Abstract
Objective
Insulin regulates mitochondrial function, thereby propagating an efficient metabolism. Conversely, diabetes and insulin resistance are linked to mitochondrial dysfunction with a decreased expression of the mitochondrial chaperone HSP60. The aim of this investigation was to determine the effect of a reduced HSP60 expression on the development of obesity and insulin resistance.
Methods
Control and heterozygous whole-body HSP60 knockout (Hsp60+/−) mice were fed a high-fat diet (HFD, 60% calories from fat) for 16 weeks and subjected to extensive metabolic phenotyping. To understand the effect of HSP60 on white adipose tissue, microarray analysis of gonadal WAT was performed, ex vivo experiments were performed, and a lentiviral knockdown of HSP60 in 3T3-L1 cells was conducted to gain detailed insights into the effect of reduced HSP60 levels on adipocyte homeostasis.
Results
Male Hsp60+/− mice exhibited lower body weight with lower fat mass. These mice exhibited improved insulin sensitivity compared to control, as assessed by Matsuda Index and HOMA-IR. Accordingly, insulin levels were significantly reduced in Hsp60+/− mice in a glucose tolerance test. However, Hsp60+/− mice exhibited an altered adipose tissue metabolism with elevated insulin-independent glucose uptake, adipocyte hyperplasia in the presence of mitochondrial dysfunction, altered autophagy, and local insulin resistance.
Conclusions
We discovered that the reduction of HSP60 in mice predominantly affects adipose tissue homeostasis, leading to beneficial alterations in body weight, body composition, and adipocyte morphology, albeit exhibiting local insulin resistance.
Keywords: Mitochondria, Stress response, Obesity, Glucose homeostasis, Insulin resistance, Adipose tissue
Highlights
-
•
Mice with reduced HSP60 levels are protected from diet-induced obesity.
-
•
Hsp60+/− mice exhibit altered adipose tissue energy metabolism.
-
•
WAT of Hsp60+/− mice exhibit elevated insulin-independent glucose uptake.
-
•
Hsp60+/− mice show improved global, but impaired WAT insulin action.
1. Introduction
The current obesity pandemic is a consequence of an imbalance of nutrient intake and energy expenditure. The resulting positive energy balance leads to obesity and can cause insulin resistance, a feature of metabolic syndrome and type 2 diabetes (T2D). Mitochondria are essential for metabolism, as they convert nutrients and metabolites into energy in the form of ATP. Mitochondrial dysfunction and the concomitant release of reactive oxygen species (ROS) are linked to insulin resistance and obesity development [1]. To ensure proper mitochondrial function, mitochondria possess a protein quality control system consisting of mitochondrial chaperones and proteases. An integral part of this mitochondrial quality control system represents the mitochondrial chaperone heat shock protein 60 (HSP60). HSP60 interacts with its co-chaperone HSP10 and enables the proper folding of more than 300 mitochondrial matrix proteins and, thereby, mitochondrial function [2,3]. Important folding substrates of this complex include the antioxidative enzyme SOD2 or acyl-CoA dehydrogenases (e.g., medium-chain acyl-CoA dehydrogenase), suggesting that HSP60 influences obesity development by affecting oxidative stress and lipid utilization [4,5]. Indeed, reports show that patients deficient in HSP60 die early and exhibit marked alterations in fatty acid metabolism [6,7]. In rodents, HSP60 deficiency is embryonically lethal [8], and reduced expression of HSP60 is present in metabolic disorders ([9,10], Attie lab diabetes database http://diabetes.wisc.edu/).
HSP60 is also part of mitochondrial stress responses (MSR), such as the mitochondrial unfolded protein response (UPRmt), a mitochondria-to-nuclear signaling pathway to control mitochondrial proteostasis by regulating expression of mitochondrial proteins, chaperones, and proteases (e.g., SIRT3, HSP10, ClpP, or LONP1 [11]). Interestingly, insulin signaling regulates mitochondrial function via HSP60 in neurons [10], showing the close interaction between UPRmt and metabolic pathways.
Though a dysregulation of the mitochondrial quality control has been associated with metabolic diseases [12,13], the deletion of different UPRmt genes results in various, even opposing, metabolic outcomes. Thus, SIRT3 deficiency accelerates the development of the metabolic syndrome, whereas ClpP deletion protects against diet-induced obesity [[14], [15], [16]]. Controversial data about the influence of mitochondrial function on metabolism also exist, especially regarding adipose tissue biology. Here, both protection from diet-induced insulin resistance (TFAM deletion [17]) and induction of insulin resistance (PGC1-alpha deletion [18]) are described. Conversely, enhancing mitochondrial function by the overexpression of MitoNEET preserves insulin sensitivity under obese conditions [19].
These data reveal that increased mitochondrial activity in adipose tissue cannot be generally linked to improved metabolic outcomes. It even suggests that a mild decrease in adipose tissue mitochondrial function can be beneficial for metabolism. This finding is of interest, as proper mitochondrial function is a prerequisite of adipocyte differentiation, and impaired differentiation capacity impairs metabolism [20]. Accordingly, low steady-state oxidative stress, often a by-product of mitochondrial dysfunction, impairs adipocyte differentiation [21].
Mice heterozygous for the Hspd1 gene, encoding HSP60, (Hsp60+/− mice) can serve as a genetic model for diabetes-related mitochondrial dysfunction, as db/db mice exhibit an approximately 50% reduction in HSP60 ([9,22,23], Attie Lab diabetes database http://diabetes.wisc.edu/). This model allows for an answer to the question of whether a mild generalized impairment in mitochondrial function due to decreased chaperone activity, as seen in diabetes, affects insulin action and obesity development. Thus, we investigated the metabolism of Hsp60+/− mice in diet-induced obesity.
In the present study, we show that the reduction of HSP60 protects against diet-induced obesity in male mice, with a pronounced reduction in adipocyte size. The adipose tissue exhibits altered energy metabolism with decreased mitochondrial function, altered autophagy, and increased insulin-independent glucose uptake. In summary, the mild global impairment of mitochondrial function protects against diet-induced obesity by modulating adipose tissue homeostasis.
2. Results
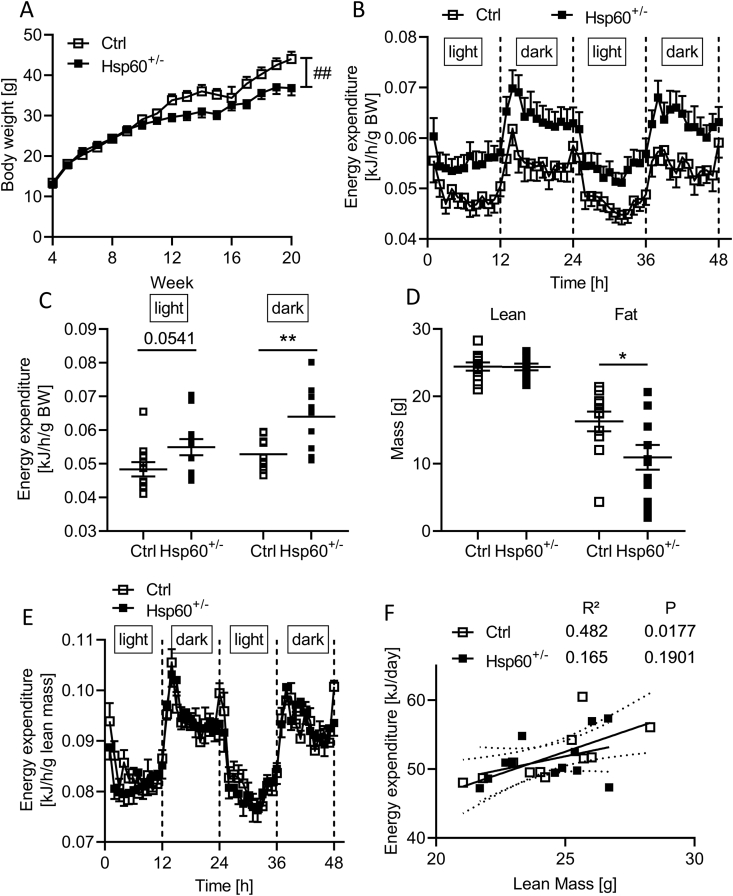
To understand the effect of global impaired mitochondrial protein homeostasis on obesity development, we fed control and Hsp60+/− mice on a C57BL/6N background (B6N), displaying an average 50% reduction in HSP60 expression across all tissues (Suppl. Fig. 1A, B), a normal chow diet (NCD, 10% kcal from fat) and a high-fat diet (HFD, 60% of kcal from fat). Feeding control and Hsp60+/− mice an NCD did not cause gross metabolic alterations. Here, heterozygous mice exhibited unaltered body weight, lean mass, fat mass, and unchanged insulin and glucose tolerance (Suppl. Fig. 1C-G). However, male Hsp60+/− mice exhibited increased energy expenditure, indicating mild effects on energy metabolism (EE, Suppl. Fig.1H). Interestingly, HFD-fed Hsp60+/− males exhibited reduced weight gain compared to the control, with a 16% reduction in body weight at 20 weeks of age (Figure 1A). The reduction in body weight was not due to altered food intake or fecal energy content as measurements of energy intake and clearance (Suppl. Fig. 2A, B) nor due to differences in body temperature between control and Hsp60+/− mice (Suppl. Fig. 2C).
Figure 1.
Male Hsp60+/−mice are protected from DIO and insulin resistance. A: Body weight development of male Ctrl and Hsp60+/− mice fed an HFD for 16 weeks (N = 12). B, C: Energy expenditure relative to body weight of male Ctrl and Hsp60+/− mice as a function of time (B) or divided into light and dark phase (C) after 14 weeks of HFD (N = 12). D: Body composition of male Ctrl and Hsp60+/− mice measured via NMR after 14 weeks of HFD (N = 12). E: Energy expenditure relative to lean mass of male Ctrl and Hsp60+/− mice as a function of time (N = 12) F: Correlation of energy expenditure and lean mass of male Ctrl and Hsp60+/− from (F). ∗P < 0.05, and ∗∗P < 0.01, after two-tailed Student's t-test. ##P < 0.01 after two-way ANOVA. All data are presented as mean ± SEM.
The observed decreased body weight may be a consequence of increased EE, as observed in Hsp60+/− males fed an NCD. Indeed, EE relative to body weight was elevated in Hsp60+/− mice, with a more pronounced effect during the dark phase (Figure 1B,C). Usually, skeletal muscle is the driving factor of EE. To this end, we determined body weight composition in control and Hsp60+/− mice and related EE to lean mass. This analysis revealed unaltered lean mass but decreased fat mass (by 33%) in heterozygous compared to control mice (Figure 1D). However, regression adjustment for either body weight or lean mass using ANCOVA [24] revealed no significant difference between the genotypes (Table 1, Table 2), thus questioning whether an elevated EE is indeed responsible for lower body weight in male Hsp60+/− mice. There was no difference between animal groups at light or dark phases when relating lean mass to EE (Figure 1E), confirming the previous finding. The positive correlation usually seen between lean mass and EE was absent in Hsp60+/− compared to control mice (Ctrl: R2 = 0.482, P = 0.0177, Hsp60+/−: R2 = 0.165, P = 0.1901; Figure 1F), hinting at potential physiological alterations of adipose tissue.
Table 1.
Energy expenditure from male mice fed HFD, adjusted for body weight using ANCOVA.
| Variables | Estimate | StdError | P value |
|---|---|---|---|
| Intercept | 0.2556 | 0.0847 | 0.0068 |
| Body weight | 0.0105 | 0.0035 | 0.0065 |
| Genotype | 0.0036 | 0.0127 | 0.7770 |
Table 2.
Energy expenditure from male mice fed HFD, adjusted for lean mass using ANCOVA.
| Variables | Estimate | StdError | P value |
|---|---|---|---|
| Intercept | 0.4810 | 0.0551 | 2.964E-08 |
| Lean | 0.0008 | 0.0015 | 0.5807 |
| Genotype | −0.0003 | 0.0173 | 0.9871 |
This metabolic phenotype was sex-specific, as female HFD-fed Hsp60+/− mice exhibited increased body weight compared to the control, with a significant increase in lean mass and unaltered fat mass assessed by NMR (Suppl. Fig. 3A, B). Heterozygous females displayed a specific increase in heart mass and an overall 10% increase in combined muscle mass (Suppl. Fig. 3C, D). Female Hsp60+/− mice showed a decreased EE relative to lean mass, but adjusting for either body weight or lean mass using an ANCOVA showed no significant genotype effect (Suppl. Fig. 3E, Supplementary Tables 1 and 2). However, the increase in body weight could be linked to decreased body temperature in female Hsp60+/− mice (Suppl. Fig. 3F).
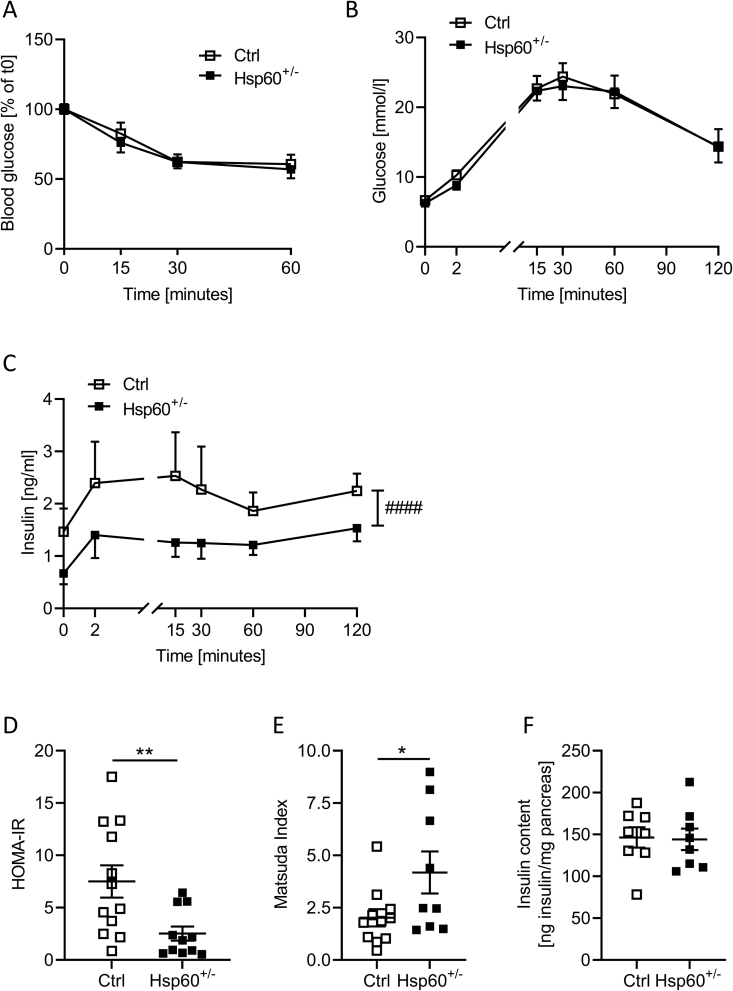
As determined by NMR in male Hsp60+/− mice, a decrease in fat mass is often linked to improved glucose tolerance and insulin action. Male Hsp60+/− mice showed unaltered insulin sensitivity and glucose tolerance compared to controls, assessed by insulin and glucose tolerance tests (Figure 2A,B). However, when we measured insulin levels during a glucose tolerance test, Hsp60+/− mice secreted (and thus needed) less insulin to maintain comparable blood glucose levels throughout the test (Figure 2C). Determining insulin resistance and sensitivity by calculating the HOMA-IR (Homeostatic Model of Insulin Resistance) and the Matsuda insulin sensitivity index revealed significantly lower insulin resistance (HOMA-IR, Ctrl = 7.51 vs. Hsp60+/− = 2.52, P = 0.0091) and higher insulin sensitivity scores (Matsuda Index, Ctrl = 2.02 vs. Hsp60+/− = 4.18, P = 0.0462) in heterozygous than control mice (Figure 2D,E). The decreased insulin release was not due to decreased insulin content in islets, as Hsp60+/− mice showed similar total pancreatic insulin content to control mice (Figure 2F). Again, this phenotype was sex-specific, as female Hsp60+/− mice showed unaltered insulin sensitivity compared to control mice (Suppl. Fig. 4A-E).
Figure 2.
Male Hsp60+/−mice show increased whole-body insulin sensitivity in DIO. A: Blood glucose values of male Ctrl and Hsp60+/− mice during an i.p.ITT after 10 weeks of HFD (N = 12). B: Blood glucose values of male Ctrl and Hsp60+/− mice during an oGTT after 11 weeks of HFD (N = 10). C: Plasma insulin values of male Ctrl and Hsp60+/− mice during the oGTT from (B) after 11 weeks of HFD (N = 10) D: HOMA-IR values calculated from (B) and (C) (N = 12): E: Matsuda insulin sensitivity index values calculated from (B) and (C) (N = 10). F: Pancreatic insulin content of male Ctrl and Hsp60+/− mice after 16 weeks of HFD (N = 8). ∗P < 0.05, and ∗∗P < 0.01, after two-tailed Student's t-test. All data are presented as mean ± SEM.
To assess organ-specific insulin sensitivity in male mice, we performed vena cava insulin injections in control and Hsp60+/− mice and evaluated insulin signaling via western blot analyses. The insulin-induced serine 473 phosphorylation of AKT in the liver, hypothalamus, and quadriceps was unaltered (Suppl. Fig. 5A-C). In line with this effect, metabolic biomarkers, such as adiponectin, glucagon, triglyceride, and non-esterified fatty acid levels in plasma, showed no differences between Hsp60+/− and control mice (Suppl. Fig. 5D-G).
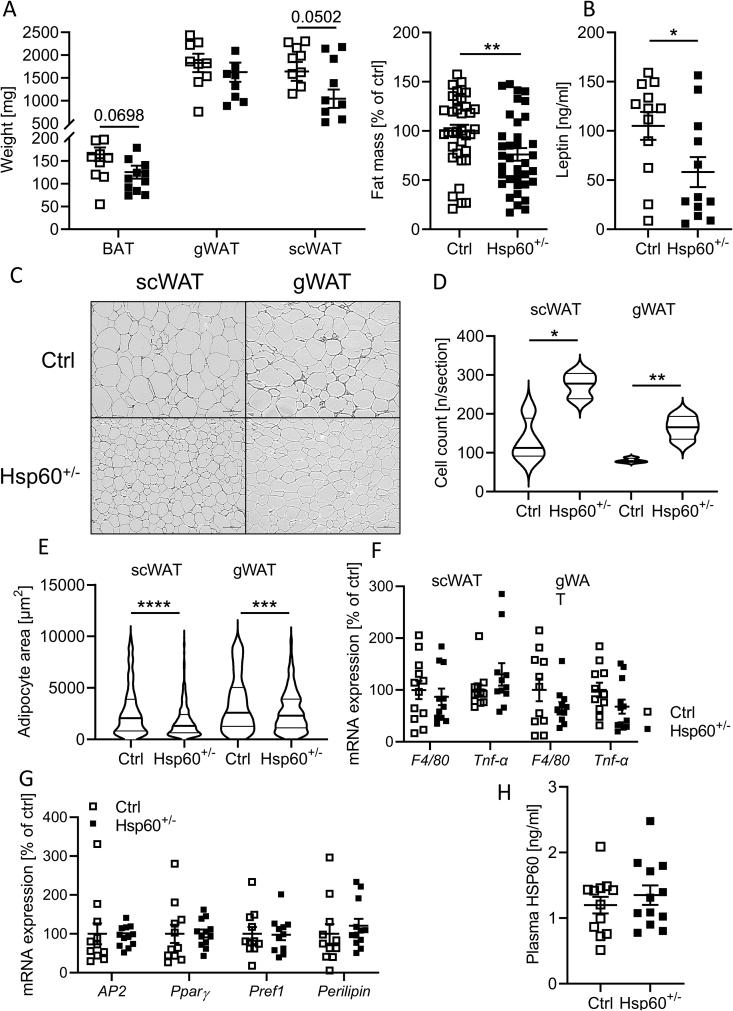
As total fat mass was decreased in male Hsp60+/− mice, we analyzed different adipose depots in more detail. Though subcutaneous and gonadal white adipose tissue (scWAT and gWAT) and brown adipose tissue (BAT) revealed only minor changes in each weight, the combination resulted in a significant, global 25% decrease in fat mass per mouse (Figure 3A), concurring with our NMR data (Figure 1D). Accordingly, leptin levels were decreased by 45 ± 20% (Figure 3B), indicating enhanced leptin sensitivity.
Figure 3.
White adipose tissue hyperplasia in male Hsp60+/−mice fed an HFD. A: Adipose tissue weights (N = 12) and combined adipose tissue weights of male Ctrl and Hsp60+/− mice after 16 weeks of HFD (N = 36, 12 animals and 3 depots). B: Plasma leptin content of male Ctrl and Hsp60+/− mice after 16 weeks of HFD (N = 12). C: Hematoxylin and eosin stain of histological sections of the scWAT and gWAT of male Ctrl and Hsp60+/− mice after 16 weeks of HFD. D, E: Adipocyte cell count (D) and average area (E) from histological sections of scWAT and gWAT of male Ctrl and Hsp60+/− mice after 16 weeks of HFD (N = 6). F: mRNA expression of inflammatory markers in scWAT and gWAT of male Ctrl and Hsp60+/− mice after 16 weeks of HFD (N = 12). G: mRNA expression of adipocyte markers in gWAT of male Ctrl and Hsp60+/− mice after 16 weeks of HFD (N = 12). H: Plasma HSP60 content of male Ctrl and Hsp60+/− mice after 16 weeks of HFD (N = 12). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001 after two-tailed Student's t-test. All data are presented as mean ± SEM.
To further delineate the effect of reduced HSP60 on adipose tissue development, we analyzed BAT, scWAT, and gWAT in detail. However, BAT did not show major differences in neither morphology, expression of brown adipocyte markers, except for a minor increase in P2RX5, nor in insulin sensitivity (Suppl. Fig. 6). Though not significantly different in weight, subcutaneous and gonadal adipocytes of Hsp60+/− mice were smaller and more numerous than control mice, with an increased frequency of small compared to large adipocytes, (Figure 3C–E), a phenotype preserved in the female sex (Suppl. Fig. 4F). Gene expression analysis of these depots showed an unchanged expression of proinflammatory markers Tnfα or F4/80 and adipocyte differentiation markers (Figure 3F,G). HSP60 can also be released from adipocytes and cause inflammation [25]. Unexpectedly, plasma HSP60 protein levels were not markedly different between groups either, despite the expected reduction in the tissue expression of 50% of Hsp60+/− mice (Figure 3H, Suppl. Fig. 1A, B).
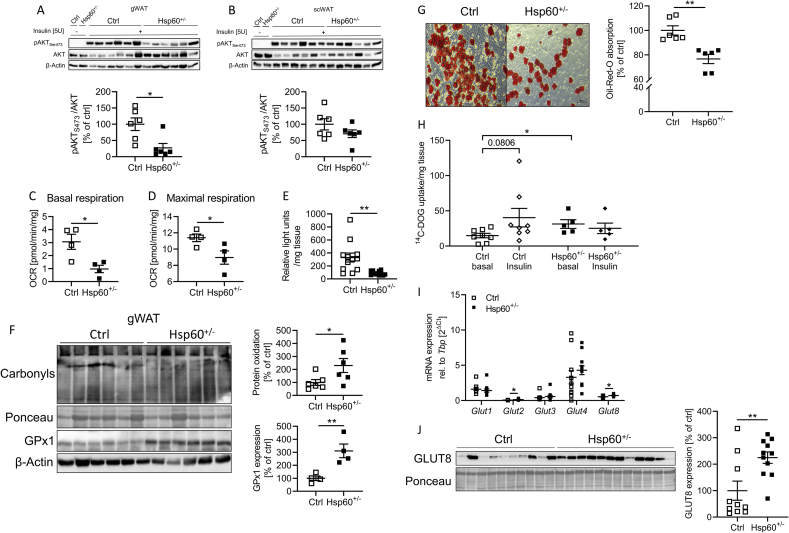
Next, we investigated adipose tissue insulin sensitivity after vena cava insulin injections in control and Hsp60+/− mice. In contrast to other tissues (Suppl. Fig. 5)A-C and 6D), the gWAT of Hsp60+/− mice exhibited insulin resistance compared to control mice, evidenced by a 73 ± 23% reduction in insulin-induced AKT S473 phosphorylation (Figure 4A), whereas insulin action in scWAT was indistinguishable between both groups (Figure 4B). Therefore, we focused on gWAT homeostasis. As HSP60 controls mitochondrial function and insulin resistance is linked to mitochondrial dysfunction, we next assessed mitochondrial respiration. Explants from the gWAT of male mice fed an HFD for 16 weeks were analyzed in the Seahorse Flux Analyzer, and clear signs of deteriorated mitochondrial function were revealed. Basal and maximal respiration were significantly reduced in heterozygous mice by 68 ± 2% and 21 ± 8%, respectively (Figure 4C,D). Following the decreased mitochondrial respiration in the gonadal adipose tissue from Hsp60+/− mice, the gWAT showed 73 ± 22% lower ATP content, measured by ATP turnover by the firefly luciferase in gWAT lysates (Figure 4E). Interestingly, the reduction of HSP60 did not lead to differences in mitochondrial DNA content, suggesting a specific defect in mitochondrial function rather than mitochondrial number (Suppl. Fig. 7A). However, the reduction of HSP60 did not induce the MSR in gWAT (Suppl. Fig. 7B).
Figure 4.
The gWAT of HFD-fed male Hsp60+/−mice is insulin resistant but shows increased basal glucose uptake. A: Western Blot and densitometric analysis of vena cava insulin-stimulated phosphorylation of AKT1S473 in the gWAT of male Ctrl and Hsp60+/− mice after 16 weeks of HFD. B: Western Blot and densitometric analysis of vena cava insulin-stimulated phosphorylation of AKT1S473 in the scWAT of male Ctrl and Hsp60+/− mice after 16 weeks of HFD. C, D: Basal (C) and maximal (D) respiration of gWAT explants of male Ctrl and Hsp60+/− mice after 16 weeks of HFD. E: Relative light unit detection after ATP-dependent Firefly luciferase assay of lysates from the gWAT of male Ctrl and Hsp60+/− mice after 16 weeks of HFD (N = 12). F: Western Blot and densitometric analysis of GPx1 expression and total protein carbonylation in the gWAT of male Ctrl and Hsp60+/− mice after 16 weeks of HFD. ∗P < 0.05, and ∗∗P < 0.01 after two-tailed Student's t-test. All data are presented as mean ± SEM. The gWAT of HFD-fed male Hsp60+/− mice is insulin resistant but shows increased basal glucose uptake. G: Representative Oil-Red-O staining of differentiated stromal vascular cells derived from the gWAT of male Ctrl and Hsp60+/− mice after 16 weeks of HFD (N = 6). H:14C-Deoxy-d-glucose uptake of gWAT explants from male Ctrl and Hsp60+/− mice after 16 weeks of HFD (N = 5–8). I: mRNA expression of different glucose transporters in the gWAT of male Ctrl and Hsp60+/− mice after 16 weeks of HFD (N = 12). J: Western Blot and densitometric analysis of GLUT8 expression in the gWAT of male Ctrl and Hsp60+/− mice after 16 weeks of HFD. ∗P < 0.05, and ∗∗P < 0.01 after two-tailed Student's t-test. All data are presented as mean ± SEM.
Another critical metabolism effector up-regulated during mitochondrial stress is fibroblast growth factor 21 (FGF-21) [26], primarily expressed in the liver and secreted into the circulation. However, neither liver or gWAT mRNA expression of FGF-21, its important co-receptor β-klotho, nor circulating FGF-21 levels were markedly different between groups (Suppl. Fig. 7C, D). Extended mitochondrial dysfunction with concomitant oxidative stress is linked to insulin resistance and can lead to irreversible protein modifications in the form of carbonylated proteins. Indeed, Hsp60+/− mice displayed a 130 ± 58% increase in cellular protein carbonylation in gWAT, whereas scWAT did not exhibit any alterations (Figure 4F, Suppl. Fig. 7E). Supporting this finding, Hsp60+/− mice exhibited a 93 ± 25% increase in 3-Nitrotyrosine level, another biomarker of oxidative stress, specifically in gWAT (Suppl. Fig. 7F). Furthermore, the antioxidative enzyme glutathione peroxidase 1 (GPx1) expression was significantly increased in the gWAT of Hsp60+/− mice compared to control mice, revealing a potential compensatory mechanism (Figure 4F). Insulin signaling and mitochondrial function are essential for proper adipocyte differentiation. Thus, we isolated the stromal vascular fraction (SVF) from the gWAT of male mice fed an HFD and performed an adipocyte differentiation protocol. After 10 days of differentiation, Hsp60+/− samples had a significant 23% reduction in Oil Red O absorption compared to the control group, indicating decreased adipocyte differentiation capacity (Figure 4G). This finding seemed to be contradictory to the unaltered expression of adipocyte markers (Figure 3G). However, it may point to a specific deficiency in differentiation capacity, resulting in a decreased number of adipocytes (Figure 3C).
We analyzed insulin-dependent glucose uptake to investigate the physiological consequences of decreased HSP60 expression on carbohydrate metabolism further. Here, gWAT explants from HFD-fed and insulin-resistant control mice showed a trend toward increased glucose uptake after insulin stimulation (P = 0.08). In contrast, explants from Hsp60+/− mice exhibiting enhanced global insulin sensitivity but impaired insulin response in gWAT (Figure 4A) were unresponsive to insulin-induced glucose uptake (Figure 4H). However, explants from Hsp60+/− gWAT showed a significant 111 ± 42% higher basal glucose uptake (Figure 4H). Analysis of the gene expression patterns of glucose transporters revealed an increased mRNA expression of insulin-independent GLUT2 and GLUT8 (Figure 4I), though GLUT2 values were close to the detection limit and probably due to the expression in endothelial (rather than adipose) cells in gWAT. Furthermore, an increased GLUT8 expression was confirmed on the protein level (Figure 4J). This effect was sex-specific, as female Hsp60+/− mice did not show an increase in the GLUT8 expression in the gWAT (Suppl. Fig. 4G). Intriguingly, a siRNA-mediated knockdown of GLUT8 in hepatocytes was recently shown to decrease basal glucose uptake [27].
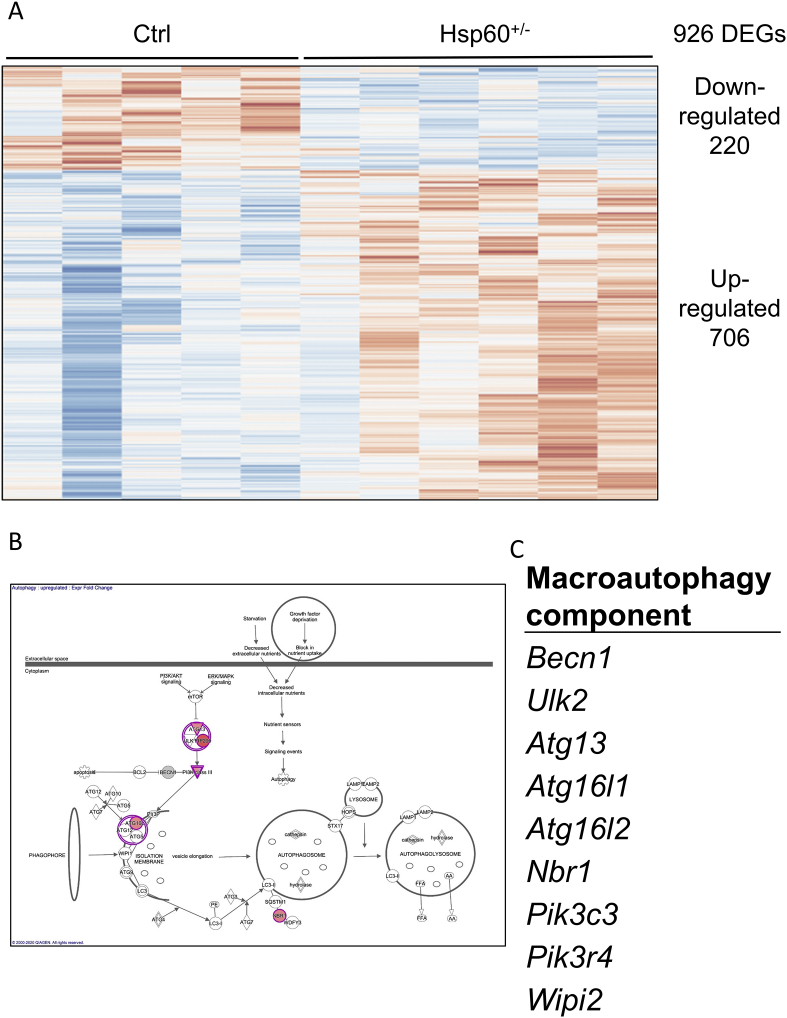
The heterozygosity for HSP60 had a considerable effect on gWAT morphology and functionality, with an apparent dissociation of adipocyte hyperplasia and cellular metabolic health. To gain additional insights into the underlying mechanisms of this phenotype, RNA samples from the gWAT of control and Hsp60+/− mice fed an HFD for 16 weeks were submitted to microarray transcriptomic analysis. In total, 926 differentially expressed genes were found. Of these, 220 were down-, and 706 were up-regulated in Hsp60+/− mice (Figure 5A). The down-regulated genes showed no enrichment in specific pathways or gene ontology terms (data not shown). However, in the subset of up-regulated genes, the Ingenuity Pathway Analysis and the gene ontology analysis showed an enrichment of genes involved in the cellular process of autophagy (Figure 5B,C, Supplementary Table 3). Autophagy represents a process by which damaged organelles can be degraded and their components recycled. In addition, it is a vital process in nutrient-depleted conditions or decreased energy production due to organelle dysfunction, allowing for survival by recycling intracellular components [28].
Figure 5.
Transcriptomic analysis of the gWAT from HFD-fed male mice indicates increased autophagy. A: Expression pattern of 926 differentially expressed genes (DEGs) after analysis of the microarray transcriptomic data. Figure was generated with the online tool ClustVis (https://biit.cs.ut.ee/clustvis/). B: Result of the ingenuity pathway analysis of up-regulated genes generated with Ingenuity Pathway Analysis (IPA) from QIAGEN. C: Genes belonging to the macroautophagy process, enriched in the subset of up-regulated genes, generated with the Ontology enRIchment anaLysis and visuaLizAtion tool Gorilla (http://cbl-gorilla.cs.technion.ac.il/).
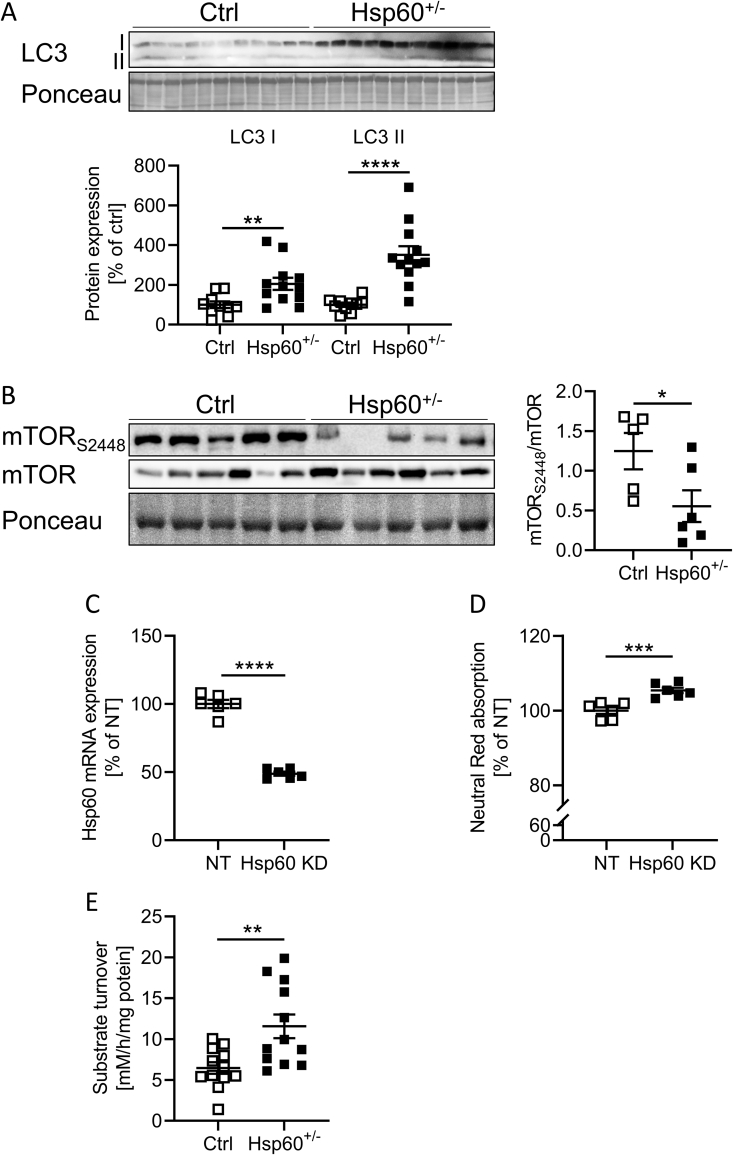
Cell components targeted for autophagy are sequestered in a membrane vesicle, the autophagosome, and directed to the lysosomes to form autolysosomes. The induction of autophagy involves the recruitment of microtubule-associated protein 1A/1B light chain 3B (LC3) and its subsequent conversion from the cytosolic form LC3-I to lipidated LC3-II at the autophagosomal membranes [29]. gWAT samples of Hsp60+/− mice showed a significant increase in LC3-I and LC3-II levels by 105 ± 37% and 251 ± 49%, respectively, in the gWAT (Figure 6A), indicating overall enhanced autophagosome presence and activity, in line with our microarray results. Activation of mammalian target of rapamycin (mTOR), an important regulator of cellular growth, is regulated by nutrient availability and inhibits autophagy. Insulin phosphorylates mTOR at S2448, leading to its activation [30]. Interestingly, S2448 phosphorylated mTOR was reduced by 55% in gWAT samples of Hsp60+/− mice, indicating decreased energy availability and an altered regulation of autophagy and insulin resistance (Figure 6B).
Figure 6.
The gWAT of HFD-fed male Hsp60+/−mice shows signs of increased autophagy and lysosomal activity. A: Western Blot and densitometric analysis of LC3 I/II in the gWAT of male Ctrl and Hsp60+/− mice after 16 weeks of HFD. B: Western Blot and densitometric analysis of phosphorylated and total mTOR protein levels in the gWAT of male Ctrl and Hsp60+/− mice after 16 weeks of HFD. C: mRNA expression of HSP60 in 3T3-L1 fibroblasts infected with siRNA targeting either non-target (NT) or Hsp60 (Hsp60 KD). D: Neutral Red absorption in NT or Hsp60 KD 3T3-L1 cells incubated with neutral red for 2 h. E: Lysosomal activity measured via fluorogenic substrate turnover in lysates of gWAT from male Ctrl and Hsp60+/− mice after 16 weeks of HFD (N = 12). ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001 after two-tailed Student's t-test. All data are presented as mean ± SEM.
However, the sole presence of the transcripts or proteins involved in autophagy is insufficient to draw conclusions about its activation. To investigate this aspect, 3T3-L1 fibroblasts were transfected with either a non-target (NT) siRNA or an siRNA directed against Hsp60 mRNA (Hsp60 KD). This approach resulted in a 51 ± 3% decrease of Hsp60 mRNA expression of Hsp60 KD compared to control cells (Figure 6C). Hsp60 KD cells displayed no differences in cell viability, as determined by an MTT assay, but reduced uptake of glucose from the insulin/IGF-containing medium, confirming our hypothesis of a different mechanism of energy supply (Suppl. Fig. 8A, B). Thus, we assessed lysosomal activity using a Neutral Red assay. This assay revealed slightly increased lysosomal activity in Hsp60 KD cells than control cells, as evidenced by enhanced Neutral Red incorporation into active lysosomes (Figure 6D). Finally, to gain further insight into the autophagic processes in vivo, we investigated this pathway in gWAT samples of Ctrl and Hsp60+/− HFD-fed mice. Protein lysates were prepared under non-denaturing conditions, and the lysosomal and proteasomal activities were measured by substrate turnover. Here, the gWAT of Hsp60+/− mice showed unaltered proteasomal activity (Suppl. Fig. 8C, D), but exhibited an almost two-fold increase in lysosomal activity, an indication of increased lysosomal/autophagic activity and a potentially altered energy supply (Figure 6E). The combined evidence from our in vitro and ex vivo analysis indicate that Hsp60+/− mice are protected against diet-induced obesity by an unexpectedly increased basal glucose uptake in adipose tissue and enhanced energy turnover due to insufficient mitochondrial energy production in the presence of local gWAT insulin resistance.
3. Discussion
Mitochondrial function and insulin sensitivity are crucial signaling nodes to engage a healthy metabolism. In general, the healthy expansion of adipocytes is characterized by elevated proliferation, known as adipocyte hyperplasia. The unhealthy expansion of adipose tissue is characterized by adipocyte hypertrophy, the enlargement of adipocytes. Hypertrophic adipocytes often exhibit mitochondrial dysfunction and secrete TNFα, causing insulin resistance [31,32]. The healthy expansion of adipose tissue depends on proper insulin action and mitochondria. Therefore, it was unexpected that a reduction of HSP60 in our study led to decreased fat mass and adipocyte hyperplasia in adipose tissue, albeit with the presence of mitochondrial dysfunction and insulin resistance in gWAT, though male Hsp60+/− mice were protected against diet-induced obesity. The question of why insulin resistance was only present in adipose tissue remains to be answered. Gonadal adipose tissue is insulin resistant, yet Hsp60+/− mice exhibit unaltered glucose tolerance, showing that local insulin resistance does not need to impact overall metabolic health. One explanation for this unexpected phenotype includes the elevated insulin-independent glucose uptake under basal conditions, which is comparable to insulin-induced glucose uptake rates and may allow normoglycemia (Figure 2, Figure 4H).
An important consequence of adipose tissue insulin action is the inhibition of lipolysis and induction of lipogenesis. Here, our transcriptomic data shows that genes for beta oxidation were significantly increased or showed a strong tendency, whereas the expression of the lipogenic transcription factor Srebf2 was decreased (Acads P = 0.07, Acadm P = 0.1, Acadl P = 0.013, Acadvl P = 0.054, Acad8 P = 0.07, Acad11 P = 0.054, Srebf2 P = 0.03, Supplementary Table 4), also suggesting an impairment of lipogenesis. Accordingly, lipid accumulation into adipocytes was decreased (Figure 4G), further confirming that the reduction of HSP60 causes gonadal adipose tissue insulin resistance. Why this insulin resistant phenotype is specific for gonadal but not subcutaneous adipose tissue is still unclear but might relate to the differences in the occurrence of oxidative stress, which was absent in the scWAT of Hsp60+/− mice (Figure 4F, Suppl. Fig. 7E and F) [33,34]. Thus, oxidative stress is a prerequisite for insulin resistance induced by decreased HSP60 levels, explaining the unaltered insulin sensitivity phenotype in scWAT [9]. This observation might also clarify the difference between the insulin-sensitive phenotype in Hsp60+/−mice fed an NCD on C57BL/6N (Suppl. Fig. 1F, G) compared to C57BL/6J background [9], as only C57BL/6J mice exhibited increased oxidative stress [35]. In addition, C57BL/6J displayed insulin resistance and alterations in their metabolic response to diet-induced obesity compared to C57BL/6N mice, further highlighting the importance of the genetic background when assessing metabolism [[36], [37], [38]].
The decreased mitochondrial function and reduced ATP production contradict our observation of potentially altered energy expenditure driven by adipose tissue. A compensation for decreased mitochondrial function can be facilitated by elevated autophagy [39], which is not as energy efficient as mitochondrial energy production [28] but might partially account for alterations in body weight development. Accordingly, gWAT from Hsp60+/− mice exhibited decreased ATP levels, suggesting partial compensation through autophagy (Figure 4E). It remains unclear why Hsp60+/− mice exhibited decreased body weight (Figure 1A and Table 1, Table 2). We have examined other parameters of energy balance but did not find any difference in food intake, fecal energy content, or body temperature. As such, it seems likely that the difference in final body weight stems from accumulating minor differences of the parameters mentioned above, which cannot be captured by our methodology. Another possibility includes alterations in housing during the measurement of energy expenditure. Mice were single-housed for the measurement of energy expenditure, which alters social interaction. It has been shown that alterations in housing can influence metabolism [24,40]. Of note, ClpP−/− mice showed a similar decreased body weight and energy expenditure but no difference in EE after ANCOVA adjustment [41]. It remains unclear why an alteration of the mitochondrial unfolded protein response (ClpP and HSP60 are members of this response) leads to reduced body weight. Further research is required to address this vital aspect.
In obese human conditions, increased autophagy in adipose tissue correlates with the degree of obesity and adipocyte hypertrophy [42]. Therefore, increased autophagy due to mitochondrial dysfunction during obesity development could represent a protective mechanism to reduce the metabolic burden. This mechanism is specific to mitochondrial dysfunction-induced autophagy, as HFD-fed mice with hyperactive autophagy due to a Beclin-1 mutation had normal adiposity and even exhibited enhanced insulin sensitivity in adipose tissue [43]. As mentioned earlier, a siRNA-mediated knockdown of GLUT8 in hepatocytes was recently shown to reduce basal glucose uptake and impair sucralose-induced autophagy [27]. This observation could help explain the discrepancy between local adipose tissue insulin resistance and unaltered blood glucose levels, which is compensated by basal, insulin-independent glucose uptake.
The decrease in HSP60 levels did not induce MSR activation (Suppl. Fig. 7B), indicating that the inability to cleave unfolded or misfolded proteins primarily causes induction of MSR in adipose tissue as evidenced by ClpP deficiency [15]. Indeed, in C. elegans, it has been shown that the accumulation of mitochondrial peptide fragments is a major activator of this response [44,45].
It is important to note that a reduction of HSP60 sex-specifically affects diet-induced obesity. Overall, both animals showed an improved metabolism with smaller adipocytes and increased insulin sensitivity. However, female Hsp60+/− mice exhibited increased body weight compared to control, with a significant increase in lean mass and unaltered fat mass. The increase in body weight was independent of food intake but was a result of decreased body temperature. It remains unknown why these discrepancies are present but imply an interaction of estrogens with mitochondrial function. It has been shown that the estrogen receptor α mediated a distinct mitochondrial unfolded protein response (UPR), an inter-membrane space UPR, which might interact with the classical MSR when HSP60 is involved. Furthermore, obese female mice showed significantly lower levels of inflammation and oxidative stress in the WAT than weight-matched males, which is a prerequisite for the induction of insulin resistance through HSP60 reduction [46]. Further studies are required to decipher the sex-dependent interaction between reduced mitochondrial function and insulin action.
In summary, our data reveal that local insulin resistance due to a reduction of HSP60 does not necessarily result in overt glucose intolerance with deteriorated metabolism. We discovered a novel gene–nutrient interaction, where the reduction of HSP60 in males fed an HFD protected against diet-induced obesity, whereas the gonadal adipose tissue revealed a region-specific insulin resistance with mitochondrial dysfunction and potential compensatory autophagy (Graphical abstract).
4. Methods
4.1. Animal studies
C57BL/6N wild-type mice (used as the control group, designated “Ctrl”) and Hsp60+/− mice were bred in-house, and littermates were group-housed in a temperature-controlled room (22 +/− 1 °C) on a 12 h light/dark cycle with free access to food and water. Animals were kept on a normal chow diet (NCD, ssniff Spezialdiäten GmbH) or a 60% high-fat diet (HFD ssniff Spezialdiäten GmbH) for 16 weeks, starting at 4 weeks of age, and the proportion of HFD was only slowly raised over a 2-week acclimatization period. An insulin tolerance test was performed with an intraperitoneal injection of insulin (0.75 U/kg body weight). Animals were starved for 16 h, and 2 g glucose/kg body weight was applied orally for the glucose tolerance test. Energy expenditure and the respiratory quotient were measured via indirect calorimetry using the PhenoMaster™ (TSE Systems GmbH). Body composition was determined via nuclear magnetic resonance spectroscopy using the EchoMRI™ (EchoMRI LLC). To assess local insulin sensitivity, mice were kept on HFD for 16 weeks and then anaesthetized. Then, 5 U of insulin was injected directly into the vena cava. Organs were harvested after the time points quoted in parentheses: Liver (2 min), quadriceps (5 min), gonadal and subcutaneous white adipose tissue (5 min), and brain and brown adipose tissue (10 min). Mice were individually caged for 48 h with free access to food and water for fecal energy content and body temperature measurements. Body temperature was measured, and feces were collected twice daily. Fecal energy content was measured in a caloric bomb.
4.2. Genotyping
PCR Typing for heterozygosity was performed using specific primers for the deleted part of the Hsp60 gene: Hsp60 P1 (5′-TAAGACAGCATTTCTCCGGTAG-3′), Hsp60 P2 (5′- CTGAGTGTTGGGATTATGCAG-3′), Hsp60 P3 (5′- GCCAGTCCTCCGATTGAC-3′).
4.3. Isolation of stroma vascular fraction
The gonadal adipose tissue of C57BL/6N Ctrl and Hsp60+/− mice fed an HFD for 16 weeks was collected, digested with Collagenase II, filtered through a 50 μm mesh, and the resulting SVF cells plated at a density of 2,5 ∗10ˆ4 cells/cm2. Differentiation was induced as described below.
4.4. Cell culture
White 3T3-L1 preadipocyte cells were cultured Gibco DMEM GlutaMAX™ (Thermo Fisher Scientific Inc., USA) culture medium supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin (Thermo Fisher Scientific Inc.), and 1% pyruvate (Thermo Fisher Scientific Inc.) at 37 °C and 5% CO2. 3T3-L1 differentiation and Oil-Red-O staining were performed as previously described [47].
4.5. Ex vivo 14C-deoxy-D-glucose uptake
Glucose uptake was measured using 14C-labeled deoxy-D-glucose (14C-DOG). Tissue explants from mice fed an HFD for 16 weeks were taken and weighed and incubated in Krebs-Ringer-Hepes buffer (KRH) with 1% BSA for 45 min with either 250 nM insulin or 10 μM Cytochalasin B before the addition of 2 mM 14C-DOG for 10 min. The tissue was then lysed and incubated with Ultima GoldTM scintillation liquid (Perkin Elmer Inc.). Radioactivity in samples was measured with the Beckman Coulter LS6500 Liquid Scintillation Counter, and the resulting values were related to explant weight.
4.6. Ex vivo seahorse assay
Tissue explants from mice fed an HFD for 16 weeks were taken, weighed, and incubated in Agilent Seahorse media. Mitochondrial respiration was monitored using the Seahorse XF Mito Stress Test Kit and the Seahorse XF24 Extracellular Flux Analyzer, measuring the oxygen consumption rate (OCR) of tissue explants. Final compound concentrations were 2 μM oligomycin, 1 μM FCCP, and 1 μM/2 μM rotenone/antimycin A.
4.7. Analytical procedures
Blood glucose was measured with a Glucometer – Contour XT (Bayer). Plasma Insulin and Adiponectin were measured with ELISAs from Alpco (Alpco Salem, BioCatGmbH). Plasma HSP60 was measured with an ELISA from Elabscience (Elabscience Inc.). Leptin was measured using an ELISA from R & D Systems (R & D Systems/Bio-Techne GmbH) and Plasma Glucagon levels with an ELISA from Yanaihara (Yanaihara Institute Inc.). FGF-21 was measured using an ELISA from Biovendor (BioVendor R&D). All ELISAs were performed according to the manufacturer's guidelines. Triacylglycerols were measured with Triglyceride Reagent ABX, and NEFAs were measured with the NEFA-HR assay according to manufacturer's guidelines (FUJIFILM Wako Chemicals Europe GmbH). The glucose in the supernatants of cultured 3T3-L1 cells was determined by the hexokinase method (Roche Diagnostics GmbH).
4.8. Immunohistochemistry
Fresh tissue samples were submerged in 4% paraformaldehyde for 24 h at 4 °C, washed in PBS, and embedded in paraffin. Slices were fixed on glass slides and stained with hematoxylin and eosin. Calculations of adipocyte size were performed with ImageJ Fiji version 1.53c.
4.9. RNA isolation from tissue
Total RNA was extracted from 5 to 20 mg tissue using the RNeasy Mini Kit (Qiagen) following the manufacturer's manual.
4.10. Analysis of gene expression by quantitative real-time PCR
RNA was reverse transcribed using oligo(dT)15 primers, random primers, Thermo Scientific dNTP-Set, and M-MLV reverse transcriptase (Promega Corporation). Real-time quantitative PCR was performed with 10 ng cDNA using GoTaq qPCR master mix (Promega Corporation) and gene-specific primers (Supplementary Table 5). Fluorescence was analyzed in the ViiA-7-Real-Time-PCR System (Applied Biosystems, Thermo Fisher Scientific Inc.). Gene expression was calculated applying the ΔΔCT method using Tbp (TATA binding protein) as a reference gene for mRNA analysis or Chdh1 for mitochondrial DNA content analysis.
4.11. Western blot analysis
Western Blots were performed on 10–15 μg total protein lysates loaded on 10% SDS-PAGE gels and transferred to PVDF membranes (GE HealthCare Life Science) for 3 h at 90 V. After the transfer, membranes were blocked in Starting Block (StartingBlockTM T20 (TBS) Blocking Buffer Thermo Scientific #37543) for 1 h. Following blocking, membranes were probed with the primary antibody (Supplementary Table 6) overnight at 4 °C. Membranes were then incubated with peroxidase-conjugated secondary antibodies (anti-rabbit Dianova #711-065-152, 1:10000, anti-mouse Dianova #715-065-150, 1:10000) at room temperature for 1h. Specific bands were detected with chemiluminescence assay (WesternBright ECL Biozym 541005X) using the ChemiDoc Touch Imaging System (BioRad). Ponceau staining or β-Actin were used as loading controls. To remove the phospho-epitope of a protein and run the total antibody, membranes were incubated in stripping buffer (Restore™ PLUS Western Blot Stripping Buffer Thermo Scientific #46430) for 20 min at room temperature, re-blocked for 30 min and re-probed. Band intensities were quantified via densitometric analysis using Image Lab 5.2.1 software and ImageJ Fiji version 1.53c.
4.12. Protein carbonylation
Carbonylated proteins were detected as previously described [48]. Briefly, after western blot transfer membranes were incubated for derivatization with 2 mM 2,4-dinitrophenylhydrazine (DNPH) in 2 M HCl and probed afterward with an anti-DNP primary antibody (Merck KGaA).
4.13. Infection with lentiviral particles
3T3-L1 cells were infected using lentiviral transduction particles with pLKO.1 plasmid containing shRNA, targeting either the Hsp60 gene (HSPD1 MISSION shRNA Lentiviral Transduction Particles, SHCLNV-NM_010477; Merck KG) or control transduction particles targeting no known mammalian genes (MISSION pLKO.1-puro Non-Mammalian shRNA Control Transduction Particles, SHC902V; Merck KG) according to manufacturer's instructions. 12 μg/ml hexadimethrine bromide (polybrene) was added for incubation overnight. For the antibiotic selection of infected cells, 5 μg/ml of puromycin was added to the media. Selection continued for at least one week.
4.14. Neutral red assay
The compound Neutral Red (3-Amino-7-dimethylamino-2-methylphenazine hydrochloride) incorporates into active lysosomes. Cells were incubated with 40 μg/ml Neutral Red for 2 h at 37 °C. After cell lysis with 1% acetic acid in 50% ethanol for 5 min, absorption was measured at 540 nm in a microplate reader.
4.15. MTT assay
The tetrazolium dye MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) was reduced by NAD(P)H dependent cellular oxidoreductases to Formazan. In a 96-well plate, 20 μl of a 5 mg/ml MTT solution was applied and incubated for 3 h at 37 °C, then washed with PBS. Cells were lysed with DMSO, and Formazan absorption was measured at 560 nm.
4.16. Luciferase assay
Homogenized tissue was lysed in a lysis buffer (Promega Dual Luciferase Assay Kit) and briefly centrifuged to assess ATP content in target tissues. The supernatant was mixed with a reaction mix containing Firefly Luciferase and Luciferin (Thermo Scientific). The measured luminescence is a direct result of the presence of free ATP, which fuels the conversion of Luciferin to Oxyluciferin by the Luciferase. Luminescence was determined in the Promega GlowMAX.
4.17. Transcriptome analysis of gWAT
To determine changes in the gene expression patterns in the gWAT between Ctrl and Hsp60+/− mice fed an HFD for 16 weeks, a transcriptomic analysis using a microarray approach was employed. RNA was isolated as described above, and RNA quality control was performed with the Agilent 2100 Bioanalyzer. The microarray was performed by OakLabs GmbH on an Agilent 8 × 60K chip. Visualization was performed with Pathway Analysis (IPA) software from Qiagen, the online tools ClustVis, and the Gene Ontology enRIchment anaLysis and visuaLizAtion tool GOrilla.
4.18. Lysosome activity (cysteine cathepsins activity)
Adipose tissue samples were homogenized with a ball mill in 500 μl of 1 mM DTT/PBS and shaken in a thermomixer for 1 h at 4 °C. Afterward, samples were sonicated on ice for 2 min at 50% amplitude and centrifuged at 14,000 rpm for 20 min at room temperature. Fat tissue lysates (0.25 μg/well) were incubated with a lysosome activity incubation buffer (containing 24 mM Cystein∗HCL, 150 mM Na-Acetate, 3 mM EDTA Dihydrate, pH 4.0) for 10 min at room temperature. To measure cysteine cathepsins, OmniCathepsin fluorogenic substrate Z-FR-AMC, Z-Phe-Arg-AMC (Enzo #BML-p-139) was used, and the final substrate concentration was 166 μM/well. AMC liberation was measured in a black 96-well plate and monitored every 180 s for 90 min at 37 °C using a fluorescence microplate plate reader (excitation: 360 nm, emission: 460 nm). Proteolytic cathepsin activity was calculated using free 7-amino-4-methylcoumarin (AMC) as fluorogenic calibration standard. The protein concentration of fat lysates was determined using a Bradford protein assay, and proteolytic activity was verified using a protease inhibitor cocktail (Sigma–Aldrich, P8340, diluted according to manufacturer's instructions).
4.19. Proteasome activity (chymotrypsin-like activity)
Fat tissue samples were homogenized with a ball mill in 150 μl lysis buffer (containing 250 mM sucrose, 25 mM HEPES, 1 mM EDTA, 10 mM magnesium chloride, and freshly added 1.7 mM DTT, pH 7.8), followed by passing lysates through a 27-gauge needle attached to a 1 ml syringe 20 times, freeze–thaw cycles 3 times, and centrifugation at 13,400 rpm, for 10 min at 4 °C. Supernatants were used for the determination of protein, using a Bradford assay, and proteasome activity. For the activity assay, samples were adjusted to 1 mg/ml protein (final protein concentration was 10 μg/well) and incubated with a proteasome activity incubation buffer (containing 225 mM Tris buffer (pH 7.8), 7.5 mM magnesium acetate, 45 mM potassium chloride, 7.5 mM magnesium chloride, and freshly added 1 mM DTT). ATP was depleted, and 15 mM 2-deoxyglucose and 0.1 mg/ml hexokinase were added to the incubation buffer to measure 20S proteasome activity. Additional ATP was added to the incubation buffer to determine 26S proteasome activity, with a final ATP concentration of 2 mM. Samples were shaken in the respective incubation buffers for 2 min at 100 rpm, followed by a 10 min incubation at room temperature. Chymotrypsin-like proteasome activity was measured using fluorogenic peptide suc-Leu-Leu-Val-Tyr-7-AMC (Enzo, #BML-P802-0005, final concentration 166 μM/well). 7-amino-4-methylcoumarin (AMC) liberation was determined in a black 96-well plate and monitored every 180 s for 90 min at 37 °C using a fluorescence microplate plate reader (excitation: 360 nm, emission: 460 nm). Proteolytic activity was calculated using free AMC as fluorogenic calibration standard and verified using proteasome inhibitor lactacystin (Enzo, BML-PI104-1000).
4.20. Statistical analysis
All data are presented as mean ± SEM. Groups were compared using an unpaired two-tailed Student's t-test. A two-way ANOVA was performed to detect interactions (e.g., between sex and treatment [diet]), and Sidak's post hoc analysis was performed when appropriate. Correlations in mice were conducted using a linear regression model. The EE ANCOVA analysis performed for this work was provided by the NIDDK Mouse Metabolic Phenotyping Centers (MMPC, www.mmpc.org) using their Energy Expenditure Analysis page (http://www.mmpc.org/shared/regression.aspx) and supported by grants DK076169 and DK115255. All in vitro experiments were performed in independent triplicates. The statistical analyses and generation of graphs were performed using GraphPad Prism 7 Software (GraphPad Software Inc.). The graphical Abstract was generated in Inkscape version 1.01.
Declaration of approval
All animal procedures were conducted according to German laws for the protection of animals and in compliance with protocols approved by local government authorities (State Agency of Environment, Health and Consumer Protection, State of Brandenburg, Germany).
Data and resource availability
All data generated or analyzed during this study are included in the published article (and its online supplementary files). No applicable resources were generated during the current study.
Author contributions
R.H. designed the study, researched data, and wrote the manuscript. M.R., M.S., K.R., K.K., S.D., C.O., M.J., W.J., and A.S. researched data and helped design experiments. A.K. designed the study, supervised all work, and wrote the manuscript. A.K. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Material and correspondence
Material request and correspondence should be addressed to Dr. André Kleinridders.
Acknowledgments
We thank Christine Gums and Annett Holms for technical assistance. We thank Peter Bross for generating and gernerously providing the Hsp60+/− mouse model. We acknowledge the support of the Deutsche Forschungsgemeinschaft and Open Access Publishing Fund of the University of Potsdam. This work was supported by the Deutsche Forschungsgemeinschaft (DFG) grant project KL 2399/4-1 (to A.K.) and by a grant from the German Ministry of Education and Research (BMBF) and the State of Brandenburg (DZD grant 82DZD00302).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101276.
Conflict of interest
The authors declare no conflict of interest.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Montgomery M.K., Turner N. Mitochondrial dysfunction and insulin resistance: an update. Endocrine Connections. 2015;4(1):R1–R15. doi: 10.1530/EC-14-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kerner M.J., Naylor D.J., Ishihama Y., Maier T., Chang H.C., Stines A.P. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell. 2005;122(2):209–220. doi: 10.1016/j.cell.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 3.Bie A.S., Comert C., Korner R., Corydon T.J., Palmfeldt J., Hipp M.S. An inventory of interactors of the human HSP60/HSP10 chaperonin in the mitochondrial matrix space. Cell Stress & Chaperones. 2020;25(3):407–416. doi: 10.1007/s12192-020-01080-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corydon T.J., Hansen J., Bross P., Jensen T.G. Down-regulation of Hsp60 expression by RNAi impairs folding of medium-chain acyl-CoA dehydrogenase wild-type and disease-associated proteins. Molecular Genetics and Metabolism. 2005;85(4):260–270. doi: 10.1016/j.ymgme.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Magnoni R., Palmfeldt J., Hansen J., Christensen J.H., Corydon T.J., Bross P. The Hsp60 folding machinery is crucial for manganese superoxide dismutase folding and function. Free Radical Research. 2014;48(2):168–179. doi: 10.3109/10715762.2013.858147. [DOI] [PubMed] [Google Scholar]
- 6.Agsteribbe E., Huckriede A., Veenhuis M., Ruiters M.H., Niezen-Koning K.E., Skjeldal O.H. A fatal, systemic mitochondrial disease with decreased mitochondrial enzyme activities, abnormal ultrastructure of the mitochondria and deficiency of heat shock protein 60. Biochemical and Biophysical Research Communications. 1993;193(1):146–154. doi: 10.1006/bbrc.1993.1602. [DOI] [PubMed] [Google Scholar]
- 7.Briones P., Vilaseca M.A., Ribes A., Vernet A., Lluch M., Cusi V. A new case of multiple mitochondrial enzyme deficiencies with decreased amount of heat shock protein 60. Journal of Inherited Metabolic Disease. 1997;20(4):569–577. doi: 10.1023/a:1005303008439. [DOI] [PubMed] [Google Scholar]
- 8.Christensen J.H., Nielsen M.N., Hansen J., Fuchtbauer A., Fuchtbauer E.M., West M. Inactivation of the hereditary spastic paraplegia-associated Hspd1 gene encoding the Hsp60 chaperone results in early embryonic lethality in mice. Cell Stress & Chaperones. 2010;15(6):851–863. doi: 10.1007/s12192-010-0194-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleinridders A., Lauritzen H.P., Ussar S., Christensen J.H., Mori M.A., Bross P. Leptin regulation of Hsp60 impacts hypothalamic insulin signaling. Journal of Clinical Investigation. 2013;123(11):4667–4680. doi: 10.1172/JCI67615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wardelmann K., Blumel S., Rath M., Alfine E., Chudoba C., Schell M. Insulin action in the brain regulates mitochondrial stress responses and reduces diet-induced weight gain. Molecular Metabolism. 2019 doi: 10.1016/j.molmet.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munch C. The different axes of the mammalian mitochondrial unfolded protein response. BMC Biology. 2018;16(1):81. doi: 10.1186/s12915-018-0548-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castro J.P., Wardelmann K., Grune T., Kleinridders A. Mitochondrial chaperones in the brain: safeguarding brain Health and metabolism? Frontiers in Endocrinology (Lausanne) 2018;9:196. doi: 10.3389/fendo.2018.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jukarainen S., Heinonen S., Ramo J.T., Rinnankoski-Tuikka R., Rappou E., Tummers M. Obesity is associated with low NAD(+)/SIRT pathway expression in adipose tissue of BMI-discordant monozygotic twins. The Journal of Cinical Endocrinology and Metabolism. 2016;101(1):275–283. doi: 10.1210/jc.2015-3095. [DOI] [PubMed] [Google Scholar]
- 14.Beck K.D., Powell-Braxton L., Widmer H.R., Valverde J., Hefti F. Igf1 gene disruption results in reduced brain size, CNS hypomyelination, and loss of hippocampal granule and striatal parvalbumin-containing neurons. Neuron. 1995;14(4):717–730. doi: 10.1016/0896-6273(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 15.Bhaskaran S., Pharaoh G., Ranjit R., Murphy A., Matsuzaki S., Nair B.C. Loss of mitochondrial protease ClpP protects mice from diet-induced obesity and insulin resistance. EMBO Reports. 2018;19(3) doi: 10.15252/embr.201745009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirschey M.D., Shimazu T., Jing E., Grueter C.A., Collins A.M., Aouizerat B. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Molecular Cell. 2011;44(2):177–190. doi: 10.1016/j.molcel.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vernochet C., Mourier A., Bezy O., Macotela Y., Boucher J., Rardin M.J. Adipose-specific deletion of TFAM increases mitochondrial oxidation and protects mice against obesity and insulin resistance. Cell Metabolism. 2012;16(6):765–776. doi: 10.1016/j.cmet.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleiner S., Mepani R.J., Laznik D., Ye L., Jurczak M.J., Jornayvaz F.R. Development of insulin resistance in mice lacking PGC-1alpha in adipose tissues. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(24):9635–9640. doi: 10.1073/pnas.1207287109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kusminski C.M., Holland W.L., Sun K., Park J., Spurgin S.B., Lin Y. MitoNEET-driven alterations in adipocyte mitochondrial activity reveal a crucial adaptive process that preserves insulin sensitivity in obesity. Nature Medicine. 2012;18(10):1539–1549. doi: 10.1038/nm.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Pauw A., Tejerina S., Raes M., Keijer J., Arnould T. Mitochondrial (dys)function in adipocyte (de)differentiation and systemic metabolic alterations. American Journal Of Pathology. 2009;175(3):927–939. doi: 10.2353/ajpath.2009.081155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernando R., Wardelmann K., Deubel S., Kehm R., Jung T., Mariotti M. Low steady-state oxidative stress inhibits adipogenesis by altering mitochondrial dynamics and decreasing cellular respiration. Redox Biology. 2020;32:101507. doi: 10.1016/j.redox.2020.101507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan S.S., Sheu M.L., Yang R.S., Chan D.C., Wu C.T., Yang T.H. The pathological role of advanced glycation end products-downregulated heat shock protein 60 in islet beta-cell hypertrophy and dysfunction. Oncotarget. 2016;7(17):23072–23087. doi: 10.18632/oncotarget.8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okamoto Y., Higashiyama H., Rong J.X., McVey M.J., Kinoshita M., Asano S. Comparison of mitochondrial and macrophage content between subcutaneous and visceral fat in db/db mice. Experimental and Molecular Pathology. 2007;83(1):73–83. doi: 10.1016/j.yexmp.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 24.Tschop M.H., Speakman J.R., Arch J.R., Auwerx J., Bruning J.C., Chan L. A guide to analysis of mouse energy metabolism. Nature Methods. 2011;9(1):57–63. doi: 10.1038/nmeth.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marker T., Sell H., Zillessen P., Glode A., Kriebel J., Ouwens D.M. Heat shock protein 60 as a mediator of adipose tissue inflammation and insulin resistance. Diabetes. 2012;61(3):615–625. doi: 10.2337/db10-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forsstrom S., Jackson C.B., Carroll C.J., Kuronen M., Pirinen E., Pradhan S. Fibroblast growth factor 21 drives dynamics of local and systemic stress responses in mitochondrial myopathy with mtDNA deletions. Cell Metabolism. 2019;30(6):1040–1054. doi: 10.1016/j.cmet.2019.08.019. e1047. [DOI] [PubMed] [Google Scholar]
- 27.Mayer A.L., Higgins C.B., Heitmeier M.R., Kraft T.E., Qian X., Crowley J.R. SLC2A8 (GLUT8) is a mammalian trehalose transporter required for trehalose-induced autophagy. Scientific Reports. 2016;6:38586. doi: 10.1038/srep38586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh R., Cuervo A.M. Autophagy in the cellular energetic balance. Cell Metabolism. 2011;13(5):495–504. doi: 10.1016/j.cmet.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parzych K.R., Klionsky D.J. An overview of autophagy: morphology, mechanism, and regulation. Antioxidants and Redox Signaling. 2014;20(3):460–473. doi: 10.1089/ars.2013.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moschella P.C., Rao V.U., McDermott P.J., Kuppuswamy D. Regulation of mTOR and S6K1 activation by the nPKC isoforms, PKCepsilon and PKCdelta, in adult cardiac muscle cells. Journal of Molecular and Cellular Cardiology. 2007;43(6):754–766. doi: 10.1016/j.yjmcc.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hotamisligil G.S., Shargill N.S., Spiegelman B.M. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science. 1993;259(5091):87–91. doi: 10.1126/science.7678183. [DOI] [PubMed] [Google Scholar]
- 32.Kusminski C.M., Scherer P.E. Mitochondrial dysfunction in white adipose tissue. Trends in Endocrinology and Metabolism. 2012;23(9):435–443. doi: 10.1016/j.tem.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bjorndal B., Burri L., Staalesen V., Skorve J., Berge R.K. Different adipose depots: their role in the development of metabolic syndrome and mitochondrial response to hypolipidemic agents. Journal of Obesity. 2011;2011:490650. doi: 10.1155/2011/490650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jankovic A., Korac A., Srdic-Galic B., Buzadzic B., Otasevic V., Stancic A. Differences in the redox status of human visceral and subcutaneous adipose tissues--relationships to obesity and metabolic risk. Metabolism. 2014;63(5):661–671. doi: 10.1016/j.metabol.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 35.Morales-Hernandez A., Martinat A., Chabot A., Kang G., McKinney-Freeman S. Elevated oxidative stress impairs hematopoietic progenitor function in C57BL/6 substrains. Stem Cell Reports. 2018;11(2):334–347. doi: 10.1016/j.stemcr.2018.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fisher-Wellman K.H., Ryan T.E., Smith C.D., Gilliam L.A., Lin C.T., Reese L.R. A direct comparison of metabolic responses to high-fat diet in C57BL/6J and C57BL/6NJ mice. Diabetes. 2016;65(11):3249–3261. doi: 10.2337/db16-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicholson A., Reifsnyder P.C., Malcolm R.D., Lucas C.A., MacGregor G.R., Zhang W. Diet-induced obesity in two C57BL/6 substrains with intact or mutant nicotinamide nucleotide transhydrogenase (Nnt) gene. Obesity (Silver Spring) 2010;18(10):1902–1905. doi: 10.1038/oby.2009.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon M.M., Greenaway S., White J.K., Fuchs H., Gailus-Durner V., Wells S. A comparative phenotypic and genomic analysis of C57BL/6J and C57BL/6N mouse strains. Genome Biology. 2013;14(7):R82. doi: 10.1186/gb-2013-14-7-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haeussler S., Kohler F., Witting M., Premm M.F., Rolland S.G., Fischer C. Autophagy compensates for defects in mitochondrial dynamics. PLoS Genetics. 2020;16(3) doi: 10.1371/journal.pgen.1008638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun M., Choi E.Y., Magee D.J., Stets C.W., During M.J., Lin E.J. Metabolic effects of social isolation in adult C57BL/6 mice. International Scholarly Research Notices. 2014;2014:690950. doi: 10.1155/2014/690950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Becker C., Kukat A., Szczepanowska K., Hermans S., Senft K., Brandscheid C.P. CLPP deficiency protects against metabolic syndrome but hinders adaptive thermogenesis. EMBO Reports. 2018 doi: 10.15252/embr.201745126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kovsan J., Bluher M., Tarnovscki T., Kloting N., Kirshtein B., Madar L. Altered autophagy in human adipose tissues in obesity. The Journal of Cinical Endocrinology and Metabolism. 2011;96(2):E268–E277. doi: 10.1210/jc.2010-1681. [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto S., Kuramoto K., Wang N., Situ X., Priyadarshini M., Zhang W. Autophagy differentially regulates insulin production and insulin sensitivity. Cell Reports. 2018;23(11):3286–3299. doi: 10.1016/j.celrep.2018.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haynes C.M., Petrova K., Benedetti C., Yang Y., Ron D. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Developmental Cell. 2007;13(4):467–480. doi: 10.1016/j.devcel.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 45.Haynes C.M., Yang Y., Blais S.P., Neubert T.A., Ron D. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Molecular Cell. 2010;37(4):529–540. doi: 10.1016/j.molcel.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nickelson K.J., Stromsdorfer K.L., Pickering R.T., Liu T.W., Ortinau L.C., Keating A.F. A comparison of inflammatory and oxidative stress markers in adipose tissue from weight-matched obese male and female mice. Experimental Diabetes Research. 2012;2012:859395. doi: 10.1155/2012/859395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ussar S., Bezy O., Bluher M., Kahn C.R. Glypican-4 enhances insulin signaling via interaction with the insulin receptor and serves as a novel adipokine. Diabetes. 2012;61(9):2289–2298. doi: 10.2337/db11-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Castro J.P., Ott C., Jung T., Grune T., Almeida H. Carbonylation of the cytoskeletal protein actin leads to aggregate formation. Free Radical Biology and Medicine. 2012;53(4):916–925. doi: 10.1016/j.freeradbiomed.2012.06.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.