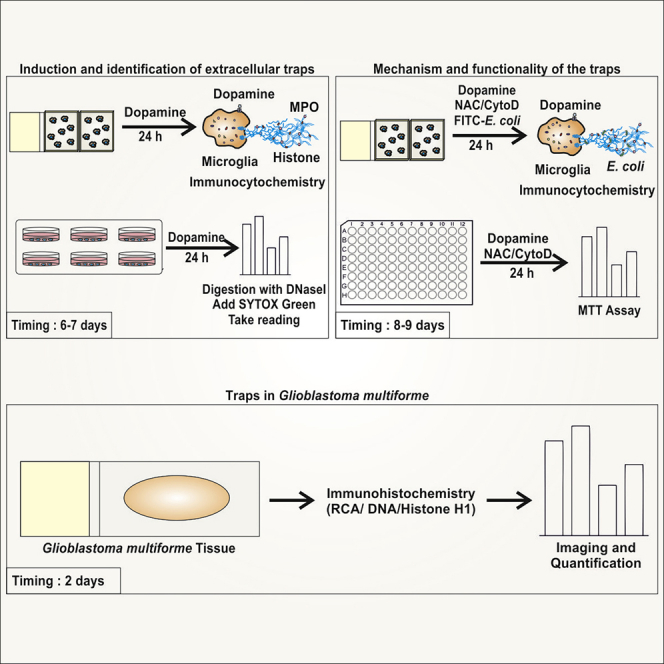
Summary
Extracellular traps (ETs) are composed of decondensed chromatin and are embedded with various antimicrobial proteins like myeloperoxidase and histones. Recently, we reported that dopamine (DA) induces ETs in BV2 microglia cell line and primary adult human microglia in a manner independent of cell death, reactive oxygen species, and actin polymerization. This protocol details how to characterize DA-induced ETs in BV2 microglia and human microglia. The protocols for characterization of ETs may also be used for other adherent cell lines.
For complete details on the use and execution of this protocol, please refer to Agrawal et al. (2021).
Subject areas: Cell-based Assays, Immunology, Microscopy, Molecular Biology, Neuroscience
Graphical abstract
Highlights
-
•
Induction and visualization of microglia ETs in cell lines and primary microglia cells
-
•
Quantification of microglia ETs formed by cell lines and present in glioblastoma tissues
-
•
Protocol for investigating the mechanism and functionality of microglia ETs
Extracellular traps (ETs) are composed of decondensed chromatin and are embedded with various antimicrobial proteins like myeloperoxidase and histones. Recently, we reported that dopamine (DA) induces ETs in BV2 microglia cell line and primary adult human microglia in a manner independent of cell death, reactive oxygen species, and actin polymerization. This protocol details how to characterize DA-induced ETs in BV2 microglia and human microglia. The protocols for characterization of ETs may also be used for other adherent cell lines.
Before you begin
Cell culture
-
1.
A T-25 flask of BV2 cell (Passage 18–43) with 80% confluency is needed. The protocol below describes the steps to be used for Myeloperoxidase staining of BV2 cells. This can also be used for primary human adult microglia by standardizing antibody dilutions. DNA/Histone H1 and RCA staining protocol for primary human adult microglia have been described which can also be used for BV2 microglia cells. The protocol for isolation of primary human adult microglia has been explained in the article published by Agrawal et al. (Agrawal et al., 2020).
Paraffin-embedded glioblastoma multiforme tissue
-
2.
For Immunohistochemistry, pathologist graded, paraffin embedded paraformaldehyde fixed glioblastoma multiforme (GBM) tissues are needed.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-DNA/Histone H1 (1:1500) | Merck | MAB3864 |
| Anti-Neutrophil Myeloperoxidase antibody (1:250) | Sigma-Aldrich | N5787 |
| Goat Anti-Rabbit IgG, Alexa Fluor 488 (1:1000) | Invitrogen | A-11034 |
| Goat Anti-Rabbit IgG, Alexa Fluor 594 (1:1000) | Invitrogen | A-11012 |
| Goat Anti-Mouse IgG, Alexa Fluor 594 (1:500) | Invitrogen | A-11005 |
| Goat anti-mouse IgG-CFL 488 (1:500) | Santa Cruz Biotechnology | sc-362257 |
| Biological samples | ||
| Paraffin-embedded paraformaldehyde-fixed glioblastoma multiforme tissues and control normal non-tumor brain tissue | Tata Memorial Cancer Hospital, Mumbai, India. All India Institute of Medical Sciences Jodhpur, India |
NA |
| Chemicals, peptides, and recombinant proteins | ||
| DMEM medium with glutamine | Himedia | AL007S |
| Dopamine | Sigma-Aldrich | H8502 |
| Fetal bovine serum | Himedia | RM9955 |
| Antibiotic-Antimycotic Solution | Himedia | A002 |
| Trypsin-EDTA (0.25%) | Gibco | 25200-056 |
| Fluoroshield with DAPI | Sigma-Aldrich | F6057 |
| SYTOX™ Green Nucleic Acid Stain | Invitrogen | S7020 |
| 2’,7’-Dichlorofluorescin diacetate | Sigma-Aldrich | D6883 |
| Xylene | Himedia | AS078 |
| Methylthiazolyldiphenyl-tetrazolium bromide | Sigma-Aldrich | M5655 |
| Isopropanol | Himedia | MB063 |
| Fluorescein Ricinus communis agglutinin-1 lectin | Vector Labs | FL-1081 |
| Ethanol | Changshu Hongsheng Fine Chemical Co. Ltd. | 1170 |
| N-acetyl-L-cysteine | Sigma-Aldrich | A9165 |
| Cytochalasin D | Sigma-Aldrich | C2618 |
| DNase I | Himedia | ML068 |
| Triton X-100 | Sigma-Aldrich | T8787 |
| Fluoroshield™ with DAPI | Sigma-Aldrich | F6057 |
| Ethylenediaminetetraacetic acid (EDTA) | Himedia | MB011 |
| Formaldehyde | Himedia | MB059 |
| Experimental models: cell lines | ||
| BV2 microglia cell line | Dr. Anirban Basu, National Brain Research Center (NBRC) | NA |
| Software and algorithms | ||
| ImageJ | Schneider et al., 2012 | https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5554542/ |
| Algorithm code for ET quantification in GBM tissues | Agrawal et al., 2021 | https://www.cell.com/iscience/fulltext/S2589-0042(20)31165-2 |
| Other | ||
| FITC tagged Escherichia coli | Vybrant Phagocytosis Assay Kit | V-6694 |
| Fluorescent microscope | Leica | Leica DM2000 LED |
| Plate reader | BioTek | Synergy H1 |
| 6-Well plates | LabWare | LW-TC006 |
| Coplin jars | Thermo Scientific | 1001363 |
Materials and equipment
| 10 X PBS – 1 L | Final concentration | Amount |
|---|---|---|
| NaCl | 1.37 M | 80 g |
| KCl | 27 mM | 2 g |
| Na2HPO4 | 10 mM | 14.4 g |
| KH2PO4 | 18 mM | 2.4 g |
| Culture Medium – 100mL | ||
| DMEM | 90% | 90mL |
| Fetal Bovine Serum | 10% | 10mL |
| Antibiotic-Antimycotic solution | 1% | Volume depends upon the amount of media added in culture flask |
Note: 10× PBS can be stored at 20°C–25°C for up to 1 year. Fresh buffer should be prepared if it is turbid. It is recommended to prepare culture medium in small volumes (50ml × 2) as needed. It can be stored at 4°C for a month. Antibiotic-Antimycotic solution should be added in the culture flask instead of adding it to the stock culture medium.
Step-by-step method details
Induction and identification of extracellular traps
Timing: Visualization of extracellular traps will take 4–5 days from seeding of the BV2 cells. Quantification of traps will take 3 days from the seeding of cells.
This step explains the induction of extracellular traps by dopamine (DA) and their identification by staining with 4′, 6-Diamidine-2′-phenylindole dihydrochloride (DAPI) and myeloperoxidase (MPO) antibody. It also explains the identification of extracellular traps in culture supernatant.
-
1.
Plate 2 × 104 BV2 cells into two well chamber slide or 2 × 105 cells in 6-well plate as needed for the experiment. Use 1 mL culture medium for two well chamber slides and 2 mL culture medium for 6-well plate. Incubate the cells 12 h–16 h at 37°C and 5% CO2.
CRITICAL: Remember to keep untreated wells as experimental secondary antibody control for Immunocytochemistry (ICC). For 6-well plate only untreated well is needed as experimental control.
-
2.
Treat cells with 250 μM of dopamine dissolved in water (Stock concentration = 20 mM) and incubate for 24 h at 37°C and 5% CO2.
-
3.Perform ICC on cells in two well chamber slides for staining of extracellular traps.Note: This step helps in visualization of the ETs produced.
-
a.Discard the medium and wash the cells two times with 1 × PBS for 5 min.
-
b.Fix the cells with 500 μL 4% PFA (prepared in 1 × PBS) for 10 min at room temperature (RT; 20°C–25°C).
 CRITICAL: Formaldehyde is hazardous. Use gloves and protective glasses while preparing.
CRITICAL: Formaldehyde is hazardous. Use gloves and protective glasses while preparing. -
c.Wash the cells two times with 1 × PBS for 5 min.
-
d.Permeabilize cells with 500 μL of 0.1% Triton X-100 (prepared in 1 × PBS) for 15 min at RT.
-
e.Block with 500 μL of 5% FBS in 0.1% Triton X-PBS (Blocking Buffer) for 1 h at 4°C in a humidified chamber.
 Pause point: At this step the protocol can be paused for longer if needed. Overnight blocking (12 h) can also be done.
Pause point: At this step the protocol can be paused for longer if needed. Overnight blocking (12 h) can also be done. -
f.Add 500 μL of 1:250 dilution of MPO primary antibody prepared in blocking buffer in the marked chambers and incubate 12 h–16 h at 4°C in a humidified chamber.
-
g.Discard primary antibody and wash the cells two times with 1 × PBS for 5 min.
 CRITICAL: Discard the supernatant in beaker containing 10% bleach. The solution should be further discarded as per the guidelines for discarding biological waste materials. Carefully tilt the chamber slides and aspirate the supernatant slowly from a corner using a pipette to avoid detachment of cells.
CRITICAL: Discard the supernatant in beaker containing 10% bleach. The solution should be further discarded as per the guidelines for discarding biological waste materials. Carefully tilt the chamber slides and aspirate the supernatant slowly from a corner using a pipette to avoid detachment of cells. -
h.Add 500 μL of 1:1000 dilution of anti-rabbit secondary antibody prepared in blocking buffer in each chamber and incubate at room temperature for 1 h. From here all the steps will be performed in dark.
-
i.Discard secondary antibody and wash the cells two times with 1 × PBS for 5 min.
-
j.Dry the slides and mount them with VECTASHIELD mounting medium containing DAPI.
-
k.Keep the slides at 4°C for 10 min to allow coverslips to settle properly. Slides are ready to be imaged (Figure 1).
-
a.
-
4.Quantify the amount of extracellular traps present in the culture supernatant.Note: This protocol was adopted from Robledo-Avila et al., Yoo et al. and Joshi et al. with modifications (Robledo-Avila et al., 2018, Yoo et al., 2014, Joshi et al., 2013). While ICC helps in visualization of the ETs, this step helps in quantifying the amount of traps produced. This determines, in an unbiased way, whether traps production is significant or not.
-
a.Add 10 U/mL DNase I to media in the wells to digest the traps. Mix well and incubate at RT for 15 min.
-
b.Stop the reaction by adding 5 nM EDTA. Mix well and collect supernatants in clean microfuge tubes.
-
c.Centrifuge at 300 × g for 5 min at RT to get rid of cell debris. Transfer supernatant into clean microfuge tubes. Keep them on ice until ready.
-
d.Add 200 μL of collected supernatant in 96-well plate.
-
e.Add 5 μM SYTOX Green in the wells.
-
f.Incubate for 15 min at room temp in dark.
-
g.Measure fluorescence using a plate reader at excitation/emission = 485 nm/530 nm.
-
a.
Note: Blank will be the culture supernatant from untreated well without SYTOX Green.
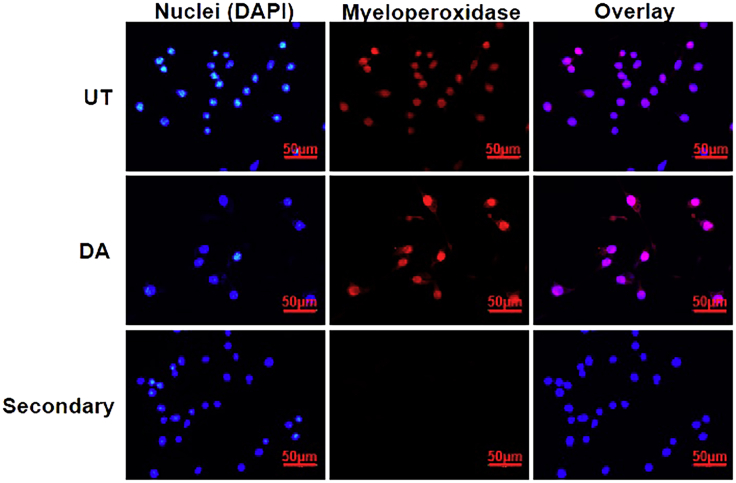
Figure 1.
DA-induced extracellular traps in BV2 microglia
BV2 microglia were incubated with 250 mM of DA for 24 h. ETs were stained with DAPI (blue) and MPO (Red). The images are representative of three experiments. At least 7 frames were imaged per well in a two well chamber slide. Scale bars 50 μm.
Mechanism and functionality of the traps
Timing: Protocol for checking dopamine induced reactive oxygen species (ROS) production will take 3 days from the seeding of BV2 cells. Protocol for checking the role of ROS in generation of traps and the functionality of traps will take 4–5 days from seeding of the BV2 cells. MTT assay will take 3 days from the seeding of cells.
This step will determine whether the extracellular traps formation is dependent or independent of cell death, reactive oxygen species (ROS) and actin polymerization. The step will also check the functionality of the traps by investigating their ability to tap FITC tagged Escherichia coli.
-
5.Check whether Dopamine induces ROS production or not.
-
a.Seed 10,000 cells per well in duplicates in 96-well microtiter plate and incubate 12h–16 h at 37°C in 5% CO2
-
b.Add 20 μM of (2′,7′-Dichlorofluorescin diacetate) DCF-DA (prepared in DMSO, Stock concentration = 2 mM) and incubate at 37°C in 5% CO2 for 30 min.
-
c.Discard the culture media and wash cells once with 1 × PBS. Be gentle with pipetting.
-
d.Add 100 μL of fresh culture media to each well.
-
e.Treat the cells with 10 mM NAC for 3 h at 37°C in 5% CO2.
-
f.Treat cells with 250 μM of dopamine for 24 h at 37°C in 5% CO2.
-
g.Measure fluorescence at excitation/emission =485 nm/Em=535 nm.
-
a.
Note: cells can also be treated with other compounds as required by the experiment and the ROS production can be measured.
-
6.Treat the cells with N-acetyl-L-cysteine (NAC) or cytochalasin D (CytoD) to check whether extracellular traps formation is dependent of independent of ROS or actin polymerization.
-
a.Plate 2 × 104 BV2 cells into two well chamber slide or 2 × 105 cells in 6-well plate as needed for the experiment. Use 1 mL culture medium for two well chamber slides and 2 mL culture medium for 6-well plate. Incubate the cells 12h–16 h at 37°C and 5% CO2.
-
b.Treat the cells with 10 mM NAC for 3 h or 10 μM CytoD for 20 min. Incubate at 37°C in 5% CO2.
-
c.Treat the cells with 250 μM dopamine and incubate at 37°C in 5% CO2 for 24 h.
-
d.Perform ICC on cells in two well chamber slides for staining of the traps as explained in step 3 of induction and identification of extracellular traps section, and take images.
-
a.
-
7.Incubate the cells with FITC tagged Escherichia coli to check the functionality of the traps.
-
a.Plate 2 × 104 BV2 cells into two well chamber slide or 2 × 105 cells in 6 well plate as needed for the experiment. Use 1 mL culture medium for two well chamber slides and 2 mL culture medium for 6 well plate. Incubate the cells 12h–16 h at 37°C and 5% CO2.
-
b.Treat the cells with 10 mM NAC for 3 h or 10 μM CytoD for 20 min. Incubate at 37°C in 5% CO2.
-
c.Treat the cells with 250 μM dopamine and incubate at 37°C in 5% CO2 for 3 h.
-
d.Add 40 μL of FITC tagged E. coli from Vybrant Phagocytosis Assay Kit to the wells and incubate for additional 21 h at 37°C in 5% CO2.Alternatives: FITC tagged E. coli can be used from any source. We used the Vybrant Phagocytosis Assay kit as it was available.
-
e.Perform ICC on cells in two well chamber slides for staining of the traps as explained in step 3 of induction and identification of extracellular traps section, and take images.
-
a.
Note: The steps for staining with primary and secondary antibody can be skipped as the ETs will be stained with DAPI.
-
8.Perform MTT assay to check for the cytotoxicity of dopamine. This step will help in determine whether dopamine induced trap formation are cell death dependent or independent.
-
a.Seed 10,000 cells per well in duplicates in 96-well microtiter plate and incubate 12h–16 h at 37°C in 5% CO2.
-
b.Treat the cells with 250 μM of dopamine and incubate at 37°C and 5% CO2 for 24 h.Note: Cells can be treated with other compounds as required by the experiment.
-
c.Remove the media carefully from the wells.
-
d.Add 100 μL of fresh serum free media to wells
-
e.Add 10 μL of 5 mg/mL MTT (prepared in 1 × PBS) to each well.
-
f.Incubate the plate at 37°C and 5% CO2 for 2 h.
-
g.Add 100 μL of acidic isopropanol to each well and mix properly.
-
h.Measure the absorbance at 570 nm using a plate reader.
-
a.
Identification and quantification of traps in glioblastoma multiforme tissues
Timing: The protocol will take 2 days.
This step will confirm the presence of extracellular traps in GBM tissues. Paraffin embedded, pathologist graded, GBM tissues are required for this step. Use tissue sections from normal brain cerebrum as reference and secondary control. The human tissue sections were prepared and section as per protocol explained by Fischer et al. (Fischer et al., 2008b, Fischer et al., 2008a).
Note: This protocol is adapted from Jha et al. with slight modifications (Jha et al., 2010).
-
9.Wash the 5 micron thick paraffin embedded tissue section slides with xylene and rehydrate slides through decreasing concentration of alcohol.
-
a.2 washes with 100% xylene for 20 min in 2 different coplin jars.
-
b.2 washes with 100% ethanol for 8 min in 2 different coplin jars.
-
c.2 washes with 95% ethanol for 8 min in 2 different coplin jars.
-
d.2 washes with 70% ethanol for 8 min in 2 different coplin jars.
-
e.2 washes with 1 × PBS for 8 min in 2 different coplin jars.
-
a.
CRITICAL: Follow the wash timings strictly.
-
10.
Mark around the tissue using ImmEdge Hydrophobic Barrier PAP Pen.
-
11.
Permeabilize the tissue with permeabilizing buffer (0.1% Triton X-100 in 1 × PBS) for 15 min at RT.
-
12.
Remove the permeabilization buffer and stain with Ricinus communis agglutinin-1 (RCA) (1:500 prepared in permeabilizing buffer) at 37°C for 1 h.
CRITICAL: Further steps of the protocol must be performed in dark.
-
13.
Block with 5% FBS in 0.1% TritonX-PBS (Blocking Buffer) for 1 h at 4°C in a humidified chamber.
Pause point: At this step the protocol can be halted if needed. Blocking can be done for up to 2 h.
-
14.
Add 1:1500 dilution of Anti DNA/Histone H1 primary antibody prepared in blocking buffer in the marked chambers and incubate 12h–16 h at 4°C in humidified chamber.
-
15.
Remove primary antibody and wash the slides 3 times in 1× PBS for 5 min in 3 different coplin jars.
-
16.
Add 1:500 dilution of anti-mouse secondary antibody and incubate the slides 37°C for 1 h in dark.
-
17.
Remove secondary antibody and was the slides 3 times in 1× PBS for 5 min in 3 different coplin jars.
-
18.
Dry the slides and mount them with Fluoroshield with DAPI .
-
19.
Keep the slides at 4°C for 10 min to allow coverslips to settle properly. Slides are ready to be imaged.
-
20.
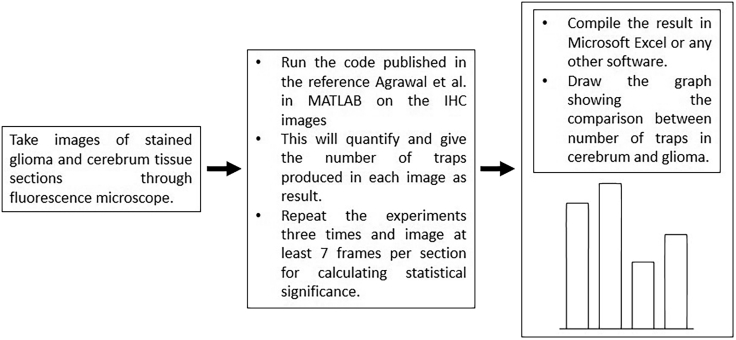
Quantify the traps using the MATLAB code published by Agrawal et al. (Agrawal et al., 2021) (Figure 2).
Figure 2.
Flow chart for the quantification protocol to be followed
Expected outcomes
This protocol explains characterization of extracellular traps by staining them with DAPI, MPO and DNA/Histone H1. ETs induced by dopamine or other stimuli can be stained and identified using this protocol. The fluorescence absorbance of SYTOX Green in the culture supernatant will quantify the traps present in the culture supernatant. Further this protocol allows quantification of traps formed in ROS, actin polymerization or cell death dependent or independent manner. The presence of ETs in GBM tissues can also be investigated and quantified using the code published (Agrawal et al., 2020).
Quantification and statistical analysis
ImageJ was used for processing of all the images (Schneider et al., 2012). Microsoft excel was used for statistical analyses. Data significance was calculated using Student’s t-test. Repeat all the experiments thrice for calculating significance.
Limitations
The protocol investigates whether extracellular traps are formed in a cell death dependent or independent manner. However, in case of CytoD treatment some cells may die. The protocol does not identify whether dying cells are also forming ETs. In case of GBM staining the protocol only identifies microglia extracellular traps as RCA stains microglia. For identifying ETs formed by other cells present in GBM other cellular markers are needed.
Troubleshooting
Problem 1
Secondary signal in Immunocytochemistry and immunohistochemistry (step 3, 14 and 16).
Potential solution
Primary and secondary antibody concentration has to be standardized. Experiments can be done initially starting with the concentration recommended in the antibody catalog. Then different concentrations of antibody should be used to establish ideal working concentration for optimized results. Working concentrations of antibodies from different companies will need to be standardized.
Problem 2
PFA fixation may lead to high background in Immunocytochemistry (step 3).
Potential solution
If the background is too high number of washes should be increased. However too many washes might also lead to washing of extracellular traps. Number of washing steps should be optimized carefully so there will be no loss of signal.
Problem
Nonspecific random signals while checking the functionality of the traps (step 7).
Potential solution
Proper washing of the slides is critical while checking the functionality of the traps. If not washed properly, FITC tagged E. coli might adhere nonspecifically on the slides giving random signals. However it should be kept in mind that the washing should not be very harsh as it may wash the traps too. Increase one more wash with the shaker on moderate speed if random signals are observed.
Problem 4
Washing of the tissue sections (step 9).
Potential solution
It is possible that tissue sections made for IHC may get washed away while processing. Use of charged slides is recommended as it helps in better attachment of the tissue to the slides. If the sections made are too old then fresh sections should be made.
Problem 5
Old dopamine solution (steps 2, 5, 6, 7 and 8).
Potential solution
Dopamine can degrade with time in solution. It is recommended to prepare fresh dopamine solution every time to maintain consistency in the results.
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Dr. Sushmita Jha (sushmitajha@iitj.ac.in).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Data and code availability
The algorithm and code generated during the study have been published in iScience by Agrawal et al.
Acknowledgments
S.J.’s laboratory was established with institutional grants from IITJ and is funded by grants from the Department of Biotechnology (BT/PR12831/MED/30/1489/2015) and the Ministry of Electronics and Information Technology, Government of India (No.4 (16)/2019-ITEA). We thank Dr. Anirban Basu’s Lab at the National Brain Research Centre (NBRC), India, for their kind gift of the BV2 microglia cell line. Modified histone antibodies were a kind gift from Dr. Vijayalakshmi Mahadevan from the Institute of Bioinformatics and Applied Biotechnology. Human brain tissue sections were obtained from the All India Institute of Medical Sciences (AIIMS), Jodhpur, and Tata Memorial Hospital, Tata Memorial Centre, Mumbai, Maharashtra, India. We thank Dr. Pushpa Potaliya and Dr. Vineet Jain, Department of Anatomy, AIIMS, Jodhpur, for valuable assistance in histological processing of the tissues.
Author contributions
I.A. designed and performed the experiments (data analysis, standardized primary microglia isolation, immunofluorescence), as well as prepared and edited the initial manuscript draft. N.S. performed BV2 microglia cell culture. S.S. did some immunocytochemistry experiments. D.B.C. wrote the code for ET quantification, and D.C. and S.A. helped with quantification. D.J. provided brain tissues for microglia isolation. S.G., S.E., and T.G. provided tissue sections for IHC studies. S.J. conceptualized the study, designed experiments, wrote the initial draft, and edited and reviewed the manuscript. All authors reviewed the manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Ishan Agrawal, Email: agrawal.5@iitj.ac.in.
Sushmita Jha, Email: sushmitajha@iitj.ac.in.
References
- Agrawal I., Saxena S., Nair P., Jha D., Jha S. Obtaining human microglia from adult human brain tissue. J. Vis. Exp. 2020 doi: 10.3791/61438. [DOI] [PubMed] [Google Scholar]
- Agrawal I., Sharma N., Saxena S., Arvind S., Chakraborty D., Chakraborty D.B., Jha D., Ghatak S., Epari S., Gupta T. Dopamine induces functional extracellular traps in microglia. iScience. 2021;24:101968. doi: 10.1016/j.isci.2020.101968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A.H., Jacobson K.A., Rose J., Zeller R. Cutting sections of paraffin-embedded tissues. Cold Spring Harb. Protoc. 2008;2008 doi: 10.1101/pdb.prot4987. pdb.prot4987. [DOI] [PubMed] [Google Scholar]
- Fischer A.H., Jacobson K.A., Rose J., Zeller R. Paraffin embedding tissue samples for sectioning. Cold Spring Harb. Protoc. 2008;2008 doi: 10.1101/pdb.prot4989. pdb.prot4989. [DOI] [PubMed] [Google Scholar]
- Jha S., Srivastava S.Y., Brickey W.J., Iocca H., Toews A., Morrison J.P., Chen V.S., Gris D., Matsushima G.K., Ting J.P. The inflammasome sensor, NLRP3, regulates CNS inflammation and demyelination via caspase-1 and interleukin-18. J. Neurosci. 2010;30:15811–15820. doi: 10.1523/JNEUROSCI.4088-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi M.B., Lad A., Bharath Prasad A.S., Balakrishnan A., Ramachandra L., Satyamoorthy K. High glucose modulates IL-6 mediated immune homeostasis through impeding neutrophil extracellular trap formation. FEBS Lett. 2013;587:2241–2246. doi: 10.1016/j.febslet.2013.05.053. [DOI] [PubMed] [Google Scholar]
- Robledo-Avila F.H., Ruiz-Rosado J.D.D., Brockman K.L., Kopp B.T., Amer A.O., Mccoy K., Bakaletz L.O., Partida-Sanchez S. Dysregulated calcium homeostasis in cystic fibrosis neutrophils leads to deficient antimicrobial responses. J. Immunol. 2018;201:2016–2027. doi: 10.4049/jimmunol.1800076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo D.-G., Floyd M., Winn M., Moskowitz S.M., Rada B. NET formation induced by Pseudomonas aeruginosa cystic fibrosis isolates measured as release of myeloperoxidase–DNA and neutrophil elastase–DNA complexes. Immunol. Lett. 2014;160:186–194. doi: 10.1016/j.imlet.2014.03.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The algorithm and code generated during the study have been published in iScience by Agrawal et al.