Abstract
Background and Objective
Coronavirus disease 2019 is a novel disease caused by the severe acute respiratory syndrome coronavirus (SARS-CoV)-2 virus. It was first detected in December 2019 and has since been declared a pandemic causing millions of deaths worldwide. Therefore, there is an urgent need to develop effective therapeutics against coronavirus disease 2019. A critical step in the crosstalk between the virus and the host cell is the binding of the receptor-binding domain (RBD) of the SARS-CoV-2 spike protein to the peptidase domain of the angiotensin-converting enzyme 2 (ACE2) receptor present on the surface of host cells.
Methods
An in silico approach was employed to design a 13-amino acid peptide inhibitor (13AApi) against the RBD of the SARS-CoV-2 spike protein. Its binding specificity for RBD was confirmed by molecular docking using pyDockWEB, ClusPro 2.0, and HDOCK web servers. The stability of 13AApi and the SARS-CoV-2 spike protein complex was determined by molecular dynamics simulation using the GROMACS program while the physicochemical and ADMET (absorption, distribution, metabolism, excretion, and toxicity) properties of 13AApi were determined using the ExPASy tool and pkCSM server. Finally, in vitro validation of the inhibitory activity of 13AApi against the spike protein was performed by an enzyme-linked immunosorbent assay.
Results
In silico analyses indicated that the 13AApi could bind to the RBD of the SARS-CoV-2 spike protein at the ACE2 binding site with high affinity. In vitro experiments validated the in silico findings, showing that 13AApi could significantly block the RBD of the SARS-CoV-2 spike protein.
Conclusions
Blockage of binding of the SARS-CoV-2 spike protein with ACE2 in the presence of the 13AApi may prevent virus entry into host cells. Therefore, the 13AApi can be utilized as a promising therapeutic agent to combat coronavirus disease 2019.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40268-021-00357-0.
Key Points
| Coronavirus disease 2019 is an ongoing pandemic caused by severe acute respiratory syndrome coronavirus (SARS-CoV)-2. The spike protein of SARS-CoV-2 interacts with the angiotensin-converting enzyme 2 receptor present on the host cell surface, leading to viral entry and infection. |
| A 13-amino acid novel peptide inhibitor (FLDKFNHNFKDLF) has been designed to block the receptor-binding domain of the SARS-CoV-2 spike protein, which impedes its interaction with the host angiotensin-converting enzyme 2 receptor. |
| The 13-amino acid peptide inhibitor is expected to restrict virus interaction and fusion with the host cells, thereby making it a promising therapeutic candidate to combat coronavirus disease 2019. |
Introduction
The pandemic of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), with more than 180 million infections and close to four million deaths, can be designated as one of the greatest threats of the century to public health. It has not only posed a significant challenge to human health globally but also caused socio-economic crises for many countries [1, 2]. The clinical spectrum of coronavirus disease 2019 (COVID-19) ranges from mild fever, cough, and shortness of breath, to severe clinical conditions characterized by respiratory failure and other long-term associated conditions [3–7]. Old age, together with pre-existing conditions such as lung or heart disease, diabetes mellitus, or a compromised immune system are known to expedite the infection time and severity [8, 9].
Structurally, the coronavirus is a single-stranded, non-segmented, positive-sense RNA virus that consists of the largest known RNA genome of 26–32 kB compared to other known viruses [10, 11]. The four major structural proteins encoded by its genome include the spike (S) protein, envelope (E) protein, membrane (M) protein, and nucleocapsid (N) protein as well as several non-structural proteins including viral proteases [12, 13]. The surface-bound spike protein, suggested to be highly susceptible to mutations, plays a crucial role in facilitating virus entry by mediating its interaction with transmembrane surface receptors on host cells. The receptor-binding domain (RBD) of the SARS-CoV-2 spike protein interacts with the peptidase domain (PD) of the angiotensin-converting enzyme 2 (ACE2) receptor on host cells, which technically marks virus entry inside the cells [14–18]. The S1 and S2 subunits in the spike protein of SARS-CoV-2 are mainly responsible for the interaction and fusion with the host cells for virus entry. The RBD in the S1 subunit initiates direct binding with the ACE2 PD, whereas the S2 subunit contains essential elements needed for membrane fusion [16, 19–22].
In humans, ACE2 is a type I, transmembrane endothelium-bound metallo-carboxypeptidase, with homology to the angiotensin-converting enzyme, well known for its role in the renin-angiotensin system [23–26]. ACE2 is a target for the treatment of hypertension [27, 28]. It is mainly expressed on endothelial cells of several organs, particularly the cardiovascular system, renal tubular epithelium, Leydig cells in the testes, and alveolar epithelial type II cells in the lungs and brain [23, 29–31]. The full-length structure of ACE2 consists of two main domains: the PD at the N-terminus and the collectrin-like domain at the C-terminus [32–34]. The spike glycoprotein of SARS-CoV-2 is known to bind to a homodimer of ACE2 [16, 17, 32].
In the current SARS-CoV-2 pandemic, a vaccine is rightfully anticipated as the most effective therapy. However, in an ongoing pandemic, vaccine development and production may not be equally efficacious in all countries. Therefore, potent, easy to generate, and cheap therapeutic agents that can effectively curb infection at the early stages also need to be developed. Several approaches, such as decoy-soluble ACE2 proteins, antibodies from the serum of infected patients, epitope-based vaccines, repurposing of drugs, and designing peptide inhibitors have been reported [35–44]. Peptides possess several attractive features compared with small-molecule and protein therapeutics, including high structural compatibility with target proteins, and the ability to specifically disrupt protein–protein interfaces [45, 46].
Effective use of computational tools can ease the way to rapidly reach a therapeutic solution for COVID-19. In the current study, we used computational biology tools to develop a therapeutic strategy utilizing a novel peptide that can inhibit the SARS-CoV-2/ACE2 interaction. We have designed a short and efficient peptide that imitates the binding of ACE2 with RBD of the SARS-CoV-2 spike protein and could significantly block their interaction in an in vitro assay system.
Materials and Methods
Structure Retrieval, Interaction Study, Peptide Design, Structure Preparation, and Molecular Docking Studies
The experimentally solved x-ray crystallographic structure of the SARS-CoV-2 spike protein and human ACE2 protein complex with PDB ID 6M17 was retrieved from the RCSB PDB database (https://www.rcsb.org/). The interaction between the SARS-CoV-2 spike protein and the ACE2 receptor protein was identified and analyzed using the Chimera Version 1.13.1 tool [47]. We focused on the interface residues of both proteins that are involved in the interaction within the 3Å region.
To target the RBD residues of the SARS-CoV-2 spike protein, a decoy peptide from the region of its interaction with ACE2 was designed. A 13-amino acid (13AA) stretch from F28 to L40 (FLDKFNHEAEDLF) from the PD of ACE2 was selected to design a short and effective peptide inhibitor. Modifications of the 13AA peptide sequence were carried out to improve its binding affinity without disturbing its physiochemical properties. Substitutions of residues at the eighth, ninth, and tenth positions gave us a 13AA peptide inhibitor (13AApi) with sequence FLDKFNHNFKDLF, which showed higher binding affinity and specificity. The 13AApi sequence was modeled in a three-dimensional structure and energetically minimized on Chimera using the Amber ff14SB force field [47].
The SARS-CoV-2 spike RBD structure obtained from PDB 6M17 was prepared for docking experiments by removing water, heteroatoms, and ligand groups, followed by adding polar hydrogen atoms to the structure using the Discovery Studio Version 21.1.0 tool [48]. Molecular docking of the 13AApi with SARS-CoV-2 spike RDB was carried out in pyDockWEB [49] and cross-checked with the HDOCK [50] and ClusPro 2.0 [51] web server tools. The docking results were visualized and analyzed in Discovery Studio Visualizer Version 21.1.0 and Chimera Version 1.13.1 tools, respectively [47, 48].
Molecular Dynamics (MD)
The system for MD simulation was prepared by soaking in a dodecahedron box filled with TIP3P water molecules. The simulation system was neutralized by adding Na+ or Cl− ions. The protein, solvent, and ion parameters were assigned using the Amber99SB-ILDN force field [52]. The initial configuration of the system was relaxed through the steepest descent energy minimization using a maximum number of steps value of 50,000. After energy minimization, all simulation systems were equilibrated in two phases: first under NVT ensemble for 1 ns to stabilize the temperature of the system and then under NPT ensemble for another 1 ns to stabilize the pressure of the simulation system. Finally, production MD runs were carried out at a constant temperature of 300 K for 500 ns using GROMACS Version 5.0.4. [53]. A time step of 2 fs was used for the simulations. A cut-off of 10 Å was used for short-range interactions while the particle-mesh Ewald method was used to handle long-range interactions. Coordinates and velocities were saved every 10 ps. The MD trajectory was processed and analyzed using MDAnalysis [54], MDTraj [55], and scikit-learn [56] python libraries. Principal component analysis was performed using MODE-TASK [57]. The MM-PBSA method was employed to evaluate the 13AApi binding energies. MM-PBSA energy calculations were performed using the g_mmpbsa program [58, 59]. Graphics were prepared using Matplotlib python library [60] and R [61].
Physicochemical and ADMET (Adsorption, Distribution, Metabolism, Excretion, and Toxicity) Analysis of 13AApi
The physicochemical and ADMET properties of the 13AApi were analyzed from ExPasy ProtParam (https://web.expasy.org/protparam/) [62] and pkCSM (http://biosig.unimelb.edu.au/pkcsm/prediction) [63] tools available online. For ADMET analysis, the pkCSM tool requires a SMILES file format of the 13AApi, which was generated using the pepSMI (https://www.novoprolabs.com/tools/convert-peptide-to-smiles-string) web tool.
Enzyme-Linked Immunosorbent Assay (ELISA)
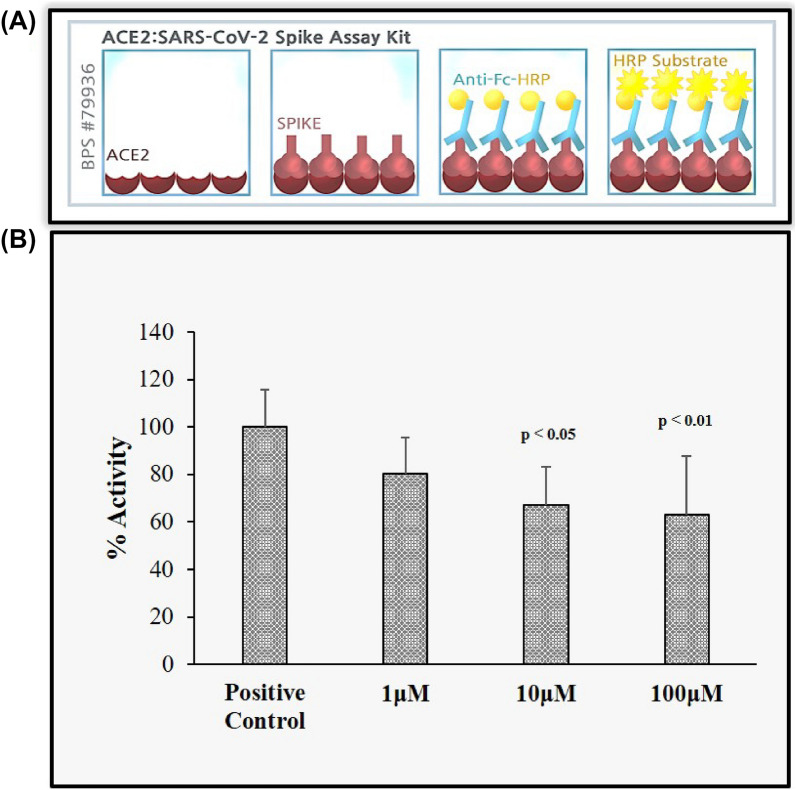
An in vitro assay was performed to validate the in silico interaction data with 13AApi. The percentage binding of the SARS-CoV-2 spike protein to ACE2 in the presence of the 13AApi was determined with an ACE2:SARS-CoV-2 spike inhibitor screening assay kit (BPS Bioscience #79936; San Diego, CA, USA) according to the protocol described in the manufacturer’s datasheet. A stock solution of the 13AApi was prepared in distilled water and the indicated working concentrations were used in the assay. A chemiluminescent reading from three biological replicates was used to determine the 13AApi activity graph.
Results and Discussion
Structural Analysis of SARS-CoV-2 Spike Protein Interaction with Host Receptor ACE2 and Design of the Novel Peptide Inhibitor
A crucial step in SARS-CoV-2 infection is the attachment of its surface spike protein with the ACE2 surface receptor on the host cell; this binding has now been widely studied and delineated [16, 17]. In terms of therapeutic strategies whereby antiviral drugs can block this interaction, previous work has mainly demonstrated the host ACE2 domain as a target; very few studies have aimed to target the SARS-CoV-2 spike protein for drug development [64–68]. Targeting the SARS-CoV-2 spike protein instead of the host ACE2 protein ensures direct action on the virus. Of note, no major therapeutic breakthroughs have been reported in targeting the SARS-CoV-2 spike protein to disrupt this primary step of interaction and disable fusion of the virus with the host cell. Several peptide-based therapeutic options to target SARS-CoV-2 and inhibit its interaction with ACE2 receptors have been reported by us and many other laboratories [69–73]. However, these peptide inhibitors are long-stretch peptides that are mainly derived from ACE2 receptors; moreover, very few have been validated in a cell-based assay.
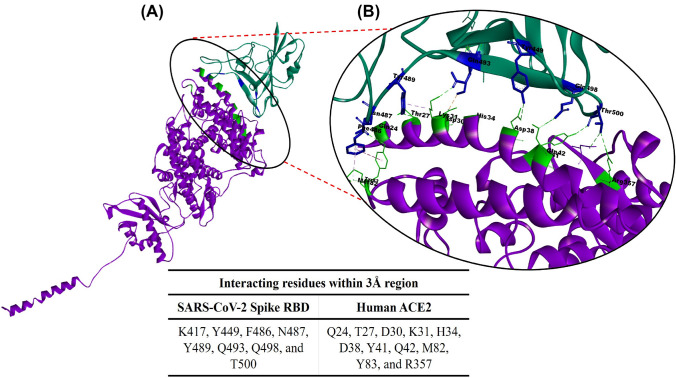
We have taken this opportunity to further explore and develop a short novel peptide inhibitor that can specifically and significantly target the RBD residues of the SARS-CoV-2 spike protein and block its interaction with the host, the ACE2 protein. The N-terminal residues of the ACE2 PD are mainly involved in its interaction with the spike protein. Therefore, a similar stretch of peptide that can mimic this binding was designed with the aim to block the RBD of the SARS-CoV-2 spike protein. The available crystal structure complex of the SARS-CoV-2 spike protein with human ACE2 (PDB ID 6M17) was retrieved from PDB database and explored as the basis of the current study. We analyzed the interaction interface and identified crucial amino acids involved in protein–protein interactions (Fig. 1A, B).
Fig. 1.
A Illustration of the interacting interface (encircled) of the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) spike receptor-binding domain [RBD] (green) and human angiotensin-converting enzyme 2 [ACE2] (purple) from the crystal structure (PDB ID 6M17). B Identification of the interacting residues within the 3 Å region. The highlighted residues in the SARS-CoV-2 spike RBD (blue) interact with human ACE2 peptidase domain residues (bright green) as demonstrated with the Discovery studio Visualizer tool. The Table shows the interacting residues within a 3 Å region that was analyzed using the Chimera tool
We next prepared a set of peptide sequences similar to the N-terminal region of the ACE2 PD that is known to interact with SARS-CoV-2 within a 3–5 Å region and tested their binding affinity and specificity with the RBD of the spike protein. The residues within the 3 Å region are crucial for a complex formation and blocking such residues could interfere with the interaction of the virus with the host cell. A 13AA stretch from F28 to L40 (FLDKFNHEAEDLF) in the PD of ACE2 was found to effectively bind to the spike protein RBD and was suitable as a template to design a short and effective peptide inhibitor. We performed modifications of the 13AA peptide sequence to improve its binding affinity without disturbing the RBD binding site and physiochemical properties. Substitution of the amino acid residues at the eighth, ninth, and tenth positions (E8N, A9F, and E10K) gave us a 13AA peptide inhibitor with sequence FLDKFNHNFKDLF, which showed higher binding affinity and specificity towards SARS-CoV-2 RBD. This peptide inhibitor was named 13AApi.
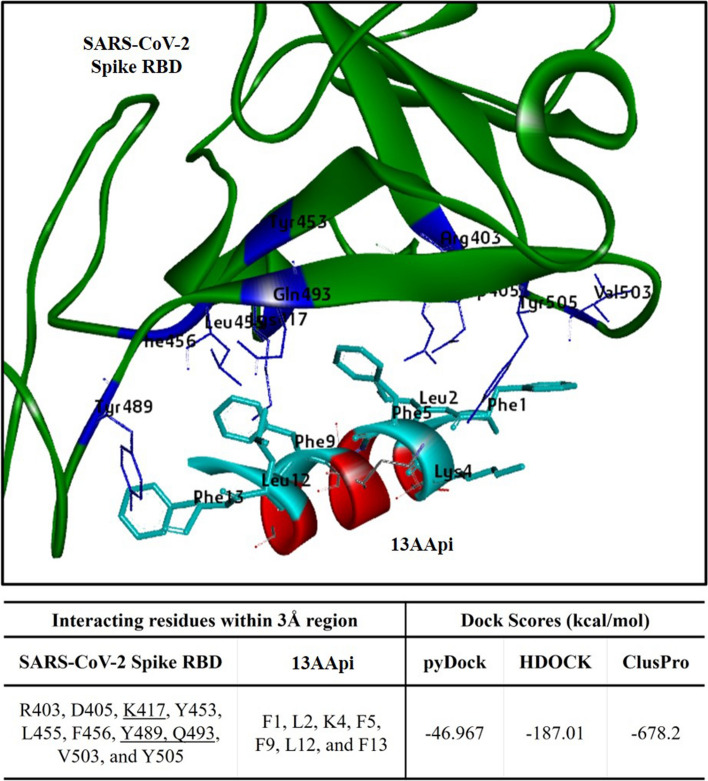
A protein-peptide blind docking study indicated that the 13AApi best-fit docking solution could bind to the residues of the spike protein RBD in the same pocket where the ACE2 PD binds (Fig. 2A). The binding energy of the initial peptide inhibitor 13AA (FLDKFNHEAEDLF) was obtained as − 42.177 kcal/mol in pyDockWEB while the docking score for the final peptide inhibitor 13AApi was increased to − 46.967 kcal/mol (Fig. 2A). We next cross-checked the docking on two other platforms: HDOCK and ClusPro 2.0. The dock score for 13AApi was − 187.01 kcal/mol in HDOCK and − 678.2 kcal/mol in ClusPro 2.0 while the scores of 13AA were − 168.91 kcal/mol in HDOCK and − 634.2 kcal/mol in ClusPro 2.0 (data not shown). Therefore, the results from HDOCK and ClusPro 2.0 are in good agreement with the pyDockWEB data, corroborating an enhancement in the docking score of 13AApi in comparison to the initial peptide. The binding position of the 13AApi was also in accord with the pyDockWEB result. The docking complex from pyDockWEB was employed for further analyses on interaction and representation.
Fig. 2.
Interaction of the novel 13-amino acid peptide inhibitor (13AApi) with the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) spike receptor-binding domain (RBD) restricts its interaction with host cell receptor angiotensin-converting enzyme 2 (ACE2) peptidase domain. Molecular docking complex of the 13AApi (red) with SARS-CoV-2 spike RBD (green) obtained with pyDockWEB. The interacting residues within the 3 Å region are highlighted in cyan and blue, respectively, while the dock scores (kcal/mol) from all three docking suits (pyDockWEB, HDOCK, and ClusPro 2.0) are listed in the table below
Three residues K417, Y489, and Q493 of the spike protein RBD within the 3 Å region were found to interact with the novel 13AApi; these residues also interact with the ACE2 receptor (Fig. 2A). The docking position of 13AApi in the RBD pocket ensured a high probability to block the interaction with the ACE2 receptor. It is also imperative to understand the stability of a complex at an atomic level. Hence, we proceeded with an MD simulation study to analyze the dynamics and stability components.
MD Simulation Study of 13AApi and the SARS-CoV-2 Spike Protein
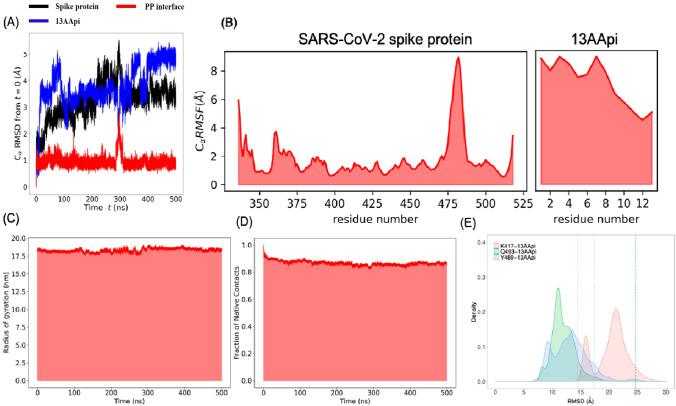
The docking complex between the SARS-CoV-2 spike protein and the 13AApi was next subjected to an MD simulation study to determine the stability of the complex. An MD simulation of 500 ns was performed using the GROMACS program. Analysis of the root mean squared deviation and the root mean squared fluctuation of the SARS-CoV-2 spike protein throughout the MD trajectory suggested no significant fluctuations in the ACE2 binding interface of the SARS-CoV-2 spike protein (Fig. 3A, B). Moreover, no significant changes in the radius of gyration and fraction of native contacts were observed, suggesting an overall stability of the SARS-CoV-2 spike protein and the 13AApi protein-peptide complex (Fig. 3C, D).
Fig. 3.
MD simulation to study the dynamics of the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) spike protein and 13-amino acid peptide inhibitor (13AApi) complex. A Root mean squared deviation (RMSD) of the Cα atoms of the SARS-CoV-2 spike protein (black), 13AApi (blue), and protein-peptide interface (red) plotted against total simulation time. B Root mean squared fluctuation of SARS-CoV-2 spike protein (black) and 13AApi. C The radius of gyration of the whole protein-peptide complex. D Fraction of native contacts of the whole protein-peptide complex. E Distance between Cα atom of K417, Y489, and Q493 with the center of mass of 13AApi at t = 0 ns is shown as dashed lines
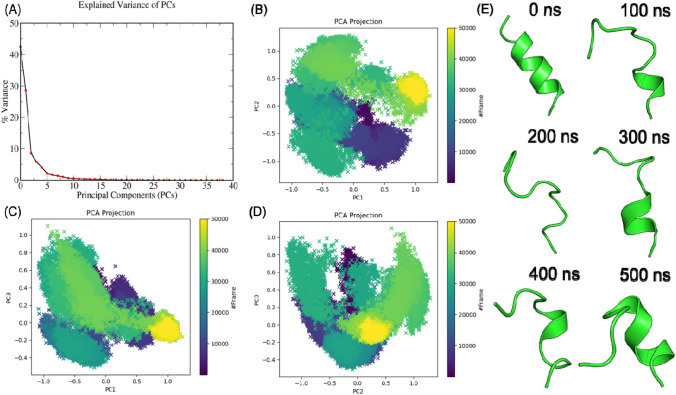
The 13AApi initially showed significant fluctuations from the docking predicted binding mode, which was stabilized after 400 ns (Fig. 3B). Conformational and positional rearrangement accounted for most of the deviations and fluctuations as 13AApi remained bound to the SARS-CoV-2 spike protein for the entire 500 ns MD trajectory (Fig. 3E). To further study the dynamics of 13AApi within the SARS-CoV-2 spike protein receptor-binding interface, a principal component analysis using cartesian coordinates and singular value decomposition approach was performed. As shown in Fig. 4A, the first three principal components explained about 85% of the variance. Analysis of the first three principal components (Fig. 4C–E) and snapshots sampled at intervals of 100 ns (Fig. 4E) revealed several conformational states. The 13AApi did not maintain a stable helical conformation as in the case of an ACE2 fragment (FLDKFNHEAEDLF), possibly because of the disruption of intramolecular contacts. Instead, 13AApi exhibited frequent unfolding and folding events (Fig. 4). A figure showing the change in the secondary structure throughout the 500 ns MD simulation is given as supporting information (Fig. S1 of the Electronic Supplementary Material). At the end of the MD simulation, i.e., at around 500 ns, 13AApi appeared to adopt a helical conformation that was necessary for its interaction with the SARS-CoV-2 spike protein. However, it seems that further optimization of 13AApi is needed to identify a sequence with stable helical conformation. To further estimate the energetics of 13AApi binding to the SARS-CoV-2 spike protein, MM-PBSA analysis was performed that revealed favorable binding energy (Table 1).
Fig. 4.
A Principal component analysis (PCA) to study the dynamics of 13-amino acid peptide inhibitor (13AApi) within the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) spike protein receptor-binding interface. B–E Screen plots showing percent variance explained by a PCA
Table 1.
13-Amino acid peptide inhibitor binding energies calculated using the MM-PBSA approach. The molecular dynamics trajectory of the last 100 ns was used for the MM-PBSA binding energy calculations
| Energy type | Energy (kJ/mol) |
|---|---|
| Van der Waal () | − 189.89 ± 31.24 |
| Electrostatic () | − 397.57 ± 88.48 |
| Polar solvation energy () | 445.82 ± 90.61 |
| Nonpolar energy () | − 25.74 ± 2.70 |
| Binding energy () | − 167.38 ± 45.08 |
In Silico Analysis of Physicochemical and ADMET Properties of 13AApi
To further understand the properties of the 13AApi in the host system, we performed an in silico prediction of its physicochemical and ADMET properties (Table 2). In brief, the 13AApi with an average molecular weight of 1684.91 g/mol and molecular formula C82H113N19O20 contains two each of the positively charged residues (K4 and K10) and negatively charged residues (D3 and D11) with a theoretical isoelectric point (pI) of 7.55. The measure of the instability index suggests that the 13AApi is in the category of a stable peptide with an estimated half-life of 1.1 h. Additionally, the hydropathy data indicated that it has suitable water-solubility properties.
Table 2.
In-silico analysis of physicochemical and ADMET (absorption, distribution, metabolism, excretion and toxicity) properties of 13-amino acid peptide inhibitor (13AApi) from ExPasy ProtParam (ExPASy—ProtParam tool) and pkCSM (pkCSM [unimelb.edu.au]) tools
| Peptide sequence | FLDKFNHNFKDLF |
|---|---|
| Physicochemical analysis | |
| Properties | |
| Molecular weight (g/mol) | 1684.91 |
| Theoretical pI | 7.55 |
| Net charge at pH 7 | 0.1 |
| Negative + positive residues | 2 +2 |
| Molecular formula | C82H113N19O20 |
| Number of atoms | 234 |
| Instability Index | − 2.21 |
| Half-life (in h) | 1.1 |
| Hydrophobicity (%) | 46.15 |
| Acidic + Basic + neutral ratio (%) | 15.38 + 23.08 + 15.38 |
| GRAVY | − 0.477 |
| ADMET analysis | |
| Absorption | |
| Water solubility (log mol/L)_ | − 2.892 |
| Skin perm (log Kp) | − 2.735 |
| Distribution | |
| Fraction unbound (human) (Fu) | 0.369 |
| BBB permeability | − 2.493 |
| CNS permeability (logPS) | − 7.089 |
| Metabolism | |
| Cytochrome P substrate | No |
| Cyctochrome P inhibitor | No |
| Excretion | |
| Total clearance (log mL/min/kg) | − 1.052 |
| Toxicity | |
| AMES toxicity | No |
| Skin sensitisation | No |
| MRTD human (log mg/kg/day) | 0.438 |
| Rat oral LD50 (mol/kg) | 2.482 |
For the ADMET analysis in pkCSM, the required SMILES file format of the 13AApi was created using pepSMI tool (PepSMI: Convert Peptide to SMILES string [novoprolabs.com])
BBB Permeability logBB < − 1 indicates poor distribution to the brain, CNS permeability logPS > − 2 classifies central nervous system penetration and logPS < − 3 classifies no central nervous system penetration, Fraction Unbound defines amount that remains unbound to plasma protein for pharmacological action, GRAVY Grand Average of Hydropathy, Instability Index value below 40 classifies stable protein/peptide, MRTD maximum recommended tolerated dose (should be less than 0.477 mg/kg/day), pI isoelectric point, Skin Perm logkp > − 2.5 classifies low skin permeability, Total Clearance includes both hepatic and renal clearance, Water Solubility logS defines solubility in water at 25 °C
Evaluation of ADMET properties suggested good water solubility with low skin permeability of 13AApi. The ADMET analyses also suggested that 13AApi was unlikely to have blood–brain barrier and central nervous system permeability. In terms of toxicity, the prediction suggested that 13AApi was safe and bore no skin toxicity or carcinogenicity (Ames test). Furthermore, the predicted maximum recommended tolerated dose for humans was below the maximum set value of 0.477 mg/kg/day (Table 2). Other properties of 13AApi are shown in Table 2.
13AApi Binds to SARS-CoV-2 and Blocks the ACE2 interaction
To validate the in silico findings of an efficient inhibition in the interaction between the SARS-CoV-2 spike and ACE2 receptor protein by 13AApi, we used an ELISA-based ACE2: SARS-CoV-2 spike inhibitor screening assay. Our data suggest that 13AApi could significantly inhibit the interaction between the SARS-CoV-2 spike and immobilized ACE2 in a concentration-dependent manner. Greater than 40% inhibition of interaction was observed at a 13AApi concentration of 100µM (Fig. 5A, B). This strongly suggests that the 13AApi can be considered for further investigation as a new modality for the therapeutic intervention of SARS-CoV-2 infection.
Fig. 5.
In vitro validation of the inhibitory effect of the 13-amino acid peptide inhibitor (13AApi) on severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) spike receptor-binding domain (RBD) was examined with an enzyme-linked immunosorbent assay (ELISA) test kit. A The image reproduced from the BP Biosciences ELISA kit demonstrates the steps in the assay. B Percent inhibitory activity of the 13AApi at the indicated concentrations on the binding of SARS-CoV-2 spike RBD to angiotensin-converting enzyme 2 (ACE2). The plotted graph represents the mean of three biological replicates
Conclusions
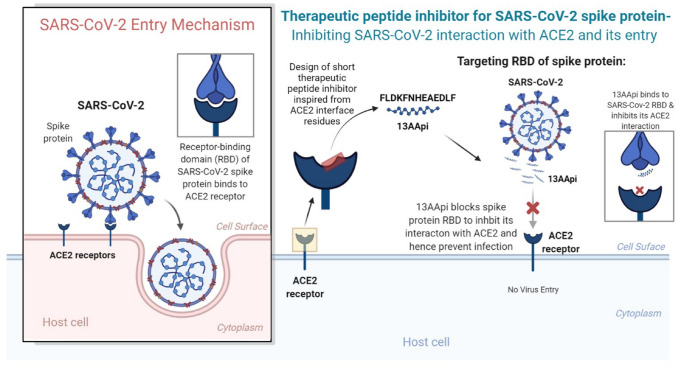
Several advantages of peptides, including ease of synthesis and modification, low toxicity, and high target specificity and selectivity, motivated us to design a potential therapeutic peptide candidate against COVID-19. The 13AApi is a novel potential therapeutic peptide that could inhibit the interaction of the SARS-CoV-2 spike protein with ACE2, thereby blocking cellular entry of the virus (Fig. 6). Our findings suggest that computationally designed inhibitory peptides may be developed as an anti-SARS-CoV-2 agent. Inhibitory activity of the designed 13AApi could be successfully validated with an ELISA. The 13AApi also displayed multiple drug-like properties such as good solubility, stability, and selectivity towards the RBD of the SARS-CoV-2 spike protein and can be further developed as a selective therapeutic candidate for COVID-19. The present study is the first step towards developing candidate drugs that can block SARS-CoV-2 entry into the host cell and further studies are warranted to determine the real-time therapeutic application of these candidate drugs.
Fig. 6.
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) and host angiotensin-converting enzyme 2 (ACE2) receptor interaction and mechanism of virus entry (left panel). A novel therapeutic peptide 13-amino acid peptide inhibitor (13AApi) blocks the receptor-binding domain (RBD) of the SARS-CoV-2 spike protein from interacting with the host cell surface ACE2 (right panel). The 13AApi is a promising therapeutic candidate to prevent virus entry and infection in the host. The image was created with BioRender
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgments
The authors acknowledge the Indian Institute of Technology Indore (IITI) for providing facilities and other support. This work was supported by Cumulative Professional Development Allowance (CPDA) from IITI to MSB.
Declarations
Funding
This work was supported by Cumulative Professional Development Allowance (CPDA) from Indian Institute of Technology Indore (IITI) to MSB.
Conflicts of Interest
Sajjan Rajpoot, Tomokazu Ohishi, Ashutosh Kumar, Qiuwei Pan, Sreeparna Banerjee, Kam Y.J. Zhang, and Mirza S. Baig have no conflicts of interest that are directly relevant to the content of this article.
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Availability of Data and Material
All the data and material have been provided in the article and/or supplementary data.
Code Availability
Not applicable.
Authors’ Contributions
MSB conceived, designed, and executed the research. AK, TO, and SR compiled and analyzed the data. MSB has written and reviewed the manuscript. QP, SB, and KYZ reviewed and edited the manuscript.
Footnotes
Sajjan Rajpoot, Tomokazu Ohishi, and Ashutosh Kumar contributed equally to the article.
References
- 1.Wang C, Wang Z, Wang G, Lau JY-N, Zhang K, Li WJST, et al. COVID-19 in early 2021: current status and looking forward. Signal Transduct Target Ther. 2021;6(1):1–14. doi: 10.1038/s41392-020-00451-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nicola M, Alsafi Z, Sohrabi C, Kerwan A, Al-Jabir A, Iosifidis C, et al. The socio-economic implications of the coronavirus and COVID-19 pandemic: a review. J Surg. 2020;78:185–193. doi: 10.1016/j.ijsu.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu YC, Chen CS, Chan YJ. The outbreak of COVID-19: an overview. J Chin Med Assoc. 2020;83(3):217–220. doi: 10.1097/JCMA.0000000000000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakraborty C, Sharma AR, Sharma G, Bhattacharya M, Lee SS. SARS-CoV-2 causing pneumonia-associated respiratory disorder (COVID-19): diagnostic and proposed therapeutic options. Eur Rev Med Pharmacol Sci. 2020;24(7):4016–4026. doi: 10.26355/eurrev_202004_20871. [DOI] [PubMed] [Google Scholar]
- 5.Leung T, Chan A, Chan E, Chan V, Chui C, Cowling B, et al. Short-and potential long-term adverse health outcomes of COVID-19: a rapid review. Emerg Microb Infect. 2020;9(1):2190–2199. doi: 10.1080/22221751.2020.1825914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yelin D, Wirtheim E, Vetter P, Kalil AC, Bruchfeld J, Runold M, et al. Long-term consequences of COVID-19: research needs. Lancet Infect Dis. 2020;20(10):1115–1117. doi: 10.1016/S1473-3099(20)30701-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nalbandian A, Sehgal K, Gupta A, Madhavan MV, McGroder C, Stevens JS, et al. Post-acute COVID-19 syndrome. Nat Med. 2021;27(4):1–15. doi: 10.1038/s41591-021-01283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cascella M, Rajnik M, Cuomo A, Dulebohn SC, Di Napoli R. Features, evaluation and treatment coronavirus (COVID-19) Treasure Island: StatPearls; 2020. [PubMed] [Google Scholar]
- 9.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi Z-L, Guo D, Rottier PJ. Coronavirus: epidemiology, genome replication and the interactions with their hosts. Virol Sin. 2016;31(1):1–2. doi: 10.1007/s12250-016-3746-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khan MI, Khan ZA, Baig MH, Ahmad I, Farouk A-E, Song YG, et al. Comparative genome analysis of novel coronavirus (SARS-CoV-2) from different geographical locations and the effect of mutations on major target proteins: an in silico insight. PLoS ONE. 2020;15(9):e0238344. doi: 10.1371/journal.pone.0238344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li G, Fan Y, Lai Y, Han T, Li Z, Zhou P, et al. Coronavirus infections and immune responses. J Med Virol. 2020;92(4):424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 15.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–80.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181(4):894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ali A, Vijayan RJS. Dynamics of the ACE2–SARS-CoV-2/SARS-CoV spike protein interface reveal unique mechanisms. Sci Rep. 2020;10(1):1–12. doi: 10.1038/s41598-019-56847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Padhi AK, Tripathi T. Can SARS-CoV-2 accumulate mutations in the S-protein to increase pathogenicity? ACS Pharmacol Transl Sci. 2020;3(5):1023–1026. doi: 10.1021/acsptsci.0c00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tai W, He L, Zhang X, Pu J, Voronin D, Jiang S, et al. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol. 2020;17(6):613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–92.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, et al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci USA. 2020;117(21):11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chakraborty C, Sharma A, Mallick B, Bhattacharya M, Sharma G, Lee S-S, et al. Evaluation of molecular interaction, physicochemical parameters and conserved pattern of SARS-CoV-2 spike RBD and hACE2: in silico and molecular dynamics approach. Eur Rev Med Pharmacol Sci. 2021;25(3):1708–1723. doi: 10.26355/eurrev_202102_24881. [DOI] [PubMed] [Google Scholar]
- 23.Zipeto D, da Palmeira JF, Argañaraz GA, Argañaraz ER. ACE2/ADAM17/TMPRSS2 interplay may be the main risk factor for COVID-19. Front Immunol. 2020;11:576745. doi: 10.3389/fimmu.2020.576745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46(4):586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu C-H, Mohammadmoradi S, Chen JZ, Sawada H, Daugherty A, Lu HS. Renin-angiotensin system and cardiovascular functions. Arterioscler Thromb Vasc Biol. 2018;38(7):e108–e116. doi: 10.1161/ATVBAHA.118.311282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ingelfinger JR. ACE2: a new target for prevention of diabetic nephropathy? J Am Soc Nephrol. 2006;17(11):2957–2959. doi: 10.1681/ASN.2006090986. [DOI] [PubMed] [Google Scholar]
- 27.Bosso M, Thanaraj TA, Abu-Farha M, Alanbaei M, Abubaker J, Al-Mulla FJMT-M, et al. The two faces of ACE2: the role of ACE2 receptor and its polymorphisms in hypertension and COVID-19. Mol Ther Methods Clin Dev. 2020;18:321–327. doi: 10.1016/j.omtm.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shenoy V, Qi Y, Katovich MJ, Raizada MK. ACE2, a promising therapeutic target for pulmonary hypertension. Curr Opin Pharmacol. 2011;11(2):150–155. doi: 10.1016/j.coph.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao, Y., Zhao, Z., Wang, Y., Zhou, Y., Ma, Y., & Zuo, W. Single-Cell RNA Expression Profiling of ACE2, the Receptor of SARS-CoV-2. American journal of respiratory and critical care medicine. 2020;202(5):756–59. [DOI] [PMC free article] [PubMed]
- 30.Uhal BD, Dang M, Dang V, Llatos R, Cano E, Abdul-Hafez A, et al. Cell cycle dependence of ACE-2 explains downregulation in idiopathic pulmonary fibrosis. Eur Respir J. 2013;42(1):198–210. doi: 10.1183/09031936.00015612. [DOI] [PubMed] [Google Scholar]
- 31.Crackower MA, Sarao R, Oudit GY, Yagil C, Kozieradzki I, Scanga SE, et al. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature. 2002;417(6891):822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- 32.Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444-8. [DOI] [PMC free article] [PubMed]
- 33.Zhang H, Wada J, Hida K, Tsuchiyama Y, Hiragushi K, Shikata K, et al. Collectrin, a collecting duct-specific transmembrane glycoprotein, is a novel homolog of ACE2 and is developmentally regulated in embryonic kidneys. J Biol Chem. 2001;276(20):17132–17139. doi: 10.1074/jbc.M006723200. [DOI] [PubMed] [Google Scholar]
- 34.Zisman LS, Keller RS, Weaver B, Lin Q, Speth R, Bristow MR, et al. Increased angiotensin-(1–7)-forming activity in failing human heart ventricles: evidence for upregulation of the angiotensin-converting enzyme homologue ACE2. Circulation. 2003;108(14):1707–1712. doi: 10.1161/01.CIR.0000094734.67990.99. [DOI] [PubMed] [Google Scholar]
- 35.Monteil V, Kwon H, Prado P, Hagelkrüys A, Wimmer RA, Stahl M, et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181(4):905–913. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Z, Yi Y, Luo X, Xiong N, Liu Y, Li S, et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol. 2020;92(9):1518–1524. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou Y, Hou Y, Shen J, Huang Y, Martin W, Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen YW, Yiu CB, Wong KY. Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CL (pro)) structure: virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Res. 2020;9:129. doi: 10.12688/f1000research.22457.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rajpoot S, Alagumuthu M, Baig MS. Dual targeting of 3clpro and Plpro of SARS-Cov-2: a novel structure-based design approach to treat Covid-19. Curr Res Struct Biol. 2021;3:9–18. doi: 10.1016/j.crstbi.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang J, Petitjean SJL, Koehler M, Zhang Q, Dumitru AC, Chen W, et al. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat Commun. 2020;11(1):4541. doi: 10.1038/s41467-020-18319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Padhi AK, Shukla R, Saudagar P, Tripathi TJI. High-throughput rational design of the remdesivir binding site in the RdRp of SARS-CoV-2: implications for potential resistance. iScience. 2021;24(1):101992. doi: 10.1016/j.isci.2020.101992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Padhi AK, Tripathi TJB. Targeted design of drug binding sites in the main protease of SARS-CoV-2 reveals potential signatures of adaptation. Biochem Biophys Res Commun. 2021;555:147–153. doi: 10.1016/j.bbrc.2021.03.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chakraborty C, Bhattacharya M, Mallick B, Sharma AR, Lee S-S, Agoramoorthy GJ. SARS-CoV-2 protein drug targets landscape: a potential pharmacological insight view for the new drug development. Expert Rev Clin Pharmacol. 2021;14(2):225–237. doi: 10.1080/17512433.2021.1874348. [DOI] [PubMed] [Google Scholar]
- 44.Huang Y, Yang C, Xu X-F, Xu W, Liu S-w. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharm Sinica. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ali AM, Atmaj J, Van Oosterwijk N, Groves MR, Dömling AJ. Stapled peptides inhibitors: a new window for target drug discovery. Comput Struct Biotechnol J. 2019;17:263–281. doi: 10.1016/j.csbj.2019.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsomaia N. Peptide therapeutics: targeting the undruggable space. Eur J Med Chem. 2015;94:459–470. doi: 10.1016/j.ejmech.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 47.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 48.Dassault Systèmes . BIOVIA, discovery studio visualizer, release 2019. San Diego: Dassault Systèmes; 2020. [Google Scholar]
- 49.Jiménez-García B, Pons C, Fernández-Recio JJB. pyDockWEB: a web server for rigid-body protein–protein docking using electrostatics and desolvation scoring. Bioinformatics. 2013;29(13):1698–1699. doi: 10.1093/bioinformatics/btt262. [DOI] [PubMed] [Google Scholar]
- 50.Yan Y, Zhang D, Zhou P, Li B, Huang S-Y. HDOCK: a web server for protein–protein and protein–DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017;45(1):365–373. doi: 10.1093/nar/gkx407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, Yueh C, et al. The ClusPro web server for protein–protein docking. Nat Protoc. 2017;12(2):255. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78(8):1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 54.Michaud-Agrawal N, Denning EJ, Woolf TB, Beckstein O. MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. J Comput Chem. 2011;32(10):2319–2327. doi: 10.1002/jcc.21787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McGibbon RT, Beauchamp KA, Harrigan MP, Klein C, Swails JM, Hernández CX, et al. MDTraj: a modern open library for the analysis of molecular dynamics trajectories. Biophys J. 2015;109(8):1528–1532. doi: 10.1016/j.bpj.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 57.Ross C, Nizami B, Glenister M, Sheik Amamuddy O, Atilgan AR, Atilgan C, et al. MODE-TASK: large-scale protein motion tools. Bioinformatics. 2018;34(21):3759–3763. doi: 10.1093/bioinformatics/bty427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumari R, Kumar R, Open Source Drug Discovery Consortium, Lynn A. g_mmpbsa- A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014;54(7):1951–62. [DOI] [PubMed]
- 59.Kumari R, Kumar R, Open Source Drug Discovery Consortium. Lynn A. g_mmpbsa: a GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model. 2014;54(7):1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 60.Hunter JD. Matplotlib: a 2D graphics environment. Comput Sci Eng. 2007;9(03):90–95. doi: 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- 61.Team RC . R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2013. [Google Scholar]
- 62.Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, Bairoch AJT. The proteomics protocols handbook. Springer; 2005. Protein identification and analysis tools on the ExPASy server; pp. 571–607. [Google Scholar]
- 63.Pires DE, Blundell TL, Ascher DB. pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J Med Chem. 2015;58(9):4066–4072. doi: 10.1021/acs.jmedchem.5b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Benítez-Cardoza CG, Vique-Sánchez JL. Potential inhibitors of the interaction between ACE2 and SARS-CoV-2 (RBD), to develop a drug. Life Sci. 2020;256:117970. doi: 10.1016/j.lfs.2020.117970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Day CJ, Bailly B, Guillon P, Dirr L, Jen FE-C, Spillings BL, et al. Multidisciplinary approaches identify compounds that bind to human ACE2 or SARS-CoV-2 spike protein as candidates to block SARS-CoV-2–ACE2 receptor interactions. MBio. 2021;12(2):e03681–e3720. doi: 10.1128/mBio.03681-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang G, Yang ML, Duan ZL, Liu FL, Jin L, Long CB, et al. Dalbavancin binds ACE2 to block its interaction with SARS-CoV-2 spike protein and is effective in inhibiting SARS-CoV-2 infection in animal models. Cell Res. 2021;31(1):17–24. doi: 10.1038/s41422-020-00450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brogi S, Calderone V. Off-target ACE2 ligands: possible therapeutic option for COVID-19? Br J Clin Pharmacol. 2020;86(6):1178–1179. doi: 10.1111/bcp.14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khelfaoui H, Harkati D, Saleh BA. Molecular docking, molecular dynamics simulations and reactivity, studies on approved drugs library targeting ACE2 and SARS-CoV-2 binding with ACE2. J Biomol Struct Dyn. 2020;5:1–17. doi: 10.1080/07391102.2020.1803967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baig MS, Alagumuthu M, Rajpoot S, Saqib U. Identification of a potential peptide inhibitor of SARS-CoV-2 targeting its entry into the host cells. Drugs R D. 2020;20(3):161–169. doi: 10.1007/s40268-020-00312-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang J, Petitjean SJ, Koehler M, Zhang Q, Dumitru AC, Chen W, et al. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat Commun. 2020;11(1):1–10. doi: 10.1038/s41467-019-13993-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Han Y, Král PJ. Computational design of ACE2-based peptide inhibitors of SARS-CoV-2. ACS Nano. 2020;14(4):5143–5147. doi: 10.1021/acsnano.0c02857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maas MN, Hintzen JC, Löffler PM, Mecinović JJCC. Targeting SARS-CoV-2 spike protein by stapled hACE2 peptides. Chem Commun. 2021;57(26):3283–3286. doi: 10.1039/D0CC08387A. [DOI] [PubMed] [Google Scholar]
- 73.Ling R, Dai Y, Huang B, Huang W, Yu J, Lu X, et al. In silico design of antiviral peptides targeting the spike protein of SARS-CoV-2. Peptides. 2020;130:170328. doi: 10.1016/j.peptides.2020.170328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.