Abstract
The oldest and most wide-ranging signal of biological activity (biosignature) on our planet is the carbon isotope composition of organic materials preserved in rocks. These biosignatures preserve the long-term evolution of the microorganism-hosted metabolic machinery responsible for producing deviations in the isotopic compositions of inorganic and organic carbon. Despite billions of years of ecosystem turnover, evolutionary innovation, organismic complexification, and geological events, the organic carbon that is a residuum of the global marine biosphere in the rock record tells an essentially static story. The ~25‰ mean deviation between inorganic and organic 13C/12C values has remained remarkably unchanged over >3.5 billion years. The bulk of this record is conventionally attributed to early-evolved, RuBisCO-mediated CO2 fixation that, in extant oxygenic phototrophs, produces comparable isotopic effects and dominates modern primary production. However, billions of years of environmental transition, for example, in the progressive oxygenation of the Earth’s atmosphere, would be expected to have accompanied shifts in the predominant RuBisCO forms as well as enzyme-level adaptive responses in RuBisCO CO2-specificity. These factors would also be expected to result in preserved isotopic signatures deviating from those produced by extant RuBisCO in oxygenic phototrophs. Why does the bulk carbon isotope record not reflect these expected environmental transitions and evolutionary innovations? Here, we discuss this apparent discrepancy and highlight the need for greater quantitative understanding of carbon isotope fractionation behavior in extant metabolic pathways. We propose novel, laboratory-based approaches to reconstructing ancestral states of carbon metabolisms and associated enzymes that can constrain isotopic biosignature production in ancient biological systems. Together, these strategies are crucial for integrating the complementary toolsets of biological and geological sciences and for interpretation of the oldest record of life on Earth.
Subject terms: Bacterial evolution, Applied microbiology, Biogeochemistry
Introduction
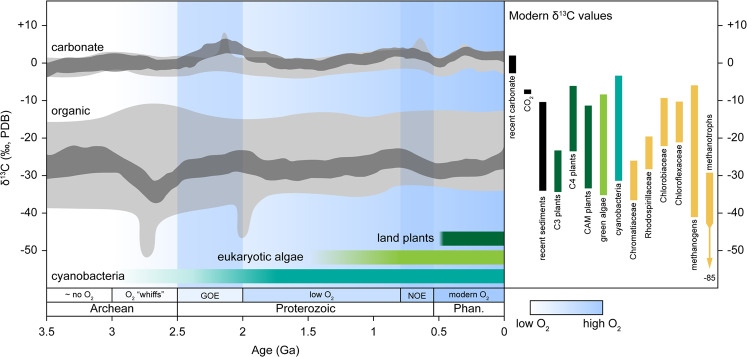
Life on Earth has generated two main repositories of information with which to reconstruct its past states: first, the genetic diversity of extant organisms, and second, the physical remnants of past life preserved in the geologic record, or biosignatures [1]. By far the most extensive biosignature record—providing the earliest potential evidence of life >3 billion years old [2–5]—is constructed from 13C/12C isotopic compositions of preserved carbonaceous material, expressed as a normalized value, δ13C, typically in units of per mil (‰) [6, 7] (Fig. 1; Box 1).
Fig. 1. The geologic record of δ13C values has remained essentially constant over ~3.5 billion years.
Geologic carbonate and organic δ13C record (left) and modern δ13C values of inorganic carbon and biomass from diverse taxa (right). Light gray fields represent the range of geologic δ13C measurements from Schidlowski [6]. Dark gray fields represent 95% confidence intervals for smoothing analyses of geologic δ13C data from Krissansen-Totton et al. [7] and references therein. Modern δ13C values from Schidlowski [6]. Bars are colored as follows: black, geologic reservoirs; dark green, land plants; light green, green algae; teal, cyanobacteria; other taxa, yellow. Phases of atmospheric oxygen are labeled at the bottom, from Lyons et al. [24] (“~ no O2” indicates <10−5 present atmospheric level (PAL), “low O2” indicates ~10−1 to 10−4 PAL, and “modern O2” indicates PAL). Qualitative O2 levels are indicated by shades of blue, with lighter shades indicating lower O2 levels and darker shades indicating higher O2 levels (also noted by the scale). The Great Oxidation Event (GOE) and Neoproterozoic Oxidation Event (NOE) are indicated by darker blue bars. The earliest potential appearance of cyanobacteria is interpreted from the oldest known oxidized sediments [24, 59, 60]; of eukaryotic algae, from oldest interpreted algal fossils [139] and molecular clock dating [140, 141]; of land plants, from the oldest interpreted pollen fossils [142] and molecular clock dating [143]. PDB Pee Dee Belemnite δ13C standard, Phan. Phanerozoic (color figure online).
This carbon isotope record is interpreted to have primarily been shaped by the biological isotopic discrimination of enzymatically driven carbon metabolism. Thus, concerted efforts have been dedicated toward disentangling this record and identifying signals potentially attributable to the metabolic innovations, ecosystem turnover, and global environmental changes that have characterized the history of life [6–10]. Such changes would be expected to manifest in variations to the carbon isotopic record over geologic time. However, deviations in inorganic and organic δ13C over the last ~3.5 billion years consistently average ~25‰, creating a largely static trend across the entirety of the record [6, 7]. Only two isolated negative excursions in organic δ13C have been resolved, one centered at ~2.7 billion years and the other at ~2 billion years. These excursions have been interpreted to reflect increased methanotrophic and/or methanogenic activity [7, 9, 11, 12]. Why are similar variations not known for other time intervals?
This curious consistency of carbon isotope biosignatures presents an ongoing challenge for interpretation of the most extensive record of life. It is an enormously complex problem, as several factors are known to affect the magnitude of biological isotope discrimination, including diversity in metabolic pathways (Table 1) as well as environmental parameters and host organism physiologies (Table 2). The overlaying of these factors serves to obfuscate individual contributions to preserved carbon biosignatures that might be of interest in understanding the early evolution of life. Further, one must consider the fidelity of the record itself, as the isotopic compositions of poorly preserved geologic samples may be affected by post-depositional abiotic processes that erase primary biogenic signatures [6, 10].
Table 1.
Isotopic discrimination, productivity, and O2 sensitivity of extant carbon fixation pathways.
| Pathway | Associated taxaa | Modern productivity (Pg C/yr)b | 13C/12C isotope discrimination Δδ13C (‰) | Isotope discrimination references | Pathway O2 sensitive?b |
|---|---|---|---|---|---|
| CBB |
Cyanobacteria [97] photoautotrophs Proteobacteria [100] autotrophic Alpha-, Beta-, Gammaproteobacteria Chloroflexi [101] Oscillochloridaceae Firmicutes [102] Sulfobacillus spp. |
~100 | ~10 to 35 | [26, 44, 45, 103–105] | Yes |
| rTCA |
Aquificae [106] Aquificales Chlorobi [107] Chlorobiales Nitrospirae [108] autotrophs Proteobacteria Magnetococcus sp. MC-1 Desulfobacter hydrogenophilus |
~1 | ~2 to 23 | [44, 45, 103–105] | Yes |
| roTCA |
Aquificae Thermosulfidibacter takaii Proteobacteria sulfur-reducing Deltaproteobacteria |
Unknown | Unknown | N/A | Unknown |
| HP/HB |
Crenarchaeota [109] Sulfolobales Marine group I [110] Thaumarchaeota [111] |
~0.7 | ~0 to 4 | [44] | No |
| Wood–Ljungdahl |
Euryarchaeota [112] Archaeoglobales methanogens Firmicutes acetogens Planctomycetes [113] anaerobic ammonium-oxidizers Proteobacteria autotrophic Deltaproteobacteria Spirochaetes [113] Treponema primitia |
~0.1 | ~5 to 80 | [45, 46, 50, 51, 53, 114] | Yes |
| DC/HB |
Crenarchaeota Thermoproteales Desulfurococcales |
<0.1 | ~0 to 4 | [44, 45] | Yes |
| 3HP |
Chloroflexi Chloroflexaceae |
<0.1 | ~0 to 14 | [20, 44, 115] | No |
| Reductive glycinec |
Proteobacteria Candidatus Phosphitivorax anaerolimi |
<0.1 | Unknown | N/A | Unknown |
Isotopic discrimination reported from literature, calculated as Δδ13C = δ13Cproduct − δ13Creactant, where product is biomass (or acetate/methane for acetogens/methanogens, respectively, utilizing the Wood–Ljungdahl pathway) and reactant is source carbon in growth media.
CBB Calvin–Benson–Bassham cycle, DC/HB dicarboxylate–4-hydroxybutyrate cycle, HP/HB 3-Hydroxyproprionate/4-hydroxybutyrate cycle, roTCA reverse oxidative tricarboxylic acid cycle, rTCA reductive citric acid cycle, 3HP 3-hydroxyproprionate bicycle.
aAssociated taxa from Berg et al. [37], Hugler and Sievert [116], Ward and Shih [25], and references therein. References for definitions of informal taxonomic groups are listed within the table at first instance. Listed taxa are not necessarily diagnostic of each pathway, but rather describe major groups of organisms where pathway can be found.
bModern productivity values from [38, 117–121], and O2 sensitivity data from Berg [122].
cRecently proposed pathway for the Deltaproteobacterium Candidatus Phosphitivorax anaerolimi [36].
Table 2.
Examples of environmental and physiological factors that affect autotrophic carbon fractionation.
| Variable | Taxa | 13C/12C isotope discrimination change (↑ or ↓) with increase in variable | Reference |
|---|---|---|---|
| Temperature | Land plants (varied) | ↓ (≤ ~4‰) | [123] |
| Diatoms (varied) | ↓ (≤~7‰; less change at high [CO2]) | [124] | |
| Marine plankton (varied) | ↑ or ↓, dependent on taxa (≤ ~4‰) | [125] | |
| pH | Spinacea oleracea (land plant) | ↓ (≤ ~3‰) | [15] |
| Skeletonema costatum (diatom), Emiliania huxleyi (coccolithophore) | ↑ or ↓, dependent on pH range (≤ ~9‰) | [47] | |
| CO2 concentration | Land plants (varied) | ↑ (≤ ~7‰) | [126] |
| Skeletonema costatum (diatom), Emiliania huxleyi (coccolithophore) | ↑ (≤ ~7‰) | [47] | |
| Emiliania huxleyi (coccolithophore) | ↑ (≤ ~7‰) | [127] | |
| Emiliania huxleyi (coccolithophore) | ↑ (≤ ~7‰) | [128] | |
| Growth rate | Phaeodactylum tricornutum (diatom) | ↓ (≤ ~20‰) | [129] |
| Emiliania huxleyi (coccolithophore) | ↓ (≤ ~7‰) | [128] | |
| Marine plankton (field samples) | ↓ (≤ ~9‰) | [130] | |
| Marine plankton (field samples) | ↓ (≤ ~8‰) | [131] | |
| Cell surface area:volume | Marine plankton (varied) | ↑ (≤ ~20‰) | [132] |
| H2 concentration | Methanothermobacter marburgensis (methanogen) | ↓ (≤ ~30‰) | [133] |
| Methanocaldococcus jannaschii (methanogen) | ↓ (≤ ~16‰) | [134] | |
| Pressure | Methanopyrus kandleri (methanogen) | ↓ (≤ ~22‰) | [135] |
Here, in addition to surveying several biological and environmental factors that quantitatively affect carbon biosignatures, we contend with an important aspect that is not typically considered—the potential role of subcellular evolution in shaping the carbon isotope record. We discuss in particular the evolution of the CO2-fixing enzyme RuBisCO (ribulose 1,5-bisphosphate carboxylase/oxygenase, EC 4.1.1.39) [13, 14], which produces comparable isotope effects in extant oxygenic phototrophs (ε ~20 to 30‰; see Box 1 for a discussion of isotope effects) to the ~25‰ mean isotopic difference between preserved inorganic and organic carbon [15–19]. RuBisCO is the catalytic bottleneck of the Calvin–Benson–Bassham (CBB) cycle used primarily by oxygenic phototrophs, though also by certain Proteobacteria, Gram-positive bacteria, and Chloroflexi [20–22] (Table 1). The CBB cycle facilitated by oxygenic phototrophs evolved early in Earth history, at least by 2.4 billion years as evidenced by broadly accepted geochemical signatures of atmospheric oxygen [23, 24]. This, in addition to the predominance of oxygenic phototrophy in modern primary production, suggests that RuBisCO has been the most important driver of carbon fixation for much of Earth history [6, 25].
There are several reasons to expect that molecular-level changes to RuBisCO enzymes over geologic history may have been imprinted upon the carbon isotope record. The range of isotope effects for differing forms of RuBisCO can extend outside that associated with well-studied oxygenic phototrophs (i.e., ε ~< 20‰; Table 3). Furthermore, extant RuBisCO carbon uptake efficiency varies as a function of external CO2 levels and protein sequence variation, which subsequently affects the degree of carbon isotope fractionation [16, 19, 26–28]. Because atmospheric CO2 levels have changed markedly over Earth history [29], one would expect molecular adaptations in RuBisCO CO2 specificity to thus be expressed in carbon biosignatures.
Table 3.
Available measurements of 13C/12C isotope effects (ε) from diverse forms of purified RuBisCO enzyme, measured under saturating CO2 levels.
| Group | Species | RuBisCO form | 13C/12C isotope effect, ε (‰) | Isotope effect reference |
|---|---|---|---|---|
| Proteobacteria | Solemya velum bivalve symbiont | IA | 24.4 | [19] |
| Cyanobacteriaa | Prochlorococcus marinus MIT9313 | IA | 24.0 | [16] |
| Cyanobacteriaa | Synechococcus elongatus PCC6301 | IB | 22.0 | [17] |
| Land plant | Flaveria bidentis | IB | 27.8 | [18] |
| Land plant | Flaveria floridana | IB | 28.6 | [18] |
| Land plant | Nicotiana tabacum | IB | 29 | [18] |
| Land plant | Spinacia oleracea | IB | 28.2–30.3 | [15, 17, 19] |
| Proteobacteria | Ralstonia eutropha | IC | 19.0 | [136] |
| Proteobacteria | Rhodobacter sphaeroides | IC | 22.4 | [136] |
| Coccolithophore | Emiliana huxleyi | ID | 11.1 | [137] |
| Diatom | Skeletonema costatum | ID | 18.5 | [27] |
| Proteobacteria | Rhodospirillum rubrum | II | 17.8–23.8 | [17, 18, 26, 73] |
| Proteobacteria | Riftia pachyptila symbiont | II | 19.5 | [74] |
An advantage of this molecular perspective is that the expectations for ancient variation in RuBisCO isotopic fractionation can be experimentally tested. Recently, molecular paleobiology has been recruited to reconcile independent biological and geological records of life by the laboratory reconstruction of ancestral enzymes and metabolic systems responsible for producing preserved biosignatures [30–32]. A fundamental issue with the interpretation of carbon isotope biosignatures is that it is not known to what extent the isotope discrimination behavior or modern biology can serve as a proxy for past life. These paleogenetic tools instead leverage modern genomic information and phylogenetic models to infer the molecular sequences of ancestral enzymes prior to their experimental synthesis and characterization [32, 33]. By this approach, the isotopic effects of inferred ancestral enzymes can be compared directly with preserved carbon isotope biosignatures, thereby reconciling biological and geological records of life [30]. Such an approach is not itself meant to be a complete solution to understanding the consistency of the carbon isotope record. Rather, these strategies can help constrain the set of contributing factors and complement further characterization of extant biological fractionation processes and the geological samples themselves. Together, these efforts provide an empirical strategy to interrogate the oldest physical remnants of ancient life.
Box 1. Mechanism and measurement of biological carbon isotope discrimination.
Measurements of carbon isotope compositions are relative, normalized to an international standard (Vienna Pee Dee Belemnite), and expressed as:
Materials enriched in 13C relative to the standard (defined as δ13C = 0‰) are thus expressed as positive δ13C values, whereas those depleted in 13C, like biogenic carbon compounds, are expressed as negative δ13C values.
In autotrophic organisms, CO2 fixing enzymes typically discriminate against the heavier 13C isotope as a result of an enzymatic kinetic isotope effect that leaves the resulting product fractionated, or depleted, in 13C relative to the CO2 source. The heavier 13C isotope requires greater activation energy to reach the transitional state, resulting in a slower reaction rate. The degree to which a given carboxylase discriminates is measured as a ratio of the reaction rate constants for each isotope (k12:k13), which is converted to an epsilon value, ε = 1000 [(kl2/k13) − 1]. For example, for RubisCO, this is directly proportional to the discrimination by the enzyme against 13C that occurs between the substrate pool (CO2) and the product (phosphoglycerate; [144]).
Enzyme fractionation has been approximated in whole cells in laboratory experiments where the δ13C of the source CO2 in the growth media is known (Table 1). This is calculated as Δδ13C = δ13Cbiomass − δ13CCO2. Since Δδ13C values encompass other physiological factors also influencing carbon isotope ratios of samples (see Table 2), ε as defined above is restricted here for carbon fractionation measured for purified enzymes.
The production and preservation of carbon isotopic biosignatures
Several biotic and abiotic factors are known to influence the magnitude of isotopic fractionation as carbon is assimilated into biomass. At the heart of carbon fixation pathways, enzyme fractionation associated with the production of biological carbon is the result of an enzymatic kinetic isotope effect that produces differences in the δ13C compositions of substrates versus products (see Box 1 for a discussion on fractionation mechanism as well as a description of notation used here). These effects arise from the isotope mass difference between 13C and 12C [34, 35], and result in a slight preference for the conversion of 12C-containing compounds to organic biomass. In addition to this enzymatic effect, environmental and physiological factors can additionally modulate the isotopic composition of fixed carbon.
There are seven known pathways of carbon fixation utilized by autotrophs, including the aforementioned CBB cycle and the recently proposed reductive glycine pathway [36] (reviewed in [25, 37]) (Table 1). These different autotrophic pathways vary in their taxonomic distributions, oxygen sensitivities, and contributions to total modern primary productivity. Though the CBB pathway is today responsible for the bulk of total fixed carbon [38–40], it is not considered to be the oldest carbon fixation mechanism. Instead, it has been suggested that the Wood–Ljungdahl pathway, today utilized by acetogenic and anaerobic ammonium oxidizing bacteria as well as methanogenic archaea, is the oldest mechanism and is proposed to be associated with the last universal common ancestor [25, 41–43].
The Δδ13C values associated with different carbon fixation pathways are calculated as the difference between δ13C of biomass and source dissolved inorganic carbon or CO2 (and thus reflect a combination of enzymatic, physiological, and environmental effects). Δδ13C measured for the rTCA, HP/HB, DC/HB, and 3HP pathways indicate smaller isotopic discriminations (Δδ13C = 0–15‰) relative to those produced by the CBB pathway (Δδ13C = 10–30‰) (Table 1). The Wood–Ljungdahl pathway can produce discrimination in excess of that of the CBB pathway (i.e., Δδ13C = 30–40‰) [44, 45], with the greatest discrimination (Δδ13C = 65‰) measured from acetogens [46]. Δδ13C values have not yet been determined for the roTCA and recently described reductive glycine pathways.
Environmental and physiological components of Δδ13C values have themselves also been investigated (Table 2). Changes in physical environmental factors, including temperature, pH, and CO2 and H2 concentrations (the latter in methanogenic organisms) all have significant effects, typically imparting between a 5 and 30‰ change in isotope discrimination. Decreased external CO2 concentrations in particular appear to reliably result in decreased isotopic fractionations. This is likely due to a Rayleigh distillation effect in which, at low CO2 concentrations, organisms will use intracellular CO2 faster than can be diffusively exchanged with external source CO2, thereby minimizing isotopic discrimination as well [47]. This relationship has been leveraged in an effort to use carbon isotopic compositions of preserved organic matter as a proxy for ancient atmospheric CO2 levels [8, 48, 49]. For variables that have been tested using methanogen cultures, including H2 concentration and pressure, fractionation can vary up to 30‰. Physiological factors, including growth rate and cell shape, can result in up to ~20‰ variability in isotope discrimination. In sum, these environmental and physiological factors can produce variability in fractionation that can meet or exceed variability attributed to differences in autotrophic metabolic pathways. However, the majority of these studies have been conducted on organisms such as eukaryotes that likely only evolved after the first 1–2 billion years of Earth history (Table 2). The autotrophic organisms more likely to have contributed to the first half of the geologic carbon isotope record are significantly underrepresented in studies of carbon isotopic fractionation.
The fate of fixed carbon is further biological recycling and/or burial. The former can result in significant carbon isotopic fractionation effects that can overprint autotrophic signatures. For example, methylotrophic methanogens (i.e., utilizing single carbon substrates other than CO2) can produce Δδ13C values as great as ~80‰ between product CH4 and source inorganic carbon [50, 51]. Further, methanotrophy, itself isotopically discriminating by ~30‰ [52], can result in an even greater depletion of 13C in biomass given an initially 13C-depleted CH4 substrate (δ13C = ~60‰) produced by methanogens. Abiotic, post-depositional processes can further alter primary biogenic isotope signatures. Thermal alteration associated with metamorphism, for example, results in preferential loss of 12C from preserved organic material [10]. Thus, care must be taken in interpreting carbon biosignatures from potentially altered samples. Independent methods for assessing thermal alteration, such as H/C content, can be used to quantify the degree of thermal maturation in filtering the carbon biosignature dataset [10].
In sum, the variability between isotope fractionation among different carbon metabolisms permits isotope compositions to not only be used broadly as a biosignature of life but can generally be used to fingerprint particular metabolic processes and the taxa associated with them. The portion of carbon that is subsequently preserved in the geologic record thus forms a remnant signature of these ancient carbon cycling processes.
Features of the carbon isotope record
The isotopic compositions of organic and inorganic carbon preserved in the geologic record, spanning >3.8 billion years, together provide the oldest forms of preserved ancient biosignatures [6, 7] (Fig. 1). Scales of isotopic measurement can range from bulk rock characterizations to those of individual, microscopic, organic fossils (e.g. [5, 53–55],). The absolute difference between inorganic and organic δ13C values in the geologic record is interpreted to reflect contemporaneous, biological isotopic fractionating processes including carbon fixation [6, 7, 10].
To date, the purportedly oldest biogenic carbon isotope measurements have been obtained from submicron graphitic inclusions in a ~4.1-billion-year-old zircon. These measurements yield a δ13C value of −24 ± 5‰, falling within the range of biological fractionation (Fig. 1) providing the earliest potential evidence of life [2]. The biogenicity of such ancient measurements is subject to controversy, owing primarily to the potential for more recent alteration of these isotopic signatures and the influence of comparable abiotic fractionation processes on the early Earth. Unambiguous assignment of these isotopic values to any particular metabolic process has not yet been achieved. For the more recent <3.5 billion years of the carbon isotope record, biogenicity of such signatures is less contentious due to a nearly coincident morphological fossil record [56, 57]. Furthermore, organic matter in more recent sediments is typically found as amorphous kerogen rather than graphite, the latter of which is more likely to have been produced abiotically and/or potentially indicative of high thermal alteration [10, 58].
Efforts have been made by geochemists and paleobiologists to filter the carbon isotope dataset to minimize representation of samples more likely to have been affected by post-depositional alteration, as well as statistically evaluate trends in the record [7]. After such treatments, a largely static isotopic trend remains but is punctuated by a significant negative excursion in organic δ13C coinciding with the late Archean to Proterozoic transition, previously noted by Hayes [9]. This excursion has been interpreted to represent the increased activity of oxygen-requiring methanotrophy. Methanotrophic recycling of buried organic material may have accompanied the initial accumulation of free oxygen following the evolution of oxygenic phototrophs. Since methanotrophy can result in exceedingly 13C-depleted carbon as described above [52], the influence of this metabolic process is a reasonable explanation for the identified excursion. Another negative isotopic anomaly, though not identified by Krissansen-Totton et al. [7], has been noted at ~2 billion years [12]. This excursion may have similarly resulted from the contributions of methanotrophs or methanogens that both produce large isotopic discriminations in excess of that typically observed for oxygenic phototrophs. On more recent geologic timescales, finer trends in the last 100 million years have been attributed to changes in atmospheric CO2 concentrations due to the empirical relationship between CO2 concentration and isotopic discrimination, as described above [8, 48, 49].
A molecular perspective on the role of rubisco evolution in shaping the carbon isotope record
Though compelling, the few identifiable signals serve to heighten the curious consistency in the remainder of the carbon isotope record, particularly considering the dynamic, early Earth biogeochemical environment. One of the most fundamental shifts in the biosphere over Earth’s 4.5-billion-year history has been the progressive oxygenation of the surface environment, mediated by the origin of oxygenic photosynthesis [24]. This process may have begun with early “whiffs” of oxygen by ~3 to 2.5 billion years ago [59, 60], but unambiguous signatures of atmospheric oxygen are not known before 2.4 billion years ago [23]. The isotopic excursion in the late Archean-early Proterozoic is likely related to these changes in environmental oxygen. However, it is still unclear why other deviations are not present due to the expected breadth of biological consequences from such a significant shift in atmospheric composition [57, 61, 62]. Furthermore, oxygen levels likely remained exceedingly low through the Proterozoic until ~0.5 billion years ago [24], yet biological isotopic trends associated with later shifts in atmospheric composition are not readily identifiable.
The first 3 billion years—the vast majority—of the carbon isotope record, produced primarily by the ancient microbial organisms that dominated the Precambrian Era (~0.54 to 4.5 billion years ago), likely requires a different level of analysis than that of the more recent geologic past. In the absence of the later-evolved, multicellular eukaryotes that typify the Phanerozoic Era (present to ~0.54 billion years ago), the early evolution of life and the major biological innovations that occurred through the Precambrian have frequently been considered rather at the subcellular level. For example, focus has been drawn to the molecular machines, enzymes, that catalyze crucial biogeochemical transformations and shaped primary productivity for billions of years [30, 39, 63, 64]. Comparatively, little attention has been given to how the molecular evolution of carbon fixation enzymes may have impacted and/or constrained features of the carbon isotope record.
This perspective can be illustrated in the evolution of the early-evolved RuBisCO enzyme, which plays a critical role in the modern biosphere and is proposed to be one of the most abundant proteins on Earth [65–67]. This enzyme is thought to have evolved more than 3 billion years ago in the anoxic environment preceding the Great Oxidation Event [13, 14, 68, 69]. Today, three forms (I–III) of RuBisCO catalyze carbon uptake in the CBB cycle; a fourth form (IV), a “RuBisCO-like” enzyme, is homologous but does not perform a carboxylase function [21, 70]. Cyanobacteria, green algae, and land plants utilize Form IA and IB RuBisCO, suggesting that the evolution of these forms are linked to that of Earth’s dominant phototrophs [71]. However, previous phylogenetic analyses indicate that other forms of RuBisCO diverged earlier than Form IA and IB homologs [13, 72]. It is then possible that ancestral enzymes preceding the evolution of oxygenic phototrophy shared greater similarity to the catalytic properties, and thus, isotopic effects, of earlier diverged forms.
However, isotopic fractionation measurements of purified RuBisCO enzymes are few, even for the better-characterized Form IA and IB enzymes. Isotope effects have been measured for certain Form IC and ID enzymes associated with coccolithophores, diatoms, and proteobacteria (ε ~11 to 22‰), which are generally distinguishable from effects for Form IA and IB (ε ~20 to 30‰) (Table 3). Isotope effects from only two organisms have been measured for Form II RuBisCO [17, 18, 26, 73, 74], and no fractionation values have been measured from Form III, which phylogenetic analysis indicates diverged earlier than Form I and II RuBisCO homologs [72]. Thus, characterization of Form III, as well as other underrepresented forms (e.g., Form IC, ID, and II), are necessary to evaluate their potential impact on the early carbon isotope record.
An anaerobic origin of RuBisCO is also of interest in the context of its substrate specificity. In addition to CO2 assimilation, RuBisCO catalyzes a competing oxygenation reaction in which RuBP is combined with O2, which in turn reduces the overall metabolic efficiency of carbon fixation. RuBisCO specificity inversely correlates with enzyme activity [75–78]. It has been argued that the balance between specificity and enzyme activity is achieved by the RuBisCO transition state, which accentuates the structural differences between otherwise similar CO2 and O2 molecules at the cost of slowed catalysis [77]. For an organism that makes its living by RuBisCO-catalyzed CO2 fixation, an atmosphere with significant amounts of O2—as well as cellular O2 produced by oxygenic photosynthesis—presents a serious hindrance [79, 80]. Extant organisms compensate for this inefficiency by various strategies, including active CO2-concentration mechanisms [81, 82]. However, such strategies would have been unnecessary during the early evolution of RuBisCO, prior to the evolution of oxygenic phototrophs when atmospheric CO2 concentrations may have been up to 2500 times higher than today [24, 28, 29, 83]. This suggests that the O2/CO2-specificity problem may only be significant in the O2-rich atmosphere that has characterized the latter half of Earth’s history [24].
Analyses of extant RuBisCO isotope effects show that changes in O2/CO2-specificity and catalytic efficiency manifest in changes to isotope effects [16, 19, 26, 27, 77]. A study of more than 100 diverse Form II and III RuBisCOs only recently found that the range of carboxylation rates extends to more than twofold that of plant RuBisCOs [84]. This may suggest that the true diversity of RuBisCO fractionation behavior is similarly not captured by existing measurements. If the specificity of RuBisCO adapted because of secular trends in environmental O2/CO2 levels, these changes would be expected to manifest in the carbon isotope record.
Future directions—paleogenetic reconstruction of ancestral carbon fixation pathways
There are several reasons why molecular-level adaptations to RuBisCO (as well as other carbon fixation enzymes) to the changing early Earth environment would leave discernible features in the carbon isotope record, as described above. These expectations warrant empirical testing. However, the use of extant biology as a proxy for ancient life is fundamentally limited. It is not known to what degree isotopic fractionation of modern organisms and their subcellular components resemble that for the enzymes, metabolic networks, and organisms that existed billions of years ago. For instance, the landscape of early carbon metabolic networks, which would have manifested from predecessor prebiotic chemical networks [85], may have been fundamentally different during and immediately after the origin of life [86, 87]. Thus, isotopic fractionation expectations derived from features of modern biology may be inherently limited in their scope.
We propose that this challenge can be met by combining the complementary strengths of geological and biological datasets. This can be accomplished through the integration of molecular paleobiology and synthetic biology tools to reconcile ancestral enzyme behaviors with the geochemical record of biological activity. This strategy applies phylogenetic models to extant genomic data to reconstruct the molecular sequences of ancestral enzymes [32, 33]. These sequences can then be synthesized in the laboratory and experimentally characterized for properties of interest. Though inferred sequences are probabilistic, they can serve to constrain the molecular sequence space that results in a particular phenotype. In vitro assessments of enzymatic properties can be conducted in addition to in vivo studies that consider the impact of ancestral enzymes on the physiology of the host organism [30, 88]. Such an approach has had prior success in elucidating fundamental features of molecular evolution. These include the evolution of enzymatic specificity [89], origins of novel functions [90, 91], and ancient enzyme promiscuity [92, 93]. However, its application to biogeochemical questions, in particular those related to the generation of ancient biosignatures, is in its infancy [30].
This strategy, for example, could be leveraged to experimentally test the relationship between CO2 specificities and isotopic effect in ancestral RuBisCO. This work could confirm expectations for deviations in carbon isotope biosignatures due to observed fractionation effects that do not conform to that observed in the geologic record. Thus, the consistency of the record would then require explanation by other factors that would balance this deviation. Alternatively, ancestral RuBisCOs might fractionate carbon much like their extant counterparts. This result would support the possibility that the molecular evolution of RuBisCO has been fundamentally constrained with regard to isotopic fractionation behavior despite long-term adaptations to the Earth’s atmosphere. In vivo experiments as described above can help determine to what extent physiological properties can overprint enzyme-level isotopic effects.
A comprehensive approach to molecular paleobiology could in the future be expanded to other carbon fixation enzymes in, for example, anaerobic taxa that might have been predominant prior to the origin of oxygenic phototrophy. Other analyses might incorporate compound-specific or site-specific isotopic investigations to work in concert with their increased use as geochemical proxies [94, 95]. These strategies would thus bridge molecular, organism, and environmental factors in disentangling the contributions to carbon isotope biosignatures. At an even broader level, molecular paleobiology techniques may also be applied to reconcile other enzyme-implicated signals in sulfur or nitrogen isotopic systems [96] or to investigate a more expansive array of ancient organic molecular biosignatures that changed over macroevolutionary timescales.
The appeal of developing new paleogenetic techniques is found in the integration of biological and geological records of life, and the recruitment of molecular biology communities toward longstanding challenges in ancient microbial ecology and biogeochemistry. We propose that this strategy, molecular paleobiology approaches used in concert with expanding microbiological and geochemical toolsets to characterize extant taxa and refine the carbon isotope dataset, will rapidly advance resolving the long-term conundrum of carbon biosignatures observed in the fossil record. What is at stake is the fundamental notion of the pervasiveness and universality of carbon isotope biosignatures, particularly as such analyses extend beyond Earth. Future work will expose the extent to which paleobiologists, microbiologists, geochemists, and planetary scientists understand the carbon isotope record, one of the foundational interpretive tools to reconstruct past biological activity.
Acknowledgements
AKG acknowledges support from the NASA Postdoctoral Fellowship in Astrobiology. BK acknowledges support from the National Science Foundation (#1724090), the NASA Early Career Faculty Award (80NSSC19K1617) and a University of Arizona Foundation Award. BK and CMC acknowledge the support of the Harvard Origins Initiative. We thank Andy Knoll for his comments and suggestions on an earlier draft of this manuscript, as well as the recommendations of three anonymous reviewers.
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Des Marais DJ, Walter MR. Astrobiology: exploring the origins, evolution, and distribution of life in the Universe. Annu Rev Ecol Syst. 1999;30:397–420. doi: 10.1146/annurev.ecolsys.30.1.397. [DOI] [PubMed] [Google Scholar]
- 2.Bell EA, Boehnke P, Harrison TM, Mao WL. Potentially biogenic carbon preserved in a 4.1 billion-year-old zircon. Proc Natl Acad Sci USA. 2015;112:14518–21. doi: 10.1073/pnas.1517557112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tashiro T, Ishida A, Hori M, Igisu M, Koike M, Mejean P, et al. Early trace of life from 3.95 Ga sedimentary rocks in Labrador, Canada. Nature. 2017;549:516–8. doi: 10.1038/nature24019. [DOI] [PubMed] [Google Scholar]
- 4.Dodd MS, Papineau D, Grenne T, Slack JF, Rittner M, Pirajno F, et al. Evidence for early life in Earth’s oldest hydrothermal vent precipitates. Nature. 2017;543:60–64. doi: 10.1038/nature21377. [DOI] [PubMed] [Google Scholar]
- 5.Schopf JW, Kitajima K, Spicuzza MJ, Kudryavtsev AB, Valley JW. SIMS analyses of the oldest known assemblage of microfossils document their taxon-correlated carbon isotope compositions. Proc Natl Acad Sci USA. 2018;115:53–58. doi: 10.1073/pnas.1718063115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schidlowski M. Carbon isotopes as biogeochemical recorders of life over 3.8 Ga of Earth history: evolution of a concept. Precambrian Res. 2001;106:117–34. doi: 10.1016/S0301-9268(00)00128-5. [DOI] [Google Scholar]
- 7.Krissansen-Totton J, Buick R, Catling DC. A statistical analysis of the carbon isotope record from the Archean to Phanerozoic and implications for the rise of oxygen. Am J Sci. 2015;315:275–316. doi: 10.2475/04.2015.01. [DOI] [Google Scholar]
- 8.Freeman KH, Hayes JM. Fractionation of carbon isotopes by phytoplankton and estimates of ancient CO2 levels. Glob Biogeochem Cycles. 1992;6:185–98. doi: 10.1029/92GB00190. [DOI] [PubMed] [Google Scholar]
- 9.Hayes JM. Global methanotrophy at the Archean-Proterozoic transition. In: Bengtson S, editor. Early life on earth. New York: Columbia University Press; 1994. p. 220–36.
- 10.Des Marais DJ. Isotopic evolution of the biogeochemical carbon cycle during the Precambrian. Rev Miner Geochem. 2001;43:555–78. doi: 10.2138/gsrmg.43.1.555. [DOI] [Google Scholar]
- 11.Schoell M, Wellmer FW. Anomalous C-13 depletion in early Precambrian graphites from Superior-Province, Canada. Nature. 1981;290:696–9. doi: 10.1038/290696a0. [DOI] [Google Scholar]
- 12.Sekine Y, Tajika E, Ohkouchi N, Ogawa NO, Goto K, Tada R, et al. Anomalous negative excursion of carbon isotope in organic carbon after the last Paleoproterozoic glaciation in North America. Geochem Geophy Geosy. 2010;11:Q08019. doi: 10.1029/2010gc003210. [DOI] [Google Scholar]
- 13.Erb TJ, Zarzycki J. A short history of RubisCO: the rise and fall (?) of Nature’s predominant CO2 fixing enzyme. Curr Opin Biotechnol. 2018;49:100–7. doi: 10.1016/j.copbio.2017.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nisbet EG, Grassineau NV, Howe CJ, Abell PI, Regelous M, Nisbet RER. The age of Rubisco: the evolution of oxygenic photosynthesis. Geobiology. 2007;5:311–35. doi: 10.1111/j.1472-4669.2007.00127.x. [DOI] [Google Scholar]
- 15.Roeske CA, O’Leary MH. Carbon isotope effects on the enzyme-catalysed carboxylation of ribulose bisphosphate. Biochemistry. 1984;23:6275–84. doi: 10.1021/bi00320a058. [DOI] [PubMed] [Google Scholar]
- 16.Scott KM, Henn-Sax M, Harmer TL, Longo DL, Frame CH, Cavanaugh CM. Kinetic isotope effect and biochemical characterization of form IA RubisCO from the marine cyanobacterium Prochlorococcus marinus MIT9313. Limnol Oceanogr. 2007;52:2199–204. doi: 10.4319/lo.2007.52.5.2199. [DOI] [Google Scholar]
- 17.Guy RD, Fogel ML, Berry JA. Photosynthetic fractionation of the stable isotopes of oxygen and carbon. Plant Physiol. 1993;101:37–47. doi: 10.1104/pp.101.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.von Caemmerer S, Tazoe Y, Evans JR, Whitney SM. Exploiting transplastomically modified Rubisco to rapidly measure natural diversity in its carbon isotope discrimination using tuneable diode laser spectroscopy. J Exp Bot. 2014;65:3759–67. doi: 10.1093/jxb/eru036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott KM, Schwedock J, Schrag DP, Cavanaugh CM. Influence of form IA RubisCO and environmental dissolved inorganic carbon on the δ13C of the clam-chemoautotroph symbiosis Solemya velum. Environ Microbiol. 2004;6:1210–9. doi: 10.1111/j.1462-2920.2004.00642.x. [DOI] [PubMed] [Google Scholar]
- 20.Ivanovsky RN, Fal YI, Berg IA, Ugolkova NV, Krasilnikova EN, Keppen OI, et al. Evidence for the presence of the reductive pentose phosphate cycle in a filamentous anoxygenic photosynthetic bacterium, Oscillochloris trichoides strain DG-6. Microbiology. 1999;145:1743–8. doi: 10.1099/13500872-145-7-1743. [DOI] [PubMed] [Google Scholar]
- 21.Tabita FR, Satagopan S, Hanson TE, Kreel NE, Scott SS. Distinct form I, II, III, and IV Rubisco proteins from the three kingdoms of life provide clues about Rubisco evolution and structure/function relationships. J Exp Bot. 2008;59:1515–24. doi: 10.1093/jxb/erm361. [DOI] [PubMed] [Google Scholar]
- 22.Caldwell PE, MacLean MR, Norris PR. Ribulose bisphosphate carboxylase activity and a Calvin cycle gene cluster in Sulfobacillus species. Microbiology. 2007;153:2231–40. doi: 10.1099/mic.0.2007/006262-0. [DOI] [PubMed] [Google Scholar]
- 23.Farquhar J. Atmospheric influence of Earth’s earliest sulfur cycle. Science. 2000;289:756–8. doi: 10.1126/science.289.5480.756. [DOI] [PubMed] [Google Scholar]
- 24.Lyons TW, Reinhard CT, Planavsky NJ. The rise of oxygen in Earth’s early ocean and atmosphere. Nature. 2014;506:307–15. doi: 10.1038/nature13068. [DOI] [PubMed] [Google Scholar]
- 25.Ward LM, Shih PM. The evolution and productivity of carbon fixation pathways in response to changes in oxygen concentration over geological time. Free Radic Biol Med. 2019;140:188–99. doi: 10.1016/j.freeradbiomed.2019.01.049. [DOI] [PubMed] [Google Scholar]
- 26.McNevin DB, Badger MR, Whitney SM, von Caemmerer S, Tcherkez GG, Farquhar GD. Differences in carbon isotope discrimination of three variants of D-ribulose-1,5-bisphosphate carboxylase/oxygenase reflect differences in their catalytic mechanisms. J Biol Chem. 2007;282:36068–76. doi: 10.1074/jbc.M706274200. [DOI] [PubMed] [Google Scholar]
- 27.Boller AJ, Thomas PJ, Cavanaugh CM, Scott KM. Isotopic discrimination and kinetic parameters of RubisCO from the marine bloom-forming diatom, Skeletonema costatum. Geobiology. 2015;13:33–43. doi: 10.1111/gbi.12112. [DOI] [PubMed] [Google Scholar]
- 28.Young JN, Rickaby RE, Kapralov MV, Filatov DA. Adaptive signals in algal Rubisco reveal a history of ancient atmospheric carbon dioxide. Philos Trans R Soc B. 2012;367:483–92. doi: 10.1098/rstb.2011.0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Catling DC, Zahnle KJ. The Archean atmosphere. Sci Adv. 2020;6:eaax1420. doi: 10.1126/sciadv.aax1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia AK, Kacar B. How to resurrect ancestral proteins as proxies for ancient biogeochemistry. Free Radic Biol Med. 2019;140:260–9. doi: 10.1016/j.freeradbiomed.2019.03.033. [DOI] [PubMed] [Google Scholar]
- 31.Kacar B, Guy L, Smith E, Baross J. Resurrecting ancestral genes in bacteria to interpret ancient biosignatures. Philos Trans R Soc A. 2017;375:20160352. doi: 10.1098/rsta.2016.0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benner S. Uniting natural history with the molecular sciences. The ultimate multidisciplinarity. Acc Chem Res. 2017;50:498–502. doi: 10.1021/acs.accounts.6b00496. [DOI] [PubMed] [Google Scholar]
- 33.Thornton JW. Resurrecting ancient genes: experimental analysis of extinct molecules. Nat Rev Genet. 2004;5:366–75. doi: 10.1038/nrg1324. [DOI] [PubMed] [Google Scholar]
- 34.Nier AO, Gulbransen EA. Variations in the relative abundance of the carbon isotopes. J Am Chem Soc. 1939;61:697–8. doi: 10.1021/ja01872a047. [DOI] [Google Scholar]
- 35.Wickman FE. Variations in the relative abundance of the carbon isotopes in plants. Nature. 1952;169:1051–1051. doi: 10.1038/1691051a0. [DOI] [PubMed] [Google Scholar]
- 36.Figueroa IA, Barnum TP, Somasekhar PY, Carlstrom CI, Engelbrektson AL, Coates JD. Metagenomics-guided analysis of microbial chemolithoautotrophic phosphite oxidation yields evidence of a seventh natural CO2 fixation pathway. Proc Natl Acad Sci USA. 2018;115:E92–E101. doi: 10.1073/pnas.1715549114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berg IA, Kockelkorn D, Ramos-Vera WH, Say RF, Zarzycki J, Hugler M, et al. Autotrophic carbon fixation in archaea. Nat Rev Microbiol. 2010;8:447–60. doi: 10.1038/nrmicro2365. [DOI] [PubMed] [Google Scholar]
- 38.Raven JA. Contributions of anoxygenic and oxygenic phototrophy and chemolithotrophy to carbon and oxygen fluxes in aquatic environments. Aquat Microb Ecol. 2009;56:177–92. doi: 10.3354/ame01315. [DOI] [Google Scholar]
- 39.Falkowski PG, Fenchel T, Delong EF. The microbial engines that drive Earth’s biogeochemical cycles. Science. 2008;320:1034–9. doi: 10.1126/science.1153213. [DOI] [PubMed] [Google Scholar]
- 40.Tabita FR, Hanson TE, Li H, Satagopan S, Singh J, Chan S. Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs. Microbiol Mol Biol Rev. 2007;71:576–99. doi: 10.1128/MMBR.00015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Braakman R, Smith E. The emergence and early evolution of biological carbon-fixation. PLoS Comput Biol. 2012;8:e1002455. doi: 10.1371/journal.pcbi.1002455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fuchs G. Alternative pathways of carbon dioxide fixation: insights into the early evolution of life? Annu Rev Microbiol. 2011;65:631–58. doi: 10.1146/annurev-micro-090110-102801. [DOI] [PubMed] [Google Scholar]
- 43.Weiss MC, Sousa FL, Mrnjavac N, Neukirchen S, Roettger M, Nelson-Sathi S, et al. The physiology and habitat of the last universal common ancestor. Nat Microbiol. 2016;1:16116. doi: 10.1038/nmicrobiol.2016.116. [DOI] [PubMed] [Google Scholar]
- 44.House CH, Schopf JW, Stetter KO. Carbon isotopic fractionation by Archaeans and other thermophilic prokaryotes. Org Geochem. 2003;34:345–56. doi: 10.1016/S0146-6380(02)00237-1. [DOI] [Google Scholar]
- 45.Preuss A, Schauder R, Fuchs G, Stichler W. Carbon isotope fractionation by autotrophic bacteria with three different CO2 fixation pathways. Z Naturforsch C. 1989;44:397–402. doi: 10.1515/znc-1989-5-610. [DOI] [Google Scholar]
- 46.Freude C, Blaser M. Carbon isotope fractionation during catabolism and anabolism in acetogenic bacteria growing on different substrates. Appl Environ Microbiol. 2016;82:2728–37. doi: 10.1128/AEM.03502-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hinga KR, Arthur MA, Pilson MEQ, Whitaker D. Carbon isotope fractionation by marine phytoplankton in culture: the effects of CO2 concentration, pH, temperature, and species. Glob Biogeochem Cycles. 1994;8:91–102. doi: 10.1029/93GB03393. [DOI] [Google Scholar]
- 48.Schubert BA, Jahren AH. Global increase in plant carbon isotope fractionation following the Last Glacial Maximum caused by increase in atmospheric pCO2. Geology. 2015;43:435–8. doi: 10.1130/G36467.1. [DOI] [Google Scholar]
- 49.Pagani M, Gröcke DR, Kucera M. The alkenone-CO2 proxy and ancient atmospheric carbon dioxide. Philos Trans R Soc A. 2002;360:609–32. doi: 10.1098/rsta.2001.0959. [DOI] [PubMed] [Google Scholar]
- 50.Londry KL, Dawson KG, Grover HD, Summons RE, Bradley AS. Stable carbon isotope fractionation between substrates and products of Methanosarcina barkeri. Org Geochem. 2008;39:608–21. doi: 10.1016/j.orggeochem.2008.03.002. [DOI] [Google Scholar]
- 51.Penger J, Conrad R, Blaser M. Stable carbon isotope fractionation by methylotrophic methanogenic archaea. Appl Environ Microbiol. 2012;78:7596–602. doi: 10.1128/AEM.01773-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Summons RE, Jahnke LL, Roksandic Z. Carbon isotopic fractionation in lipids from methanotrophic bacteria: relevance for interpretation of the geochemical record of biomarkers. Geochim Cosmochim Acta. 1994;58:2853–63. doi: 10.1016/0016-7037(94)90119-8. [DOI] [PubMed] [Google Scholar]
- 53.House CH, Schopf JW, McKeegan KD, Coath CD, Harrison TM, Stetter KO. Carbon isotopic composition of individual Precambrian microfossils. Geology. 2000;28:707–10. doi: 10.1130/0091-7613(2000)28<707:CICOIP>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 54.Ueno Y, Isozaki Y, Yurimoto H, Maruyama S. Carbon isotopic signatures of individual Archean microfossils(?) from Western Australia. Int Geol Rev. 2001;43:196–212. doi: 10.1080/00206810109465008. [DOI] [Google Scholar]
- 55.Williford KH, Ushikubo T, Schopf JW, Lepot K, Kitajima K, Valley JW. Preservation and detection of microstructural and taxonomic correlations in the carbon isotopic compositions of individual Precambrian microfossils. Geochim Cosmochim Acta. 2013;104:165–82. doi: 10.1016/j.gca.2012.11.005. [DOI] [Google Scholar]
- 56.Schopf JW, Kudryavtsev AB, Czaja AD, Tripathi AB. Evidence of Archean life: stromatolites and microfossils. Precambrian Res. 2007;158:141–55. doi: 10.1016/j.precamres.2007.04.009. [DOI] [Google Scholar]
- 57.Knoll AH, Nowak MA. The timetable of evolution. Sci Adv. 2017;3:e1603076. doi: 10.1126/sciadv.1603076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schopf JW, Kudryavtsev AB, Agresti DG, Czaja AD, Wdowiak TJ. Raman imagery: a new approach to assess the geochemical maturity and biogenicity of permineralized Precambrian fossils. Astrobiology. 2005;5:333–71. doi: 10.1089/ast.2005.5.333. [DOI] [PubMed] [Google Scholar]
- 59.Crowe SA, Dossing LN, Beukes NJ, Bau M, Kruger SJ, Frei R, et al. Atmospheric oxygenation three billion years ago. Nature. 2013;501:535–8. doi: 10.1038/nature12426. [DOI] [PubMed] [Google Scholar]
- 60.Anbar AD, Duan Y, Lyons TW, Arnold GL, Kendall B, Creaser RA, et al. A whiff of oxygen before the great oxidation event? Science. 2007;317:1903–6. doi: 10.1126/science.1140325. [DOI] [PubMed] [Google Scholar]
- 61.Fischer WW, Valentine JS. How did life come to tolerate and thrive in an oxygenated world? Free Radic Biol Med. 2019;140:1–3. doi: 10.1016/j.freeradbiomed.2019.07.021. [DOI] [PubMed] [Google Scholar]
- 62.Lenton TM, Boyle RA, Poulton SW, Shields-Zhou GA, Butterfield NJ. Co-evolution of eukaryotes and ocean oxygenation in the Neoproterozoic era. Nat Geosci. 2014;7:257–65. doi: 10.1038/ngeo2108. [DOI] [Google Scholar]
- 63.Moore EK, Jelen BI, Giovannelli D, Raanan H, Falkowski PG. Metal availability and the expanding network of microbial metabolisms in the Archaean eon. Nat Geosci. 2017;10:629–36. doi: 10.1038/ngeo3006. [DOI] [Google Scholar]
- 64.Nitschke W, McGlynn SE, Milner-White EJ, Russell MJ. On the antiquity of metalloenzymes and their substrates in bioenergetics. Biochim Biophys Acta. 2013;1827:871–81. doi: 10.1016/j.bbabio.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 65.Ellis RJ. The most abundant protein in the world. Trends Biochem Sci. 1979;4:241–4. doi: 10.1016/0968-0004(79)90212-3. [DOI] [Google Scholar]
- 66.Raven JA. Rubisco: still the most abundant protein of Earth? N Phytol. 2013;198:1–3. doi: 10.1111/nph.12197. [DOI] [PubMed] [Google Scholar]
- 67.Bar-On YM, Milo R. The global mass and average rate of rubisco. Proc Natl Acad Sci USA. 2019;10:4738–43. doi: 10.1073/pnas.1816654116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tabita FR, Hanson TE, Satagopan S, Witte BH, Kreel NE. Phylogenetic and evolutionary relationships of RubisCO and the RubisCO-like proteins and the functional lessons provided by diverse molecular forms. Philos Trans R Soc B. 2008;363:2629–40. doi: 10.1098/rstb.2008.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ashida H, Danchin A, Yokota A. Was photosynthetic RuBisCO recruited by acquisitive evolution from RuBisCO-like proteins involved in sulfur metabolism? Res Microbiol. 2005;156:611–8. doi: 10.1016/j.resmic.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 70.Kim SM, Lim HS, Lee SB. Discovery of a RuBisCO-like protein that functions as an oxygenase in the novel d-hamamelose pathway. Biotechnol Bioprocess Eng. 2018;23:490–9. doi: 10.1007/s12257-018-0305-6. [DOI] [Google Scholar]
- 71.Schopf JW. The paleobiological record of photosynthesis. Photosynth Res. 2011;107:87–101. doi: 10.1007/s11120-010-9577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kacar B, Hanson-Smith V, Adam ZR, Boekelheide N. Constraining the timing of the Great Oxidation Event within the Rubisco phylogenetic tree. Geobiology. 2017;15:628–40. doi: 10.1111/gbi.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roeske CA, O’Leary MH. Carbon isotope effect on carboxylation of ribulose bisphosphate catalyzed by ribulosebisphosphate carboxylase from Rhodospirillum rubrum. Biochemistry. 1985;24:1603–7. doi: 10.1021/bi00328a005. [DOI] [PubMed] [Google Scholar]
- 74.Robinson JJ, Scott KM, Swanson ST, O’Leary MH, Horken K, Tabita FR, et al. Kinetic isotope effect and characterization of form II RubisCO from the chemoautotrophic endosymbionts of the hydrothermal vent tubeworm Riftia pachyptila. Limnol Oceanogr. 2003;1:48–54. doi: 10.4319/lo.2003.48.1.0048. [DOI] [Google Scholar]
- 75.Savir Y, Noor E, Milo R, Tlusty T. Cross-species analysis traces adaptation of Rubisco toward optimality in a low-dimensional landscape. Proc Natl Acad Sci USA. 2010;107:3475–80. doi: 10.1073/pnas.0911663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Studer RA, Christin PA, Williams MA, Orengo CA. Stability-activity tradeoffs constrain the adaptive evolution of RubisCO. Proc Natl Acad Sci USA. 2014;111:2223–8. doi: 10.1073/pnas.1310811111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tcherkez GG, Farquhar GD, Andrews TJ. Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc Natl Acad Sci USA. 2006;103:7246–51. doi: 10.1073/pnas.0600605103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Flamholz AI, Prywes N, Moran U, Davidi D, Bar-On YM, Oltrogge LM, et al. Revisiting trade-offs between Rubisco kinetic parameters. Biochemistry. 2019;58:3365–76. doi: 10.1021/acs.biochem.9b00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Leegood RC. A welcome diversion from photorespiration. Nat Biotechnol. 2007;25:539–40. doi: 10.1038/nbt0507-539. [DOI] [PubMed] [Google Scholar]
- 80.Griffiths H. Plant biology: designs on Rubisco. Nature. 2006;441:940–1. doi: 10.1038/441940a. [DOI] [PubMed] [Google Scholar]
- 81.Badger MR, Price GD. CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J Exp Bot. 2003;54:609–22. doi: 10.1093/jxb/erg076. [DOI] [PubMed] [Google Scholar]
- 82.Scott KM, Leonard JM, Boden R, Chaput D, Dennison C, Haller E, et al. Diversity in CO2-concentrating mechanisms among Chemolithoautotrophs from the genera Hydrogenovibrio, Thiomicrorhabdus, and Thiomicrospira, ubiquitous in sulfidic habitats worldwide. Appl Environ Microbiol. 2019;85:e02096–18. doi: 10.1128/aem.02096-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rickaby REM, Eason Hubbard MR. Upper ocean oxygenation, evolution of RuBisCO and the Phanerozoic succession of phytoplankton. Free Radic Biol Med. 2019;140:295–304. doi: 10.1016/j.freeradbiomed.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Davidi D, Shamshoum M, Guo Z, Bar‐On YM, Prywes N, Oz A et al. Highly active rubiscos discovered by systematic interrogation of natural sequence diversity. EMBO J. 2020: e104081; 10.15252/embj.2019104081. [DOI] [PMC free article] [PubMed]
- 85.Adam ZR, Fahrenbach AC, Jacobson SM, Kacar B, Zubarev DY. Radiolysis generates a complex organosynthetic chemical network. Sci Rep. 2021;11:1743. doi: 10.1038/s41598-021-81293-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tian T, Chu XY, Yang Y, Zhang X, Liu YM, Gao J, et al. Phosphates as energy sources to expand metabolic networks. Life. 2019;9:43. doi: 10.3390/life9020043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Goldford JE, Hartman H, Smith TF, Segre D. Remnants of an ancient metabolism without phosphate. Cell. 2017;168:1126–34. doi: 10.1016/j.cell.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 88.Siddiq MA, Loehlin DW, Montooth KL, Thornton JW. Experimental test and refutation of a classic case of molecular adaptation in Drosophila melanogaster. Nat Ecol Evol. 2017;1:0025. doi: 10.1038/s41559-016-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thornton JW, Need E, Crews D. Resurrecting the ancestral steroid receptor: ancient origin of estrogen signaling. Science. 2003;301:1714–7. doi: 10.1126/science.1086185. [DOI] [PubMed] [Google Scholar]
- 90.Hochberg GKA, Thornton JW. Reconstructing ancient proteins to understand the causes of structure and function. Annu Rev Biophys. 2017;46:247–69. doi: 10.1146/annurev-biophys-070816-033631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ohno S. Ancient linkage groups and frozen accidents. Nature. 1973;244:259–62. doi: 10.1038/244259a0. [DOI] [PubMed] [Google Scholar]
- 92.Risso VA, Gavira JA, Mejia-Carmona DF, Gaucher EA, Sanchez-Ruiz JM. Hyperstability and substrate promiscuity in laboratory resurrections of Precambrian beta-lactamases. J Am Chem Soc. 2013;135:2899–902. doi: 10.1021/ja311630a. [DOI] [PubMed] [Google Scholar]
- 93.Copley SD. An evolutionary biochemist’s perspective on promiscuity. Trends Biochem Sci. 2015;40:72–78. doi: 10.1016/j.tibs.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Eiler JM, Bergquist B, Bourg I, Cartigny P, Farquhar J, Gagnon A, et al. Frontiers of stable isotope geoscience. Chem Geol. 2014;372:119–43. doi: 10.1016/j.chemgeo.2014.02.006. [DOI] [Google Scholar]
- 95.Close HG. Compound-specific isotope geochemistry in the ocean. Ann Rev Mar Sci. 2019;11:27–56. doi: 10.1146/annurev-marine-121916-063634. [DOI] [PubMed] [Google Scholar]
- 96.Garcia AK, McShea H, Kolaczkowski B, Kacar B. Reconstructing the evolutionary history of nitrogenases: Evidence for ancestral molybdenum-cofactor utilization. Geobiology. 2020;18:394–411. doi: 10.1111/gbi.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Woese CR, Stackebrandt E, Macke TJ, Fox GE. A phylogenetic definition of the major eubacterial taxa. Syst Appl Microbiol. 1985;6:143–51. doi: 10.1016/S0723-2020(85)80047-3. [DOI] [PubMed] [Google Scholar]
- 98.Bhattacharya D, Yoon HS, Hackett JD. Photosynthetic eukaryotes unite: endosymbiosis connects the dots. Bioessays. 2004;26:50–60. doi: 10.1002/bies.10376. [DOI] [PubMed] [Google Scholar]
- 99.Adl SM, Simpson AG, Lane CE, Lukes J, Bass D, Bowser SS, et al. The revised classification of eukaryotes. J Eukaryot Microbiol. 2012;59:429–93. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Garrity G, Bell J, Lilburn T. Phylum XIV. Proteobacteria phyl. nov. In: Brenner D, Krieg N, Staley J, Garrity G (eds). Bergey’s manual of systematic bacteriology, 2nd edn, vol. 2 (The Proteobacteria), part B (The Gammaproteobacteria). Springer: New York; 2005. p. 1.
- 101.Garrity G, Holt J. Phylum BVI. Chloroflexi phy. nov. In: Boone D, Castenholz R, Garrity G, editors. Bergey’s manual of systematic bacteriology, 2nd ed. vol. 1 (The Archaea and the deeply branching and phototrophic Bacteria). New York: Springer; 2001. p. 427–46.
- 102.Gibbons NE, Murray RGE. Proposals concerning the higher taxa of bacteria. Int J Syst Bacteriol. 1978;28:1–6. doi: 10.1099/00207713-28-1-1. [DOI] [Google Scholar]
- 103.Quandt L, Gottschalk G, Ziegler H, Stichler W. Isotope discrimination by photosynthetic bacteria. Fems Microbiol Lett. 1977;1:125–8. doi: 10.1111/j.1574-6968.1977.tb00596.x. [DOI] [Google Scholar]
- 104.Sirevåg R, Buchanan BB, Berry JA, Troughton JH. Mechanisms of CO2 fixation in bacterial photosynthesis studied by the carbon isotope fractionation technique. Arch Microbiol. 1977;112:35–38. doi: 10.1007/BF00446651. [DOI] [PubMed] [Google Scholar]
- 105.Zyakun AM, Lunina ON, Prusakova TS, Pimenov NV, Ivanov MV. Fractionation of stable carbon isotopes by photoautotrophically growing anoxygenic purple and green sulfur bacteria. Microbiology. 2009;78:757–68. doi: 10.1134/S0026261709060137. [DOI] [Google Scholar]
- 106.Reysenbach A. Phylum BI. Aquificae phy. nov. In: Boone D, Castenholz R, Garrity G, editors. Bergey’s manual of systematic bacteriology, 2nd ed. vol. 1 (The Archaea and the deeply branching and phototrophic Bacteria). New York: Springer; 2001. p. 359–67.
- 107.Garrity G, Holt J. Phylum BXI. Chlorobi phy. nov. In: Boone D, Castenholz R, Garrity G, editors. Bergey’s manual of systematic bacteriology, 2nd ed. vol. 1 (The Archaea and the deeply branching and phototrophic Bacteria). New York: Springer; 2001. p. 601–23.
- 108.Garrity G, Holt J. Phylum BVlll. Nitrospirae phy. nov. In: Boone D, Castenholz R, Garrity G (eds). Bergey’s Manual of Systematic Bacteriology, 2nd ed. vol. 1 (The Archaea and the deeply branching and phototrophic Bacteria). New York: Springer; 2001. p. 451–64.
- 109.Garrity G, Holt J. Phylum AI. Crenarchaeota phy. nov. In: Boone D, Castenholz R, Garrity G, editors. Bergey’s manual of systematic bacteriology, 2nd ed. vol. 1 (The Archaea and the deeply branching and phototrophic Bacteria). New York: Springer-Verlag; 2001. p. 169–210.
- 110.Takai K, Oida H, Suzuki Y, Hirayama H, Nakagawa S, Nunoura T, et al. Spatial distribution of marine crenarchaeota group I in the vicinity of deep-sea hydrothermal systems. Appl Environ Microbiol. 2004;70:2404–13. doi: 10.1128/AEM.70.4.2404-2413.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol. 2008;6:245–52. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- 112.Garrity G, Holt J (2001). Phylum AII. Euryarchaeota phy. nov. In: Boone D, Castenholz R, Garrity G, editors. Bergey’s manual of systematic bacteriology, 2nd ed. vol. 1 (The Archaea and the deeply branching and phototrophic Bacteria). New York: Springer; 2001. p. 211–355.
- 113.Garrity G, Holt J. The Road Map to the Manual. In: Boone D, Castenholz R, Garrity G, editors. Bergey’s manual of systematic bacteriology, 2nd ed. vol. 1 (The Archaea and the deeply branching and phototrophic Bacteria). Springer: New York; 2001. p. 119–66.
- 114.Gelwicks JT, Risatti JB, Hayes JM. Carbon isotope effects associated with autotrophic acetogenesis. Org Geochem. 1989;14:441–6. doi: 10.1016/0146-6380(89)90009-0. [DOI] [PubMed] [Google Scholar]
- 115.Holo H, Sirevåg R. Autotrophic growth and CO2 fixation of Chloroflexus aurantiacus. Arch Microbiol. 1986;145:173–80. doi: 10.1007/BF00446776. [DOI] [Google Scholar]
- 116.Hugler M, Sievert SM. Beyond the Calvin cycle: autotrophic carbon fixation in the ocean. Ann Rev Mar Sci. 2011;3:261–89. doi: 10.1146/annurev-marine-120709-142712. [DOI] [PubMed] [Google Scholar]
- 117.Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. Primary production of the biosphere: Integrating terrestrial and oceanic components. Science. 1998;281:237–40. [DOI] [PubMed]
- 118.Pachiadaki MG, Sintes E, Bergauer K, Brown JM, Record NR, Swan BK et al. Major role of nitrite-oxidizing bacteria in dark ocean carbon fixation. Science. 2017;358:1046–51. [DOI] [PubMed]
- 119.Ingalls AE, Shah SR, Hansman RL, Aluwihare LI, Santos GM, Druffel ERM et al. Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. Proc Natl Acad Sci USA. 2006;103:6442–7. [DOI] [PMC free article] [PubMed]
- 120.Wuchter C, Abbas B, Coolen MJL, Herfort L, van Bleijswijk J, Timmers P et al. Archaeal nitrification in the ocean. Proc Natl Acad Sci USA. 2006;103:12317–22. [DOI] [PMC free article] [PubMed]
- 121.Liu Y, Whitman WB. Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann NY Acad Sci. 2008;1125:171–89. [DOI] [PubMed]
- 122.Berg IA. Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl Environ Microbiol. 2011;77:1925–36. [DOI] [PMC free article] [PubMed]
- 123.Deleens E, Treichel I, O’Leary MH. Temperature dependence of carbon isotope fractionation in CAM plants. Plant Physiol. 1985;79:202–6. doi: 10.1104/pp.79.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Degens ET, Guillard RRL, Sackett WM, Hellebust JA. Metabolic fractionation of carbon isotopes in marine plankton—I. Temperature and respiration experiments. Deep Sea Res Oceanogr Abstr. 1968;15:1–9. doi: 10.1016/0011-7471(68)90024-7. [DOI] [Google Scholar]
- 125.Wong WW, Sackett WM. Fractionation of stable carbon isotopes by marine phytoplankton. Geochim Cosmochim Acta. 1978;42:1809–15. doi: 10.1016/0016-7037(78)90236-3. [DOI] [Google Scholar]
- 126.Schubert BA, Jahren AH. The effect of atmospheric CO2 concentration on carbon isotope fractionation in C3 land plants. Geochim Cosmochim Acta. 2012;96:29–43. doi: 10.1016/j.gca.2012.08.003. [DOI] [Google Scholar]
- 127.Riebesell U, Revill AT, Holdsworth DG, Volkman JK. The effects of varying CO2 concentration on lipid composition and carbon isotope fractionation in Emiliania huxleyi. Geochim Cosmochim Acta. 2000;64:4179–92. doi: 10.1016/S0016-7037(00)00474-9. [DOI] [Google Scholar]
- 128.Wilkes EB, Lee RBY, McClelland HLO, Rickaby REM, Pearson A. Carbon isotope ratios of coccolith–associated polysaccharides of Emiliania huxleyi as a function of growth rate and CO2 concentration. Org Geochem. 2018;119:1–10. doi: 10.1016/j.orggeochem.2018.02.006. [DOI] [Google Scholar]
- 129.Laws EA, Bidigare RR, Popp BN. Effect of growth rate and CO2 concentration on carbon isotopic fractionation by the marine diatom Phaeodactylum tricornutum. Limnol Oceanogr. 1997;42:1552–60. doi: 10.4319/lo.1997.42.7.1552. [DOI] [Google Scholar]
- 130.Fry B, Wainright SC. Diatom sources of 13C-rich carbon in marine food webs. Mar Ecol Prog Ser. 1991;76:149–57. doi: 10.3354/meps076149. [DOI] [Google Scholar]
- 131.Bidigare RR, Fluegge A, Freeman KH, Hanson KL, Hayes JM, Hollander D, et al. Consistent fractionation of 13C in nature and in the laboratory: growth-rate effects in some haptophyte algae. Glob Biogeochem Cycles. 1997;11:279–92. doi: 10.1029/96GB03939. [DOI] [PubMed] [Google Scholar]
- 132.Popp BN, Laws EA, Bidigare RR, Dore JE, Hanson KL, Wakeham SG. Effect of phytoplankton cell geometry on carbon isotopic fractionation. Geochim Cosmochim Acta. 1998;62:69–77. doi: 10.1016/S0016-7037(97)00333-5. [DOI] [Google Scholar]
- 133.Valentine DL, Chidthaisong A, Rice A, Reeburgh WS, Tyler SC. Carbon and hydrogen isotope fractionation by moderately thermophilic methanogens. Geochim Cosmochim Acta. 2004;68:1571–90. doi: 10.1016/j.gca.2003.10.012. [DOI] [Google Scholar]
- 134.Topçuoğlu BD, Meydan C, Nguyen TB, Lang SQ, Holden JF, Kelly RM. Growth kinetics, carbon isotope fractionation, and gene expression in the hyperthermophile Methanocaldococcus jannaschii during hydrogen-limited growth and interspecies hydrogen transfer. Appl Environ Microbiol. 2019;85:e00180–19. 10.1128/AEM.00180-19. [DOI] [PMC free article] [PubMed]
- 135.Takai K, Nakamura K, Toki T, Tsunogai U, Miyazaki M, Miyazaki J, et al. Cell proliferation at 122 °C and isotopically heavy CH4 production by a hyperthermophilic methanogen under high-pressure cultivation. Proc Natl Acad Sci USA. 2008;105:10949–54. doi: 10.1073/pnas.0712334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Thomas PJ, Boller AJ, Satagopan S, Tabita FR, Cavanaugh CM, Scott KM. Isotope discrimination by form IC RubisCO from Ralstonia eutropha and Rhodobacter sphaeroides, metabolically versatile members of ‘Proteobacteria’ from aquatic and soil habitats. Environ Microbiol. 2019;21:72–80. doi: 10.1111/1462-2920.14423. [DOI] [PubMed] [Google Scholar]
- 137.Boller AJ, Thomas PJ, Cavanaugh CM, Scott KM. Low stable carbon isotope fractionation by coccolithophore RubisCO. Geochim Cosmochim Acta. 2011;75:7200–7. doi: 10.1016/j.gca.2011.08.031. [DOI] [Google Scholar]
- 138.Jaffe AL, Castelle CJ, Dupont CL, Banfield JF. Lateral gene transfer shapes the distribution of RuBisCO among Candidate Phyla Radiation bacteria and DPANN archaea. Mol Biol Evol. 2019;36:435–46. doi: 10.1093/molbev/msy234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Butterfield NJ. Modes of pre-Ediacaran multicellularity. Precambrian Res. 2009;173:201–11. doi: 10.1016/j.precamres.2009.01.008. [DOI] [Google Scholar]
- 140.Hedges SB, Blair JE, Venturi ML, Shoe JL. A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol Biol. 2004;4:2. doi: 10.1186/1471-2148-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yang EC, Boo SM, Bhattacharya D, Saunders GW, Knoll AH, Fredericq S, et al. Divergence time estimates and the evolution of major lineages in the florideophyte red algae. Sci Rep. 2016;6:21361. doi: 10.1038/srep21361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rubinstein CV, Gerrienne P, de la Puente GS, Astini RA, Steemans P. Early Middle Ordovician evidence for land plants in Argentina (eastern Gondwana) N Phytol. 2010;188:365–9. doi: 10.1111/j.1469-8137.2010.03433.x. [DOI] [PubMed] [Google Scholar]
- 143.Morris JL, Puttick MN, Clark JW, Edwards D, Kenrick P, Pressel S, et al. The timescale of early land plant evolution. Proc Natl Acad Sci USA. 2018;115:E2274–E2283. doi: 10.1073/pnas.1719588115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hayes JM. Fractionation of carbon and hydrogen isotopes in biosynthetic processes. Rev Miner Geochem. 2001;43:225–77. doi: 10.2138/gsrmg.43.1.225. [DOI] [Google Scholar]