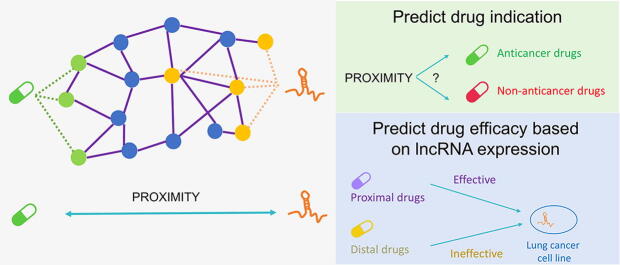
Graphical abstract
Keywords: Long non-coding RNA-based therapy, Network proximity, Drug repurposing
Abstract
Despite the fact that an increased amount of survival-related lncRNAs have been found in cancer, few drugs that target lncRNAs are approved for treatment. Here, we developed a network-based algorithm, LncTx, to repurpose the medications that potentially act on survival-related lncRNAs in lung cancer. We used eight survival-related lncRNAs derived from our previous study to test the efficacy of this method. LncTx calculates the shortest path length (proximity) between the drug targets and the lncRNA-correlated proteins in the protein–protein interaction network (interactome). LncTx contains seven different proximity measures, which are calculated in the unweighted or weighted interactome. First, to test the performance of LncTx in predicting correct indication of drugs, we benchmarked the proximity measures based on the accuracy of differentiating anticancer drugs from non-anticancer drugs. The closest proximity weighted by clustering coefficient (closestCC) has the best performance (AUC around 0.8) compared to other proximity measures across all survival-related lncRNAs. The majority of the other six proximity measures have decent performance as well, with AUC greater than 0.7. Second, to evaluate whether LncTx can repurpose the drugs effectively acting on the lncRNAs, we clustered the drugs according to their proximities by hierarchical clustering. The drugs with smaller proximity (proximal drugs) were proved to be more effective than the drugs with larger proximity (distal drugs). In conclusion, LncTx enables us to accurately identify anticancer drugs and can potentially be an index to repurpose effective agents acting on survival-related lncRNAs in lung cancer.
1. Introduction
Non-small cell lung cancer (NSCLC) is a heterogenous disease with different molecular drivers and genetic aberrations [1], [2]. Surveying the druggable targets in each patient can provide better care and prolong the survival time [3], [4], [5]. For instance, patients having EGFR mutation, one of the most common genetic aberration in lung adenocarcinoma [6], can benefit from gefitinib, an EGFR tyrosine kinase inhibitor (TKI), and had favorable treatment response compared to those without the drug target [3], [7], [8], [9]. Other examples include alectinib and crizotinib in ALK-positive NSCLC [10]; atezolizumab for the first-line treatment of PD-L1-selected NSCLC patients [11]. However, only certain population can benefit from the precision therapy. The discovery of more therapeutic target is thus necessary. Recent years have shown the explosion of studies on non-coding RNAs [12]. Whether non-coding RNAs can become therapeutic targets is worth further investigation.
Long non-coding RNAs (lncRNAs) are expected to be promising drug targets due to characters of tissue specificity, rapid turnover and low expression abundance [13], [14]. These features may be associated with lower dose of medication and lesser adverse effects in other organs [14]. Antisense oligonucleotide (ASO) has been used to target lncRNAs for years [15]. But since ASO is a large and highly-charged molecule, its drug-likeness makes the delivery ASO into human body a challenging task [16]. Few, if any, ASO-based therapies were approved in treating lung cancer patients. Considering the complex structure of lncRNAs, it was believed that a sophisticated pocket structure which is suitable for binding drug-like molecules can be found [16]; these enable small molecule to target the special domains of the lncRNAs [17]. However, because of the dynamic structure of lncRNA, effective small molecules targeting lncRNA have still been limited [18].
Given that roles of most lncRNAs in lung cancer have not been fully discovered, numerous published literatures have revealed crucial lncRNAs that are clearly associated with pathogenesis and prognosis of lung cancer, such as MALAT1 [19], [20], [21] and HOTAIR [22], [23], [24]. In one of our recent works [25], we constructed lncRNA association networks to investigate functionally similar and co-regulated lncRNAs in lung cancer. Some lncRNA modules within the lncRNA association networks were correlated with the overall survival of the patients. In addition, we proved that the modular signature could be used as a novel prognostic biomarker in lung cancer. In terms of the biological functions, we found that the survival-related lncRNA modules were significantly associated with cancer hallmarks and pathways. However, it was uncertain whether these modules can become therapeutic targets as well. Although it is not denying that few drugs can directly target lncRNAs [18], the rationale of this study is based on the hypothesis that drugs can indirectly act on a lncRNA through influencing proteins that are highly correlated with that lncRNA.
With the availability of public pharmacogenomic data [26], [27], [28], the predictive modeling techniques, particularly machine learning [29], [30], [31] and deep learning [32], [33], [34], have been largely applied in the prediction of drug response. Combining in vitro datasets and patients’ samples is even more powerful in analyzing pharmacogenomic data [35]. Network science is another pivotal discipline dealing with complex biological systems [36], [37], [38], [39]. The application of network biology in pharmacology brings new insights into early drug discovery, optimal drug combination regimen [40] and drug repurposing [41], [42]. The advantages of using network pharmacology include unraveling disease mechanisms through topology-based pathways, and prioritizing the candidate targets considering their network effects [43], [44], [45], [46], [47]. Network proximity was proved to be an effective measure for predicting drug efficacy, particularly in Parkinson’s disease and several inflammatory disorders [41]. It was revealed that protein targets of the effective drugs tend to localize close to the disease-related proteins in the interactome [41]. Novel drug-disease association can be predicted considering the proximity as well [41].
The network-based drug repurposing strategy is expected to identify potentially effective drugs acting on cancer-related lncRNAs. Meanwhile, it is also questioned that whether properties of the network (e.g., node degree) are important factors in predicting the effectiveness of a drug. Therefore, we developed a network-based method, LncTx, which measures the proximity between the drug targets and lncRNA-correlated proteins in the interactome. We accentuated the effects of the network properties by weighting the interactome with various different network parameters. Eight survival-related lncRNAs discovered in our previous work [25] were selected as the therapeutic targets. LncTx was used to repurpose the drugs that are potentially effective at targeting these lncRNAs.
2. Materials and methods
2.1. Data sources and data pre-processing
The interactome was derived from the open-access data provided by Guney et al [41] who collected the experimentally validated protein–protein interactions from various databases [48], [49], [50], [51], [52], [53], [54], [55].
Two drug lists were used in this study. One was adapted from the public data created by Guney et al [41], which included 237 drugs. Since both anticancer and non-anticancer drugs were included in this list, it was used to assess the performance of predicting the indication of anticancer drug. The other drug list was derived from Genomics of Drug Sensitivity in Cancer (GDSC) [27], [56], which consists of two datasets (GDSC1 and GDSC2). All drugs within this list are anticancer drugs, either clinically approved or under pre-clinical assessment. Because the improved screening techniques and procedures were used in GDSC2, we removed the duplicates shown in GDSC1, combined the two datasets, and selected the drug response of the non-small cell lung cancer cell lines. There are 429 anticancer drugs in total and their protein targets. We referred to DRUGBANK [57] for protein targets of chemotherapy agents because such information was not revealed in GDSC.
The basal gene expression of the cancer cell lines was derived from the Cancer Cell Line Encyclopedia (CCLE) [26]. Reads per kilobase million (RPKM) of the cancer cell lines was retrieved. The pre-processing process was detailed in our previous work [58]. In brief, genes with empty expression value across more than 20% of the samples were removed. The remaining empty expression was imputed with the minimal expression value in the dataset. RPKM was then log2 transformed. Finally, 188 lung cancer cell lines were selected for subsequent analysis.
The analyses were implemented with R packages dplyr and ggplot2 [59].
2.2. Survival analysis and functional enrichment analysis
The clinical data of lung cancer patients were downloaded from the lung adenocarcinoma (LUAD) project in the Cancer Genome Atlas (TCGA-LUAD) [60]. Log rank test was used to assess the effect of lncRNA expression on the survival probability in the subgroups (lncRNA = high v.s. lncRNA = low) of TCGA-LUAD patients. The median of the lncRNA expression was used to define the patient subgroup. The univariate survival analysis was visualized with a Kaplan-Meier plot. Cox proportional hazard (Cox PH) model [61] was used for multivariable analysis to adjust the factors that potentially influence the survival, including age and cancer stage. We used binary classification in cancer stage where patients with stage I and II were assigned to early stage, and stage III and IV were assigned to late stage. The hazard ratio of the effect of high lncRNA expression on overall survival was calculated and visualized with a forest plot.
The biological functions of the lncRNAs were deduced from the function enrichment analysis. Under the norm of “guilt by association” [62], which is commonly used in inferring the functions of unknown genes [63], top 200 lncRNA-correlated mRNAs were selected to predict functions of the lncRNAs. Hypergeometric test was used for hypothesis testing. We implemented the analysis on a web-based platform, The Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 [64]. Concerning the multiple hypothesis testing, Benjamini-Hochberg procedure were used to correct p-value. The significant level of the adjusted p-value was set at 0.05.
2.3. Calculation of the edge weights
The following network properties were used in the calculation of edge weights in the interactome, including degree, betweenness centrality, and clustering coefficient. In addition, we integrated information from differential expressed analysis (DEA) into edge weights. DEA was conducted by comparing the gene expression of tumor and adjacent normal tissues in TCGA-LUAD. The analysis was implemented by limma [65] and edgeR [66]. The HTSeq counts were downloaded from TCGA-LUAD by TCGAbiolinks [67]. The fold change and p-value derived from DEA were used to calculate the edge weights. We used as the p-value (PVal) weight and as the fold change (LogFC) weight. The edge weights (w) between node i and node j (Eij(w)) were defined as follow:
-
•
Degree weight: Eij(Degree) =
-
•
Betweenness centrality (BC) weight: Eij(BC) =
-
•
Clustering coefficient (CC) weight: Eij(CC) =
-
•
P-value (PVal) weight: Eij(PVal) =
-
•
Fold change (LogFC) weight: Eij(LogFC) =
where i and j are nodes in the interactome.
2.4. Calculation of the proximity in the interactome
The proximity was defined as the shortest path length between the protein target(s) of a drug and the proteins of interest in the interactome. To deduce the proximity between the drug targets and the survival-related lncRNAs, we used top 20 correlated proteins of each survival-related lncRNA. Specifically, the choice of selecting negatively- or positively- correlated proteins is based on the association network from which the lncRNAs were derived [25]. Seven different proximity measures are defined as below. Of note, the shortest proximity and the closest proximity were adapted from Guney et al [41].
Let T= {t|t ∈ drug targets}; G={g|g ∈ lncRNA correlated proteins}; W = {w|w ∈ weighting methods (BC, CC, PValue, LogFC, Degree)}; l (a,b)w = the proximity betweeb node a and node b under w weighing method.
-
•
Shortest proximity =
-
•
Closest proximity =
-
•
ClosestBC proximity =
-
•
ClosestDegree proximity =
-
•
ClosestPVal proximity =
-
•
ClosestLogFC proximity =
-
•
ClosestCC proximity =
where closestBC is the closest proximity weighted by betweenness centrality; closestDegree is the closest proximity weighted by degree of nodes; closestCC is the closest proximity weighted by clustering coefficient; closestPVal is the closest proximity weighted by p-value from DEA; closestFC is the closest proximity weighted by fold-change from DEA.
The network analysis was implemented with pandas and networkX [68] in Python.
2.5. Evaluation of the proximity as a predictor of anticancer drugs
The drug list from Guney et al [41] contains 237 drugs with diverse indications. In this study, the indications were grouped into two categories (anticancer or non-anticancer drugs) or three categories (anti-lung cancer, anti-other cancer or non-anticancer drugs) by the consensus of two licensed medical doctors based on the latest clinical guidelines [57]. We then built the receiver operating characteristic (ROC) curve by examining different proximity threshold and calculated area under the receiver operating characteristic (AUC) curve. The analysis was implemented with R packages pROC [69] and ROSE [70].
The proximity is correlated with the mean degree of drug targets (Supplementary Fig. S2A–H). To reduce the effect of the degree on the proximity, we transformed the proximity into z-score. To begin with, we randomly selected nodes with similar degree as the drug targets, and calculated the proximity between these nodes and lncRNA-correlated proteins. More specifically, to derive the nodes with similar degree as the drug targets, the nodes in the interactome were sorted on the basis of the node degree, and were collected in the bins where the number of nodes within one bin did not exceed 100. We then randomly selected the nodes from the bins that the drug targets belong to. By doing so, the degree of the selected nodes will be very close (or equal) to the drug targets. We repeated the procedure 100 times and calculated the mean and standard deviation of these degree-adjusted proximities. A z-score of the proximity can then be derived by
-
•
where p is the proximity of interest; p(rand) is the proximity derived from the randomly selected nodes in similar degree; is the mean of the samples; is the standard deviation of the samples.
To visualize the relation between seven proximity measures and drug categories, we reduced the seven dimensions (proximities) into three dimensions by principal component analysis (PCA). Top three principal components were selected. The 3-D PCA analysis was performed with R packages rgl and plot3D [71].
2.6. Evaluation of the proximities as a predictor of drug effectiveness
Drug effectiveness based on lncRNA expression was quantified by the correlation between lncRNA expression and drug IC50 or AUC under IC50. We took the absolute value to Spearman’s correlation coefficient (SCC) to model two possible biological mechanisms. 1) if SCC is positive (i.e., lncRNA expression is positively correlated with drug IC50 or AUC under IC50), the drugs may act on the proteins that are negatively correlated with these lncRNAs; and 2) if SCC is negative, the drugs might potentially act on these lncRNAs via off-target effects since no known lncRNAs are the direct targets of the drugs when we conducted this study. Therefore, the absolute value of SCC between drug IC50 and lncRNA expression (absIC50SCC) as well as the absolute value of the SCC between drug AUC under IC50 (absAUCSCC) were used to quantify the sensitivity of the drug toward certain lncRNA.
To categorize the drugs, we conducted hierarchical clustering (HC) to identify drug clusters based on seven proximity measures. Specifically, Euclidian distance between the proximities of drugs was used as distance measure, and complete linkage was used as the agglomerative method. Drugs within the clusters that have smaller proximities are defined as proximal drugs, while those with larger proximities are defined as distal drugs.
To test whether the proximities can predict drug sensitivity, we arranged drug clusters as the same order in hierarchical clustering and visualized the absIC50SCC or absAUCSCC in each cluster with box plots. A cubic polynomial was fitted to observe the trend of proximity and drug sensitivity. Next, we compared the absIC50SCC or absAUCSCC between proximal drugs (clusters with lower proximities) and distal drugs (clusters with higher proximities) by Wilcoxon rank-sum test. Significant level was set at p ≤ 0.05.
3. Results
3.1. An overview of the analytical pipeline
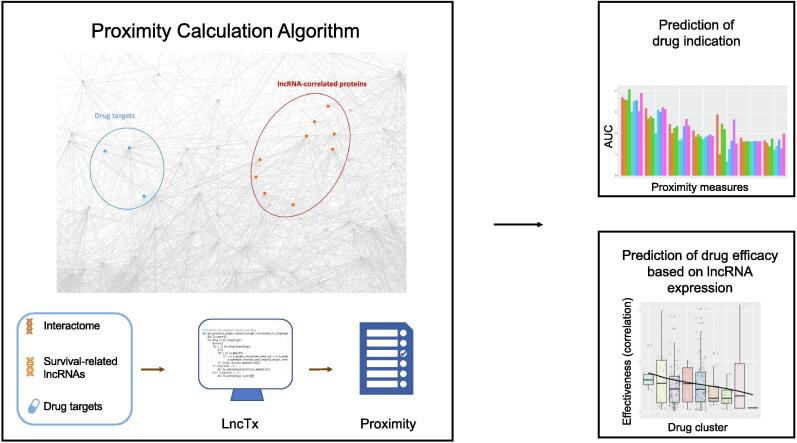
The flowchart of this study was shown in Fig. 1. To predict effective drugs acting on the survival-related lncRNAs in lung cancer, we derived eight lncRNAs candidates from our previous study [25], basal gene expression of cancer cell lines from CCLE [26], and two public drug lists from Guney et al [41] and GDSC [27]. We first examined the accuracy of proximity in predicting the correct drug indication (e.g., anticancer or non-anticancer drugs), and benchmarked seven different proximity measures. Second, we used hierarchical clustering to categorize drug clusters into proximal drugs and distal drugs. Finally, we validated the results by analyzing the correlation between the drug response and lncRNA expression.
Fig. 1.
The workflow of the study. Seven proximity measures were used to calculate the shortest path length between the protein targets of a drug and the lncRNA-correlated proteins in the interactome. Performance of the drug indication prediction was compared among different proximity measures. An unsupervised learning method was used to cluster drugs on the basis of seven proximity measures. The proximal drugs were validated by the correlation of the response of the cell lines and the lncRNA gene expression.
3.2. The survival-related lncRNAs are associated with cancer hallmarks
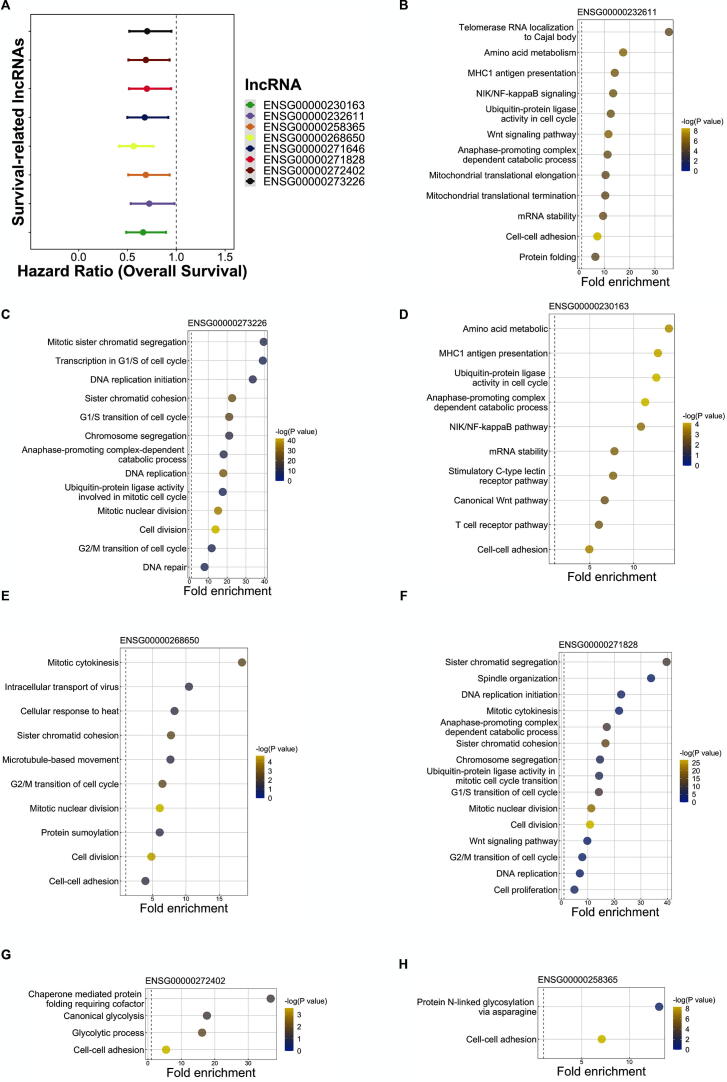
We selected eight lncRNAs from the modular signature of lung adenocarcinoma in our previous study [25], and examined the association between lncRNA expression and patients’ overall survival (Fig. 2A, Supplementary Fig. S1). The results showed that higher expression of lncRNAs is associated with favorable prognosis. We also investigated roles of lncRNAs in lung cancer with functional enrichment analysis which revealed crucial cancer hallmarks such as cell cycle and cell–cell adhesion. (Fig. 2B–H).
Fig. 2.
The roles of eight survival-related lncRNAs in lung cancer. (A) The patients from TCGA-LUAD project were divided into two subgroups according to the median expression of each survival-related lncRNA. Cox PH model was used to conduct multivariable survival analysis, adjusting cancer stage and patients’ age. Hazard ratio (HR) of high lncRNA expression and the confidence interval of HR were calculated. (B-H) Functional enrichment analysis was conducted by querying top 200 lncRNA-correlated mRNAs. Significantly enriched biological processes and their fold enrichment were shown in the plots. The color key represents the Benjamin-corrected p-value.
3.3. The weighted proximity can accurately predict the indication of anticancer drugs
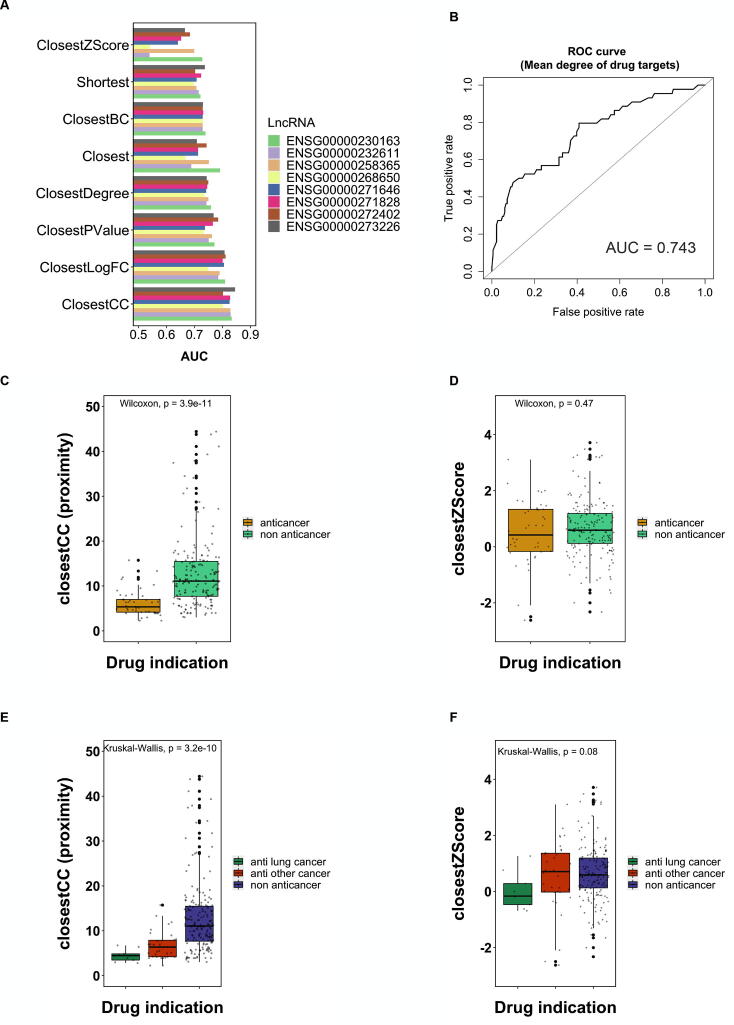
We calculated the proximity by seven different measures (Shortest, Closet, ClosestBC, ClosestDegree, ClosestPValue, ClosestLogFC, and ClosestCC). When categorizing drugs into anticancer or non-anticancer drugs, closestCC had the best performance compared to other proximity measures, with AUC greater than 0.8 across eight survival-related lncRNAs. The result implied that closestCC may be an optimal proximity measure to predict anti-neoplastic agents (Fig. 3A).
Fig. 3.
Prediction of the indication of the anticancer drugs using seven different proximity measures across eight survival-related lncRNAs. (A) The proximity is the shortest path length between survival-related lncRNAs and the drug targets. The label of the drug is dichotomous (i.e., anticancer or non-anticancer). A receiver operating characteristic (ROC) curve was constructed by setting different threshold of the proximity to assess the diagnostic accuracy. Area under the receiver operating characteristic curve (AUC) was then calculated by summing the area under the ROC curve. The AUC of different proximity measures were compared across eight survival-related lncRNAs. (B) ROC curve was constructed considering the mean degree of the drug targets. (C) Comparison of the ClosestCC between anticancer drugs and non-anticancer drugs in ENSG00000232611. (D) Comparison of the ClosestZScore between anticancer drugs and non-anticancer drugs in ENSG00000232611. (E) Comparison of the ClosestCC between anti-lung cancer drugs, anti-other cancer drugs and non-anticancer drugs in ENSG00000232611. (F) Comparison of the ClosestZScore between anti-lung cancer drugs, anti-other cancer drugs and non-anticancer drugs in ENSG00000232611. closestBC: the closest proximity weighted by betweenness centrality; closestDegree: the closest proximity weighted by degree of nodes; closestCC: the closest proximity weighted by clustering coefficient; closestPVal: the closest proximity weighted by p-value from DEA; closestFC: the closest proximity weighted by fold-change from DEA. closestZScore: the closest proximity transformed to z-score.
Degree of nodes in the interactome is an important factor in cancer network biology. Nodes with high degree tend to be hubs and play pivotal roles in the network [36], [37], [38], [63]. Considering the importance of hub genes in cancer, it was speculated that the degree of drug targets may influence the prediction of anti-neoplastic drugs. In other words, the drug that can target hub proteins in the interactome may be a more potent anti-neoplasm agent than the drug that target low-degree proteins. In fact, we found that the diagnostic accuracy of the mean degree of drug targets was acceptable (AUC = 0.743) (Fig. 3B). Further, degree was found being negatively correlated with proximity (Supplementary Fig. S2). To investigate the net effect of proximity on diagnostic accuracy of drug indication without the influence of degree, we transformed the proximity into the z-score (ClosestZScore) (See Materials and Methods). We then re-examined the correlation between ClosestZScore and the mean degree of drug targets (Supplementary Fig. S2). It was found that the degree effect in all lncRNAs diminishes. Particularly, in lncRNAs ENSG00000232611, the degree effect can be completely adjusted (Supplementary Fig. S2J). Compared to other proximity measures, ClosestZScore proximity has the lowest AUC in predicting drug indication, particularly in ENSG00000232611 whose degree effect was completely removed (Fig. 3A). We further compared the ClosestCC proximity and ClosestZScore proximity in anticancer and non-anticancer drugs. In ENSG00000232611, the anticancer drugs had significantly lower closestCC proximity than the non-anticancer drugs (Wilcoxin P = 3.9e−11) (Fig. 3C), but showed no significant difference in the ClosestZScore proximity (Fig. 3D). Similar results were found as well when classifying the drug indications into three categories (i.e., anti-lung cancer, anti-other cancer, or non-anticancer drugs) (Fig. 3E and F). 3D-PCA also revealed distinct distribution of the drugs in these three categories (Supplementary Fig. S3). The above findings suggest that the degree may interact with the proximity and have an effect on predicting the indication of anticancer drugs.
3.4. Drugs with low proximity tend to be effective
To quantify the effectiveness of the drugs that act on lncRNAs, we computed the absolute value of Spearman correlation coefficient (SCC) between the lncRNA expression and the cell response to drugs (absIC50SCC or absAUCSCC). Here, we assume that the drug response will be significantly correlated with the lncRNA expression if that drug is acting (either directly or indirectly) on that lncRNA. To prove that the proximity can be an index in surveying the drugs acting on a lncRNA, we examined the relation between the proximity and the absIC50SCC or absAUCSCC of eight survival-related lncRNAs across seven proximity measures with scatter plots. A cubic polynomial was fitted to reveal the trend (Supplementary Fig. S4–11). We found that the drugs with higher absIC50SCC or absAUCSCC tend to have smaller proximity, while the drugs with larger proximity tend to have lower absIC50SCC or absAUCSCC. In other words, many drugs with low proximity tend to be effective, while few drugs with high proximity are effective. Most of the fitted lines showed a decreasing trend, suggesting that there may exist a negative correlation between proximity and drug efficacy.
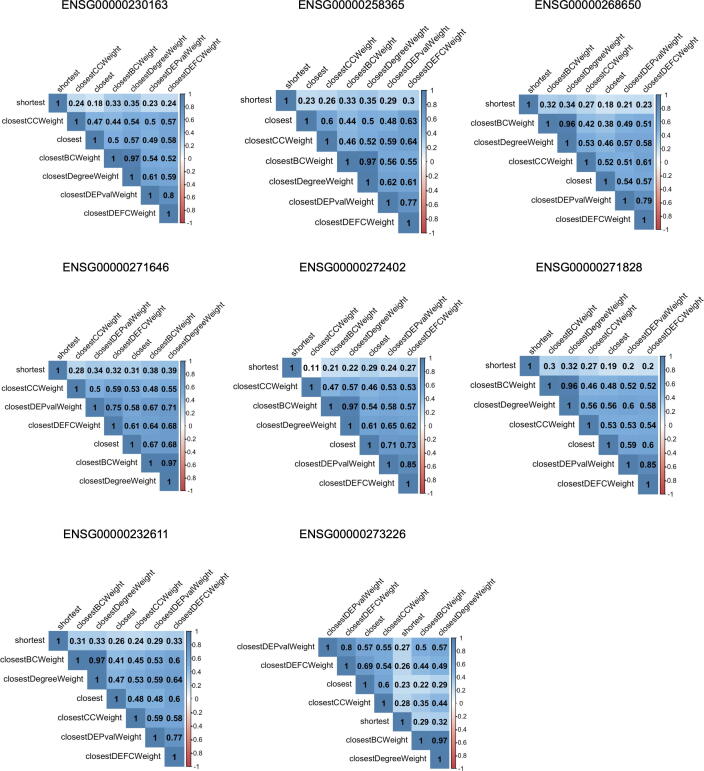
However, it was also noted that not all proximity measures have the same trend. For instance, the fitted line in the shortest proximity is concave up (Supplementary Fig. S6G). Hence, to analyze the similarity of the proximities calculated by different measures, the correlation between proximity measure was compared (Fig. 4). The results showed that the shortest proximity have relatively low correlation with other measures, implying the dissimilarity of shortest proximity.
Fig. 4.
Correlation of the seven proximity measures. Spearman correlation coefficient (SCC) was calculated between proximities from different proximity measures. The values in heatmaps represent SCC.
3.5. Proximal drugs are more effective than distal drugs
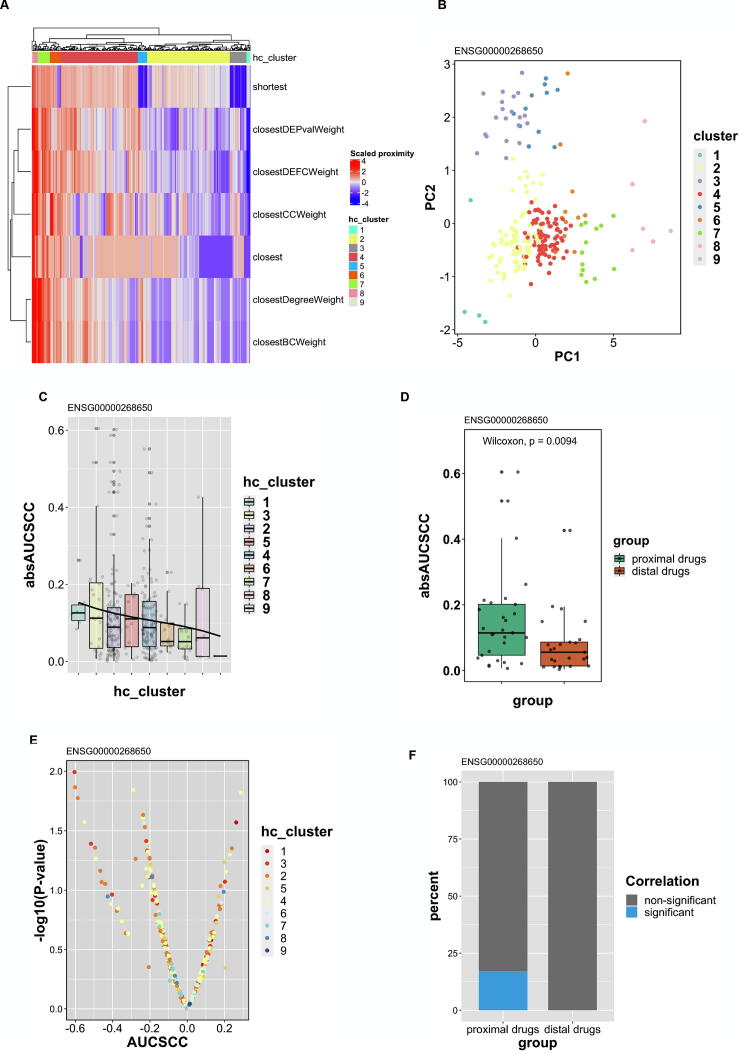
According to the above findings, the drugs with higher absAUCSCC or absIC50SCC tend to have lower proximity. However, whether proximity can be a predictor of drug sensitivity based on lncRNA expression still need to be proved. In ENSG00000268650, we used hierarchical clustering (HC) to define new drug clusters according to their proximities derived from seven different measures (Fig. 5A). The scaled proximities and clusters were shown in the heatmap. Cluster 1 and 3 had smaller proximities and were defined as proximal drugs, while cluster 7, 8, and 9 were defined as distal drugs due to larger proximities. Since the proximities calculated from the seven measures are highly collinear (Fig. 4), we used principal component analysis (PCA) to reduce seven proximity measures (dimensions) into two main principal components. We found that all clusters identified by HC can be clearly distinguished. Specifically, proximal drugs (cluster 1 and 3) localize in the area with smaller value in both PC1 and PC2, while distal drugs (cluster 7 to 9) have larger values (Fig. 5B).
Fig. 5.
Prediction and evaluation of the drugs acting on ENSG00000268650. (A) Seven measures were used to calculate the proximities between drug targets and lncRNA (ENSG00000268650)-correlated proteins. The proximities were scaled to conduct hierarchical clustering where nine drug clusters were identified. (B) Proximities derived from seven measures were dimensionally reduced to two main components by PCA. The identified nine clusters were plotted on PC1 and PC2. (C) The absAUCSCCs of each cluster were shown in a box plot. A fitted cubic polynomial line was added to observe the trend. (D) Wilcoxin rank sum test was used to compare the absAUCSCC of proximal drugs (clusters 1 and 3) and distal drugs (cluster 7–9). (E) The correlation between lncRNA expression and drug response (AUC) in different drug clusters were shown on a volcano plot by plotting -log(p-value) against AUCSCC. The percentage of significant drugs (p ≤ 0.05) in proximal and distal drug clusters was shown in (F). hc_cluster: clusters identified by hierarchical clustering; AUCSCC: The spearman correlation coefficient between lncRNA expression and AUC of drug IC50; absAUCSCC: the absolute value of spearman correlation coefficient between lncRNA expression and AUC of drug IC50.
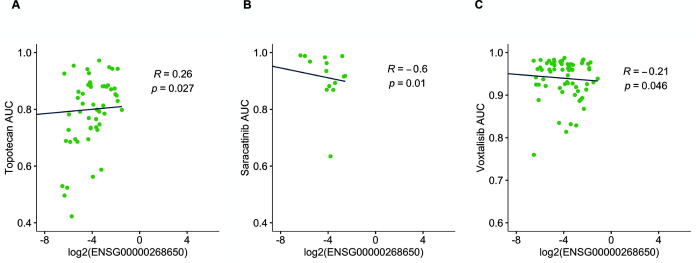
We next examined the distribution of absAUCSCC in different drug clusters identified by HC (Fig. 5C). We arranged the clusters as the same order in HC and found a clear decreasing trend from cluster 1 through cluster 9 (Fig. 5C). This result suggests that drug clusters with lower proximity tend to be more effective in treating cell lines with aberrant expression of the lncRNA (ENSG00000268650). To validate whether proximal drugs are more effective than distal drugs, we further compared the absAUCSCC of these drugs clusters. We discovered that proximal drugs have significantly higher correlation with lncRNA expression (absAUCSCC) than distal drugs (Wilcoxin p = 0.0094) (Fig. 5D). We also noticed that a larger proportion of proximal drugs have significant correlation between AUC of IC50 and lncRNA gene expression (Fig. 5E and F). Within the significant proximal drugs, some of the drugs have been currently used in treating lung cancer, such as topotecan [72], [73], [74] (Fig. 6A); the others are not currently available in treating lung cancer but have potential to be applied in the future, such as saracatinib [75] and voxtalisib [76] (Fig. 6B and C). The results from the other survival-related lncRNAs were shown in Supplementary Fig. S12–15 and Supplementary Table 1. In some lncRNAs, such as ENSG00000271646, proximal drugs do not have higher absAUCSCC or absIC50SCC compared to distal drugs (data not shown), implying that not all survival-related lncRNAs in our study are ideal therapeutic targets. All in all, the above results suggest that some, but not all, prognostic lncRNAs can be the therapeutic target as well. Furthermore, drug candidates within proximal clusters may be effective medications acting on the lncRNA (Table 1).
Fig. 6.
The correlation between the expression of the survival-related lncRNA and the cell response to the repurposed drugs. Three drugs were selected from the proximal clusters (cluster 1 and 3). The expression of ENSG00000268650 was plotted against the AUC of the IC50curve (AUC) of topotecan (G), saracatinib (H), and voxtalisib (I). Linear regression was used to calculate the fitted line. R: The Spearman correlation coefficient (SCC); p: the p-value of SCC.
Table 1.
Summary of the results in this study.
| lncRNA | Biomarker |
Accuracy of the drug indication prediction (AUC) | |
|---|---|---|---|
| Prognostic | Therapeutic | ||
| ENSG00000268650 | Yes | Yes | High (0.801) |
| ENSG00000272402 | Yes | Yes | High (0.802) |
| ENSG00000258365 | Yes | Yes | High (0.828) |
| ENSG00000273226 | Yes | Yes | High (0.845) |
| ENSG00000271828 | Yes | Equivocal | High (0.827) |
| ENSG00000232611 | Yes | No | High (0.829) |
| ENSG00000230163 | Yes | No | High (0.833) |
| ENSG00000271646 | Yes | No | High (0.825) |
4. Conclusions and discussion
With the accessibility of next-generation sequencing technology in clinical practice, it is expected that more and more genes, including lncRNAs, will be found to be involved in cancer progression [77], patients’ prognosis [78], pathological subgroups [79], and drug resistance [80]. It is also clear that the speed of new anticancer drugs approved annually would be far less than the new targets or biomarkers being discovered. Therefore, drug repurposing is of importance to provide more treatment opportunities to patients.
A study by Guney et al [41] in 2016 revealed that proximity between drug targets and disease-related proteins can accurately predict drug indication across different diseases and provide a new insight into drug repurposing. We adapted and applied their method to discover drug targets and the lncRNA-related proteins in the interactome in order to reveal effective drugs acting on the survival-related lncRNAs in lung cancer.
Previous studies showed that the hub genes in biological networks may be important targets in cancer [36], [37]. In this study, we found that the proximity is negatively correlated with the degree. Further, both proximity and degree of drug targets involved in the prediction of drug indication, providing another clue that degree is an important network property in cancer biology. Apart from degree, parameters in the networks, including clustering coefficient and betweenness centrality, quantify the characteristics of the nodes or the neighbors of the nodes. Under the assumption that network structure plays important roles in cancer biology, we integrated these parameters by adding weights in the interactome. In terms of drug indication prediction, we found that the weighted proximity measures outperformed the non-weighted measures, particularly the closest proximity weighted with clustering coefficient (closestCC).
When assessing the performance of proximity in predicting effective drugs targeting lncRNA expression (data not shown), however, we found ambiguous results. First, given the same proximity measure, the value of AUC varies across different lncRNAs, and most of the AUCs are close to 0.5. Second, closestCC proximity was not better than other proximity measures. The above findings suggest that, when predicting drug sensitivity toward lncRNA, it may be less proper to simply consider the proximity calculated from a single measure, because the results may not be robust. Therefore, considering seven different proximity measures, we performed hierarchical clustering to define clusters of proximal and distal drugs.
The design of this study was not to systematically reveal novel lncRNA therapeutic biomarkers, where comparing the genome between responsive and non-responsive cohorts may be necessary. In fact, we were more interested in investigating whether the known survival-related lncRNAs can be the therapeutic targets as well. Considering the dynamic structure and the metabolism of lncRNAs, it is not expected that all prognostic lncRNA biomarker can be therapeutic biomarkers.
We conducted literature search for some of the identified proximal drugs. For example, six effective drugs within the proximal drugs (cluster 1 and 3) were revealed acting on ENSG00000268650. Among them, topotecan in cluster 1 was found having positive results in treating NSCLC [74]. Being a cytotoxic agent, topotecan was revealed to kill cancer cells by interrupting the cell cycle through upregulating CDKN1A-encoded p21 in the lung cancer cell line [81]. Given the low objective response rate with single-agent, topotecan still showed high stable disease rate and, when in combination with other chemotherapy agents and radiotherapy, showed encouraging results [82]. Hence, according to Vennepureddy et al [74], topotecan in combination with other agents was recommended as the first-line advanced NSCLC therapy if the patient cannot tolerate the standard platinum-based therapy. Saracatinib is a Src-kinase inhibitor used to treat NSCLC in vivo [83]. Saracatinib seemed to be able to restore the epithelial-mesenchymal transition and disrupt spheroidogenesis in ovarian cancer cell lines and the subcutaneous xenograft model [84]. A phase II trial showed that 4 out of 37 patients had stable disease for at least four months, and an additional two patients had partial response. This result suggested that a subset of NSCLC patients may have benefitted from saracatinib given that the molecular subtypes have been unclear [75]. Therefore, using ENSG00000268650 as a saracatinib therapeutic response biomarker may be worth further investigations in the future. Voxtalisib is another drug in the proximal drug cluster acting on ENSG00000268650. It is a dual-PI3K/mTOR kinase inhibitor and showed promising results in follicular lymphoma [76]. In glioblastoma multiform, voxtalisib combined with temozolomide can significantly decrease the intracranial xenograft tumor size compared with temozolomide alone [85]. Although limited anti-solid-tumor activity was revealed in a phase Ib trial [86], its brain-penetrant character [87] may still be promising in treating lung cancer with brain metastasis.
In ENSG00000272402, we revealed four effective drugs within the proximal drug cluster acting on this lncRNA (Supplementary Fig. S12G-J). Vinblastine and vinorelbine are chemotherapy originally used in NSCLC [88]. Meanwhile, we revealed two potential agents targeting ENSG00000272402 in lung cancer. Serdemetan, a novel tryptamine derivative, showed potent in vitro and in vivo antiproliferative activity. A phase I clinical trial [89] suggested its association with p53 induction and modest clinical activity despite the side effect of exposure-related QTc prolong [89]. AZD7969 is a potent inhibitor of glycogen synthase kinase 3 (GSK3β) undergoing preclinical toxicity assessment [90]. The above results showed that some of the proximal drugs are currently used to treat NSCLC, while many of them are under investigation. Our findings can provide even more insight to these drugs, particularly in cell lines/patients with over- or under-expression in these survival-related lncRNAs. More detail regarding the drug clusters in each survival-related lncRNA can be found in Supplementary Table 1.
In conclusion, LncTx is a network-based method to repurpose drugs acting on the survival-related lncRNAs. In this study, we revealed that the proximity between drug targets and the lncRNA-correlated proteins can be a decent predictor of anticancer drug (Table 1). Furthermore, we found that some of the survival-related lncRNAs are more susceptible to proximal drugs, suggesting that proximity can be used to predict the treatment response (Table 1). This result, as far as we know, is the first study using weighted biological network to repurpose the drugs targeting the survival-related lncRNAs. Given the limitation that our results were only validated on GDSC/CCLE datasets, it is still expected that the method can be applied to other cancers and diseases to select effective drugs for lncRNA-based treatment in the future.
Funding
This work was supported by the Ministry of Science and Technology, Taiwan (MOST 109–2320-B-002–017-MY3, MOST 109–2327-B-006–004, MOST 109–2221-E-010–012-MY3, MOST 109–2221-E-002–161-MY3 and MOST 109–2221-E-010–011-MY3), the Higher Education Sprout Project (NTU-109L8837A and NTU-110L8808), and Emerging Infectious and Major Disease Research Program and Taiwan Biotech Innovation Academy (AS-KPQ-110-EIMD). The funding agencies had no role in the design, analysis, and interpretation of the data or writing of the manuscript.
CRediT authorship contribution statement
Albert Li: Data curation, Investigation, Formal analysis, Methodology, Validation, Visualization, Writing - original draft. Hsuan-Ting Huang: Formal analysis, Methodology, Validation. Hsuan-Cheng Huang: Conceptualization, Methodology, Writing - original draft, Supervision, Project administration, Funding acquisition. Hsueh-Fen Juan: Conceptualization, Methodology, Writing - original draft, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.07.007.
Contributor Information
Hsuan-Cheng Huang, Email: hsuancheng@ym.edu.tw.
Hsueh-Fen Juan, Email: yukijuan@ntu.edu.tw.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Duma N., Santana-Davila R., Molina J.R. Non-small cell lung cancer: Epidemiology, screening, diagnosis, and treatment. Mayo Clin Proc. 2019;94(8):1623–1640. doi: 10.1016/j.mayocp.2019.01.013. [DOI] [PubMed] [Google Scholar]
- 2.Longo D.L., Reck M., Rabe K.F. Precision diagnosis and treatment for advanced non-small-cell lung cancer. N Engl J Med. 2017;377(9):849–861. doi: 10.1056/NEJMra1703413. [DOI] [PubMed] [Google Scholar]
- 3.Arbour K.C., Riely G.J. Systemic therapy for locally advanced and metastatic non-small cell lung cancer: A Review. JAMA. 2019;322(8):764. doi: 10.1001/jama.2019.11058. [DOI] [PubMed] [Google Scholar]
- 4.Elkrief A., Kazandjian S., Bouganim N. Changes in lung cancer treatment as a result of the coronavirus Disease 2019 Pandemic. JAMA Oncol. 2020;6(11):1805. doi: 10.1001/jamaoncol.2020.4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirsch F.R., Scagliotti G.V., Mulshine J.L., Kwon R., Curran W.J., Wu Y.-L. Lung cancer: Current therapies and new targeted treatments. Lancet. 2017;389(10066):299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 6.Howlader N., Forjaz G., Mooradian M.J., Meza R., Kong C.Y., Cronin K.A. The effect of advances in lung-cancer treatment on population mortality. N Engl J Med. 2020;383(7):640–649. doi: 10.1056/NEJMoa1916623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobayashi S., Boggon T.J., Dayaram T., Jänne P.A., Kocher O., Meyerson M. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352(8):786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 8.Lynch T.J., Bell D.W., Sordella R., Gurubhagavatula S., Okimoto R.A., Brannigan B.W. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 9.Mok T.S., Wu Y.-L., Thongprasert S., Yang C.-H., Chu D.-T., Saijo N. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 10.Peters S., Camidge D.R., Shaw A.T., Gadgeel S., Ahn J.S., Kim D.-W. Alectinib versus Crizotinib in Untreated ALK-Positive Non-Small-Cell Lung Cancer. N Engl J Med. 2017;377(9):829–838. doi: 10.1056/NEJMoa1704795. [DOI] [PubMed] [Google Scholar]
- 11.Herbst R.S., Giaccone G., de Marinis F., Reinmuth N., Vergnenegre A., Barrios C.H. Atezolizumab for First-Line Treatment of PD-L1-Selected Patients with NSCLC. N Engl J Med. 2020;383(14):1328–1339. doi: 10.1056/NEJMoa1917346. [DOI] [PubMed] [Google Scholar]
- 12.Gates A.J., Gysi D.M., Kellis M., Barabási A.-L. A wealth of discovery built on the Human Genome Project – by the numbers. Nature. 2021;590(7845):212–215. doi: 10.1038/d41586-021-00314-6. [DOI] [PubMed] [Google Scholar]
- 13.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81(1):145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Statello L., Guo C.-J., Chen L.-L., Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22(2):96–118. doi: 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amodio N., Stamato M.A., Juli G., Morelli E., Fulciniti M., Manzoni M. Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity. Leukemia. 2018;32(9):1948–1957. doi: 10.1038/s41375-018-0067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warner K.D., Hajdin C.E., Weeks K.M. Principles for targeting RNA with drug-like small molecules. Nat Rev Drug Discov. 2018;17(8):547–558. doi: 10.1038/nrd.2018.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smola M.J., Christy T.W., Inoue K., Nicholson C.O., Friedersdorf M. SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the <em>Xist</em> lncRNA in living cells. Proc Natl Acad Sci. 2016;113:10322–10327. doi: 10.1073/pnas.1600008113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsui M., Corey D.R. Non-coding RNAs as drug targets. Nat Rev Drug Discov. 2017;16(3):167–179. doi: 10.1038/nrd.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amodio N., Raimondi L., Juli G., Stamato M.A., Caracciolo D., Tagliaferri P. MALAT1: A druggable long non-coding RNA for targeted anti-cancer approaches. J Hematol Oncol. 2018;11(1) doi: 10.1186/s13045-018-0606-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji P., Diederichs S., Wang W., Böing S., Metzger R. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 21.Schmidt L.H., Spieker T., Koschmieder S., Humberg J., Jungen D., Bulk E. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol. 2011;6(12):1984–1992. doi: 10.1097/JTO.0b013e3182307eac. [DOI] [PubMed] [Google Scholar]
- 22.Loewen G., Jayawickramarajah J., Zhuo Y., Shan B. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol. 2014;7:90. doi: 10.1186/s13045-014-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren M.-M., Xu S., Wei Y.-B., Yang J.-J., Yang Y.-N. Roles of HOTAIR in lung cancer susceptibility and prognosis. Mol Genet Genomic Med. 2020;8:e1299. doi: 10.1002/mgg3.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woo C.J., Kingston R.E. HOTAIR lifts noncoding RNAs to new levels. Cell. 2007;129(7):1257–1259. doi: 10.1016/j.cell.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 25.A. Li, W.-H. Yu, C.-L. Hsu, H.-C. Huang and H.-F. Juan (2021) Modular signature of long non-coding RNA association networks as a prognostic biomarker in lung cancer. In Proceedings of the 19th Asia Pacific Bioinformatics Conference, 3-5 February 2021, Taiwan. [DOI] [PMC free article] [PubMed]
- 26.Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M. A landscape of pharmacogenomic interactions in cancer. Cell. 2016;166(3):740–754. doi: 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seashore-Ludlow B., Rees M.G., Cheah J.H., Cokol M., Price E.V., Coletti M.E. Harnessing connectivity in a large-scale small-molecule sensitivity dataset. Cancer Discov. 2015;5(11):1210–1223. doi: 10.1158/2159-8290.CD-15-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ali M., Aittokallio T. Machine learning and feature selection for drug response prediction in precision oncology applications. Biophys Rev. 2019;11(1):31–39. doi: 10.1007/s12551-018-0446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang C.-T., Hsieh C.-H., Lee W.-C., Liu Y.-L., Yang T.-S., Hsu W.-M. Therapeutic targeting of non-oncogene dependencies in high-risk neuroblastoma. Clin Cancer Res. 2019;25(13):4063–4078. doi: 10.1158/1078-0432.CCR-18-4117. [DOI] [PubMed] [Google Scholar]
- 31.Adam G., Rampášek L., Safikhani Z., Smirnov P., Haibe-Kains B., Goldenberg A. Machine learning approaches to drug response prediction: challenges and recent progress. NPJ Precis Oncol. 2020;4(1) doi: 10.1038/s41698-020-0122-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.D. Baptista, P. G. Ferreira and M. Rocha (2021) Deep learning for drug response prediction in cancer. Brief Bioinform 22: 360-379. [DOI] [PubMed]
- 33.Chiu Y.-C., Chen H.-I., Zhang T., Zhang S., Gorthi A., Wang L.-J. Predicting drug response of tumors from integrated genomic profiles by deep neural networks. BMC Med Genomics. 2019;12(S1) doi: 10.1186/s12920-018-0460-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuenzi B.M., Park J., Fong S.H., Sanchez K.S., Lee J., Kreisberg J.F. Predicting drug response and synergy using a deep learning model of human cancer cells. Cancer Cell. 2020;38(5):672–684.e6. doi: 10.1016/j.ccell.2020.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nath A., Lau E.Y.T., Lee A.M., Geeleher P., Cho W.C.S., Huang R.S. Discovering long noncoding RNA predictors of anticancer drug sensitivity beyond protein-coding genes. Proc Natl Acad Sci U S A. 2019;116(44):22020–22029. doi: 10.1073/pnas.1909998116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barabási A.-L., Gulbahce N., Loscalzo J. Network medicine: A network-based approach to human disease. Nat Rev Genet. 2011;12(1):56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barabási A.-L., Oltvai Z.N. Network biology: Understanding the cell's functional organization. Nat Rev Genet. 2004;5(2):101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 38.Boguñá M., Bonamassa I., De Domenico M., Havlin S., Krioukov D., Serrano M.Á. Network geometry. Nat Rev Phys. 2021;3(2):114–135. [Google Scholar]
- 39.Liu C., Ma Y., Zhao J., Nussinov R., Zhang Y.-C., Cheng F. Computational network biology: Data, models, and applications. Phys Rep. 2020;846:1–66. [Google Scholar]
- 40.Cheng F., Kovács I.A., Barabási A.L. Network-based prediction of drug combinations. Nat Commun. 2019;10:1197. doi: 10.1038/s41467-019-09186-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guney E., Menche J., Vidal M., Barábasi A.L. Network-based in silico drug efficacy screening. Nat Commun. 2016;7:10331. doi: 10.1038/ncomms10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeng X., Zhu S., Lu W., Liu Z., Huang J., Zhou Y. Target identification among known drugs by deep learning from heterogeneous networks. Chem Sci. 2020;11(7):1775–1797. doi: 10.1039/c9sc04336e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fotis C., Antoranz A., Hatziavramidis D., Sakellaropoulos T., Alexopoulos L.G. Network-based technologies for early drug discovery. Drug Discov Today. 2018;23(3):626–635. doi: 10.1016/j.drudis.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 44.Quinn R.A., Nothias L.-F., Vining O., Meehan M., Esquenazi E., Dorrestein P.C. Molecular networking as a drug discovery, drug metabolism, and precision medicine strategy. Trends Pharmacol Sci. 2017;38(2):143–154. doi: 10.1016/j.tips.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 45.Hopkins A.L. Network pharmacology: The next paradigm in drug discovery. Nat Chem Biol. 2008;4(11):682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 46.Gao S., Tibiche C., Zou J., Zaman N., Trifiro M., O’Connor-McCourt M. Identification and construction of combinatory cancer hallmark-based gene signature sets to predict recurrence and chemotherapy benefit in stage II Colorectal Cancer. JAMA Oncol. 2016;2(1):37. doi: 10.1001/jamaoncol.2015.3413. [DOI] [PubMed] [Google Scholar]
- 47.Li J., Lenferink A.E.G., Deng Y., Collins C., Cui Q., Purisima E.O. Identification of high-quality cancer prognostic markers and metastasis network modules. Nat Commun. 2010;1(1) doi: 10.1038/ncomms1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matys V., Fricke E., Geffers R., Gößling E., Haubrock M. TRANSFAC ® : Transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aranda B., Achuthan P., Alam-Faruque Y., Armean I., Bridge A. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2009;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ceol A., Chatr Aryamontri A., Licata L., Peluso D., Briganti L. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2009;38:D532–D539. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stark C., Breitkreutz B.-J., Chatr-aryamontri A., Boucher L., Oughtred R. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 2010;39:D698–D704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keshava Prasad T.S., Goel R., Kandasamy K., Keerthikumar S., Kumar S. Human Protein Reference Database—2009 update. Nucleic Acids Res. 2008;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kanehisa M., Goto S., Hattori M., Aoki-Kinoshita K.F., Itoh M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duarte N.C., Becker S.A., Jamshidi N., Thiele I., Mo M.L., Vo T.D. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci. 2007;104(6):1777–1782. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruepp A., Brauner B., Dunger-Kaltenbach I., Frishman G., Montrone C., Stransky M. CORUM: The comprehensive resource of mammalian protein complexes. Nucleic Acids Res. 2007;36(Database):D646–D650. doi: 10.1093/nar/gkm936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang W., Soares J., Greninger P., Edelman E.J., Lightfoot H. Genomics of Drug Sensitivity in Cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:D955–961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wishart D.S., Knox C., Guo A.C., Shrivastava S., Hassanali M. DrugBank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu C.-H., Hsu C.-L., Lu P.-C., Lin W.-C., Juan H.-F., Huang H.-C. Identification of lncRNA functions in lung cancer based on associated protein-protein interaction modules. Sci Rep. 2016;6(1) doi: 10.1038/srep35939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wickham H. Springer-Verlag; New York: 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- 60.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Breslow N.E. Analysis of Survival Data under the Proportional Hazards Model. Int Stat Rev/Revue Internationale de Statistique. 1975;43(1):45. doi: 10.2307/1402659. [DOI] [Google Scholar]
- 62.Aravind L. Guilt by association: contextual information in genome analysis. Genome Res. 2000;10:1074–1077. doi: 10.1101/gr.10.8.1074. [DOI] [PubMed] [Google Scholar]
- 63.Bass J.I.F., Diallo A., Nelson J., Soto J.M., Myers C.L., Walhout A.J.M. Using networks to measure similarity between genes: association index selection. Nat Methods. 2013;10(12):1169–1176. doi: 10.1038/nmeth.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dennis G., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4(5) doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 65.M. E. Ritchie, B. Phipson, D. Wu, Y. Hu, C. W. Law, et al. (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research 43: e47-e47. [DOI] [PMC free article] [PubMed]
- 66.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (Oxford, England) 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Colaprico A., Silva T.C., Olsen C., Garofano L., Cava C. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2015;44:e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.A. A. Hagberg, D. A. Schult and P. J. Swart (2008) Exploring Network Structure, Dynamics, and Function using NetworkX. Proceedings of the 7th Python in Science Conference (SciPy 2008).
- 69.Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.-C. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinf. 2011;12(1) doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lunardon N., Menardi G., Torelli N. ROSE: A package for binary imbalanced learning. R J. 2014;6(1):79. doi: 10.32614/RJ-2014-008. [DOI] [Google Scholar]
- 71.Feng D., Tierney L. Computing and Displaying Isosurfaces in R. J Stat Softw. 2008;28(1) doi: 10.18637/jss.v028.i01. [DOI] [Google Scholar]
- 72.Horita N., Yamamoto M., Sato T., Tsukahara T., Nagakura H., Tashiro K. Topotecan for relapsed small-cell lung cancer: Systematic Review and Meta-Analysis of 1347 Patients. Sci Rep. 2015;5(1) doi: 10.1038/srep15437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stewart D.J. Update on the role of topotecan in the treatment of non-small cell lung cancer. Oncologist. 2004;9(S6):43–52. doi: 10.1634/theoncologist.9-90006-43. [DOI] [PubMed] [Google Scholar]
- 74.Vennepureddy A., Atallah J.-P., Terjanian T. Role of topotecan in non-small cell lung cancer: a review of literature. World J Oncol. 2015;6(5):429–436. doi: 10.14740/wjon950e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Laurie S.A., Goss G.D., Shepherd F.A., Reaume M.N., Nicholas G., Philip L. A phase II trial of saracatinib, an inhibitor of src kinases, in previously-treated advanced non-small-cell lung cancer: the princess margaret hospital phase II consortium. Clin Lung Cancer. 2014;15(1):52–57. doi: 10.1016/j.cllc.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 76.Brown J.R., Hamadani M., Hayslip J., Janssens A., Wagner-Johnston N., Ottmann O. Voxtalisib (XL765) in patients with relapsed or refractory non-Hodgkin lymphoma or chronic lymphocytic leukaemia: an open-label, phase 2 trial. Lancet Haematol. 2018;5(4):e170–e180. doi: 10.1016/S2352-3026(18)30030-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Silva-Fisher J.M., Dang H.X., White N.M., Strand M.S., Krasnick B.A., Rozycki E.B. Long non-coding RNA RAMS11 promotes metastatic colorectal cancer progression. Nat Commun. 2020;11(1) doi: 10.1038/s41467-020-15547-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li J., Chen Z., Tian L., Zhou C., He M.Y., Gao Y. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut. 2014;63(11):1700–1710. doi: 10.1136/gutjnl-2013-305806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Belorkar A., Vadigepalli R., Wong L. SPSNet: subpopulation-sensitive network-based analysis of heterogeneous gene expression data. BMC Syst Biol. 2018;12:28. doi: 10.1186/s12918-018-0538-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu K., Gao L., Ma X., Huang J.-J., Chen J., Zeng L. Long non-coding RNAs regulate drug resistance in cancer. Mol Cancer. 2020;19(1) doi: 10.1186/s12943-020-01162-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Y. M. Kang, A. Lan, Y. H. Huang, K. M. Hsu, Y. Chao, et al. (2020) Identification of key genes and pathways associated with topotecan treatment using multiple bioinformatics tools. J Chin Med Assoc 83: 446-453. [DOI] [PubMed]
- 82.Patel S.H., Ajlouni M., Chapman R., Lu M., Movsas B., Kim J.H. A prospective phase II study of induction carboplatin and vinorelbine followed by concomitant topotecan and accelerated radiotherapy (ART) in locally advanced non-small cell lung cancer (NSCLC) J Thorac Oncol. 2007;2(9):831–837. doi: 10.1097/JTO.0b013e318145b2e5. [DOI] [PubMed] [Google Scholar]
- 83.Chiu L.-Y., Hsin I.-L., Tsai J.-N., Chen C.-J., Ou C.-C., Wu W.-J. Combination treatment of Src inhibitor Saracatinib with GMI, a Ganoderma microsporum immunomodulatory protein, induce synthetic lethality via autophagy and apoptosis in lung cancer cells. J Cell Physiol. 2021;236(2):1148–1157. doi: 10.1002/jcp.29924. [DOI] [PubMed] [Google Scholar]
- 84.Huang R Y.-J., Wong M.K., Tan T.Z., Kuay K.T., Ng A.H.C., Chung V.Y. An EMT spectrum defines an anoikis-resistant and spheroidogenic intermediate mesenchymal state that is sensitive to e-cadherin restoration by a src-kinase inhibitor, saracatinib (AZD0530) Cell Death Dis. 2013;4(11):e915. doi: 10.1038/cddis.2013.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Prasad G., Sottero T., Yang X., Mueller S., James C.D., Weiss W.A. Inhibition of PI3K/mTOR pathways in glioblastoma and implications for combination therapy with temozolomide. Neuro-Oncology. 2011;13(4):384–392. doi: 10.1093/neuonc/noq193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schram A.M., Gandhi L., Mita M.M., Damstrup L., Campana F., Hidalgo M. A phase Ib dose-escalation and expansion study of the oral MEK inhibitor pimasertib and PI3K/MTOR inhibitor voxtalisib in patients with advanced solid tumours. Br J Cancer. 2018;119(12):1471–1476. doi: 10.1038/s41416-018-0322-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wen P.Y., Cloughesy T.F., Olivero A.G., Morrissey K.M., Wilson T.R., Lu X. First-in-Human Phase I Study to Evaluate the Brain-Penetrant PI3K/mTOR Inhibitor GDC-0084 in patients with progressive or recurrent high-grade glioma. Clin Cancer Res. 2020;26(8):1820–1828. doi: 10.1158/1078-0432.CCR-19-2808. [DOI] [PubMed] [Google Scholar]
- 88.Kosty M.P., Fleishman S.B., Herndon J.E., Coughlin K., Kornblith A.B., Scalzo A. Cisplatin, vinblastine, and hydrazine sulfate in advanced, non-small-cell lung cancer: a randomized placebo-controlled, double-blind phase III study of the Cancer and Leukemia Group B. J Clin Oncol. 1994;12(6):1113–1120. doi: 10.1200/JCO.1994.12.6.1113. [DOI] [PubMed] [Google Scholar]
- 89.Tabernero J., Dirix L., Schöffski P., Cervantes A., Lopez-Martin J.A., Capdevila J. A phase I first-in-human pharmacokinetic and pharmacodynamic study of serdemetan in patients with advanced solid tumors. Clin Cancer Res. 2011;17(19):6313–6321. doi: 10.1158/1078-0432.CCR-11-1101. [DOI] [PubMed] [Google Scholar]
- 90.Hall A.P., Escott K.J., Sanganee H., Hickling K.C. Preclinical toxicity of AZD7969: Effects of GSK3β inhibition in adult stem cells. Toxicol Pathol. 2015;43(3):384–399. doi: 10.1177/0192623314544468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.