Abstract
F. nucleatum is an anaerobic bacterium that is associated with several tumor entities and promotes tumorigenesis. Recent evidence suggests that F. nucleatum binds the inhibitory receptor carcinoembryonic antigen cell adhesion molecule 1 (CEACAM1) via the trimeric autotransporter adhesin CbpF. However, whether this binding is functional or whether other fusobacterial trimeric autotransporter adhesins are involved in CEACAM1 activation is unknown. In this study, using F. nucleatum mutants lacking the type 5c trimeric autotransporter adhesins fvcA (CbpF), fvcB, fvcC, and fvcD, we show that F. nucleatum CbpF binds and activates CEACAM1 and also binds carcinoembryonic antigen (CEA), a tumor-associated protein. We further find that CEACAM antibodies directed against the CEACAM N-terminal domain block the CbpF-CEACAM1 interaction. In functional assays, we demonstrate CbpF-dependent inhibition of CD4+ T cell response. Thus, we characterize an immune evasion mechanism in which F. nucleatum uses its surface protein CbpF to inhibit T cell function by activating CEACAM1.
Keywords: F. nucleatum, CbpF, trimeric autotransporter adhesins, CEACAM1, CEA
Introduction
Tumors harbor a diverse microbiome that impacts carcinogenesis, cancer progression and therapy (Javaheri et al., 2016; Koniger et al., 2016; Gur et al., 2019a; Nejman et al., 2020). Among tumor-associated bacteria, Fusobacterium nucleatum has attracted increasing attention in recent years. F. nucleatum is a gram-negative anaerobic bacterium found in the oral cavity and associated with periodontal disease. Initially discovered to be enriched in colorectal cancer, F. nucleatum was since shown to be associated with esophageal (Yamamura et al., 2016), breast (Nejman et al., 2020; Parhi et al., 2020) and pancreatic cancer (Mitsuhashi et al., 2015; Nejman et al., 2020), and to promote both tumor growth and metastasis (Mima et al., 2016; Bullman et al., 2017; Parhi et al., 2020).
Besides contributing to a pro-inflammatory tumor microenvironment, F. nucleatum protects tumor cells from killing by NK cells and tumor infiltrating T cells. Mechanistically, we previously found that the F. nucleatum adhesion protein Fap2 engages TIGIT, an inhibitory receptor expressed on NK cells and T cells (Gur et al., 2015). Furthermore, we and others showed that F. nucleatum specifically targets carcinoembryonic antigen cell adhesion molecule 1 (CEACAM1) (Brewer et al., 2019; Gur et al., 2019b), an inhibitory receptor expressed on endothelial, epithelial, and immune cells.
CEACAM1 mediates cell adhesion via homophilic binding (CEACAM1-CEACAM1) or heterophilic binding to carcinoembryonic antigen (CEA), a tumor-associated adhesion molecule (Gray-Owen and Blumberg, 2006). Additionally, proteins of several bacteria have been identified as ligands for CEACAM1: Helicobacter pylori HopQ (Javaheri et al., 2016; Koniger et al., 2016; Gur et al., 2019a) Neisseria ssp. Opa proteins (Boulton and Gray-Owen, 2002), Haemophilus influenza P5 (Hill et al., 2001), group B Streptococcus β protein (van Sorge et al., 2021), Escherichia coli Afa/Dr adhesins (Berger et al., 2004), Moraxella catarrhalis UspA1 (Hill and Virji, 2003), and a yet unidentified ligand on Acinetobacter baumanii (Ambrosi et al., 2020). Besides bacteria, Candida albicans was found to bind CEACAM1 (Klaile et al., 2017).
Recently, the F. nucleatum type 5c trimeric autotransporter CbpF (CEACAM binding protein of Fusobacterium) was discovered to bind CEACAM1 and CEA using matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry and N-terminal sequencing (Brewer et al., 2019). However, whether binding of F. nucleatum CbpF to CEACAM1 is functional or whether other fusobacterial trimeric autotransporter adhesins are involved in CEACAM1 activation remains unknown. In this study, using F. nucleatum deletion mutants of four fusobacterial 5c trimeric autotransporter proteins including CbpF, we studied the role of these proteins in CEACAM1 binding and activation.
Materials and Methods
Ethics
The collection of blood samples from healthy donors was approved by the Institutional Review Board of Hadassah Medical Center (HMO-0030-12).
Primary Human T Cells and Cell Lines
To obtain primary human CD4+ CEACAM1-positive T cells we first isolated PBMCs from peripheral blood of healthy donors by centrifugation on Lymphoprep (StemCells Technologies). We then seeded single cells together with irradiated (6000 rad) feeder cells (50,000 allogeneic PBMCs from two donors and 5,000 RPMI 8866 cells) and 0.2 µl PHA (Sigma-Aldrich) per well in 96-well U-bottom plates. After a week, the same numbers of irradiated feeder cells were added again. Cultures were maintained in DMEM:F-12 Nutrient Mixture (70:30) supplemented with 10% human serum (Sigma Aldrich), 1% each of non-essential amino acids (Biological Industries, BI), L-glutamine (BI), sodium pyruvate (BI), and penicillin-streptomycin (BI), as well as recombinant human IL-2 (500 IU/ml, Peprotech). Following expansion, clones were stained for CD4, CD8, and CEACAM1 (Biolegend) and CEACAM1-positive CD4+ T cell clones were pooled together. Cells were consistently assessed for their expression of CD4, CD8, and CEACAM1 throughout the experiments.
All cell lines used in the study were originally obtained from the ATCC: human EBV-transformed 721.221 cells, mouse mastocytoma P815 cells, and mouse thymoma BW cells. Cell lines were grown in RPMI supplemented with 10% heat-inactivated fetal calf serum (FCS), 1% each of non-essential amino acids, L-glutamine, sodium pyruvate and penicillin-streptomycin (all from Biological Industries). Cells were grown at 37°C in a humidified 5% CO2 incubator.
Bacteria
The generation of F. nucleatum ATCC 23726 gene deletion mutants is described elsewhere (Casasanta et al., 2020). All mutants were generated in the F. nucleatum 23726 ∆galKT background that is subsequently referred to as Fnn. Bacteria were grown at 37°C on anaerobic blood agar plates (Novamed) or chocolate agar plates (Novamed) under anaerobic conditions generated using Oxoid AnaeroGen 2.5L Jars and Sachets (Thermo Fisher).
Antibodies and Fusion Proteins
The following antibodies were used: purified α-mouse IL-2 (JES6-1A12) and biotinylated α-mouse IL-2 (JES6-5H4) from Biolegend; purified α-human IFN-γ (Nib42) and biotinylated α-human IFN-γ (4S.B3) from Biolegend; purified α-human CD3 (Hit3a) from Biolegend. For flow cytometry, the following antibodies were used in blocking experiments: α-PVR (in-house antibody) used as a negative control; CEACAM antibodies 18/20 binding to the N-terminal domain of CEACAM1, 3, 5 and 6 and CC1/3/5-Sab binding to the N-terminal domain of CEACAM1, 3, 5 (B. B. Singer), C5-1X/8/8 and B3-17 binding to the A1/B1 domain of CEACAM1 (B. B. Singer), 6G5j binding to the N-terminal domain of CEACAM1, 3, 5, 6, 8 (B. B. Singer), and ASL-32 binding to CEACAM1, 5, 6 (Biolegend).
The fusion proteins CEACAM1-Fc, CEACAM3-Fc, CEACAM5-Fc, CEACAM6-Fc, CEACAM8-Fc, mouse sCEACAM1-Fc, and macaque CEACAM1-Fc were generated in HEK293T cells and purified using Protein A/G-Sepharose affinity Chromatography, as previously described (Singer et al., 2014; Javaheri et al., 2016; Moonens et al., 2018).
FITC-Labeling of Bacteria and Flow Cytometry
Bacteria were washed twice and incubated with 0.1 mg/ml FITC (Sigma-Aldrich) in PBS at room temperature in the dark for 30 minutes. Subsequently, bacteria were washed three times in PBS to remove unbound FITC. For flow cytometry, 721.221 cells were used as carrier cells to facilitate gating. To this end, bacteria were divided into 96-well plates and incubated with 721.221 cells for 30 minutes on ice to allow for bacterial adhesion to the cells (60 million bacteria were placed together with 100,000 721.221 cells per well). Next, cells were washed and incubated with the indicated amount of CEACAM fusion proteins for 1 hour on ice followed by a 30-minute incubation with Alexa Fluor 647-conjugated donkey anti-human IgG (Jackson Immunoresearch). Histograms of cell-bound bacteria stained with CEACAM fusion proteins were gated on FITC-positive cells.
For blocking experiments, 2 µg of CEACAM1-Ig were preincubated with 1 µg of the respective antibodies for 1 hour on ice. Values obtained without blocking antibodies were arbitrarily set to 1 and all other values were normalized accordingly.
IL-2 Release Assay From Mouse Thymoma BW Cell Transfectants
Bacteria were divided into 96 well plates (30 µl of bacteria at an OD600 of 1 per well) and incubated for one hour at 37°C in complete RPMI. Subsequently, BW CEACAM1 cells were added at 50,000 per well and incubated with the bacteria for 48 hours at 37°C. Next, supernatants were collected and mouse IL-2 levels were quantified by a sandwich ELISA. The generation of BW cells expressing chimeric CEACAM1 (composed of the extracellular portion of human CEACAM1 fused to the mouse CD3ζ chain) was previously described (Markel et al., 2002).
Redirected Cytokine Secretion Assay
Irradiated mouse mastocytoma P815 cells that express the Fcγ receptor were coated with anti-human CD3 antibodies (25,000 P815 cells and 0.1 µg anti-CD3 per well) for 1 hour on ice. Subsequently, bacteria were added (15 million bacteria per well; 1:600). Finally, 25,000 CEACAM1-positive CD4+ T cells were added to each well and the plates were incubated for 48 hours at 37°C. Levels of IFN-γ in supernatants were quantified by a sandwich ELISA.
Statistical Analysis
All statistical analyses were performed using GraphPad Prism 6. Data were statistically analyzed with one-way ANOVA followed by Dunnett’s multiple comparison test. A two-tailed unpaired t test was used to determine statistical significance of differences between two groups. p-values less than 0.05 were considered statistically significant (* p ≤ 0.05; ** p ≤ 0.01; *** p ≤ 0.001).
Results
F. nucleatum CbpF Binds and Activates CEACAM1
In this study, we sought to comprehensively investigate the role of fusobacterial 5c trimeric autotransporter adhesins in CEACAM1 binding. To this end, we characterized F. nucleatum mutants generated in the F. nucleatum 23726 ∆galKT strain, which allows for targeted gene deletion (Casasanta et al., 2020). In line with previously established nomenclature (Casasanta et al., 2020), we refer to this strain as Fnn throughout the manuscript and figures.
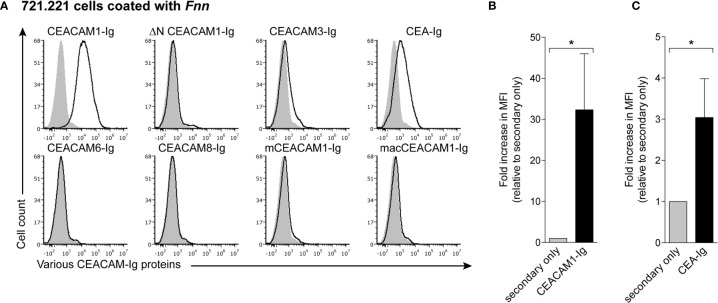
To assess binding of Fnn to CEACAM proteins, we first stained bacteria with several different CEACAM fusion proteins in which the extracellular domain of the respective CEACAM is fused to the Fc portion of human IgG1. An overview of features of the CEACAM fusion proteins tested in this study is given in Supplemental Table 1 . We used 721.221 cells as carrier cells for the FITC-labeled bacteria to facilitate efficient gating in flow cytometry. While 721.221 cells alone did not bind human CEACAM1-Ig, ∆N CEACAM1-Ig that lacks the N-terminal domain, CEACAM3-Ig, CEA-Ig, CEACAM6-Ig, CEACAM8-Ig, mouse CEACAM1-Ig (mCEACAM1-Ig) or rhesus macaque CEACAM1-Ig (macCEACAM1-Ig) ( Supplemental Figure 1A ), we observed staining of cell-bound Fnn by CEACAM1-Ig and CEA-Ig ( Figures 1A–C ), consistent with previous findings (Brewer et al., 2019; Gur et al., 2019b). In contrast, we observed no binding when bacteria were stained with ∆N-CEACAM1-Ig, indicating that the CEACAM1 N-terminal domain is critical for this interaction. Similarly, no binding of mCEACAM1-Ig and macCEACAM1-Ig homologues as well as CEACAM3-Ig, CEACAM6-Ig, and CEACAM8-Ig was detected ( Figure 1A ).
Figure 1.
Fnn specifically binds to the N-terminal domain of human CEACAM1. (A) 721.221 cells coated with FITC-labeled Fnn were stained with 2 μg of human CEACAM1-Ig, ∆N CEACAM1-Ig that lacks the N-terminal domain, CEACAM3-Ig, CEA-Ig, CEACAM6-Ig, CEACAM8-Ig, mouse CEACAM1-Ig (mCEACAM1-Ig) or rhesus macaque CEACAM1-Ig (macCEACAM1-Ig). Filled grey histograms represent staining with secondary antibody only. Representative histograms from three independent repeats are shown. (B) CEACAM1-Ig and (C) CEA-Ig staining quantified and shown as fold increase in MFI relative to secondary only. Bars represent means of three independent experiments ± SD. Statistical significance was assessed using a two-tailed unpaired t test. *p ≤ 0.05.
The trimeric autotransporter adhesin CbpF was previously suggested to be the fusobacterial ligand of CEACAM1. Therefore, we next examined binding of CEACAM1-Ig to Fnn single mutants each lacking one of the type 5c trimeric autotransporter adhesins fvcA (CbpF), fvcB, fvcC, fvcD, as well as a quadruple mutant deficient for all four adhesins (fvcABCD). Whereas binding of CEACAM1-Ig to the ∆fvcB, ∆fvcC and ∆fvcD mutants was similar to Fnn, little or no staining was observed for both Fnn ∆fvcA (∆CbpF) and Fnn ∆fvcABCD ( Figure 2A ). To corroborate these results, we used a reporter system previously generated in our lab (Markel et al., 2002). In this system, mouse thymoma BW cells expressing chimeric CEACAM1 composed of the extracellular portion of human CEACAM1 fused to the mouse CD3ζ chain secrete mouse IL-2 upon binding and activation of CEACAM1 by specific ligands (illustrated in Figure 2B ). Whereas mouse IL-2 levels detected for Fnn ∆fvcB, ∆fvcC and ∆fvcD mutants were similar to Fnn, both the Fnn ∆fvcA (∆CbpF) and ∆fvcABCD mutants failed to activate CEACAM1 ( Figure 2C ). Taken together, these assays demonstrate that CbpF both binds to and activates human CEACAM1.
Figure 2.
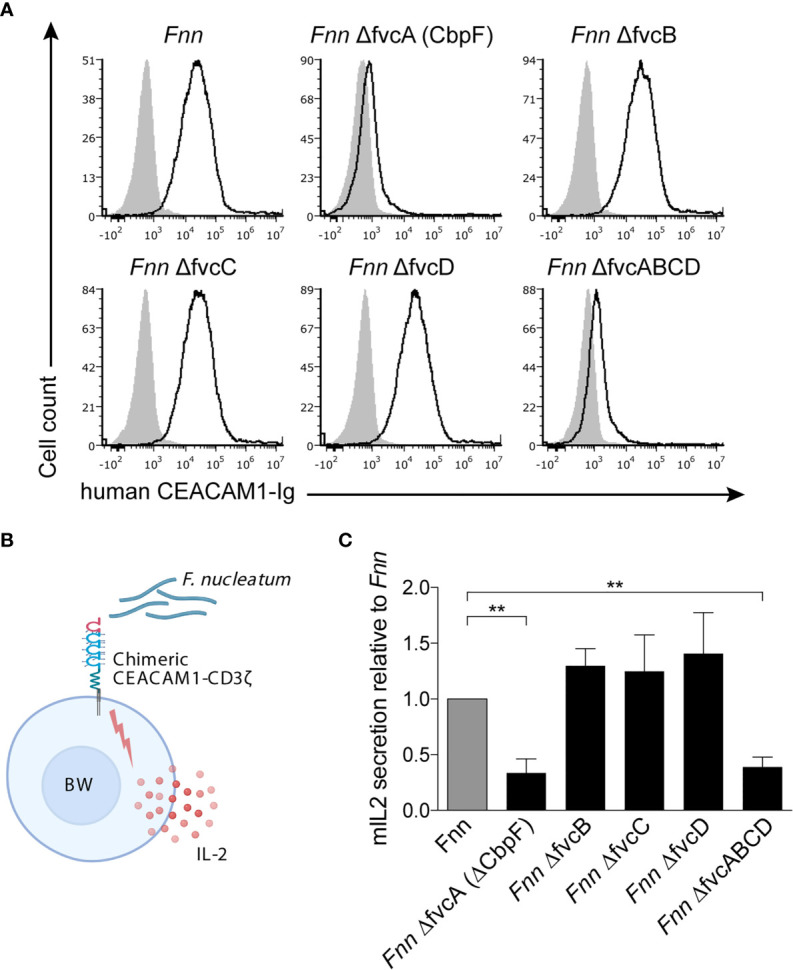
The Fnn ∆CbpF mutant fails to bind CEACAM1. (A) FITC-labeled Fnn and single mutants of the trimeric autotransporter adhesins fvcA (cbpF), fvcB, fvcC, fvcD, as well as a mutant lacking all four adhesins (Fnn fvcABCD) were stained with 3 μg of human CEACAM1-Ig. 721.221 cells were used as carrier cells. Filled gray histograms represent staining with secondary antibody only. One representative staining out of two is shown. (B) Schematic representation of the BW reporter assay. BW5147 cells stably transfected with human CEACAM1 fused to the transmembrane and cytoplasmic domain of mouse CD3ζ chain (chimeric recptor-CD3ζ) are incubated with above-mentioned Fnn strains. Activation of CEACAM1 by a ligand results in mIL-2 quantified by ELISA. (C) CEACAM1-expressing BW cells were incubated with Fnn strains at a ratio of 1:300. Mouse IL-2 in the supernatants was determined by ELISA 48 hours later. Bars represent means of three independent experiments performed in triplicates ± SD. Fnn bacteria were arbitrarily set to 1 and other values were normalized accordingly. Statistical significance was assessed using one-way ANOVA with Dunnett’s multiple comparison test. **p ≤ 0.01.
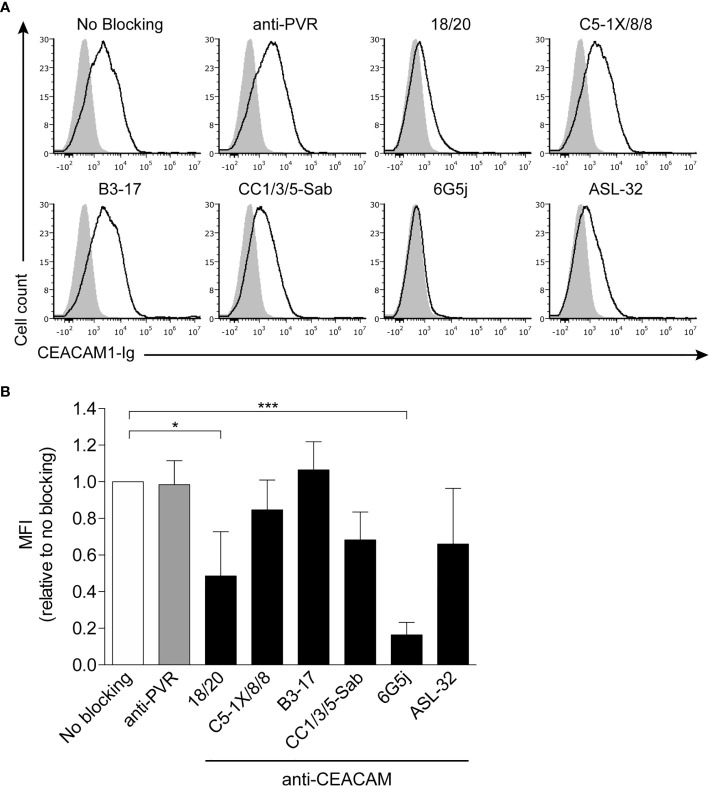
The CEACAM Antibodies 18/20 and 6G5j That Target the N-Terminal Domain Block F. nucleatum CbpF Binding to CEACAM1
Next, to further investigate the CEACAM1 binding site that mediates interaction with CbpF, we used different anti-CEACAM1 antibodies to attempt to block the interaction: 18/20, CC1/3/5-Sab, 6G5j, and ASL-32 that bind to the CEACAM1 N-domain; C5-1X/8/8 and B3-17 that bind to the A1-B1 domains (Javaheri et al., 2016; van Sorge et al., 2021). To assess blocking, 721.221 cell-bound FITC-labeled Fnn were stained with CEACAM1-Ig, following preincubation with the respective antibodies. Anti-PVR antibodies served as a negative control, since PVR is not expressed on 721.221 cells (Stanietsky et al., 2009). Both the C5-1X/8/8 and B3-17 antibodies did not significantly alter binding of CEACAM1 to CbpF, further corroborating that it is the CEACAM1 N-domain that mediates binding to CbpF ( Figure 3A , quantification in Figure 3B ). In contrast, blocking with 18/20 and 6G5j CEACAM antibodies led to a significant reduction in MFI (to 48.6% and 16.4%, respectively, relative to no blocking) ( Figure 3A , quantification in Figure 3B ). The decreases in MFI observed for CC1/3/5-Sab and ASL-32 were not statistically significant.
Figure 3.
Binding of CEACAM1 to Fnn is blocked by anti-CEACAM1 18/20 and 6G5J. (A) FITC-labeled Fnn were stained with 2 μg of human CEACAM1-Ig preincubated with 1 µg of anti-PVR or different anti-CEACAM1 antibodies. 721.221 cells were used as carrier cells. Filled grey histograms represent staining with secondary antibody only. (B) Quantification of three independent experiments. Bars represent means of three independent experiments ± SD. Values obtained without blocking antibody were arbitrarily set to 1 and all other values were normalized accordingly. Statistical significance was assessed using one-way ANOVA with Dunnett’s multiple comparison test. *p ≤ 0.05; ***p ≤ 0.001.
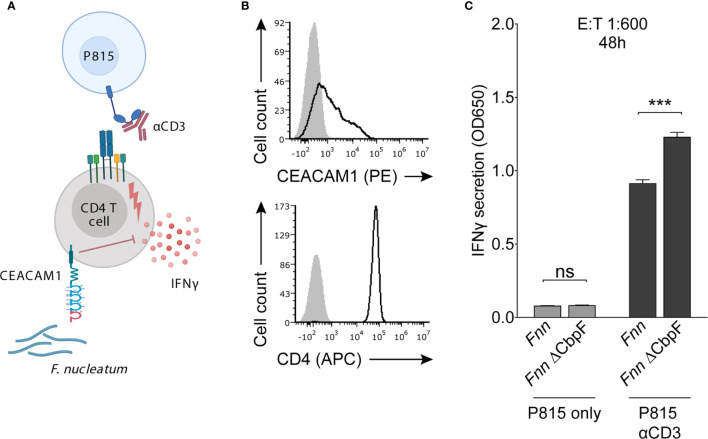
CbpF Binding to CEACAM1 Leads to Inhibition of the CD4+ T Cell Response
To assess the effect of CbpF on CD4+ T cell function, we used a redirected cytokine secretion assay. Mouse mastocytoma P815 cells expressing the Fcγ receptor were coated with an anti-CD3 antibody to enable activation of T cells ( Figure 4A ) and were further incubated with and without bacteria. The cells were then incubated with CEACAM1-positive CD4+ T cells that were obtained from peripheral blood of healthy donors ( Figure 4B ). The P815 cells coated with anti-CD3, CEACAM1-positive CD4+ T cells and Fnn or the Fnn ∆CbpF were incubated for 48 hours, and IFN-γ secretion was assessed by ELISA. In the absence of anti-CD3 antibody, little IFN-γ secretion was observed ( Figure 4C ). In contrast, in the presence of anti-CD3, IFN-γ levels were significantly higher when CEACAM1-positive CD4+ T cells were incubated with Fnn ∆CbpF as compared to Fnn ( Figure 4C ). These results indicate that CbpF inhibits IFN-γ secretion from CEACAM1-positive CD4+ T cells.
Figure 4.
Fnn inhibits CD4+ T cell IFN-γ secretion by activating CEACAM1. (A) Schematic representation of the redirected cytokine assay. Mouse mastocytoma P815 cells expressing FcγR were coated with anti-CD3 to enable activation of T cells and subsequent IFN-γ secretion. Cells were incubated with Fnn or the Fnn ∆CbpF mutant at a E:T ratio of 1:600 and IFN-γ secretion was quantified by ELISA after 48 hours of incubation. (B) Flow cytometry staining of primary IL-2 activated human CD4+ T cells with anti-CEACAM1 and anti-CD4. Filled grey histograms represent staining with secondary antibody only. (C) Quantification of IFN-γ secretion determined by ELISA. Bars represent means of triplicates ± SD. The graphs represent data collected from two independent experiments. Statistical significance was assessed using a two-tailed unpaired t test. ***p ≤ 0.001. NS, not significant.
Discussion
In this study, we used single mutants of the type 5c trimeric autotransporter adhesins fvcA (CbpF), fvcB, fvcC, and fvcD, as well as a quadruple mutant lacking all four genes, to investigate the role of these proteins in CEACAM1 binding and activation. Using both flow cytometry and functional assays, we demonstrated that CbpF binds and activates CEACAM1. The interaction was significantly blocked by the CEACAM 18/20 and 6G5j antibodies that bind the N-terminal domain. In line with these results, CEACAM1-Ig lacking the N-terminal domain failed to interact with F. nucleatum. Furthermore, using a redirected cytokine secretion assay, we showed that CbpF inhibits the IFN-γ response in activated CEACAM1-positive CD4+ T cells.
Members of the CEACAM family are known to interact with many bacteria such as Helicobacter pylori (Javaheri et al., 2016; Koniger et al., 2016; Gur et al., 2019a), Haemophilus influenza (Hill et al., 2001), and group B Streptoccocus (van Sorge et al., 2021). These bacterial ligands share little structural homology (Brewer et al., 2019), strongly suggesting they evolved independently. Furthermore, not only bacteria but also Candida albicans specifically binds to CEACAM1 (Klaile et al., 2017). This notion implies a critical importance for the interaction of pathogens with CEACAM1 to allow for cellular adhesion, invasion, as well as for immune evasion.
Here we assessed the impact of CbpF-dependent CEACAM1 activation on the CD4+ T cell response. However, CEACAM1 is expressed not only on immune cells, but also on certain endothelial cells and epithelial cells, including cancer cells (Gray-Owen and Blumberg, 2006). The impact of F. nucleatum CbpF binding to CEACAM1 on these cells in the tumor microenvironment is unknown. Depending on the isoform expression profile, CEACAM1 activation can result in growth inhibition or allow for cell proliferation (Singer et al., 2000). Moreover, CEACAM1 expression on several tumor entities such as colorectal cancer (Ieda et al., 2011) and malignant melanoma (Thies et al., 2002; Thies et al., 2007) is associated with increased metastatic potential. Since F. nucleatum is maintained in distant metastases of CRC (Bullman et al., 2017), it will be of interest to investigate the impact of the interaction between F. nucleatum CbpF and CEACAM1-positive cancer cells on both cancer cell proliferation and metastasis.
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by The Institutional Review Board of Hadassah Medical Center (HMO-0030-12). Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
JG and OM conceived the study. JG, AS, AU, CC, LA, BI, and OB performed the experiments. JG and OM analyzed the data. DJS contributed F. nucleatum mutants. BS contributed CEACAM fusion proteins and antibodies. JG drafted the manuscript. OM and GB supervised the project. All authors contributed to the article and approved the submitted version.
Funding
JG is supported by the German Research Foundation (DFG; project number 429842436) with a postdoctoral research fellowship. This work was supported by the Israel Science Foundation (Moked grant), the GIF Foundation, the ICRF professorship grant, the ISF Israel-China grant, the MOST-DKFZ grant, and by the ERC Marie Curie grant.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Schematic figures were created with BioRender (BioRender.com).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.692544/full#supplementary-material
References
- Ambrosi C., Scribano D., Sarshar M., Zagaglia C., Singer B. B., Palamara A. T. (2020). Acinetobacter baumannii Targets Human Carcinoembryonic Antigen-Related Cell Adhesion Molecules (Ceacams) for Invasion of Pneumocytes. mSystems 5 (6), e00604–20. 10.1128/mSystems.00604-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger C. N., Billker O., Meyer T. F., Servin A. L., Kansau I. (2004). Differential Recognition of Members of the Carcinoembryonic Antigen Family by Afa/Dr Adhesins of Diffusely Adhering Escherichia coli (Afa/Dr DAEC). Mol. Microbiol. 52 (4), 963–983. 10.1111/j.1365-2958.2004.04033.x [DOI] [PubMed] [Google Scholar]
- Boulton I. C., Gray-Owen S. D. (2002). Neisserial Binding to CEACAM1 Arrests the Activation and Proliferation of CD4+ T Lymphocytes. Nat. Immunol. 3 (3), 229–236. 10.1038/ni769 [DOI] [PubMed] [Google Scholar]
- Brewer M. L., Dymock D., Brady R. L., Singer B. B., Virji M., Hill D. J. (2019). Fusobacterium Spp. Target Human CEACAM1 Via the Trimeric Autotransporter Adhesin Cbpf. J. Oral. Microbiol. 11 (1):1565043. 10.1080/20002297.2018.1565043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullman S., Pedamallu C. S., Sicinska E., Clancy T. E., Zhang X., Cai D., et al. (2017). Analysis of Fusobacterium Persistence and Antibiotic Response in Colorectal Cancer. Science 358 (6369), 1443–1448. 10.1126/science.aal5240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casasanta M. A., Yoo C. C., Udayasuryan B., Sanders B. E., Umaña A., Zhang Y., et al. (2020). Fusobacterium nucleatum Host-Cell Binding and Invasion Induces IL-8 and CXCL1 Secretion That Drives Colorectal Cancer Cell Migration. Sci. Signal 13 (641), eaba9157. 10.1126/scisignal.aba9157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray-Owen S. D., Blumberg R. S. (2006). CEACAM1: Contact-Dependent Control of Immunity. Nat. Rev. Immunol. 6 (6), 433–446. 10.1038/nri1864 [DOI] [PubMed] [Google Scholar]
- Gur C., Ibrahim Y., Isaacson B., Yamin R., Abed J., Gamliel M., et al. (2015). Binding of the Fap2 Protein of Fusobacterium Nucleatum to Human Inhibitory Receptor TIGIT Protects Tumors From Immune Cell Attack. Immunity 42 (2), 344–355. 10.1016/j.immuni.2015.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur C., Maalouf N., Gerhard M., Singer B. B., Emgård J., Temper V., et al. (2019. a). The Helicobacter pylori HopQ Outermembrane Protein Inhibits Immune Cell Activities. Oncoimmunology 8 (4), e1553487. 10.1080/2162402x.2018.1553487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur C., Maalouf N., Shhadeh A., Berhani O., Singer B. B., Bachrach G., et al. (2019. b). Fusobacterium nucleatum Supresses Anti-Tumor Immunity by Activating CEACAM1. Oncoimmunology 8 (6), e1581531. 10.1080/2162402x.2019.1581531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill D. J., Toleman M. A., Evans D. J., Villullas S., Van Alphen L., Virji M. (2001). The Variable P5 Proteins of Typeable and Non-Typeable Haemophilus influenzae Target Human CEACAM1. Mol. Microbiol. 39 (4), 850–862. 10.1046/j.1365-2958.2001.02233.x [DOI] [PubMed] [Google Scholar]
- Hill D. J., Virji M. (2003). A Novel Cell-Binding Mechanism of Moraxella catarrhalis Ubiquitous Surface Protein UspA: Specific Targeting of the N-domain of Carcinoembryonic Antigen-Related Cell Adhesion Molecules by Uspa1. Mol. Microbiol. 48 (1), 117–129. 10.1046/j.1365-2958.2003.03433.x [DOI] [PubMed] [Google Scholar]
- Ieda J., Yokoyama S., Tamura K., Takifuji K., Hotta T., Matsuda K., et al. (2011). Re-expression of CEACAM1 Long Cytoplasmic Domain Isoform is Associated With Invasion and Migration of Colorectal Cancer. Int. J. Cancer 129 (6), 1351–1361. 10.1002/ijc.26072 [DOI] [PubMed] [Google Scholar]
- Javaheri A., Kruse T., Moonens K., Mejias-Luque R., Debraekeleer A., Asche C. I., et al. (2016). Helicobacter pylori Adhesin HopQ Engages in a Virulence-Enhancing Interaction With Human Ceacams. Nat. Microbiol. 2, 16189. 10.1038/nmicrobiol.2016.189 [DOI] [PubMed] [Google Scholar]
- Klaile E., Müller M. M., Schäfer M. R., Clauder A. K., Feer S., Heyl K. A., et al. (2017). Binding of Candida albicans to Human CEACAM1 and CEACAM6 Modulates the Inflammatory Response of Intestinal Epithelial Cells. mBio 8 (2), e02142–16. 10.1128/mBio.02142-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koniger V., Holsten L., Harrison U., Busch B., Loell E., Zhao Q., et al. (2016). Helicobacter pylori Exploits Human CEACAMs Via HopQ for Adherence and Translocation of Caga. Nat. Microbiol. 2, 16188. 10.1038/nmicrobiol.2016.188 [DOI] [PubMed] [Google Scholar]
- Markel G., Wolf D., Hanna J., Gazit R., Goldman-Wohl D., Lavy Y., et al. (2002). Pivotal Role of CEACAM1 Protein in the Inhibition of Activated Decidual Lymphocyte Functions. J. Clin. Invest. 110 (7), 943–953. 10.1172/jci15643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mima K., Nishihara R., Qian Z. R., Cao Y., Sukawa Y., Nowak J. A., et al. (2016). Fusobacterium nucleatum in Colorectal Carcinoma Tissue and Patient Prognosis. Gut 65 (12), 1973–1980. 10.1136/gutjnl-2015-310101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhashi K., Nosho K., Sukawa Y., Matsunaga Y., Ito M., Kurihara H., et al. (2015). Association of Fusobacterium Species in Pancreatic Cancer Tissues With Molecular Features and Prognosis. Oncotarget 6 (9), 7209–7220. 10.18632/oncotarget.3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moonens K., Hamway Y., Neddermann M., Reschke M., Tegtmeyer N., Kruse T., et al. (2018). Helicobacter pylori Adhesin HopQ Disrupts Trans Dimerization in Human Ceacams. EMBO J. 37 (13), e98665. 10.15252/embj.201798665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejman D., Livyatan I., Fuks G., Gavert N., Zwang Y., Geller L. T., et al. (2020). The Human Tumor Microbiome Is Composed of Tumor Type-Specific Intracellular Bacteria. Science 368 (6494), 973–980. 10.1126/science.aay9189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parhi L., Alon-Maimon T., Sol A., Nejman D., Shhadeh A., Fainsod-Levi T., et al. (2020). Breast Cancer Colonization by Fusobacterium nucleatum Accelerates Tumor Growth and Metastatic Progression. Nat. Commun. 11 (1), 3259. 10.1038/s41467-020-16967-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer B. B., Opp L., Heinrich A., Schreiber F., Binding-Liermann R., Berrocal-Almanza L. C., et al. (2014). Soluble CEACAM8 Interacts With CEACAM1 Inhibiting TLR2-Triggered Immune Responses. PloS One 9 (4), e94106. 10.1371/journal.pone.0094106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer B. B., Scheffrahn I., Obrink B. (2000). The Tumor Growth-Inhibiting Cell Adhesion Molecule CEACAM1 (C-CAM) Is Differently Expressed in Proliferating and Quiescent Epithelial Cells and Regulates Cell Proliferation. Cancer Res. 60 (5), 1236–1244. [PubMed] [Google Scholar]
- Stanietsky N., Simic H., Arapovic J., Toporik A., Levy O., Novik A., et al. (2009). The Interaction of TIGIT With PVR and PVRL2 Inhibits Human NK Cell Cytotoxicity. Proc. Natl. Acad. Sci. U.S.A. 106 (42), 17858–17863. 10.1073/pnas.0903474106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thies A., Berlin A., Brunner G., Schulze H. J., Moll I., Pfüller U., et al. (2007). Glycoconjugate Profiling of Primary Melanoma and its Sentinel Node and Distant Metastases: Implications for Diagnosis and Pathophysiology of Metastases. Cancer Lett. 248 (1), 68–80. 10.1016/j.canlet.2006.05.020 [DOI] [PubMed] [Google Scholar]
- Thies A., Moll I., Berger J., Wagener C., Brümmer J., Schulze H. J., et al. (2002). CEACAM1 Expression in Cutaneous Malignant Melanoma Predicts the Development of Metastatic Disease. J. Clin. Oncol. 20 (10), 2530–2536. 10.1200/jco.2002.05.033 [DOI] [PubMed] [Google Scholar]
- van Sorge N. M., Bonsor D. A., Deng L., Lindahl E., Schmitt V., Lyndin M., et al. (2021). Bacterial Protein Domains With a Novel Ig-Like Fold Target Human CEACAM Receptors. EMBO J., 40 (7), e106103. 10.15252/embj.2020106103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamura K., Baba Y., Nakagawa S., Mima K., Miyake K., Nakamura K., et al. (2016). Human Microbiome Fusobacterium nucleatum in Esophageal Cancer Tissue is Associated With Prognosis. Clin. Cancer Res. 22 (22), 5574–5581. 10.1158/1078-0432.ccr-16-1786 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.