control).
Author contributions
J.Z. and Q.H. wrote the main manuscript text and Z.L. prepared figures. J.J. and Y.L. isolated and identified the bacterial strain. The methods were verified by W.Y. All authors reviewed the manuscript.
Funding
This work was supported by the National Key Research Project of China (grant number 2016YFD0500906), Innovative Research Team for Fujian Academy of Agriculture Science (FAAS) (grant numbers STIT2017-1-5; STIT2017-3-10), Innovative Research Project of FAAS (grant numbers PC-2018-6 and PC-2018-8 and the Public Scientific Research Institution fund (grant number 2019R1026-17).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Macowan KJ, Minette JE. The role of mycoplasma strain f38 in contagious caprine pleuropneumonia (cccp) in Kenya. Vet. Rec. 1977;101(19):380–381. doi: 10.1136/vr.101.19.380. [DOI] [PubMed] [Google Scholar]
- 2.Macowan KJ, Minette JE. A mycoplasma from acute contagious caprine pleuropneumonia in Kenya. Trop. Anim. Health Prod. 1976;8(1):91–95. doi: 10.1007/BF02383376. [DOI] [PubMed] [Google Scholar]
- 3.Rurangirwa, et al. Treatment of contagious caprine pleuropneumonia. Trop. Anim. Health Prod. 1981;13(3):177–182. doi: 10.1007/BF02237919. [DOI] [PubMed] [Google Scholar]
- 4.Oie. Contagious Caprine Pleuropneumonia. In Manual of diagnostic tests and vaccines for terrestrial animals (mammals, birds and bees), 1000–1012 (World Organization for Animal Health, 2008).
- 5.Adehan RK, Ajuwape ATP, Adetosoye AI, Alaka OO. Characterization of mycoplasmas isolated from pneumonic lungs of sheep and goats. Small Rumin. Res. 2006;63(1–2):44–49. doi: 10.1016/j.smallrumres.2005.01.014. [DOI] [Google Scholar]
- 6.Chu Y, Zhao P, Gao P, He Y, Lu Z. Mycoplasmas and mycoplasmosis in sheep and goats. Anim. Husb. Feed Sci. 2010;2(3):128–131. doi: 10.19578/j.cnki.ahfs.2010.03.009. [DOI] [Google Scholar]
- 7.Hernandez, et al. Mycoplasma mycoides subsp. capri associated with goat respiratory disease and high flock mortality. Can. Vet. J. 2006;47(4):366–369. doi: 10.1080/03079450600598244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thiaucourt, et al. Mycoplasma mycoides, from "mycoides small colony" to "capri": A microevolutionary perspective. BMC Genomics. 2011;12(1):114. doi: 10.1186/1471-2164-12-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thiaucourt F, Bölske G. Contagious caprine pleuropneumonia and other pulmonary mycoplasmoses of sheep and goats. Rev. Sci. Tech. 1996;15(4):1397–1414. doi: 10.1111/j.1439-0531.1996.tb01445.x. [DOI] [PubMed] [Google Scholar]
- 10.Bölske, et al. Diagnosis of contagious caprine pleuropneumonia by detection and identification of Mycoplasma capricolum subsp. capripneumoniae by pcr and restriction enzyme analysis. J. Clin. Microbiol. 1996;34(4):785–791. doi: 10.1002/(SICI)1097-4660(199604)65:4<380::AID-JCTB452>3.0.CO;2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hotzel H, Sachse K, Pfutzner H. A pcr scheme for differentiation of organisms belonging to the mycoplasma mycoides cluster. Vet. Microbiol. 1996;49(1–2):31. doi: 10.1016/0378-1135(95)00176-X. [DOI] [PubMed] [Google Scholar]
- 12.Kumar P, Roy A, Bhanderi BB, Pal BC. Isolation, identification and molecular characterization of Mycoplasma isolates from goats of Gujarat state, India. Veterinarski Arhiv. 2011;81(4):443–458. doi: 10.1258/la.2011.010112. [DOI] [Google Scholar]
- 13.Righter DJ, Rurangirwa FR, Call DR, Mcelwain TF. Development of a bead-based multiplex pcr assay for the simultaneous detection of multiple mycoplasma species. Vet. Microbiol. 2011;153(3–4):246–256. doi: 10.1016/j.vetmic.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 14.Zheng JQ, et al. Establishment and application of a multiplex PCR assay for simultaneous detection of Mycoplasma ovipneumoniae, Mycoplasma mycoides subsp. capri and Mycoplasma arginini. Chin J. Vet. Sci. 2016;36(7):1131–1134. doi: 10.16303/j.cnki.1005-4545.2016.07.10. [DOI] [Google Scholar]
- 15.Yu-Sheng L, Jin-Xiu J, Bin J, Wei Y, Qi-Lin HU. Development of a triplex PCR assay for detection of Mycoplasma ovipneumoniae, M. mycoides subsp. capri and M. capricolum subsp. capripneumoniae. Chin. J. Prev. Vet. Med. 2018 doi: 10.3969/j.issn.1008-0589.201710026. [DOI] [Google Scholar]
- 16.Chu YF, Lu ZX, Zhao P, Peng-cheng G, Ying HE. Development of nested PCR assay for detecting Mycoplasma mycoides subsp. capri. Vet. Sci. Chin. 2007 doi: 10.16656/j.issn.1673-4696.2007.11.003. [DOI] [Google Scholar]
- 17.Fitzmaurice, et al. Real-time polymerase chain reaction assays for the detection of members of the mycoplasma mycoides cluster. N. Z. Vet. J. 2008;56(1):40–47. doi: 10.1080/00480169.2008.36803. [DOI] [PubMed] [Google Scholar]
- 18.Yu-Sheng L, Jin-Xiu J, Jing-Peng Z. Establishment of a SYBR Green I qRT-PCR for rapid detection of Mycoplasma mycoides subsp. capri. J. Agric. Biotechnol. 2017;2017(11):171–178. doi: 10.3969/j.issn.1674-7968.2017.11.017. [DOI] [Google Scholar]
- 19.Lin YS, Jiang JX, Zhang JP, Wei Y, Hu QL. Establishment of a SYBR green I qRT-PCR for rapid detection of Mycoplasma capricolum subsp. capripneumoniae. J. Agric. Biotechnol. 2018;26(2):339–345. doi: 10.3969/j.issn.1674-7968.2018.02.017. [DOI] [Google Scholar]
- 20.Wittwer CT, Reed GH, Gundry CN, Pryor RJ. High-resolution genotyping by amplicon melting analysis using LC Green. Clin. Chem. 2003;49(6):853–860. doi: 10.1373/49.6.853. [DOI] [PubMed] [Google Scholar]
- 21.Reed GH, Wittwer CT. Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin. Chem. 2004;50(10):1748–1754. doi: 10.1373/clinchem.2003.029751. [DOI] [PubMed] [Google Scholar]
- 22.Lin Y, Jiang J, Zhang J, et al. Development of a TaqMan real-time PCR for detection of the Mycoplasma mycoides subsp. capri. Small Rumin. Res. 2019;175:31–36. doi: 10.1016/j.smallrumres.2019.02.014. [DOI] [Google Scholar]
- 23.Li Y, Zhang JH, Hu SP, Wang L, Xin JQ. Reclassification of the four china isolated strains of the pathogen for contagious caprine pleuropneumonia. Acta Microbiol. Sinica. 2007;47(5):769–773. doi: 10.3321/j.issn:0001-6209.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 24.Wang D, Zhang RT. Study on the pathogen of contagious caprine pleuropneumonia in China. Chin. Vet. Sci. 1988;9:5–7. [Google Scholar]
- 25.Zhou LB, Guo JG, Liu Q. Isolation and identification of the pathogen of Contagious caprine pleuropneumonia in Guangxi goats, Shanghai. J. Anim. Husb. Vet. Med. 2007;4:16–19. [Google Scholar]
- 26.Amores J, et al. Comparison of culture and pcr to detect mycoplasma agalactiae and Mycoplasma mycoides subsp. capri in ear swabs taken from goats. Vet. Microbiol. 2010;140(1–2):105–108. doi: 10.1016/j.vetmic.2009.06.036. [DOI] [PubMed] [Google Scholar]
- 27.Landolt P, Stephan R, Scherrer S. Development of a new high resolution melting (HRM) assay for identification and differentiation of Mycobacterium tuberculosis complex samples. Sci. Rep. 2019 doi: 10.1038/s41598-018-38243-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murillo E, Muskus C, Agudelo LA, Vélez ID, Ruiz-Lopez F. A new high-resolution melting analysis for the detection and identification of plasmodium in human and anopheles vectors of malaria. Sci. Rep. 2019;9(1):1–9. doi: 10.1038/s41598-018-36515-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chibssa TR, et al. An HRM assay to differentiate sheeppox virus vaccine strains from sheeppox virus field isolates and other Capripoxvirus species. Sci. Rep. 2019;9:6646. doi: 10.1038/s41598-019-43158-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woubit S, et al. A PCR for the detection of mycoplasmas belonging to the mycoplasma mycoides cluster: Application to the diagnosis of contagious agalactia. Mol. Cell. Probes. 2007;21(5):391–399. doi: 10.1016/j.mcp.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 31.Douarre PE, Cashman W, Buckley J, Coffey A, O'Mahony JM. High resolution melting PCR to differentiate Mycobacterium avium subsp. paratuberculosis "cattle type" and "sheep type". J. Microbiol. Methods. 2012;88(1):172–174. doi: 10.1016/j.mimet.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 32.Gurtler V, Grando D, Mayall BC, Wang J, Ghaly-Derias S. A novel method for simultaneous enterococcus species identification/typing and van genotyping by high resolution melt analysis. J. Microbiol. Methods. 2012;90(3):167–181. doi: 10.1016/j.mimet.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, Singh P, Mustapha A. Multiplex high resolution melt-curve real-time pcr assay for reliable detection of salmonella. Food Contr. 2018;91(1):225–230. doi: 10.1016/j.foodcont.2018.03.043. [DOI] [Google Scholar]
- 34.Jiang JX, Lin YS, You W, Hu QL. Isolation and identification of Mycoplasma mycoides subsp. capri FJ-GT strain. Chin. Agric. Sci. Bull. 2016;32(29):11–16. doi: 10.11924/j.issn.1000-6850.casb16030148. [DOI] [Google Scholar]