Abstract
Prostate cancer resistance to next-generation hormonal treatment with enzalutamide is a major problem and eventuates into disease lethality. Biologically active glucocorticoids that stimulate glucocorticoid receptor (GR) have an 11β-OH moiety and resistant tumors exhibit loss of 11β-HSD2, the oxidative (11β-OH → 11-keto) enzyme that normally inactivates glucocorticoids, allowing elevated tumor glucocorticoids to drive resistance by stimulating GR. Here, we show up-regulation of hexose-6-phosphate dehydrogenase (H6PD) protein occurs in prostate cancer tissues of men treated with enzalutamide, human-derived cell lines, and patient-derived prostate tissues treated ex vivo with enzalutamide. Genetically silencing H6PD blocks NADPH generation, which inhibits the usual reductive directionality of 11β-HSD1, to effectively replace 11β-HSD2 function in human-derived cell line models, suppress the concentration of biologically active glucocorticoids in prostate cancer and reverse enzalutamide resistance in mouse xenograft models. Similarly, pharmacologic blockade of H6PD with rucaparib normalizes tumor glucocorticoid metabolism in human cell lines and reinstates responsiveness to enzalutamide in mouse xenograft models. Our data show that blockade of H6PD, which is essential for glucocorticoid synthesis in humans, normalizes glucocorticoid metabolism and reverses enzalutamide resistance in mouse xenograft models. We credential H6PD as a pharmacologic vulnerability for treatment of next-generation androgen receptor antagonist-resistant prostate cancer by depleting tumor glucocorticoids.
One Sentence Summary:
Inhibiting H6PD restores prostate cancer response to AR antagonists by normalizing glucocorticoid metabolism in mouse xenograft models.
INTRODUCTION
Next-generation androgen receptor (AR) antagonists such as enzalutamide (enz), that potently suppress the androgen axis, have transformed modern therapy for advanced prostate cancer (1–3). However, pathways that bypass AR have emerged as major mechanisms of enz resistance. The glucocorticoid signaling axis has been implicated in the resistance mechanism. Evidence includes tumor up-regulation of glucocorticoid receptor (GR) expression (4, 5), along with concurrent loss of 11β-hydroxysteroid dehydrogenase-2 (11β-HSD2) protein (6), which is the major enzyme that inactivates biologically active glucocorticoids (cortisol in human and corticosterone in mouse). Systemic loss of 11β-HSD2 biochemical activity is also evident in multiple clinical trials of enz and apalutamide, including, for example, biochemically and clinically phenocopying germline loss-of-function mutations in 11β-HSD2 (7, 8). Together, these data suggest that an adaptation in glucocorticoid metabolism may occur under conditions of enzalutamide treatment.
Oxidative inactivation of (11β-OH) cortisol by 11β-HSD2 to (11-keto) cortisone is directionally opposed by 11β-HSD1, the alternative isoenzyme which is usually reductive and NADPH dependent (fig. S1). Loss of 11β-HSD2 expression, as occurs in enz-resistant prostate cancer, shifts the balance toward 11β-HSD1 and the reductive direction without any change in 11β-HSD1 expression, increasing cortisol and engendering enz resistance (6). Located inside the inner leaflet of the endoplasmic reticulum, 11β-HSD1 interacts with hexose-6-phosphate dehydrogenase (H6PD), the major generator of endoplasmic reticulum NADPH, which in turn drives 11β-reduction. Loss of H6PD reverses the reaction so that 11β-HSD1 instead drives 11β-oxidation (9–12). Therefore, we hypothesized that blockade of H6PD, which supplies NADPH for 11β-HSD1, could restore the oxidative directionality of glucocorticoid metabolism, normalize tumor glucocorticoid concentrations and reverse enz resistance.
RESULTS
H6PD up-regulation occurs with antiandrogen resistance and enables a metabolic shift toward the active glucocorticoid, cortisol
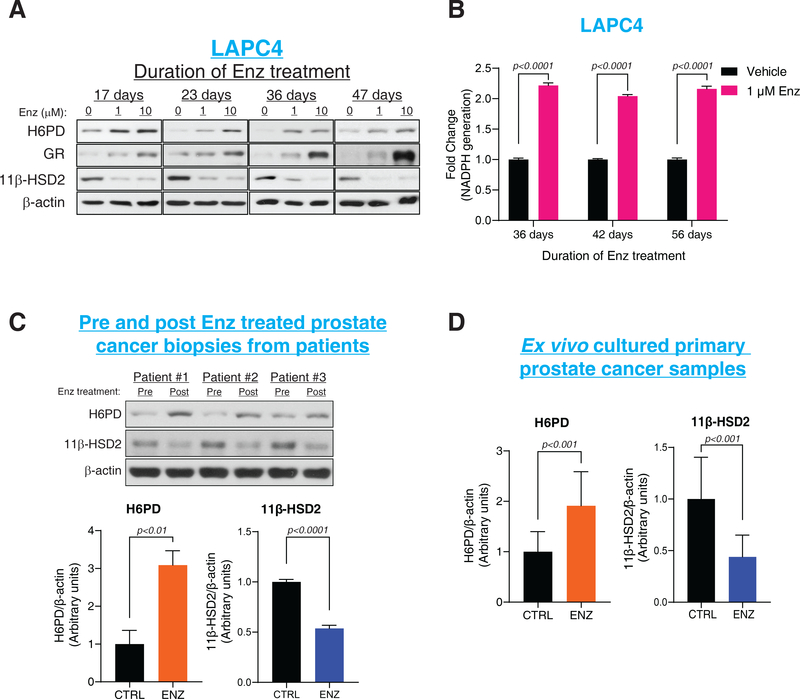
To determine the effects of enzalutamide on H6PD, we treated human prostate cancer cell line models and obtained tissues from men with prostate cancer. H6PD protein (but not transcript) was increased with Enz treatment in vitro in 3 human cell line models (LAPC4, VCaP and MDA-Pca-2b) of castration-resistant prostate cancer (CRPC) (Fig. 1A and fig. S2AȓE). Sustained H6PD protein expression required continuous enz treatment (fig. S2F) and led to greater than a 2-fold increase in NADPH generation (p<0.0001) in the endoplasmic reticulum, where glucocorticoid-metabolizing 11β-HSD isoenzymes reside (Fig. 1B). Compared with pre-treatment patient tissues, an increase in H6PD was also detectable post-treatment in men with prostate cancer treated with enz and prostate tissues treated with enz ex vivo (Fig. 1C–D and fig. S2G). Glucocorticoid metabolic alterations that sustain elevated tumor cortisol are also commonly induced by other next-generation AR antagonists, such as apalutamide (fig. S3AȓC).
Figure 1. H6PD and regulation of glucocorticoid metabolism with enzalutamide resistance..
(A) Effects of Enz treatment on H6PD, GR and 11β-HSD2 in the LAPC4 CRPC model as assessed by immunoblot. (B) NADPH generation in LAPC4 cells with enz treatment. NADPH was assessed using a bioluminescence assay in endoplasmic reticulum extracted from control and enz-treated cells. (C) H6PD and 11β-HSD2 protein in tissues from enz-treated patients with prostate cancer. Bar graphs represent band intensity; 3 paired tissues from 3 patients with prostate cancer were analyzed. (D) Effects of enz on H6PD and 11β-HSD2 protein with ex vivo treatment of 12 clinical tissues from prostate cancer patients. For all panels unless otherwise noted, error bars represent the SEM, and p values were calculated using unpaired 2-tailed t-tests.
H6PD regulates NADPH generation, active glucocorticoid synthesis and glucocorticoid receptor (GR) signaling in enzalutamide-resistant prostate cancer models
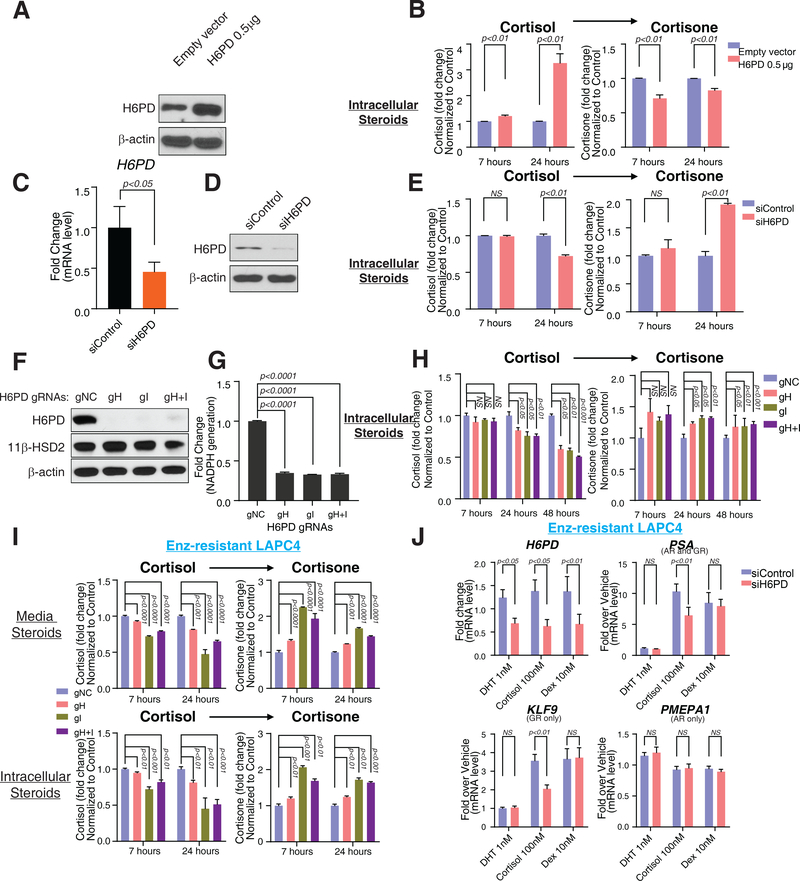
The existence of humans with germline loss-of-function mutations in H6PD, which impairs cortisol activation and causes apparent cortisone reductase deficiency, concomitantly demonstrates the essentiality of H6PD for the reductive directionality of 11β-HSD1 and that absence of H6PD is compatible with survival to adulthood, although endocrine anomalies including ACTH up-regulation do occur (13, 14). In prostate cancer, reversal of sustained cortisol in enz resistance with H6PD loss might be expected to reverse drug resistance (fig. S1). Similar to increased endogenous H6PD seen with enz treatment, forced expression of H6PD in vitro allows cortisol to persist by retarding conversion to cortisone (Fig. 2A–B and fig. S4A). In contrast, transient or stable H6PD loss hastens cellular conversion from cortisol to cortisone (Fig. 2C–H and fig. S4BȓC, fig. S5AȓB). Moreover, as the enzyme which activates glucocorticoid with NAPDH provided by H6PD, 11β-HSD1 expression shifts the balance toward cortisol metabolism and 11β-HSD1 knockdown reverses the favored reductive directionality (fig. S6). Similarly, cortisol inactivation is accelerated by H6PD loss in enz-resistant prostate cancer in vitro (Fig. 2I, fig. S5C) and results in specific reversal of cortisol-induced GR signaling (Fig. 2J, fig. S7).
Figure 2. Requirement of H6PD for NADPH generation, glucocorticoid metabolism and glucocorticoid receptor (GR) signaling.
(A, B) H6PD overexpression and conversion from cortisol to cortisone in LAPC4 cells. Cells were transfected with 0.5 μg H6PD expression plasmid, and subsequently treated with [3H]-cortisol (100 nM) for the indicated times, followed by steroid extraction from cells, steroid separation and quantitation with HPLC. (C, D and E) Cortisol inactivation and siRNA knockdown of H6PD in LAPC4. Cells were transfected with H6PD targeted siRNA smart-pool for 48 hours, followed by treatment with [3H]-cortisol (100 nM) for the indicated times; then steroid was extracted from cells and separated and quantitated with HPLC. (F, G and H) H6PD CRISPR/Cas9 KO and cortisol-to-cortisone conversion in LAPC4. Cells were infected with a lentivirus containing H6PD guide RNAs (gRNAs), followed by selection with puromycin. (I) Effects of H6PD loss on enz-impeded conversion from cortisol to cortisone. (J) Cortisol-induced GR signaling and reversibility with siRNA-mediated H6PD knockdown. Dexamethasone (dex), is used as a control that is impervious to 11β-HSD metabolism. LAPC4 cells were treated with Enz for 36 days, starved with phenol-red-free medium containing 5% charcoal:dextran-stripped FBS for 48 hours and transfected with a H6PD targeted siRNA smart-pool and treated with the indicated conditions for 24 hours to test H6PD requirement. Induction of PSA expression, is GR- and metabolism-dependent. Expression of KLF9, is regulated only by GR. Expression of PMEPA1, is regulated only by AR, is induced with dihydrotestosterone (DHT) only. Expression is normalized to vehicle-treated cells (not shown) and RPLP0 expression. For all panels unless otherwise noted, error bars represent the SEM; p values were calculated using unpaired 2-tailed t-tests.
Genetic loss of H6PD reinstates enzalutamide responsiveness by specifically suppressing tumor corticosterone in mouse xenograft models
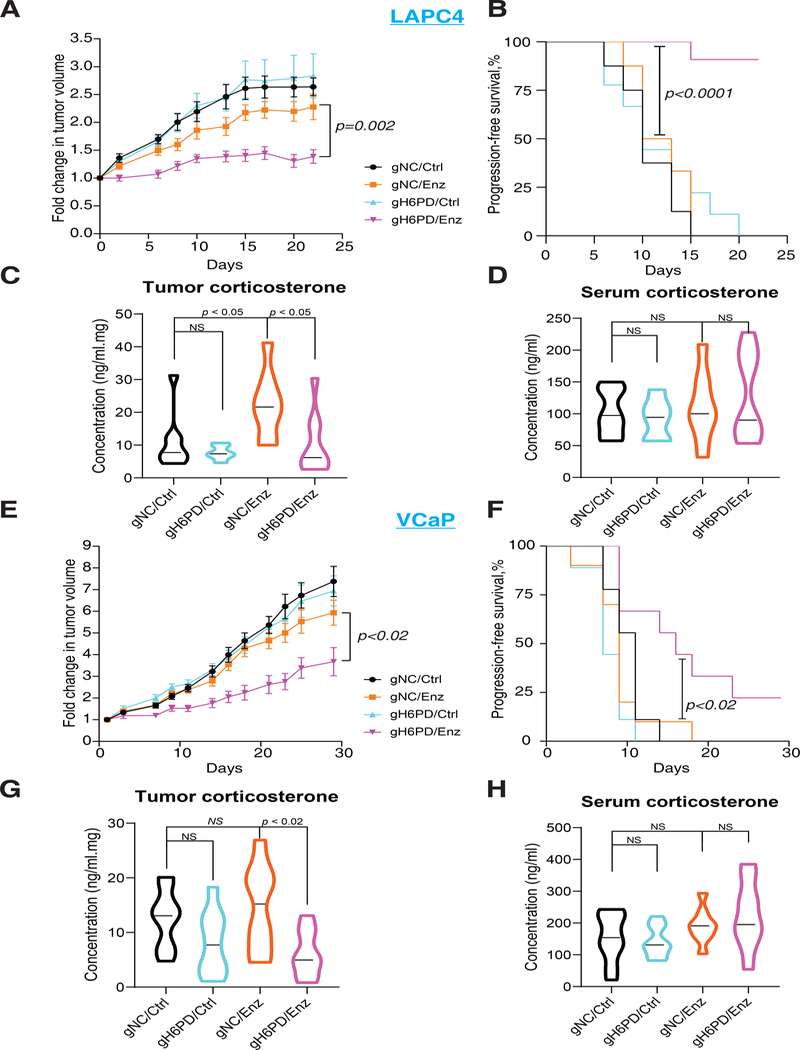
To determine the necessity of H6PD for enz resistance in vivo, we conducted mouse xenograft studies in 2 models with and without H6PD knockout. The absence of H6PD expression specifically and significantly increased the inhibitory effect of enz on xenograft tumor growth in both LAPC4 and VCaP models, as assessed by changes in tumor volume (p=0.002 and p<0.02) and progression-free survival (p<0.0001 and p<0.02) (Fig. 3A–B and 3E–F). In contrast, H6PD loss has no effect on tumor growth or progression in the absence of enz treatment, thus showing that H6PD loss by itself does not have non-specific toxic effects. We next assessed the in vivo effect of H6PD blockade on mouse glucocorticoids (corticosterone) in enz-treated xenografts. In both models, H6PD loss significantly suppressed corticosterone concentrations (p=0.02 for LAPC4 and p<0.02 for VCaP) as assessed by mass spectrometry (Fig. 3C and G). In contrast, there was no effect on serum corticosterone (Fig. 3D and H). Loss of H6PD expression (fig. S8A and C) and reversal of enz-induced glucocorticoid metabolism by H6PD loss with ex vivo radiolabeled steroid studies were confirmed in xenografts (fig. S8B).
Figure 3. Effects of H6PD loss on sensitivity to enzalutamide therapy and tumor corticosterone concentrations in mouse xenograft models.
(A and E) H6PD loss and the inhibitory effect of enz on A, LAPC4 and E, VCaP xenograft tumor growth. (B and F) H6PD loss and progression-free survival in enz-treated B, LAPC4 and F, VCaP xenografts. For both xenograft studies, cells with H6PD gRNA (gH6PD) or control gRNA (gNC) were grown in orchiectomized mice supplemented with DHEA and arbitrarily assigned to enz or chow (Ctrl). Tumor volume was compared between gH6PD/Enz and gNC/Enz with an unpaired two-tailed t-test on day 20 (LAPC4) or day 29 (VCaP). Progression-free survival was compared between gH6PD/Enz and gNC/Enz with a log-rank test. The number of mice in the LAPC4 gNC/Ctrl, gNC/Enz, gH6PD/Ctrl and gH6PD/Enz groups were 8, 8, 9, and 11, respectively. The number of mice in the VCaP gNC/Ctrl, gNC/Enz, gH6PD/Ctrl and gH6PD/Enz groups were 9, 10, 9, and 9, respectively. (C and G) Effects of H6PD knockout with and without enz treatment on the absolute concentration of corticosterone (active glucocorticoid in mice) in C, LAPC4 and G, VCaP xenograft tumors. (D and H) The absolute concentration of corticosterone in sera by treatment group, with and without H6PD loss in mice with D, LAPC4 and H, VCaP xenografts. For all panels unless otherwise noted, error bars represent the SEM; p values were calculated using unpaired 2-tailed t tests.
Pharmacologic blockade of H6PD suppresses active tumor glucocorticoids and restores enzalutamide responsiveness
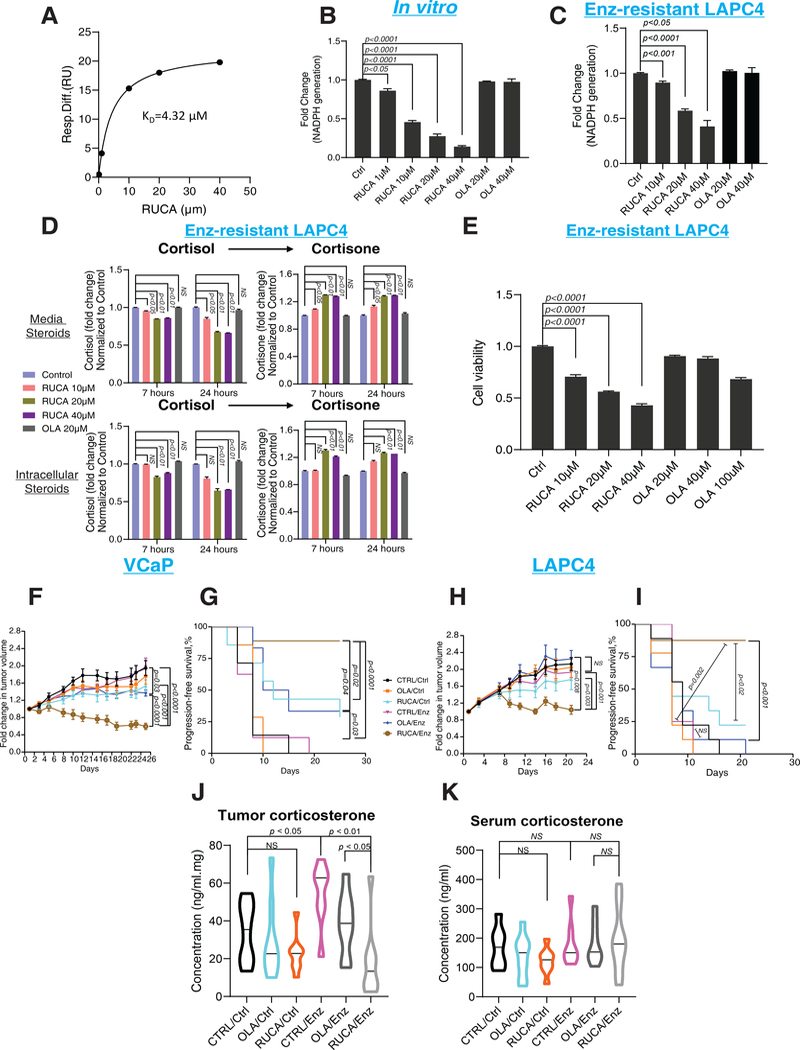
We next wished to determine whether H6PD pharmacologic blockade would similarly ablate sustained tumor glucocorticoids and reverse enz resistance. To our knowledge, no drug has been developed for the specific purpose of H6PD inhibition. However, of four PARP inhibitors, off-target H6PD inhibition was specifically identified only with rucaparib (15). We confirmed that rucaparib but not olaparib (another PARP inhibitor) specifically bound H6PD (Fig. 4A and fig. S9), rucaparib but not olaparib inhibited H6PD activity and neither inhibited G6PD activity (fig. S10). Additionally, we determined that rucaparib inhibited H6PD generation of NADPH in vitro and in enz-resistant prostate cancer (Fig 4B–C). Pharmacologic blockade of H6PD by rucaparib and inhibition of endoplasmic reticulum NADPH synthesis restored the glucocorticoid metabolic balance toward the oxidative directionality of inactive 11-keto glucocorticoids (Fig 4D), which in turn limited cell viability in vitro (Fig 4E). H6PD overexpression rescued the loss of enz-resistant LAPC4 cell viability induced by rucaparib, further confirming an H6PD-dependent mechanism for rucaparib (fig S11). Chemical modifications of the rucaparib azepinone indole ring system were conducted to understand the basic H6PD pharmacophore units and how they contrasted and compared to published PARP structure-activity-relationships. Our initial efforts led to the synthesis of compounds 2–5 (fig S12A). In general, changes within the indole core led to steep losses in activity as both the des-fluoro compound (2) and the benzimidazole (3) resulted in a greater than 20x loss of H6PD inhibitory potency. Capping the rucaparib lactam NH resulting in methyl congener 4 led to a modest ~10x loss of inhibition. Modification of the terminal aminomethyl to the primary hydroxyl analog 5 was reasonably well tolerated with an H6PD IC50 of 27 μM. The structural changes within compounds 2–4, which are known molecules studied in the PARP field, (16, 17) were well tolerated as PARP-1 inhibitors; however, the hydroxyl group modification in compound 5 (16) led to a compound with poor PARP inhibition activity, as a basic amine is typically required in this position of the rucaparib scaffold for good PARP inhibition. Overall, these data suggest that most of the basic pharmacophore features found in rucaparib are essential to inhibit H6PD enzyme activity (fig S12B).
Figure 4. Pharmacologic inhibition of H6PD and effects on enzalutamide resistance.
(A) A steady-state affinity model for rucaparib and H6PD was obtained by surface plasmon resonance (SPR). Purified H6PD protein was covalently immobilized, rucaparib was injected, and the data were fit into a steady-state affinity model. (B) Effects of rucaparib andolaparib on H6PD production by NADPH in vitro. Rucaparib or olaparib was added to a reaction cocktail with H6PD protein, D-glucosamine 6-phosphate and NADP, and after 1 hour incubation, NADPH production was measured using Glo Detection Reagent. (C) Effects of rucaparib or olaparib on H6PD activity in enz-resistant LAPC4 cells. Cells were treated with rucaparib for 24 hours, then microsomal NADPH was measured. (D) Cortisol metabolism in enz-resistant LAPC4 cells treated with rucaparib or olaparib. Cells were pretreated with rucaparib or olaparib for 1 hour, then [3H]-cortisol (100 nM) was added; the steroids were extracted, separated and quantitated by HPLC after the indicated incubation times. (E) Viability of enz-resistant LAPC4 cells treated with rucaparib. Cells were treated with 100 nM cortisol with 10 μM enz combined with the indicated drugs for 5 days and assayed using CellTiter-Glo. Viability is normalized to Ctrl. (F and H) Rucaparib and the inhibitory effect of enz on xenograft growth in the F, VCaP and H, LAPC4 mouse xenograft models. (G and I) Effects of rucaparib on progression-free survival in enz-treated xenograft models. G, VCaP or I, LAPC4 xenografts were grown in orchiectomized mice supplemented with DHEA and arbitrarily divided among 3 groups each for control or enz diet: control, olaparib or rucaparib. Tumor volume was compared between CTRL/Enz and RUCA/Enz or between RUCA/Ctrl and RUCA/Enz with an unpaired two-tailed t-test on day 21 (LAPC4) or 25 (VCaP). To compare progression-free survival, the difference between CTRL/Enz and RUCA/Enz or between RUCA/Ctrl and RUCA/Enz was calculated with a log-rank test. The number of mice in the LAPC4 CTRL/Ctrl, OLA/Ctrl, RUCA/Ctrl, CTRL/Enz, OLA/Enz and RUCA/Enz groups were 9, 9, 9, 8, 9 and 8, respectively. The number of mice in the VCaP CTRL/Ctrl, OLA/Ctrl, RUCA/Ctrl, CTRL/Enz, OLA/Enz and RUCA/Enz groups were 7, 7, 7, 8, 6 and 9, respectively. (J) Rucaparib treatment and the absolute concentration of corticosterone in LAPC4 xenograft tumors and (K) in sera. For all panels unless otherwise noted, error bars represent the SEM; p values were calculated using unpaired 2-tailed t-tests.
Finally, VCaP xenografts were grown and arbitrarily assigned to treatment in 1 of 6 groups: Ctrl, rucaparib alone, olaparib alone, enz alone, rucaparib + enz, olaparib + enz (Fig 4F). Fold change in tumor volume was significantly decreased in rucaparib + enz compared with rucaparib alone (p<0.001) as well as compared with enz alone (p<0.0001). Differences in progression-free survival were similarly significant in these comparisons for rucaparib + enz (p<0.0001) (Fig. 4G). Results for the comparison between rucaparib versus rucaparib + enz were essentially identical in a second xenograft model (all p values <0.02) (Fig. 4H and 4I). Consistent with the H6PD inhibition activity of rucaparib (without suppression of H6PD expression; fig. S13), corticosterone was significantly suppressed in rucaparib + enz treated tumors compared with enz alone (p=0.002) and olaparib + enz (p<0.05) treated xenografts (Fig. 4J). Serum corticosterone did not differ among treatment groups (Fig. 4K).
DISCUSSION
Accumulating evidence implicates a role for GR in driving treatment-resistant prostate cancer, as well as in other steroid-dependent diseases, including breast cancer (18–21). The interpretation of the data on GR as a potential driver has understandably been met with some controversy (22, 23). Some of the debate is attributable to tumor responses that can occur with exogenous glucocorticoid administration, which may appear to be in contradistinction to tumor GR and local glucocorticoid metabolism as driver of treatment resistance (24). However, those responses are probably mainly due to adrenal androgen suppression by exogenous glucocorticoids (25), an indirect effect, which differs from direct effects of glucocorticoids on GR in the tumor (26).
Our data suggest that just as circulating and tumor tissue androgen concentrations are disassociated in prostate cancer, which is particularly apparent in the context of medical castration because of local tumor metabolism (27–29), this circulating/tumor steroid concentration discordance also occurs with tumor glucocorticoids because of local glucocorticoid metabolism (30). In other words, tumor metabolic regulation causes tissue glucocorticoid concentrations to differ from circulating concentrations. Previously, we showed that loss of 11β-HSD2 co-occurs along with GR up-regulation with enzalutamide resistance (6). The loss of 11β-HSD2 results in an increase in tumor tissue glucocorticoids, which is required for GR stimulation and enzalutamide resistance. From a therapeutic standpoint, replacement of 11β-HSD2 expression is challenging. Instead, we exploit sustained tumor expression of the remaining isoenzyme, 11β-HSD1, and modify its preferred directionality (31). The existence of humans with apparent cortisone reductase deficiency, who have loss-of-function mutations in H6PD, validates the requirement of H6PD for reductive 11β-HSD1 directionality, establishes the biochemical requirement of this enzyme for synthesis of biologically active glucocorticoids and demonstrates that the absence of H6PD does not have overtly toxic effects (14). Our data genetically validate H6PD as a metabolic vulnerability of prostate cancer that is resistant to next generation hormonal therapy and demonstrate the feasibility of pharmacologic targeting to reverse drug resistance in human cell lines and mouse xenograft models. Such a strategy targeting H6PD circumvents the requirement for complete and systemic GR antagonism that may be associated with adrenal insufficiency. Humans with genetic loss of H6PD may have androgen excess; however, in the context of prostate cancer treatment with potent AR antagonists, these androgens would be blocked.
Although our major focus here was on enzalutamide resistance, we also found that apalutamide, another potent AR antagonist, induces similar changes in glucocorticoid metabolism. Inhibition of extragonadal androgen synthesis with the CYP17A1 inhibitor abiraterone is yet another widely utilized hormonal therapy for prostate cancer. A neoadjuvant clinical study of abiraterone showed that increased prostate tissue cortisol concentrations accompany GR up-regulation, which is consistent with engagement of a similar resistance mechanism occurring with abiraterone therapy (32). Together, this evidence suggests that H6PD pharmacologic blockade may have a role in reversing resistance to multiple next-generation hormonal therapies for the treatment of prostate cancer.
To our knowledge, the protein structure of H6PD has not been resolved. Therefore, although our structure activity relationship studies of rucaparib identify chemical moieties that are necessary for H6PD inhibition, the precise molecular basis for the interaction between drug and enzyme has yet to be determined. The protein structural basis for the selectivity of rucaparib for H6PD, which provides NADPH in the endoplasmic reticulum, over G6PD which generates NADPH in the cytosol, similarly remains to be resolved.
Limitations of our study include the possibility that there may be some contributing effect of PARP inhibition in the xenograft studies combining rucaparib with enzalutamide. However, the absence of such an effect with olaparib, along with the xenograft studies with genetic H6PD KO together indicate that H6PD inhibition is a critical pharmacologic target. Another limitation is the uncertainty of the relationship between rucaparib concentrations achieved in our mouse xenograft studies which were sufficient to reverse enz resistance compared with those that are clinically achievable in men with prostate cancer. In other words, it is possible that H6PD inhibition with clinically achievable concentrations of rucaparib is insufficient to restore glucocorticoid inactivation. Furthermore, although the existence of humans apparent cortisone reductase deficiency who have H6PD loss-of-function mutations suggests the tolerability of H6PD loss in humans, acute loss with pharmacologic inhibition might have additional adverse effects.
Finally, both rucaparib and olaparib are PARP inhibitors recently FDA-approved for the treatment of metastatic CRPC (33, 34). These clinical indications are specifically for patients who have homologous recombination repair gene mutations and for the PARP inhibitor to be given singly. Although this was not the primary purpose of our study, our results raise distinguishing features between these drugs that can be rapidly translated and tested in the clinic in combination with next-generation hormonal therapies, including enzalutamide and apalutamide. An ongoing phase Ib clinical trial combining rucaparib with either enzalutamide or abiraterone will assess the safety and tolerability of these combinations (ClinicalTrials.gov #NCT04179396). However, any clinical benefit associated with this combination will probably have to be assessed in a subsequent clinical study with a comparator arm.
In conclusion, H6PD blockade reverses enzalutamide resistance by normalizing glucocorticoid metabolism in human cell lines and mouse xenograft models of prostate cancer. Rucaparib is a modest H6PD inhibitor that might be exploited for this effect clinically. Clinical studies are necessary to determine the effects of rucaparib on tumor glucocorticoid metabolism and enzalutamide responsiveness in men with prostate cancer. However, more potent H6PD inhibitors are probably necessary.
MATERIALS AND METHODS
Study Design
In general, all studies were predesigned to test the hypothesis that H6PD loss or inhibition reinstates glucocorticoid metabolism and downstream effects including reversal of drug resistance. Mouse xenograft studies were designed with sample size estimates based on tumor take rate and effects on tumor growth using genetic or pharmacologic inhibitors of steroidogenesis (35–38). Xenograft studies were performed with predefined endpoints and two experimenters such that the tumor measurements were done by a person blinded to treatment group. Mice were arbitrarily (not by formal randomization) assigned to treatment groups in drug treatment studies. The number of replicates is defined for each study individually. No outliers were excluded in any studies.
Cell lines, chemicals and plasmids
LAPC4 cells were a generous gift from Dr. Charles Sawyers (Memorial Sloan Kettering Cancer, New York, NY) and were maintained in Iscove’s modified Dulbecco’s medium (IMDM) with 10% fetal bovine serum (Gemini) and 1% pen-strep (Gibco). VCaP cells were purchased from American Type Culture Collection (ATCC) and cultured in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% fetal bovine serum and 1% pen-strep. MDA-PCa-2b cells were purchased from ATCC and cultured in BRFF-HPC1 (Athena ES) containing 20% fetal bovine serum and 1% pen-strep.
Control gRNA (5’-ATCTGCCATGGCGTCCTGGC-3’) and H6PD gRNAs (gH: 5’-CAGAGGGGCAGTGCCGTCGT-3’ and gI: 5’-GGGTGAGGACCTCCGTCAGA-3’) were inserted into a CRISPR plasmid backbone, lentiCRISPR v2, [a generous gift from Dr. Feng Zhang (Addgene Plasmid #52961)] according to the protocol they provided (39). Then these constructs were used to generate the H6PD knock out LAPC4 and VCaP stable cell lines by using a lentiviral system. The viral packaging and infection were performed as previously described (6, 37). Briefly, 293T cells (ATCC) were cotransfected with 10 μg each of constructed plasmid (containing gRNAs), pMD2.G, and psPAX2 vector for 48 hours to package the virus. Then the virus was concentrated by using PEG-it Virus Precipitation Solution (System Biosciences) according to the provided protocol. Next, LAPC4 and VCaP cells were infected with the concentrated virus for 24 hours with addition of polybrene (10 μg/ml), followed by selection with 2 μg/ml puromycin for ~2 weeks. Enzalutamide (Enz) was obtained courtesy of Medivation; rucaparib and olaparib were purchased from MedChemExpress. All Enz- and vehicle-treated cells were maintained in medium containing 10 nM DHEA. Cell lines were authenticated by DDC Medical and routinely screened for mycoplasma contamination using primers 5’-ACACCATGGGAGCTGGTAAT-3’ and 5’-GTTCATCGACTTTCAGACCCAAGGCAT-3’ (3, 4).
Gene expression and immunoblot
Total RNA was extracted with a GenElute Mammalian Total RNA miniprep kit (Sigma-Aldrich) and 1μg RNA was reverse-transcribed to cDNA with the iScript cDNA Synthesis Kit (Bio-Rad). An ABI 7500 Real-Time PCR instrument (Applied Biosystems) was used to perform the qPCR analysis, using iTaq Fast SYBR Green Supermix with ROX (Bio-Rad) in 96-well plates at a final reaction volume of 10 μl. The qPCR analysis was carried out in triplicate with the following primer sets:
H6PD
Forward: 5’-CTGCAGCACGTCCGGATCCC-3’;
Reverse: 5’- TGGCGCGGTTGATGAGAGGC-3’
PSA
Forward: 5’-GCATGGGATGGGGATGAAGTAAG-3’;
Reverse: 5’-CATCAAATCTGAGGGTTGTCTGGA-3’
KLK4
Forward: 5’-GGAACTCTTGCCTCGTTTCTGG-3’;
Reverse: 5’-AGCGGGTCATAGAGCTTACTGC-3’
FKBP5
Forward: 5’-CCCCCTGGTGAACCATAATACA-3’;
Reverse: 5’-AAAAGGCCACCTAGCTTTTTGC-3’)
KLF9
Forward: 5’-AACACGCCTCCGAAAAGAGG-3’;
Reverse: 5’- CGTCTGAGCGGGAGAACTTT-3’
PPAP2A
Forward: 5’-TGGAGCGATGTGTTGACTGGAC-3’;
Reverse: 5’-GCAGAGTTGTATGAGAGTCCTCC-3’
BCL6
Forward: 5’-CATGCAGAGATGTGCCTCCACA-3’;
Reverse: 5’-TCAGAGAAGCGGCAGTCACACT-3’
PMEPA1
Forward: 5’-GTGCAACTGCAAACGCTCTT-3’;
Reverse: 5’-AGCTTGTAGTGGCTCAGCAG-3’
ENDOD1
Forward: 5’-GCTTCGCCACCCTCTACAGCA-3’;
Reverse: 5’-AATCGCCTCCTCAAGGTTGCTG-3’
MRP4
Forward: 5’-CTGTTGGAGGATGGTGATCTGAC-3’;
Reverse: 5’-CTGCTAACTTCCGCATCTACTGC-3’
RPLP0 (large ribosomal protein P0, a housekeeping gene)
Forward: 5’-CGAGGGCACCTGGAAAAC-3’;
Reverse: 5’-CACATTCCCCCGGATATGA-3’.
For steroid-treated cells, each mRNA transcript was quantitated by normalizing the sample values to RPLP0 and to vehicle-treated cells. All gene expression studies were repeated in at least three independent experiments.
For protein analysis, immunoblots were performed as described previously (6). Briefly, total cellular protein was extracted with ice-cold RIPA lysis buffer (Sigma-Aldrich) containing protease inhibitors (Roche). Protein concentration was determined using a BCA protein assay (Pierce Protein Research Products, Thermo Scientific). Protein, 30–50 μg, was separated by 8% SDS-PAGE and then transferred to a nitrocellulose membrane (Millipore). After incubating with the anti-11β-HSD2 antibody (Santa Cruz; 1:3000), anti-H6PD antibody (Origene; 1:6000) overnight at 4°C, the appropriate secondary antibody was incubated for 1 hour at room temperature. A chemiluminescent detection system (Thermo Scientific) was used to detect the bands with peroxidase activity. An anti-β-actin antibody (Sigma-Aldrich; 1:5000) was used as a control for sample loading.
H6PD and G6PD protein purification
H6PD and G6PD overexpressing plasmids were purchased from OriGene (RC209890L1, RC220625L1) and used to generate a 293T cell line stably expressing human H6PD or G6PD by using a lentiviral system. Briefly, 293T cells (ATCC) were cotransfected with 10 μg each of H6PD or G6PD overexpressing plasmid, pMD2.G, and psPAX2 vector for 48–72 hours. Then, these 293T cells were infected with the virus for 24 hours followed by selection with 2 μg/ml puromycin for ~2 weeks. Then ~108 of the stable cells were used for H6PD or G6PD protein purification with the FLAG M Purification Kit (Sigma, Cat # CELLMM2) according to the protocol they provided. Simply, cell pellets were washed with 10 volumes of PBS and centrifuged. Packed cells were suspended in CelLytic M reagent and incubated for 20 minutes on ice. After being centrifuged, the supernatant was loaded into the prepared column, which includes ANTI-FLAG M2 affinity gel under gravity flow. The column was then washed with 10 column volumes of 1 × wash buffer, which removed any proteins that were not bound to the M2 antibody, and then H6PD or G6PD protein was eluted with 1 ml 1X wash buffer containing 200 ng/ml 3X FLAG peptide.
In vitro and vivo H6PD activity assay
In vitro H6PD activity assay. A reaction cocktail (pH: 7.4) was prepared with the following reagents: 21 ml purified water, 5 ml 250 mM glycylglycine buffer (pH: 7.4), 1ml 60 mM D-glucosamine 6-phosphate solution, 1ml 20 mM β-nicotinamide adenine dinucleotide phosphate solution, 1ml 300 mM magnesium chloride solution. NADP/NADPH-Glo Detection Reagent was purchased from Promega (Cat # G9061) and prepared according to the protocol. Briefly, Luciferin Detection Reagent was reconstituted with Reconstitution Buffer, the NADP/NADPH-Glo Detection Reagent was formed by adding the Reductase and Reductase Rubstrate to reconstituted Luciferin Detection Buffer. 29 μl reaction cocktail and 1μl enzyme were added to 96-well plates in triplicate, and then incubated for 1 hour at room temperature. 30 μl NADP/NADPH-Glo Detection Reagent were added to each well and the mixture incubated with shaking for 1 hour at room temperature. The luminescence of each well was recorded with a luminometer (NADPH generation represents H6PD activity).
In vivo H6PD activity assay. Endoplasmic reticulum (ER) was extracted from ~2×108 cells by using Endoplasmic Reticulum Isolation Kit (Sigma, Cat # ER0100) according to the protocol they provided. Cell pellets were washed with 10 volumes of PBS and centrifuged. Packed cells were suspended in 1x Hypotonic Extraction Buffer and incubated for 20 minutes on ice to allow the cells to swell. After being centrifuged, the cells were broken in 1x Isotonic Extraction Buffer with 10 strokes of a Dounce homogenizer. The homogenate was centrifuged at 1000 x g for 10 minutes at 4°C, then the floating lipid layer was aspirated without aspirating the post-nuclear supernatant. The supernatant was then centrifuged at 12000 x g for 15 minutes at 4°C, and the floating lipid layer was aspirated without aspirating the post-mitochondrial supernatant. The supernatant fraction was ultracentrifuged at 100,000 x g for 1 hour at 4°C, and then the pellet was solubilized in 400 μl cold 250 mM glycylglycine buffer. Protein concentration was determined as described below (Gene expression and immunoblot). Equal amounts of ER were used for the H6PD activity assay. Briefly, 100 μl ER (100 μg protein) was incubated with NADP/NADPH-Glo Detection Reagent for 1 hour at room temperature in 96-well plates I n triplicate. The luminescence of each well was recorded with luminometer.
Cortisol metabolism
Cells (~106 cells per well) were seeded and maintained in 12-well plates that were coated with poly-DL-ornithine (Sigma-Aldrich) for ~24 hours and then treated with [3H]-cortisol (1,000,000 counts per minute (c.p.m.) per well; PerkinElmer) and non-radiolabeled cortisol (100 nM final concentration). Both media and cells were collected at the indicated time points. Briefly, 300 μl media was collected, then the cells were scraped and centrifuged at 10,000 x g for 2 minutes twice to remove all the media without disturbing the pellet. The cell pellets were resuspended in 300 μl PBS. Collected media and cells were incubated with 300 units of β-glucuronidase (Helix pomatia; Novoprotein) at 37°C for at least 2 hours, extracted with 600 μl 1:1 ethyl acetate : isooctane, and concentrated under a nitrogen stream.
For HPLC analysis, the concentrated samples were dissolved in 50% methanol and injected on a Breeze 1525 system equipped with model 717 plus autoinjector (Waters Corp.). Steroid metabolites were separated on a Luna 150 × 4.6 mm, 3 μM C18 reverse-phase column (Phenomenex) using methanol/water gradients at 30°C. The column effluent was analyzed using a β-RAM model 3 in-line radioactivity detector (IN/US Systems, Inc.) using Liquiscint scintillation mixture (National Diagnostics). All metabolism studies were performed in triplicate and repeated in independent experiments.
Gene reinstatement and knockdown
Gene expression. LAPC4 cells were plated and maintained in 12-well plates coated with poly-DL-ornithine (~7 × 105 cells/well) for overnight, then an H6PD-expressing plasmid, H6PD-Myc-DDK-tagged (OriGene) or 11β-HSD1-overexpressing plasmid (Sinobio) was introduced into the cells with Lipofectamine 3000 Reagent (Life Technology). After 48 hours transfection, the cells were used to determine the cortisol metabolism by HPLC as described above.
Gene knockdown. LAPC4 cells were seeded in 12-well plates coated with poly-DL-ornithine at 60%−80% confluence. After incubation overnight, the cells were transfected with siRNA following the Lipofectamine RNAiMAX Reagent (Life Technology) protocol provided by the manufacturer for 48 hours. Then cells were used for evaluating cortisol metabolism and gene expression. The experiments were performed with Dhamarcon SMARTpool: ON-TARGETplus H6PD siRNA, L-004692–01-0005, 11β-HSD1 siRNAs (ThermoFisher, 107742 and 107743) or ON-TARGET plus Non-targeting Pool, #D-001810–10-05 with a final concentration of 25 nM siRNA.
Cell viability assay
~104 long-term enzalutamide (Enz)-treated LAPC4 cells were plated in triplicate in 96-well plates coated with poly-DL-ornithine and incubated overnight, then treated with 10 μM enz and 100 nM cortisol combined with indicated drug treatments for 5 days and assayed using CellTiter-Glo (Promega). Viability was normalized to control.
Mouse xenograft studies
All mouse studies were performed under a protocol approved by the Institutional Animal Care and Use Committee (IACUC) of the Cleveland Clinic Lerner Research Institute. All NSG male mice (6–8 weeks old) were purchased from the Jackson Laboratory and the number of mice used in this study was based on previously published mouse xenograft studies by our lab that determined effects of steroid pathway inhibition/augmentation on xenograft growth (37, 38, 40). Mice were surgically orchiectomized and implanted with DHEA pellets to mimic human adrenal DHEA production in patients with CRPC. 1 week later, mice were prepared for cell injections.
For the evaluation of the H6PD role in reversing enzalutamide resistance, either 107 vector control (gNC) or 107 H6PD knockout (gH6PD) LAPC4 or VCaP cells (100 μl in 50% matrigel and 50% growth media) were subcutaneously injected into mice. When tumors reached 100 or 150 mm3 (length × width × width × 0.52), for LAPC4 and VCaP xenografts, respectively, the mice were arbitrarily divided into 2 groups each for control and H6PD knockout cells: Enz diet 62.5 mg/kg and [as described in (1)] or chow alone groups. Based on the daily chow consumption, approximately 0.3125 mg Enz was consumed per mouse per day. Enz in chow and chow alone were obtained from Medivation. Tumor volume was measured every other day, and progression-free survival was assessed as time to 1.5-fold (LAPC4) or 2.0-fold (VCaP) increase in tumor volume (a baseline of 100 or 150 mm3) from the time Enz or chow alone was initiated. The number of mice in the LAPC4 gNC/Ctrl, gNC/Enz, gH6PD/Ctrl and gH6PD/Enz groups were 8, 8, 9, and 11, respectively. The number of mice in the VCaP gNC/Ctrl, gNC/Enz, gH6PD/Ctrl and gH6PD/Enz groups were 9, 10, 9, and 9, respectively. The number of mice in each treatment group was determined by those that survived surgical procedures and had reached a tumor volume to initiate treatment. Protein levels of H6PD in the gNC/Enz and gH6PD/Enz xenografts were analyzed by immunoblot. Briefly, ~40–50 mg xenograft tissue was minced and added into soft tissue homogenizing CK14 tubes (Betin Technologies) with 150 μl RIPA buffer containing protease inhibitors. Xenograft tissues were homogenized with a homogenizer (Minilys, Betin Technologies) 6 times (40 seconds each time) at the highest speed. Tubes were incubated on ice for 5–10 minutes between each homogenization. The lysates were then centrifuged for 30 minutes at 15,000 x g and the supernatants were used for immunoblot analysis.
For evaluation of whether rucaparib (RUCA) or olaparib (OLA) reverses enzalutamide resistance, 107 LAPC4 or VCaP cells (100 μl in 50% matrigel and 50%growth media) were subcutaneously injected into mice. When tumors reached 100 or 150 mm3 (length × width × width × 0.52), the mice were arbitrarily divided into 3 groups each for control and Enz diet: control [safflower seed oil (Sigma-Aldrich) with 10% DMSO], OLA (MedChemExpress, 150 mg/kg in safflower seed oil with 10% DMSO) or RUCA (MedChemExpress, 150 mg/kg in safflower seed oil with 10% DMSO). The mice were given control, OLA or RUCA by oral gavage BID, 5 days on, 2 days off. The remaining procedures were performed as described above. The number of mice in the LAPC4 CTRL/Ctrl, OLA/Ctrl, RUCA/Ctrl, CTRL/Enz, OLA/Enz and RUCA/Enz groups were 9, 9, 9, 8, 9 and 8, respectively. The number of mice in the VCaP CTRL/Ctrl, OLA/Ctrl, RUCA/Ctrl, CTRL/Enz, OLA/Enz and RUCA/Enz groups were 7, 7, 7, 8, 6 and 9, respectively.
Human tissue studies
All deidentified human tissues were obtained under institutional review board (IRB)-approved protocols. Patients #1–3 were samples from the same patients used in our previous study (3). Paired pre-Enz treatment and post-Enz treatment tissues from Patient #1 and Patient #2 were obtained from patients with localized prostate cancer treated with Enz plus ADT for two months prior to the second biopsy in a clinical trial (NCT02064582) at the University of Texas Southwestern Medical Center. Biopsies were obtained using image-guidance with a Koelis Urostation. Paired pre- and post-Enz treatment tissues from Patient #3 were lymph node tissues which were obtained from CT-guided biopsy of metastatic CRPC from Cleveland Clinic, pathologic identification of tumor was done by an expert prostate cancer pathologist.
12 fresh prostate tissue cores (60–80 mg) from Patients #5-#16 were obtained from the peripheral zone of radical prostatectomy specimens at Cleveland Clinic, confirmed to have tumor in or in close proximity to cores by an expert prostate cancer pathologist, minced and aliquoted to two equal portions. One was treated with 10 nM DHEA plus vehicle, and the other was treated with 10 nM DHEA plus 10 μM Enz. Both tissues were maintained in 3 ml DMEM containing 10% fetal bovine serum and incubated in a 5% CO2 humidified incubator. After 4 days of culture, 2 more ml medium with DHEA plus either vehicle or Enz was added to each part. The tissues were collected after 7–8 days treatment. The same procedures were performed as described above for protein extraction and immunoblot.
Mass spectrometry
Xenograft analysis. At least 24 mg tumor tissue was homogenized with 1 ml LC-MS grade water (Fisher) by using homogenizer. The mixture was then centrifuged. 800 μl supernatant was transferred to a glass tube, followed by the addition of 80 μl 10 ng/ml internal standard (corticosterone-d8) (Steraloids). The steroids and the internal standard were extracted with methyl tert butyl ether (Across) evaporated to dryness under N2 then reconstituted with 500 μl of 50% methanol.
Mouse serum analysis. At the endpoint of the xenograft study, mouse serum was collected. 20 μl serum and internal standard (corticosterone-d8) were precipitated with 200 μl methanol. After centrifugation, the supernatant was transferred to HPLC vials prior to mass spectrometry analysis.
The LC-MS/MS system has an ultra-pressure liquid chromatography system (Shimadzu Corporation) which is consists of two LC-30AD pumps, a DGU-20A5R degasser, a CTO-30A column oven, SIL-30AC autosampler, and a system controller CBM-20A and coupled with a Qtrap 5500 mass spectrometer (AB Sciex). Data acquisition and processing were performed using Analyst software (version 1.6.2) from ABSciex.
Steroids were ionized using electrospray ionization in positive mode. Analytes were quantified using multiple reaction monitoring. The mass transitions for corticosterone, 11-dehydrocorticosterone, and internal standard are 347.3/121.0, 345.3/121.0, and 355.3/125.0, respectively. Separation of steroids was achieved using a Zorbax Eclipse plus C18 column (Agilent) using mobile phase consisting of (A) 0.2% formic acid in water and (B) 0.2% formic acid in (methanol:acetonitrile, 60:40) with a gradient program at a flow rate of 0.2 ml/min. Sample injection volume was 10 μl.
Surface plasmon resonance (SPR) technology based binding assay
The binding of RUCA to H6PD was analyzed using an SPR technology-based Biacore 3000 instrument (GE Lifescience). All the experiments were carried out using 1 × PBS plus 0.1% DMSO as running buffer with a constant flow rate of 10 μL/min at 25°C. H6PD protein, which was diluted in 10 mM sodium acetate buffer (pH 5.0) to a final concentration of 3 μg/μl, was covalently immobilized on the hydrophilic carboxymethylated dextran matrix of the CM5 sensor chip (GE Lifescience) using standard primary amine coupling procedure. RUCA was dissolved in the running buffer with different concentrations ranging from 0.01 to 100 μM. All data were analyzed by BIAevaluation software, and the sensorgrams were processed by automatic correction for nonspecific bulk refractive index effects. The kinetic analyses of the RUCA/H6PD binding were performed based on the 1:1 Langmuir binding fit model according to the procedures described in the software manual.
H6PD IC50 assay protocol
All compounds were set to 5 mM in DMSO and serially diluted using a 3-fold scheme, yielding a 10-point IC50 curve ranging from 200 μM to 10 nM. Compound or DMSO was titrated (4% v/v final) into 33.3 ng recombinant human H6PD, 2 mM D-glucosamine 6-phosphate (Sigma #G5509), and 10 mM MgCl2 (Alfa Aesar #12288) in a 40 mM Gly-Gly (Sigma #50200) pH 7.2 buffer and allowed to incubate at room temperature for 15 minutes. Reactions were initiated by the titration of 20 μM NADP+ (Sigma #N0505) and incubated at room temperature for 1 hour. Reactions were then quenched by adding an equal volume of Promega NAD(P)H-Glo detection reagent (Promega #G9062) and incubating at room temperature for 1 hour. The resulting luminescence was quantified on a BioTek Synergy 4 plate reader. Following background subtraction (DMSO reaction without H6PD present), the average uninhibited signal was calculated (DMSO reaction with H6PD present), and the % inhibition calculated by dividing the signal of wells with compound present with the average uninhibited signal and multiplying by 100. Data was plotted using PRISM 8.1.1 and fit with a log(inhibitor) vs response – four parameter variable slope equation to determine IC50 values. All compounds were tested in duplicate on the same 384-well assay plate, twice (Corning #3574).
Statistical analysis
Statistical data analyses were performed in GraphPad Prism software and Microsoft Excel. In general, for mouse xenograft studies, progression-free survival was compared among groups with a log-rank test. For other comparative analyses, unpaired 2-tailed t test was used unless otherwise noted. P < 0.05 was considered statistically significant.
Supplementary Material
Fig. S1. Enzalutamide treatment promotes H6PD protein expression in prostate cancer.
Fig. S2. Long-term apalutamide treatment sustains cortisol by increasing H6PD protein and suppressing 11β-HSD2 protein in prostate cancer cells.
Fig. S3. H6PD regulates glucocorticoid metabolism directionality in prostate cancer.
Fig. S4. H6PD loss with CRISPR reverses enzalutamide-sustained cortisol in the VCaP model.
Fig. S5. 11β-HSD1 expression regulates cortisol metabolism.
Fig. S6. Cortisol-induced GR signaling is specifically reversible with siRNA-mediated H6PD knockdown.
Fig. S7. H6PD loss of expression and glucocorticoid metabolism normalization in xenograft tissues.
Fig. S8. Rucaparib, but not olaparib, binds to H6PD protein in vitro.
Fig. S9. The effects of rucaparib or olaparib on H6PD and G6PD activity.
Fig. S10. H6PD expression rescues rucaparib-induced loss of LAPC4 cell viability.
Fig. S11. Structural modification of rucaparib and H6PD structure-activity relationships using a biochemical-based NADPH detection enzymatic assay.
Fig. S12. Neither rucaparib nor olaparib treatment affect H6PD or GR protein expression in either LAPC4 or VCaP xenograft tumors.
Acknowledgments
Funding: This work was supported by grants from the National Cancer Institute (R01CA172382 to NS, R01CA190289 to NS, and R01CA236780 to NS) and the Prostate Cancer Foundation (to NS).
Footnotes
Competing interests: NS has been a consultant for Janssen, Pfizer and Celgene.
Data and materials availability:
All data are available in the main text or supplementary data.
REFERENCES AND NOTES
- 1.Tran C et al. , Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science 324, 787–790 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dai C, Heemers H, Sharifi N, Androgen Signaling in Prostate Cancer. Cold Spring Harb Perspect Med 7, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watson PA, Arora VK, Sawyers CL, Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer 15, 701–711 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arora VK et al. , Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell 155, 1309–1322 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Isikbay M et al. , Glucocorticoid receptor activity contributes to resistance to androgen-targeted therapy in prostate cancer. Horm Cancer 5, 72–89 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J et al. , Aberrant corticosteroid metabolism in tumor cells enables GR takeover in enzalutamide resistant prostate cancer. eLife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alyamani M et al. , Deep androgen receptor suppression in prostate cancer exploits sexually dimorphic renal expression for systemic glucocorticoid exposure. Ann Oncol 31, 369–376 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaochar S, Mitsiades N, Glucocorticoids mediate adverse events of deep androgen receptor axis inhibition in prostate cancer patients. Ann Oncol 31, 323–325 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Atanasov AG et al. , Direct protein-protein interaction of 11beta-hydroxysteroid dehydrogenase type 1 and hexose-6-phosphate dehydrogenase in the endoplasmic reticulum lumen. Biochim Biophys Acta 1783, 1536–1543 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Bujalska IJ et al. , Hexose-6-phosphate dehydrogenase confers oxo-reductase activity upon 11 beta-hydroxysteroid dehydrogenase type 1. J Mol Endocrinol 34, 675–684 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Lavery GG et al. , Hexose-6-phosphate dehydrogenase knock-out mice lack 11 beta-hydroxysteroid dehydrogenase type 1-mediated glucocorticoid generation. J Biol Chem 281, 6546–6551 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Banhegyi G, Csala M, Benedetti A, Hexose-6-phosphate dehydrogenase: linking endocrinology and metabolism in the endoplasmic reticulum. J Mol Endocrinol 42, 283–289 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Lavery GG et al. , Steroid biomarkers and genetic studies reveal inactivating mutations in hexose-6-phosphate dehydrogenase in patients with cortisone reductase deficiency. J Clin Endocrinol Metab 93, 3827–3832 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lavery GG et al. , Novel H6PDH mutations in two girls with premature adrenarche: ‘apparent’ and ‘true’ CRD can be differentiated by urinary steroid profiling. Eur J Endocrinol 168, K19–26 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knezevic CE et al. , Proteome-wide Profiling of Clinical PARP Inhibitors Reveals Compound-Specific Secondary Targets. Cell Chem Biol 23, 1490–1503 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas HD et al. , Preclinical selection of a novel poly(ADP-ribose) polymerase inhibitor for clinical trial. Mol Cancer Ther 6, 945–956 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Thorsell AG et al. , Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors. J Med Chem 60, 1262–1271 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kach J, Conzen SD, Szmulewitz RZ, Targeting the glucocorticoid receptor in breast and prostate cancers. Science translational medicine 7, 305ps319 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Obradovic MMS et al. , Glucocorticoids promote breast cancer metastasis. Nature 567, 540–544 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Sharifi N, Steroid receptors aplenty in prostate cancer. The New England journal of medicine 370, 970–971 (2014). [DOI] [PubMed] [Google Scholar]
- 21.West DC et al. , Discovery of a Glucocorticoid Receptor (GR) Activity Signature Using Selective GR Antagonism in ER-Negative Breast Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 24, 3433–3446 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narayanan S, Srinivas S, Feldman D, Androgen-glucocorticoid interactions in the era of novel prostate cancer therapy. Nat Rev Urol 13, 47–60 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Montgomery B, Cheng HH, Drechsler J, Mostaghel EA, Glucocorticoids and prostate cancer treatment: friend or foe? Asian journal of andrology 16, 354–358 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Bono JS et al. , Abiraterone and increased survival in metastatic prostate cancer. The New England journal of medicine 364, 1995–2005 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zein J et al. , HSD3B1 genotype identifies glucocorticoid responsiveness in severe asthma. Proceedings of the National Academy of Sciences of the United States of America 117, 2187–2193 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Claessens F, Joniau S, Helsen C, Comparing the rules of engagement of androgen and glucocorticoid receptors. Cell Mol Life Sci 74, 2217–2228 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Desai K, McManus JM, Sharifi N, Hormonal Therapy for Prostate Cancer. Endocrine Reviews (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montgomery RB et al. , Maintenance of intratumoral androgens in metastatic prostate cancer: a mechanism for castration-resistant tumor growth. Cancer research 68, 4447–4454 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai C et al. , Direct Metabolic Interrogation of Dihydrotestosterone Biosynthesis from Adrenal Precursors in Primary Prostatectomy Tissues. Clinical cancer research : an official journal of the American Association for Cancer Research 23, 6351–6362 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Auchus RJ, Sharifi N, Sex Hormones and Prostate Cancer. Annu Rev Med 71, 33–45 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Chapman K, Holmes M, Seckl J, 11beta-hydroxysteroid dehydrogenases: intracellular gate-keepers of tissue glucocorticoid action. Physiol Rev 93, 1139–1206 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Efstathiou E et al. , Clinical and Biological Characterisation of Localised High-risk Prostate Cancer: Results of a Randomised Preoperative Study of a Luteinising Hormone-releasing Hormone Agonist with or Without Abiraterone Acetate plus Prednisone. European urology 76, 418–424 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Bono J et al. , Olaparib for Metastatic Castration-Resistant Prostate Cancer. 382, 2091–2102 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Abida W et al. , Non-BRCA DNA Damage Repair Gene Alterations and Response to the PARP Inhibitor Rucaparib in Metastatic Castration-Resistant Prostate Cancer: Analysis From the Phase II TRITON2 Study. 26, 2487–2496 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z, Alyamani M, Li J, Upadhyay U, Balk SP, Taplin M -E., Auchus, R.J., Sharifi, N., Redirecting abiraterone metabolism to biochemically fine tune prostate cancer anti-androgen therapy. Nature 533, 547–551 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alyamani M et al. , Steroidogenic Metabolism of Galeterone Reveals a Diversity of Biochemical Activities. Cell Chem Biol 24, 825–832 e826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang KH et al. , A gain-of-function mutation in DHT synthesis in castration-resistant prostate cancer. Cell 154, 1074–1084 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang KH et al. , Dihydrotestosterone synthesis bypasses testosterone to drive castration-resistant prostate cancer. Proceedings of the National Academy of Sciences of the United States of America 108, 13728–13733 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanjana NE, Shalem O, Zhang F, Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Z et al. , Conversion of abiraterone to D4A drives anti-tumour activity in prostate cancer. Nature, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Enzalutamide treatment promotes H6PD protein expression in prostate cancer.
Fig. S2. Long-term apalutamide treatment sustains cortisol by increasing H6PD protein and suppressing 11β-HSD2 protein in prostate cancer cells.
Fig. S3. H6PD regulates glucocorticoid metabolism directionality in prostate cancer.
Fig. S4. H6PD loss with CRISPR reverses enzalutamide-sustained cortisol in the VCaP model.
Fig. S5. 11β-HSD1 expression regulates cortisol metabolism.
Fig. S6. Cortisol-induced GR signaling is specifically reversible with siRNA-mediated H6PD knockdown.
Fig. S7. H6PD loss of expression and glucocorticoid metabolism normalization in xenograft tissues.
Fig. S8. Rucaparib, but not olaparib, binds to H6PD protein in vitro.
Fig. S9. The effects of rucaparib or olaparib on H6PD and G6PD activity.
Fig. S10. H6PD expression rescues rucaparib-induced loss of LAPC4 cell viability.
Fig. S11. Structural modification of rucaparib and H6PD structure-activity relationships using a biochemical-based NADPH detection enzymatic assay.
Fig. S12. Neither rucaparib nor olaparib treatment affect H6PD or GR protein expression in either LAPC4 or VCaP xenograft tumors.
Data Availability Statement
All data are available in the main text or supplementary data.