Abstract
The role of the long noncoding RNA CCAT1 NC_000008.10:g.128220661C > T (rs67085638) in the development of colon cancer has been reported. Therefore, we assessed the prevalence of rs67085638 in patients with gastric cancer (GC). We also evaluated the effect of rs67085638 on B-cell-specific Moloney leukaemia virus insertion site 1 (BMI1) transcripts in primary GC and counterpart histopathologically confirmed disease-free margin tissue. Using high-resolution melting analysis, we evaluated rs67085638 frequency in patients with the GC genotype (n = 214) and controls (n = 502) in a Polish Caucasian population. qRT-PCR was used to determine BMI1 transcripts. We observed the trend of rs67085638 association in all patients with GC (ptrend = 0.028), a strong risk of the GC genotype in male (ptrend = 0.035) but not female (ptrend = 0.747) patients, and the association with non-cardia GC (ptrend = 0.041), tumour stages T3 (ptrend = 0.014) and T4 (ptrend = 0.032), differentiation grading G3 (ptrend = 0.009), lymph node metastasis stage N3 (ptrend = 0.0005) and metastasis stage M0 (ptrend = 0.027). We found that significantly increased BMI1 transcripts were associated with the primary GC genotype classified as grade G3 (p = 0.011) and as lymph node metastasis N3 (p = 0.010) and counterpart marginal tissues (p = 0.026, p = 0.040, respectively) from carriers of the T/T versus C/C genotypes. rs67085638 may contribute to increased BMI1 transcripts and the progression and rapid growth of GC.
Subject terms: Cancer, Risk factors
Introduction
Gastric cancer (GC) is a deadly disease resulting in more than 841,000 deaths each year1. The development of GC is due to the interaction of environmental and genetic factors. Environmental carcinogenic factors encompass low socioeconomic status, tobacco smoking, radiation, Helicobacter pylori infection, low consumption of fruits and vegetables and high intake of salty and smoked food, male sex, obesity and older age2–6. However, not everyone is exposed to the environmental carcinogenic factors that develop into GC, which suggests a strong genetic background contributing to its occurrence6–8. A genetic study demonstrated the different variants of genes that contribute to the development of GC8. Genome-wide association studies (GWAS) have revealed many loci as major genetic components in the development of GC9–11. Many previously studied single nucleotide polymorphisms (SNPs) did not reach statistical significance in GWAS, but they may force the development and progression of GC6–8.
Long noncoding RNAs (lncRNAs) are a class of transcripts that are longer than 200 nucleotides in length and are not translated to proteins12. In the nuclei and cytosol, lncRNAs play critical roles in various mechanisms regulating gene expression, which can be related to development, progression and drug resistance in cancer13–15. Abnormal lncRNA expression has been demonstrated in many malignancies, including GC15.
Recently, studies have suggested that colon cancer-associated transcript-1 (CCAT1), also designated LOC100507056 or the cancer-associated region lncRNA-5, plays a role in the growth and invasion of GC16–19. CCAT1 includes a region 2,628 base pairs in length, and its expression results in two isoforms of lncRNAs: CCAT1-L and CCAT1-S. CCAT1-L is exclusively situated in the nucleus, whereas the short isoform, CCAT1-S, is positioned in the cytoplasm20.
The CCAT1 NC_000008.10:g.128220661C > T (rs67085638) polymorphism is associated with the development of colon cancer21. However, little is known about the contribution of rs67085638 in the development of GC. In our study, we assessed the prevalence of the CCAT1 rs67085638 polymorphism in patients with GC in a Polish Caucasian population. We also evaluated the distribution of rs67085638 in different clinicopathological characteristics of GC.
Recent findings demonstrated that the presence of the minor allele of rs67085638 increased the expression of CCAT122. Moreover, lncRNA CCAT1 knockdown significantly downregulated B-cell-specific Moloney leukaemia virus insertion site 1 (BMI1) mRNA in GC23. Therefore, we also assessed the effect of CCAT1 rs67085638 on the transcript levels of BMI1 in primary GC tissue and counterpart histopathologically confirmed disease-free margin tissue.
Materials and methods
Study population
The studied subjects included 214 patients with gastric adenocarcinoma (GC, 54 cardia and 160 non-cardia) who were diagnosed by oncologists. All cases were gastric adenocarcinoma and were confirmed by pathological examination with routine evaluation of differentiation stage and grade according to the staging system of the tumour node metastasis (TNM) classification system and the World Health Organization (Table 1)24. Patient data and primary GC tissue samples were obtained from subjects enrolled between July 2016 and January 2019 at the Department of Oncological Gastroenterology, Maria Sklodowska-Curie Oncology Center, Warsaw, Poland. The inclusion criteria were age over 18 years, new diagnosis of gastric adenocarcinoma, and ability to endure surgical treatment or chemotherapy. The exclusion criteria encompassed patients with a non-adenocarcinoma and the presence of an extragastric tumour. Subjects with severe systemic disorders were also excluded from the group of patients with GC.
Table 1.
Available clinicopathological characteristics of patients with gastric cancer.
|
Sex No. of patients |
Male 139 |
Female 75 |
||
| aMean age (years) ± SD | 56.1 ± 11.8 | 57.5 ± 12.1 | ||
|
GC localization No. of patients |
Cardia 54 |
Non-cardia 160 |
||
|
Histological type No. of patients |
Diffuse 111 |
Intestinal 79 |
Mixed 18 |
Undetermined 6 |
|
Tumour stage No. of patients |
T1 24 |
T2 32 |
T3 86 |
T4 72 |
|
Lymph node metastasis stage No. of patients |
N0 78 |
N1 35 |
N2 42 |
N3 59 |
|
Metastasis stage No. of patients |
M0 192 |
M1 22 |
||
|
Histological grading No. of patients |
G1 7 |
G2 66 |
G3 141 |
T tumour, N node, M metastasis, G grade22.
aAge at first diagnosis.
The control group was matched to patients by age and sex and included 175 healthy females with a mean age of 56.9 ± 12.3 years and 327 healthy males with a mean age of 55. 8 ± 11.8 years randomly selected during routine medical examinations at the Department of Radiotherapy of the Greater Poland Cancer Center in Poznań, Poland, Subjects who were pregnant, had a history of cancer or had blood relatives with GC going back two generations were excluded from the control group. We also obtained body mass index (BMI) data for the controls and patients with GC as well as the presence of diabetes in the GC patients (Supplementary Table 125).
Tissue samples
We found the greatest association of rs67085638 with G3 differentiation grading and with N3 lymph node metastasis in patients with GC. Therefore primary GC tissue and counterpart histopathologically confirmed disease-free margin tissue from these patients were collected to determine the influence of rs67085638 on the level of BMI1 transcript. Primary GC tissue samples and distal surgical resection margin histopathologically confirmed disease-free tissue samples were obtained from 42 patients with a mean age of 51.3 ± 7.6 years and classified as grade G3 at the time of surgery and from 42 patients with a mean age of 52.4 ± 6.6 years and classified as N3 at the time of surgery at the Department of Oncological Gastroenterology, Maria Sklodowska-Curie Oncology Center, Warsaw. A portion of the tissue sample was immediately snap-frozen in liquid nitrogen and stored at − 80 °C until RNA isolation was performed.
Genetic analysis
DNA was isolated from peripheral blood cells via a salting-out procedure. The primers were designated using Oligo 7.6 software (DBA Oligo, Inc., Colorado Springs, CO). The NC_000008.10:g.128220661C > T (rs67085638) polymorphism DNA fragment (140 bp) was amplified using the following primers: forward 5′ GCTGTAAATAACGCTGAT 3′ and reverse 5′AACTGAATGAGATGAAGG 3′. The rs67085638 SNP was then genotyped via high-resolution melting (HRM) curve analysis previously described26 using HOT FIREPol EvaGreen (Solis BioDyne, Tartu, Estonia) with a LightCycler 480 system (Roche Diagnostics, Mannheim, Germany). The presence of this SNP was reanalysed by Sanger sequencing analysis of arbitrarily chosen samples, comprising 10% of the samples from both cases and controls. The concordance rate between HRM and sequencing was 100%.
Reverse transcription and quantitative real-time PCR (qRT-PCR) analysis of BMI1 transcript levels in GC tissue and histopathologically confirmed disease-free margin tissue
Frozen primary GC and distal surgical resection counterpart margin histopathologically confirmed disease-free tissues were homogenized, and total RNA was isolated according to the method of Chomczyński and Sacchi27. RNA quality was determined spectrophotometrically and by agarose gel electrophoresis. RNA samples were treated with DNase I, quantified, and reverse-transcribed into complimentary DNA (cDNA) (Supplementary Table 2). Quantitative analysis of BMI1 cDNA (Supplementary data 1) was performed by the Light Cycler480 II Real-Time PCR System (Roche Diagnostics GmbH, Mannheim, Germany) with SYBR Green I as the detection dye. BMI1 cDNA was quantified using the relative quantification method with a calibrator (Supplementary Table 2). The quantity of the BMI1 transcript in each sample was standardized by the geometric mean of reference transcript levels: hydroxymethylbilane synthase (HMBS) and beta-2-microglobulin (B2M) (Supplementary Table 2). The BMI1 mRNA levels were expressed as multiples of these cDNA concentrations in the calibrator.
Statistical analysis
The distinction in genotypic prevalence between the patients and controls and their genotype deviation from Hardy–Weinberg (HW) equilibrium were evaluated using a χ2 test. The rs67085638 SNP was tested for association with GC using the Cochran–Armitage p-trend test (ptrend). The χ2 and Fisher exact tests were used to determine the differences in genotypic and allelic distributions between the patients and controls. The odds ratios (ORs) and 95% confidence intervals (95% CIs) were also calculated. Logistic regression analysis was used to adjust for the effect of age, BMI and the presence of diabetes. A p-value of < 0.05 was considered statistically significant. Statistical analysis comparing BMI1 transcript levels between the T/T versus C/C and C/T versus C/C genotype carriers was evaluated using the Kruskal–Wallis test with Dunn's post hoc test. Statistical analyses were conducted using Statistica version 10, 2011 (Stat Soft, Inc., Tulsa, USA), as previously described28.
Ethical approval
The study procedures were approved by the Local Ethical Committee of the Poznań University of Medical Sciences (reference number of ethical approval: 673/15 and 190/19). The study was carried out in accordance with the approved guidelines. Informed consent was obtained from all individual participants included in the study.
Results
Distribution of the CCAT1 rs67085638 SNP between the group of patients with GC and the control group
The χ2 test of HW equilibrium showed values of 0.973 and 0.949 for the patients with GC and the control group, respectively. The genotype distribution of the rs67085638 SNP in the group of patients is presented in Table 2. We found a significant association of the rs67085638 SNP with all the patients with GC, and the p-trend value calculated for the rs67085638 polymorphism was ptrend = 0.028. The logistic regression analysis, which was adjusted for the effects of age, BMI and presence of diabetes, demonstrated an association of the rs6983267 SNP with T/T as well as the T/T + C/T genotype and T allele for all the patients with GC. For the T/T versus C/C genotypes, the adjusted OR was 1.360 (95% CI 1.011–1.828, p = 0.041); for the C/T + T/T versus C/C genotypes, the adjusted OR was 1.423 (95% CI 1.025–1.977, p = 0.035); and for T versus C, the adjusted OR was 1.373 (95% CI 1.056–1.784, p = 0.018). However, there was no significant association of the rs6983267 SNP for C/T versus C/C, where the adjusted OR was 1,430 (95% CI 0.981–2.085, p = 0.063).
Table 2.
Prevalence of the CCAT1 rs67085638 polymorphism among patients with GC and controls.
| Genotype | Patients (frequency) | Controls (frequency) | Odds ratio (95%CI) | pa | Adjusted odds ratio (95%CI)b | p | ptrend |
|---|---|---|---|---|---|---|---|
| All | |||||||
| C/C | 85(0.40) | 239 (0.48) | Referent | Referent | 0.028 | ||
| C/T | 101 (0.47) | 217 (0.43) | 1.309 (0.9295 –1.843) | 0.123 | 1.430 (0.981–2.085) | 0.063 | |
| T/T | 28 (0.13) | 46 (0.09) | 1.712 (1.006–2.911) | 0.0458 | 1.360 (1.011–1.828) | 0.041 | |
| C/T + T/T | 129 (0.60) | 263 (0.52) | 1.379 (0.9964–1.909) | 0.0522 | 1.423 (1.025–1.977) | 0.035 | |
| MAFc | 0.37 | 0.31 | 1.303 (1.027–1.653) | 0.029 | 1.373 (1.056–1.784) | 0.018 | |
| Sex | |||||||
|---|---|---|---|---|---|---|---|
| Male | |||||||
| C/C | 49 (0.35) | 158 (0.48) | Referent | – | Referent | – | 0.035 |
| C/T | 68 (0.49) | 139 (0.43) | 1.577 (1.024–2.431) | 0.0381 | 1.640 (1.059–2.540) | 0.026 | |
| T/T | 22 (0.16) | 30 (0.9) | 2.365 (1.251–4.470) | 0.0071 | 1.554 (1.124–2.147) | 0.007 | |
| C/T + T/T | 90 (0.65) | 169 (0.52) | 1.717 (1.139–2.588) | 0.0094 | 1.789 (1.181–2.712) | 0.001 | |
| MAFd | 0.40 | 0.30 | 1.543 (1.152–2.066) | 0.0035 | 1.610 (1.169–2.216) | 0.003 | |
| Female | |||||||
| C/C | 36 (0.48) | 81 (0.46) | Referent | – | Referent | – | 0.747 |
| C/T | 33 (0.44) | 78 (0.44) | 0.952 (0.5407–1.676) | 0.864 | 0.958 (0.539–1.702) | 0.896 | |
| T/T | 6 (0.08) | 16 (0.09) | 0.844 (0.3051–2.334) | 0.743 | 0.931 (0.520–1.667) | 0.807 | |
| C/T + T/T | 39 (0.52) | 94 (0.54) | 0.934 (0.5430–1.605) | 0.803 | 0.935 (0.540–1.620) | 0.809 | |
| MAFc | 0.30 | 0.31 | 0.935 (0.617–1.417) | 0.752 | 0.973 (0.611–1.551) | 0.909 | |
Significant results are highlighted in bold font.
aχ2. bORs were adjusted by age, BMI and presence of diabetes. cMinor allele frequency.
The division of the patients based on sex revealed a significant contribution of rs67085638 to GC in the male patients (ptrend = 0.035). For T/T versus C/C, the adjusted OR was 1.554 (95% CI 1.124–2.147, p = 0.007); for C/T versus C/C, the adjusted OR was 1.640 (95% CI 1.059–2.540, p = 0.026); for C/T + T/T versus C/C, the adjusted OR was 1.789 (95% CI 1.181–2.712, p = 0.001); and for T versus C, the adjusted OR was 1.610 (95% CI 1.169–2.216, p = 0.003). However, there was no contribution of rs67085638 to GC in the female patients (ptrend = 0.747). For T/T versus C/C, the adjusted OR was 0.931 (95% CI 0.520–1.667, p = 0.807); for C/T versus C/C, the adjusted OR was 0.958 (95% CI 0.539–1.702, p = 0.896); for C/T + T/T versus C/C, the adjusted OR was 0.935 (95% CI 0.540–1.620, p = 0.809); and for T versus C, the adjusted OR was 0.973 (95% CI 0.611–1.551, p = 0.909).
Prevalence of the CCAT1 rs67085638 SNP among patients with cardia and non-cardia localization of GC
In the patients with cardia localization of GC, ptrend = 0.253, and for T/T versus C/C, the adjusted OR was 1.334 (95% CI 0.839–2.123, p = 0.222); for C/T versus C/C, the adjusted OR was 1.257 (95% CI 0.664–2.377, p = 0.481); for C/T + T/T versus C/C, the adjusted OR was 1.411 (95% CI 0.792–2.512, p = 0.241); and for T versus C, the adjusted OR was 1.279 (95% CI 0.820–1.993, p = 0.277).
In the patients with non-cardia localization of GC, we found a significant association of the rs67085638 SNP with GC, and the p-trend value calculated for the rs67085638 polymorphism was ptrend = 0.041. The logistic regression analysis demonstrated an association of the rs6983267 SNP with the C/T + T/T genotype as well as the T allele. For C/T + T/T versus C/C, the adjusted OR was 1.576 (95% CI 1.051–2.364, p = 0.028), and for T versus C, the adjusted OR was 1.404 (95% CI 1.047–1.881, p = 0.023). However, there was no significant association of the rs6983267 SNP with T/T versus C/C; the adjusted OR was 1.320 (95% CI 0.982–1.773, p = 0.065), and for C/T versus C/C, the adjusted OR was 1.252 (95% CI 0.922–1.985, p = 0.122).
Prevalence of the CCAT1 rs67085638 SNP in patients with diffuse, mixed and intestinal histological types of GC
There was no association of the rs67085638 polymorphism with diffuse, mixed and intestinal histological types of GC (Table 3). In the patients with diffuse histological types of GC, ptrend = 0.073, and for T/T versus C/C, the adjusted OR was 1.353 (95% CI 0.965–1.897, p = 0.078); for C/T versus C/C, the adjusted OR was 1.348 (95% CI 0.863–2.106, p = 0.189); for C/T + T/T versus C/C, the adjusted OR was 1.440 (95% CI 0.943–2.199, p = 0.091); and for T versus C, the adjusted OR was 1.402 (95% CI 0.999–1.966, p = 0.049). In the patients with mixed histological types of GC, ptrend = 0.493, and for T/T versus C/C, the adjusted OR was 1.238 (95% CI 0.552–2.774, p = 0.603); for C/T versus C/C, the adjusted OR was 1.810 (95% CI 0.579–5.657, p = 0.306); for C/T + T/T versus C/C, the adjusted OR was 1.427 (95% CI 0.541–3.759, p = 0.471); and for T versus C, the adjusted OR was 1.267 (95% CI 0.632–2.540, p = 0.503). In the patients with intestinal histological types of GC, ptrend = 0.306, and for T/T versus C/C, the adjusted OR was 1.206 (95% CI 0.804–1.809, p = 0.363); for C/T versus C/C, the adjusted OR was 1.278 (95% CI 0.767–2.128, p = 0.345); for C/T + T/T versus C/C, the adjusted OR was 1.314 (95% CI 0.781–2.211, p = 0.303); and for T versus C, the adjusted OR was 1.172 (95% CI 0.795–1.727, p = 0.422).
Table 3.
Prevalence of the CCAT1 rs67085638 polymorphism among localization of GC and histological type of GC.
| GC localization | |||||||
|---|---|---|---|---|---|---|---|
| Genotype | Patients (frequency) | Controls (frequency) | Odds ratio (95%CI) | pa | Adjusted odds ratio (95%CI)c | p | ptrend |
| Cardia | |||||||
| C/C | 22 (0.41) | 239 (0.48) | Referent | – | Referent | – | 0.253 |
| C/T | 25 (0.46) | 217 (0.43) | 1.252 (0.686–2.285) | 0.464 | 1.257 (0.664–2.377) | 0.481 | |
| T/T | 7 (0.13) | 46 (0.09) | 1.653 (0.667–4.096) | 0.273 | 1.334 (0.839–2.123) | 0.222 | |
| C/T + T/T | 32 (0.59) | 263 (0.52) | 1.322 (0.747–2.339) | 0.337 | 1.411 (0.792–2.512) | 0.241 | |
| MAFd | 0.36 | 0.31 | 1.271 (0.840–1.925) | 0.256 | 1.279 (0.820–1.993) | 0.277 | |
| Non-cardia | |||||||
| C/C | 63 (0.39) | 239 (0.48) | Referent | – | Referent | – | 0.041 |
| C/T | 76 (0.48) | 217 (0.43) | 1.329 (0.908–1.945) | 0.143 | 1.252 (0.922–1.985) | 0.122 | |
| T/T | 21 (0.13) | 46 (0.09) | 1.732 (0.964–3.113) | 0.064 | 1.320 (0.982–1.773) | 0.065 | |
| C/T + T/T | 97 (0.61) | 263 (0.52) | 1.399 (0.974–2.010) | 0.069 | 1.576 (1.051–2.364) | 0.028 | |
| MAFd | 0.37 | 0.31 | 1.314 (1.009–1.710) | 0.0421 | 1.404 (1.047–1.881) | 0.023 | |
| Histological type | |||||||
|---|---|---|---|---|---|---|---|
| Diffuse | |||||||
| C/C | 44 (0.40) | 239 (0.48) | Referent | – | Referent | – | 0.073 |
| C/T | 52 (0.47) | 217 (0.43) | 1.302 (0.837–2.024) | 0.241 | 1.348 (0.863–2.106) | 0.189 | |
| T/T | 15 (0.13) | 46 (0.09) | 1.771 (0.910–3.447) | 0.0893 | 1.353 (0.965–1.897) | 0.078 | |
| C/T + T/T | 67(0.60) | 263 (0.52) | 1.384 (0.910–2.104) | 0.128 | 1.440 (0.943–2.199) | 0.091 | |
| MAFd | 0.37 | 0.31 | 1.317 (0.972–1.785) | 0.0747 | 1.402 (0.999–1.966) | 0.052 | |
| Mixed | |||||||
| C/C | 7 (0.39) | 239 (0.48) | Referent | Referent | – | 0.493 | |
| C/T | 9 (0.50) | 217 (0.43) | 1.416 (0.5184–3.868) | 0.495 | 1.810 (0.579–5.657) | 0.306 | |
| T/T | 2 (0.11) | 46 (0.09) | 1.484 (0.2987–7.376) | 0.644b | 1.238 (0.552–2.774) | 0.603 | |
| C/T + T/T | 11 (0.61) | 263 (0.52) | 1.428 (0.5446–3.744) | 0.467 | 1.427 (0.541–3.759) | 0.471 | |
| MAFd | 0.36 | 0.31 | 1.271(0.636–2.543) | 0.4703b | 1.267 (0.632–2.540) | 0.503 | |
| Intestinal | |||||||
| C/C | 33 (0.42) | 239 (0.48) | Referent | – | Referent | – | 0.306 |
| C/T | 37 (0.47) | 217 (0.43) | 1.235 (0.746–2.044) | 0.411 | 1.278 (0.767–2.128) | 0.345 | |
| T/T | 9 (0.11) | 46 (0.09) | 1.417 (0.636–3.160) | 0.392 | 1.206 (0.804–1.809) | 0.363 | |
| C/T + T/T | 46 (0.58) | 263 (0.52) | 1.267 (0.784–2.048) | 0.334 | 1.314 (0.781–2.211) | 0.303 | |
| MAFd | 0.35 | 0.31 | 1.201(0.8432–1.711) | 0.310 | 1.172 (0.795–1.727) | 0.422 | |
The number of patients with undetermined histological type GC [(C/C-1 (0.17); C/T-3 (0.50): T/T-2 (0.33)] was too small; therefore, we did not apply statistical logistic regression.
Significant results are highlighted in bold font.
aχ2 or bFisher’s exact test. cORs were adjusted by age, BMI and presence of diabetes. dMinor allele frequency.
Prevalence of the CCAT1 rs67085638 SNP in patients in various tumour stages of GC
We found an association of the rs67085638 polymorphism with T3 and T4 tumour stages in the patients with GC (Table 4). In the patients with T3 tumour stage GC, ptrend = 0.014, and for T/T versus C/C, the adjusted OR was 1.522 (95% CI 1.039–2.230, p = 0.031); for C/T versus C/C, the adjusted OR was 1.807 (95% CI 1.087–3.006, p = 0.022); for C/T + T/T versus C/C. The adjusted OR was 1.716 (95% CI 1.022–2.879, p = 0.041), and for T versus C, the adjusted OR was 1.565 (95% CI 1.116–2.194, p = 0.009). In patients with T4 tumour stage GC, ptrend = 0.032, and for C/T versus C/C, the adjusted OR was 2.190 (95% CI 1.205–3.979, p = 0.010); for C/T + T/T versus C/C, the adjusted OR was 2.164 (95% CI (1.215–3.855, p = 0.009); and for T versus C, the adjusted OR was 1.505 (95% CI 1.048–2.159, p = 0.027). However, we did not observe an association of rs67085638 with T4 tumour stage; for T/T versus C/C, the adjusted OR was 1.403 (95% CI 0.924–2.133, p = 0.111).
Table 4.
Prevalence of the CCAT1 rs67085638 polymorphism among various tumour stages and differentiation grades of GC.
| Tumor stage | |||||||
|---|---|---|---|---|---|---|---|
| Genotype | Patients (frequency) | Controls (frequency) | Odds ratio (95%CI) | pa | Adjusted odds ratio (95%CI)c | p | ptrend |
| T1 | |||||||
| C|C | 14 (0.58) | 239 (0.48) | Referent | – | Referent | – | 0.244 |
| C|T | 9 (0.37) | 217 (0.43) | 0.708 (0.300–1.669) | 0.428 | 0.649 (0.233–1.805) | 0.406 | |
| T/T | 1 (0.04) | 46 (0.09) | 0.371 (0.476–2.89) | 0.481b | 0.615 (0.219–1.727) | 0.355 | |
| C/T + T|T | 10 (0.42) | 263 (0.52) | 0.649 (0.283–1.489) | 0.304 | 0.655 (0.284–1.511) | 0.320 | |
| MAFd | 0.23 | 0.31 | 0.669 (0337–1.328) | 0.335b | 0.512 (0.209–1.257) | 0.144 | |
| T2 | |||||||
| C|C | 18 (0.56) | 239 (0.48) | Referent | – | Referent | – | 0.937 |
| C|T | 8 (0.25) | 217 (0.43) | 0.490 (0.209–1.149) | 0.0945 | 0.653 (0.253–1.686) | 0.378 | |
| T/T | 6 (0.19) | 46 (0.09) | 1.732 (0.652–4.599) | 0.265 | 1.329 (0.813– 2.173) | 0.254 | |
| C/T + T|T | 14 (0.44) | 263 (0.52) | 0.707 (0.344–1.452) | 0.343 | 1.001 (0.457–2.211) | 0.989 | |
| MAFd | 0.31 | 0.31 | 1.022 (0.593–1.764) | 0.937 | 1.389 (0.771–2.504) | 0.273 | |
| T3 | |||||||
| C|C | 29 (0.34) | 239 (0.48) | Referent | – | Referent | – | 0.014 |
| C|T | 45 (0.52) | 217 (0.43) | 1.709 (1.035–2.823) | 0.0348 | 1.807 (1.087–3.006) | 0.022 | |
| T/T | 12 (0.14) | 46 (0.09) | 2.150 (1.022–4.521) | 0.0399 | 1.522 (1.039–2.230) | 0.031 | |
| C/T + T|T | 57 (0.66) | 263 (0.52) | 1.786 (1.105–2.887) | 0.0169 | 1.716 (1.022–2.879) | 0.041 | |
| MAFd | 0.40 | 0.31 | 1.507 (1.080–2.102) | 0.0154 | 1.565 (1.116–2.194) | 0.009 | |
| T4 | |||||||
| C|C | 24 (0.33) | 239 (0.48) | Referent | – | Referent | – | 0.032 |
| C|T | 39 (0.54) | 217 (0.43) | 1.790 (1.042–3.074) | 0.0331 | 2.190 (1.205–3.979) | 0.010 | |
| T/T | 9 (0.13) | 46 (0.09) | 1.948 (0.8507–4.463) | 0.1094 | 1.403 (0.924–2.133) | 0.111 | |
| C/T + T|T | 48 (0.67) | 263 (0.52) | 1.817 (1.080–3.059) | 0.0230 | 2.164 (1.215–3.855) | 0.009 | |
| MAFd | 0.39 | 0.31 | 1.784 (1.266–2.514) | 0.0009 | 1.505 (1.048–2.159) | 0.027 | |
| Histological grading | |||||||
|---|---|---|---|---|---|---|---|
| G1 | |||||||
| C|C | 2 (0.28) | 239 (0.48) | Referent | – | Referent | – | 0.328 |
| C|T | 4 (0.57) | 217 (0.43) | 2.203 (0.399–12.151) | 0.422b | 1.144 (0.158–8.279) | 0.893 | |
| T/T | 1 (0.14) | 46 (0.09) | 2.598 (0.231–29.265) | 0.415b | 1.540 (0.451–5.260) | 0.489 | |
| C/T + T|T | 5 (0.71) | 263 (0.52) | 2.272 (0.437–11.824) | 0.455b | 0.946 (0.131–6.835) | 0.956 | |
| MAFd | 0.43 | 0.31 | 1.687 (0.5802–4.904 | 0.384b | 1.659 (0.569–4.839) | 0.354 | |
| G2 | |||||||
| C|C | 31 (0.47) | 239 (0.48) | Referent | – | Referent | – | 0.947 |
| C|T | 29 (0.44) | 217 (0.43) | 1.030 (0.6011–1.766) | 0.0913 | 1.057 (0.614–1.818) | 0.841 | |
| T/T | 6 (0.09) | 46 (0.09) | 1.006 (0.3969–2.548) | 0.9906 | 1.013 (0.634–1.618) | 0.957 | |
| C/T + T|T | 35 (0.53) | 263 (0.52) | 1.026 (0.6135–1.716) | 0.9220 | 1.332 (0.754–2.354) | 0.322 | |
| MAFd | 0.31 | 0.31 | 1.013 (0.685–1500) | 0.947 | 1.034 (0.697–1.536) | 0.866 | |
| G3 | |||||||
| C|C | 52 (0.37) | 239 (0.48) | Referent | – | Referent | – | 0.009 |
| C|T | 68 (0.48) | 217 (0.43) | 1.440 (0.960–2.160) | 0.0767 | 1.494 (0.992–2.250) | 0.054 | |
| T/T | 21 (0.15) | 46 (0.09) | 2.098 (1.155–3.812) | 0.0136 | 1.468 (1.084–1.990) | 0.013 | |
| C/T + T|T | 89 (0.63) | 263 (0.52) | 1.555 (1.059–2.284) | 0.0237 | 1.616 (1.096–2.383) | 0.015 | |
| MAFd | 0.39 | 0.31 | 1.438 (1.093–1.892) | 0.0092 | 1.438 (1.091–1.893) | 0.009 | |
Significant results are highlighted in bold font.
aχ2 or bFisher’s exact test. cORs were adjusted by age, BMI and presence of diabetes. dMinor allele frequency.
We did not find a significant association of rs67085638 with T1 and T2 tumour stages of GC (Table 4). In the patients with T1 tumour stage GC, ptrend = 0.244, and for T/T versus C/C, the adjusted OR was 0.615 (95% CI 0.219–1.727, p = 0.355); for C/T versus C/C, the adjusted OR was 0.615 (95% CI 0.219–1.727, p = 0.406); for C/T + T/T versus C/C, the adjusted OR was 0.655 (95% CI 0.284–1.511, p = 0.320); and for T versus C, the adjusted OR was 0.512 (95% CI 0.209–1.257, p = 0.144). In the patients with T2 tumour stage GC, ptrend = 0.937, and for T/T versus C/C, the adjusted OR was 1.329 (95% CI 0.813– 2.173, p = 0.254); for C/T versus C/C, the adjusted OR was 0.653 (95% CI 0.253–1.686, p = 0.378); for C/T + T/T versus C/C, the adjusted OR was 1.001 (95% CI 0.457–2.211, p = 0.989); and for T versus C, the adjusted OR was 1.389 (95% CI 0.771–2.504, p = 0.273).
Prevalence of the CCAT1 rs67085638 SNP in patients with differentiation grading of GC
We observed an association of rs67085638 with G3 differentiation grading in the patients with GC (Table 4). In the patients with G3 differentiation grading of GC, ptrend = 0.009, and for T/T versus C/C, the adjusted OR was 1.468 (95% CI 1.084–1.990, p = 0.013); for C/T + T/T versus C/C, the adjusted OR was 1.616 (95% CI 1.096–2.383, p = 0.015); and for T versus C, the adjusted OR was 1.438 (95% CI 1.091–1.893, p = 0.009). However, we did not observe an association between rs67085638 and G3 differentiation grading; for C/T versus C/C, the adjusted OR was 1.494 (95% CI 0.992–2.250, p = 0.054). We did not find a significant association of rs67085638 with G1 and G2 differentiation grading of GC in patients with GC (Table 4). In the patients with G1 differentiation grading of GC, ptrend = 0.328, and for T/T versus C/C, the adjusted OR was 1.540 (95% CI 0.451–5.260, p = 0.489); for C/T versus C/C, the adjusted OR was 1.144 (95% CI 0.158–8.279, p = 0.893); for C/T + T/T versus C/C, the adjusted OR was 0.946 (95% CI 0.131–6.835, p = 0.956); and for T versus C, the adjusted OR was 1.659 (95% CI 0.569–4.839, p = 0.354). In the patients with G2 differentiation grading of GC, we found that ptrend = 0.947, and for T/T versus C/C, the adjusted OR was 1.013 (95% CI 0.634–1.618, p = 0.957); for C/T versus C/C, the adjusted OR was 1.057 (95% CI 0.614–1.818, p = 0.841); for C/T + T/T versus C/C, the adjusted OR was 1.332 (95% CI 0.754–2.354, p = 0.322); and for T versus C, the adjusted OR was 1.034 (95% CI 0.697–1.536, p = 0.866).
Prevalence of the CCAT1 rs67085638 SNP in patients with various lymph node and GC metastasis stages
We found a significant association of rs67085638 with N3 lymph node metastasis in GC (Table 5). In the patients with N3 lymph node metastasis of GC, ptrend = 0.0005, and for T/T versus C/C, the adjusted OR was 9.134 (95% CI 1.250–2.929, p = 0.003); for C/T versus C/C, the adjusted OR was 3.081 (95% CI 1.492–6.362, p = 0.002); for C/T + T/T versus C/C, the adjusted OR was 2.703 (95% CI 1.462–4.997, p = 0.001); and for T versus C, the adjusted OR was 1.980 (95% CI 1.345–2.915, p = 0.001). However, we did not observe an association between rs67085638 and the patients with N0, N1 and N2 lymph node metastasis (Table 5). In the patients with N0 lymph node metastasis of GC, ptrend = 0.626, and for T/T versus C/C, the adjusted OR was 1.120 (95% CI 0.639–1.808, p = 0.642); for C/T versus C/C, the adjusted OR was 1.224 (95% CI 0.739–2.027, p = 0.431); for C/T + T/T versus C/C, the adjusted OR was 1.319 (95% CI 0.769–2.262, p = 0.314); and for T versus C, the adjusted OR was 1.192 (95% CI 0.798–1.781, p = 0.390). In the patients with N1 lymph node metastasis, ptrend = 0.909, and for T/T versus C/C, the adjusted OR was 1.152 (95% CI 0.648–2.048, p = 0.629); for C/T versus C/C, the adjusted OR was 0.895 (95% CI 0.410–1.953, p = 0.780); for C/T + T/T versus C/C, the adjusted OR was 1.029 (95% CI 0.514–2.059, p = 0.935); and for T versus C, the adjusted OR was 1.082 (95% CI 0.639–1.830, p = 0.769). In the patients with N2 lymph node metastasis, ptrend = 0.475, and for T/T versus C/C, the adjusted OR was 1.324 (95% CI 0.810–2.167, p = 0.261); for C/T versus C/C, the adjusted OR was 0.998 (95% CI 0.448–2.222, p = 0.997); for C/T + T/T versus C/C, the adjusted OR was 1.118 (95% CI 0.562–2.225, p = 0.750); and for T versus C, the adjusted OR was 1.204 (95% CI 0.719–2.017, p = 0.480).
Table 5.
Prevalence of the CCAT1 rs67085638 polymorphism among various lymph nodes and stages of metastasis of GC.
| Lymph node metastasis stage | |||||||
|---|---|---|---|---|---|---|---|
| Genotype | Patients (frequency) | Controls (frequency) | Odds ratio (95%CI) | pa | Adjusted odds ratio (95%CI)c | p | ptrend |
| N0 | |||||||
| C|C | 34 (0.43) | 239 (0.48) | Referent | – | Referent | – | 0.626 |
| C|T | 37 (0.47) | 217 (0.43) | 1.199 (0.727–1.977) | 0.478 | 1.224 (0.739–2.027) | 0.431 | |
| T/T | 7 (0.09) | 46 (0.09) | 1.070 (0.447 –2.560) | 0.880 | 1.120 (0.639–1.808) | 0.642 | |
| C/T + T|T | 44 (0.56) | 263 (0.52) | 1.176 (0.727–1.902) | 0.508 | 1.319 (0.769–2.262) | 0.314 | |
| MAFd | 0.33 | 0.31 | 1.092 (0.762–1.567) | 0.631 | 1.192 (0.798–1.781) | 0.390 | |
| N1 | |||||||
| C|C | 17 (0.48) | 239 (0.48) | Referent | – | Referent | – | 0.909 |
| C|T | 14 (0.40) | 217 (0.43) | 0.907 (0.437–1.88) | 0.794 | 0.895 (0.410–1.953) | 0.780 | |
| T/T | 4 (0.11) | 46 (0.09) | 1.223 (0.393–3.801) | 0.759b | 1.152 (0.648–2.048) | 0.629 | |
| C/T + T|T | 18 (0.51) | 263 (0.52) | 0.962 (0.485–1.910) | 0.912 | 1.029 (0.514–2.059) | 0.935 | |
| MAFd | 0.31 | 0.31 | 1.031 (0.612–1.738) | 0.894b | 1.082 (0.639–1.830) | 0.769 | |
| N2 | |||||||
| C|C | 19 (0.45) | 239 (0.48) | Referent | – | Referent | – | 0.475 |
| C|T | 17 (0.40) | 217 (0.43) | 0.985 (0.499–1.945) | 0.966 | 0.998 (0.448–2.222) | 0.997 | |
| T/T | 6 (0.14) | 46 (0.09) | 1.641 (0.622–4.33) | 0.313 | 1.324 (0.810–2.167) | 0.261 | |
| C/T + T|T | 23 (0.55) | 263 (0.52) | 1.100 (0.584–2.071) | 0.768 | 1.118 (0.562–2.225) | 0.750 | |
| MAFd | 0.34 | 0.31 | 1.186 (0.743–1.896) | 0.476 | 1.204 (0.719–2.017) | 0.480 | |
| N3 | |||||||
| C|C | 15 (0.25) | 239 (0.48) | Referent | – | Referent | – | 0.0005 |
| C|T | 33 (0.56) | 217 (0.43) | 2.423 (1.281–4.584) | 0.0053 | 3.081 (1.492–6.362) | 0.002 | |
| T/T | 11 (0.19) | 46 (0.09) | 3.810 (1.645–8.824) | 0.001 | 9.134 (1.250–2.929) | 0.003 | |
| C/T + T|T | 44 (0.74) | 263 (0.52) | 2.666 (1.446–4.915) | 0.0012 | 2.703 (1.462–4.997) | 0.001 | |
| MAFd | 0.47 | 0.31 | 1.964 (1.335–2.88) | 0.0005 | 1.980 (1.345–2.915) | 0.001 | |
| Metastasis stage | |||||||
|---|---|---|---|---|---|---|---|
| M0 | |||||||
| C|C | 76 (0.39) | 239 (0.48) | Referent | – | Referent | – | 0.027 |
| C|T | 90 (0.47) | 217 (0.43) | 1.304 (0.9132–1.863) | 0.1435 | 1.348 (0.940–1.934) | 0.104 | |
| T/T | 26 (0.14) | 46 (0.09) | 1.777 (1.030–3.068) | 0.0373 | 1.349 (1.023–1.778) | 0.033 | |
| C/T + T|T | 116 (0.60) | 263 (0.52) | 1.387 (0.989–1.945) | 0.0575 | 1.438 (1.021– 2.025) | 0.037 | |
| MAFd | 0.37 | 0.31 | 1.320 (1.031–1.689) | 0.0273 | 1.357 (1.033–1.782) | 0.028 | |
| M1 | |||||||
| C|C | 9 (0.41) | 239 (0.48) | Referent | – | Referent | – | 0.638 |
| C|T | 11 (0.50) | 217 (0.43) | 1.346 (0.547–3.311) | 0.516 | 1.175 (0.608–5.060) | 0.297 | |
| T/T | 2 (0.09) | 46 (0.09) | 1.155 (0.242–5.520) | 0.695 | 1.019 (0.458–2.267) | 0.962 | |
| C/T + T|T | 13 (0.59) | 263 (0.52) | 1.313 (0.5510–3.127) | 0.538 | 1.729 (0.624–4.794) | 0.291 | |
| MAFd | 0.34 | 0.31 | 1.163 (0.615–2.201) | 0.621b | 1.431 (0.703–2.913) | 0.322 | |
Significant results are highlighted in bold font.
aχ2 or bFisher’s exact test. cORs were adjusted by age, BMI and presence of diabetes. dMinor allele frequency.
We also found a contribution of the rs67085638 SNP to the M0 metastasis stage of GC (Table 5). In the patients with M0 metastasis stage GC, we observed ptrend = 0.027, and for T/T versus C/C, the adjusted OR was 1.349 (95% CI 1.023–1.778, p = 0.033); for C/T + T/T versus C/C, the adjusted OR was 1.438 (95% CI 1.021– 2.025, p = 0.037); and for T versus C, the adjusted OR was 1.357 (95% CI 1.033–1.782, p = 0.028). However, we did not observe an association of rs67085638 with M0 metastasis stage; for C/T versus C/C, the adjusted OR was 1.348 (95% CI 0.940–1.934, p = 0.104). We did not find a contribution of the rs67085638 SNP to the M1 metastasis stage of GC (Table 5). In the patients with M1 metastasis stage, we observed ptrend = 0.638, and for T/T versus C/C, the adjusted OR was 1.019 (95% CI 0.458–2.267, p = 0.962); for C/T versus C/C, the adjusted OR was 1.175 (95% CI 0.608–5.060, p = 0.297); for C/T + T/T versus C/C, the adjusted OR was 1.729 (95% CI 0.624–4.794, p = 0.291); and for T versus C, the adjusted OR was 1.431 (95% CI 0.703–2.913, p = 0.322).
Influence of the rs67085638 polymorphism on BMI1 transcript levels in GC tissue and counterpart histopathologically confirmed disease-free margin tissue
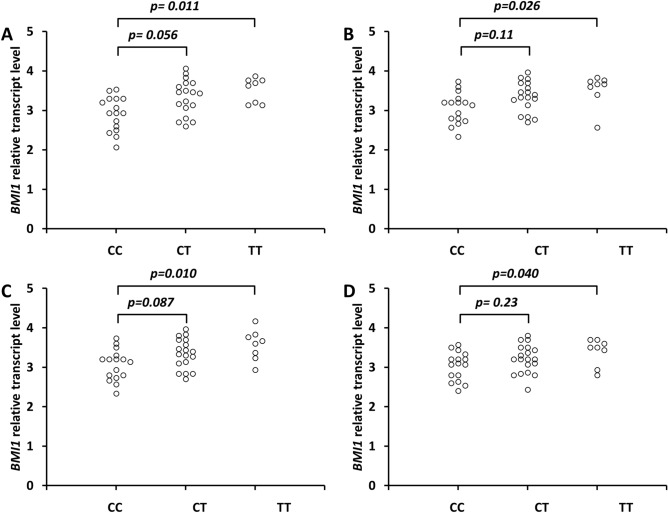
We found significantly increased BMI1 transcript levels in the primary GC tissue classified as grade G3 (p = 0.011) and counterpart histopathologically confirmed disease-free margin tissue (p = 0.026, respectively) in the carriers of the T/T versus C/C genotypes but not for the carriers of the C/T versus C/C genotypes (p = 0.056, p = 0.11, respectively) (Fig. 1A, B). There was also a statistically significant increase in the BMI1 transcript levels in the primary GC tissue classified as lymph node metastasis N3 (p = 0.010) and counterpart histopathologically confirmed disease-free margin tissue (p = 0.040) in the carriers of the T/T versus C/C genotypes but not in the carriers of the C/T versus C/C genotypes (p = 0.087, p = 0.23, respectively) (Fig. 1C,D).
Figure 1.
Effect of the CCAT1 rs67085638 SNP on BMI1 transcript levels in primary GC tissue samples classified as grade G3 (A) and those classified as N3 lymph nodes (C) and distal counterpart surgical resection margins histopathologically confirmed disease-free for grade G3 (B) and N3 lymph nodes (D). Frozen tissue was homogenized, followed by total RNA isolation. Quantitative analyses of BMI1 transcript levels were performed by qRT-PCR using the SYBR Green I system (Supplementary Table 2). The quantity of BMI1 transcript levels in each sample was standardized by the geometric mean of references using HMBS and B2M cDNA levels. Kruskal–Wallis test with aDunn’s post hoc test (Supplementary Table 2).
Discussion
CCAT1 and CCAT2 are located in the 8q24.21 region, which is frequently amplified in colorectal cancer. In the 8q24 region, several complex molecular interactions are tissue-specific29 between CCAT1 and MYC, which regulate CCAT1 and MYC gene expression20,30–32. CCAT1 is situated inside a super‐enhancer region of the MYC promotor32. CCAT1-L protects against interactions between MYC and its enhancers, recruiting CCCTC‐binding factor or CTCF20. Moreover, CCAT1 lncRNA functions as a sponge for some tumour suppressor miRNAs to titrate the MYC protein33. In this context, MYC binds to the promoter of CCAT1 and increases its transcription16.
Overexpression of CCAT1 has been observed in various malignancies compared to normal counterpart tissue, including GC15. CCAT1 displays oncogenic properties in various cancers, and in the cytoplasm or nucleus, it supports biological processes in cancer, such as proliferation, migration, cell cycle progression, apoptosis, and chemoresistance15. Moreover, CCAT1 positively correlated with the clinicopathological characteristics, progression and treatment outcomes of various cancers, including advanced TNM stage, vascular invasion, overall survival and recurrence-free survival, among others15.
In our studies, we observed a trend of rs67085638 SNP association in all patients with GC. rs67085638 was a strong risk factor for GC in male but not female patients. To date, the rs67085638 and rs7013433 polymorphisms have been demonstrated as risk factors for colorectal cancer21. However, we did not find an association between the rs7013433 polymorphism and either the occurrence of GC or the clinicopathological characteristics of GC (data not shown). The increased risk of rs67085638 in males can be due to the additive effect of environmental factors, including tobacco smoking, regular alcohol consumption, and limited consumption of fruits and vegetables, which are common in males in our country. Our observation is consistent with findings in other populations in which male sex is an important factor contributing to the development of GC6,34.
To date, the role of CCAT1 lncRNA in the development of GC has been intensively studied. CCAT1 was significantly elevated in primary GC tissue compared with normal gastric tissue and significantly correlated with the progression of GC and supports the proliferation and migration of GC cells16–19. A detailed study revealed that CCAT1 is involved in the negative regulation of the miR-219-1 and miR-490/hnRNPA1 axes, contributing to malignant transformation, progression and migration of GC cells19,23,35. CCAT1 expression was associated with larger gastric tumour size, lymphatic metastasis and advanced TNM stage19. CCAT1-L is also involved in the epithelial-mesenchymal transition and metastasis of gastric adenocarcinoma18.
We found that rs67085638 was associated with tumour stages T3 and T4, histological differentiation G3, lymph node metastasis stage N3 and metastasis stage M0. In addition, we determined that homozygous carriers of the CCAT1 rs67085638 T allele exhibited increased BMI1 transcript levels compared to homozygous carriers of the CCAT1 rs67085638 C allele from primary GC tissues as well as in histopathologically confirmed cancer-free margin tissue. Our findings suggest that the CCAT1 rs67085638 T allele elevating BMI1 expression might support increased rapid growth compared to lower-grade tumour cells and the expansion of cancer cells into the outer lining of the stomach or other organs and lymph node metastasis.
BMI1 is an oncogene and catalytic component of polycomb group proteins involved in epigenetic gene silencing. BMI1displays a critical function in tissue-specific regulation of gene expression and consequently several elementary cellular processes36. SNP rs6983267 has been shown to be located in the enhancer region of MYC, which plays a role in the regulation of CCAT1 expression21. Recently, it was demonstrated that the presence of the T allele of rs67085638 increased CCAT1 expression22. Moreover, CCAT1 expression correlated with BMI1 mRNA and protein levels in both GC cells in vitro and in vivo in murine tumour models23. CCAT1 knockdown repressed the proliferation and invasion of gallbladder cancer cells via miR-218-5p controlling BMI1 transcript translation37. In cigarette smoke extract-exposed human bronchial epithelial cells, CCAT1 negatively regulates miR-218, which modulates BMI1 expression and cell cycle progression38. BMI1 has been found to be a characteristic marker of poor prognosis in patients with breast cancer, nasopharyngeal carcinoma, oesophageal squamous cell carcinoma and gastric carcinoma36,39,40. Several studies have shown that BMI1 is associated with the progression, epithelial-mesenchymal transition and metastasis of GC41–43. The reduced expression of BMI1 inhibits epithelial-mesenchymal transition and spreading of melanoma cells44.
Conclusion
Our study is the first to demonstrate that the CCAT1 rs67085638 SNP is a risk factor for gastric carcinogenesis in Caucasian Polish individuals. We also found that homozygous carriers of CCAT1 rs67085638 T were associated with increased BMI1 transcript levels and increased rapid growth compared to lower-grade tumour cells, spread of cancer cells to neighbouring tissues and lymph node metastasis. However, our studies have some limitations. In patients with tumour stage T4, we observed the greatest contribution of C/T but not T/T to GC, which may be due to our relatively small group of patients with GC. This study should be repeated in other independent cohorts. Moreover, we need precisely explain the mechanism by which rs67085638 may regulate the transcription of BMI1.
Supplementary Information
Author contributions
T.O. Research idea, wrote the main manuscript text, Development of research method, prepared Fig. 1, supervisor moleculr biology technician, performing experiments. A.L. Collection data, supervisor moleculr biology technician. A.B. Performing experiments. A.S. Statistics, P.P. Collection data, description of the material section. T.T. Prepared supplementary data, supervisor moleculr biology technician. T.M. Collection data, description of the material section P.H. Collection data, description of the material section. P.P.J Wrote the main manuscript text, approval of manuscript. All authors reviewed the manuscript.
Funding
This study was funded by Grant No. 502-01-01124182-07474 from Poznań University of Medical Sciences and by Maria Sklodowska-Curie National Research Institute of Oncology subsidy from Ministry of Science and Higher Education. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-94576-9.
References
- 1.Global Burden of Disease Cancer Collaboration. Fitzmaurice C, et al. The global burden of cancer 2013. JAMA Oncol. 2015;1(4):505–527. doi: 10.1001/jamaoncol.2015.0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: Descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol. Biomark. Prev. 2014;23:700–713. doi: 10.1158/1055-9965.EPI-13-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma SH, et al. Impact of alcohol drinking on gastric cancer development according to Helicobacter pylori infection status. Br. J. Cancer. 2015;113:1381–1388. doi: 10.1038/bjc.2015.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nomura AM, Wilkens LR, Henderson BE, Epplein M, Kolonel LN. The association of cigarette smoking with gastric cancer: The multiethnic cohort study. Cancer Causes Control. 2012;23:51–58. doi: 10.1007/s10552-011-9854-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camargo MC, et al. Improved survival of gastric cancer with tumour Epstein–Barr virus positivity: An international pooled analysis. Gut. 2014;63:236–243. doi: 10.1136/gutjnl-2013-304531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang X, et al. Landscape of dietary factors associated with risk of gastric cancer: A systematic review and dose-response meta-analysis of prospective cohort studies. Eur. J. Cancer. 2015;51:2820–2832. doi: 10.1016/j.ejca.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Choi YJ, Kim N. Gastric cancer and family history. Korean J. Intern. Med. 2016;31:1042–1053. doi: 10.3904/kjim.2016.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gigek CO, et al. Genetic variants in gastric cancer: Risks and clinical implications. Exp. Mol. Pathol. 2017;103:101–111. doi: 10.1016/j.yexmp.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 9.Helgason H, et al. Loss-of-function variants in ATM confer risk of gastric cancer. Nat. Genet. 2015;47:906–910. doi: 10.1038/ng.3342. [DOI] [PubMed] [Google Scholar]
- 10.Cai M, et al. Environmental factors, seven GWAS-identified susceptibility loci, and risk of gastric cancer and its precursors in a Chinese population. Cancer Med. 2017;6:708–720. doi: 10.1002/cam4.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hishida A, et al. GWAS analysis reveals a significant contribution of PSCA to the risk of Heliobacter pylori-induced gastric atrophy. Carcinogenesis. 2019;40:661–668. doi: 10.1093/carcin/bgz016. [DOI] [PubMed] [Google Scholar]
- 12.Akhade VS, Pal D, Kanduri C. Long noncoding RNA: Genome organization and mechanism of action. Adv. Exp. Med. Biol. 2017;1008:47–74. doi: 10.1007/978-981-10-5203-3_2. [DOI] [PubMed] [Google Scholar]
- 13.Qi F, et al. Long noncoding AGAP2-AS1 is activated by SP1 and promotes cell proliferation and invasion in gastric cancer. J. Hematol. Oncol. 2017;10:48. doi: 10.1186/s13045-017-0420-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malek E, Jagannathan S, Driscoll JJ. Correlation of long non-coding RNA expression with metastasis, drug resistance and clinical outcome in cancer. Oncotarget. 2014;5:8027–8038. doi: 10.18632/oncotarget.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang N, et al. Pivotal prognostic and diagnostic role of the long non-coding RNA colon cancer-associated transcript 1 expression in human cancer (review) Mol. Med. Rep. 2019;19:771–782. doi: 10.3892/mmr.2018.9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang F, et al. Long noncoding RNA CCAT1, which could be activated by c-Myc, promotes the progression of gastric carcinoma. J. Cancer Res. Clin. Oncol. 2013;139:437–445. doi: 10.1007/s00432-012-1324-x. [DOI] [PubMed] [Google Scholar]
- 17.Shan T, Chen YG, Hong B, Zhou H, Xia JZ. Expression and clinical significance of long non-coding RNA CCAT1 in gastric cancer. Zhonghua Yi Xue Za Zhi. 2017;97:1411–1414. doi: 10.3760/cma.j.issn.0376-2491.2017.18.012. [DOI] [PubMed] [Google Scholar]
- 18.Fang H, et al. Upregulation of long noncoding RNA CCAT1-L promotes epithelial-mesenchymal transition in gastric adenocarcinoma. Onco Targets Ther. 2018;11:5647–5655. doi: 10.2147/OTT.S170553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X, Zhou Y, Qian H. CCAT1 expressed in malignant and pre-malignant human gastric tissues. Cell. Mol. Biol. (Noisy-le-grand) 2017;63:89–93. doi: 10.14715/cmb/2017.63.5.16. [DOI] [PubMed] [Google Scholar]
- 20.Xiang JF, et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24:513–531. doi: 10.1038/cr.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, et al. Long noncoding RNA CCAT1 polymorphisms are associated with the risk of colorectal cancer. Cancer Genet. 2018;222–223:13–19. doi: 10.1016/j.cancergen.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 22.Xiao ZS, Zhao L, Zhang XN, Li HX, Yin ZH. Effect of rs67085638 in long non-coding RNA (CCAT1) on colon cancer chemoresistance to paclitaxel through modulating the microRNA-24-3p and FSCN1. Cell. Mol. Med. 2021 doi: 10.1111/jcmm.16210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li N, Jiang K, Fang LP, Yao LL, Yu Z. Knockdown of long noncoding RNA CCAT1 inhibits cell growth, invasion and peritoneal metastasis via downregulation of Bmi-1 in gastric cancer. Neoplasma. 2018;65:736–744. doi: 10.4149/neo_2018_171206N801. [DOI] [PubMed] [Google Scholar]
- 24.Zhao B, et al. Assessment of the 8th edition of TNM staging system for gastric cancer: The results from the SEER and a single-institution database. Future Oncol. 2018;14:3023–3035. doi: 10.2217/fon-2018-0299. [DOI] [PubMed] [Google Scholar]
- 25.Hebebrand J, et al. A proposal of the European Association for the study of obesity to improve the ICD-11 diagnostic criteria for obesity based on the three dimensions etiology, degree of adiposity and health risk. Obes. Facts. 2017;10:284–307. doi: 10.1159/000479208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rolf HA, Vossen M. Genotyping DNA variants with high-resolution melting analysis. Methods Mol. Biol. 2017;1492:17–28. doi: 10.1007/978-1-4939-6442-0_2. [DOI] [PubMed] [Google Scholar]
- 27.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- 28.Tybura P, Grzywacz A, Samochowiec A, Samochowiec J. Associations between candidate genes with schizophrenia susceptibility and the treatment efficiency. Psychiatr. Pol. 2011;45:811–823. [PubMed] [Google Scholar]
- 29.Ahmadiyeh N, et al. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc. Natl. Acad. Sci. U. S. A. 2010;107:9742–9746. doi: 10.1073/pnas.0910668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim T, et al. Long-range interaction and correlation between MYC enhancer and oncogenic long noncoding RNA CARLo-5. Proc. Natl. Acad. Sci. U. S. A. 2014;111:4173–4178. doi: 10.1073/pnas.1400350111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sur IK, et al. Mice lacking a Myc enhancer that includes human SNP rs6983267 are resistant to intestinal tumors. Science. 2012;338:1360–1363. doi: 10.1126/science.1228606. [DOI] [PubMed] [Google Scholar]
- 32.Hamilton MJ, Young MD, Sauer S, Martinez E. The interplay of long non-coding RNAs and MYC in cancer. AIMS Biophys. 2015;2:794–809. doi: 10.3934/biophy.2015.4.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng L, Yang SB, Xu FF, Zhang JH. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J. Exp. Clin. Cancer Res. 2015;34:18. doi: 10.1186/s13046-015-0136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song M, Rabkin CS, Camargo MC. Gastric cancer: An evolving disease. Curr. Treat. Options Gastroenterol. 2018;16:561–569. doi: 10.1007/s11938-018-0203-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou B, et al. The long noncoding RNA colon cancer-associated transcript-1/miR-490 axis regulates gastric cancer cell migration by targeting hnRNPA1. IUBMB Life. 2016;68:201–210. doi: 10.1002/iub.1474. [DOI] [PubMed] [Google Scholar]
- 36.Guo BH, et al. Bmi-1 promotes invasion and metastasis, and its elevated expression is correlated with an advanced stage of breast cancer. Mol. Cancer. 2011;10:10. doi: 10.1186/1476-4598-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma MZ, et al. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death Dis. 2015;6:e583. doi: 10.1038/cddis.2014.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu L, et al. Epigenetic silencing of miR-218 by the lncRNA CCAT1, acting via BMI1, promotes an altered cell cycle transition in the malignant transformation of HBE cells induced by cigarette smoke extract. Toxicol. Appl. Pharmacol. 2016;304:30–41. doi: 10.1016/j.taap.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 39.Song LB, et al. Bmi-1 is a novel molecular marker of nasopharyngeal carcinoma progression and immortalizes primary human nasopharyngeal epithelial cells. Cancer Res. 2006;66:6225–6232. doi: 10.1158/0008-5472.CAN-06-0094. [DOI] [PubMed] [Google Scholar]
- 40.Ji H, et al. Expression and clinicopathological significance of Mel-18 and Bmi-1 in esophageal squamous cell carcinoma. Technol. Cancer Res. Treat. 2017;16:828–834. doi: 10.1177/1533034617705055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu C, et al. Reduction of gastric cancer proliferation and invasion by miR-15a mediated suppression of Bmi-1 translation. Oncotarget. 2016;7:14522–14536. doi: 10.18632/oncotarget.7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu WW, Jiang H, Zhang CT, Peng Y. The SNAIL/miR-128 axis regulated growth, invasion, metastasis, and epithelial-to-mesenchymal transition of gastric cancer. Oncotarget. 2017;8:39280–39295. doi: 10.18632/oncotarget.16849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao FL, Li WS, Liu CL, Zhao GQ. Silencing Bmi-1 enhances the senescence and decreases the metastasis of human gastric cancer cells. World J. Gastroenterol. 2013;19:8764–8769. doi: 10.3748/wjg.v19.i46.8764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y, et al. Downregulation of Bmi-1 suppresses epithelial-mesenchymal transition in melanoma. Oncol. Rep. 2017;37:139–146. doi: 10.3892/or.2016.5244. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.