Abstract
Gastric cancer (GC) is the second leading cause of cancer deaths around the world. Chemoresistance is an important reason for poor prognosis of GC. Saikosaponin D (SSD) is a natural constituent from Radix Bupleuri and exhibits various activities including antitumors. This study investigated the effects and the mechanisms of SSD on cisplatin (cis-diamminedichloroplatinum, DDP) sensitivity of GC cells. Findings suggested that SSD could promote the inhibitory effect of DDP on proliferation and invasion and increase DDP-induced apoptosis in SGC-7901 and DDP-resistant cell line SGC-7901/DDP. We further identified that SSD increased levels of LC3 B and cleaved caspase 3 and decreased levels of p62, IKK β, p-IκB α, and NF-κB p65, suggesting that SSD might inhibit the IKK β/NF-κB pathway and induce both cell autophagy and apoptosis in SGC-7901 and SGC-7901/DDP. A further study indicated that SSD enhanced the effect of DDP-induced cleaved caspase 3 level rise and NF-κB pathway suppression, especially in SGC-7901/DDP cells. Conclusively, SSD enhanced DDP sensitivity of GC cells; the potential molecular mechanisms were that SSD-induced apoptosis and autophagy and inhibited the IKK β/NF-κB pathway in GC cells. These findings suggested that SSD might contribute to overcoming DDP resistance in GC treatment.
Introduction
Saikosaponin D (SSD) is an active constituent from the traditional Chinese medicine Radix Bupleuri. SSD possesses various specific pharmacological activities, such as anti-inflammatory,1 antitumor,2 immunoregulation,3 anti-allergic,4 and anti-apoptosis activities.5 Interestingly, SSD was also proved to significantly induce apoptosis and increase the radiosensitivity of liver cancer cells and suppress the growth of liver tumors.6,7 SSD might trigger autophagic death in liver cancer cells via the AMPK–mTOR pathway.8 Additionally, SSD could enhance chemosensitivity of cancer cells. For example, SSD could enhance the sensitivity of human non-small cell lung cancer cells to gefitinib by inhibiting the STAT3/Bcl-2 signaling pathway.9 Zhang and colleagues found that SSD suppressed the malignant phenotype of Hep3B cells and increased their chemosensitivity in vitro and in vivo.10 Shi et al. proved that SSD could enhance the sensitivity of MCF-7/adriamycin cells toward adriamycin by down-regulating MDR1 and P-gp expression.11 To date, few literatures reported the effects of SSD on gastric cancer (GC).
GC is the second most common cause of cancer deaths around the world.12 The incidence of GC is higher in Eastern Asia, European, and South America Countries than in North America and Africa regions.13 According to statistics, in 70–90% of cases, the chemotherapy is not effective enough, and drug resistance occurs.14 The accepted mechanisms of chemoresistance of tumor cells could be divided into two categories: intrinsic and acquired resistance.15−17 Nevertheless, the mechanisms of chemoresistance in GC, such as tumor microenvironment characteristics,18 genetic and non-genetic factors,19,20 and mutations in drug targets,21 are still poorly understood. Recently, some natural compounds showed activities of reducing drug-resistant of GC. For example, α-hederin is one of the main components from Nigella sativa seed, which is a common Chinese traditional medicine. Liu and colleagues found that α-hederin might inhibit the proliferation and induce the apoptosis of HGC-27/DDP cells by increasing intracellular reactive oxygen species levels and activating mitochondrial pathway.22 β-Elemene, which is a natural novel plant-derived drug with anticancer activities, showed inhibitory effects on the metastasis of SGC-7901/ADR cells by modulating the miR-1323/Cbl-b/EGFR pathway.23 Cardamonin, a natural chalcone, might reduce 5-fluorouracil resistance of GC cells by regulating Wnt/β-catenin pathway.24
In this study, we evaluated the effects of SSD on several GC cell lines, including SGC-7901, MGC-803, and HGC-27. SGC-7901/DDP, the cisplatin (cis-diamminedichloroplatinum, DDP)-resistant cellular model, was established using SGC-7901 under elevated concentrations of DDP. The potential molecular mechanisms of SSD enhancing DDP sensitivity of SGC-7901/DDP were investigated. Additionally, the sensitivity of SGC-7901/DDP cells to DDP was confirmed by the increased DDP-induced apoptosis in the presence of cardamonin. We also confirmed that SSD could induce cell autophagy of SGC-7901/DDP cells. Furthermore, we explored the relationship between apoptosis and autophagy processes induced by SSD.
Results
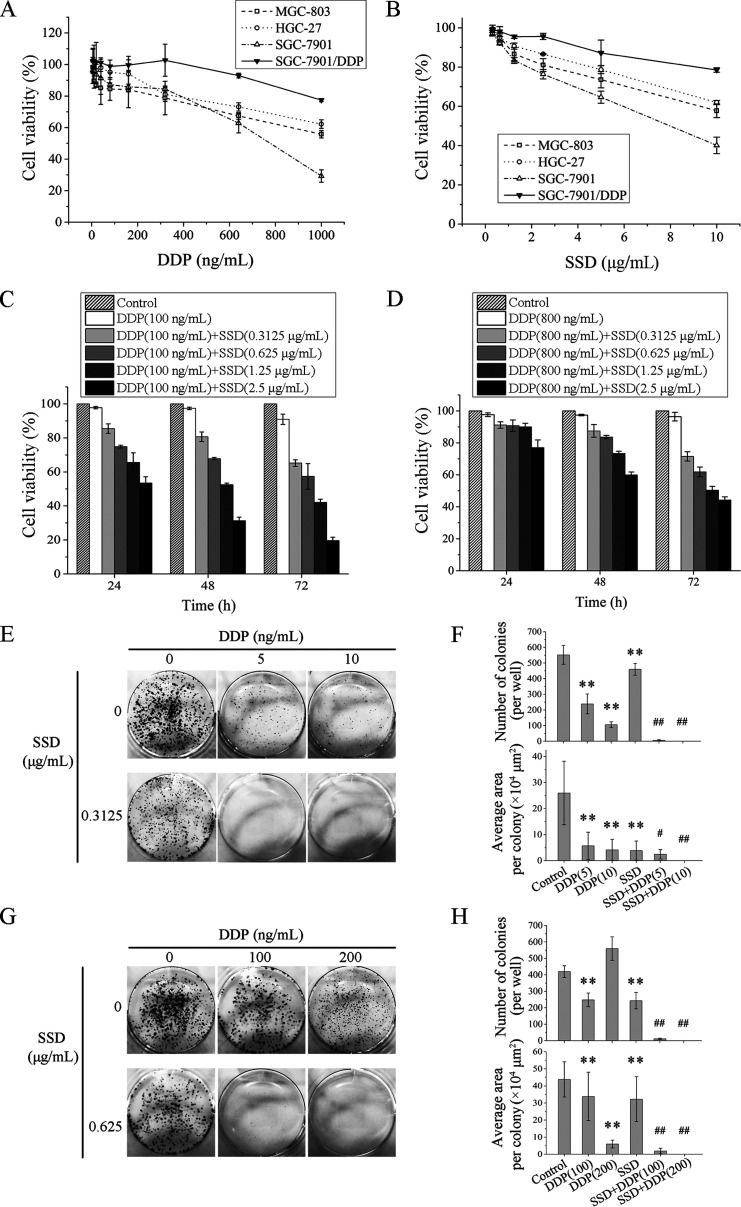
SSD Strengthens the Inhibitory Effects of DDP on Cell Proliferation
To examine the roles of SSD in GC DDP-resistant and -sensitive cell lines, MGC-803, HGC-27 and SGC-7901, and DDP-resistant cells SGC-7901/DDP were all treated with different concentrations of SSD for 48 h. As shown in Figure 1A, the most sensitive cell line to DDP was SGC-7901, followed by MGC-803 and HGC-27, and SGC-7901/DDP was the least sensitive. Subsequently, we exposed all the cells above to different concentrations of SSD for 48 h, and the results showed that SSD could reduce the cell growth in a dose-dependent manner (Figure 1B). The efficacy of DDP modulated by SSD against SGC-7901 and SGC-7901/DDP cells are, respectively, shown in Figure 1C,D. Treatment with SSD significantly enhanced the sensitivities of SGC-7901 and SGC-7901/DDP to DDP in a dose- and time-dependent manner. Additionally, 100 ng/mL DDP alone or combined with 0.3125 μg/mL SSD could not affect cell growth of GES-1 for 24, 48, or 72 h. But coadministration of 2.5 μg/mL SSD and 100 ng/mL DDP could suppress GES-1 viability (Figure S1).
Figure 1.
SSD augments the inhibitory effect of DDP in DDP-sensitive MGC-803, HGC-27 and SGC-7901, and DDP-resistant SGC-7901/DDP cells. Effects of DDP (A) and SSD (B) alone on proliferation of four GC cells were detected by 1-(4,5-demethylthiazol-2-yl)-3,5-diphenyltetrazoliumbromide (MTT) assays. Effects of SSD on DDP-induced SGC-7901 (C) and SGC-7901/DDP (D) cell growth inhibition were also determined by MTT assays. Photomicrographs of SGC-7901 (E) and SGC-7901/DDP (G) were shown, and the number of colonies per well and average area of each colony were analyzed in SGC-7901 (F) and SGC-7901/DDP (H) cells. **p < 0.01, compared with control cells; #p < 0.05, ##p < 0.01, compared with DDP alone.
To further identify effects of SSD on DDP-induced growth suppression of GC cells, colony formation assays were conducted with SGC-7901 and SGC-7901/DDP cells after treatment with SSD or/and DDP for 72 h. As shown in Figure 1E,F, DDP or SSD alone could significantly reduce the number and the average area of cell colonies, but the combination of DDP and SSD is more effective (SSD combinated with 5 ng/mL DDP, p < 0.05; SSD combinated with 10 ng/mL DDP, p < 0.01). In SGC-7901/DDP cells, treatment with 100 ng/mL DDP could decrease colony number, but 200 ng/mL DDP not. However, 100 and 200 ng/mL DDP could both inhibit the colony area, especially the high concentration. Few cell colonies grew when DDP and SSD were combined (Figure 1G,H).
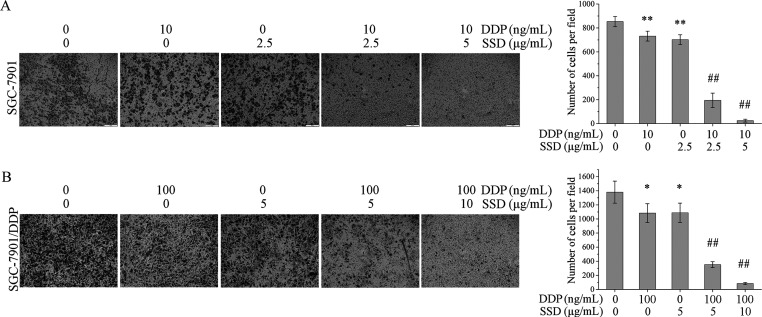
SSD Inhibits GC Cell Invasion and Migration
To investigate the effects of SSD on DDP-induced invasion and migration suppression of GC cells, the Boyden chamber assay and wound-healing assays were performed in vitro. In order to avoid the adverse effects of too many dead cells on the invasion and migration assay, low concentrations of SSD or DDP were used to treat the cells. As shown in Figure 2A, DDP or SSD alone could inhibit invasion of SGC-7901 (**p < 0.01), and SSD further increased DDP-induced invasion suppression (##p < 0.01). The similar results were observed in SGC-7901/DDP cells (Figure 2B), too. Additionally, DDP combined with SSD could also inhibit cell migration of SGC-7901 and SGC-7901/DDP more significantly than DDP or SSD alone did (Figure S2).
Figure 2.
SSD increased DDP-induced invasion suppression. The Boyden chamber assay in SGC-7901 (A) and SGC-7901/DDP (B). Cells that crossed the membrane were monitored by microscopy and counted. *p < 0.05, and **p < 0.01, compared with the control group; #p < 0.05, and ##p < 0.01, compared with the group treated with DDP alone.
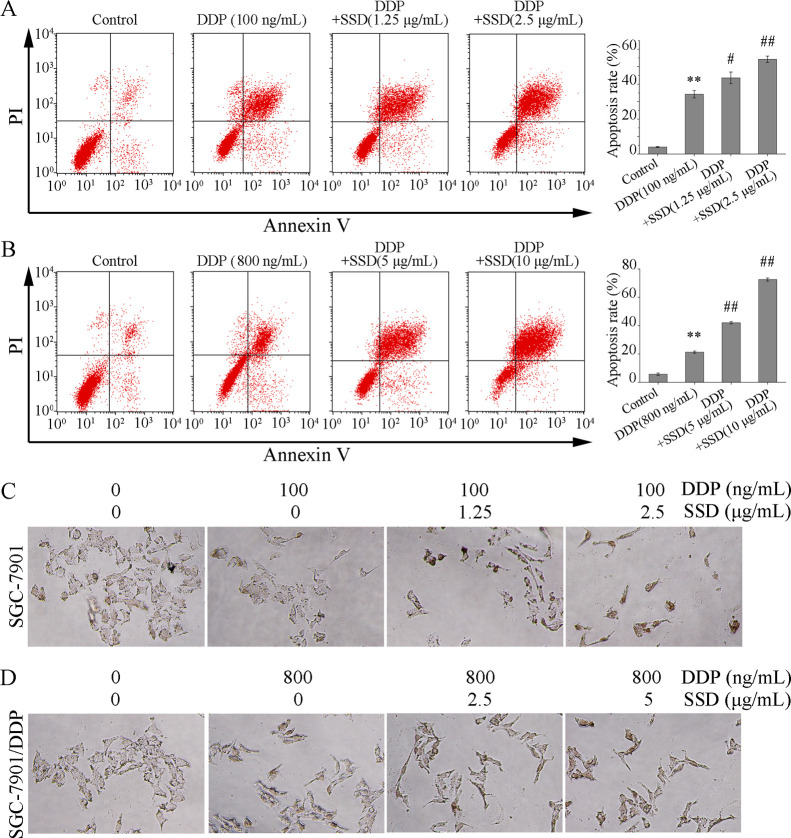
SSD Increases DDP-Induced Cell Apoptosis
To clarify the effects of SSD on DDP-induced cell apoptosis, double dyes of Annexin V-FITC and propidium iodide were used to stain the apoptotic cells. As illustrated in Figure 3A, DDP (100 ng/mL) could clearly induce SGC-7901 apoptosis (##p < 0.01) and SSD significantly strengthened the apoptosis-inducing effect of DDP (**p < 0.01). Similar results were also observed in SGC-7901/DDP (Figure 3B).
Figure 3.
Effects of DDP combined with SSD on apoptosis of SGC-7901 and SGC-7901/DDP cells. (A) Flow cytometric analysis for cell apoptosis in SGC-7901 cells; (B) flow cytometric analysis for cell apoptosis in SGC-7901/DDP cells. **p < 0.01, compared with control; #p < 0.05, ##p < 0.01, compared with DDP alone. (C,D) TUNEL assay of DDP or/and SSD in SGC-7901 or SGC-7901/DDP cells for 48 h.
To confirm the FACS results, TdT-UTP nick end labeling (TUNEL) assay was performed. As shown in Figure 3C, most nuclei were stained as a discernible brown in the treatment groups with DDP combined with SSD (Figure 3C,D), compared with the control group.
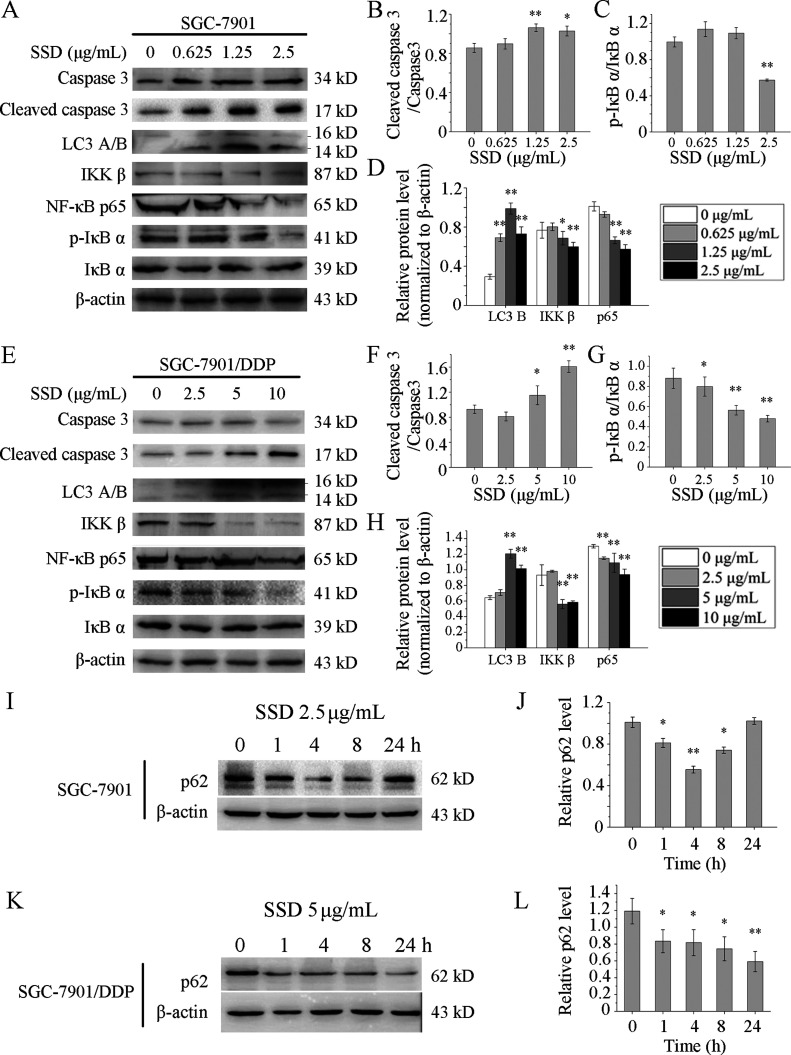
SSD Alone Modulates Expression of Apoptosis-Related Proteins
Since high concentration of DDP or SSD would weaken the cell adhesion capability and affect the cell harvest rate and protein extraction rate, so low concentrations were selected to treat SGC-7901 and SGC-7901/DDP cells. Expressions of apoptosis-related proteins were determined by Western blot. Cleaved caspase 3 was up-regulated remarkably by SSD in SGC-7901 (Figure 4A,B) and SGC-7901/DDP (Figure 4E,F) cells. These results partly reveal the cause of SSD enhancing DDP-induced GC cell apoptosis. Additionally, expressions of IKK β, one of the phosphorylase subunits of IκB, and its downstream protein NF-κB p65 were both suppressed by SSD in SGC-7901 (Figure 4A,D) and SGC-7901/DDP (Figure 4E,H) cells. Accordingly, the phosphorylation level of IκB α, one of the inhibitors of NF-κB, was also decreased in SGC-7901 (Figure 4A,C) and SGC-7901/DDP (Figure 4E,G) cells. These results indicated that SSD might inhibit the NF-κB pathway, and it may be part of the reason why SSD suppresses GC cell proliferation.
Figure 4.
SSD alone increased expressions of several apoptosis- and autophagy-related proteins. SGC-7901 (A) and SGC-7901/DDP cells (E) were treated with SSD at the indicated concentrations for 48 h, and the expression levels of proteins were detected by Western blot. Lanes of Western blot were measured by the densitometer. Level of cleaved caspase 3 was normalized to the level of pro-caspase 3 in SGC-7901 (B) and SGC-7901/DDP (F), respectively. Expression of p-IκB α was normalized to the expression of total IκB α in SGC-7901 (C) and SGC-7901/DDP (G), respectively. Levels of LC3 B, IKK β, and NF-κB p65 were normalized to β-actin levels in SGC-7901 (D) and SGC-7901/DDP (H), respectively. SGC-7901 (I) and SGC-7901/DDP (K) cells were treated with SSD at a concentration of 2.5 and 5 μg/mL, respectively, for the indicated time. Western blot images were quantified by densitometric analysis, and levels of p62 were normalized to β-actin levels in SGC-7901 (J) and SGC-7901/DDP (L), respectively. *p < 0.05, **p < 0.01, compared with control.
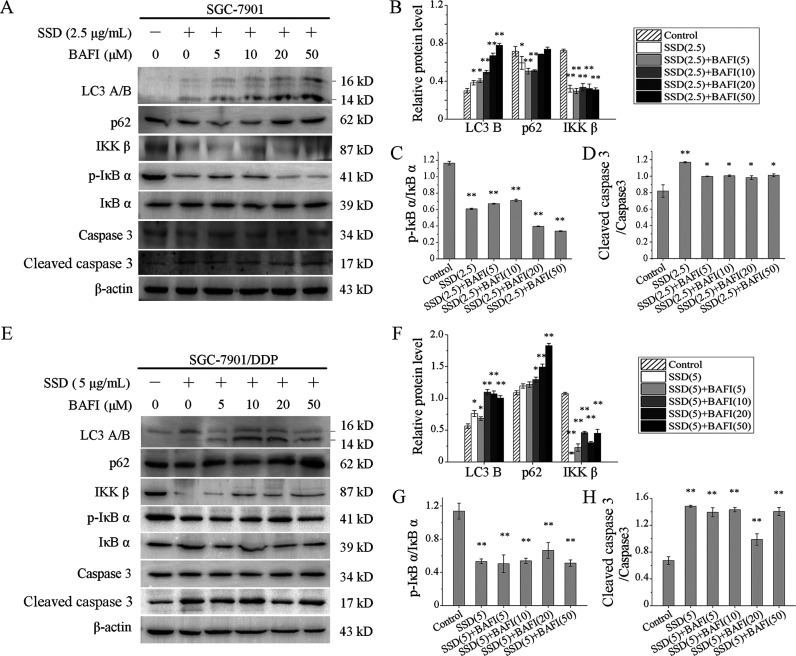
To further identify effects of SSD on cell apoptosis, bafilomycin A1, an inhibitor of late-stage autophagy was used to treat SGC-7901 and SGC-7901/DDP in the presence or absence of SSD. As shown in Figure 5, levels of cleaved caspase 3 were still increased in both SGC-7901 (Figure 5A,D) and SGC-7901/DDP (Figure 5E,H) cells. In addition, levels of p-IκB α and IKK β were still suppressed by SSD in the two cells above and not affected by bafilomycin A1. Interestingly, the expression level of p62 in SGC-7901 cells showed an initial decrease followed by a rise change with increasing bafilomycin A1 concentration (Figure 5A,B). However, bafilomycin A1 upregulated p62 expression significantly in SGC-7901/DDP cells (Figure 5E,F). Besides, levels of LC3 B in both SGC-7901 and SGC-7901/DDP were clearly increased at the presence of bafilomycin A1. Increasing evidence has shown that bafilomycin A1 could hinder the degradation of p6225,26 and potentially elevate LC3 B accumulation.27,28 These were consistent with the results above. Therefore, p62 degradation was inhibited in SGC-7901 and SGC-7901/DDP cells, indicating that bafilomycin A1 had suppressed cell autophagy effectively. In conclusion, SSD could induce gastric cell apoptosis.
Figure 5.
SSD induced autophagy-independent apoptosis. SGC-7901 (A) and SGC-7901/DDP (E) cells were treated by bafilomycin A1 at the indicated concentrations in the presence or absence of SSD, and several apoptosis- and autophagy-related proteins were detected by Western blot. Levels of LC3 B, p62, and IKK β were normalized to β-actin levels in SGC-7901 (B) and SGC-7901/DDP (F), respectively. Expression of p-IκB α was normalized to the expression of total IκB α in SGC-7901 (C) and SGC-7901/DDP (G), respectively. Level of cleaved caspase 3 was normalized to the level of pro-caspase 3 in SGC-7901 (D) and SGC-7901/DDP (H), respectively. *p < 0.05, **p < 0.01, compared with control.
SSD Alone Modulates Expression of Autophagy-Related Proteins
The level of LC3 B, indicators of autophagy, was increased by SSD in a dose-dependent manner both in SGC-7901 (Figure 4A,D) and SGC-7901/DDP (Figure 4E,H) cells. Furthermore, as shown in Figure 4I, decreased protein levels of p62, at least over the first 4 h of exposure, were observed in SGC-7901 (Figure 4I,J). And the p62 expression was suppressed over 24 h in SGC-7901/DDP (Figure 4K,L). These results suggest that SSD might induce autophagic flux.
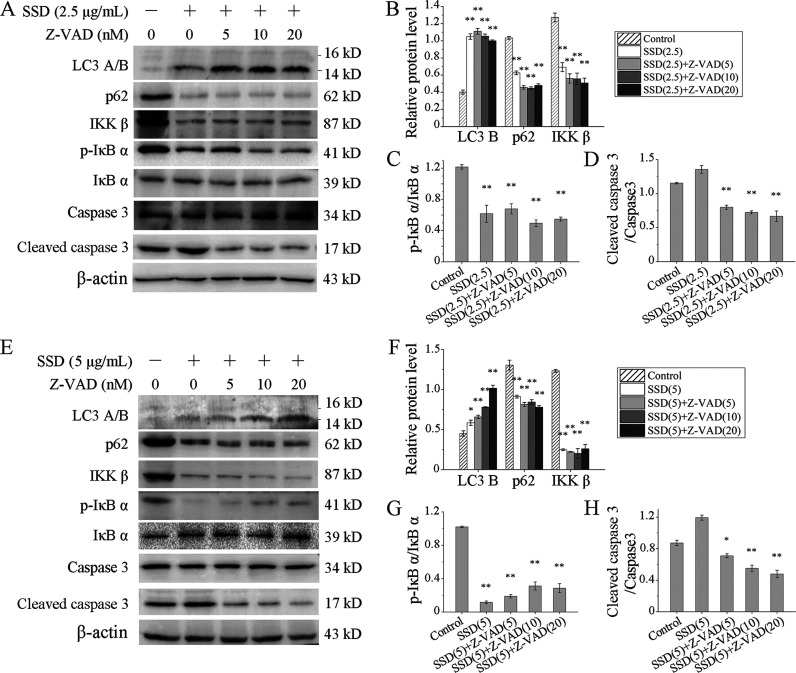
To further identify whether cell apoptosis affected SSD-induced autophagy, SGC-7901 and SGC-7901/DDP were treated with Z-VAD-FMK, a pan-caspase inhibitor, in the presence or absence of SSD. Levels of LC3 B were still increased by SSD, and expressions of p62 still decreased in both SGC-7901 (Figure 6A,B) and SGC-7901/DDP (Figure 6E,F) cells. Levels of p-IκB α were still down-regulated by SSD and not affected by Z-VAD-FMK (Figure 6A,C,E,G). As a control, cleaved caspase 3 was also detected, and its levels were up-regulated by SSD in the absence of Z-VAD-FMK. Once Z-VAD-FMK involved, levels of cleaved caspase 3 extremely decreased, indicating the Z-VAD-FMK working well.
Figure 6.
Apoptosis suppression did not affect the expression regulation of SSD on autophagy-related proteins. SGC-7901 (A) and SGC-7901/DDP (E) cells were treated by Z-VAD-FMK at the indicated concentrations in the presence or absence of SSD, and several apoptosis- and autophagy-related proteins were detected by Western blot. Levels of LC3 B, p62, and IKK β were normalized to β-actin levels in SGC-7901 (B) and SGC-7901/DDP (F), respectively. Expression of p-IκB α was normalized to the expression of total IκB α in SGC-7901 (C) and SGC-7901/DDP (G), respectively. Level of cleaved caspase 3 was normalized to the level of pro-caspase 3 in SGC-7901 (D) and SGC-7901/DDP (H), respectively. *p < 0.05, **p < 0.01, compared with control.
Combination of SSD and DDP Promotes Cell Apoptosis and Autophagy in GC Cells
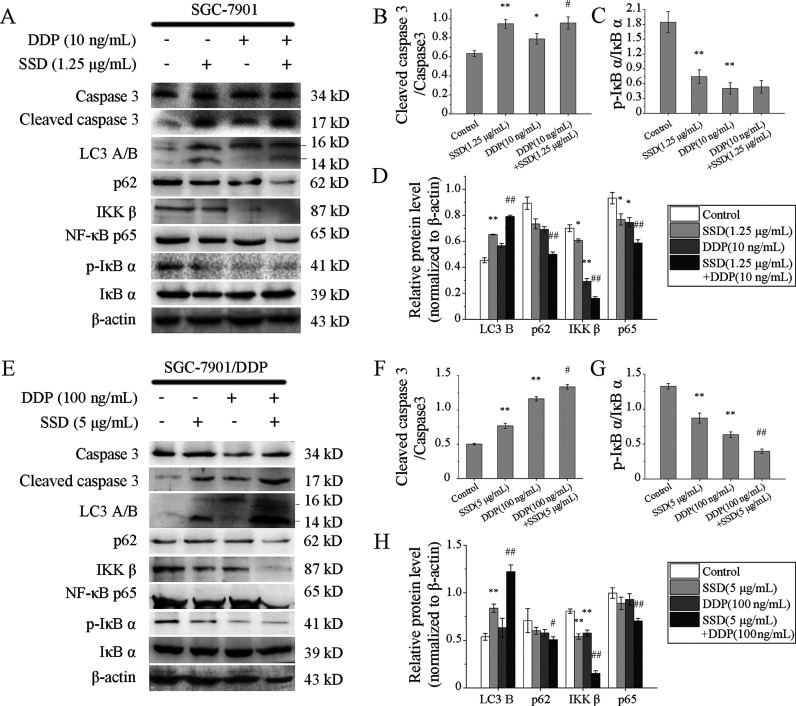
To investigate the effects of the combination of SSD and DDP on GC cells, several apoptosis- and autophagy-related proteins were detected by Western blot. DDP alone could not affect the expressions of LC3 B and p62 in SGC-7901 (Figure 7A,D), as well as in SGC-7901/DDP (Figure 7E,H). But once SSD involved, the level of LC3 B increased, while that of p62 decreased in the cells above. The results suggested that SSD indeed induced GC cell autophagy, but DDP alone did not.
Figure 7.
Effects of DDP combined with SSD on expressions of apoptosis- and autophagy-related proteins in GC cells. Protein levels in SGC-7901 (A) and SGC-7901/DDP (E) cells were analyzed by Western blots. Lanes were measured by a densitometer. Level of cleaved caspase 3 was normalized to the level of pro-caspase 3 in SGC-7901 (B) and SGC-7901/DDP (F), respectively. Expression of p-IκB α was normalized to the expression of total IκB α in SGC-7901 (C) and SGC-7901/DDP (G), respectively. Levels of LC3 B, p62, IKK β, and NF-κB p65 were normalized to β-actin levels in SGC-7901 (D) and SGC-7901/DDP (H), respectively. β-Actin was used as an internal control. *p < 0.05, **p < 0.01, compared with control; #p < 0.05, ##p < 0.01, compared with DDP alone.
DDP-induced expression suppressions of p-IκB (Figure 7C,G), IKK β, and NF-κB p65 (Figure 7A,D,E,H) were all promoted by SSD. The results indicated that combination of SSD and DDP could more effectively inhibit NF-κB pathway activities than SSD or DDP alone did. Furthermore, SSD enhanced the effect of DDP-induced protein level rise of cleaved caspase 3 both in SGC-7901 (Figure 7A,B) and SGC-7901/DDP (Figure 7E,F) cells.
Discussion
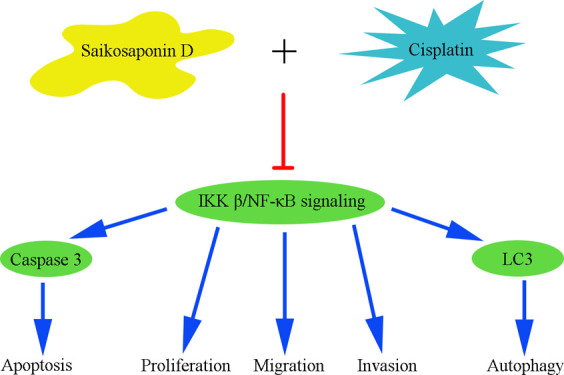
GC is a high-recurrence-rate tumor. The NF-κB pathway is a key step of many aberrantly expressed proteins with tumor-promoting or -suppressing properties promoting the pathology of GC. For instance, NF-κB could affect expressions of a number of genes including cyclin D1 and COX-2,29 and levels of VEGF and E-cadherin indirectly in GC.30,31 Hence, NF-κB might be a useful therapeutic target contributing to conventional therapy and avoiding drug-tolerance in GC patients. Parthenolide was proved to inhibit NF-κB effectively and then suppress GC cell growth in vitro and in vivo.32 Some natural components such as resveratrol and curcumin have showed inhibitory effects of NF-κB activities and exhibited anti-tumor properties.33−36 In this study, we found SSD decreased the NF-κB p65 expression (Figure 4), especially when SSD and DDP were combined (Figure 7). The results imply that NF-κB might be the potential target for SSD to inhibit GC cell proliferation and migration and promote DDP sensitivity of SGC-7901/DDP cells. However, how SSD affects downstream factors of NF-κB needs further confirmation.
Autophagy is a critical process involved in various cellular processes, such as cell survival, proliferation, differentiation and so on. Dysregulation of autophagy has been reported in the occurrence of multiple diseases, including cancer. Regulation of SSD on autophagy is quite diverse in different conditions. Wang et al. showed that SSD could increase LC3 B levels and induce autophagic cell death in apoptosis-defective cell8. Wang and colleagues found SSD-induced LC3 puncta formation in U2OS cells.37 SSD was also proved to increase autophagy puncta formation in ADPKD cells.38 However, in the case of EV-A71 virus infection, SSD may play a cytoprotective role by inhibiting autophagy.39 In another study, authors also found that SSD suppressed pancreatic stellate cell autophagy via the PI3K/Akt/mTOR pathway to prevent pancreatic fibrosis.40 Here, we found that SSD increased LC3 B levels in SGC-7901 and SGC-7901/DDP cells (Figures 4–7). Accordingly, levels of p62 in these two cells were extremely decreased by SSD in the absence of bafilomycin A1 (Figures 4, 6, and 7). Thus, we believe that SSD induces autophagy in SGC-7901 and SGC-7901/DDP cells.
However, autophagy’s roles are very complicated in cancer. Usually, a physiological level of autophagy contributes to cellular survival responding to various adverse conditions, such as starvation, mitochondrial damage, and pathogen infection.41−43 Excessive level of autophagy was proved related to cell death.44 For example, autophagy may promote Fas-induced but not TRAIL-induced apoptosis.45 Yin and colleagues identified that withaferin-A inhibited cell growth of cisplatin-resistant human oral cancer cells by inducing both apoptosis and autophagic cell death via the MAPK/RAS/RAF pathway.46 In this study, we proved that SSD could increase LC3 B levels and decrease p62 levels in the presence of caspase inhibitor Z-VAD-FMK (Figure 6). Considering that SSD could induce cell apoptosis and enhance DDP-induced apoptosis (Figure 4), we speculated that SSD might trigger autophagic cell death in GC cells. But, more evidence is needed to identify this hypothesis.
Conclusions
In summary, SSD inhibited GC cell proliferation and migration, induced apoptosis and autophagy. Furthermore, SSD enhanced DDP-induced proliferation and migration suppression and apoptosis. The potential molecular mechanism might be that SSD inhibited IKK β/NF-κB signaling in GC cells.
Materials and Methods
Reagents
SSD [≥95%, high-performance liquid chromatography (HPLC)] was purchased from Sigma-Aldrich (St. Louis, MO, USA), with a molecular formula of C42H68O13 and molecular weight of 780.98. DDP [Pt(NH3)2Cl2, ≥99.9%], MTT (98%), mitomycin C (pharmaceutical primary standard), bafilomycin A1 (≥90%, HPLC), and Z-VAD-FMK (≥98%, HPLC) were all purchased from Sigma-Aldrich (St. Louis, MO, USA).
Cell Culture
To access the effects of SSD on GC, the human gastric epithelial cell line GES-1 and three typical GC cell lines MGC-803, HGC-27, and SGC-7901 were selected. Both MGC-803 and SGC-7901 are poorly differentiated GC cell lines and HGC-27 is an undifferentiated cell line. Both SGC-7901 and HGC-27 were derived from lymph node metastasis. The cell lines above were all purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). All the GC cells were cultured in RPMI 1640 (Gibco, USA) supplemented with 10% fetal bovine serum (Gibco, USA). SGC-7901/DDP (#GF466) was obtained from GeFan Biotech (Shanghai, China) and maintained in RPMI 1640 (Gibco, USA) containing 10% fetal bovine serum and 1000 ng/mL DDP. GES-1 cells were growing in Dulbecco’s modified Eagle’s medium (Gibco, USA) containing 10% fetal bovine serum. To avoid contamination, all cell culture media were supplemented with 100 μg/mL streptomycin and 100 U/mL penicillin.
MTT Assay
Cells (5 × 103 cells/well) were seeded in 96-well culture plates and incubated overnight. DDP or SSD was added in the medium at the indicated concentrations for 24, 48, or 72 h. Then, MTT solution (5 mg/mL) was added to each well at a volume ratio of 1/10 and incubated for another 4 h at 37 °C. The supernatant was discarded, and 150 μL of dimethyl sulfoxide (Sigma) was added to dissolve the precipitate. Absorbance at 570 nm was measured by SpectraMax M2 Multimode Microplate Readers (Molecular Devices, USA).
Colony Formation Assay
After treating with DDP or/and SSD for 48 h, a total of 500 cells were seeded in each well of a 12-well plate and incubated in media for 10 days. Cells were fixed with 4% paraformaldehyde at room temperature, followed by staining with crystal violet for 15 min. Colony numbers were calculated using Image-Pro Plus 6.0.
Wound-Healing Assay
Cells were seeded in a 12-well culture plate and grown to 80% of confluence. Then, the plate was scratched across the surface of the cell monolayer with a sterile pipette tip after the treatment with 0.5 μM mitomycin C, an inhibitor of cell proliferation, for 1 h. Immediately after wounding (0 h), and up to 12 and 24 h, wounds were captured in five different fields by phase-contrast microscopy. The wound width was compared with the initial width at 0 h time point in the different fields.
Boyden Chamber Assay
Standard 24-well chemotaxis chambers (Millipore, USA) were used in this study. The upper chamber wells, of which the bottom was coated with Matrigel, were filled with cultures of cells with or without a 48 h pre-treatment with SSD or/and DDP at indicated concentrations. After incubation at 37 °C for 12 h, the cells on the upper surface that did not invade were wiped off with a cotton swab. The cells that crossed the membrane were fixed with methanol, stained with 0.1% crystal violet for 10 min, and then photographed using an optical microscope (Olympus, Japan).
Cell Apoptosis Assay
Cells (5 × 105 cells/well) were seeded in Φ 35 mm plates and then treated with different concentrations of SSD or DDP. After incubation for 48 h, cells were harvested and washed twice with ice-cold 1× phosphate-buffered saline (PBS). Apoptosis-mediated cell death was analyzed with a FITC-Annexin V Apoptosis Detection Kit according to the manufacturer’s instructions. Data analysis was carried out using FACScan flow cytometry (BD, USA).
TUNEL Assay
The TUNEL assay kit (Beyotime, China) was used to access cell apoptosis. Cells treated with SSD or/and DDP for 48 h were washed with PBS and fixed with 4% paraformaldehyde for 30 min. After washing once with PBS, cells were incubated in PBS containing 0.3% Triton X-100 for 5 min at room temperature. And then, cells were washed again followed by incubation in 0.3% H2O2 with PBS for 20 min at room temperature. Thereafter, according to the instructions, biotin-labeled solution was added, and cells were incubated in dark at 37 °C for 60 min. Then, labeled reaction termination solution was added, and cells were incubated at room temperature for 10 min. After PBS washing thrice, streptavidin horseradish peroxidase (HRP) solution was added, and cells were incubated at room temperature for 10 min. After washing with PBS thrice again, DAB was added and cells were incubated at room temperature for 30 min. Cells were imaged under an optical microscope (Olympus, Japan).
Western Blot
Cells were washed twice with ice-cold 1× PBS and lysed with RIPA lysis buffer containing 1 mM phenylmethylsulfonyl fluoride, 1 μg/mL aprotinin, 1 μg/mL leupeptin, and 1 μg/mL pepstatin for 30 min on ice. The whole cell lysates were centrifuged at 12,000g for 15 min at 4 °C, and then, total protein concentration was determined using a BCA Protein Assay Kit (Solarbio, China). The protein samples were electrophoresed in 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to poly(vinylidene difluoride) membranes. After blocked with 5% non-fat dry milk in PBS-T for 1 h at room temperature, the membrane was incubated with the primary antibody overnight. After three 10 min washes in PBS-T, the membranes were incubated with HRP-conjugated secondary antibodies for 1 h at room temperature. After another three 10 min washes in PBS-T, immunoreactive proteins were detected with ECL Western blotting analysis system (Solarbio, China) and captured using ImageQuant LAS 500 imager (GE, USA).
The primary antibodies used were as follows: rabbit anti-LC3 A/B (Cell Signaling Technology, USA), rabbit anti-p62 (Abcam, USA), rabbit anti-IKK β (Abcam, USA), rabbit anti-IκB α (Abcam, USA), rabbit anti-p-IκB α (Cell Signaling Technology, USA), mouse anti-β-actin (Santa Cruz, USA), rabbit anti-NF-κB p65 (Abcam, USA), mouse anti-caspase 3 (Santa Cruz, USA), and rabbit anti-cleaved caspase 3 (Abcam, USA).
Statistical Analysis
If not otherwise indicated, data were obtained from three independent experiments. Mean values were calculated and used for analysis of standard deviation or standard error. Statistical analyses were performed using the Student’s two-tailed t-tests. p-Values < 0.05 were considered as statistically significant. Data analysis was performed with Prism 5.0 software (GraphPad, USA).
Acknowledgments
This work was supported by grant from the National Natural Science Foundation of China (31701125), Shanxi Undergraduate Training Programs for Innovation and Entrepreneurship (2018595), and the Fund for Shanxi 1331 Project Key Subjects Construction (1331KSC).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c01795.
Effects of SSD combined with DDP on GES-1 and effects of SSD combined with DDP on cell migration (PDF)
Author Contributions
Conceptualization: J.H. and P.L. Data curation: J.H., P.L., and B.S. Funding acquisition: J.H., P.L., and J.T. Investigation: J.H., P.L., and B.S. Methodology: J.H. and P.L. Resources: J.H. and P.L. Visualization: J.H., P.L. and B.S. Writing original draft: J.H. Writing review and editing: P.L.
The authors declare no competing financial interest.
Supplementary Material
References
- Lu C.-N.; Yuan Z.-G.; Zhang X.-L.; Yan R.; Zhao Y.-Q.; Liao M.; Chen J.-X. Saikosaponin a and its epimer saikosaponin d exhibit anti-inflammatory activity by suppressing activation of NF-κB signaling pathway. Int. Immunopharmacol. 2012, 14, 121–126. 10.1016/j.intimp.2012.06.010. [DOI] [PubMed] [Google Scholar]
- Zhao L.; Li J.; Sun Z. B.; Sun C.; Yu Z. H.; Guo X. Saikosaponin D inhibits proliferation of human osteosarcoma cells via the p53 signaling pathway. Exp. Ther. Med. 2019, 17, 488–494. 10.3892/etm.2018.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Z.-L.; Li X.-J.; Dang H.; Wang F.; Xu X.-Y. Saikosaponin-d affects the differentiation, maturation and function of monocyte-derived dendritic cells. Exp. Ther. Med. 2014, 7, 1354–1358. 10.3892/etm.2014.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y.; Piao X.; Piao X. Saikosaponin-d inhibits β-conglycinin induced activation of rat basophilic leukemia-2H3 cells. Int. Immunopharmacol. 2012, 13, 257–263. 10.1016/j.intimp.2012.04.021. [DOI] [PubMed] [Google Scholar]
- Lixing X.; Zhouye J.; Liting G.; Ruyi Z.; Rong Q.; Shiping M. Saikosaponin-d-mediated downregulation of neurogenesis results in cognitive dysfunction by inhibiting Akt/Foxg-1 pathway in mice. Toxicol. Lett. 2018, 284, 79–85. 10.1016/j.toxlet.2017.11.009. [DOI] [PubMed] [Google Scholar]
- Wang B.-F.; Wang X.-j.; Kang H.-F.; Bai M.-H.; Guan H.-T.; Wang Z.-W.; Zan Y.; Song L.-Q.; Min W.-L.; Lin S.; et al. Saikosaponin-D enhances radiosensitivity of hepatoma cells under hypoxic conditions by inhibiting hypoxia-inducible factor-1α. Cell. Physiol. Biochem. 2014, 33, 37–51. 10.1159/000356648. [DOI] [PubMed] [Google Scholar]
- Wang B.-F.; Dai Z.-J.; Wang X.-J.; Bai M.-H.; Lin S.; Ma H.-B.; Wang Y.-L.; Song L.-Q.; Ma X.-L.; Zan Y.; et al. Saikosaponin-d increases the radiosensitivity of smmc-7721 hepatocellular carcinoma cells by adjusting the g0/g1 and g2/m checkpoints of the cell cycle. BMC Complementary Altern. Med. 2013, 13, 263. 10.1186/1472-6882-13-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong V. K.; Li T.; Law B. Y.; Ma E. D.; Yip N. C.; Michelangeli F.; Law C. K.; Zhang M. M.; Lam K. Y.; Chan P. L.; et al. Saikosaponin-d, a novel SERCA inhibitor, induces autophagic cell death in apoptosis-defective cells. Cell Death Dis. 2013, 4, e720 10.1038/cddis.2013.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J.-c.; Long F.; Zhao J.; Hang J.; Ren Y.-g.; Chen J.-y.; Mu B. The Effects and Mechanisms by which Saikosaponin-D Enhances the Sensitivity of Human Non-small Cell Lung Cancer Cells to Gefitinib. J. Cancer 2019, 10, 6666–6672. 10.7150/jca.30361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.-Y.; Jiang Z.-M.; Ma X.-F.; Li Y.; Liu X.-Z.; Li L.-L.; Wu W.-H.; Wang T. Saikosaponin-d Inhibits the Hepatoma Cells and Enhances Chemosensitivity Through SENP5-Dependent Inhibition of Gli1 SUMOylation Under Hypoxia. Front. Pharmacol. 2019, 10, 1039. 10.3389/fphar.2019.01039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.; Guan X.; Xue H.; Wang P.; Wang M.; Gai X. Reversal of P-glycoprotein-mediated multidrug resistance is induced by saikosaponin D in breast cancer MCF-7/adriamycin cells. Pathol., Res. Pract. 2017, 213, 848–853. 10.1016/j.prp.2017.01.022. [DOI] [PubMed] [Google Scholar]
- Torre L. A.; Siegel R. L.; Ward E. M.; Jemal A. Global Cancer Incidence and Mortality Rates and Trends--An Update. Cancer Epidemiol., Biomarkers Prev. 2016, 25, 16–27. 10.1158/1055-9965.EPI-15-0578. [DOI] [PubMed] [Google Scholar]
- Casamayor M.; Morlock R.; Maeda H.; Anjani J. Targeted literature review of the global burden of gastric cancer. Ecancermedicalscience 2018, 12, 883. 10.3332/ecancer.2018.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimondi A.; Nichetti F.; Peverelli G.; Bartolomeo M. D.; Braud F. D.; Pietrantonio F. Genomic markers of resistance to targeted treatments in gastric cancer: potential new treatment strategies. Pharmacogenomics 2018, 19, 1047–1068. 10.2217/pgs-2018-0077. [DOI] [PubMed] [Google Scholar]
- Gottesman M. M. Mechanisms of cancer drug resistance. Annu. Rev. Med. 2002, 53, 615–627. 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- Shi W.-J.; Gao J.-B. Molecular mechanisms of chemoresistance in gastric cancer. World J. Gastrointest. Oncol. 2016, 8, 673–681. 10.4251/wjgo.v8.i9.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. K.; Choi I. J.; Kim C. G.; Kim H. S.; Oshima A.; Michalowski A.; Green J. E. A gene expression signature of acquired chemoresistance to cisplatin and fluorouracil combination chemotherapy in gastric cancer patients. PLoS One 2011, 6, e16694 10.1371/journal.pone.0016694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testerman T. L.; Morris J. Beyond the stomach: an updated view of Helicobacter pylori pathogenesis, diagnosis, and treatment. World J. Gastroenterol. 2014, 20, 12781–12808. 10.3748/wjg.v20.i36.12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y.; Dang S.; Hou P. Gene methylation in gastric cancer. Clin. Chim. Acta 2013, 424, 53–65. 10.1016/j.cca.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Selaru F. M.; David S.; Meltzer S. J.; Hamilton J. P. Epigenetic events in gastrointestinal cancer. Am. J. Gastroenterol. 2009, 104, 1910–1912. 10.1038/ajg.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q.; Zhu C.; Zhang Y.; Wang Y.; Wang Y.; Zhu L.; Yang X.; Li J.; Nie H.; Jiang S.; et al. Molecular analysis of gastric cancer identifies genomic markers of drug sensitivity in Asian gastric cancer. J. Cancer 2018, 9, 2973–2980. 10.7150/jca.25506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Lei H.; Ma J.; Deng H.; He P.; Dong W. α-Hederin Increases The Apoptosis Of Cisplatin-Resistant Gastric Cancer Cells By Activating Mitochondrial Pathway In Vivo And Vitro. OncoTargets Ther. 2019, 12, 8737–8750. 10.2147/OTT.S221005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M.; Liu B.; Song H.; Yu R.; Zou D.; Chen Y.; Ma Y.; Lv F.; Xu L.; Zhang Z.; et al. β-Elemene inhibits the metastasis of multidrug-resistant gastric cancer cells through miR-1323/Cbl-b/EGFR pathway. Phytomedicine 2020, 69, 153184. 10.1016/j.phymed.2020.153184. [DOI] [PubMed] [Google Scholar]
- Hou G.; Yuan X.; Li Y.; Hou G.; Liu X. Cardamonin, a natural chalcone, reduces 5-fluorouracil resistance of gastric cancer cells through targeting Wnt/β-catenin signal pathway. Invest. New Drugs 2020, 38, 329–339. 10.1007/s10637-019-00781-9. [DOI] [PubMed] [Google Scholar]
- Wang R.; Wang J.; Hassan A.; Lee C.-H.; Xie X.-S.; Li X. Molecular basis of V-ATPase inhibition by bafilomycin A1. Nat. Commun. 2021, 12, 1782. 10.1038/s41467-021-22111-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A.; Tagawa Y.; Yoshimori T.; Moriyama Y.; Masaki R.; Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct. Funct. 1998, 23, 33–42. 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- Song J.; Zhang Z.; Hu Y.; Li Z.; Wan Y.; Liu J.; Chu X.; Wei Q.; Zhao M.; Yang X. An aqueous extract of Prunella vulgaris L. inhibits the growth of papillary thyroid carcinoma by inducing autophagy in vivo and in vitro. Phytother. Res. 2021, 35, 2691–2702. 10.1002/ptr.7015. [DOI] [PubMed] [Google Scholar]
- Xiao Q.; Chen X.-H.; Jiang R.-C.; Chen S.-Y.; Chen K.-F.; Zhu X.; Zhang X.-l.; Huang J.-j.; Qin Y.; Zhang G.-P.; et al. Ubc9 Attenuates Myocardial Ischemic Injury Through Accelerating Autophagic Flux. Front. Pharmacol. 2020, 11, 561306. 10.3389/fphar.2020.561306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manu K. A.; Shanmugam M. K.; Li F.; Chen L.; Siveen K. S.; Ahn K. S.; Kumar A. P.; Sethi G. Simvastatin sensitizes human gastric cancer xenograft in nude mice to capecitabine by suppressing nuclear factor-kappa B-regulated gene products. J. Mol. Med. 2014, 92, 267–276. 10.1007/s00109-013-1095-0. [DOI] [PubMed] [Google Scholar]
- Nam S. Y.; Ko Y. S.; Jung J.; Yoon J.; Kim Y. H.; Choi Y. J.; Park J. W.; Chang M. S.; Kim W. H.; Lee B. L. A hypoxia-dependent upregulation of hypoxia-inducible factor-1 by nuclear factor-κB promotes gastric tumour growth and angiogenesis. Br. J. Cancer 2011, 104, 166–174. 10.1038/sj.bjc.6606020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z.; Liu X.; Tang Z.; Zhou Y.; Qiao L. Possible regulatory role of Snail in NF-κB-mediated changes in E-cadherin in gastric cancer. Oncol. Rep. 2013, 29, 993–1000. 10.3892/or.2012.2200. [DOI] [PubMed] [Google Scholar]
- Jiang Y.; Zhang Y.; Chu F.; Xu L.; Wu H. Circ_0032821 acts as an oncogene in cell proliferation, metastasis and autophagy in human gastric cancer cells in vitro and in vivo through activating MEK1/ERK1/2 signaling pathway. Cancer Cell Int. 2020, 20, 74. 10.1186/s12935-020-1151-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Jiang A.; Qi B.; Ma Z.; Xiong Y.; Dou J.; Wang J. Resveratrol Protects against Helicobacter pylori-Associated Gastritis by Combating Oxidative Stress. Int. J. Mol. Sci. 2015, 16, 27757–27769. 10.3390/ijms161126061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P.-M.; Cheng Y.-W.; Wu T.-C.; Chen C.-Y.; Lee H. MnSOD overexpression confers cisplatin resistance in lung adenocarcinoma via the NF-κB/Snail/Bcl-2 pathway. Free Radical Biol. Med. 2015, 79, 127–137. 10.1016/j.freeradbiomed.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Imran M.; Ullah A.; Saeed F.; Nadeem M.; Arshad M. U.; Suleria H. A. R. Curcumin, anticancer; antitumor perspectives: A comprehensive review. Crit. Rev. Food Sci. Nutr. 2018, 58, 1271–1293. 10.1080/10408398.2016.1252711. [DOI] [PubMed] [Google Scholar]
- Jin R.; Xia Y.; Chen Q.; Li W.; Chen D.; Ye H.; Zhao C.; Du X.; Shi D.; Wu J.; et al. Da0324, an inhibitor of nuclear factor-κB activation, demonstrates selective antitumor activity on human gastric cancer cells. Drug Des., Dev. Ther. 2016, 10, 979–995. 10.2147/DDDT.S90081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.; Zhang Y.; Zhang Q.; Peng S.; Shen C.; Yu Y.; Zhang M.; Yang W.; Wu Q.; Zhang Y.; et al. Content decline of SERCA inhibitors saikosaponin a and d attenuates cardiotoxicity and hepatotoxicity of vinegar-baked Radix bupleuri. Environ. Toxicol. Pharmacol. 2017, 52, 129–137. 10.1016/j.etap.2017.04.002. [DOI] [PubMed] [Google Scholar]
- Shi W.; Xu D.; Gu J.; Xue C.; Yang B.; Fu L.; Song S.; Liu D.; Zhou W.; Lv J.; et al. Saikosaponin-d inhibits proliferation by up-regulating autophagy via the CaMKKβ-AMPK-mTOR pathway in ADPKD cells. Mol. Cell. Biochem. 2018, 449, 219–226. 10.1007/s11010-018-3358-0. [DOI] [PubMed] [Google Scholar]
- Li C.; Huang L.; Sun W.; Chen Y.; He M.-L.; Yue J.; Ballard H. Saikosaponin D suppresses enterovirus A71 infection by inhibiting autophagy. Signal Transduction Targeted Ther. 2019, 4, 4. 10.1038/s41392-019-0037-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L.-H.; Li C.-X.; Zhuo Y.-Z.; Yang L.; Cui N.-Q.; Zhang S.-K. Saikosaponin d ameliorates pancreatic fibrosis by inhibiting autophagy of pancreatic stellate cells via PI3K/Akt/mTOR pathway. Chem.-Biol. Interact. 2019, 300, 18–26. 10.1016/j.cbi.2019.01.005. [DOI] [PubMed] [Google Scholar]
- Green D. R.; Levine B. To be or not to be? How selective autophagy and cell death govern cell fate. Cell 2014, 157, 65–75. 10.1016/j.cell.2014.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C.; Bassik M. C.; Moresi V.; Sun K.; Wei Y.; Zou Z.; An Z.; Loh J.; Fisher J.; Sun Q.; et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature 2012, 481, 511–515. 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi A. M.; Ryter S. W.; Levine B. Autophagy in human health and disease. N. Engl. J. Med. 2013, 368, 1845–1846. 10.1056/NEJMc1303158. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Levine B. Autosis and autophagic cell death: the dark side of autophagy. Cell Death Differ. 2015, 22, 367–376. 10.1038/cdd.2014.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gump J. M.; Staskiewicz L.; Morgan M. J.; Bamberg A.; Riches D. W. H.; Thorburn A. Autophagy variation within a cell population determines cell fate through selective degradation of Fap-1. Nat. Cell Biol. 2014, 16, 47–54. 10.1038/ncb2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X.; Yang G.; Ma D.; Su Z. Inhibition of cancer cell growth in cisplatin-resistant human oral cancer cells by withaferin-A is mediated via both apoptosis and autophagic cell death, endogenous ROS production, G2/M phase cell cycle arrest and by targeting MAPK/RAS/RAF signalling pathway. J. BUON. 2020, 25, 332–337. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.