Abstract
Bioactive constituents from natural sources are of great interest as alternatives to synthetic compounds for the treatment of various diseases, including diabetes mellitus. In the present study, phytochemicals present in Leucaena leucocephala (Lam.) De Wit leaves were identified by gas chromatography–mass spectrometry and further examined by qualitative and quantitative methods. α-Amylase enzyme activity assays were performed and revealed that L. leucocephala (Lam.) De Wit leaf extract inhibited enzyme activity in a dose-dependent manner, with efficacy similar to that of the standard α-amylase inhibitor acarbose. To determine which phytochemicals were involved in α-amylase enzyme inhibition, in silico virtual screening of the absorption, distribution, metabolism, excretion, and toxicity properties was performed and pharmacophore dynamics were assessed. We identified hexadecenoic acid and oleic acid ((Z)-octadec-9-enoic acid) as α-amylase inhibitors. The binding stability of α-amylase to those two fatty acids was confirmed in silico by molecular docking and a molecular dynamics simulation performed for 100 ns. Together, our findings indicate that L. leucocephala (Lam.) De Wit-derived hexadecanoic acid and oleic acid are natural product-based antidiabetic compounds that can potentially be used to manage diabetes mellitus.
Introduction
Diabetes mellitus (DM) is a metabolic disease that results in hyperglycemia. The hormone insulin, produced by β cells in pancreatic islets, is required for uptake of glucose from the blood into cells for energy or for storage in the liver as glycogen. In DM, the body either does not produce enough or cannot effectively use insulin.1−4 In type 2 DM (T2DM), the body becomes resistant to insulin owing to disturbed insulin receptor cell signaling, and glucose builds up in circulation, whereas cells are glucose-starved. This forces the pancreas to work harder to produce more insulin. In later stages of the disease, cells in the pancreas become damaged and do not produce enough insulin.5 DM-related disease complications include congestive heart failure, atherosclerosis, cardiovascular disease, peripheral vascular disease, chronic kidney disease, neuropathy, and retinopathy.6−13 A potential strategy for people with DM is to adjust glucose absorption from the intestine into the bloodstream.14 Retardation of glucose absorption by inhibiting carbohydrate hydrolyzing enzymes is one therapeutic approach to hyperglycemia.15,16
Currently, several synthetic antidiabetic drugs are available to lower blood glucose level: metformin, sulfonylureas, meglitinides, thiazolidinediones, dipeptidyl peptidase-4 inhibitors, glucagon-like peptide-1 receptor agonists, and sodium–glucose cotransporter-2 inhibitors.17,18
Natural products and traditional herbal medicines also have potential antidiabetic efficacy.19−21 Naturally occurring chemical compounds in plants, called phytoconstituents, can inhibit α-amylase and be used to manage blood glucose level in T2DM with fewer side effects than those caused by synthetic agents.19,22,23 Pancreatic α-amylase plays a key role in the digestive system,24 and inhibition of α-amylase activity decreases the glucose level in the bloodstream.15,22,25−28 However, synthetic α-amylase inhibitors have been reported to have various side effects, such as diarrhea, nausea, dyspepsia, myocardial infarction, hypoglycemia, liver damage, flatulence, abdominal pain, dropsy, and heart failure.22,23,29
Leucaena leucocephala (Lam.) De Wit is a fast-growing tree (angiosperm, Fabaceae) indigenous to tropical countries near the equator.30−34 Its leaves are frequently eaten by ruminants33,35,36 and are traditionally used as an anthelmintic therapy,37−40 though the leaves can potentially cause alimentary toxicosis.41 The antioxidative and antidiabetic properties of L. leucocephala (Lam.) De Wit leaves have been investigated.42−45 We designed the present study to identify and investigate the phytochemical constituents of local L. leucocephala (Lam.) leaves (Nagapattinam district in Tamil Nadu, India) and their potential antidiabetic properties.
We identified phytochemicals by gas chromatography–mass spectrometry (GC–MS), performed in silico screening of those phytochemicals to identify their biophysical properties, and characterized their interactions with α-amylase using molecular dynamics simulations. We identified two fatty acids as antidiabetic drug candidates because of their ability to inhibit α-amylase activity.
Results and Discussion
Phytochemical Analysis: Qualitative Histochemical Analysis
The phytochemical content of L. leucocephala leaves was histochemically analyzed, and the results are presented in Table S1. Leaf samples contained large amounts of tannins, terpenoids, polyphenols, and flavonoids and smaller quantities of saponins.
Qualitative Phytochemical Analysis
We qualitatively identified the phytochemical constituents in the alcohol and aqueous extracts of L. leucocephala leaves, such as tannins, saponins, flavonoids, steroids, terpenoids, triterpenoids, anthraquinones, polyphenols, glycosides, and coumarins (Table S2). Alkaloids were not detected in either extract type, whereas flavonoids were abundantly detected in both. Aqueous extracts were enriched in saponins, anthraquinones, polyphenols, and coumarins.
Quantitative Analysis
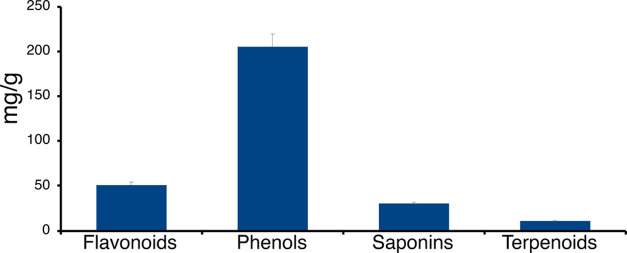
The 70% ethanol leaf extract of L. leucocephala was quantitatively analyzed for its phytochemical constituents (Table S3 and Figure 1). L. leucocephala leaf extract contained many phenolic constituents as well as flavonoids and saponins, but only small amounts of terpenoids were detected, which is consistent with other studies describing the metabolic profile of L. leucocephala (Figure 1).46−48
Figure 1.
Quantitative analysis of phytochemicals in L. leucocephala leaf powder. Values (mg of phytochemical per gram of leaf powder) are expressed as mean ± standard deviation (SD) based on experiments performed in triplicate.
In Vitro Anti-α-Amylase Enzyme Activity of L. leucocephala Leaf Extracts
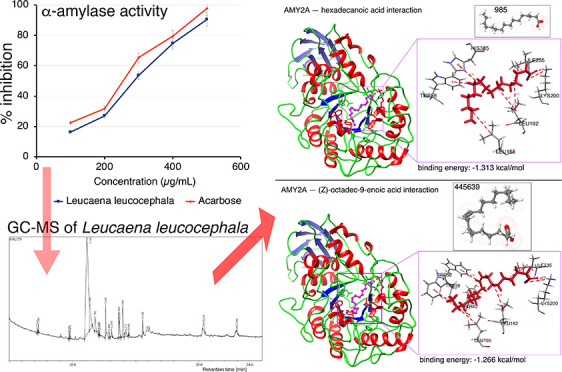
The in vitro α-amylase inhibitory activity of the 70% ethanol L. leucocephala leaf extract was investigated. The L. leucocephala leaf extract inhibited α-amylase activity in a dose-dependent manner (Table S4 and Figure 2). Acarbose, a clinically approved anti-α-amylase drug, was used as a comparative standard. The L. leucocephala leaf extract inhibited α-amylase activity to an extent similar (within a difference of ∼±5%) to that of acarbose (Figure 2).
Figure 2.
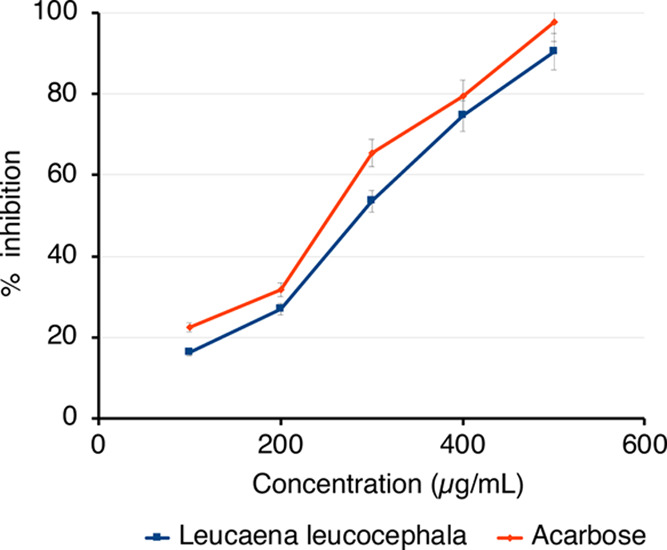
Efficacy of anti-α-amylase enzyme activity of L. leucocephala leaf extract (70% ethanol) compared with that of acarbose. Mean values are from independent experiments performed in triplicate (maximum mean deviation ±5%).
GC–MS Analysis
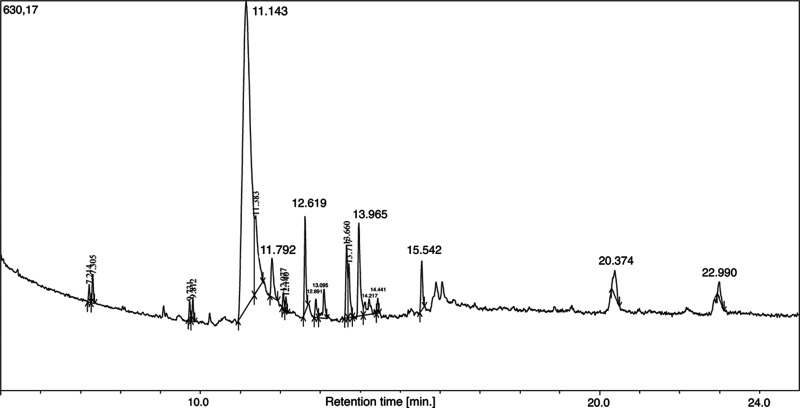
We next analyzed the 70% ethanol extract using GC–MS analysis and unambiguously identified 17 specific phytochemical compounds; cyclohexane-1,2,3,4,5,6-hexol was particularly abundant (peak 5) (Table 1 and Figure 3).43,49,50
Table 1. Phytocomponents Identified in the Ethanolic (70%) Leaf Extract of L. leucocephala by GC–MSa.
| peak | RT | area | area (%) | height | height (%) | A/H | compound IUPAC name |
|---|---|---|---|---|---|---|---|
| 1 | 7.214 | 56 975 | 0.53 | 28 986 | 1.75 | 1.97 | 6-methylheptan-1-ol, CAS number: 1653-40-3 |
| 2 | 7.305 | 97 848 | 0.90 | 45 863 | 2.77 | 2.13 | 3,7-dimethylnonane, CAS number: 17302-32-8 |
| 3 | 9.731 | 62 260 | 0.58 | 30 452 | 1.84 | 2.04 | 6-methylheptan-1-ol, CAS number: 1653-40-3 |
| 4 | 9.812 | 80 458 | 0.74 | 35 780 | 2.16 | 2.25 | 3,7-dimethyldecane, CAS number: 17312-54-8 |
| 5 | 11.143 | 6 692 445 | 61.84 | 496 677 | 29.98 | 13.47 | cyclohexane-1,2,3,4,5,6-hexol, CAS number: 87-89-8 |
| 6 | 11.383 | 760 814 | 7.03 | 124 537 | 7.52 | 6.11 | 4-bromo-2-[(2-phenylbenzotriazol-5-yl)iminomethyl]phenol, NIST number: 223598 |
| 7 | 11.792 | 272 398 | 2.52 | 62 726 | 3.79 | 4.34 | hexadecanoic acid, CAS number: 57-10-3 |
| 8 | 12.077 | 60 474 | 0.56 | 27 190 | 1.64 | 2.22 | tridecan-1-ol, CAS number: 112-70-9 |
| 9 | 12.140 | 50 414 | 0.47 | 23 921 | 1.44 | 2.11 | 2,3,6-trimethyloctane, CAS number: 62016-33-5 |
| 10 | 12.619 | 397 897 | 3.68 | 154 947 | 9.35 | 2.57 | 2,6,10-trimethylpentadecane, CAS number: 3892-00-0 |
| 11 | 12.891 | 91 527 | 0.85 | 29 926 | 1.81 | 3.06 | tetradecanal, CAS number: 124-25-4 |
| 12 | 13.095 | 180 050 | 1.66 | 46 539 | 2.81 | 3.87 | tetradecanal, CAS number: 124-25-4 |
| 13 | 13.660 | 345 690 | 3.19 | 117 115 | 7.07 | 2.95 | 2-(diethylamino)-N-(2,6-dimethylphenyl)acetamide, CAS number: 137-58-6 |
| 14 | 13.717 | 267 510 | 2.47 | 87 110 | 5.26 | 3.07 | N-trimethylsilyl aniline, CAS number: 3768-55-6 |
| 15 | 13.965 | 573 744 | 5.30 | 149 994 | 9.05 | 3.83 | (Z)-octadec-9-enoic acid, CAS number: 112-80-1 |
| 16 | 14.217 | 198 276 | 1.83 | 23 725 | 1.43 | 8.36 | undecan-1-ol, CAS number: 112-42-5 |
| 17 | 14.441 | 51 164 | 0.47 | 24 316 | 1.47 | 2.10 | not identified |
| 18 | 15.542 | 233 427 | 2.16 | 79 035 | 4.77 | 2.95 | (E)-3,7,11,15-tetramethyl-2- hexadecen-1-ol, CAS number: 7541-49-3 |
| 19 | 20.374 | 236 481 | 2.19 | 40 896 | 2.47 | 5.78 | 2,6-dimethylhepta-1,5-diene, CAS number: 6709-39-3 |
| 20 | 22.990 | 111 694 | 1.03 | 26 867 | 1.62 | 4.16 | spiro[4.4]nona-3,8-diene, CAS number: 6569-94-4 |
Note: RT, retention time; A/H, area/height.
Figure 3.
GC chromatogram of the ethanolic (70%) L. leucocephala leaf extract. Further details are provided in Table S5. The top 20 GC peaks (based on A/H values) (Table 1) were selected for MS analysis, which unambiguously revealed 17 chemicals (Table 1).
In Silico Virtual Screening: In Silico Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) Property Analysis
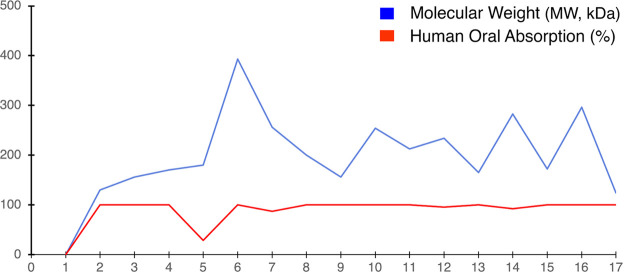
To assess the possible clinical potential of the 17 phytochemicals identified by GC–MS in the ethanolic (70%) leaf extract of L. leucocephala, we determined their ADMET properties and physicochemical parameters in silico (Table S5 and Figure 4). The percentage of human oral absorption was predicted based on a quantitative multiple linear regression model (>80% indicates high absorption and <25% poor absorption). Overall, predicted qualitative human oral absorption was assessed on a scale from 1 to 3 (1, 2, or 3 for low, medium, or high absorption, respectively). The ADMET assessment uses a knowledge-based set of rules, including assessment for suitable values of % human oral absorption, number of metabolites, number of rotatable bonds, log P, solubility, MW, and cell permeability.51 Hexadecanoic acid and oleic acid met the physicochemical and drug-likeness criteria best (≥80% human oral absorption; MW 250–500 (Table S5 and Figure 4)), also known as the Rule of 5.52,53
Figure 4.
In silico ADMET analyses. (1) 6-Methylheptan-1-ol; (2) 3,7-dimethylnonane; (3) 3,7-dimethyldecane; (4) cyclohexane-1,2,3,4,5,6-hexol; (5) 4-bromo-2-[(2-phenylbenzotriazol-5-yl)iminomethyl]phenol; (6) hexadecanoic acid; (7) tridecan-1-ol; (8) 2,3,6-trimethyloctane; (9) 2,6,10-trimethylpentadecane; (10) tetradecanal; (11) 2-(diethylamino)-N-(2,6-dimethylphenyl)acetamide; (12) N-trimethylsilyl aniline; (13) (Z)-octadec-9-enoic acid; (14) undecan-1-ol; (15) (E)-3,7,11,15-tetramethyl-2- hexadecen-1-ol; (16) 2,6-dimethylhepta-1,5-diene; and (17) spiro[4.4]nona-3,8-diene.
Molecular Docking
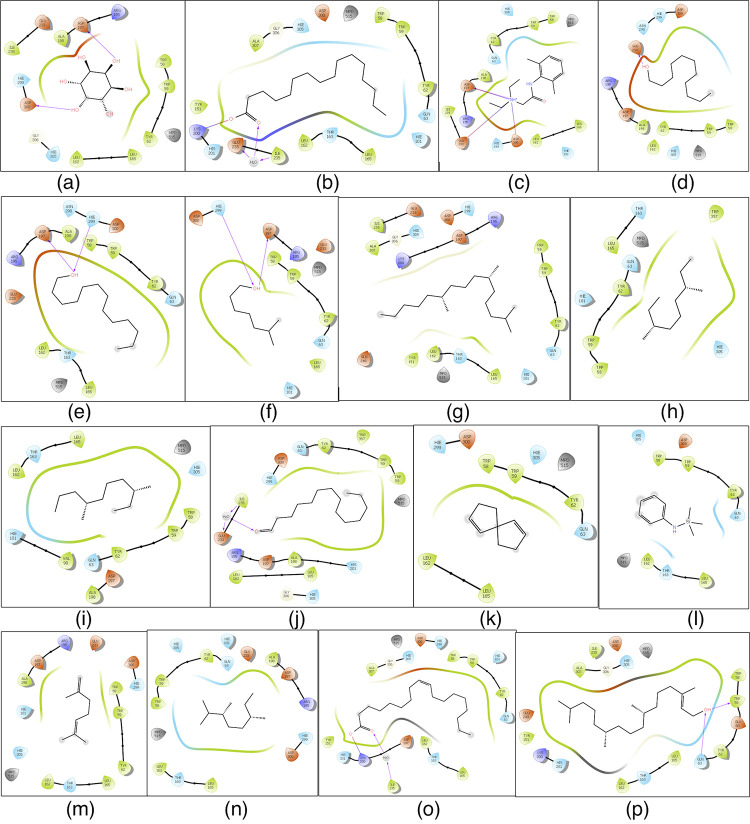
Active-site α-amylase amino acid (AA) residues were identified as TRP58, TRP59, TYR62, GLN63, LEU65, TYR151, LEU162, THR163, LEU165, LYS200, Glu233, ILE235, ASP300, and HIS305.16 We used the Glide 4.0 XP extra precision scoring function and docking protocol to determine the binding affinities of the 17 selected phytochemicals as ligands for α-amylase (Table S6). α-Amylase–ligand interactions are presented in two dimensions in Figure 5. α-Amylase affinity for the ligands hexadecanoic acid and (Z)-octadec-9-enoic acid was −1.313 and −1.266 kcal/mol, respectively.
Figure 5.
Schematic representation of 2D interactions of α-amylase with select phytochemicals identified in the ethanolic (70%) leaf extract of L. leucocephala. (a) Cyclohexane-1,2,3,4,5,6-hexol; (b) hexadecanoic acid; (c) 2-(diethylamino)-N-(2,6-dimethylphenyl)acetamide; (d) undecan-1-ol; (e) tridecan-1-ol; (f) 6-methylheptan-1-ol; (g) 2,6,10-trimethylpentadecane; (h) 3,7-dimethylnonane; (i) 3,7-dimethyldecane; (j) tetradecanal; (k) spiro[4.4]nona-3,8-diene; (l) N-trimethylsilyl aniline; (m) 2,6-dimethylhepta-1,5-diene; (n) 2,3,6-trimethyloctane; (o) (Z)-octadec-9-enoic acid; and (p) (E)-3,7,11,15-tetramethylhexadec-2-en-1-ol. Black lines indicate C–C (chemicals) or peptide bonds (protein). Amino acid color code indicates hydrophobicity and polarity. Van der Waals forces/hydrogen bonds are indicated by colored arrows. Hexadecanoic acid and (Z)-octadec-9-enoic acid interact with key AAs in the active site of α-amylase.16
Highest Occupied Molecular Orbital (HOMO)–Lowest Unoccupied Molecular Orbital (LUMO) Energy Gap Analysis
Because higher gap energies between molecules indicate lower stability and reactivity, we focused on the low HOMO–LUMO energy gaps of select phytochemicals present in the ethanolic (70%) leaf extract of L. leucocephala. The fatty acids hexadecanoic acid (−1.583; very low) and (Z)-octadec-9-enoic acid (−13.161; low) had reasonable low energy gaps (Table S7). Those properties assisted us in defining the frontier molecular orbitals of the molecules (Figure 6).
Figure 6.
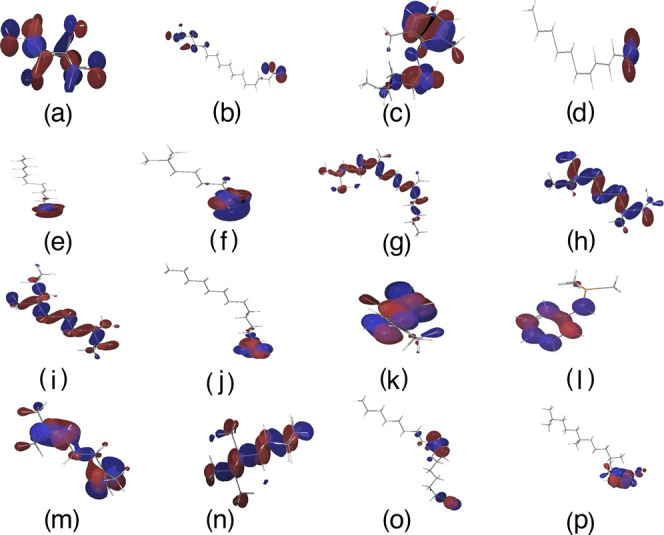
Schematic diagram of the HOMO–LUMO energy gap preferences of select phytochemicals identified from the ethanolic (70%) leaf extract of L. leucocephala. Frontier energies of HOMO and LUMO and intrinsic electronic proprieties of select phytochemicals are shown. (a) Cyclohexane-1,2,3,4,5,6-hexol; (b) hexadecanoic acid; (c) 2-(diethylamino)-N-(2,6-dimethylphenyl)acetamide; (d) undecan-1-ol; (e) tridecan-1-ol; (f) 6-methylheptan-1-ol; (g) 2,6,10-trimethylpentadecane; (h) 3,7-dimethylnonane; (i) 3,7-dimethyldecane; (j) tetradecanal; (k) spiro[4.4]nona-3,8-diene; (l) N-trimethylsilyl aniline; (m) 2,6-dimethylhepta-1,5-diene; (n) 2,3,6-trimethyloctane; (o) (Z)-octadec-9-enoic acid; and (p) (E)-3,7,11,15-tetramethylhexadec-2-en-1-ol. The positive electron density is shown in red color, whereas negative is shown in blue.
Molecular Dynamics
Based on our analyses of the ADMET properties and the HOMO–LUMO data, we further focused on hexadecanoic acid and (Z)-octadec-9-enoic acid. Although cyclohexane-1,2,3,4,5,6-hexol (better known as inositol) was the most abundant compound in the ethanolic (70%) leaf extract of L. leucocephala (Figure 3 and Table 1) and is known as a hypoglycemic agent,49,50 hexadecanoic acid and (Z)-octadec-9-enoic acid met the ADMET and HOMO–LUMO criteria best. Thus, we proceeded to analyze the stability of these α-amylase protein–fatty acid complexes by calculating the root-mean-square deviation (RMSD) of atomic positions and root-mean-square fluctuation (RMSF) values and determining the α-amylase protein contact map (Figures 7, 8, 9, and 10). The RMSD plots distinguished conformational changes caused by binding between the fatty acids and α-amylase. The hexadecanoic acid−α-amylase complex structure exhibited an energy difference (protein vs protein-bound ligand) of ∼3.6 Å (less than 4 Å for hydrogen bonding) at 40 ns and after 80 ns. The (Z)-9-octadecanoic acid−α-amylase complex showed stabilized energy differences at <4 Å after 70 ns. The RMSF plots show residue fluctuations during the molecular dynamics simulations (Figures 9 and 10).
Figure 7.
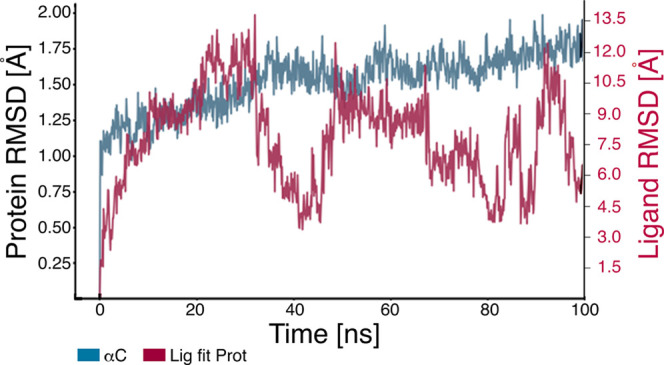
RMSD plot of α-amylase protein with hexadecanoic acid. The plot shows the RMSD evolution of α-amylase (left Y-axis). The ligand RMSD (right Y-axis) indicates the stability of the ligand hexadecanoic acid with respect to α-amylase and its binding pocket. In the plot, “Lig fit Prot” shows the RMSD of hexadecanoic acid when the α-amylase–hexadecanoic acid complex is aligned to the protein backbone of the reference, followed by measurement of the RMSD of hexadecanoic acid heavy atoms. Values significantly larger than the RMSD of α-amylase indicate that hexadecanoic acid diffused from its initial binding site. Good binding was observed at 40 ns and after 80 ns.
Figure 8.
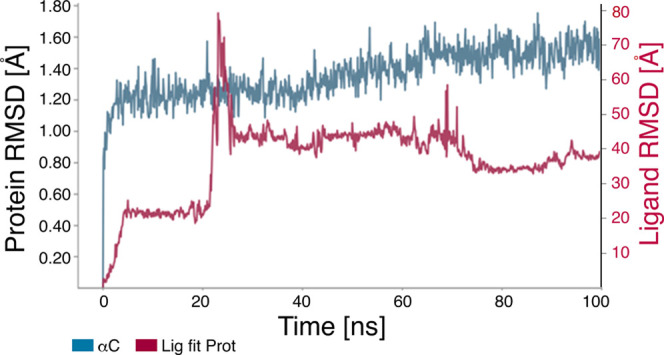
RMSD plot of α-amylase protein with (Z)-octadec-9-enoic acid. The plot shows the RMSD evolution of α-amylase (left Y-axis). The ligand RMSD (right Y-axis) indicates the stability of the ligand (Z)-octadec-9-enoic acid with respect to α-amylase and its binding pocket. In the plot, Lig fit Prot shows the RMSD of (Z)-octadec-9-enoic acid when the α-amylase–(Z)-octadec-9-enoic acid complex is aligned to the protein backbone of the reference, followed by measurement of the RMSD of the (Z)-octadec-9-enoic acid heavy atoms. Values significantly larger than the RMSD of α-amylase indicate that (Z)-octadec-9-enoic diffused from its initial binding site. Stable binding was observed after 70 ns.
Figure 9.
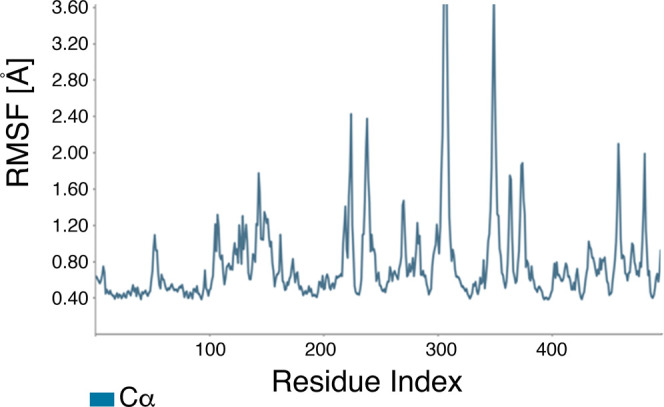
RMSF plot of the α-amylase protein with hexadecanoic acid. The X-axis shows the AA residue number of the α-amylase protein. On this plot, peaks indicate areas of α-amylase that fluctuated most during the simulation.
Figure 10.
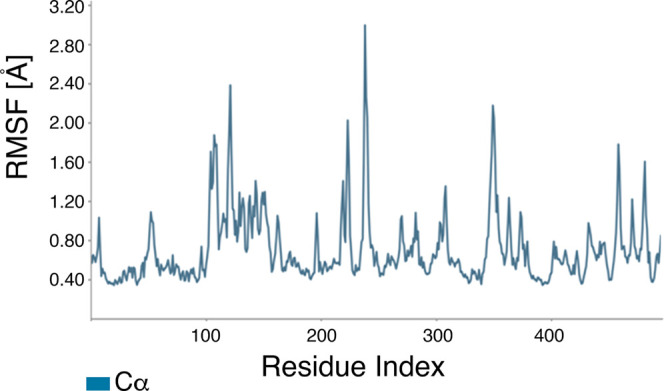
RMSF plot of the α-amylase protein with (Z)-octadec-9-enoic acid. The X-axis shows the AA residue numbers of α-amylase. On this plot, peaks indicate areas of α-amylase that fluctuated most during the simulation.
Figures 11 and 12 show the specific AA residues involved in fluctuations and interactions with these two phytochemicals.
Figure 11.
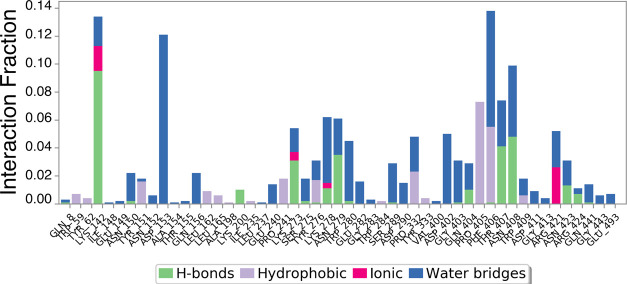
Bar diagram of α-amylase protein contacts with hexadecanoic acid showing the specific AAs involved in fluctuations and interactions. The X-axis indicates the AA residue number of the α-amylase protein. Protein interactions with the ligand were monitored throughout the simulation. Interactions were categorized by type and summarized in the plot above. α-Amylase protein–hexadecanoic acid interactions (or “contacts”) were categorized into four types: hydrogen bonds, hydrophobic interactions, ionic interactions, and water bridges. The stacked bar charts were normalized over the course of the trajectory: for example, a value of 0.7 suggests that the specific interaction was observed during 70% of the simulation time. Values over 1.0 are possible as some protein residue may make multiple contacts of the same subtype with the ligand.
Figure 12.
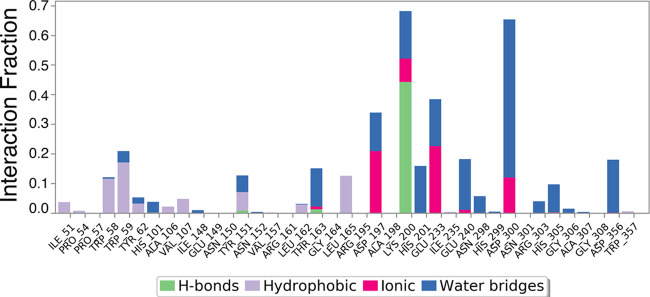
Bar diagram of α-amylase protein contacts with (Z)-octadec-9-enoic acid showing the specific AAs involved in fluctuations and interactions. The X-axis shows the AA residue number of the α-amylase protein. Protein interactions with the ligand were monitored throughout the simulation. These interactions were categorized by type and are summarized in the plot above. α-Amylase protein–(Z)-octadec-9-enoic acid interactions (or contacts) were categorized into four types: hydrogen bonds, hydrophobic interactions, ionic interactions, and water bridges. The stacked bar charts were normalized over the course of the trajectory: for example, a value of 0.7 indicates that the specific interaction was maintained for 70% of the simulation time.
An in silico α-amylase docking analysis with hexadecanoic acid and (Z)-octadec-9-enoic acid confirmed the specific AA residue binding sites and binding energy (Figure 13). Interacting AA residues bound within 4 Å radius of ligands were chosen for visualization and prediction by molecular rendering. Both fatty acids interact with the active site of α-amylase via AA residues TRP58, LEU162 and 165, Lys200, Glu233, and ILE235. Lys200 interacts with the ligands by hydrogen bonds for activity, and it is common for both fatty acids (Figure 13). AA residues TRP58, LEU162 and 165, Glu233, and ILE235 were identified as hydrophobic/π–π contacts in the protein binding site, and these AA residues are common binding partners for both fatty acids and are also key AA residues interacting with the standard α-amylase inhibitor acarbose (Figure 13).129 Glu233 is widely regarded as a key AA in the active site of α-amylase, where it acts as a general acid/base catalyst.16 Differences between Figures 11 and 12 on the one hand and Figure 13 on the other hand are based on the time-dependent dynamic nature of the analyses shown in Figures 7–12.
Figure 13.
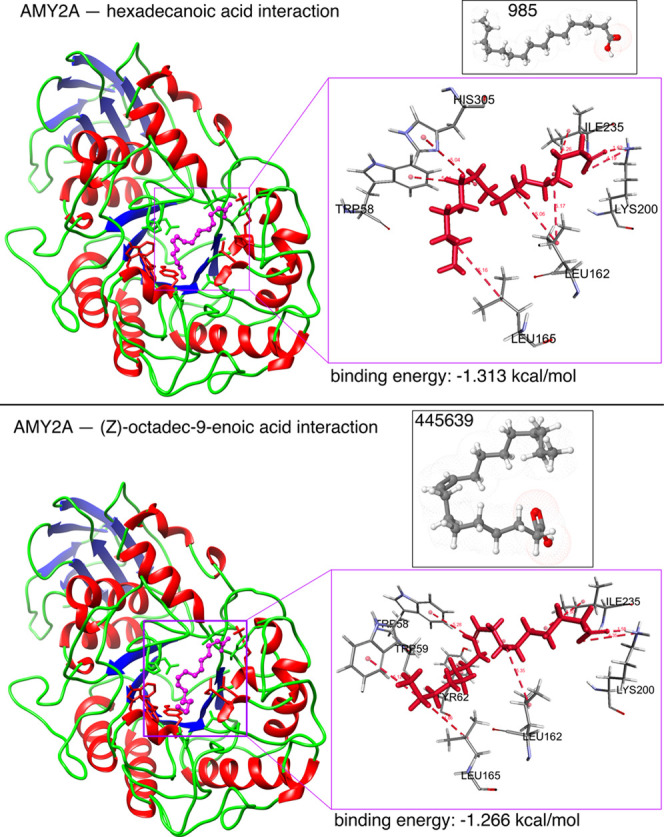
In silico α-amylase docking analyses with hexadecanoic acid and (Z)-octadec-9-enoic acid. The α-amylase protein structure is shown as a brown backbone ribbon, and the compounds are indicated with sticks. Interacting residues were determined using BIOVIA Discovery Studio Visualizer (Dassault Systems K.K., Tokyo, Japan);54 hydrogen bonds are represented by pale red dotted lines. Docking was performed using Glide 4.0 (XP) extra precision (Schrödinger) to depict the binding mode and calculate binding energies as described previously.55−57 Hexadecanoic acid interacted with surrounding residues through hydrophobic interactions with TRP59, TYR62, GLN63, TYR151, THR163, HIS201, GLU233, and GLY306; H-bonds with LYS200; and π–π interactions with TRP58, LEU162, and LEU165. (Z)-Octadec-9-enoic acid interacted with surrounding residues through hydrophobic interactions with TRP59, TYR62, GLN63, TYR151, THR163, HIS201, GLU233, ASP300, and GLY306; H-bonds with LYS200; and π–π interactions with TRP58, LEU162, and LEU165.
We performed phytochemical analyses of L. leucocephala leaf extracts using various solvents (methanol and ethanol for quantitative and qualitative analyses, respectively; 70% ethanol for GC–MS analysis). Our phytochemical studies revealed that the L. leucocephala leaf extract contains many phenolic constituents and is rich in flavonoids, coumarins, and saponins but not alkaloids.
L. leucocephala leaves have been reported to function as α-amylase inhibitors to reduce blood sugar level and aid in starch digestion.42−44 We found that an ethanol extract of L. leucocephala leaves had significant α-amylase inhibition activity, comparable to that of the standard anti-diabetes drug acarbose.58−61 Our GC–MS analysis of L. leucocephala leaf extract revealed the presence of 17 phytochemicals, including hexadecanoic acid and oleic acid ((Z)-octadec-9-enoic acid). In silico molecular docking and dynamics studies indicated that hexadecanoic acid and (Z)-octadec-9-enoic acid are potential α-amylase enzyme inhibitors.
Our data strongly indicate these two fatty acids as natural drugs for the treatment of T2DM. Accordingly, further in vitro and in vivo experimental studies62,63 are required to confirm and validate the possible α-amylase enzyme inhibitory activity of hexadecanoic acid and oleic acid. As nutraceuticals, hexadecanoic acid and oleic acid could serve for nonpharmacological strategies for treating T2DM.64−67 However, possible detrimental side effects (e.g., fatty liver, insulin resistance, atherosclerosis, or other cardiovascular diseases) of long-term treatment at higher doses must be considered and would require further animal and human studies.67−74 Moreover, although L. leucocephala leaves could be a reasonable alternative medicine in the form of a food supplement for relief of T2DM,75,76 isolated and purified fatty acids from different plant origins and with proven inhibitory activity of key enzymes related to T2DM77−82 would require optimized production processes at an industrial scale and would have to pass through the entire drug development process, including clinical trials.83−85
Conclusions
We examined the phytochemical composition and potential antidiabetic activity of L. leucocephala leaves. Two potential α-amylase inhibitors were identified through in vitro and in silico analyses. The fatty acids hexadecanoic acid and (Z)-octadec-9-enoic acid from L. leucocephala leaves had significant α-amylase enzyme inhibitory activity and are potential alternative natural product-based drugs for the treatment of T2DM.
Experimental Section
Materials
All chemicals used in this research were of analytical grade and were purchased from Sigma-Aldrich (St. Louis, MO).
Origin of L. leucocephala (Lam.) De Wit
L. leucocephala (Lam.) De Wit was collected from the Nagapattinam district in Tamil Nadu, India (10.7906° N, 79.8428° E), in December 2019 and authenticated (voucher specimen no. 375 deposited in the herbarium of the Department of Botany, Annamalai University, Chidambaram, Tamil Nadu) by Professor Mullainathan. Plant material was washed with normal and distilled water, dried in the dark, and ground to a fine powder, as described previously.62,63,86−88
Preparation and Qualitative Phytochemical Analysis of Leaf Extracts
Ground L. leucocephala leaf powder was soaked in 70% ethanol for 24 h with mild shaking at room temperature. After 24 h, the sample was filtered using Whatman grade 1 filter paper (Sigma-Aldrich) and concentrated by a rotary vacuum evaporator to 1 mg/mL.62,63,86−88 Concentrated extract was stored at 4 °C until further use.89,90 Preliminary qualitative phytochemical characterization of the L. leucocephala leaf extract was performed to identify and characterize phytochemical constituents such as anthraquinones, coumarins, polyphenol, terpenoids, saponins, tannins, steroids, alkaloids, flavonoids, glycosides, triterpenoids, and terpenoids following the standard protocols of Harborne.91
Quantitative Analysis of Phytochemicals
Leaf powder was quantitatively analyzed using standard procedures.91−99
Determination of Total Phenols by UV/VIS Spectrophotometry
Total phenol content was estimated using the method of Mbaebie et al.92L. leucocephala leaf powder (250 mg) was soaked in 10 mL of ether for 15 min to extract phenolic components. Approximately 2.5 mL of ether extract was transferred to a 50 mL conical flask, and 5 mL of sterile distilled water was added. Ammonium hydroxide solution (1 mL) and amyl alcohol (2.5 mL) were added. Sterile distilled water was added to a volume of 13 mL, followed by a 30 min incubation for color development. Optical density (OD) at a wavelength of 565 nm was measured using a UV/Vis spectrophotometer (Lambda 265, PerkinElmer Health Sciences Pvt. Ltd., Chennai, Tamil Nadu, India).
Determination of Flavonoid Content
L. leucocephala leaf powder (250 mg) was extracted with 10 mL of 80% aqueous methanol and incubated at room temperature for 30 min. The methanol extract was filtered through Whatman grade 1 filter paper (Sigma-Aldrich). The filtrate was transferred into a crucible and kept in a water bath to facilitate evaporation; the amount of sample left after evaporation was weighed. Total flavonoid content was determined using previously described standard procedures.91,93−95,100
Estimation of Total Terpenoid Content
L. leucocephala leaf powder (250 mg) was soaked in methanol (100%, 10 mL) for 24 h. One milliliter of filtrate (crude extract) was extracted with 3 mL of petroleum ether (1:3 ratio). The ether extract was evaporated under reduced pressure. The dried ether extract was collected to determine the total terpenoid content of the leaves using standard procedures.91,96,97
Estimation of Saponin Content
L. leucocephala leaf powder (250 mg) was mixed in 10 mL of 80% aqueous ethanol. The ethanol extract was heated at 55 °C for 1 h in a water bath. The filtrate was transferred to 10 mL of ethanol, and the volume of this mixture was reduced to 5 mL by boiling in a hot water bath. Diethyl ether was added, and the concentrated solution was shaken vigorously in a separating funnel; the aqueous layer was removed for purification by repeating the steps described above. Five milliliters of butanol was added to the filtrate, followed by evaporation of the mixture by immersion in hot water. Total saponin content in the leaf extracts was assessed by standard procedures.91,98,99,101
Qualitative Histochemical Tests
Qualitative phytochemical analyses were performed using the protocols of Adetuyi and Popoola,102 Trease and Evans,103 and Sofowora104 with slight modifications. Briefly, a small quantity of dried and finely powdered leaf sample was placed on a grease-free microscopic slide and treated with specific chemicals and reagents, followed by a 2 min incubation step. Light microscopy (pathology microscope BLS 111, BLISCO, Haryana, India) was used to observe and record color changes over time. Development of a black color after FeCl3 treatment indicated the presence of tannins, whereas the development of a yellow color after treatment with H2SO4 indicated the presence of saponins. Development of an orange color after dinitrophenol hydrazine treatment indicated the presence of terpenoids, and a green color after toluidine blue treatment indicated the presence of polyphenols. Finally, development of a yellow color after treatment with diluted ammonia and H2SO4 indicated the presence of flavonoids.91,105,106
In Vitro α-Amylase Inhibitory Activity
To determine the ability of the leaf extract to inhibit α-amylase activity, the 3,5-dinitrosalicylic acid (DNSA) method was used.107−109 Briefly, 100 mg of L. leucocephala leaf powder extract (80% ethanol) was dissolved in 100 mL of 80% ethanol, and serial dilutions (100, 200, 300, 400, and 500 μg/mL) were prepared. Five hundred microliters of α-amylase solution (HiMedia, Mumbai, Maharashtra, India) was mixed with 500 μL of leaf extract, and tubes were incubated for 10 min at 25 °C. Five hundred microliters of starch solution (0.5% in water (w/v); Nice Chemicals Pvt. Ltd., Kochi, Kerala, India) was added to each tube, followed by an additional 10 min incubation. The reaction was terminated by adding 1 mL of DNSA and boiling in a water bath at 85–90 °C for 5 min. The reaction mixture was cooled to ambient temperature, and absorbance was measured at 540 nm using a UV/Vis spectrophotometer (Lambda 265, PerkinElmer). Acarbose (an antidiabetic drug used to treat T2DM)58−61 was used as the positive control. The α-amylase enzyme inhibitory activity of the leaf extract is expressed as inhibition percentage and was calculated using the following equation
Percentage of α-amylase inhibition was plotted against extract concentration, and half maximal inhibitory concentration (IC50) was calculated.
Gas Chromatography–Mass Spectrometry Analysis
The GC–MS analysis was carried out using a Shimadzu QP2010PLUS (Shimadzu Analytical India Pvt. Ltd., Chennai) system comprising an AOC-20i autosampler and GC interfaced to an MS instrument with an Rtx-5Ms column (Restek, Bellefonte, PA; column diameter: 0.32 mm, column length: 30 m, column thickness: 0.50 mm) operated in electron impact mode at 70 eV. Helium gas (99.99%) was used as the carrier gas at a constant flow of 1.73 mL/min. An injection volume of 5 mL (split ratio of 10:1), injector temperature of 270 °C, and ion source temperature of 200 °C were used. The Turbo Mass Ver. 5.2.0 software supplied with the device was used. Interpretation of the GC–MS results was conducted using the database of the National Institute of Standards and Technology (NIST), which contains more than 62,000 MS patterns. The spectra of unknown components were identified based on comparison with spectra of known components stored in the NIST library, and the chemicals with their structures were unambiguously identified in the PubChem database.110−112
Computational In Silico Studies: Data Collection
The crystallographic 3-dimensional (3D) protein structure of the α-amylase protein (PDB ID: 3OLE) was retrieved from the Protein Data Bank113 at a resolution of 1.55 Å. Compound structures were downloaded from the PubChem database114 as described previously.55−57
Ligand Preparation
The LigPrep module of the Schrödinger software package (Schrödinger, LLC, New York, NY) was used to generate accurate and energy-minimized 3D molecular structures of the ligands. All compound structures were imported into the workspace, and the LigPrep module created 3D conformational structures and possible combinations of ligand compounds by adding potential missing atoms. The OPLS3E force field was used to minimize the energy of the ligands.115
Absorption, Distribution, Metabolism, Excretion, and Toxicity Properties of the Ligands
For all compounds of interest, ADMET properties and physicochemical parameters affecting ADMET, including solubility, lipophilicity, and permeability, as well as hydrogen bonding parameters,116−119 were determined using the Qikprop module of the Schrödinger software package (Schrödinger, LLC).120 Based on compound information, ligand properties were calculated. Molecular weight (MW), solvent-accessible surface area, partition coefficient (QP log Po/w), pharmacokinetic properties (blood–brain barrier penetration (QP log BB), gut–blood barrier penetration (QPP MDCK), and serum binding protein (QP log Khsa)), and pharmacological properties were determined.121
Highest Occupied Molecular Orbital–Lowest Unoccupied Molecular Orbital
The frontier molecular orbitals of the compounds were calculated using density functional theory.122,123 The HOMO and LUMO gap energy predictions were executed using the jaguar module of the Schrödinger software package (Schrödinger, LLC).124,125 Various parameters, such as dipole movement, chemical hardness, and chemical softness, were determined. The HOMO–LUMO gap energies were determined using the equation KE gap = EHOMO – ELUMO, and the calculated HOMO–LUMO gap energy value was reported as described previously.121
Molecular Docking and Dynamics: Protein Preparation
In this study, the α-amylase 3D protein crystal structure was prepared using the protein preparation wizard panel of the Schrödinger software package (Schrödinger, LLC). The 3D protein crystal structure of α-amylase was imported into the workspace and preprocessed, and the missing amino acid residues were filled.126 Water molecules were removed from the ligand-binding domain. H-bonds were optimized using the hydrogen bond optimizer, and the α-amylase protein structure was subjected to a minimization process to obtain the lowest-energy conformational structure.121
Receptor Grid Generation
Grids were generated using the receptor grid generation module of the Schrödinger software package (Schrödinger, LLC). This module generates a grid box in cocrystallized acarviostatins and compares it with the ligand acarbose of human α-amylase to define the center of the grid box for optimal inhibitory ligand position.127−129
Molecular Docking
Default parameters were used for molecular docking, which was performed using the Glide 4.0 XP extra precision module of Schrödinger software package (Schrödinger, LLC). Interacting AA residues bound within 4 Å radius of ligands were chosen for visualization and prediction by molecular rendering. The binding affinity between each ligand and α-amylase was calculated, and phytochemical ligands were ranked by scoring function.16,55−57,126,129
Molecular Dynamics Simulations of Protein and Ligand Complexes
Based on the docking scores, ADMET properties, and HOMO–LUMO gaps, 17 phytochemical compounds were subjected to molecular dynamics simulations using the Desmond module of the Schrödinger software package (Schrödinger, LLC).130 α-Amylase protein–phytochemical compound ligand complexes were adjusted in the cubic box, and the solvation model used was a simple point charge model. System equilibration was set to a temperature of 300 K and a pressure of 1 bar. A graphic processing unit was used for production and trajectory preparation. Each trajectory was generated and recorded for up to 100 ns. Calculations of the RMSD, RMSF,131 radius of gyration, and torsional studies were performed using the trajectory files of the protein–ligand complexes.121,132,133
Statistical Analysis
Data obtained in this study were analyzed by Student’s t test using SPSS software (IBM SPSS Statistics; Armonk, NY). Experiments were performed in triplicate, and data are presented as mean ± SD. The significance of differences between groups was determined using unpaired Student’s t test, and the significance of differences within groups was assessed using paired Student’s t test.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c02350.
Additional qualitative and quantitative experimental results are presented in supplementary Tables S1-S7. Qualitative histochemical analysis of L. leucocephala leaf powder (Table S1); qualitative phytochemical analysis of L. leucocephala leaf extracts (Table S2); quantitative analysis of phytochemicals in L. leucocephala leaf extracts (70% ethanol) (Table S3); in vitro anti α-amylase enzyme activity of L. leucocephala leaf extract (70% ethanol) compared with that of acarbose (Table S4); ADMET properties of phytochemicals identified by GC–MS from ethanolic (70%) leaf extracts of L. leucocephala (Table S5); HOMO–LUMO energy gap calculation of all of the phytochemicals identified by GC–MS from the ethanolic (70%) leaf extracts of L. leucocephala (Table S6); predicted docking score of α-amylase with select phytochemicals identified by GC–MS from the ethanolic (70%) leaf extract of L. leucocephala (Table S7) (PDF)
Author Contributions
Conceptualization: S.R. and K.H.; methodology: S.R., S.M., P.V., and U.S.; validation: S.R., K.H., P.-S.K., and A.K.; formal analysis: S.R., S.M., P.V., U.S., and K.H.; investigation: S.R., S.M., P.V., U.S., and K.H.; resources: S.R., U.S., and K.H.; data curation: S.R. and K.H.; writing—original draft preparation: S.R. and K.H.; writing—review and editing; A.K., P.-S.K., and K.H.; visualization: S.R., P.-S.K., and A.K.; supervision: S.R. and K.H.; project administration: S.R. and K.H.; and funding acquisition: S.R., A.K., P.-S.K., and K.H. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Khan R. M. M.; Chua Z. J. Y.; Tan J. C.; Yang Y.; Liao Z.; Zhao Y. From pre-diabetes to diabetes: diagnosis, treatments and translational research. Medicina 2019, 55, 546. 10.3390/medicina55090546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molitch M. E.Diabetes Mellitus. In Clinical Methods: The History, Physical, and Laboratory Examinations; Walker H. K.; Hall W. D.; Hurst J. W., Eds.; Butterworth Publishers: Boston, 1990. [PubMed] [Google Scholar]
- Sapra A.; Bhandari P.. Diabetes Mellitus. In StatPearls; StatPearls Publishing: Treasure Island (FL), 2020. [Google Scholar]
- Wikipedia Diabetes. https://en.wikipedia.org/wiki/Diabetes (accessed January 14, 2021).
- Goyal R.; Jialal I.. Diabetes Mellitus Type 2. In StatPearls; StatPearls Publishing: Treasure Island (FL), 2020. [Google Scholar]
- Zhang B.; Zhao W.; Tu J.; Wang X.; Hao Y.; Wang H.; Zhao Y.; Mizuno K.; Tseng Y.; Bu H. The relationship between serum 25-hydroxyvitamin D concentration and type 2 diabetic peripheral neuropathy: a systematic review and a meta-analysis. Medicine 2019, 98, e18118 10.1097/MD.0000000000018118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.; Balan P.; Popovich D. G. Review of ginseng anti-diabetic studies. Molecules 2019, 24, 4501. 10.3390/molecules24244501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. A.; Choi D. W.; Kwon J.; Lee D. W.; Park E. C. Association between continuity of care and type 2 diabetes development among patients with thyroid disorder. Medicine 2019, 98, e18537 10.1097/MD.0000000000018537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez M. L.; Perez S.; Mena-Molla S.; Desco M. C.; Ortega A. L. Oxidative stress and microvascular alterations in diabetic retinopathy: future therapies. Oxid. Med. Cell. Longevity 2019, 2019, 4940825 10.1155/2019/4940825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z.; Zhu X.; Kaminga A. C.; Xu H. Associated risk factors and their interactions with type 2 diabetes among the elderly with prediabetes in rural areas of Yiyang City: A nested case-control study. Medicine 2019, 98, e17736 10.1097/MD.0000000000017736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari U.; Heese K. Cardiovascular dementia - a different perspective. Open. Biochem. J. 2010, 4, 29–52. 10.2174/1874091X01004010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese K.; Beck K. F.; Behrens M. H.; Pluss K.; Fierlbeck W.; Huwiler A.; Muhl H.; Geiger H.; Otten U.; Pfeilschifter J. Effects of high glucose on cytokine-induced nerve growth factor (NGF) expression in rat renal mesangial cells. Biochem. Pharmacol. 2003, 65, 293–301. 10.1016/S0006-2952(02)01487-9. [DOI] [PubMed] [Google Scholar]
- Banday M. Z.; Sameer A. S.; Nissar S. Pathophysiology of diabetes: an overview. Avicenna J. Med. 2020, 10, 174–188. 10.4103/ajm.ajm_53_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sintsova O.; Gladkikh I.; Kalinovskii A.; Zelepuga E.; Monastyrnaya M.; Kim N.; Shevchenko L.; Peigneur S.; Tytgat J.; Kozlovskaya E.; Leychenko E. Magnificamide, a beta-defensin-like peptide from the mucus of the sea anemone heteractis magnifica, is a strong inhibitor of mammalian alpha-amylases. Mar. Drugs 2019, 17, 542. 10.3390/md17100542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proença C.; Ribeiro D.; Freitas M.; Fernandes E. Flavonoids as potential agents in the management of type 2 diabetes through the modulation of α-amylase and α-glucosidase activity: a review. Crit. Rev. Food Sci. Nutr. 2021, 1–71. 10.1080/10408398.2020.1862755. [DOI] [PubMed] [Google Scholar]
- Teng H.; Chen L. α-Glucosidase and α-amylase inhibitors from seed oil: a review of liposoluble substance to treat diabetes. Crit. Rev. Food Sci. Nutr. 2017, 57, 3438–3448. 10.1080/10408398.2015.1129309. [DOI] [PubMed] [Google Scholar]
- Singhal S.; Kumar S. Current perspectives on management of type 2 diabetes in youth. Children 2021, 8, 37. 10.3390/children8010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhlaghi F.; Matson K. L.; Mohammadpour A. H.; Kelly M.; Karimani A. Clinical pharmacokinetics and pharmacodynamics of antihyperglycemic medications in children and adolescents with type 2 diabetes mellitus. Clin. Pharmacokinet. 2017, 56, 561–571. 10.1007/s40262-016-0472-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S.; Mittal A.; Babu D.; Mittal A. Herbal medicines for diabetes management and its secondary complications. Curr. Diabetes Rev. 2021, 17, 437–456. 10.2174/1573399816666201103143225. [DOI] [PubMed] [Google Scholar]
- Jirapure K.; Undale V. Antidiabetics with herbs: a compressive review on interactions. Curr. Diabetes Rev. 2021, e011221190237 10.2174/1573399817999210112191718. [DOI] [PubMed] [Google Scholar]
- Atanasov A. G.; Zotchev S. B.; Dirsch V. M.; Supuran C. T. Natural products in drug discovery: advances and opportunities. Nat. Rev. Drug Discovery 2021, 20, 200–216. 10.1038/s41573-020-00114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahy S. K.; Kumar A.; Bhatt R. Hedychium coronarium rhizomes: promising antidiabetic and natural inhibitor of alpha-amylase and alpha-glucosidase. J. Diet. Suppl. 2020, 17, 81–87. 10.1080/19390211.2018.1483462. [DOI] [PubMed] [Google Scholar]
- Etsassala N.; Badmus J. A.; Waryo T. T.; Marnewick J. L.; Cupido C. N.; Hussein A. A.; Iwuoha E. I. Alpha-glucosidase and alpha-amylase inhibitory activities of novel abietane diterpenes from salvia africana-lutea. Antioxidants 2019, 8, 421. 10.3390/antiox8100421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikipedia Alpha-Amylase. https://en.wikipedia.org/wiki/Alpha-amylase (accessed January 14, 2021).
- Lachowicz S.; Wisniewski R.; Ochmian I.; Drzymala K.; Pluta S. Anti-microbiological, anti-hyperglycemic and anti-obesity potency of natural antioxidants in fruit fractions of Saskatoon berry. Antioxidants 2019, 8, 397. 10.3390/antiox8090397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeed M.; Majeed S.; Mundkur L.; Nagabhushanam K.; Arumugam S.; Beede K.; Ali F. Standardized Emblica officinalis fruit extract inhibited the activities of alpha-amylase, alpha-glucosidase, and dipeptidyl peptidase-4 and displayed antioxidant potential. J. Sci. Food Agric. 2020, 100, 509–516. 10.1002/jsfa.10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong L.; Feng D.; Wang T.; Ren Y.; Liu Y.; Wang J. Inhibitors of α-amylase and α-glucosidase: potential linkage for whole cereal foods on prevention of hyperglycemia. Food Sci. Nutr. 2020, 8, 6320–6337. 10.1002/fsn3.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papoutsis K.; Zhang J.; Bowyer M. C.; Brunton N.; Gibney E. R.; Lyng J. Fruit, vegetables, and mushrooms for the preparation of extracts with α-amylase and α-glucosidase inhibition properties: a review. Food Chem. 2021, 338, 128119 10.1016/j.foodchem.2020.128119. [DOI] [PubMed] [Google Scholar]
- Fei Y.; Tsoi M. F.; Cheung B. M. Y. Cardiovascular outcomes in trials of new antidiabetic drug classes: a network meta-analysis. Cardiovasc. Diabetol. 2019, 18, 112. 10.1186/s12933-019-0916-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikipedia Leucaena leucocephala. https://en.wikipedia.org/wiki/Leucaena_leucocephala (accessed January 13, 2021).
- Tropical Forages. Leucaena leucocephala. https://web.archive.org/web/20150304155421/http://www.tropicalforages.info/key/Forages/Media/Html/Leucaena_leucocephala.htm (accessed January 13, 2021).
- Hughes C. E.Leucaena, A Genetic Resources Handbook.; Oxford University Press: U.K., 1998. [Google Scholar]
- Jones R. J.; Brewbaker J. L.; Sorensson C. T.. Leucaena leucocephala (Lamk) de Wit. In Plant Resources of South-East Asia; Mannetje L.; Jones R. M., Eds.; Pudoc Scientific Publishers: Wageningen, the Netherlands, Vol. 199, pp 150–154. [Google Scholar]
- Parrotta J. A.SO-ITF-SM-52: Leucaena leucocephala (Lam.) de Wit Leucaena, tantan Leguminosae (Mimosoideae) Legume family; United States Department of Agriculture: USA, 1992; pp 1–8.
- Clément C.; Witschi U.; Kreuzer M. The potential influence of plant-based feed supplements on sperm quantity and quality in livestock: a review. Anim. Reprod. Sci. 2012, 132, 1–10. 10.1016/j.anireprosci.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Shelton H. M.; Gutteridge R. C.; Mullen B. F.; Bray R. A.. Leucaena – Adaptation, Quality and Farming Systems; ACIAR: Canberra, Australia, 1989. [Google Scholar]
- Alonso-Díaz M. A.; Torres-Acosta J. F.; Sandoval-Castro C. A.; Aguilar-Caballero A. J.; Hoste H. In vitro larval migration and kinetics of exsheathment of Haemonchus contortus larvae exposed to four tropical tanniniferous plant extracts. Vet. Parasitol. 2008, 153, 313–319. 10.1016/j.vetpar.2008.01.042. [DOI] [PubMed] [Google Scholar]
- Alonso-Díaz M. A.; Torres-Acosta J. F. J.; Sandoval-Castro C. A.; Hoste H. Tannins in tropical tree fodders fed to small ruminants: a friendly foe?. Small Ruminant Res. 2010, 89, 164–173. 10.1016/j.smallrumres.2009.12.040. [DOI] [Google Scholar]
- von Son-de Fernex E.; Alonso-Diaz M. A.; Mendoza-de Gives P.; Valles-de la Mora B.; Gonzalez-Cortazar M.; Zamilpa A.; Castillo Gallegos E. Elucidation of Leucaena leucocephala anthelmintic-like phytochemicals and the ultrastructural damage generated to eggs of Cooperia spp. Vet. Parasitol. 2015, 214, 89–95. 10.1016/j.vetpar.2015.10.005. [DOI] [PubMed] [Google Scholar]
- Ademola I. O.; Akanbi A. I.; Idowu S. O. Comparative nematocidal activity of chromatographic fractions of Leucaena leucocephala. Seed against gastrointestinal sheep nematodes. Pharm. Biol. 2005, 43, 599–604. 10.1080/13880200500301761. [DOI] [Google Scholar]
- Hammond A. C. Leucaena toxicosis and its control in ruminants. J. Anim. Sci. 1995, 73, 1487–1492. 10.2527/1995.7351487x. [DOI] [PubMed] [Google Scholar]
- Chowtivannakul P.; Srichaikul B.; Talubmook C. Antidiabetic and antioxidant activities of seed extract from Leucaena leucocephala (Lam.) de Wit. Agric. Nat. Resour. 2016, 50, 357–361. 10.1016/j.anres.2016.06.007. [DOI] [Google Scholar]
- Nurmaylindha V.; Widodo G. P.; Herowati R. Molecular docking analysis of Leucaena leucocephala and Trigonella foenum-graecum chemical constituents on antidiabetic macromolecular targets and prediction of the pharmacokinetic profiles. AIP Conf. Proc. 2020, 2243, 020015 10.1063/5.0006279. [DOI] [Google Scholar]
- Wan-Nadilah W. A.; Khozirah S.; Khatib A.; Hamid A. A.; Hamid M. Evaluation of the α-glucosidase inhibitory and free radical scavenging activities of selected traditional medicine plant species used in treating diabetes. Int. Food Res. J. 2019, 26, 75–85. [Google Scholar]
- Elya B.; Handayani R.; Sauriasari R.; Azizahwati; Hasyyati U. S.; Permana I. T.; Permatasari Y. I. Antidiabetic activity and phytochemical screening of extracts from Indonesian plants by inhibition of alpha amylase, alpha glucosidase and dipeptidyl peptidase IV. Pak. J. Biol. Sci. 2015, 18, 279–284. 10.3923/pjbs.2015.279.284. [DOI] [Google Scholar]
- Chen C. Y.; Wang Y. D. Secondary metabolites from Leucaena leucocephala. Chem. Nat. Compd. 2011, 47, 145–146. 10.1007/s10600-011-9862-z. [DOI] [Google Scholar]
- Herrera R.; Verdecia D. M.; Ramírez J. L.; García M.; Cruz A. M. Secondary metabolites of Leucaena leucochephala. Their relationship with some climate elements, different expressions of digestibility and primary metabolites. Cuba J. Agric. Sci. 2017, 51, 107–116. [Google Scholar]
- Zayed M. Z.; Samling B. Phytochemical constituents of the leaves of Leucaena leucocephala from malaysia. Int. J. Pharm. Pharm. Sci. 2016, 8, 174–179. 10.22159/ijpps.2016v8i12.11582. [DOI] [Google Scholar]
- Werner E. F.; Froehlich R. J. The potential role for myoinositol in the prevention of gestational diabetes mellitus. Am. J. Perinatol. 2016, 33, 1236–1241. 10.1055/s-0036-1584273. [DOI] [PubMed] [Google Scholar]
- Andersen D. B.; Holub B. J. The relative response of hepatic lipids in the rat to graded levels of dietary myo-inositol and other lipotropes. J. Nutr. 1980, 110, 496–504. 10.1093/jn/110.3.496. [DOI] [PubMed] [Google Scholar]
- Ji D.; Xu M.; Udenigwe C. C.; Agyei D. Physicochemical characterisation, molecular docking, and drug-likeness evaluation of hypotensive peptides encrypted in flaxseed proteome. Curr. Res. Food Sci. 2020, 3, 41–50. 10.1016/j.crfs.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benet L. Z.; Hosey C. M.; Ursu O.; Oprea T. I. BDDCS, the Rule of 5 and drugability. Adv. Drug Delivery Rev. 2016, 101, 89–98. 10.1016/j.addr.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski C. A.; Lombardo F.; Dominy B. W.; Feeney P. J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Delivery Rev. 2001, 46, 3–26. 10.1016/S0169-409X(00)00129-0. [DOI] [PubMed] [Google Scholar]
- Puranik N. V.; Srivastava P.; Bhatt G.; John Mary D. J. S.; Limaye A. M.; Sivaraman J. Determination and analysis of agonist and antagonist potential of naturally occurring flavonoids for estrogen receptor (ERα) by various parameters and molecular modelling approach. Sci. Rep. 2019, 9, 7450 10.1038/s41598-019-43768-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramanik S.; Kutzner A.; Heese K. Lead discovery and in silico 3D structure modeling of tumorigenic FAM72A (p17). Tumor Biol. 2015, 36, 239–249. 10.1007/s13277-014-2620-7. [DOI] [PubMed] [Google Scholar]
- Pramanik S.; Kutzner A.; Heese K. 3D structure, dimerization modeling, and lead discovery by ligand-protein interaction analysis of p60 transcription regulatorprotein (p60TRP). Mol. Inf. 2016, 35, 99–108. 10.1002/minf.201500035. [DOI] [PubMed] [Google Scholar]
- Pramanik S.; Thaker M.; Perumal A. G.; Ekambaram R.; Poondla N.; Schmidt M.; Kim P. S.; Kutzner A.; Heese K. Proteomic atomics reveals a distinctive uracil-5-methyltransferase. Mol. Inf. 2020, 39, 1900135 10.1002/minf.201900135. [DOI] [PubMed] [Google Scholar]
- Zhang W.; Kim D.; Philip E.; Miyan Z.; Barykina I.; Schmidt B.; Stein H. A multinational, observational study to investigate the efficacy, safety and tolerability of acarbose as add-on or monotherapy in a range of patients: the Gluco VIP study. Clin. Drug Invest. 2013, 33, 263–274. 10.1007/s40261-013-0063-3. [DOI] [PubMed] [Google Scholar]
- Zhu Q.; Tong Y.; Wu T.; Li J.; Tong N. Comparison of the hypoglycemic effect of acarbose monotherapy in patients with type 2 diabetes mellitus consuming an Eastern or Western diet: a systematic meta-analysis. Clin. Ther. 2013, 35, 880–899. 10.1016/j.clinthera.2013.03.020. [DOI] [PubMed] [Google Scholar]
- Hedrington M. S.; Davis S. N. Considerations when using alpha-glucosidase inhibitors in the treatment of type 2 diabetes. Expert Opin. Pharmacother. 2019, 20, 2229–2235. 10.1080/14656566.2019.1672660. [DOI] [PubMed] [Google Scholar]
- Wikipedia Acarbose. https://en.wikipedia.org/wiki/Acarbose#::text=Acarbose%20(INN)%20is%20an%20anti,as%20Prandase%20(Bayer%20AG (accessed January 18, 2021).
- Mishra M.; Huang J.; Lee Y. Y.; Chua D. S.; Lin X.; Hu J. M.; Heese K. Gastrodia elata modulates amyloid precursor protein cleavage and cognitive functions in mice. Biosci. Trends 2011, 5, 129–138. 10.5582/bst.2011.v5.3.129. [DOI] [PubMed] [Google Scholar]
- Feng L.; Manavalan A.; Mishra M.; Sze S. K.; Hu J. M.; Heese K. Tianma modulates blood vessel tonicity. Open Biochem. J. 2012, 6, 56–65. 10.2174/1874091X01206010056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza C. O.; Vannice G. K.; Rosa Neto J. C.; Calder P. C. Is palmitoleic acid a plausible nonpharmacological strategy to prevent or control chronic metabolic and inflammatory disorders?. Mol. Nutr. Food Res. 2018, 62, 1700504 10.1002/mnfr.201700504. [DOI] [PubMed] [Google Scholar]
- Mozaffarian D.; Cao H.; King I. B.; Lemaitre R. N.; Song X.; Siscovick D. S.; Hotamisligil G. S. Trans-palmitoleic acid, metabolic risk factors, and new-onset diabetes in U.S. adults: a cohort study. Ann. Intern. Med. 2010, 153, 790–799. 10.7326/0003-4819-153-12-201012210-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman K.; Haider K.; Jabeen K.; Akash M. S. H. Current perspectives of oleic acid: Regulation of molecular pathways in mitochondrial and endothelial functioning against insulin resistance and diabetes. Rev. Endocr. Metab. Disord. 2020, 21, 631–643. 10.1007/s11154-020-09549-6. [DOI] [PubMed] [Google Scholar]
- López-Gómez C.; Santiago-Fernández C.; García-Serrano S.; García-Escobar E.; Gutiérrez-Repiso C.; Rodríguez-Díaz C.; Ho-Plágaro A.; Martín-Reyes F.; Garrido-Sánchez L.; Valdés S.; Rodríguez-Cañete A.; Rodríguez-Pacheco F.; García-Fuentes E. Oleic acid protects against insulin resistance by regulating the genes related to the PI3K signaling pathway. J. Clin. Med. 2020, 9, 2615. 10.3390/jcm9082615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua M. C.; Su H. M.; Lai M. W.; Yao T. C.; Tsai M. H.; Liao S. L.; Lai S. H.; Huang J. L. Palmitoleic and dihomo-γ-linolenic acids are positively associated with abdominal obesity and increased metabolic risk in children. Front. Pediatr. 2021, 9, 628496 10.3389/fped.2021.628496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes E. A.; Rafacho A. Implications of palmitoleic acid (palmitoleate) on glucose homeostasis, insulin resistance and diabetes. Curr. Drug. Targets 2017, 18, 619–628. 10.2174/1389450117666151209120345. [DOI] [PubMed] [Google Scholar]
- Tardy A. L.; Morio B.; Chardigny J. M.; Malpuech-Brugère C. Ruminant and industrial sources of trans-fat and cardiovascular and diabetic diseases. Nutr. Res. Rev. 2011, 24, 111–117. 10.1017/S0954422411000011. [DOI] [PubMed] [Google Scholar]
- Sobczak A. I. S.; Blindauer C. A.; Stewart A. J. Changes in plasma free fatty acids associated with type-2 diabetes. Nutrients 2019, 11, 2022. 10.3390/nu11092022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauquier F.; Léotoing L.; Philippe C.; Spilmont M.; Coxam V.; Wittrant Y. Pros and cons of fatty acids in bone biology. Prog. Lipid Res. 2015, 58, 121–145. 10.1016/j.plipres.2015.03.001. [DOI] [PubMed] [Google Scholar]
- Palomer X.; Pizarro-Delgado J.; Barroso E.; Vázquez-Carrera M. Palmitic and oleic acid: the yin and yang of fatty acids in type 2 diabetes mellitus. Trends Endocrinol. Metab. 2018, 29, 178–190. 10.1016/j.tem.2017.11.009. [DOI] [PubMed] [Google Scholar]
- Mary Ann Liebert, Inc. 3: Final report on the safety assessment of oleic acid, lauric acid, palmitic acid, myristic acid, and stearic acid. J. Am. Coll. Toxicol. 1987, 6, 321–401.. 10.3109/10915818709098563. [DOI] [Google Scholar]
- McKennon S. A.Non-Pharmaceutical Intervention Options for Type 2 Diabetes: Diets and Dietary Supplements (Botanicals, Antioxidants, and Minerals). In Endotext; Feingold K. R.; Anawalt B.; Boyce A.; Chrousos G.; de Herder W. W.; Dhatariya K.; Dungan K.; Grossman A.; Hershman J. M.; Hofland J.; Kalra S.; Kaltsas G.; Koch C.; Kopp P.; Korbonits M.; Kovacs C. S.; Kuohung W.; Laferrere B.; McGee E. A.; McLachlan R.; Morley J. E.; New M.; Purnell J.; Sahay R.; Singer F.; Stratakis C. A.; Trence D. L.; Wilson D. P., Eds.; South Dartmouth: MA, 2000. [Google Scholar]
- Forouhar E.; Sack P. Non-traditional therapies for diabetes: fact or fiction. J. Community Hosp. Intern. Med. Perspect. 2012, 2, 18447 10.3402/jchimp.v2i2.18447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C. H.; Hsu C. H.; Ng L. T. Inhibitory potential of fatty acids on key enzymes related to type 2 diabetes. Biofactors 2013, 39, 415–421. 10.1002/biof.1082. [DOI] [PubMed] [Google Scholar]
- Mahmood N. A review of α-amylase inhibitors on weight loss and glycemic control in pathological state such as obesity and diabetes. Comp. Clin. Pathol. 2016, 25, 1253–1264. 10.1007/s00580-014-1967-x. [DOI] [Google Scholar]
- Anh L. H.; Xuan T. D.; Dieu Thuy N. T.; Quan N. V.; Trang L. T. Antioxidant and a-amylase inhibitory activities and phytocompounds of Clausena indica fruits. Medicines 2020, 7, 10. 10.3390/medicines7030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T. H.; Kim S. M. Alpha-glucosidase inhibitory activities of fatty acids purified from the internal organ of sea cucumber Stichopus japonicas. J. Food Sci. 2015, 80, H841–847. 10.1111/1750-3841.12810. [DOI] [PubMed] [Google Scholar]
- Liu Q. Z.; Zhang H.; Dai H. Q.; Zhao P.; Mao Y. F.; Chen K. X.; Chen Z. X. Inhibition of starch digestion: the role of hydrophobic domain of both α-amylase and substrates. Food Chem. 2021, 341, 128211 10.1016/j.foodchem.2020.128211. [DOI] [PubMed] [Google Scholar]
- Nehdi I. A.; Sbihi H.; Tan C. P.; Al-Resayes S. I. Leucaena leucocephala (Lam.) de Wit seed oil: characterization and uses. Ind. Crops Prod. 2014, 52, 582–587. 10.1016/j.indcrop.2013.11.021. [DOI] [Google Scholar]
- Vasquez-Rios G.; Nadkarni G. N. SGLT2 inhibitors: emerging roles in the protection against cardiovascular and kidney disease among diabetic patients. Int. J. Nephrol. Renovasc. Dis. 2020, 13, 281–296. 10.2147/IJNRD.S268811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar V. S.; Dubrofsky L.; Boulet J.; Cherney D. Z. Making a case for the combined use of SGLT2 inhibitors and GLP1 receptor agonists for cardiorenal protection. J. Bras. Nefrol. 2020, 42, 467–477. 10.1590/2175-8239-jbn-2020-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Cao D.; Yan M.; Liu M. The feasibility of Chinese massage as an auxiliary way of replacing or reducing drugs in the clinical treatment of adult type 2 diabetes: a systematic review and meta-analysis. Medicine 2020, 99, e21894 10.1097/MD.0000000000021894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.; Tao Z.; Jin Y.; Yuan Y.; Dong T. T. X.; Tsim K. W. K.; Zhou Z. Flavonoids, a potential new insight of Leucaena leucocephala foliage in ruminant health. J. Agric. Food Chem. 2018, 66, 7616–7626. 10.1021/acs.jafc.8b02739. [DOI] [PubMed] [Google Scholar]
- Kotchabhakdi N.; Seanjum C.; Kiwfo K.; Grudpan K. A simple extract of Leucaena leucocephala (Lam.) de Wit leaf containing mimosine as a natural color reagent for iron determination. Microchem. J. 2021, 162, 105860 10.1016/j.microc.2020.105860. [DOI] [Google Scholar]
- Manavalan A.; Ramachandran U.; Sundaramurthi H.; Mishra M.; Sze S. K.; Hu J. M.; Feng Z. W.; Heese K. Gastrodia elata Blume (tianma) mobilizes neuro-protective capacities. Int. J. Biochem. Mol. Biol. 2012, 3, 219–241. [PMC free article] [PubMed] [Google Scholar]
- Sukumaran S.; Kiruba S.; Mahesh M.; Nisha S. R.; Miller P. Z.; Ben C. P.; Jeeva S. Phytochemical constituents and antibacterial efficacy of the flowers of Peltophorum pterocarpum (DC.) Baker ex Heyne. Asian Pac. J. Trop. Med. 2011, 4, 735–738. 10.1016/S1995-7645(11)60183-1. [DOI] [PubMed] [Google Scholar]
- Ali M. S.; Amin M. R.; Kamal C. M.; Hossain M. A. In vitro antioxidant, cytotoxic, thrombolytic activities and phytochemical evaluation of methanol extract of the A. philippense L. leaves. Asian Pac. J. Trop. Biomed. 2013, 3, 464–469. 10.1016/S2221-1691(13)60097-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborne A. J.Phytochemical Methods: A Guide to Modern Techniques of Plant Analysis, 3rd ed.; Harborne A. J., Ed.; Springer: Netherlands, 1998. [Google Scholar]
- Mbaebie B. O.; Edeoga H. O.; Afolayan A. J. Phytochemical analysis and antioxidants activities of aqueous stem bark extract of Schotia latifolia Jacq. Asian Pac. J. Trop. Biomed. 2012, 2, 118–124. 10.1016/S2221-1691(11)60204-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.-Y.; Tang C.-Y. Determination of total phenolic and flavonoid contents in selected fruits and vegetables, as well as their stimulatory effects on mouse splenocyte proliferation. Food Chem. 2007, 101, 140–147. 10.1016/j.foodchem.2006.01.014. [DOI] [Google Scholar]
- da Silva L. A. L.; Pezzini B. R.; Soares L. Spectrophotometric determination of the total flavonoid content in Ocimum basilicum L. (Lamiaceae) leaves. Pharmacogn. Mag. 2015, 11, 96–101. 10.4103/0973-1296.149721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba S. A.; Malik S. A. Determination of total phenolic and flavonoid content, antimicrobial and antioxidant activity of a root extract of Arisaema jacquemontii Blume. J. Taibah Univ. Sci. 2015, 9, 449–454. 10.1016/j.jtusci.2014.11.001. [DOI] [Google Scholar]
- Harman-Ware A. E.; Sykes R.; Peter G. F.; Davis M. Determination of terpenoid content in pine by organic solvent extraction and fast-GC analysis. Front. Energy Res. 2016, 4, 2 10.3389/fenrg.2016.00002. [DOI] [Google Scholar]
- Jiang Z.; Kempinski C.; Chappell J. Extraction and analysis of terpenes/terpenoids. Curr. Protoc. Plant Biol. 2016, 1, 345–358. 10.1002/cppb.20024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kołodziej B.; Sęczyk Ł.; Sugier D.; Kędzia B.; Chernetskyy M.; Gevrenova R.; Henry M. Determination of the yield, saponin content and profile, antimicrobial and antioxidant activities of three Gypsophila species. Ind. Crops Prod. 2019, 138, 111422 10.1016/j.indcrop.2019.05.071. [DOI] [Google Scholar]
- Gurfinkel D. M.; Rao A. V. Determination of saponins in legumes by direct densitometry. J. Agric. Food Chem. 2002, 50, 426–430. 10.1021/jf010754c. [DOI] [PubMed] [Google Scholar]
- Bohm B. A.; Koupai-Abyazani M. R. Flavonoids and condensed tannins from leaves of Hawaiian Vaccinium reticulatum and V. calycinum (Ericaceae). Pac. Sci. 1994, 48, 458–463. [Google Scholar]
- Ferguson N. M. A textbook of pharmacognosy. J. Pharm. Sci. 1956, 45, 1–374. [Google Scholar]
- Adetuyi A. O.; Popoola A. V. Extraction and dyes ability potential studies of the colourant in zanthoxylum zanthoxyloides plant on cotton fabric. J. Sci. Eng. Technol. 2001, 8, 3291–3299. [Google Scholar]
- Trease G. E.Textbook of Pharmacognosy, 7th ed.; In Trease G. E., Ed.; Bailliere Tindall: Kent, U.K., 1957. [Google Scholar]
- Sofowora A. Research on medicinal plants and traditional medicine in Africa. J. Altern. Complement. Med. 1996, 2, 365–372. 10.1089/acm.1996.2.365. [DOI] [PubMed] [Google Scholar]
- Kuster V. C.; Vale F. H. A. Leaf histochemistry analysis of four medicinal species from Cerrado. Rev. Bras. Farmacogn. 2016, 26, 673–678. 10.1016/j.bjp.2016.05.015. [DOI] [Google Scholar]
- Mujeeb F.; Bajpai P.; Pathak N. Phytochemical evaluation, antimicrobial activity, and determination of bioactive components from leaves of Aegle marmelos. BioMed. Res. Int. 2014, 2014, 497606 10.1155/2014/497606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohun A. F.; Cook I. J. An improved dinitrosalicylic acid method for determining blood and cerebrospinal fluid sugar levels. J. Clin. Pathol. 1962, 15, 169–180. 10.1136/jcp.15.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S.; Jana M. L.; Sengupta D.; Naskar A. K. A note on the estimation of microbial glycosidase activities by dinitrosalicylic acid reagent. Appl. Microbiol. Biotechnol. 2000, 53, 732–735. 10.1007/s002530000327. [DOI] [PubMed] [Google Scholar]
- Sudha P.; Zinjarde S. S.; Bhargava S. Y.; Kumar A. R. Potent α-amylase inhibitory activity of Indian Ayurvedic medicinal plants. BMC Complement. Altern. Med. 2011, 11, 5. 10.1186/1472-6882-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proestos C.; Sereli D.; Komaitis M. Determination of phenolic compounds in aromatic plants by RP-HPLC and GC-MS. Food Chem. 2006, 95, 44–52. 10.1016/j.foodchem.2004.12.016. [DOI] [Google Scholar]
- Shettar A. K.; Sateesh M. K.; Kaliwal B. B.; Vedamurthy A. B. In vitro antidiabetic activities and GC-MS phytochemical analysis of Ximenia americana extracts. S. Afr. J. Bot. 2017, 111, 202–211. 10.1016/j.sajb.2017.03.014. [DOI] [Google Scholar]
- Janakiraman N.; Johnson M.; Sahaya S. S. GC–MS analysis of bioactive constituents of Peristrophe bicalyculata (Retz.) Nees. (Acanthaceae). Asian Pac. J. Trop. Biomed. 2012, 2, S46–S49. 10.1016/S2221-1691(12)60128-2. [DOI] [Google Scholar]
- Berman H. M.; Battistuz T.; Bhat T. N.; Bluhm W. F.; Bourne P. E.; Burkhardt K.; Feng Z.; Gilliland G. L.; Iype L.; Jain S.; Fagan P.; Marvin J.; Padilla D.; Ravichandran V.; Schneider B.; Thanki N.; Weissig H.; Westbrook J. D.; Zardecki C. The Protein Data Bank. Acta Crystallogr., D: Biol. Crystallogr. 2002, 58, 899–907. 10.1107/S0907444902003451. [DOI] [PubMed] [Google Scholar]
- Kim S.; Chen J.; Cheng T.; Gindulyte A.; He J.; He S.; Li Q.; Shoemaker B. A.; Thiessen P. A.; Yu B.; Zaslavsky L.; Zhang J.; Bolton E. E. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019, 47, D1102–D1109. 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder E.; Damm W.; Maple J.; Wu C.; Reboul M.; Xiang J. Y.; Wang L.; Lupyan D.; Dahlgren M. K.; Knight J. L.; Kaus J. W.; Cerutti D. S.; Krilov G.; Jorgensen W. L.; Abel R.; Friesner R. A. OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins. J. Chem. Theory Comput. 2016, 12, 281–296. 10.1021/acs.jctc.5b00864. [DOI] [PubMed] [Google Scholar]
- Chandrasekaran B.; Abed S. N.; Al-Attraqchi O.; Kuche K.; Tekade R. K.. Chapter 21 – Computer-Aided Prediction of Pharmacokinetic (ADMET) Properties. In Advances in Pharmaceutical Product Development and Research, Dosage Form Design Parameters Tekade R. K., Ed.; Academic Press: Cambridge, MA, 2018; pp 731–755. [Google Scholar]
- Lagorce D.; Douguet D.; Miteva M. A.; Villoutreix B. O. Computational analysis of calculated physicochemical and ADMET properties of protein-protein interaction inhibitors. Sci. Rep. 2017, 7, 46277 10.1038/srep46277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J.; Wang N.-N.; Yao Z.-J.; Zhang L.; Cheng Y.; Ouyang D.; Lu A.-P.; Cao D.-S. ADMETlab: a platform for systematic ADMET evaluation based on a comprehensively collected ADMET database. J. Cheminform. 2018, 10, 29. 10.1186/s13321-018-0283-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H. A.ADMET Properties: Overview and Current Topics. In Drug Design: Principles and Applications; Grover A., Ed.; Springer: Singapore, 2017. [Google Scholar]
- Ioakimidis L.; Thoukydidis L.; Mirza A.; Naeem S.; Reynisson J. Benchmarking the reliability of QikProp. Correlation between experimental and predicted values. QSAR Comb. Sci. 2008, 27, 445–456. 10.1002/qsar.200730051. [DOI] [Google Scholar]
- Senthil R.; Sakthivel M.; Usha S. Structure-based drug design of peroxisome proliferator-activated receptor gamma inhibitors: ferulic acid and derivatives. J. Biomol. Struct. Dyn. 2020, 1–17. 10.1080/07391102.2020.1740790. [DOI] [PubMed] [Google Scholar]
- Abu-Melha S. Pyridyl thiosemicarbazide: synthesis, crystal structure, DFT/B3LYP, molecular docking studies and its biological investigations. Chem. Cent. J. 2018, 12, 101. 10.1186/s13065-018-0469-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alex A. A.4.16 – Quantum Mechanical Calculations in Medicinal Chemistry: Relevant Method or a Quantum Leap Too Far?. In Comprehensive Medicinal Chemistry II; Taylor J. B.; Triggle D. J., Eds.; Elsevier: Oxford, U.K., 2007; pp 379–420. [Google Scholar]
- Bredas J.-L. Mind the gap!. Mater. Horiz. 2014, 1, 17–19. 10.1039/C3MH00098B. [DOI] [Google Scholar]
- Griffith J. S.; Orgel L. E. Ligand-field theory. Chem. Soc. Rev. 1957, 11, 381–393. 10.1039/qr9571100381. [DOI] [Google Scholar]
- Friesner R. A.; Murphy R. B.; Repasky M. P.; Frye L. L.; Greenwood J. R.; Halgren T. A.; Sanschagrin P. C.; Mainz D. T. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- Ban T.; Ohue M.; Akiyama Y. Multiple grid arrangement improves ligand docking with unknown binding sites: Application to the inverse docking problem. Comput. Biol. Chem. 2018, 73, 139–146. 10.1016/j.compbiolchem.2018.02.008. [DOI] [PubMed] [Google Scholar]
- Sirisha G. V. D.; Vijaya Rachel K.; Zaveri K.; Yarla N. S.; Kiranmayi P.; Ganash M.; Alkreathy H. M.; Rajeh N.; Ashraf G. M. Molecular docking and in vitro studies of soap nut trypsin inhibitor (SNTI) against phospholipase A2 isoforms in therapeutic intervention of inflammatory diseases. Int. J. Biol. Macromol. 2018, 114, 556–564. 10.1016/j.ijbiomac.2018.03.139. [DOI] [PubMed] [Google Scholar]
- Qin X.; Ren L.; Yang X.; Bai F.; Wang L.; Geng P.; Bai G.; Shen Y. Structures of human pancreatic α-amylase in complex with acarviostatins: Implications for drug design against type II diabetes. J. Struct. Biol. 2011, 174, 196–202. 10.1016/j.jsb.2010.11.020. [DOI] [PubMed] [Google Scholar]
- Bowers K. J.; Chow E.; Xu H.; Dror R. O.; Eastwood M. P.; Gregersen B. A.; Klepeis J. L.; Kolossvary I.; Moraes M. A.; Sacerdoti F. D.; Salmon J. K.; Shan Y.; Shaw D. E. In Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters, Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, Association for Computing Machinery: Tampa, FL, 2006; p 84.
- Margreitter C.; Oostenbrink C. MDplot: Visualise Molecular Dynamics. R J. 2017, 9, 164–186. 10.32614/RJ-2017-007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N.; Goel N.; Chand Yadav T.; Pruthi V. Quantum chemical, ADMET and molecular docking studies of ferulic acid amide derivatives with a novel anticancer drug target. Med. Chem. Res. 2017, 26, 1822–1834. 10.1007/s00044-017-1893-y. [DOI] [Google Scholar]
- Kumar N.; Gupta S.; Chand Yadav T.; Pruthi V.; Kumar Varadwaj P.; Goel N. Extrapolation of phenolic compounds as multi-target agents against cancer and inflammation. J. Biomol. Struct. Dyn. 2019, 37, 2355–2369. 10.1080/07391102.2018.1481457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.