Abstract
Epithelial cell adhesion molecule (EpCAM) is a transmembrane glycoprotein which is involved in cell signaling, proliferation, maturation, and movement, all of which are crucial for the proper development of cells and tissues. Cleavage of the EpCAM protein leads to the up-regulation of c-myc, e-fabp, and cyclins A and E which promote tumorigenesis. EpCAM can act as potential diagnostic and prognostic biomarker for different types of cancers as it is also found to be expressed in epithelia and epithelial-derived neoplasms. Hence, we aimed to analyze the EpCAM gene expression and any associated feedback in the patients of two major types of lung cancer (LC) i.e., lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC), based on the publicly available online databases. In this study, server-based gene expression analysis represents the up-regulation of EpCAM in both LUAD and LUSC subtypes as compared to the corresponding normal tissues. Besides, the histological sections revealed the over-expression of EpCAM protein in cancerous tissues by depicting strong staining signals. Furthermore, mutation analysis suggested missense as the predominant type of mutation both in LUAD and LUSC in the EpCAM gene. A significant correlation (P-value < 0.05) between the higher EpCAM expression and lower patient survival was also found in this study. Finally, the co-expressed genes were identified with their ontological features and signaling pathways associated in LC development. The overall study suggests EpCAM to be a significant biomarker for human LC prognosis.
Keywords: EpCAM gene, Lung adenocarcinoma, Lung squamous cell carcinoma, Gene expression, Biomarker, mRNA expression and regulation, Gene mutation
1. Introduction
The malady that ensues when cells adopt bizarre behaviors i.e., disregarding anti-growth signals, being indifferent to anti-apoptotic pathways, deceiving immune surveillance, and sustaining autonomous replicative potential and angiogenesis-is called cancer [1]. Such atypical cell growth is invasive and comes right after heart diseases as the leading cause of deaths worldwide. The International Agency for Research on Cancer (IARC) has reported a rise to 19.3 million cases and 10 million deaths due to cancer in 2020 [2]. Lung cancer (LC) is one of the most prominent cancers, comprising of 11.6% of the total cancer cases around the world, affecting both male and female alike [3]. According to the Global Burden of Disease Study, the five-year survival rate of LC was 17.8% in 2020 which is significantly lower than that of other types of cancers [4]. And, LCs cause 18.4% of the total cancer deaths globally [5]. Non-small cell lung cancer (NSCLC) is the most common pathological type of LC that covers about 85% of all LCs. Squamous cell lung carcinoma (LUSC) and Lung adenocarcinoma (LUAD) are two important histopathological subtypes of NSCLC [6]. LUSC is one of the most common form of NSCLC where approximately 400,000 new cases occur annually, accounting for 20–30% NSCLCs. On the other hand, LUAD occurs more in non-smoking females and the survival rate of LUAD patients is poor because of drug resistance, failure to diagnose at an early stage and lack of efficient treatments [[7], [8], [9]]. Early diagnosis, especially in the case of LUADs, can definitely reduce the mortality rate associated with LCs.
Previous studies suggest that the cell adhesion molecules like EpCAM protein have a role in cancer's intercellular and cell-extracellular matrix interactions. The differential expression i. e, upregulation or downregulation of several cell adhesion genes reveals the association between cell adhesion proteins and Differentially Expressed Genes (DEGs) [10]. For instance, CDH-1 (Cadherin-1), ITGB6 (Integrin beta 6) and DSC3 (Desmocollin-3) are found to be upregulated in breast cancer [11]. Also, CD44 and CD29 (Integrin beta-1) are downregulated in ovarian cancer whereas, ICAM-1 (Intercellular Adhesion Molecule 1) and CDH3 (Cadherin-3) are upregulated [12]. Further, colorectal cancer upregulates CDH1 and CDH3, but downregulates CDH19 (Cadherin 19) and PTPRF [13]. LC has been shown to upregulate EpCAM, ICAM-1, ITGA3 (Integrin alpha-3/beta-1), ITGB4 (Integrin alpha-6/beta-4), DSP (Desmoplakin), and DSC3 genes [[14], [15], [16]]. Thus, differential expression of DEGs could be used as possible biomarkers for detecting LCs like LUAD and LUSC [17].
A subset of cell adhesion proteins that are located on the surface of the cell and participate in cellular binding with either other cells or extracellular matrix (ECM) are called ‘cell adhesion molecules (CAMs)’ [18]. If a cell binds to another cell of the same type through these adhesion molecules, then this binding is called homotypic cell-cell adhesion [19,20]. Epithelial Cell Adhesion Molecule (EpCAM) is a trans-membrane glycoprotein of about 40 KD which consists of 314 amino acids. It is a homophilic cell-cell adhesion molecule independent of Ca2+ [21] and contains an extracellular domain (EpEX), a transmembrane domain, and an intracellular domain (EpICD) [22]. According to both in vivo and in vitro studies, EpCAM protein has pivotal roles in cell signalling, proliferation, formation, differentiation, and maintenance of organ morphology [23]. EpCAM acts as a cell surface marker on progenitor cells as well as various stem cells along with being expressed in multiple types of epithelial tissues [24,25]. During the early stages of first development such as in the morula, the EpCAM gene is expressed, although it remains tissue-specific [26]. The murine homologue of EpCAM gene shows expression on thymocytes, antigen-presenting cells, T-cells, and definitely on epithelia. The interaction between intra-epithelial lymphocytes and epithelial cells is assisted by the selective expression of the CAMs on them [27]. Therefore, the expression of murine EpCAM of non-epithelial nature may facilitate homotypic adhesive interactions between dendritic cells or epithelial cells with thymocytes. Inconsistent expression like up-regulation or down-regulation or de novo expression is demonstrated by maximum CAMs during and after a malignant transformation occurs [26]. EpCAM is overexpressed in different types of cancers including breast cancer, ovarian cancer, and head and neck squamous cell cancer [28]. Due to the overexpression of EpCAM protein molecules in cancerous cells, it appears to be important in tumorigenesis and metastasis of carcinomas, so it can act as a potential prognostic marker as well as a possible target for immunotherapeutic strategies [29]. Previous studies revealed that EpCAM acts as prognostic marker in gallbladder carcinoma, breast cancer, and colorectal cancer. Moreover, EpCAM is also referred as a universal molecular marker for the detection of circulating tumor cells (CTCs) [30]. The overexpression of EpCAM has been proved to be an indicator of lesser survival rates in breast cancer and ovarian cancer patients [31,32]. This overexpression of EpCAM can be utilized as a biomarker for detection of LUAD and LUSC as EpCAM distributes differently for squamous cell carcinomas and adenocarcinomas. This adds a good diagnostic value in the precise recognition of a certain type of carcinoma [33,34].
LUAD and LUSC are the most prevalent of NSCLCs and the disease diagnosis in the early stages is unsatisfactory. Prognostic factors are meant to determine the features of patients and the stage of the cancer before the treatment is started. Some traditional prognostic factors for survival in patients with LUAD and LUSC are performance status, stage-tumor dimension, nodal status, and weight loss [35]. Moreover, symptoms are seen in advanced stages (stage III or stage IV) in about 70% of patients diagnosed with LC [36]. The lack of symptoms in the initial stage and traditional prognosis such as determination of cancer metastasis by chest radiograph, CT scan or determining the presence of Napsin-A or TTF-1 proteins helps confirm the cancer progression. However, these methods are not satisfactory for early-prognosis and cannot lead to targeted treatment which is why survival rate is quite low [37]. Presently, there are almost no biomarkers available for detection of LC which can be applied in clinical use. This occurs either due to the lack of a robust sensitivity and specificity of the biomarkers or their functional relevance with lung carcinogenesis [36]. Thus, a biomarker is crucially required to detect LC in earlier stages, which can significantly increase the survival rate of the patients. Therefore, in this study, we hypothesize that studying gene expression of EpCAM may unveil its immunotherapy as well as prognostic value for diagnosis and treating LUAD and LUSC.
2. Materials and methods
2.1. Expression analysis of EpCAM in different types of cancerous and normal tissues
The expression pattern of EpCAM mRNA or transcript in numerous cancerous and their respective normal tissues was analyzed by three different databases i.e., Oncomine (https://www.oncomine.org) [38], GEPIA2 (http://gepia2.cancer-pku.cn) [39], and GENT2 (http://gent2.appex.kr) [40]. The Oncomine database is mainly used for analyzing the translational bioinformatics, which aids in discovering the highly ranked over-expressed genes from their 715 independent datasets, containing 86,733 clinical cancer samples and 12,764 standard tissue samples [38]. During the analysis, the threshold of p-value was kept at 1E-4 and the thresholds for fold change and gene rank were set at 2.0 and Top 10%, respectively. The Gene Expression Profiling Interactive Analysis or GEPIA2 webserver integrates the mRNA expression data of 9736 tumors and 8587 normal samples from The Cancer Genome Atlas (TCGA) program and the Genotype-Tissue Expression (GTEx) project. This server is widely recognized as the tool for differential expression analysis, patient survival analysis, detection of similar gene(s), correlation analysis etc. [39]. During analysis by GEPIA2 server, all the parameters were kept at their default values. And the Gene Expression database of Normal and Tumor tissues-2 (GENT2) server generates the result of expression analysis of a particular gene or mRNA by analyzing more than 68,000 samples stored in its database. This server has a user-friendly interface that uses the Apache Lucene indexing and Google Web Toolkit (GWT) framework to predict the results [40]. Like GEPIA2, during the experiment, all the parameters were kept default in GENT2 webserver.
2.2. Expression analysis of EpCAM in cancerous and normal lung tissues
Four different web servers were used to analyze the expression of EpCAM in normal and cancerous human lung tissues i.e., Oncomine [38], GEPIA2 [39], UALCAN (http://ualcan.path.uab.edu/) [41], and HPA (https://www.proteinatlas.org/) [42]. UALCAN is a user-friendly and interactive web server that allows the users to identify novel biomarkers and perform various in silico analyses of potential genes of interest by providing free access to the publicly available cancer OMICS data, such as the TCGA database [41]. This server was used to determine the relative expression pattern of EpCAM in both the target LUAD and LUSC samples from the TCGA database. The Human Protein Atlas (HPA) is a publicly available, Swedish-based project which was initiated in 2003. The aim of this project is to map all the human proteins in cells, tissues and organs using an integration of different types of omics technologies, for example, mass spectrometry-based proteomics, transcriptomics, antibody-based imaging etc. [42]. The HPA database is the official webserver of this project. In the HPA database, a visual comparison was made between a cancerous lung tissue (with lung-specific antibody HPA026761) and a normal lung tissue (without any lung-specific antibody CAB030012). In all these servers, default parameters were used to predict the results and the p-value less than 0.05 was considered statistically significant.
2.3. Analysis of the association of EpCAM expression with different clinical features and promoter methylation
To analyze the relationship of EpCAM gene expression with different types of clinical features, the UALCAN server [41] was again used in this step. In this study, both the LUAD and LUSC cancerous tissues were compared with normal tissues and numerous features i.e., individual cancer stages, patient's race, patient's gender, patient's age, patient's smoking habit, tumor histology, nodal metastasis status, and T53 mutation status, were noted. Thereafter, to analyze the DNA methylation pattern, the UCSC Xena Functional Genomic Explorer (https://xenabrowser.net/) was utilized which provides the users to analyze the functional genomic data for determining the correlations between numerous genomic and phenotypic variables [43]. The server generated the methylation pattern of EpCAM gene by analyzing 877 LUAD and 765 LUSC samples from GDC TCGA database. From the server, the Illumina Human Methylation 27 and Illumina Human Methylation 450 patterns were analyzed keeping all the parameters default.
2.4. Analysis of mutations and copy number alterations in the EpCAM gene
The cbioportal server (https://www.cbioportal.org/) was used to determine the pattern of the mutations and alterations in copy number of the EpCAM gene. The server is a freely available online server designed to visualize and interpret multidimensional cancer genomics datasets [44]. In our analysis, total 2983 LUAD samples and 1176 LUSC samples from different databases available in the server, were utilized to find out the anomalies or mutations and copy numbers of the EpCAM gene using the GISTIC algorithm (Genomic Identification of Significant Targets in Cancer), keeping all the parameters default. The server predicts the probable mutations in different regions along the EpCAM gene, which might be responsible for cancer development.
2.5. Determining the relationship between EpCAM gene expression and the survival of LC patients
The relationship between the expression of EpCAM gene and the survival of patients with LC was determined by the PrognoScan server (http://dna00.bio.kyutech.ac.jp/PrognoScan/). PrognoScan server is a database for meta-analysis of the prognostic values of genes. It searches the publicly available microarray datasets to establish a relationship between the expression of a target gene and the prognosis of the disease caused by that particular gene such as overall survival (OS) and disease free survival (DFS) [45]. The server plots the expression levels of a gene using the Kaplan-Meier statistical method. During the analysis, default parameters were used and any p-value <0.05 was considered as the statistically significant value.
2.6. Determination of the genes co-expressed with EpCAM in LC tissues
The profiling of genes co-expressed with EpCAM was performed by three different servers i.e., Oncomine [38], GEPIA2 [39], and UCSC Xena web browser [43]. At first, the Oncomine server was used to find out the co-expressed genes. The server generates the correlation scores and the co-expressed genes are expected to have better correlation scores. Thereafter, the GEPIA2 server was exploited to analyze the correlation between EpCAM gene and the gene that was found to be highly co-expressed with EpCAM in the Oncomine server. GEPIA2 server generated the p-values when presenting the relation of the two selected genes and any p-value with less than 0.05 value was considered as statistically significant. Finally, the UCSC Xena web browser was used to predict the gene expression pattern of the two selected genes in LC patients. The co-expression analysis was conducted for both LUAD and LUSC.
2.7. Determination of gene ontology and signaling pathways of EpCAM and its related genes in LC development
The gene ontology (GO) and cell signaling pathways of EpCAM gene were retrieved from the Enrichr server (http://amp.pharm.mssm.edu/Enrichr/). This server takes the advantage of using the enrichment analysis inferring knowledge about a target gene set by comparing it to multiple genomics datasets representing prior biological knowledge [46,47]. In this analysis, we used the EpCAM gene and all the co-expressed genes from the previous step to find out the Go terms i.e., GO biological process, GO molecular function, and GO cellular component. And the signaling pathways were also determined from BioPlanet 2019, KEGG 2019 Human, and Reactome 2016 databases via the Enrichr web server.
3. Results
3.1. Analysis of the mRNA expression of EpCAM in different types of cancerous and normal tissues
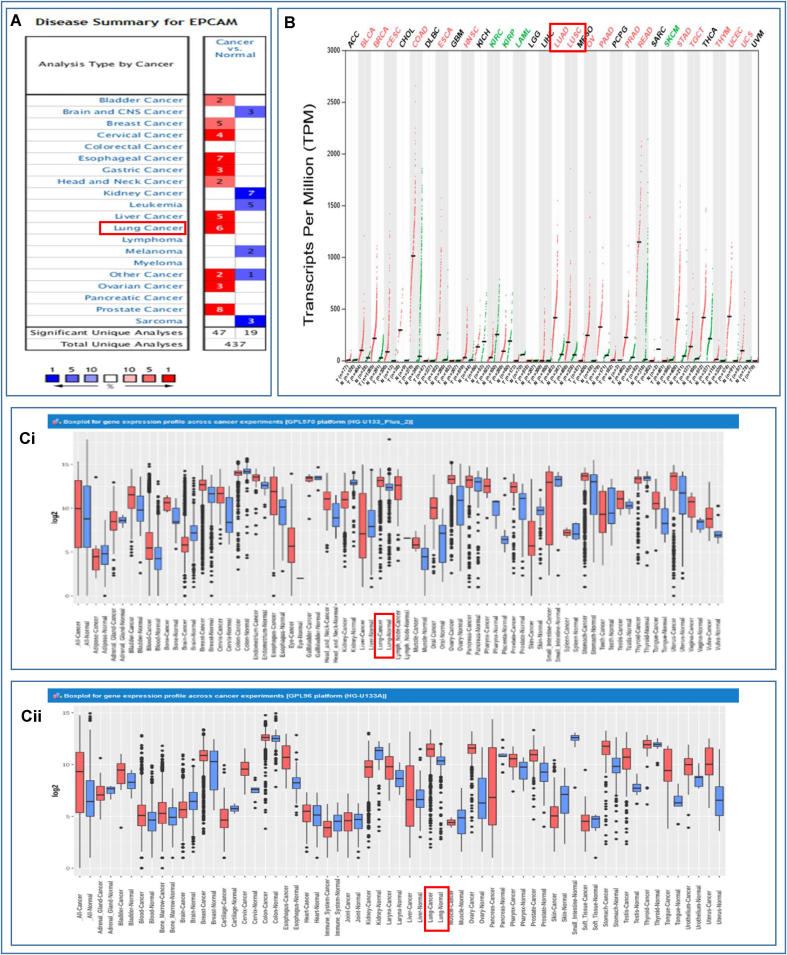
The differential expression of EpCAM transcripts or mRNAs in different cancer tissues and their respective healthy counterparts was determined using three web-based servers, i.e., Oncomine, GEPIA2, and GENT2 servers. In the Oncomine server, the statistically significant results were found in only 66 studies ranked in the top 10%, among the 437 unique datasets. The up-regulation of EpCAM was evident in numerous malignancies including lung, breast, bladder, cervical, esophageal, gastric, head and neck, liver, ovarian, prostate, and other cancers. Likewise, EpCAM was found to be highly expressed in both LUAD and LUSC patients while downregulated in the patients of Brain, CNS, Kidney Cancer, Leukemia, Melanoma, and Sarcoma (Fig. 1A). Fig. 1B illustrates the comparative expression of EpCAM in 33 different human cancers and their corresponding normal tissues. Furthermore, the overexpression of EpCAM in various cancers was revealed in another analysis using the Affymetrix HG-U133pLUS2 (Fig. 1Ci) and HG-U133A (Fig. 1Cii) platform of the GENT2 database as portrayed in Fig. 1C. Therefore, these outcomes provided strong shreds of evidence regarding the higher expression of EpCAM gene in the LC tissues when contrasted with the normal, non-cancerous tissues (Fig. 1).
Fig. 1.
Pattern of the tissue-wide expression of EpCAM in different human cancers, (A) comparison between cancer versus normal tissues in which high and low expression of mRNA has been indicated by red and blue colors, respectively, (B) the dot plot depicts the gene expression profile of the EpCAM gene in 33 different types of human cancers including tumors and normal tissue samples together. Herein, red and green dashed lines indicate the average expression value in all tumor and normal tissues, respectively, and (C) the box-plot showing the EpCAM mRNA expression in tumors and respective normal tissues using the Affymetrix HG-U133pLUS2 (Ci) and HG-U133A platforms (Cii) of the GENT2 database where the boxes indicate the median, the dots represent outliers, the red-boxes refer to the tumor tissues, whereas the blue-boxes represent the expression of normal tissues. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Expression of EpCAM transcript in human LC tissues
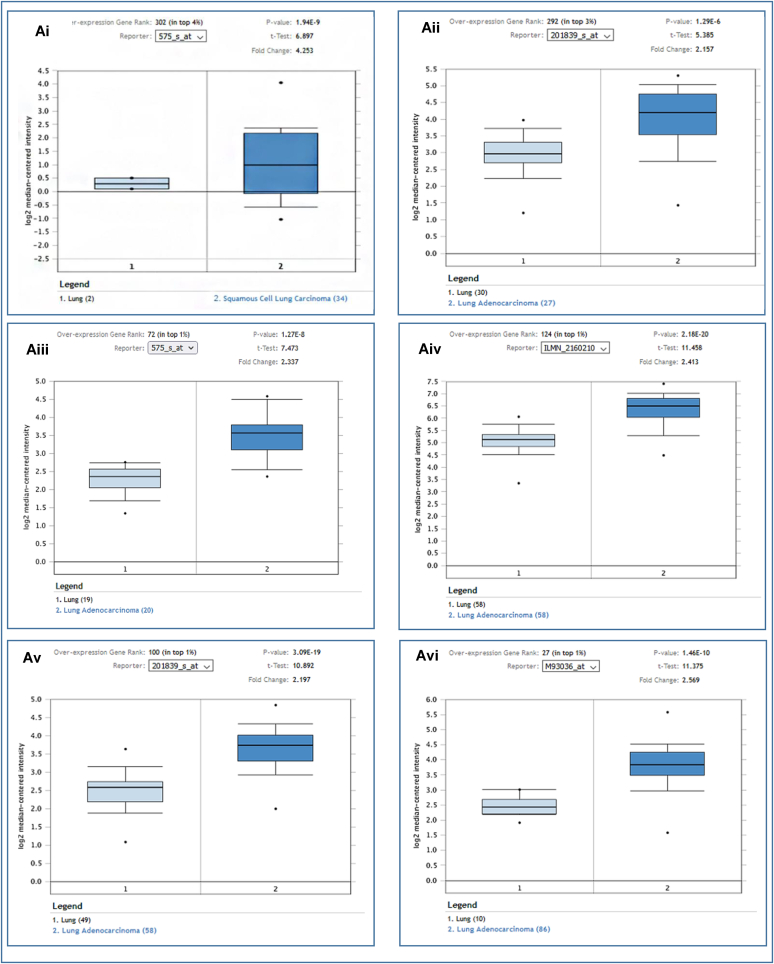
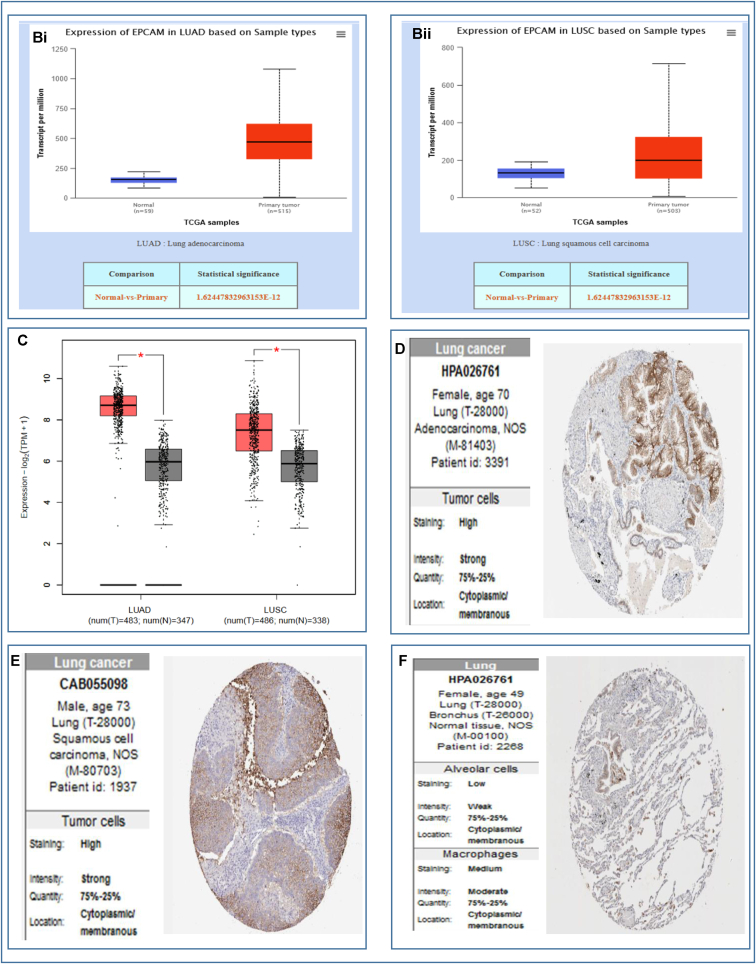
The Oncomine webserver was used to analyze the EpCAM gene expression for each subtype of LC compared to the normal tissues. The results revealed the overexpression of EpCAM in two different LCs i.e., LUAD and LUSC (Fig. 2Ai-vi and Table 1). Further assessment of TCGA datasets with UALCAN and GEPIA2 servers also showed considerable up-regulation of EpCAM in both LUAD and LUSC (Fig. 2Bi, 2Bii and 2C). In addition to that, the relative immunohistochemistry analysis was performed between normal and cancer tissues using the HPA database. While comparing the staining signals between normal and cancerous lung tissues, the signal was found to be moderate to weak in normal alveolar cells, whereas both LUAD and LUSC tissue samples showed strong staining signals as depicted in Fig. 2D, E and 2F, respectively. The results indicate that the EpCAM expression is relatively higher in cancerous tissues than the normal tissues (Fig. 2 and Table 1).
Table 1.
The expression of EpCAM in the two subtypes of LC from the Oncomine database.
| Dataset | Parameters | Samples | P- value | Gene Rank | Fold Change |
|---|---|---|---|---|---|
| Talbot Lung (n = 93) | Normal Lung | 2 | – | – | – |
| Squamous Cell Lung Carcinoma | 34 | 1.94E-9 | 302 | 4.253 | |
| Su Lung (n = 66) | Normal | 30 | – | – | – |
| Lung Adenocarcinoma | 27 | 1.29E-6 | 292 | 2.157 | |
| Strearman Lung (n = 39) | Normal | 19 | – | – | – |
| Lung Adenocarcinoma | 20 | 1.27E-8 | 72 | 2.337 | |
| Selamat Lung (n = 116) | Normal | 58 | – | – | – |
| Lung Adenocarcinoma | 58 | 2.18E-20 | 124 | 2.413 | |
| Landi Lung (n = 107) | Normal | 49 | – | – | – |
| Lung Adenocarcinoma | 58 | 3.09E-19 | 100 | 2.197 | |
| Beer Lung (n = 96) | Normal | 10 | – | – | – |
| Lung Adenocarcinoma | 86 | 1.46E-10 | 27 | 2.569 |
Fig. 2.
Analysis of the EpCAM expression in LUAD and LUSC in which (A) box-plots showing comparative expression between normal (left) and cancer tissue (right) - for LUAD and LUSC (Ai-Avi), (B–C) box-plots showing the expression of EpCAM mRNA based on sample types in tumor tissue and normal tissues for LUAD (Bi) and LUSC (Bii), using the UALCAN and GEPIA2 (C) servers, respectively, and (D–F) the immunohistochemistry images of the expression of EpCAM in LUAD (D) and LUSC (E) tissues as well as normal tissue (F) retrieved from the HPA database. The number within the parenthesis in Fig. 2 (Ai-Avi) represents the number of the samples deposited in the database.
3.3. Association of EpCAM expression with clinical characteristics of LC patients
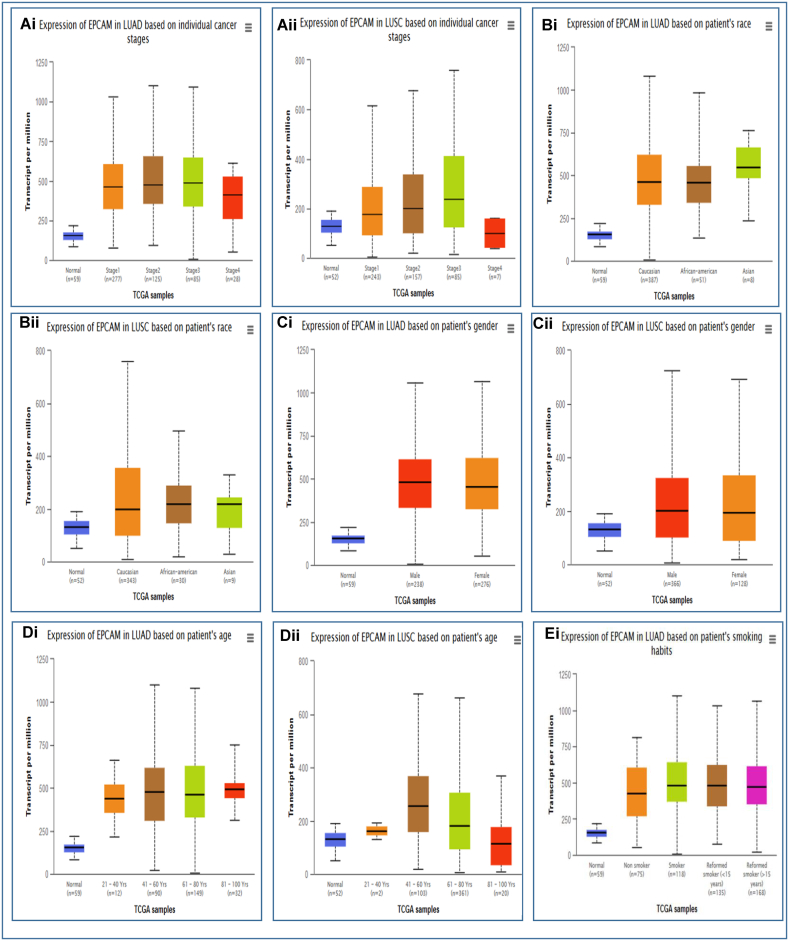
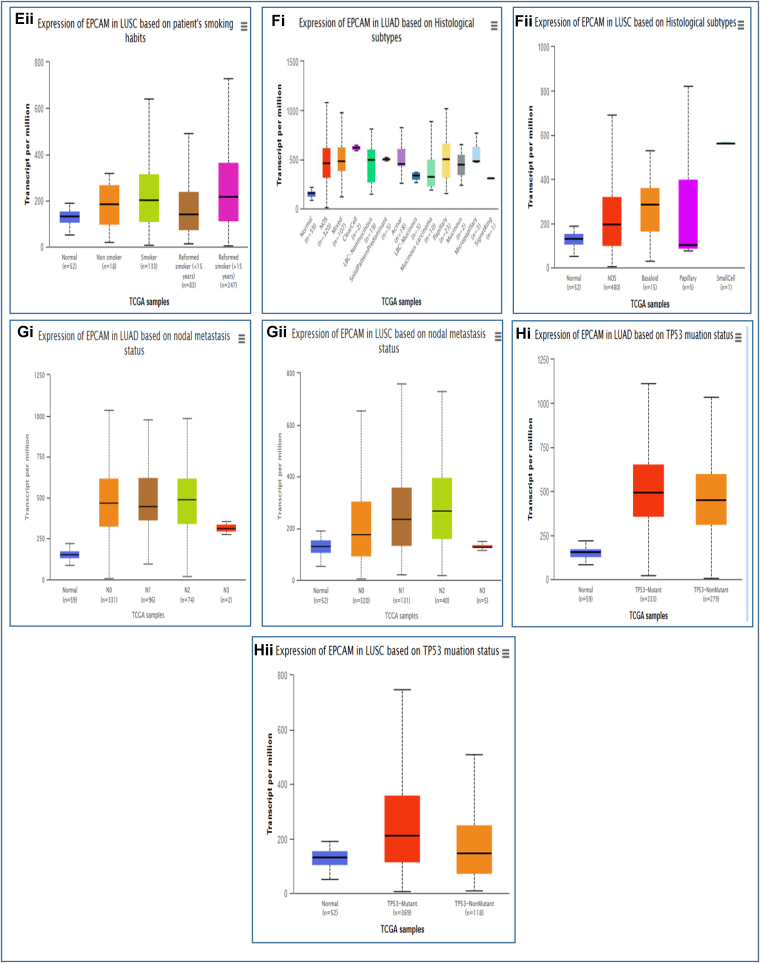
The relationship between EpCAM gene expression and the clinical representation of LUAD and LUSC patients along with the control healthy individuals was observed using the TCGA dataset via the UALCAN database. Up-regulation of EpCAM for LUAD resulted in different parameters including individual cancer stages, patient's race, gender, age, smoking habit, tumor histology, nodal metastasis, and TP53 mutation status, as depicted in Fig. 3 and listed in Table 2a. In the analysis, the increased expression of EpCAM was the highest in a fluctuating manner for LUAD (Fig. 3Ai) and in an incremental manner for LUSC (Fig. 3Aii) in the middle stages of LC. Also, the expression of EpCAM was found to be the highest among the patients from Asia for LUAD (Fig. 3Bi) and Caucasian regions for LUSC (Fig. 3Bii). The increased expression level was almost the same in male and female patients for both LUAD and LUSC (Fig. 3Ci, ii). Moreover, Fig. 3Di and ii shows higher expression in the middle age group (41–80 years old) for LUAD, whereas the expression was higher in the age group of 41–60 years old for LUSC. Up-regulation of EpCAM expression can also be seen considering other clinicopathological parameters, including smoking habit (Fig. 3Ei, ii), tumor histology (Fig. 3Fi,ii), nodal metastasis (Fig. 3Gi,ii), and TP53 mutation status (Fig. 3Hi, ii) (Table 2ab). These results indicate that the expression of EpCAM and clinical characteristics is significantly higher in LC compared to the healthy, normal individuals.
Fig. 3.
The analysis of EpCAM expression with clinical characteristics of LUAD and LUSC patients. The EpCAM mRNA expression in LUAD (Ai-Hi) and LUSC (Aii-Hii) showing individual cancer stage, patient's race, gender, age group, smoking habit, histological subtypes, nodal metastasis, and TP53 status, respectively.
Table 2.
aThe relationship between EpCAM and numerous clinicopathological features of LUAD.
| EpCAM expression based on | Features | Expression of mRNA | Number of samples | Statistical significance (p-value) |
|---|---|---|---|---|
| Individual cancer stages | Normal | Underexpression | 59 | – |
| Stage-1 | Overexpression | 277 | <1E-12 | |
| Stage-2 | Overexpression | 125 | 1.624E-12 | |
| Stage-3 | Overexpression | 85 | <1E-12 | |
| Stage-4 | Overexpression | 28 | 2.237E-06 | |
| Patient's race | Normal | Underexpression | 59 | – |
| Caucasian | Overexpression | 387 | <1E-12 | |
| African American | Overexpression | 51 | 1.728E-12 | |
| Asian | Overexpression | 8 | 2.996E-04 | |
| Patient's gender | Normal | Underexpression | 59 | – |
| Male | Overexpression | 238 | 1 .624E-12 | |
| Female | Overexpression | 276 | <1E-12 | |
| Patient's age | Normal | Underexpression | 59 | – |
| 21-40 Yrs | Overexpression | 12 | 5.362E-02 | |
| 41–60 | Overexpression | 90 | 1.624E-12 | |
| 61–80 | Overexpression | 149 | <1E-12 | |
| 81–100 | Overexpression | 32 | 2.480E-10 | |
| Patient's smoking habit | Normal | Underexpression | 59 | – |
| Non smoker | Overexpression | 75 | <1E-12 | |
| Smoker | Overexpression | 118 | <1E-12 | |
| Reformed Smoker 1 | Overexpression | 135 | <1E-12 | |
| Reformed Smoker 2 | Overexpression | 168 | 1.624E-12 | |
| Histological subtypes | Normal | Underexpression | 59 | – |
| NOS | Overexpression | 820 | 1.624E-12 | |
| Mixed | Overexpression | 107 | 1.624E-12 | |
| Clear Cell | Overexpression | 2 | <1E-12 | |
| LBC-Non Mucinous | Overexpression | 19 | 2.341E-06 | |
| Solid Pattern Predominant | Overexpression | 5 | 1.921E-03 | |
| Acinar | Overexpression | 18 | 5.900E-05 | |
| LBC-Mucinous | Overexpression | 5 | 4.740E-02 | |
| Mucinous carcinoma | Overexpression | 10 | 7.959E-03 | |
| Papillary | Overexpression | 23 | 8.074E-08 | |
| Mucinous | Overexpression | 2 | 3.905E-01 | |
| Micropapillary | Overexpression | 3 | 5.009E-02 | |
| Nodal metastasis status | Normal | Underexpression | 59 | – |
| NO | Overexpression | 331 | 1.624E-12 | |
| N1 | Overexpression | 96 | 1.624E-12 | |
| N2 | Overexpression | 74 | <1E-12 | |
| N3 | Overexpression | 2 | 3.781E-08 | |
| TP53 mutation status | Normal | Underexpression | 59 | – |
| TP53-Mutant | Overexpression | 233 | 1.624E-12 | |
| TP53-NonMutant | Overexpression | 279 | <1E-12 |
| 2b. The relationship between EpCAM and numerous clinicopathological features of LUSC. | ||||
|---|---|---|---|---|
| EpCAM expression based on | Features | Expression of mRNA | Number of samples | Statistical significance (p-value) |
| Individual cancer stages | Normal | Underexpression | 52 | – |
| Stage-1 | Overexpression | 243 | 1.3E-12 | |
| Stage-2 | Overexpression | 157 | 3E-13 | |
| Stage-3 | Overexpression | 85 | 1.232E-10 | |
| Stage 4 | Overexpression | 7 | 5.25E-01 | |
| Patient's race | Normal | Underexpression | 52 | – |
| Caucasian | Overexpression | 343 | 1.11E-16 | |
| African American | Overexpression | 30 | 2.375E-04 | |
| Asian | Overexpression | 9 | 1.351E-01 | |
| Patient's gender | Normal | Underexpression | 52 | – |
| Male | Overexpression | 366 | <1E-12 | |
| Female | Overexpression | 128 | 1.118E-10 | |
| Patient's age | Normal | Underexpression | 52 | – |
| 21-40 Yrs | Overexpression | 2 | 1.683E-01 | |
| 41–60 | Overexpression | 103 | 1.743E-12 | |
| 61–80 | Overexpression | 361 | 1.624E-12 | |
| 81–100 | Overexpression | 20 | 4.949E-01 | |
| Patient's smoking habit | Normal | Underexpression | 52 | – |
| Non smoker | Overexpression | 18 | 9.979E-03 | |
| Smoker | Overexpression | 133 | 1.36E-10 | |
| Reformed Smoker 1 | Overexpression | 83 | 6.791E-04 | |
| Reformed Smoker 2 | Overexpression | 247 | 1.11E-16 | |
| Tumor histology | Normal | Underexpression | 52 | – |
| NOS | Overexpression | 480 | <1E-12 | |
| Basaloid | Overexpression | 15 | 2.115E-03 | |
| Papillary | Overexpression | 5 | 3.078E-01 | |
| Nodal metastasis status | Normal | Underexpression | 52 | – |
| NO | Overexpression | 320 | 1.624E-12 | |
| N1 | Overexpression | 131 | 4.251E-13 | |
| N2 | Overexpression | 40 | 2.341E-06 | |
| N3 | Overexpression | 5 | 2.815E-01 | |
| TP53 mutation status | Normal | Underexpression | 52 | – |
| TP53-Mutant | Overexpression | 369 | <1E-12 | |
| TP53-NonMutant | Overexpression | 118 | 5.822E-05 | |
3.4. Analysis of promoter methylation of LC from TCGA dataset
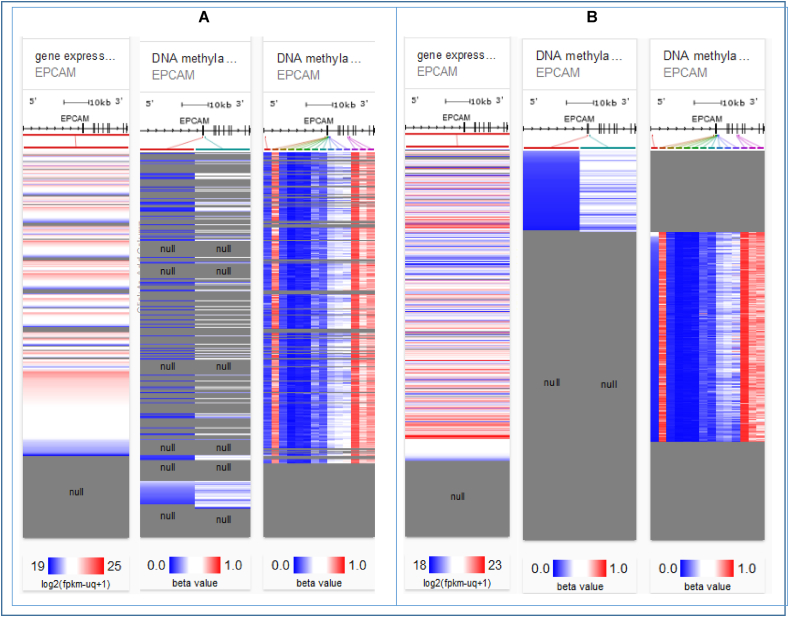
DNA methylation is crucial to understand because when it is found in a gene promoter, it acts to inhibit or lower the transcription process. Thus, promoter methylation is an important factor to be considered while studying cellular development, gene silencing, mRNA expression, tissue differentiation, genetic imprinting etc [48]. Also, hypermethylation of high-density CpG regions or genome-wide hypomethylation has been proved to be correlated with numerous types of cancer. Hence, the correlation between EpCAM expression and DNA methylation for both LUAD and LUSC was analyzed using two different methylation assays available in the server i.e., Illumina Human Methylation 27 and Illumina Human Methylation 450. The result in the heat maps revealed a likewise negative relation between the EpCAM expression and some CpG islands which indicates the overexpression of EpCAM and for both genes methylation was found to be responsible for mutation and possibly, cancer formation (Fig. 4).
Fig. 4.
Promoter methylation of the EpCAM gene in LC tissues. Heat map of EpCAM expression and DNA methylation status for LUAD (A) and LUSC (B).
3.5. Analysis of mutations, copy number alterations, and expression of mutant EpCAM transcript
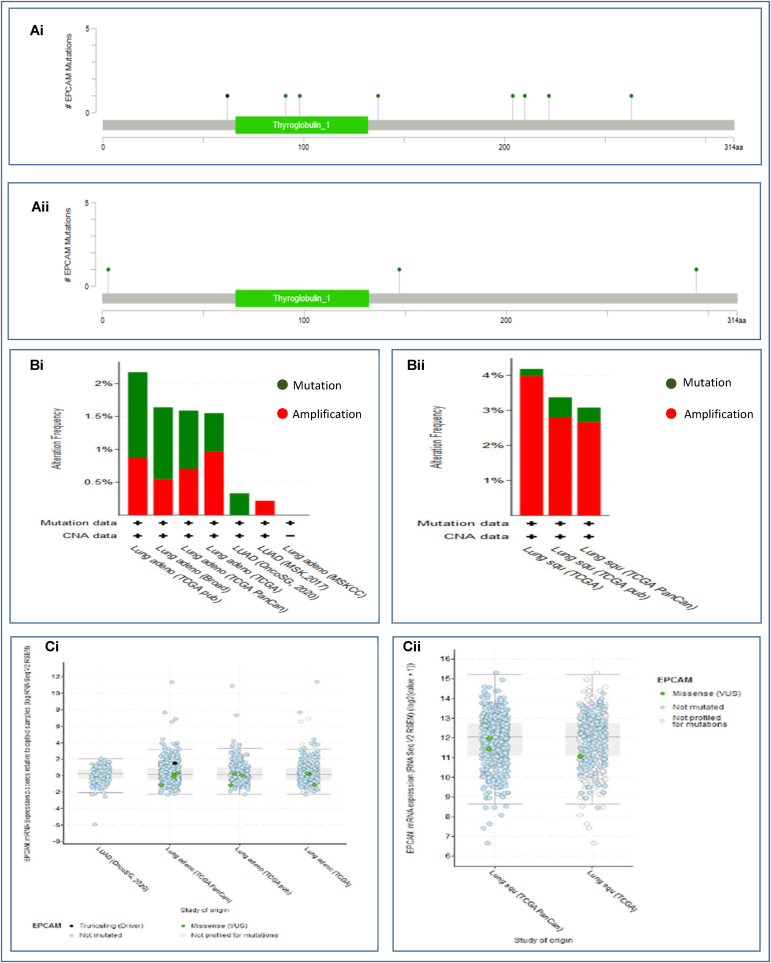
Alterations of the EpCAM gene in both types of LCs covering a total of 2983 samples from 8 LUAD studies, and 1176 samples from 3 LUSC studies were determined using cBioPortal database (Table 3). The EpCAM gene was found to be altered in 28 (<1%) among the quarried LUAD samples with an achieved somatic mutation frequency of 0.5%. A total of 14 mutations including 13 missenses and 1 splice mutations located within 1–314 residues of Thyroglobulin_1 domain were identified. All these mutations took place within 47600710 to 47607038 base pair of chromosome 2. Interestingly, the highest mutation in both LUAD and LUSC was the missense mutation, occurring in a total of 1765 (Fig. 5Ai) and 977 samples (Fig. 5Aii), respectively. Furthermore, in the alteration frequency analysis, LUAD TCGA pub occupied the highest frequency of 2.17% of 230 cases, among the six categories of the studies (Fig. 5Bi). Finally, profiling of mutated EpCAM mRNA expression was conducted where eleven cases of mutation including ten missense and one non-sense were found in LUAD (Fig. 5Ci). Hence, the results suggested that alterations found in EpCAM protein in the quarried LUAD tissues are not possibly responsible for the over expression of EpCAM. Again, the EpCAM gene was altered in 42 (4%) of quarried LUSC samples with a somatic mutation frequency of 0.3%. A total of 4 missense mutations were detected in patients with multiple samples of LUAD. In the alteration frequency analysis, the highest frequency was reported (3.37% of 178 cases) for LUSC TCGA pub among the three categories of the studies (Fig. 5Bii). Thereafter, the mutated EpCAM mRNA expression was profiled for LUSC and the result revealed three cases of analysis all of which were missense mutations (Fig. 5Cii). Therefore, these findings suggested that the overexpression of EpCAM in LUSC might not be correlated with mutations or copy number alterations found in EpCAM protein just like the LUAD samples (Fig. 5). This also provided indication that mutations or alterations in EpCAM gene might not be pivotal in causing LC even though this could be a case that is needed further studies.
Table 3.
A list of EpCAM mutational positions and types in LUAD and LUSC from the TCGA dataset.
| Sample ID | Cancer Type | Protein Change | Mutation Type | Chromosome No | Start Position | End Position | Number of Samples |
|---|---|---|---|---|---|---|---|
| TCGA-78-8662-01 | Lung Adenocarcinoma | X62_splice | Splice | 2 | 47600710 | 47600710 | 1014 |
| LUAD-B00416 | Lung Adenocarcinoma | G222V | Missense | 2 | 47606915 | 47606915 | 803 |
| LUAD-S01315 | Lung Adenocarcinoma | N91S | Missense | 2 | 47601034 | 47601034 | 1765 |
| BGI-RS55 | Lung Adenocarcinoma | Q204E | Missense | 2 | 47606146 | 47606146 | 107 |
| TCGA-44-4112-01 | Lung Adenocarcinoma | G263V | Missense | 2 | 47607038 | 47607038 | 371 |
| TCGA-44-6779-01 | Lung Adenocarcinoma | A210T | Missense | 2 | 47606164 | 47606164 | 89 |
| TCGA-78-7163-01 | Lung Adenocarcinoma | D98E | Missense | 2 | 47601056 | 47601056 | 24 |
| TCGA-44-4112-01 | Lung Adenocarcinoma | G263V | Missense | 2 | 47607038 | 47607038 | 434 |
| TCGA-44-6779-01 | Lung Adenocarcinoma | A210T | Missense | 2 | 47606164 | 47606164 | 91 |
| TCGA-78-7163-01 | Lung Adenocarcinoma | D98E | Missense | 2 | 47601056 | 47601056 | 49 |
| TCGA-86-8279-01 | Lung Adenocarcinoma | E137A | Missense | 2 | 47601172 | 47601172 | 458 |
| TCGA-44-4112-01 | Lung Adenocarcinoma | G263V | Missense | 2 | 47607038 | 47607038 | 389 |
| TCGA-44-6779-01 | Lung Adenocarcinoma | A210T | Missense | 2 | 47606164 | 47606164 | 93 |
| TCGA-78-7163-01 | Lung Adenocarcinoma | D98E | Missense | 2 | 47601056 | 47601056 | 27 |
| TCGA-37-3789-01 | Lung Squamous Cell Carcinoma | M294I | Missense | 2 | 47612328 | 47612328 | 379 |
| TCGA-98-8021-01 | Lung Squamous Cell Carcinoma | E147Q | Missense | 2 | 47602386 | 47602386 | 930 |
| TCGA-22-4604-01 | Lung Squamous Cell Carcinoma | P3S | Missense | 2 | 47596651 | 47596651 | 225 |
| TCGA-37-3789-01 | Lung Squamous Cell Carcinoma | M294I | Missense | 2 | 47612328 | 47612328 | 977 |
Fig. 5.
Genetic alteration and mutations of EpCAM in LUAD and LUSC tissues. Herein (Ai) lollipop plot shows the type of alteration in fourteen mutation spots within the peptide sequence (1–314 residues) of Thyroglobulin_1 domain in EpCAM from LUAD tissues, (Aii) depicts the alterations in only four mutation spots in EpCAM from LUSC tissues, (Bi and Bii) bar diagrams show the mutation frequencies and genome alteration in the EpCAM gene for LUAD and LUSC, respectively, and (Ci and Cii) indicate the correlation between the expression and copy number alteration of EpCAM for LUAD and LUSC in the TCGA dataset.
3.6. EpCAM expression and clinical prognosis of LC patients
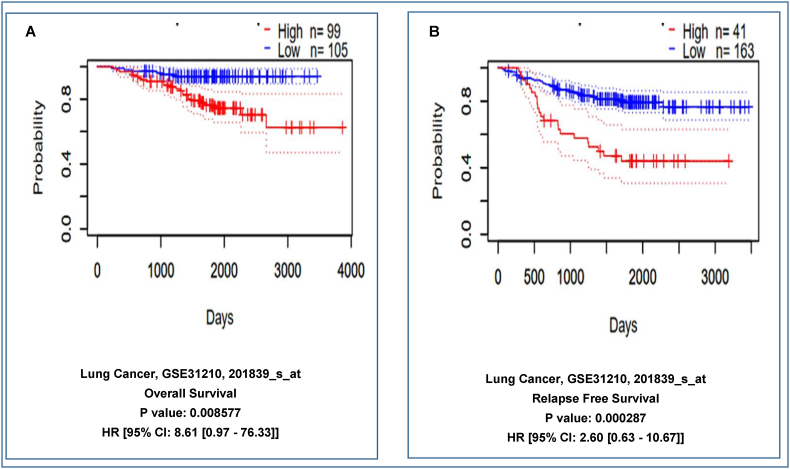
The relationship between the level of EpCAM expression and patient's survival in LC was analyzed using the PrognoScan database (significant level was kept at P-value < 0.05 and HR > 1). The analysis showed a negative correlation of EpCAM expression with the survival of the patients. According to the analysis, high expression of EpCAM might lead to lower survival rate whereas low or normal expression of the protein should enhance the survival rate of the patients. For dataset GSE31210 (Number of samples = 204), patients with low EpCAM expression (n = 105 and 163) had significantly higher survival probability whereas higher expression (n = 99 and 41) of EpCAM was responsible lower survival rate in the patients as illustrated in Fig. 6A and B. The results revealed that the enhanced expression of EpCAM could be responsible for relatively poor prognosis in LC patients.
Fig. 6.
The in Kaplan-Meier plot represents the relationship between EpCAM gene expression and survival of LC patients. The survival curves demonstrate patients' survival with the high (red) and low (blue) expression of EpCAM in the plots where (A–B) showing overall survival, and relapse-free survival, respectively. The analysis was focused on the EpCAM expression in LC patients. Here, HR, hazard ratio; CI, confidence interval. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
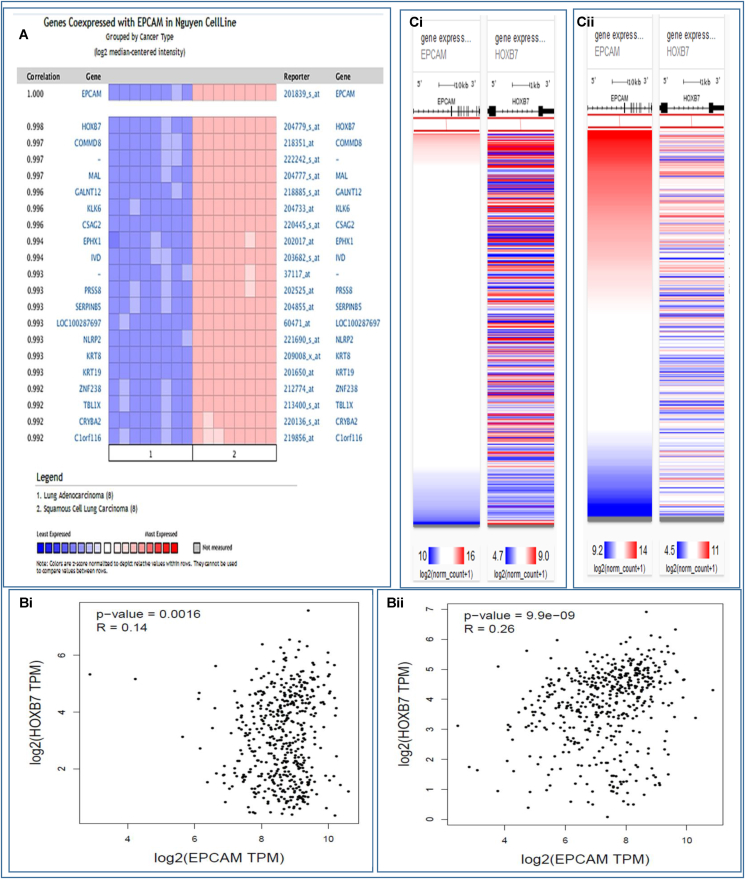
3.7. Analysis of gene signatures linked to EpCAM and human LC
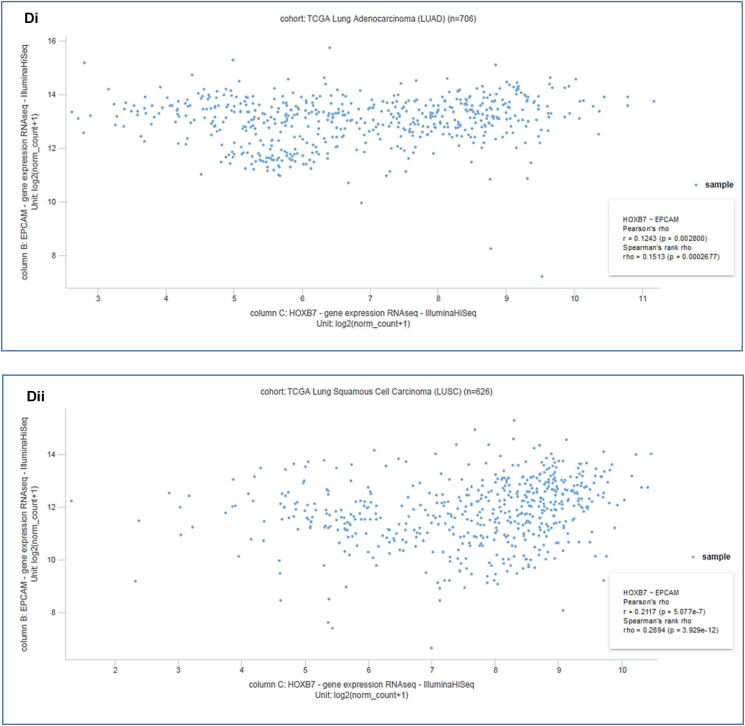
The co-expression profile of EpCAM was analyzed with 19 genes using a total of 16 samples from LUAD and LUSC patients through the Oncomine database (Fig. 7A). The HOXB7 was found to be mostly co-expressed (R = 0.998) among the total 19 genes. The EpCAM and HOXB7 were found to be positively correlated with the Spearman coefficient in both LUAD (R = 0.14) (Fig. 7Bi) and LUSC (R = 0.26) (Fig. 7Bii) using GEPIA2 server. Furthermore, the Pearson and Spearman correlation analyses were performed to ensure the positive correlation between EpCAM and HOXB7 in LC patients using TCGA data through UCSC Xena server (Fig. 7C–D). For the LUAD (Fig. 7Ci-Di) and LUSC (Fig. 7Cii-Dii) patients, the Pearson correlation values were found to be 0.1243 and 0.2117, respectively. Besides this, the Spearman correlation values were also reported to be 0.1513 and 0.2894, respectively. Therefore, these results suggest that these two genes might be interlinked in different signaling pathways in LC progression.
Fig. 7.
Co-expression profile of the EpCAM and co-expressed genes in human LC. The figure shows (A) co-expression profile of EpCAM derived from the Oncomine database, (B) correlation analysis between EpCAM and HOXB7 obtained by GEPIA2 server, (C) heatmap of mRNA expression for EpCAM and HOXB7 genes across LC in the TCGA database, and (D) co-expression analysis between EpCAM and HOXB7 genes in LC using UCSC Xena server.
3.8. Determination of gene ontologies and signaling pathways linked to EpCAM and LC progression
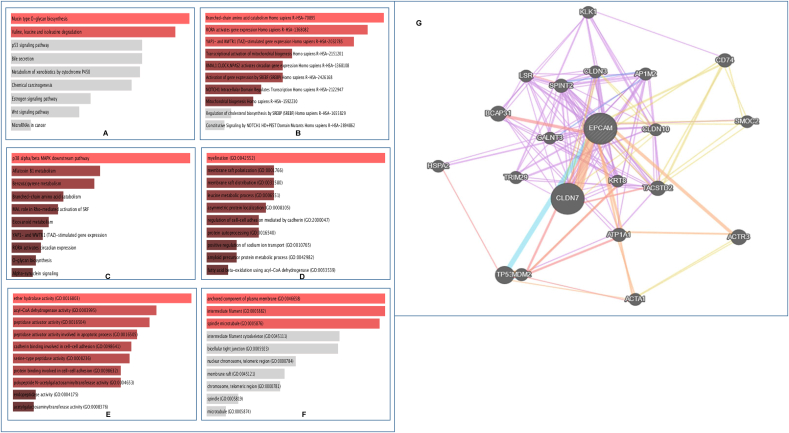
Based on the EpCAM and correlated genes, signaling pathways and gene ontological features that lead to the progression of LC in humans were identified. For pathway determination, the results from three databases depicted in Fig. 8A–C were considered. In the analysis of KEGG human 2019 database, different significant pathways were found including mucin type O-glycan biosynthesis and branched chain amino acid (BCAA) metabolism i.e., degradation of valine, leucine and isoleucine. (Fig. 8A). Furthermore, Reactome 2016 database showed pathways related to amino acid catabolism, protein metabolism, activation of gene expression by RORA, BMAL1:CLOCK, NPAS2, and SREBF (SREBP), YAP1- and WWTR1 (TAZ)- stimulated gene expression, mitochondrial biogenesis, regulation of transcription by NOTCH1 intracellular domain, etc. (Fig. 8B). Finally, the analysis of the Bioplanet 2019 database revealed the p38 alpha/beta MAPK downstream pathway as well as some other significant pathways related to aflatoxin B1 metabolism, benzo(a)pyrene metabolism, amino acid catabolism, Rho-mediated activation of SRF, eicosanoid metabolism, YAP1- and WWTR1 (TAZ)-stimulated gene expression, O-glycan biosynthesis, and alpha-synuclein signaling (Fig. 8C). These pathways above, therefore, are anticipated to be involved in the progression of LC. After that, the GO terms were also considered and determined for the corresponding genes. The suggested GO features in the analysis mainly include myelination, membrane raft polarization and distribution, regulation of cell-cell adhesion, protein autoprocessing, regulation of ion transport, protein and fat metabolic process (Fig. 8D), hydrolase, dehydrogenase, peptidase and transferase (Fig. 8E), and mitotic spindle activity (Fig. 8F). The cell signaling pathway where EpCAM gene is involved is depicted in Fig. 8G. These results suggested a possible correlation of EpCAM and its correlated genes in the development and progression of LC.
Fig. 8.
Analysis of the pathways and gene ontologies related to EpCAM expression and LC. Pathways and ontologies are achieved from (A) KEGG human 2019, (B) Reactome 2016, (C) BioPlanet 2019, (D) GO biological process 2018, (E) GO molecular function 2018, and (F) GO cellular component 2018. The length and the color gradient of the bar represent the level of significance; brighter color indicates the more significant term and vice versa. (G) Figure representing the cell signaling network where EpCAM is involved. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
LC has the highest cancer-related mortality rate worldwide and is infamously known for its appearance as one of the most frequently diagnosed malignant tumors [49,50]. NSCLC, consisting of two subtypes i.e., LUAD and LUSC, alone constitutes of an estimated 85% of all LC cases [50]. Hence, early detection and correct prognosis of LC are pivotal to understand a patient's illness circumstances and set up further appropriate treatment approaches, which is why the accurate prediction is crucial to both patients and physicians and researchers [51,52].
According to previous reports, LC in the majority of the patients (approx. 75%) seem to already reach the advanced stage (stage III/IV) during diagnosis which only puts a greater emphasis on the gravity of early diagnosis in case of LC [53]. For instance, surgical resection of NSCLC presents a favorable prognosis in case of small, localized tumors (stage I) with 5-year survival rates of 70–90% [[54], [55], [56]]. This suggests a strong correlation between the early diagnosis of LC and enhanced survival rates. Therefore, the early diagnosis of LC still remains a major deciding factor for optimal outcomes, even with the addition of contemporary advancements and breakthroughs [57]. In this study, the significance of EpCAM as a prognostic marker in the early determination of two NSCLC i.e., LUAD and LUSC using bioinformatics was evaluated [33]. Based on the multiple databases, it can be concluded that the expression level of EpCAM is positively correlated with the progression of LC. The analysis of EpCAM expression in LUAD and LUSC displayed a negative correlation with all cases of overall survival, diseases free survival and relapse-free survival with the overall HR > 1. It was also observed that high EpCAM level showed a detrimental effect on the survival rate than those with lower EpCAM levels. A previous study suggested poor prognosis in LC patients due to increased expression of EpCAM which further validated this observation [33].
Again, the expression patterns of EpCAM in the two cancer tissues were also found to be significantly associated with different clinical characteristics of LC patients including, tumor histology, patient's race, gender, age, smoking habit, nodal metastasis status, etc. Therefore, analysis of these features requires further investigation since a high expression level of EpCAM might suggest a risk of cancer transformation and progression. Computer-aided systems and digital imaging, combined with the methodological aspects of modern immunohistochemistry, provide excellent insights into immunohistochemical scoring [33,58,59]. The immunohistochemical data of EpCAM exhibited a strong nuclear immunoreactivity of EpCAM in every target LC cells. Robust and intense staining of cancer cells that distinguish them from normal alveolar cells, helped to recognize the higher levels of EpCAM expression in LUAD and LUSC tissues. Varied alterations of four categories i.e., somatically acquired genetic, epigenetic, transcriptomic, and proteomic alterations, constitute a series of histopathological process that contribute to cancer progression [58]. Any of these alterations in the genomic region (either loss or gain) can result in either suppressive or oncogenic effects [60]. The cBioPortal webserver was utilized to explore the copy number alterations, mutations, and mutant mRNA expressions of EpCAM. The EpCAM gene was found to be altered in 28 (<1%) of quarried LUAD samples with 0.5% somatic mutation frequency, whereas alteration was reported in 42 (4%) of the LUSC samples with 0.3% somatic mutation frequency. The highest mutation in both LUAD and LUSC was found to be a missense mutation, occurring in total of 1765 and 977 samples, respectively.
This study also investigated the potential correlation between DNA methylation and EpCAM expression which revealed a negative correlation between them. Transcriptional silencing of genes usually occurs as an outcome of DNA methylation that are found mainly on the CpG islands of the promoters of the genes. Following this prospect, several studies on the methylation of promoter in multiple cancer types including LC were conducted, which implied that the expression of EpCAM is inversely correlated with DNA methylation in tissues from cancer patients [61]. Through further co-expression and correlation analysis, it was found that 19 genes generated a positive correlation with the EpCAM gene and the homeobox B7 (HOXB7) was found to be positively correlated with EpCAM expression (R = 0.998). Analysis conducted by other servers also confirmed the relationship between EpCAM and HOXB7. A dysregulated HOXB7 was suggested to act as a critical player in regulating tumorigenesis and metastases of some cancers. A relevant previous study found the increased expression of HOXB7 to be associated with poor clinical outcomes in LUAD patients leading to significant correlation with short survival time [62].
Finally, the correlated genes were used to analyze the possible EpCAM related pathways responsible for the development of LC. In the KEGG pathway analysis, the correlated genes were mostly related to the O-glycan biosynthesis and BCAA metabolism [63]. The observation seems reasonable as sometimes the process of oncogenesis is dependent on cellular energy and metabolites provided by amino acids that are degraded by overexpressed enzymes in many cancer types [64,65]. In the GO analysis, the widely used ontology term was myelination. Cognitive impairment is a common consequence of chemotherapy where myelination is reportedly claimed as an underlying factor according to previous studies [66]. Furthermore, membrane raft polarization facilitates another significant correlated molecular function which includes cell adhesion and migration. The cell adhesion disorders and aggressive phenotypes of migration and invasion constitute the malignant phenotype of cancer which is modulated by the dynamic feature of the cancer cell surface. Lipid rafts are also recently found to contribute to the cancer cell adhesion and migration [67]. Overall, the pathways and GO enrichment analysis portrayed the significance of EpCAM and its correlated genes in different oncogenic processes leading to LC development.
Overall, this study supports the idea that the expression of EpCAM gene could be used as the prognostic biomarker for the early detection of human LUAD and LUSC cases. However, more in vivo and in vitro studies are needed to finally confirm the outcome of this study.
5. Conclusion
In this study, we targeted the molecular signatures that play key roles in the development and progression of LUAD and LUSC. In cancer, prognostic factors are important for early diagnosis as well as efficient treatment and thus help patients prevent the risk of overtreatments. To determine the potency of EpCAM as a potential prognostic marker in LC development, the mRNA expression, DNA methylation, mutations and CNAs, correlated genes and the prognostic features were analyzed in this study. The analyses exhibited a sharp overexpression as well as a potential correlation of EpCAM to the LC development. Furthermore, it also indicates the probable signaling pathways and gene ontological features related to EpCAM and its expression in LC progression. These pathways could be the significant target to interfere with the development of cancer. For this reason, EpCAM is suggested to be an effective biomarker as well as a potential therapeutic target in effort of preventing LC in humans.
Data availability statement
Authors made all the data generated during experiment and analysis available within the manuscript.
Funding statement
Authors received no specific funding from any external sources.
Author contribution
BS and ATM conceived the study. BS designed the analysis. ATM, MAU and BS conducted the experiments. ATM, MAU and YA wrote the paper. NA, BS and BR edited the paper. BR supervised the study. BS was the co-supervisor of the study.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Acknowledgements
The authors are thankful to Swift Integrity Computational Lab, Dhaka, Bangladesh, for providing support during the preparation of the manuscript.
Contributor Information
Bishajit Sarkar, Email: sarkarbishajit@gmail.com.
Bashudev Rudra, Email: bashudev@cu.ac.bd.
References
- 1.Harrington K.J. Biology of cancer. Medicine. 2011 Dec 1;39(12):689–692. doi: 10.1016/j.mpmed.2011.09.015. [DOI] [Google Scholar]
- 2.Globocan New global cancer data. UICC. 2020 https://www.uicc.org/news/globocan-2020-new-global-cancer-data Internet]. Uicc.org. 2021 [cited 27 January 2021] Available from: [Google Scholar]
- 3.Liu C., Li X., Shao H., Li D. Identification and validation of two lung adenocarcinoma-development characteristic gene sets for diagnosing lung adenocarcinoma and predicting prognosis. Front. Genet. 2020;11 doi: 10.3389/fgene.2020.565206. PMid:33408736 PMCid:PMC7779611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong M.C., Lao X.Q., Ho K.F., Goggins W.B., Shelly L.A. Incidence and mortality of lung cancer: global trends and association with socioeconomic status. Sci. Rep. 2017;7(1):1–9. doi: 10.1038/s41598-017-14513-7. PMid:28361988 PMCid:PMC5374459, 10.1038/s41598-017-02997-2, 10.1038/srep45846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. PMid:30207593. [DOI] [PubMed] [Google Scholar]
- 6.Anusewicz D., Orzechowska M., Bednarek A.K. Lung squamous cell carcinoma and lung adenocarcinoma differential gene expression regulation through pathways of Notch, Hedgehog, Wnt, and ErbB signalling. Sci. Rep. 2020;10(1):1–15. doi: 10.1038/s41598-020-77284-8. PMid:33273537 PMCid:PMC7713208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519. doi: 10.1038/nature11404. PMid:22960745 PMCid:PMC3466113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donner I., Katainen R., Sipilä L.J., Aavikko M., Pukkala E., Aaltonen L.A. Germline mutations in young non-smoking women with lung adenocarcinoma. Lung Canc. 2018;122:76–82. doi: 10.1016/j.lungcan.2018.05.027. PMid:30032850. [DOI] [PubMed] [Google Scholar]
- 9.Ling D.J., Chen Z.S., Liao Q.D., Feng J.X., Zhang X.Y., Yin T.Y. Differential effects of MTSS1 on invasion and proliferation in subtypes of non-small cell lung cancer cells. Experimental and therapeutic medicine. 2016;12(2):1225–1231. doi: 10.3892/etm.2016.3382. PMid:27446348 PMCid:PMC4950550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okegawa T., Pong R.C., Li Y., Hsieh J.T. The role of cell adhesion molecule in cancer progression and its application in cancer therapy. Acta Biochim. Pol. 2004;51(2):445–457. doi: 10.18388/abp.2004_3583. PMid:15218541. [DOI] [PubMed] [Google Scholar]
- 11.Calaf G.M., Roy D., Narayan G., Balajee A.S. Differential expression of cell adhesion molecules in an ionizing radiation-induced breast cancer model system. Oncol. Rep. 2013;30(1):285–291. doi: 10.3892/or.2013.2448. PMid:23670055. [DOI] [PubMed] [Google Scholar]
- 12.Xi X.W., Chen J.L., Feng Y.J., Wan X.P., Gu K.J. Differential expression of adhesion molecules (CD44, CD29, ICAM-1 and E-cadherin) in ovarian cancer SK-OV-3ip1 cells grown as monolayer and multicellular aggregates. Chin. J. Canc. Res. 2003;15(1):19–23. doi: 10.1007/s11670-003-0005-3. [DOI] [Google Scholar]
- 13.Bujko M., Kober P., Mikula M., Ligaj M., Ostrowski J., Siedlecki J.A. Expression changes of cell cell adhesion related genes in colorectal tumors. Oncology letters. 2015;9(6):2463–2470. doi: 10.3892/ol.2015.3107. PMid:26137091 PMCid:PMC4473523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boelens M.C., van den Berg A., Vogelzang I., Wesseling J., Postma D.S., Timens W., Groen H.J. Differential expression and distribution of epithelial adhesion molecules in non-small cell lung cancer and normal bronchus. J. Clin. Pathol. 2007;60(6):608–614. doi: 10.1136/jcp.2005.031443. PMid:16489176 PMCid:PMC1955047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schardt C., Heymanns J., Schardt C., Rotsch M., Havemann K. Differential expression of the intercellular adhesion molecule-1 (ICAM-1) in lung cancer cell lines of various histological types. Eur. J. Canc. 1993;29(16):2250–2255. doi: 10.1016/0959-8049(93)90217-4. [DOI] [PubMed] [Google Scholar]
- 16.Hase T., Sato M., Yoshida K., Girard L., Takeyama Y., Horio M.…Hasegawa Y. Pivotal role of epithelial cell adhesion molecule in the survival of lung cancer cells. Canc. Sci. 2011;102(8):1493–1500. doi: 10.1111/j.1349-7006.2011.01973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu Y., Tian X. Analysis of genes associated with prognosis of lung adenocarcinoma based on GEO and TCGA databases. Medicine. 2020;99(19) doi: 10.1097/MD.0000000000020183. PMid:32384511 PMCid:PMC7220259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thirion B., Pereira S., Neveol A., Dahamna B., Darmoni S. AMIA... Annual Symposium Proceedings. AMIA Symposium. 2007. October). French MeSH Browser: a cross-language tool to access MEDLINE/PubMed. 1132-1132. [PubMed] [Google Scholar]
- 19.Janiszewska M., Primi M.C., Izard T. Cell adhesion in cancer: beyond the migration of single cells. J. Biol. Chem. 2020;295(8):2495–2505. doi: 10.1074/jbc.REV119.007759. PMid:31937589 PMCid:PMC7039572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lodish H., Berk A., Zipursky S.L., Matsudaira P., Baltimore D., Darnell J. Molecular Cell Biology. fourth ed. WH Freeman; 2000. Cell-cell adhesion and communication. [Google Scholar]
- 21.Huang L., Yang Y., Yang F., Liu S., Zhu Z., Lei Z., Guo J. Functions of EpCAM in physiological processes and diseases. Int. J. Mol. Med. 2018;42(4):1771–1785. doi: 10.3892/ijmm.2018.3764. PMid:30015855 PMCid:PMC6108866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herlyn M., Steplewski Z., Herlyn D., Koprowski H. Colorectal carcinoma-specific antigen: detection by means of monoclonal antibodies. Proc. Natl. Acad. Sci. Unit. States Am. 1979;76(3):1438–1442. doi: 10.1073/pnas.76.3.1438. PMid:286328 PMCid:PMC383267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schnell U., Cirulli V., Giepmans B.N. EpCAM: structure and function in health and disease. Biochim. Biophys. Acta Biomembr. 2013;1828(8):1989–2001. doi: 10.1016/j.bbamem.2013.04.018. PMid:23618806. [DOI] [PubMed] [Google Scholar]
- 24.Kamimoto K., Kaneko K., Kok C.Y.Y., Okada H., Miyajima A., Itoh T. Heterogeneity and stochastic growth regulation of biliary epithelial cells dictate dynamic epithelial tissue remodeling. Elife. 2016;5 doi: 10.7554/eLife.15034.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmelzer E., Zhang L., Bruce A., Wauthier E., Ludlow J., Yao H.L.…Reid L.M. Human hepatic stem cells from fetal and postnatal donors. J. Exp. Med. 2007;204(8):1973–1987. doi: 10.1084/jem.20061603. PMid:17664288 PMCid:PMC2118675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trzpis M., McLaughlin P.M., de Leij L.M., Harmsen M.C. Epithelial cell adhesion molecule: more than a carcinoma marker and adhesion molecule. Am. J. Pathol. 2007;171(2):386–395. doi: 10.2353/ajpath.2007.070152. PMid:17600130 PMCid:PMC1934518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agace W.W., Higgins J.M., Sadasivan B., Brenner M.B., Parker C.M. T-lymphocyte-epithelial-cell interactions: integrin αE (CD103) β7, LEEP-CAM and chemokines. Curr. Opin. Cell Biol. 2000;12(5):563–568. doi: 10.1016/S0955-0674(00)00132-0. [DOI] [PubMed] [Google Scholar]
- 28.Moldenhauer G., Salnikov A.V., Lüttgau S., Herr I., Anderl J., Faulstich H. Therapeutic potential of amanitin-conjugated anti-epithelial cell adhesion molecule monoclonal antibody against pancreatic carcinoma. J. Natl. Cancer Inst. 2012;104(8):622–634. doi: 10.1093/jnci/djs140. PMid:22457476. [DOI] [PubMed] [Google Scholar]
- 29.Armstrong A., Eck S.L. EpCAM: a new therapeutic target for an old cancer antigen. Canc. Biol. Ther. 2003;2(4):320–326. doi: 10.4161/cbt.2.4.451. PMid:14508099. [DOI] [PubMed] [Google Scholar]
- 30.Han S., Zong S., Shi Q., Li H., Liu S., Yang W.…Hou F. Is Ep-CAM expression a diagnostic and prognostic biomarker for colorectal cancer? A systematic meta-analysis. EBioMedicine. 2017;20:61–69. doi: 10.1016/j.ebiom.2017.05.025. PMid:28558958 PMCid:PMC5478257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spizzo G., Gastl G., Obrist P., Went P., Dirnhofer S., Bischoff S.…Dietrich H. High Ep-CAM expression is associated with poor prognosis in node-positive breast cancer. Breast Canc. Res. Treat. 2004;86(3):207–213. doi: 10.1023/B:BREA.0000036787.59816.01. PMid:15567937. [DOI] [PubMed] [Google Scholar]
- 32.Spizzo G., Went P., Dirnhofer S., Obrist P., Moch H., Baeuerle P.A.…Zeimet A.G. Overexpression of epithelial cell adhesion molecule (Ep-CAM) is an independent prognostic marker for reduced survival of patients with epithelial ovarian cancer. Gynecol. Oncol. 2006;103(2):483–488. doi: 10.1016/j.ygyno.2006.03.035. PMid:16678891. [DOI] [PubMed] [Google Scholar]
- 33.Pak M.G., Shin D.H., Lee C.H., Lee M.K. Significance of EpCAM and TROP2 expression in non-small cell lung cancer. World J. Surg. Oncol. 2012 Apr 6;10:53. doi: 10.1186/1477-7819-10-53. PMid:22482828 PMCid:PMC3342122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seligson D.B., Pantuck A.J., Liu X., Huang Y., Horvath S., Bui M.H.…Figlin R.A. Epithelial cell adhesion molecule (KSA) expression: pathobiology and its role as an independent predictor of survival in renal cell carcinoma. Clin. Canc. Res. 2004;10(8):2659–2669. doi: 10.1158/1078-0432.CCR-1132-03. PMid:15102668. [DOI] [PubMed] [Google Scholar]
- 35.Crvenkova S. Prognostic factors and survival in NSCLC patients treated with chemoradiotherapy. Open access Macedonian journal of medical sciences. 2015;3(1):75. doi: 10.3889/oamjms.2015.003. PMid:27275200 PMCid:PMC4877793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lemjabbar-Alaoui H., Hassan O.U., Yang Y.W., Buchanan P. Lung cancer: biology and treatment options. Biochim. Biophys. Acta Rev. Canc. 2015;1856(2):189–210. doi: 10.1016/j.bbcan.2015.08.002. PMid:26297204 PMCid:PMC4663145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma Y., Fan M., Dai L., Kang X., Liu Y., Sun Y., Chen K. The expression of TTF-1 and Napsin A in early-stage lung adenocarcinoma correlates with the results of surgical treatment. Tumor Biol. 2015;36(10):8085–8092. doi: 10.1007/s13277-015-3478-z. [DOI] [PubMed] [Google Scholar]
- 38.Rhodes D.R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pandey A., Chinnaiyan A.M. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang Z., Kang B., Li C., Chen T., Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019 doi: 10.1093/nar/gkz430. Jul 2;47(W1):W556-60. PMid:31114875 PMCid:PMC6602440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park S.J., Yoon B.H., Kim S.K., Kim S.Y. GENT2: an updated gene expression database for normal and tumor tissues. BMC Med. Genom. 2019 Jul;12(5):101. doi: 10.1186/s12920-019-0514-7. PMid:31296229 PMCid:PMC6624177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chandrashekar D.S., Bashel B., Balasubramanya S.A., Creighton C.J., Ponce-Rodriguez I., Chakravarthi B.V., Varambally S. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017 Aug 1;19(8):649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., Olsson I. Tissue-based map of the human proteome. Science. 2015 Jan 23;(6220):347. doi: 10.1126/science.1260419. PMid:25613900. [DOI] [PubMed] [Google Scholar]
- 43.Goldman M.J., Craft B., Hastie M., Repečka K., McDade F., Kamath A., Banerjee A., Luo Y., Rogers D., Brooks A.N., Zhu J. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020 May 22:1–4. doi: 10.1038/s41587-020-0546-8. PMid:32444850 PMCid:PMC7386072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., Antipin Y. 2012. The cBio Cancer Genomics Portal: an Open Platform for Exploring Multidimensional Cancer Genomics Data. PMid:22588877 PMCid:PMC3956037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mizuno H., Kitada K., Nakai K., Sarai A. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med. Genom. 2009 Dec 1;2(1):18. doi: 10.1186/1755-8794-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma'ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013 Dec 1;14(1):128. doi: 10.1186/1471-2105-14-128. PMid:23586463 PMCid:PMC3637064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., McDermott M.G. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016 Jul 8;44(W1) doi: 10.1093/nar/gkw377. W90-7. PMid:27141961 PMCid:PMC4987924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phillips T. The role of methylation in gene expression. Nature Education. 2008;1(1):116. [Google Scholar]
- 49.Cruz C.S.D., Tanoue L.T., Matthay R.A. Lung cancer: epidemiology, etiology, and prevention. Clin. Chest Med. 2011;32(4):605–644. doi: 10.1016/j.ccm.2011.09.001. PMid:22054876 PMCid:PMC3864624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Y., Wang M., Liu B. Exploring and comparing of the gene expression and methylation differences between lung adenocarcinoma and squamous cell carcinoma. J. Cell. Physiol. 2019;234(4):4454–4459. doi: 10.1002/jcp.27240. PMid:30317601. [DOI] [PubMed] [Google Scholar]
- 51.Bernicker E.H., Allen T.C., Cagle P.T. Update on emerging biomarkers in lung cancer. J. Thorac. Dis. 2019;11(Suppl 1):S81. doi: 10.21037/jtd.2019.01.46. PMid:30775031 PMCid:PMC6353743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moons K.G.M., Altman D.G., Vergouwe Y., Royston P. Prognosis and prognostic research: application and impact of prognostic models in clinical practice. BMJ (Clinical research ed.) 2009;338 doi: 10.1136/bmj.b606. b606, b606. PMid:19502216. [DOI] [PubMed] [Google Scholar]
- 53.Walters S. Lung cancer survival and stage at diagnosis in Australia, Canada, Denmark, Norway, Sweden and the UK: a population-based study, 2004-2007. Thorax. 2013;68:551–564. doi: 10.1136/thoraxjnl-2012-202297. PMid:23399908. [DOI] [PubMed] [Google Scholar]
- 54.Goldstraw P. The IASLC lung cancer staging project: proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the TNM classification for lung cancer. J. Thorac. Oncol. 2016;11:39–51. doi: 10.1016/j.jtho.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 55.Nesbitt J.C., Putnam J.B., Jr., Walsh G.L., Roth J.A., Mountain C.F. Survival in early-stage non-small cell lung cancer. Ann. Thorac. Surg. 1995;60:466–472. doi: 10.1016/0003-4975(95)00169-L. [DOI] [PubMed] [Google Scholar]
- 56.Shah R., Sabanathan S., Richardson J., Mearns A.J., Goulden C. Results of surgical treatment of stage I and II lung cancer. J. Cardiovasc. Surg. 1996;37:169–172. [PubMed] [Google Scholar]
- 57.Bradley S.H., Kennedy M.P., Neal R.D. Recognising lung cancer in primary care. Adv. Ther. 2019;36(1):19–30. doi: 10.1007/s12325-018-0843-5. PMid:30499068 PMCid:PMC6318240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laurinavicius A., Laurinaviciene A., Ostapenko V., Dasevicius D., Jarmalaite S., Lazutka J. Immunohistochemistry profiles of breast ductal carcinoma: factor analysis of digital image analysis data. Diagn. Pathol. 2012;7 doi: 10.1186/1746-1596-7-27. 27, 27. PMid:22424533 PMCid:PMC3319425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu L., Xiao Z., Tu H., Tong B., Chen S. The expression and prognostic significance of Drp1 in lung cancer: a bioinformatics analysis and immunohistochemistry. Medicine. 2019;98(48) doi: 10.1097/MD.0000000000018228. e18228, e18228. PMid:31770286 PMCid:PMC6890372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klonowska K., Czubak K., Wojciechowska M., Handschuh L., Zmienko A., Figlerowicz M., Dams-Kozlowska H., Kozlowski P. Oncogenomic portals for the visualization and analysis of genome-wide cancer data. Oncotarget. 2016;7(1):176–192. doi: 10.18632/oncotarget.6128. PMid:26484415 PMCid:PMC4807991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keller L., Werner S., Pantel K. Biology and clinical relevance of EpCAM. Cell Stress. 2019;3(6):165. doi: 10.15698/cst2019.06.188. PMid:31225512 PMCid:PMC6558934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yuan W., Zhang X., Xu Y., Li S., Hu Y., Wu S. Role of HOXB7 in regulation of progression and metastasis of human lung adenocarcinoma. Mol. Carcinog. 2014;53(1):49–57. doi: 10.1002/mc.21947. PMid:22911672. [DOI] [PubMed] [Google Scholar]
- 63.Vazquez A., Kamphorst J.J., Markert E.K. Cancer metabolism at a glance. J. Cell Sci. 2016;129:3367–3373. doi: 10.1242/jcs.181016. PMid:27635066 PMCid:PMC6518336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ananieva E. Targeting amino acid metabolism in cancer growth and antitumor immune response. World J. Biol. Chem. 2015;6:281–289. doi: 10.4331/wjbc.v6.i4.281. PMid:26629311 PMCid:PMC4657121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.DeBerardinis R.J., Chandel N.S. Fundamentals of cancer metabolism. Sci Adv. 2016;2 doi: 10.1126/sciadv.1600200. [PMC free article] [PubMed] [Google Scholar]The recent review by DeBerardinis and Chandel provides a useful overview of the role of metabolism in cancer. PMid:27386546 PMCid:PMC4928883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Briones T.L., Woods J. Dysregulation in myelination mediated by persistent neuroinflammation: possible mechanisms in chemotherapy-related cognitive impairment. Brain Behav. Immun. 2014;35:23–32. doi: 10.1016/j.bbi.2013.07.175. PMid:23916895 PMCid:PMC3858476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Murai T. The role of lipid rafts in cancer cell adhesion and migration. International journal of cell biology. 2012 doi: 10.1155/2012/763283. PMid:22253629 PMCid:PMC3255102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Authors made all the data generated during experiment and analysis available within the manuscript.