Abstract

A leucine aminopeptidase primarily hydrolyzes amino acid leucine from the N-terminus end of proteins and is involved in free amino acid regulation, which makes it a potential therapeutic target against neglected tropical diseases including leishmaniasis. We here report the purification and characterization of the leucine aminopeptidase from Leishmania donovani (LdLAP). Using a set of biophysical and biochemical methods, we demonstrate that this enzyme was properly folded after expression in a bacterial system and catalytically active when supplemented with divalent metal cofactors with synthetic fluorogenic peptides. Subsequently, enzymatic inhibition assay denoted that LdLAP activity was inhibited by peptidomimetics, particularly actinonin, which caused potent inhibition and exhibited stronger binding association with the LdLAP. Stronger association of actinonin with the LdLAP was due to a stable complex formation mostly mediated by hydrogen bonding with catalytic and substrate-binding residues in the C-terminal catalytic domain. With molecular dynamics simulation studies, we demonstrate that peptidomimetics retain their topological space in the LdLAP catalytic pocket and form a stable complex. These results expand the current knowledge of aminopeptidase biochemistry and highlight that specific actinonin or peptidomimetic-based inhibitors may emerge as leads to combat leishmaniasis.
1. Introduction
Leishmaniasis, caused by the protozoan parasite Leishmania, is transmitted through female phlebotomine sandflies and exists in three major forms—visceral (the severest form of the disease), cutaneous, and mucocutaneous. Approximately 20 Leishmania species are thought to cause different forms of the disease affecting an estimated 0.7 to 1 million people annually (https://www.who.int/news-room/fact-sheets/detail/leishmaniasis). The existing medications present multiple limitations such as drug toxicity1 and resistance,2 necessitating the need to identify essential parasitic pathways that can be targeted to combat this neglected tropical disease.
Aminopeptidases hydrolyze amino acid residues from the amino-termini of proteins or peptide substrates3 and play crucial roles in several physiological processes like free amino acid regulation,4 cell-cycle control,5 selective protein degradation,6 protein maturation,7 signal peptide modification,8 turnover,9,10 nutrient acquisition,11 tumor growth,12 and angiogenesis.13 In fact, peptidases of different classes are fundamental enzymes identified and characterized in numerous microorganisms, plants, and animals and play vital roles in parasitic processes by either degrading peptides or interacting with peptide-dependent signaling.9 Moreover, variations in expression patterns or catalytic functions of a leucine aminopeptidase (LAP) affect peptide activation, resulting into alterations in tumor cell proliferation, angiogenesis, and invasion.9,14 The functional insights into the LAP reveal its broad functions in different living systems. For instance, in bacteria, LAPs are implicated in site-specific homologous recombination and regulation of transcription.9 In plants, LAPs work as molecular chaperones15 in addition to being important for turnover.16 On the contrary, LAPs in mammals are responsible for N-terminal processing of some proteins and determine cell redox status.17 Furthermore, LAPs are implicated in proliferation, migration, and invasion18 and provide free amino acids necessary for growth and survival.17 LAPs have been studied in several human parasites such as Trypanosoma, Plasmodium, and Leishmania and have been described as potential drug targets in disease-causing protozoa.4,17,19 For example, the LAP of the malaria-causing parasite Plasmodium falciparum is regarded as a druggable candidate to combat malaria.20 Furthermore, LAP from the carcinogenic Helicobacter pylori is highly expressed during nitric oxide (NO) stress21 suggesting its role to evade host immunity and appears to contribute to the survival of drug-resistant parasites22 proving the essentiality of LAPs throughout living systems. Additionally, the vaccination of sheep and cattle with a Fasciola hepatica LAP against fasciolosis23 and the growth inhibition of Trypanosoma brucei by the metalloprotease inhibitor arphamenine A24 further substantiate the claims of aminopeptidases particularly LAPs to be potential vaccine and druggable candidates against parasitic infections.
We here report the purification and characterization of the leucine aminopeptidase from Leishmania donovani (LdLAP) using biochemical, biophysical, and structural approaches. Our work provides key insights into the functional and structural attributes of the LdLAP and suggests that peptidomimetics or the chemical libraries synthesized from them may emerge as frontline inhibitors of the LdLAP, which may pave the way for novel therapeutics against neglected tropical diseases (NTDs) in general and leishmaniasis in particular.
2. Results
2.1. Sequence and Structural Analysis of the LdLAP
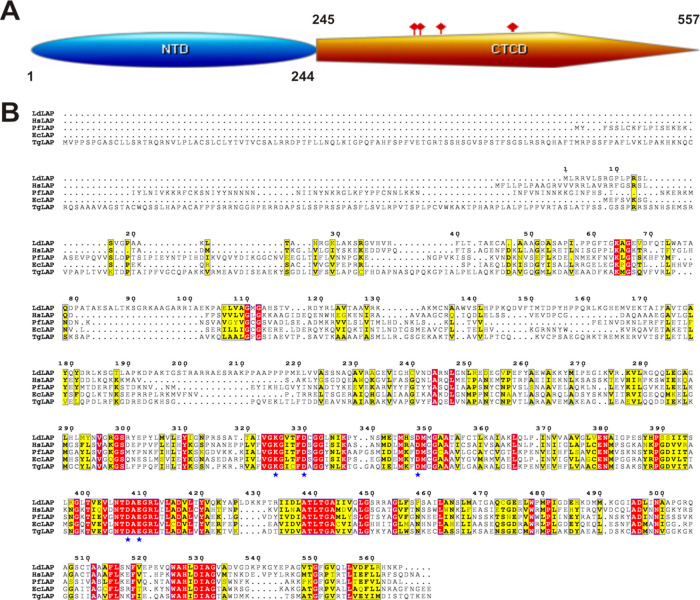
The LdLAP sequence was identified to be a cytosolic leucyl aminopeptidase involved in the processing and regular turnover by catalyzing the removal of unsubstituted N-terminal amino acids from various intracellular peptides. Moreover, the N-terminal domain (residues 1–244) was predicted to have an unspecified or unknown function, while the C-terminal domain (residues 245–557) was identified as the M17 aminopeptidase domain. Further sequence analysis of LAPs from bacteria to mammals reveals a conserved active site embedded in the C-terminal catalytic domain (Figure 1A). Five catalytic residues (K325, D330, D348, D407, and E409) make up the active site, which has interactions with the two metal cofactors, usually Mn(II), Co(II), or Zn(II) in the catalytic center of each subunit (Figure 1B). The overall sequence identity of the LdLAP (UniProt ID E9BGD0) with other LAPs was low in the N-terminal domain and high in the C-terminal domain (Figure 1B).
Figure 1.
Sequence analysis of the LdLAP. (A) Domain architecture of the LdLAP displaying its two domains, i.e., the N-terminal domain (NTD, blue) and the C-terminal catalytic domain (CTCD, orange) with active site residues as orange squares. (B) Alignment of the LdLAP sequence with various LAP sequences depicting a conserved catalytic domain and active site residues (K325, D330, D348, D407, and E409) that are presented as blue stars. Conserved and similar residues are shown in red and yellow boxes, respectively.
The N-terminal domain was found to be highly variable with amino acid insertions, whose roles are far from known. An amino acid signature motif composed of residues NTDAEGRL is highly conserved in all LAPs. It contains two catalytic residues D407 and E409 of the LdLAP, which represents a feature of all known or characterized M17 leucine aminopeptidases (LAPs).
The homology model of a full-length LdLAP could not be generated due to lack of a suitable template for the major portion of the N-terminal non-catalytic domain of the LdLAP. Like other LAPs, a large portion of the N-terminal domain of the LdLAP shows variability in amino acid composition and very little sequence conservation with existing LAPs unlike the C-terminal catalytic domain, which is not only highly conserved among other LAPs from various organisms but also possesses a conserved catalytic site. Therefore, a homology model was generated without the initial 180 residues of the N-terminal domain of the LdLAP (Figure 2A).
Figure 2.
Structural characteristics of the LdLAP. (A) Cartoon representation of the modeled structure of the LdLAP. The active site residues are depicted as sticks along with zinc ions as green spheres. (B) Secondary structure of the modeled LdLAP. Cylinders and arrows are representative of helices and sheets, respectively. (C) Ribbon representation of the hexameric structure of the LdLAP. Each monomeric unit is colored differently, whereas two metal ions of each subunit are shown as gray spheres.
Similar to other LAPs,17 the leishmanial LAP carries structural elements that make it form a biological hexamer carrying six active sites in the cavity inside the oligomer. The substrate-binding channel adjacent to the active site appears narrower and can lodge shorter peptides of up to five amino acid residues, which explain the shorter substrate preference of all known LAPs. The catalytic site contains two metal cofactors (zinc ions), which were imported during homology modeling from the template structure, i.e., the leucine aminopeptidase from Pseudomonas putida (PDB ID 3H8E). The two catalytic metal ions in each subunit of the LdLAP are coordinated by catalytic residues including three aspartates, one glutamate, and one lysine (Figure 2A). For catalytic activity, both metal binding sites must be occupied. The two metal cofactors are involved in substrate and water molecule binding and stabilize the transition state intermediate. Additionally, LAPs also contain a bicarbonate ion in the catalytic active site, which acts as a general acid/base in the catalytic reaction. The secondary structure mostly resembles the template catalytic domain (Figure 2B). The modeled LdLAP hexamer portrays a triangular shape with round corners in which the C-terminal domains of six LdLAP monomers assemble to build the core of a hexamer, while the N-terminal domains give rise to three corners (Figure 2C). The cavity encompassing six substrate-binding channels interconnects at the center of the hexamer.
The comparison of the LdLAP modeled structure with other known LAP structures from Escherichia coli (EcLAP, PDB ID 1GYT), Pseudomonas putida (PpLAP, PDB ID 3H8G), Staphylococcus aureus (SaLAP, PDB ID 3H8E), Coxiella burnetii (CbLAP, PDB ID 3IJ3), and Francisella tularensis (FtLAP, PDB ID 3PEI) highlights an identical orientation of all LAP monomers and a similar overall shape of the hexamer. The C-terminal catalytic domain of the LdLAP is highly similar to that of other LAPs as pairwise alignment shows nearly 90% Cα atoms of the C-terminal domain overlapping with their counterpart atoms in the EcLAP, PpLAP, SaLAP, CbLAP, and FtLAP with a root-mean-square deviation (RMSD) of less than 1.2 Å, demonstrating sequence identity between 34 and 38% over the corresponding positions. Higher fold organization and conservation of the C-terminal domain in leucine aminopeptidases are often linked to their identical catalytic function.25
2.2. Cloning and Purification of the LdLAP
The LdLAP gene with 1698 bp coding for 565 amino acids was cloned into pET28a vector in the frame between restriction sites NdeI and HindIII followed by its confirmation through the Sanger sequencing method. The overexpression of the LdLAP, which is an approximately 62 kDa protein, was examined on 10% (w/v) SDS-PAGE. The soluble LdLAP enzyme was purified by immobilized metal affinity chromatography (IMAC) and gel filtration chromatography. Protein purity was analyzed with an SDS-PAGE gel that indicates toward protein homogeneity (Figure 3A).
Figure 3.

Purification and biophysical characterization. (A) SDS-PAGE gel showing the purified LdLAP in lane 1, while lane M presents a prestained protein marker. (B–C) Far-UV CD spectra of the LdLAP with and without metal cofactors and at different pHs. (D) Heat denaturation plots of the LdLAP.
2.3. LdLAP Expresses as a Properly Folded Protein
With CD spectroscopy, we determined the average secondary structure of the LdLAP. The high ellipticity of the protein indicates toward its proper folding (Figure 3B). The LdLAP like its other reported counterparts is a metal-dependent protein; hence, we monitored the metal-induced changes in the LdLAP. Our experiments delineated that both Co(II) and Mn(II), which are metal cofactors of this protein, increase its ellipticity (Figure 3B). The CD result analyzed through the DichroWeb server suggests that there is an increase in the α-helix of the LdLAP when incubated with divalent metal cofactors Co(II) and Mn(II) (Table 1). It is possible that these metal-induced shifts in the secondary structure could be necessary for the protein to be catalytically active.
Table 1. Secondary Structure Contents of the LdLAP with Metal Cofactors and at Various pHs.
| condition | α-helices | β-sheets | random coils |
|---|---|---|---|
| LdLAP (apo) | 23 | 25 | 52 |
| LdLAP with Co(II) | 23 | 27 | 50 |
| LdLAP with Mn(II) | 26 | 25 | 49 |
| LdLAP at pH 6.0 | 19 | 22 | 59 |
| LdLAP at pH 8.0 | 23 | 25 | 52 |
| LdLAP at pH 10.0 | 21 | 23 | 56 |
While recording far-UV CD spectra in different chemical environments, it was observed that the LdLAP lost a significant degree of the secondary structure at acidic and highly alkaline pHs, suggesting that the protein was more stable at pH 8.0 (Figure 3C). At both high and low pHs, the ellipticity at 222 nm is significantly reduced, and there is a noticeable shift in ellipticity from 208 toward 202 nm, which indicates a significant loss of the secondary structure. This is in line with the protein activity profile at different pHs, which shows the protein to be most active at pH 8.0, implying that extreme pH distorted the secondary structure of the LdLAP. While estimating the thermal stability, it was observed that the LdLAP was stable in solution (Figure 3C). However, the protein showed precipitation upon the addition of metal cofactors when the temperature crossed 325 K.
2.4. Peptidomimetics Potently Inhibit the LdLAP
As M17 leucine aminopeptidases are optimally active at slightly alkaline pH, the catalytic activity of the LdLAP was measured against a leucine-containing fluorogenic substrate Leu-AMC (l-leucine-7-amido-4-methylcoumarin) and other substrates linked to the fluorophore AMC (7-amido-4-methylcoumarin). The determined steady-state kinetic parameters demonstrated high affinity and turnover of the LdLAP when it was supplemented with the metal cofactors (Figure 4A and Table 2). However, the LdLAP only cleaved leucine efficiently and had noticeable activity for hydrolysis of methionine from the substrate Met-AMC (Figure 4B).
Figure 4.

Enzyme activity and inhibition. (A) Michaelis–Menten fit for the amidolytic activity of the LdLAP. Data represents mean activity ± SD (N = 3). (B) Enzyme assay of the LdLAP with different fluorogenic peptides. (C, E, and G) Double-reciprocal plots demonstrating competitive inhibition of the LdLAP with peptidomimetics, while chemical structures of peptidomimetics are presented in the insets of figures. (D, F, and H) Determination of the inhibition constant for actinonin, bestatin, and amastatin.
Table 2. Steady-State Kinetic Parameters of the LdLAP for Hydrolysis of the Peptide Substrate Leu-AMC.
| metal | concentration (μM) | Km (μM) | kcat (×10–3 min–1) | kcat/Km (μM–1 min–1) |
|---|---|---|---|---|
| Co(II) | 100 | 68.11 ± 6.3 | 22 ± 1.4 | 0.323 |
| Mn(II) | 100 | 57.29 ± 5.7 | 20 ± 1.2 | 0.349 |
The activity observed for the liberation of other amino acid residues or the peptides used was negligible suggesting the LdLAP to be largely specific to leucine hydrolysis. Further, peptidomimetic aminopeptidase inhibitors actinonin, amastatin, and bestatin were examined through in vitro assay that inhibited the LdLAP potently in a dose-dependent manner. The other aminopeptidases inhibited by peptidomimetic inhibitors include M20 family aminopeptidases10 and other classes of metal-dependent aminopeptidases.26 We next performed kinetic assays with and without inhibitors to determine the mode of inhibition toward the LdLAP. It was found that all the tested inhibitors act through a competitive mode of binding, competing with the substrate for binding to the catalytic site with inhibition constant (Ki) values found to be 2.7, 4.375, and 7.18 nM for actinonin, bestatin, and amastatin, respectively (Figure 4C–H).
2.5. LdLAP Interacts with Peptidomimetics with High Affinity
As the Ki of amastatin was relatively higher than that of other peptidomimetic inhibitors, fluorescence spectroscopy was performed to calculate the binding affinity of the LdLAP with bestatin and actinonin. The LdLAP ORF encodes for five tryptophan residues positioned at 74, 138, 263, 478, and 525, which we used as intrinsic fluorophores to get the intrinsic fluorescence spectra of the LdLAP. Upon excitation at 290 nm, the LdLAP exhibited maximum fluorescence emission at 375–380 nm indicating the tryptophan residues to be positioned on the exterior of the protein and hence highly accessible to the solvent. We monitored the changes in the intrinsic fluorescence intensity of the LdLAP upon its binding with peptidomimetic inhibitors, which exhibited some fluorescence intensity and were thus properly blanked during data analysis. Moreover, the maximum emission of the LdLAP at 379 nm decreases upon binding to peptidomimetics in a concentration-dependent manner (Figure 5A–F). As all peptidomimetic compounds inhibited the LdLAP through a competitive mode, a decrease in LdLAP emission may be attributed to the interaction of the LdLAP excited-state fluorophores with their environment within the protein. This decline in LdLAP fluorescence emission is expected as proteins undergo conformational changes upon binding to ligands, which are monitored by recording the changes in tryptophan fluorophores of the protein that are sensitive to the changes in the local environment. Therefore, a decrease in LdLAP fluorescence emission when titrated with peptidomimetics can be attributed to the protein–inhibitor complex formation. Analysis of the binding association constant suggested actinonin to have a high-order binding association with the LdLAP (Figure 5C), which corroborates with its high binding affinity and the high potency observed with the biochemically determined inhibition constant.
Figure 5.
Peptidomimetics showing high binding association for the LdLAP. (A–F) Fluorescence spectra of the LdLAP with two potent inhibitors actinonin and bestatin along with the Stern–Volmer and modified Stern–Volmer plots showing binding association of peptidomimetics with the leishmanial LAP.
2.6. Peptidomimetics Bind at LdLAP Catalytic Sites
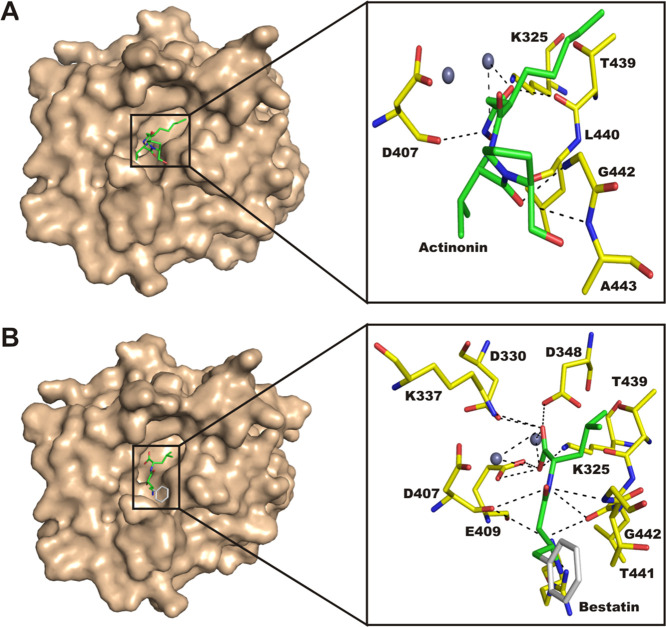
To understand the inhibition mechanism of the LdLAP with peptidomimetics, protein–ligand docking studies were performed using AutoDock Vina. Structurally, all inhibitors bound both to the metal-dependent catalytic site and its adjoining substrate-binding sites in particular the S1 and S2 sites. The major interacting force involved was hydrogen bonding mostly with the catalytic residues. Actinonin occupied the S1′ pocket and had a binding affinity of −7.2 kcal mol–1. Residues of the LdLAP like Lys325, Asp407, Glu409, Gly410, Arg411, Leu440, Gly442, and Ala443 formed hydrogen bonds with actinonin, while only Ala531 displayed hydrophobic interaction (Figure 6A). Similarly, bestatin exhibited more hydrogen bond interactions and fitted deep into the catalytic pocket with a binding affinity of −7.1 kcal mol–1. It interacted with the leishmanial LAP through hydrogen bonds with residues Asp407, Arg409, Leu440, and Gly442, whereas residues Ile445 and Ala531 showed hydrophobic interactions (Figure 6B).
Figure 6.
Molecular docking. (A,B) Surface and atomic interactions of peptidomimetic inhibitors with the LdLAP. Inhibitors and interacting residues are labeled and displayed as sticks. Metal ions and polar contacts are shown as gray spheres and black dashed lines, respectively.
2.7. Peptidomimetics Formed Stable Complexes with the LdLAP
In order to evaluate stability of the complexes of the LdLAP with peptidomimetics, molecular dynamics simulations (MDS) were performed followed by analysis of various parameters including root-mean-square deviation (RMSD), root-mean-square fluctuation (RMSF), and the radius of gyration (Rg). The RMSD demonstrated that both the LdLAP and its complexes with actinonin and bestatin converged to equilibrium toward the end of a 20 ns production run. Among the two complexes, the RMSD of the LdLAP–bestatin complex showed comparatively lesser RMS deviations and reached equilibrium early during the production run. On the other hand, the LdLAP–actinonin complex exhibited a deviation early and stabilized afterward, indicating that actinonin binding induced a conformational change and raised the RMSD but formed a very stable complex with no major deviations after 5 ns (Figure 7A). A similar pattern was demonstrated by the RMSF plot where the unbound protein and the complexes showed no major fluctuations in the catalytic and other residues, which are involved in the binding of inhibitors (Figure 7B). Thus, the RMSF profile demonstrated that the LdLAP forms highly stable complexes with peptidomimetics that may be a reason for their potency. To assess changes in the compactness, Rg was evaluated as it provides an overview of the overall mobility of atomic assembly during MDS. Both the native LdLAP and complexes displayed a steady Rg particularly after 15 ns, signifying that all systems were compact (Figure 7C). These results highlight that peptidomimetic inhibitors make a stable complex with the LdLAP, which is in agreement with the results of molecular docking and spectroscopic measurements.
Figure 7.
MD simulation. Group parameters RMSD (A), RMSF (B), and Rg (C) highlighting that peptidomimetic inhibitors form a stable complex with the LdLAP. Residues are numbered as per the full-length LdLAP sequence in Figure 7B, whereas catalytic residues are denoted by green downward arrows.
3. Discussion
Aminopeptidases utilize a metal cofactor for hydrolysis of peptide bonds.27 Among many such enzymes, the M17 leucine aminopeptidase is a major cytosolic exopeptidase, which hydrolyzes leucine residues from the amino termini of peptides.17 Such enzymes are widespread in living systems and play multiple physiological or biological roles by either degrading peptides or interacting with peptide-dependent signaling.9 Parasitic leucine aminopeptidases belonging to malarial parasites Plasmodium,4Leishmania,19 and Trypanosoma(17) are important for parasite biology due to their involvement in free amino acid regulation and have thus been investigated as therapeutic or vaccine candidates against parasitic adversaries.
We cloned, expressed, and purified the LdLAP and demonstrated with far-UV circular dichroism that it was expressed in a properly folded form in bacteria. We next found this enzyme to be catalytically active against the peptide substrate Leu-AMC and demonstrated its optimum activity at pH 8.0. Activity seemed to decrease below or beyond pH 8.0 like other members of leucine aminopeptidases.17,28 Like previously reported for protozoan LAPs,28,29 the LdLAP did not cleave residues other than leucine efficiently from peptide substrates and demonstrated optimal activity with Co(II) as a cofactor similar to other leucine aminopeptidases4,28,29 and aminopeptidases in general.10 The purified LdLAP follows Michaelis–Menten kinetics like other M17 aminopeptidases. In fact, the only known LAP that does not obey Michaelis–Menten kinetics and shows S-shaped kinetics is the LAP of H. pylori (HpLAP)30 due to a distinguishing hydrophilic pocket at the S1 subsite, which accommodates charged groups found in basic residues.31 This renders the HpLAP more efficient against basic residues. On the contrary, the LdLAP like most LAPs acts as an exopeptidase releasing only leucine with high efficiency while acting on smaller peptides like other exopeptidases, which suggests its role in the release of amino acids in the terminal stages of protein catabolism.19,28 As such, the substrates of the LdLAP may be the shorter peptides released in the last stages of proteasomal protein degradation pathways or the peptides degraded by endoproteases in the parasitophorous vacuole (PV) and sent to the cytoplasm for the generation of free amino acids by aminopeptidases. As many aminopeptidases are pivotal to parasite biology and may thus emerge as potential targets, we characterized the recombinant LdLAP expressed in E. coli and examined potent inhibitors.
All LAP structures known till date from different organisms are homologous hexamers with great structural similarities. The comparison of overall root-mean-square deviations (RMSDs) between LAP orthologs reveals an RMSD between 0.27 to 1.2 Å and tremendous sequence identity in the C-terminal catalytic domains. Moreover, M17 LAP family members are bilobal proteins in which three monomeric chains combine to form trimers, which in turn associate to form homohexamers of varying molecular weights albeit without the involvement of disulfide bridges.31,32 The analysis of the homohexamer reveals the active sites to be facing a central cavity and isolated from the bulk solvent but accessible to its substrates through solvent channels. The mechanism of aminopeptidase activity of LAPs suggests a great degree of conservation of the catalytic mechanism with the involvement of water molecules and metal ions being indispensable for catalysis. Additionally, a bicarbonate ion in the catalytic center is deemed necessary for aminopeptidase activity.31,33 Without the supplementation of a metal cofactor in assay buffer, LdLAP activity was negligible. In fact, the LdLAP demonstrated activity only with the supplementation of divalent cofactors particularly Co(II). This signifies the metal dependence of the LdLAP like other metal-dependent aminopeptidases belonging to different classes.3,34
Given that aminopeptidases are targeted for therapeutic intervention against many adversaries, we screened peptidomimetics in vitro and found the LdLAP to be potently inhibited by all tested peptidomimetic inhibitors like bestatin, actinonin, and amastatin with a competitive mode of inhibition. Notably, potent peptidomimetics showed high binding association and formed a stable complex with the LdLAP. Peptidomimetics are known to inhibit many cytosolic aminopeptidases, in particular M20 aminopeptidases10 and M17 leucine aminopeptidases.17,29 To formulate peptidomimetics as potential therapeutics, specificity remains a major challenge as such inhibitors act upon many classes of aminopeptidases. Therefore, chemical modifications that render peptidomimetics highly specific to LAPs are a need of the hour. An alternate approach could be to develop synthetic schemes to modify peptidomimetics such that specific inhibitors targeting LAPs can be synthesized. An approach employing structure-based drug design may emerge as a frontline method in this direction. Moreover, many peptide analogs may exhibit increased inhibitor potency and specificity. Such analogs may emerge as useful scaffolds to design novel small molecule inhibitors to cause potent and selective inhibition of the leishmanial leucine aminopeptidase.
4. Conclusions
In this study, we have expressed the leishmanial leucine aminopeptidase in a bacterial system and purified it to homogeneity. This aminopeptidase demonstrated structural stability and optimal activity for the excision of leucine with the supplementation of divalent metal chlorides. Structurally, the LdLAP displays a highly conserved C-terminal catalytic domain and active site surrounded by two metal cofactors. Moreover, peptidomimetic inhibitors, in particular actinonin, inhibit the LdLAP potently with high binding affinity at the catalytic center. Actinonin also retained its topological space in the catalytic center and made a stable complex with the LdLAP. Altogether, our work provides key insights into the structure and biochemistry of the LdLAP and highlights the potency of peptidomimetics as proteolytic inhibitors of this enzyme, which might have implications in the structure-based drug design against trypanosomatid parasites.
5. Methods
5.1. Protein Sequence Analysis and Homology Modeling
The amino acid sequence of the LdLAP (565 residues) was retrieved from the UniProt database (https://www.uniprot.org/) with ID E9BGD0 and analyzed with the web-based SMART server35 in a normal mode to find the domain organization and PDB homologues. The program BLASTp36 was employed to detect similar PDB structures of the LdLAP (without N-terminal 180 residues), and then, the structure of the leucine aminopeptidase from Pseudomonas putida (PDB ID 3H8E) was used as a template (percentage query coverage, 98; percentage identity, 37.8) to perform homology modeling using the program Modeller.37 The model with the lowest DOPE score was picked, and its geometry was evaluated using RAMPAGE.38 Ramachandran outliers in the model were fixed, and the energy of the structure was minimized through GROMACS39 by the steepest descent method.
5.2. Cloning and Expression of the LdLAP
The LdLAP gene of length 1.7 kb was amplified with polymerase chain reaction (PCR) using gene-specific primers and cloned into the expression vector pET28a between the restriction sites NdeI and HindIII. The cloning was confirmed by overnight double digestion with restriction enzymes NdeI and HindIII at 37 °C and DNA sequencing. The resulting pET28a–LdLAP construct encoded the full-length LdLAP protein with an N-terminal 6X-histidine tag when overexpressed in BL21(DE3) cells in LB medium supplemented with 50 μg mL–1 kanamycin. Protein expression was induced with 1 mM isopropyl-d-thiogalactopyranoside (IPTG) as reported previously.19
5.3. Purification of the LdLAP
Cells expressing the LdLAP were harvested with centrifugation at 4830g and resuspended in lysis buffer with the composition of 50 mM Tris-HCl (pH 8.0), 300 mM NaCl, 30 mM imidazole, 2 mM phenylmethylsulfonyl fluoride (PMSF), and 1 mM β-mercaptoethanol. After resuspension, the cells were treated with a lysozyme and DNase and kept on ice for 1 h and then sonicated at an amplitude of 30% with a pulse break of 9 s each for 40 min. The cell lysate was centrifuged at 26,000g for 45 min at 4 °C, and the supernatant was loaded on HisTrap HP beads (GE Healthcare) calibrated with buffer carrying 50 mM Tris-HCl (pH 8.0), 300 mM NaCl, and 40 mM imidazole. An imidazole gradient was used to ward off impurities, and the protein was eluted with buffer containing 300 mM imidazole followed by analysis of protein purity on 10% (w/v) SDS-PAGE.
5.4. Aminopeptidase Assay and Inhibition
The amidolytic activity of the LdLAP was determined in vitro in a metal cofactor-supplemented biochemical assay in 50 mM Tris-HCl (pH 8.0) buffer by measuring the liberation of l-leucine from the fluorogenic peptide substrate, Leu-AMC (Sigma-Aldrich). Release of AMC was monitored at 37 °C on a TECAN Infinite M200 Pro spectrofluorometer with a λex and λem pair of 355 and 460 nm, respectively. For steady-state kinetic parameters, fluorescence was employed to deduce product formation (AMC) by utilizing an AMC standard curve. Data was plotted, and all parameters were determined in GraphPad Prism. To study the inhibitory efficacy of peptidomimetics like bestatin, amastatin, and actinonin (Santa Cruz Biotechnology) on the amidolytic activity, the LdLAP was preincubated in assay buffer with inhibitors (0 to 20 nM) for 30 min at 37 °C before the addition of a substrate to measure the residual amidolytic activity. The mode of inhibition was deduced with double-reciprocal plots.
5.5. Circular Dichroism Spectroscopy
Circular dichroism (CD) spectroscopy was used to study the secondary structure of the LdLAP on a JASCO-J1500 CD spectrometer in 20 mM Tris-HCl (pH 8.0) and 50 mM NaCl at 25 °C in a quartz cell with a path length of 0.2 cm using a protein concentration of 2 μM. Three scans were accumulated at a scan speed of 50 nm min–1, with data being collected from 250 to 195 nm. Three replicate experiments were carried out with the average of three scans deemed as the final spectra. The data was plotted using GraphPad Prism software.
5.6. Fluorescence Measurements
The purified LdLAP (5 μM) in buffer containing 20 mM Tris-HCl (pH 8.0) and 50 mM NaCl at 25 °C was excited (λex ∼ 290 nm, λem ∼ 300–400 nm) on a Fluormax 300 fluorescence spectrophotometer (Hitachi). Since tryptophan fluorescence is strongly influenced by its local environment, we titrated the LdLAP with the aminopeptidase inhibitors and recorded fluorescence measurements by monitoring a decrease in intensity. The relative fluorescence intensity obtained by [(F0 – F)/F0], where F0 and F were the fluorescence intensities in the absence and presence of the substrates, was plotted against the inhibitor concentration as shown earlier,40 and the quenching and association constants were calculated by Stern–Volmer and modified Stern–Volmer plots, respectively.
5.7. Molecular Docking
Molecular docking was performed with the program AutoDock Vina 1.1.2 version41 to predict the binding mechanisms and atomic interactions of peptidomimetic inhibitors with the modeled LdLAP structure. The structural coordinates of all ligands including peptidomimetics were retrieved from PubChem,42 and their geometry was optimized with Discovery studio 3.5.43 The structural coordinates of all peptidomimetics were used to generate the inbuilt files in Autodock Tools (ADT). Ligand torsion angles were kept as per default settings. Hydrogen and water were removed, and the grid was set up at the catalytic pocket of the LdLAP. Polar hydrogens were added, and a grid-based procedure (grid dimensions, 29 × 24 × 38) was followed for docking while employing the Lamarckian genetic algorithm44 for docking. For every inhibitor, a total of nine poses were generated with the LdLAP, and the related poses (RMSD < 1.2 Å) were clustered. The pose with the highest binding affinity was chosen as a representative, while PyMOL (http://pymol.org) was employed for molecular visualization.
5.8. Molecular Dynamics Simulation
Molecular dynamics simulations were carried out using the GROMOS96 43a1 force field in the program package GROMACS.39 The production run was executed with SPC as a water model for the LdLAP and its complexes with peptidomimetics actinonin and bestatin. For every run, the best affinity pose was taken after molecular docking. Complex topologies were generated as reported previously.10 Every system was solvated and neutralized by the addition of monovalent ions to ensure electroneutrality. After an energy minimization step to reduce steric clashes and prepare stable systems, two canonical equilibration runs were added before a 20 ns molecular dynamics run was started to collect data.
Acknowledgments
The authors acknowledge DST-FIST- and UGC-SAP-sponsored instrument facilities of the Department of Biotechnology & Bioinformatics, School of Life Sciences as well as the Center for Modeling Simulation & Design (CMSD), University of Hyderabad, Hyderabad for computational resources for MD simulation studies. S.Y.B. is thankful to the Indian Council of Medical Research (ICMR) for the Senior Research Fellowship. Authors also acknowledge Dr. Tushar Vaidya for providing genomic DNA of Leishmania donovani and Mr. Samayaditya Singh for helping during MDS experiments.
Author Present Address
† Indian Institute of Science, CV Raman Road, Bengaluru 560012, India (S.Y.B.)
This work was funded by the Council of Scientific and Industrial Research [CSIR, project no. 37 (1686)/17/EMR-II], and Indian Council of Medical Research [ICMR, project no. 61/15/2020-IMM/BMS], Government of India as the grant to I.A.Q.
The authors declare no competing financial interest.
References
- Sundar S.; Chakravarty J. Antimony toxicity. Int. J. Environ. Res. Public Health 2010, 7, 4267–4277. 10.3390/ijerph7124267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponte-Sucre A.; Gamarro F.; Dujardin J.-C.; Barrett M. P.; López-Vélez R.; García-Hernández R.; Pountain A. W.; Mwenechanya R.; Papadopoulou B. Drug resistance and treatment failure in leishmaniasis: A 21st century challenge. PLoS Neglected Trop. Dis. 2017, 11, e0006052 10.1371/journal.pntd.0006052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A. Aminopeptidases: structure and function. FASEB J. 1993, 7, 290–298. 10.1096/fasebj.7.2.8440407. [DOI] [PubMed] [Google Scholar]
- Stack C. M.; Lowther J.; Cunningham E.; Donnelly S.; Gardiner D. L.; Trenholme K. R.; Skinner-Adams T. S.; Teuscher F.; Grembecka J.; Mucha A.; Kafarski P.; Lua L.; Bell A.; Dalton J. P. Characterization of the Plasmodium falciparum M17 leucyl aminopeptidase. A protease involved in amino acid regulation with potential for antimalarial drug development. J. Biol. Chem. 2007, 282, 2069–2080. 10.1074/jbc.M609251200. [DOI] [PubMed] [Google Scholar]
- Peer W. A. The role of multifunctional M1 metallopeptidases in cell cycle progression. Ann. Bot. 2011, 107, 1171–1181. 10.1093/aob/mcq265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecker S. H.; Goldberg A. L.; Mitch W. E. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J. Am. Soc. Nephrol. 2006, 17, 1807–1819. 10.1681/ASN.2006010083. [DOI] [PubMed] [Google Scholar]
- Frottin F.; Bienvenut W. V.; Bignon J.; Jacquet E.; Jacome A. S. V.; Van Dorsselaer A.; Cianferani S.; Carapito C.; Meinnel T.; Giglione C. MetAP1 and MetAP2 drive cell selectivity for a potent anti-cancer agent in synergy, by controlling glutathione redox state. Oncotarget 2016, 7, 63306–63323. 10.18632/oncotarget.11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varland S.; Osberg C.; Arnesen T. N-terminal modifications of cellular proteins: The enzymes involved, their substrate specificities and biological effects. Proteomics 2015, 15, 2385–2401. 10.1002/pmic.201400619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui M.; Fowler J. H.; Walling L. L. Leucine aminopeptidases: diversity in structure and function. Biol. Chem. 2006, 387, 1535–1544. 10.1515/BC.2006.191. [DOI] [PubMed] [Google Scholar]
- Bhat S. Y.; Qureshi I. A. Mutations of key substrate binding residues of leishmanial peptidase T alter its functional and structural dynamics. Biochim. Biophys. Acta, Gen. Subj. 2020, 1864, 129465. 10.1016/j.bbagen.2019.129465. [DOI] [PubMed] [Google Scholar]
- White R. C.; Gunderson F. F.; Tyson J. Y.; Richardson K. H.; Portlock T. J.; Garnett J. A.; Cianciotto N. P.; Isberg R.; Roy C. Type II Secretion-Dependent Aminopeptidase LapA and Acyltransferase PlaC Are Redundant for Nutrient Acquisition during Legionella pneumophila Intracellular Infection of Amoebas. mBio 2018, 9, e00528–18. 10.1128/mBio.00528-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman-Rojas L.; Rangel R.; Salameh A.; Edwards J. K.; Dondossola E.; Kim Y. G.; Saghatelian A.; Giordano R. J.; Kolonin M. G.; Staquicini F. I.; Koivunen E.; Sidman R. L.; Arap W.; Pasqualini R. Cooperative effects of aminopeptidase N (CD13) expressed by nonmalignant and cancer cells within the tumor microenvironment. Proc. Natl. Acad. Sci. 2012, 109, 1637–1642. 10.1073/pnas.1120790109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondossola E.; Rangel R.; Guzman-Rojas L.; Barbu E. M.; Hosoya H.; St. John L. S.; Molldrem J. J.; Corti A.; Sidman R. L.; Arap W.; Pasqualini R. CD13-positive bone marrow-derived myeloid cells promote angiogenesis, tumor growth, and metastasis. Proc. Natl. Acad. Sci. 2013, 110, 20717–20722. 10.1073/pnas.1321139110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani S.; Shibata K.; Kikkawa F.; Hattori A.; Tsujimoto M.; Ishii M.; Kobayashi H. Essential role of placental leucine aminopeptidase in gynecologic malignancy. Expert Opin. Ther. Targets 2007, 11, 453–461. 10.1517/14728222.11.4.453. [DOI] [PubMed] [Google Scholar]
- Scranton M. A.; Yee A.; Park S.-Y.; Walling L. L. Plant leucine aminopeptidases moonlight as molecular chaperones to alleviate stress-induced damage. J. Biol. Chem. 2012, 287, 18408–18417. 10.1074/jbc.M111.309500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waditee-Sirisattha R.; Hattori A.; Shibato J.; Rakwal R.; Sirisattha S.; Takabe T.; Tsujimoto M. Role of the Arabidopsis leucine aminopeptidase 2. Plant Signaling Behav. 2011, 6, 1581–1583. 10.4161/psb.6.10.17105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm J.; Valente M.; García-Caballero D.; Wilson K. S.; González-Pacanowska D. Structural Characterization of Acidic M17 Leucine Aminopeptidases from the TriTryps and Evaluation of Their Role in Nutrient Starvation in Trypanosoma brucei. mSphere 2017, 2, e00226-17. 10.1128/mSphere.00226-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.; Dai G.; Wang S.; Zhao Y.; Wang X.; Zhao X.; Zhang H.; Wei L.; Zhang L.; Guo S.; Song W.; Guo L.; Fang C. Inhibition of the proliferation, migration, and invasion of human breast cancer cells by leucine aminopeptidase 3 inhibitors derived from natural marine products. Anti-Cancer Drugs 2020, 31, 60–66. 10.1097/CAD.0000000000000842. [DOI] [PubMed] [Google Scholar]
- Morty R. E.; Morehead J. Cloning and characterization of a leucyl aminopeptidase from three pathogenic Leishmania species. J. Biol. Chem. 2002, 277, 26057–26065. 10.1074/jbc.M202779200. [DOI] [PubMed] [Google Scholar]
- Trenholme K. R.; Brown C. L.; Skinner-Adams T. S.; Stack C.; Lowther J.; To J.; Robinson M. W.; Donnelly S. M.; Dalton J. P.; Gardiner D. L. Aminopeptidases of malaria parasites: new targets for chemotherapy. Infect. Disord. Drug Targets 2010, 10, 217–225. 10.2174/187152610791163363. [DOI] [PubMed] [Google Scholar]
- Qu W.; Zhou Y.; Shao C.; Sun Y.; Zhang Q.; Chen C.; Jia J. Helicobacter pylori proteins response to nitric oxide stress. J. Microbiol. 2009, 47, 486–493. 10.1007/s12275-008-0266-0. [DOI] [PubMed] [Google Scholar]
- Kaakoush N. O.; Asencio C.; Mégraud F.; Mendz G. L. A redox basis for metronidazole resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 2009, 53, 1884–1891. 10.1128/AAC.01449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acosta D.; Cancela M.; Piacenza L.; Roche L.; Carmona C.; Tort J. F. Fasciola hepatica leucine aminopeptidase, a promising candidate for vaccination against ruminant fasciolosis. Mol. Biochem. Parasitol. 2008, 158, 52–64. 10.1016/j.molbiopara.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Knowles G. The effects of arphamenine-A, an inhibitor of aminopeptidases, on in-vitro growth of Trypanosoma brucei brucei. J. Antimicrob. Chemother. 1993, 32, 172–174. 10.1093/jac/32.1.172. [DOI] [PubMed] [Google Scholar]
- Burley S. K.; David P. R.; Lipscomb W. N. Leucine aminopeptidase: bestatin inhibition and a model for enzyme-catalyzed peptide hydrolysis. Proc. Natl. Acad. Sci. 1991, 88, 6916–6920. 10.1073/pnas.88.16.6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; Liu C.; Lin Y. L.; Li F. Structural insights into central hypertension regulation by human aminopeptidase A. J. Biol. Chem. 2013, 288, 25638–25645. 10.1074/jbc.M113.494955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat S. Y.; Dey A.; Qureshi I. A. Structural and functional highlights of methionine aminopeptidase 2 from Leishmania donovani. Int. J. Biol. Macromol. 2018, 115, 940–954. 10.1016/j.ijbiomac.2018.04.090. [DOI] [PubMed] [Google Scholar]
- Jia H.; Nishikawa Y.; Luo Y.; Yamagishi J.; Sugimoto C.; Xuan X. Characterization of a leucine aminopeptidase from Toxoplasma gondii. Mol. Biochem. Parasitol. 2010, 170, 1–6. 10.1016/j.molbiopara.2009.11.005. [DOI] [PubMed] [Google Scholar]
- Jia H.; Terkawi M. A.; Aboge G. O.; Goo Y. K.; Luo Y.; Li Y.; Yamagishi J.; Nishikawa Y.; Igarashi I.; Sugimoto C.; Fujisaki K.; Xuan X. Characterization of a leucine aminopeptidase of Babesia gibsoni. Parasitology 2009, 136, 945–952. 10.1017/S0031182009006398. [DOI] [PubMed] [Google Scholar]
- Dong L.; Cheng N.; Wang M.-W.; Zhang J.; Shu C.; Zhu D.-X. The leucyl aminopeptidase from Helicobacter pylori is an allosteric enzyme. Microbiology 2005, 151, 2017–2023. 10.1099/mic.0.27767-0. [DOI] [PubMed] [Google Scholar]
- Modak J. K.; Rut W.; Wijeyewickrema L. C.; Pike R. N.; Drag M.; Roujeinikova A. Structural basis for substrate specificity of Helicobacter pylori M17 aminopeptidase. Biochimie 2016, 121, 60–71. 10.1016/j.biochi.2015.11.021. [DOI] [PubMed] [Google Scholar]
- Cadavid-Restrepo G.; Gastardelo T. S.; Faudry E.; de Almeida H.; Bastos I. M.; Negreiros R. S.; Lima M. M.; Assumpção T. C.; Almeida K. C.; Ragno M.; Ebel C.; Ribeiro B. M.; Felix C. R.; Santana J. M. The major leucyl aminopeptidase of Trypanosoma cruzi (LAPTc) assembles into a homohexamer and belongs to the M17 family of metallopeptidases. BMC Biochem. 2011, 12, 46. 10.1186/1471-2091-12-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan S.; Oellig C. A.; Birru W. A.; Caradoc-Davies T. T.; Stack C. M.; Lowther J.; Skinner-Adams T.; Mucha A.; Kafarski P.; Grembecka J.; Trenholme K. R.; Buckle A. M.; Gardiner D. L.; Dalton J. P.; Whisstock J. C. Structure of the Plasmodium falciparum M17 aminopeptidase and significance for the design of drugs targeting the neutral exopeptidases. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 2449–2454. 10.1073/pnas.0911813107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat S. Y.; Jagruthi P.; Srinivas A.; Arifuddin M.; Qureshi I. A. Synthesis and characterization of quinoline-carbaldehyde derivatives as novel inhibitors for leishmanial methionine aminopeptidase 1. Eur. J. Med. Chem. 2020, 186, 111860. 10.1016/j.ejmech.2019.111860. [DOI] [PubMed] [Google Scholar]
- Letunic I.; Bork P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis S.; Madden T. L. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004, 32, W20–W25. 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B.; Sali A. Protein Structure Modeling with MODELLER. Methods Mol. Biol. 2017, 1654, 39–54. 10.1007/978-1-4939-7231-9_4. [DOI] [PubMed] [Google Scholar]
- Wang W.; Xia M.; Chen J.; Deng F.; Yuan R.; Zhang X.; Shen F. Data set for phylogenetic tree and RAMPAGE Ramachandran plot analysis of SODs in Gossypium raimondii and G. arboreum. Data Brief 2016, 9, 345–348. 10.1016/j.dib.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Spoel D.; Lindahl E.; Hess B.; Groenhof G.; Mark A. E.; Berendsen H. J. C. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- Bhat S. Y.; Bhandari S.; Thacker P. S.; Arifuddin M.; Qureshi I. A. Development of quinoline-based hybrid as inhibitor of methionine aminopeptidase 1 from Leishmania donovani. Chem. Biol. Drug Des. 2021, 97, 315–324. 10.1111/cbdd.13783. [DOI] [PubMed] [Google Scholar]
- Trott O.; Olson A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009, 31, 455–461. 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.; Thiessen P. A.; Bolton E. E.; Chen J.; Fu G.; Gindulyte A.; Han L.; He J.; He S.; Shoemaker B. A.; Wang J.; Yu B.; Zhang J.; Bryant S. H. PubChem Substance and Compound databases. Nucleic Acids Res. 2016, 44, D1202–D1213. 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Diego: Accelrys Software Inc. Discovery Studio Modeling Environment, Release 3.5; Accelrys Software Inc.: 2012. [Google Scholar]
- Fuhrmann J.; Rurainski A.; Lenhof H.-P.; Neumann D. A new Lamarckian genetic algorithm for flexible ligand-receptor docking. J. Comput. Chem. 2010, 31, 1911–1918. 10.1002/jcc.21478. [DOI] [PubMed] [Google Scholar]