Abstract
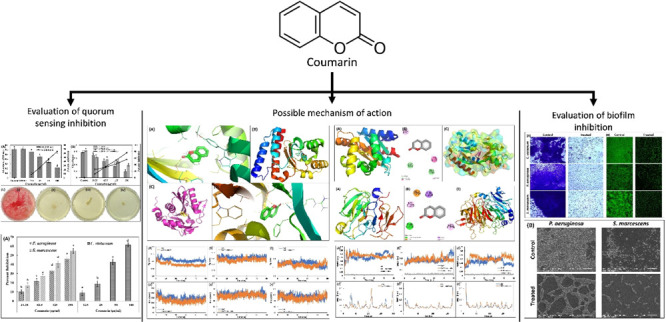
Quorum sensing (QS) and biofilm inhibition are recognized as the novel drug targets for the broad-spectrum anti-infective strategy to combat the infections caused by drug-resistant bacterial pathogens. Many compounds from medicinal plants have been found to demonstrate anti-infective activity. However, broad-spectrum anti-QS and antibiofilm efficacy and their mode of action are poorly studied. In this study, the efficacy of coumarin was tested against QS-regulated virulent traits of Gram-negative bacteria. Coumarin inhibited the production of violacein pigment in Chromobacterium violaceum 12472 by 64.21%. Similarly, there was 87.25, 70.05, 76.07, 58.64, 48.94, and 81.20% inhibition of pyocyanin, pyoverdin, and proteolytic activity, lasB elastase activity, swimming motility, and rhamnolipid production, respectively, in Pseudomonas aeruginosa PAO1. All tested virulence factors of Serratia marcescens MTCC 97 were also suppressed by more than 50% at the highest sub-minimum inhibitory concentration. Moreover, the biofilms of bacterial pathogens were also inhibited in a dose-dependent manner. Molecular docking and molecular dynamics (MD) simulation gave insights into the possible mode of action. The binding energy obtained by docking studies ranged from −5.7 to −8.1 kcal mol–1. Coumarin was found to be docked in the active site of acylhomoserine lactone (AHL) synthases and regulatory proteins of QS. MD simulations further supported the in vitro studies where coumarin formed a stable complex with the tested proteins. The secondary structure of all proteins showed a negligible change in the presence of coumarin. Computational studies showed that the possible mechanisms of anti-QS activity were the inhibition of AHL synthesis, antagonization of QS-regulatory proteins, and blocking of the receptor proteins. The findings of this study clearly highlight the potency of coumarin against the virulence factors of Gram-negative bacterial pathogens that may be developed as an effective inhibitor of QS and biofilms.
Introduction
Infectious diseases are still one of the major causes of human mortality and morbidity across the globe after cancer and cardiovascular diseases.1 In the last two decades, a global rise in the emergence and spread of antimicrobial resistance (AMR) accelerated among bacterial pathogens.2 AMR is now considered a global public health threat that warrants immediate action. The WHO (2019) reported that nearly 700,000 people die each year only because of the infections caused by drug-resistant microbes. If no action is taken, it (AMR) is expected to become a major cause of mortality by 2050, even surpassing cancer.3 The problem became more serious due to the lack of development of novel antibiotics in the recent two decades. These problems triggered urgent calls from national and international agencies to tackle the problem of AMR at various levels through integrated approaches including the development of new anti-infective agents to combat AMR. One of the promising strategies is to develop anti-infective agents targeting bacterial cell-to-cell communication, that is, quorum sensing (QS)-regulated virulence traits in pathogenic bacteria.4 QS is a global regulatory mechanism in most of the pathogenic bacteria which controls the production of various bacterial functions including virulence factors.5 The key advantage of anti-virulence drugs is that they specifically target the expression of virulence of bacteria without affecting bacterial growth, thereby reducing the chances of development of resistance against them.
Recent understanding on the role of microbial biofilms in bacterial pathogenicity and resistance to antimicrobial drugs clearly indicated that biofilms are one of the major obstacles in antimicrobial chemotherapy. Earlier, it was assumed that microbes grow in the planktonic mode only. However, it was found that microbes reside in the complex structures called biofilms. These complex structures comprise microbial cells and extracellular polymeric substances produced by bacteria. There is contrast in the expression of microbial phenotypes when they grow in the biofilm mode compared to the planktonic mode. In biofilms, microbes interact with each other and regulate the expression of certain set of important genes.6 According to National Institute of Health (NIH) estimates, more than 80% of infections are encouraged and established by biofilm development that poses heavy burden on the cost of human health.7 A vast majority of infections are associated with the biofilm development of either opportunistic or pathogenic microbes.8
In this study, we have explored the broad-spectrum antibiofilm and anti-QS activity of coumarin against Gram-negative bacteria. The anti-QS and antibiofilm studies were performed against a number of virulent traits of Chromobacterium violaceum 12472, Serratia marcescens MTCC 97, and Pseudomonas aeruginosa PAO1. Moreover, the antibiofilm activity of coumarin was validated microscopically using light microscopy, electron microscopy, and confocal microscopy. In computational studies, coumarin was docked against 10 proteins of four different targets to study their interactions. Molecular dynamics (MD) simulation studies were carried out to explore the stability of the docked complex that how they behave in the aqueous system.
Results and Discussion
Inhibition of Violacein Production in C. violaceum 12472
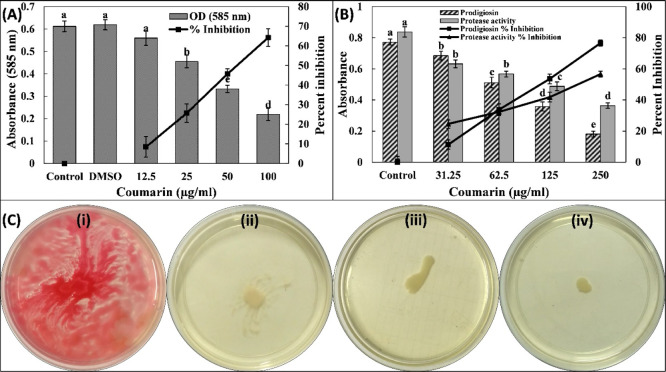
The preliminary investigation of anti-QS activity of coumarin was performed by assessing the inhibition of violacein production in C. violaceum 12472. The production of this pigment in C. violaceum 12472 is controlled by acylhomoserine lactone (AHL)-regulated QS. In this assay, any reduction in the pigment production is an indicator of anti-QS activity. The quantitative estimation of this pigment was assessed spectrophotometrically. The data clearly show that the absorbance (585 nm) of pigment in untreated control was 0.61 ± 0.02 that decreased in a dose-dependent manner by the treatment of coumarin (Figure 1A). Approximately 65% inhibition of the pigment was recorded in the presence of highest sub-minimum inhibitory concentration (MIC) (100 μg/mL) of coumarin. Results show the inhibitory potential coumarin on QS-mediated violacein production in C. violaceum 12472. Our findings are in agreement with a previous report in which structurally related coumarins inhibited violacein production in C. violaceum 12472.17
Figure 1.
(A) Effect of coumarin on the violacein production in C. violaceum 12472. The combined df and F values are 5 and 121.945, respectively. (B) Effect of coumarin on the prodigiosin production and exoprotease activity S. marcescens MTCC 97. The combined df and F values for prodigiosin production are 4 and 248.452, respectively. The combined df and F values for exoprotease activity are 4 and 145.625, respectively. Different letters above the error bars represent different significance groups by the Tukey test at p-value = 0.05. Data are represented as mean values of triplicate readings, and the bar is standard deviation. The percent inhibition is shown on the secondary y-axis. (C) Swarming motility of S. marcescens MTCC 97. (i) Untreated control, (ii) 62.5, (iii) 125, and (iv) 250 μg/mL.
Inhibition of QS-Controlled Virulence Factors of P. aeruginosa PAO1
Pyocyanin is a blue green pigment whose production is controlled by QS in P. aeruginosa. The amount of pyocyanin in the untreated control was found to be 6.58 ± 0.49 μg/mL in the cell-free supernatant (CFS). The presence of 31.25, 62.5, 125, and 250 μg/mL coumarin decreased the pyocyanin levels by 31.70, 42.79, 64.97, and 87.25% in P. aeruginosa PAO1, respectively (Table S3). However, dimethyl sulfoxide (DMSO) (0.5% v/v), the solvent control, showed an insignificant (p-value = 0.84) change in the pyocyanin production. Pyocyanin contributes in the pathogenicity of P. aeruginosa by interfering with several cellular functions of the host. In cystic fibrosis subjects, pyocyanin and its precursor have been reported to hinder the proper beating of human respiratory cilia apart from altering the expression of numerous immune modulatory proteins.18 Pyocyanin aids P. aeruginosa in biofilm establishment and suppresses the host’s defence system by enhancing the apoptosis of human neutrophils.19 Pyoverdin (a fluorescent siderophore) is another pigment produced by P. aeruginosa that plays a vital role in virulence of infections caused in host. A concentration-dependent response was found by the supplementation of coumarin in culture media, as shown in Table S3. More than 70% reduction in production of this siderophore was recorded upon treatment with coumarin (250 μg/mL). Pyoverdin contributes in the pathogenicity of infections caused by P. aeruginosa by confiscating the transferrin protein, thereby causing deficiency of iron in host’s tissues.20 In the lungs of subjects with cystic fibrosis, this siderophore helps in establishment of P. aeruginosa infection by evading the lipoplatin recognition.21 Therefore, inhibition of these pigments shows the ability of coumarin in diminishing the pathogenicity of P. aeruginosa.
The major enzymes that cause cellular damage in P. aeruginosa infections are the proteases and elastases. The effect of coumarin on exoprotease activity was assessed by azocasein degradation assay. The presence of 31.25, 62.5, 125, and 250 μg/mL coumarin inhibited the proteolytic activity of P. aeruginosa PAO1 by 12.76, 31.73, 41.27, and 76.07% in the CFS, respectively (Table S3). Similarly, lasB elastase activity was also reduced by ∼60% with coumarin treatment. The proteolytic enzymes produced by bacteria increases the bacterial invasion to overcome the host’s defence system by cleaving the proteins of host’s cells. Many other hydrolytic enzymes, such as elastases, secreted by bacteria during infection degrade the tissues of host to overpower the immune response.22 The synthesis of las proteins is controlled by QS whose expression also assists the formation of pathogenic biofilms.23 A similar finding has been reported earlier where treatment with sub-MIC of 6-gingerol resulted in downregulation of lasB gene that encodes for elastase enzyme.24 Reduced production of these enzymes in culture supernatant of P. aeruginosa PAO1 indicates the modulation of lasI-lasR QS by coumarin.
Rhamnolipids are surfactants produced by P. aeruginosa that plays a vibrant role in the attachment of bacterial cells to surfaces and in the maintenance of the biofilm architecture.25 The rhamnolipid production in P. aeruginosa PAO1 was reduced by 15.48, 26.69, 39.09, and 48.94% in the presence of 31.25, 62.5, 125, and 250 μg/mL coumarin, respectively (Table S3). These surfactants also assist in surface motility of P. aeruginosa. Previously, a study has revealed that curcumin inhibit the production of rhamnolipid in P. aeruginosa PAO1.26 The motility of P. aeruginosa is controlled by QS. The bacterial motility plays a crucial role in the pathogenicity of P. aeruginosa,27 and hence, it is also considered an important virulence factor to assess for the development of anti-QS agents. The untreated control P. aeruginosa PAO1 swam to the entire Petri plate within 18 h of incubation with an average diameter of 88.66 ± 1.15 mm. The swimming diameter decreased to 48.33, 33.00, and 21.66 mm by the addition of 31.25, 62.5, and 125 μg/mL coumarin in culture media, respectively (Table S3). More than 80% inhibition of swimming motility of P. aeruginosa was found at highest sub-MIC of coumarin (250 μg/mL). Tea polyphenols have also been found to reduce such bacterial motility in P. aeruginosa.(28)
Inhibition of QS-Controlled Virulence Factors of S. marcescens MTCC 97
The broad-spectrum anti-QS activity of coumarin was studied by evaluating the effect of coumarin on QS-controlled virulence factors of another Gram-negative bacteria, that is, S. marcescens MTCC 97. Prodigiosin is a red pigment whose production is governed by QS circuit of S. marcescens. There are minimum of four AHLs produced by S. marcescens that govern prodigiosin production, biofilm formation, motility, and carbapenem resistance. The data presented in Figure 1B clearly shows that prodigiosin production was dose-decently decreased by the treatment of coumarin. The pigment biogenesis was inhibited by >75% at 250 μg/mL coumarin. A study has found in some strains of S. marcescens that there may be a common regulatory link in the synthesis of prodigiosin, hemagglutination, and flagellar variation.29 Such bacterial pigments are known to hinder the immune responses and cause cytotoxicity to the host’s cells.30 These pigments produced by bacteria are sometimes considered essential for the survival and are also related to the pathogenicity of bacteria.31 The finding of this study is in agreement with an earlier report in which petroselinic acid inhibited the prodigiosin production in S. marcescens ATCC 14756.32 Furthermore, the azocasein-degrading exoprotease activity in S. marcescens was also evaluated. A significant reduction (p-value < 0.05) in proteolytic activity was found in the CFS of S. marcescens MTCC 97 by the treatment of coumarin. More than 50% inhibition was recorded at 250 μg/mL (Figure 1B). This is a key virulence factor of S. marcescens as secretion of proteases in infections governs the inflammatory and immune responses in host.33 The swimming motility is characteristic to many virulent strains of S. marcescens and also plays a vital role in certain nosocomial infections such as urinary tract infections associated with a catheter.34 The untreated S. marcescens MTCC 97 swarmed to the entire plate with dark red pigment production (Figure 1C). The presence of coumarin (250 μg/mL) reduced swarming motility up to 90%, in which the prodigiosin pigment was also inhibited, further validating the prodigiosin inhibition data. Such flagellar-mediated bacterial motilities regulate the adherence of S. marcescens which are required for the biofilm development.35
Broad-Spectrum Inhibition of Biofilm Development by Coumarin
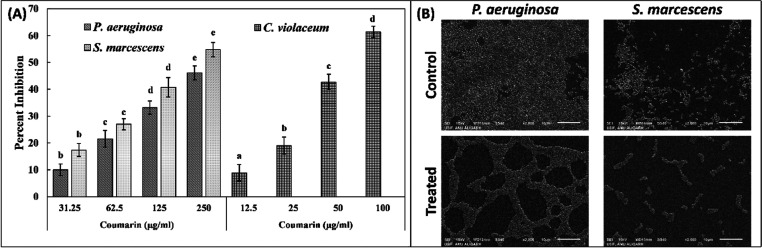
The effect of coumarin on biofilm formation of abovementioned three bacteria was also evaluated. There was concentration-dependent inhibition of biofilm formation of test bacteria (Figure 2A). The presence of 31.25, 62.5, 125, and 250 μg/mL coumarin decreased the development of the P. aeruginosa PAO1 biofilm by 09.98, 21.51, 33.16, and 46.10%, respectively, with respect to the control. Similarly, the biofilm of S. marcescens MTCC 97 was reduced by >50% at the highest sub-MIC (250 μg/mL). The biofilm of C. violaceum 12472 was maximally inhibited (>60%) by treatment of coumarin (100 μg/mL).
Figure 2.
(A) Effect of coumarin on the biofilm formation of C. violaceum 12472, P. aeruginosa PAO1, and S. marcescens MTCC 97. Different letters above the error bars represent different significance groups by the Tukey test at p-value = 0.05. Data are represented as mean values of triplicate readings, and bar is standard deviation. The combined df and F values for biofilm inhibition of P. aeruginosa are 5 and 155.128, respectively. The combined df and F values for biofilm inhibition of S. marcescens are 5 and 171.195, respectively. The combined df and F values for biofilm inhibition of C. violaceum are 5 and 226.252, respectively. (B) Scanning electron microscopic images of P. aeruginosa PAO1 and S. marcescens MTCC 97 biofilms in the absence and presence of sub-MIC (250 μg/mL) coumarin.
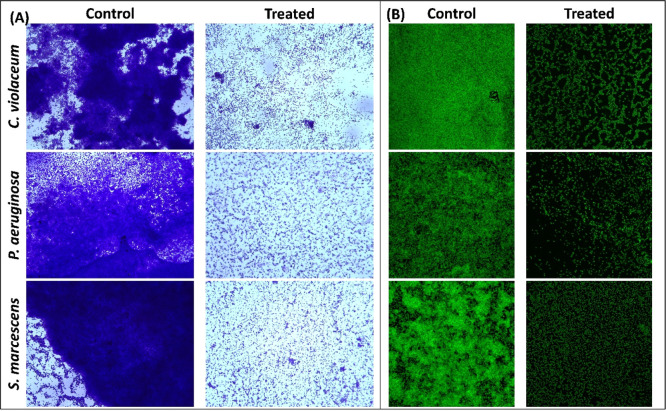
Quantitative biofilm data were further validated microscopically using light microscopy, electron microscopy, and confocal microscopy. The untreated control of all test bacteria showed a dense mat-like structure of biofilms on glass coverslip’s surface, as visualized under a light microscope (Figure 3A). Treatment with respective sub-MIC remarkably decreased the formation of aggerate-like structures of bacterial cells. A dense cluster of cells was also observed under a scanning electron microscope in the control slides, as shown in Figure 2B. The colonization of each bacterium on the glass surface was reduced in the presence of coumarin. The similar findings were also recorded by confocal laser scanning microscopy, in which there were a thick cluster of cellular mass comprising multiple layers in control slides (Figure 3B). Treatment with coumarin reduced the biofilm development where bacteria were mostly seen as single layer of cells.
Figure 3.
(A) Light microscopic images of C. violaceum 12472, P. aeruginosa PAO1, and S. marcescens MTCC 97 biofilms in the absence and presence of sub-MIC coumarin. (B) Confocal laser scanning microscopic images of C. violaceum 12472, P. aeruginosa PAO1, and S. marcescens MTCC 97 biofilms in the absence and presence of sub-MIC coumarin. Sub-MIC against C. violaceum 12472 was 100 μg/mL, and sub-MIC against P. aeruginosa PAO1 and S. marcescens MTCC 97 was 250 μg/mL.
The role of biofilms in disease development is well recognized, and about 80% of bacterial infections are associated with biofilm development.7 It has been documented that the pathogenicity of P. aeruginosa is also due to the biofilms, as bacterial cells in the biofilm mode of growth are more resistant to physical and chemical treatment.36 For example, P. aeruginosa biofilms growing on urinary catheters are approximately 1000 times more resistant to tobramycin compared to planktonic cells.37 Coumarin has also been reported previously to inhibit the biofilm of Escherichia coli and C. violaceum.(17,38) The findings of this study validate broad-spectrum inhibition of biofilm development in Gram-negative bacteria by coumarin.
Molecular Docking
To obtain a closer insight into the antivirulence potential of coumarin, molecular docking of coumarin with the protein involved in QS and biofilms was performed. The parameters used in the molecular docking were first validated. The natural ligand (N-3-oxo-dodecanoyl-l-homoserine lactone) of LasR protein was first extracted from the crystal structure and then redocked. The ligand was found to be docked in the same binding pocket as it was in the crystal structure (Figure S1), validating the applied docking procedure. Coumarin exhibited different biding affinities with different tested proteins. The binding constant and the lowest binding energies are presented in Table 1. The energies of top five poses are enlisted in Table S4.
Table 1. Binding Energies and Binding Constants for the Interaction of Coumarin with Different Proteins Obtained Using AutoDock Vina.
| S. no. | PDB ID | protein name | binding energy (kcal mol–1) | binding constant (M–1) |
|---|---|---|---|---|
| 1. | 1RO5 | LasI | –5.7 | 1.5 × 104 |
| 2. | 1KZF | EsaI | –6.6 | 6.9 × 104 |
| 3. | 2UV0 | LasR | –8.1 | 8.7 × 105 |
| 4. | 3IT7 | LasA | –7.5 | 3.1 × 105 |
| 5. | 3QP5 | CviR | –7.7 | 4.4 × 105 |
| 6. | 3QP1 | CviR′ | –7.7 | 4.4 × 105 |
| 7. | 4JVI | PqsR | –6.5 | 5.8 × 104 |
| 8. | 3JVV | PilT | –6.3 | 4.1 × 104 |
| 9. | 3HX6 | PilY1 | –6.3 | 4.1 × 104 |
| 10. | RhlR | –6.4 | 4.9 × 104 |
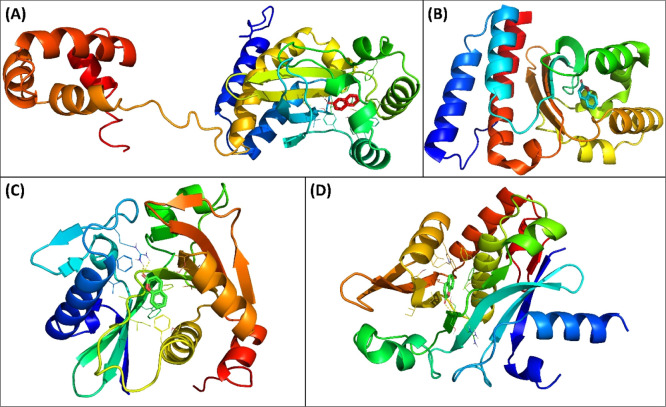
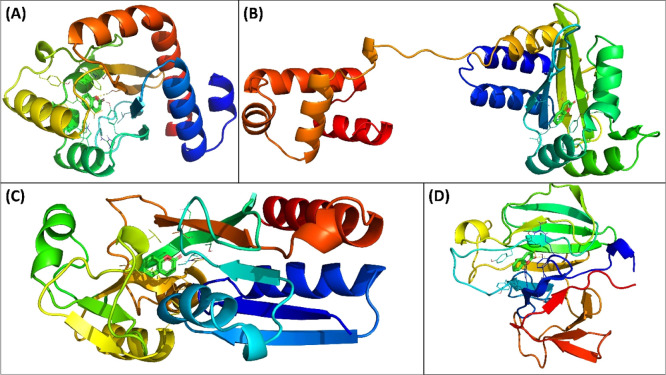
CviR is a receptor of C. violaceum ATCC 31532 which senses the QS signal molecule (C6-AHL), and the presence of sufficient amount of signal molecules activates the expression of QS-controlled genes. There are two domains of CviR which are joined together by a short flexible random coil; one is ligand-binding domain (LBD), and other is DNA-binding domain (DBD).39 Another receptor protein of C. violaceum ATCC 12472 is CviR′ that exhibits 87% sequence identity with CviR. This receptor protein senses 3-hydroxy-C10-AHL. The acyl group of C6-AHL forms one hydrogen bond with Asp97, the lactone carbonyl group attaches to Trp84 via a hydrogen bond, and carbonyl oxygen forms two hydrogen bonds with Ser155 and Tyr80 of the receptor CviR.39 Coumarin interacted in same binding cavity where its antagonist (chlorolactone) binds. The binding energy (BE) was obtained as −7.7 kcal mol–1 for coumarin–CviR interaction. Coumarin bound to Asp97 via hydrogen bonds and to Ser155 by electrostatic interactions. Moreover, coumarin also interacted with Ile99, Trp11, and Met135 of CviR by hydrophobic interactions (Figure 4A). Coumarin interacted with CviR′ with −7.7 kcal mol–1 BE. Tyr80 of CviR′ formed hydrogen bonds with coumarin, and Asp97 electrostatically interacted with the protein (Figure 4B). Coumarin also interacted with Ile99, Trp11, Ala130, and Met135 of CviR′ with hydrophobic interactions. The blocking of AHL autoinducer’s binding site by any molecule has been proposed as an effective strategy to antagonize the transcription factor CviR. The findings indicated that binding of coumarin to CviR may antagonize the expression of QS-linked traits.
Figure 4.
(A) Molecular docked complex of coumarin with CviR (3QP5). (B) Molecular docked complex of coumarin with CviR′ (3QP1). (C) Molecular docked complex of coumarin with LasI (1RO5). (D) Molecular docked complex of coumarin with EsaI (1KZF).
LasI is an HSL synthesis protein of P. aeruginosa that produces 3-oxo-C12-HSL. The protein shares 31% identity and 47% homology with RhlI, a counterpart of P. aeruginosa AHL synthase. Coumarin was found to interact with Arg30 via hydrogen bonds (Figure 4C). Moreover, coumarin also interacted with Trp69, Phe105, Ile107, Phe117, and Val148 via hydrophobic interactions. The binding constant and energy for coumarin–LasI interactions were found to be 1.5 × 104 M–1 and −5.7 kcal mol–1, respectively. The crystal structure has deciphered that N-terminal amino acids of LasI such as Phe27, Arg30, and Trp33 are crucial for the formation of S-adenosyl methionine (SAM)-binding pocket. Phe105 of LasI is a conserved residue for binding to acyl-chain.40 EsaI, AHL synthase of Pantoea stewartii, synthesizes 3-oxo-C6-HSL and has 44% homology and 23% identity with RhlI. The BE for coumarin–EsaI interactions was found to be −6.6 kcal mol–1. Coumarin interacted with Arg100 of EsaI via a hydrogen bond with 2.83 Å as the bond length (Figure 4D). Coumarin also bound to Phe101, Val142, Met146, and Leu150 by hydrophobic interactions. Hence, the interaction of coumarin with AHL synthases, such as LasI and EsaI, might be preventing the binding of precursor molecules and hence inhibiting the synthesis of functional signal molecules.
LasR is a transcriptional activator of numerous genes related to virulence and pathogenicity of P. aeruginosa. The docked-out conformation of coumarin with the lowest BE toward LasR was obtained as −8.1 kcal mol–1, corresponding to a binding constant of 8.7 × 105 M–1. It is interesting that coumarin was docked to LasR in the same binding cavity where its natural ligand (3-oxo-C12-acylhomoserine lactone) binds. Tyr56 of LasR formed the hydrogen bond with coumarin at a distance of 2.93 Å. Other amino acids of LasR such as Leu36, Tyr56, Tyr64, Asp73, Trp88, Ala105, and Leu100 were found to be interacting with coumarin via hydrophobic interactions (Figure 5A). The interaction of 3-oxo-C12-AHL to LasR induces the transcription of an array of virulent genes of P. aeruginosa.(41) It is anticipated that the binding of coumarin will compete against 3-oxo-C12-AHL for the interaction with LasR and consequently decrease the expression of QS-controlled genes. In vitro results have demonstrated that coumarin successfully inhibited the LasR-dependent QS traits of P. aeruginosa such as motility and biofilms. One of the possible mechanisms for such inhibitory activity may be the competition of coumarin for the same biding site in LasR.
Figure 5.
(A) Molecular docked complex of coumarin with LasR (2UV0). (B) Molecular docked complex of coumarin with RhlR. (C) Molecular docked complex of coumarin with PqsR (4JVI). (D) Molecular docked complex of coumarin with LasA (3IT7).
RhlR is a transcription regulator in P. aeruginosa where binding of butanoyl-homoserine lactone with the regulator activates the certain virulent gene expression. Coumarin interacted with Val60 and Tyr72 via hydrophobic interactions (Figure 5B). Moreover, Trp68 and Asp81 of RhlR were complexed to coumarin through van der Waals (vdW) forces. The binding constant and BE for coumarin–RhlR interactions were obtained as 4.9 × 104 M–1 and −6.4 kcal mol–1, respectively. Another transcriptional regulator of P. aeruginosa is PqsR that controls the expression of virulent genes. PqsR is activated by the binding of the Pseudomonas quinolone signal (PQS) and 2-heptyl-4-quinolone.42 Coumarin interacted with PqsR with a BE of −6.5 kcal mol–1 and a binding constant of 5.8 × 104 M–1. Molecular docking revealed that coumarin interacted with PqsR at the same binding site where its inhibitor binds. Coumarin interacted with Ala102, Ile149, Ala168, Leu208, Ile236, and Pro238 of PqsR via hydrophobic interactions (Figure 5C). PqsR controls the transcription of major synthase genes that are located in polycistronic operon (pqsABCDE).43 Another possible anti-QS mode of action of coumarin may be the inhibition of synthesis of AHL synthases, as revealed by molecular docking.
LasA is a gene whose product is known to be involved in the proteolytic and elastolytic activities of P. aeruginosa. LasA is considered an important virulence factor of P. aeruginosa that shows proteolytic and elastinolytic activities. Post docking analysis revealed that coumarin interacted with LasA with BE as −7.5 kcal mol–1. Arg12 and Trp 17 of LasA formed hydrogen bonds with coumarin at 2.26 and 1.89 Å bond lengths, respectively (Figure 5D). Moreover, Tyr15, Tyr39, and Tyr49 interacted with coumarin via hydrophobic interactions. LasA, a staphylolytic endopeptidase, cleaves the pentaglycine bridge of peptidoglycan and also performs elastinolytic activity.44
Furthermore, molecular docking of coumarin was also performed with proteins involved in biofilm formation viz. PilT and PilY1. The binding affinity of coumarin with PilT was found to be −6.3 kcal mol–1 that corresponds to a binding constant of 4.1 × 104 M–1. Docking results revealed that coumarin interacted with Thr231 of PilT by the hydrogen bond at 2.36 Å (Figure S2). Coumarin also formed electrostatic bonding with Glu258. Moreover, Thr230, Lys235, and Arg239 formed hydrophobic interactions with coumarin. The finding corroborates with the earlier finding where plumbagin interacted with PilT with biding affinity as −6.3 kcal mol–1.45 Similarly, the BE for coumarin-PilY1 interaction was found to be −6.3 kcal mol–1. Coumarin formed two hydrogen bonds with Arg753 and one hydrogen bond with Ser846 (Figure S3). Glu1044 of PilY1 electrostatically interacted with coumarin, and Lys790 was involved in hydrophobic interactions. It can be deduced from molecular docking analysis that the interaction of coumarin with pilus proteins may also be a possible mechanism for the inhibition of biofilms of the test bacteria.
Molecular Dynamics Simulations
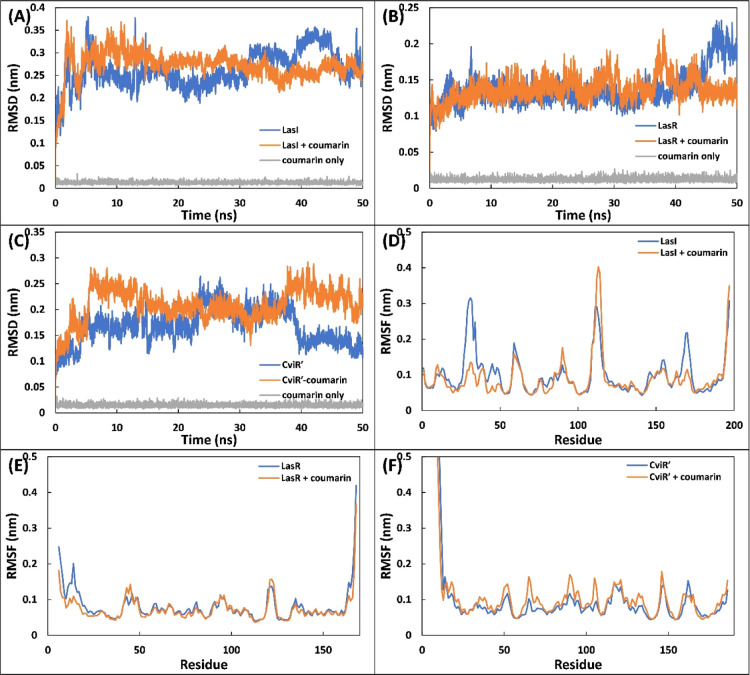
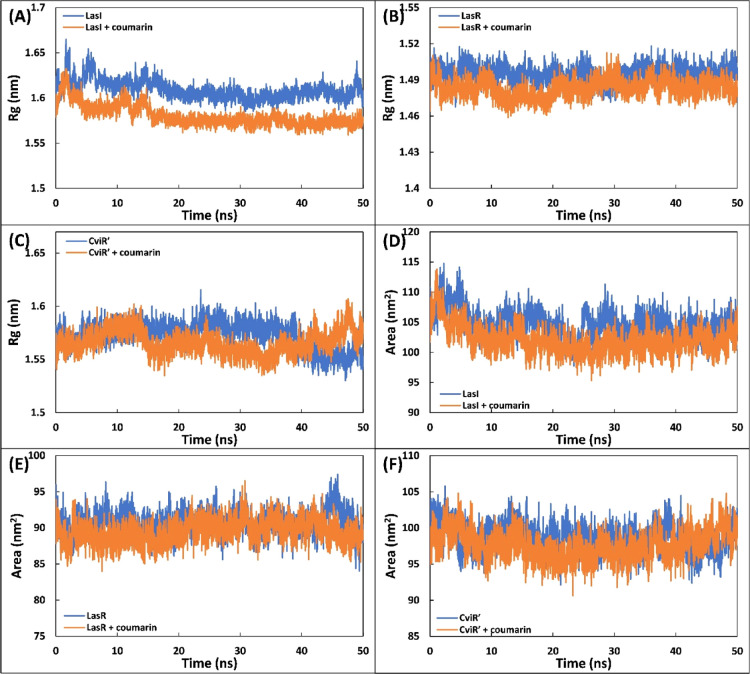
The complexes of coumarin with different target proteins of QS and biofilms were further studied using MD simulations. Three proteins (LasI, LasR, and CviR′) of different targets were selected as initial conformations for MD simulation studies. The values of root-mean-square deviation (RMSD), Rg, and solvent accessible surface area (SASA) of protein only, coumarin-protein complex, and coumarin only are enlisted in Table S5. The initial assessment of the MD simulation was carried out by calculating the RMSD of the proteins in the absence and presence of coumarin with respect to their initial backbone structures. The RMSD of the protein and their complexes is presented in Figure 6A–C. The RMSD of all proteins and complexes were below 0.4 nm, and complexes showed a similar RMSD to their respective proteins. For instance, the RMSD of LasI and LasI–coumarin complex was found to be 0.263 ± 0.039 and 0.272 ± 0.030 nm, respectively (Figure 6A). In all cases, the system (RMSD) reached equilibrium after initial few nanoseconds and becomes stable. The root-mean-square fluctuation (RMSF) of alpha carbon of individual residues of the proteins in the absence and presence of coumarin was calculated from the trajectory, as shown in Figure 6D–F. RMSF analysis shows that a similar pattern of fluctuations in amino acids of the protein was observed even in the presence of coumarin with minor variations. For instance, Lys31 of LasI showed maximum fluctuation, while in the complex form, Gly113 was the most fluctuating residue (Figure 6D). The RMSF of coumarin was also calculated and is shown in Figure S4. The atoms of coumarin showed some fluctuations with all proteins, indicating a dynamical shift from their initial positions. The shift in position of coumarin may induce different interaction modes with nearby residues as simulation progressed. This results in shift between hydrogen bonds and hydrophobic interactions.46 The radius of gyration (Rg) of three proteins (LasI, LasR, and CviR′) with and without coumarin was calculated as a function of time, and the results are presented in Figure 7A–C. The Rg of the all proteins and their complexes remained nearly the same during entire simulation period with minor variations. Similarly, Rg of coumarin also remained nearly constant over the simulation period (50 ns), indicating that coumarin did not undergo major conformational change.47 The stability of all the complexes was further validated by calculating changes in SASA as a function of time. The data clearly show that SASA of proteins and their complexes with coumarin was negligibly altered throughout simulation duration (Figure 7D–F). The above calculations, that is, RMSD, RMSF, Rg, and SASA, validate that the complexes of coumarin are well stable under solvent systems as of their respective protein counterparts.
Figure 6.
(A) RMSD of backbone of LasI, LasI–coumarin complex, and coumarin during simulation. (B) RMSD of LasR, LasR–coumarin complex, and coumarin. (C) RMSD of CviR′, CviR′–coumarin complex, and coumarin. (D) RMSF of the central carbon alpha of LasI and LasI–coumarin complex. (E) RMSF of LasR and LasR–coumarin complex. (F) RMSF of CviR′ and CviR′–coumarin complex.
Figure 7.
(A) Variation in the radius of gyration (Rg) of the LasI and LasI–coumarin complex as a function of time. (B) Variation in the radius of gyration (Rg) of LasR and the LasR–coumarin complex as a function of time. (C) Variation in the radius of gyration (Rg) of CviR′ and the CviR′–coumarin complex as a function of time. (D) SASA of LasI and the LasI–coumarin complex as a function of time. (E) SASA of LasR and the LasR–coumarin complex as a function of time. (F) SASA of CviR′ and the CviR′–coumarin complex as a function of time.
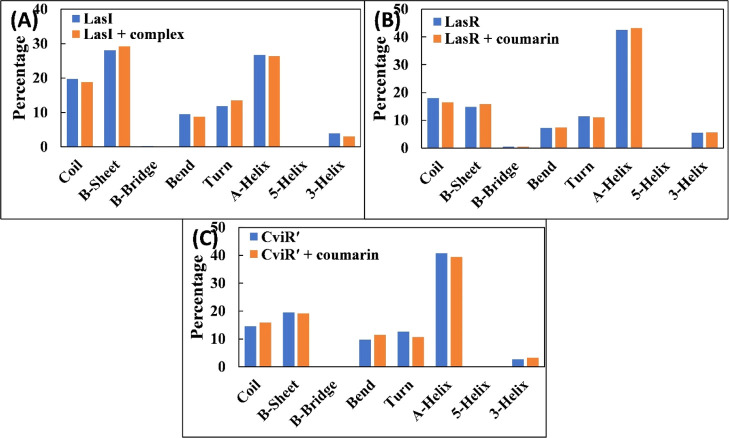
The changes in the secondary structure of proteins were calculated, and the data obtained are shown in Figure 8A–C. The percentage of α-helix and β-sheet in LasI was found to be 26.67 and 28.04, respectively. The presence of coumarin exhibited a negligible change in the secondary structure of LasI. Similar results were obtained for LasR and CviR′, in which the presence of coumarin did not alter the secondary structure of the proteins. The data also validated the structural stability of the proteins even in the presence of coumarin. The interactions between coumarin and proteins were studied by analyzing the hydrogen bonds. The hydrogen bond existence between coumarin and proteins with % existence of >1% was calculated. The existence of the hydrogen bond remained approximately the same during the simulation duration. The residues of LasI that were involved in hydrogen bond formation were Arg30, Trp33, Ile107, Ser109, and Thr145. Similarly, Tyr56 and Trp60 of LasR were involved in the hydrogen bond formation. To obtain further detailed insights regarding binding forces responsible for the interaction of coumarin with the simulated proteins, different energies were calculated using MM-PBSA (molecular mechanics Poisson–Boltzmann surface area) methods. 100 snapshots were extracted at equal time intervals from the whole trajectory and used for MM-PBSA calculations (Table 2). In protein–drug interactions, various non-covalent forces such as hydrogen bonds, electrostatic forces, hydrophobic interactions, polar forces, and so on are responsible for the binding.46 Each of these interactions either contribute positively or negatively to the overall BE. Electrostatic (Elec) and vdW forces mainly favored the binding process of coumarin with LasR and CviR′. vdW forces were the major driver for the interaction of coumarin with LasI, while there were also small contributions of Elec and SASA energies. Polar solvation energy (PSE) impaired binding of coumarin with all proteins. A small contribution of SASA energy was found in the overall binding. The average binding energy for the complexation of coumarin with LasI, LasR, and CviR′ was found to be −8.6, −14.2, and −10.3 kcal mol–1, respectively.
Figure 8.
(A) Percentage of the secondary structure in the LasI and LasI–coumarin complex. (B) Percentage of the secondary structure in the LasR and LasR–coumarin complex. (C) Percentage of the secondary structure in CviR′ and CviR′–coumarin complex.
Table 2. Binding Free Energy (kcal mol–1) Calculated by the MM-PBSA method for 100 Snapshots of MD Simulationa.
| proteins |
|||
|---|---|---|---|
| energy | LasI | LasR | CviR′ |
| ΔEvdW | –20.2 ± 0.6 | –20.6 ± 0.5 | –21.4 ± 0.2 |
| ΔEele | –1.4 ± 1.1 | –17.1 ± 0.5 | –13.6 ± 0.1 |
| ΔEPSE | 15.1 ± 0.4 | 25.7 ± 0.3 | 27.1 ± 0.1 |
| ΔESSASA | –2.1 ± 0.1 | –2.2 ± 0.1 | –2.3 ± 0.1 |
| ΔEBE | –8.6 ± 0.1 | –14.2 ± 1.4 | –10.3 ± 0.1 |
ΔEvdW: van der Waals energy, ΔEele: electrostatic energy, ΔEPSE: polar solvation energy, ΔESASA: solvent accessible surface area energy, and ΔEBE: binding energy.
Conclusions
In this study, we report the broad-spectrum anti-QS and antibiofilm activity of coumarin against Gram-negative bacteria. Nearly all virulence factors of the tested bacteria were reduced by >50% at sub-MICs. Coumarin not only inhibited the biofilm development but also altered the architecture of biofilms of the bacteria. The binding energy from molecular docking studies ranged from −5.7 to −8.1 kcal mol–1. Coumarin occupied the active site of the proteins (AHL synthases and regulatory proteins) and formed a stable complex with the tested proteins. Possible mechanisms of anti-QS activity may be the inhibition of signal molecule synthesis, antagonization of QS-regulatory proteins, and blocking of receptor proteins. Results showed the promising potency of coumarin against the virulence factors of Gram-negative bacteria that may be developed as an effective inhibitor of the QS and biofilm.
Materials and Methods
Materials and Reagents
Azocasein (A2765), coumarin (C4261, purity ≥99%), and elastin Congo red (ECR) (E0502) were purchased from Sigma Aldrich, USA. Trichloroacetic acid (RM7570) and orcinol (MB242) were procured from Hi-Media Laboratories, Mumbai, India. 2,3,5-Triphenyltetrazolium chloride (65599) was purchased from Sisco Research Laboratories (SRL) Pvt. Ltd. The details including catalogue number, purity, company name, and country of the chemical used in this study are provided in Table S1.
Bacterial Strains and Growth Conditions
C. violaceum ATCC 12472 (ATCC, USA) and P. aeruginosa PAO1 were obtained as a gift from Prof. RJC McLean, USA. S. marcescens MTCC 97 was obtain form Microbial Type Culture Collection (MTCC, India). The bacterial strains were grown in Luria–Bertani (LB) broth (0.5% yeast extract, 15.0 g of tryptone, and 0.5% NaCl), otherwise stated.
Determination of MIC
The MIC of coumarin against test bacteria was determined by microbroth dilution assay using tetrazolium chloride (TTC) as the indicator dye.9 Briefly, two-fold dilution of coumarin was made in a 96-well microtiter plate in LB broth. 10 μL of culture from the log phase of bacteria were inoculated in the wells containing varying concentrations of coumarin. No treatment with coumarin or solvent was given in the control group. Following 24 h of incubation, 10 μL of TTC (4 mg/mL) was added to each well, and the microtiter plate was incubated at 37 °C for 30 min in the dark. The wells were observed for change in color. The development of pink and/or red color is due to the metabolic activity of the actively growing cells. The minimum concentration at which no color (pink or red) was observed was considered as MIC. To further validate the bacterial growth, culture from the wells with no color change was spotted in LB agar plates. The MIC of coumarin against C. violaceum 12472 was found to be 200 μg/mL, while 500 μg/mL both against P. aeruginosa PAO1 and S. marcescens MTCC 97.
Assessment of Violacein Pigment Production in C. violaceum 12472
The relative quantification of violacein pigment production was determined spectrophotometrically following the earlier described procedure.10 Briefly, C. violaceum 12472 was grown in LB broth in the absence and presence of varying sub-inhibitory concentrations (sub-MICs) of coumarin (12.5, 25, 50, and 100 μg/mL). The cultures were allowed to grow for 18 h at 30 °C. Following incubation, 1 mL of culture from each treatment group was centrifuged for 5 min at 10,000 rpm, and supernatant was discarded. The violacein pigment present in the pellet was extracted in 1 mL DMSO by vigorous vortexing. The mixture was again centrifuged to settle down the bacterial cells. Three independent replicates of each group were used in this experiment. The optical density (OD) of the supernatant was recorded at 585 nm against DMSO as blank using an UV-2600 spectrophotometer, Shimadzu, Japan.
Quantitative Evaluation of Virulence Factors of P. aeruginosa PAO1
The method for the assessment of virulence factors of P. aeruginosa PAO1 was adopted with minor changes, as described previously.11P. aeruginosa PAO1 was cultured in the absence and presence of varying sub-MICs of coumarin (31.25, 62.5, 125, and 250 μg/mL) in LB broth (otherwise stated) for 18 h at 37 °C under shaking (250 rpm) condition. On completion of incubation, the CFS was obtained by centrifuging the cultures at 10,000 rpm for 5 min. The obtained CFS was used further for the assessment of virulence factors of P. aeruginosa PAO1, otherwise stated. Three independent replicates of each group were used, and fresh CFS was obtained for each experiment.
For the estimation of pyocyanin, P. aeruginosa PAO1 was cultured in Pseudomonas broth (PB) (20 g/L peptone, 1.4 g/L MgCl2, and 10 g/L K2SO4) as this medium maximizes the production of pyocyanin. 5 mL of CFS was extracted in chloroform (3 mL) by vigorous vortexing, and the aqueous phase was discarded. The chloroform phase was again extracted in 1.2 mL of 0.2 N HCl. The mixture was kept at room temperature to separate in layers, and the organic phase was discarded. The absorbance of the aqueous phase was recorded at 520 nm using a spectrophotometer. Pyoverdin was determined spectrofluorophotometrically. For the determination of pyoverdin, 0.1 mL of CFS was mixed with 0.9 mL of Tris–HCl (50 mM) of pH 7.4. The fluorescence intensity (460 nm) of each sample was measured by exciting at 400 nm using a spectrofluorometer (RF-5301PC, Shimadzu, Japan).
The exoprotease activity was determined using azocasein degradation assay. Briefly, 0.1 mL of CFS was mixed with 1 mL of azocasein (0.3% in 50 mM Tris–HCl containing 0.5 mM CaCl2, pH 7.5). The reaction mixture was left for incubation at 37 °C for 4 h under shaking conditions. 0.5 mL of ice cold trichloroacetic acid (10%) was added to stop the reaction, and the reaction mixture was centrifuged at 13,000 rpm for 8 min to pellet down the insoluble azocasein. The OD of the supernatant was recorded at 400 nm using a spectrophotometer. The elastase activity of P. aeruginosa PAO1 was determined in the CFS using ECR assay. Briefly, 0.1 mL of CFS was mixed with 0.9 mL of ECR buffer (5 mg/mL ECR and 1 mM CaCl2 in 100 mM Tris, pH 7.5) and incubated under shaking conditions at 37 °C for 3 h. The reaction was terminated by adding 1 mL sodium phosphate buffer (100 mM) of pH 6.0 and placing the samples at 4 °C for 30 min. The insoluble ECR was removed by centrifuging the reaction mixture at 13,000 rpm for 8 min. The absorbance of the supernatant was recorded at 495 nm using a spectrophotometer.
The relative amount of rhamnolipid in each sample was determined using the orcinol method. The CFS (300 μL) was added to diethyl ether (600 μL) and mixed by vortexing for 1 min. The mixture was kept for 30 min under static conditions for phase separation, and the organic phase was taken. The organic phase was then completely dried by evaporating at room temperature. 100 μL of deionized water was added to each sample and gently shaken to solubilize. 900 μL of orcinol solution (0.19% w/v orcinol in 53% H2SO4) was mixed to each sample and heated at 80 °C for 30 min. The samples were allowed to cool at room temperature, and absorbance was recorded at 421 nm using a spectrophotometer.
The swimming motility of P. aeruginosa PAO1 was assessed on soft agar plates (0.3% agar). Briefly, 5 μL of culture was taken from the log phase and spotted on LB soft gar plates containing varying sub-MICs of coumarin. In control plates, no treatment was given. The Petri plates were incubated at 37 °C for 18 h under static conditions. The zone of swimming was measured by a transparent ruler in millimeters (mm), and percent inhibition was calculated with respect to the control group.
Determination of Virulence Factors of S. marcescens MTCC 97
The determination of prodigiosin pigment production was performed using the standard method, as described earlier.12 Briefly, S. marcescens MTCC 97 was grown in the absence and presence of sub-MIC coumarin (31.25, 62.5, 125, and 250 μg/mL) in LB broth for 18 h at 30 °C in a shaking incubator (250 rpm). The pellet was obtained from liquid culture by centrifugation. The pellet was resuspended in 1 mL of acidified ethanol (96 mL ethanol + 4 mL 1 M HCl) by vigorous vortexing. The mixture was centrifuged (13,000 rpm for 5 min) to settle down cell debris, and the absorbance of supernatant (containing prodigiosin) was recorded at 534 nm using a spectrophotometer. Three independent replicates of each group were used for the determination of virulence factors of S. marcescens MTCC 97, and percent inhibition was calculated with respect to untreated control.
The exoprotease activity in the CFS of S. marcescens MTCC 97 was determined using azocasein degradation assay. S. marcescens MTCC 97 was cultured in the absence and presence of coumarin for 18 h at 30 °C under shaking conditions. The CFS was obtained by centrifugation. 0.1 mL of CFS was mixed with 1 mL of azocasein (0.3% w/v), and the reaction mixture was incubated at 37 °C with mild shaking. The reaction was stopped by the addition of 0.5 mL of ice-cold trichloroacetic acid (10% w/v), and insoluble azocasein was removed by centrifugation. The OD of the supernatant was recorded at 400 nm using a spectrophotometer. Percent inhibition was determined with respect to untreated control.
Swarming motility of S. marcescens MTCC 97 was studied in soft agar plates (0.5% agar). 5 μL of culture taken from actively growing log phase was spotted on soft agar LB plates containing varying sub-MICs of coumarin. For control, no treatment was given to S. marcescens MTCC 97. The swimming zone was measured by a transparent ruler in millimeters.
Biofilm Inhibition
Quantitative Evaluation of Biofilm in 96-Well Microtiter Plate
The quantification of biofilm was performed by crystal violet assay in a 96-well microtiter polystyrene plate following the standard procedure with slight modifications.13 10 μL of culture from the log phase of bacteria was seeded in the wells of the microtiter plate containing varying sub-MICs of coumarin. The sub-MICs for C. violaceum 12474 were 12.5, 25, 50, and 100 μg/mL. The sub-MICs for P. aeruginosa PAO1 and S. marcescens MTCC 97 were 31.25, 62.5, 125, and 250 μg/mL. The wells without any treatment of coumarin were taken as the control group. The plate was incubated for 24 h at their respective optimum growth temperature of each bacterium under static conditions. Following incubation, the planktonic cells in the medium were removed by discarding the growth medium and washing the wells gently with sterile phosphate buffer. The wells were allowed to air-dry at room temperature and then stained with 200 μL of crystal violet (0.1% w/v) for 15 min. The unbound dye was removed by washing with sterile phosphate buffer, and the stained biofilms were dissolved in 200 μL of ethanol (90%). The absorbance of each well was recorded at 620 nm using a microplate reader (Thermo Scientific Multiskan EX, UK). Four independent replicates of each group were taken for biofilm inhibition assay.
Microscopic Assessment of Biofilm Inhibition
The microscopic assessment of biofilm inhibition was performed on a glass surface. The glass coverslips for light and confocal microscopic analysis were prepared, as mentioned below. 50 μL of culture from the log phase of test bacteria was inoculated in 24-well tissue culture plates containing 3 mL of culture media with the respective highest sub-MICs. The highest sub-MICs for C. violaceum12474, P. aeruginosa PAO1, and S. marcescens MTCC 97 were 100, 250, and 250 μg/mL, respectively. Sterile glass coverslips of size 1 cm2 were placed in each well. The control slides were not given any treatment. On completion of incubation period (24 h), the glass slides were removed and gently washed with sterile phosphate buffer to remove loosely adhered cells. The biofilms on glass sides were stained with few drops of 0.1% w/v crystal violet solution for 15 min for light microscopy. For confocal microscopy, glass slides were stained with 0.1% acridine orange for 20 min. The excess dye was washed gently, and slides finally air-dried at room temperature for 30 min. The biofilms were visualized under a light microscope (Olympus BX60, model BX60F5, Olympus Optical Co. Ltd. Japan) equipped with a color VGA camera (Sony, Model no. SSC-DC-58AP, Japan), and images were captured at 40× magnification. Confocal microscopic analysis was carried out using Zeiss LSM780 at 63× magnification, at University Sophisticated Instrumentation Facility (USIF), AMU, Aligarh.
For scanning microscopic evaluation, the treatment was given, as mentioned above. The coverslips were removed from wells and gently washed with sterile phosphate buffer. The biofilms were fixed with glutaraldehyde (2.5% glutaraldehyde in 50 mM phosphate buffer) for 24 h at 4 °C. The biofilms were dehydrated by treating with a gradient of ethanol (20–100%) for 10 min each. The coverslips were then air dried at room temperature and coated with gold before visualization. The biofilms were visualized using a JEOL-JSM 6510 LV scanning electron microscope, at USIF, AMU, Aligarh.
Molecular Docking
The possible mechanism of the antibiofilm and anti-QS activity of coumarin was studied by molecular docking using AutoDock Vina.14 The three-dimensional (3D) structure of coumarin [CID: 323] was downloaded from PubChem in sdf format and converted to pdb format using Chimera 1.14. Using MGL Tools-1.5.6, the ligand was made flexible to obtain the best conformation, and the coordinate was saved in pdbqt format. The 3D crystal structures of receptor proteins were downloaded from Protein Data Bank. The structure of RhlR was downloaded from SWISS-MODEL Repository (ID: P54292) due to unavailability at Protein Data Bank. The water molecules in the crystal structure were deleted, and non-polar hydrogen and Kollman charges were added. The coordinates of proteins were saved in pdbqt format. The grid size and center of each receptor are enlisted in Table S2. The spacing of the grid was 1 Å. The analysis of docked complexes was performed using PyMol, LigPlot+, and Discovery Studio.
Molecular Dynamics Simulation
The complexes with lowest binding energy were further studied by MD simulation using gromacs 2018.1 with amber99sb-ILDN force field.15 The topology of coumarin was generated with amber99sb force field using Antechamber package in AmberTools19. TIP3P water model was used for the solvation in the triclinic box. Proteins were neutralized by adding sodium or chlorine counter ions. 50 ns of standard MD simulation was performed, and coordinates were saved for each trajectory. The RMSF, RMSD, SASA, and radius of gyration (Rg) were calculated using gromacs utilities. MM-PBSA analysis was performed to calculate the binding energy of coumarin with the proteins.16
Statistical Analysis
The experiments were three or four independent replicates, and data presented are mean value with standard deviation. For analysis of statistical significance, one-way ANOVA was performed using the Tukey test at a significance level of 0.05. The comparison of means was carried out using post-hoc analysis. Different letters in different treatment groups represent different significance groups at p-value = 0.05 and are in ascending order of values starting from letter “a”.
Acknowledgments
W.A. extends their appreciation to the Deanship of Scientific Research at King Khalid University to fund this work through research groups program under grant no. R.G.P.1/179/41. The authors are grateful to the Deanship of Scientific Research, King Saud University, for funding this work through the research group no. RG-1440-059. F.A.Q. is thankful to CSIR [File no. 09/112(0626)2k19 EMR] for providing SRF.
Glossary
Abbreviation
- ECR
elastin-congo red
- MD
molecular dynamics
- Rg
radius of gyration
- rmsd
root-mean-square deviation
- RMSF
root-mean-square fluctuation
- SASA
solvent accessible surface area
- Sub-MIC
sub inhibitory concentration
- TCA
trichloroacetic acid
- AHL
acyl homoserine lactone
- AI
autoinducer
- CFS
cell-free supernatant
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c02046.
Molecular docking images; RMSF; details of the chemicals; docking grid size and center; anti-QS data of P. aeruginosa PAO1; binding energies; and MD data (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- WHO . The Top 10 Causes of Death, 2018.
- Shakoor S.; Platts-Mills J. A.; Hasan R. Antibiotic-Resistant Enteric Infections. Infect. Dis. Clin. 2019, 33, 1105–1123. 10.1016/j.idc.2019.05.007. [DOI] [PubMed] [Google Scholar]
- WHO . No Time to Wait: Securing the Future from Drug-Resistant Infections, 2019.
- Kalia V. C. Quorum Sensing Inhibitors: An Overview. Biotechnol. Adv. 2013, 31, 224–245. 10.1016/j.biotechadv.2012.10.004. [DOI] [PubMed] [Google Scholar]
- Rutherford S. T.; Bassler B. L. Bacterial Quorum Sensing: Its Role in Virulence and Possibilities for Its Control. Cold Spring Harbor Perspect. Med. 2012, 2, a012427. 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Stoodley L.; Costerton J. W.; Stoodley P. Bacterial Biofilms: From the Natural Environment to Infectious Diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- Schachter B. Slimy Business—the Biotechnology of Biofilms. Nat. Biotechnol. 2003, 21, 361–365. 10.1038/nbt0403-361. [DOI] [PubMed] [Google Scholar]
- Donlan R. M. Biofilm Formation: A Clinically Relevant Microbiological Process. Clin. Infect. Dis. 2001, 33, 1387–1392. 10.1086/322972. [DOI] [PubMed] [Google Scholar]
- Eloff J. A Sensitive and Quick Microplate Method to Determine the Minimal Inhibitory Concentration of Plant Extracts for Bacteria. Planta Med. 1998, 64, 711–713. 10.1055/s-2006-957563. [DOI] [PubMed] [Google Scholar]
- Matz C.; Deines P.; Boenigk J.; Arndt H.; Eberl L.; Kjelleberg S.; Jürgens K. Impact of Violacein-Producing Bacteria on Survival and Feeding of Bacterivorous Nanoflagellates. Appl. Environ. Microbiol. 2004, 70, 1593–1599. 10.1128/AEM.70.3.1593-1599.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qais F. A.; Khan M. S.; Ahmad I. Broad-Spectrum Quorum Sensing and Biofilm Inhibition by Green Tea against Gram-Negative Pathogenic Bacteria: Deciphering the Role of Phytocompounds through Molecular Modelling. Microb. Pathog. 2019, 126, 379–392. 10.1016/j.micpath.2018.11.030. [DOI] [PubMed] [Google Scholar]
- Slater H.; Crow M.; Everson L.; Salmond G. P. Phosphate Availability Regulates Biosynthesis of Two Antibiotics, Prodigiosin and Carbapenem, in Serratia via Both Quorum-Sensing-Dependent and -Independent Pathways. Mol. Microbiol. 2003, 47, 303–320. 10.1046/j.1365-2958.2003.03295.x. [DOI] [PubMed] [Google Scholar]
- O’Toole G. A.; Kolter R. Flagellar and Twitching Motility Are Necessary for Pseudomonas Aeruginosa Biofilm Development. Mol. Microbiol. 1998, 30, 295–304. 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- Trott O.; Olson A. J. AutoDock Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2009, 31, 455–461. 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Spoel D.; Lindahl E.; Hess B.; Groenhof G.; Mark A. E.; Berendsen H. J. C. GROMACS: Fast, Flexible, and Free. J. Comput. Chem. 2005, 26, 1701–1718. 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- Kumari R.; Kumar R.; Lynn A. G_mmpbsa —A GROMACS Tool for High-Throughput MM-PBSA Calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- D’Almeida R. E.; Molina R. D. I.; Viola C. M.; Luciardi M. C.; Nieto Peñalver C.; Bardón A.; Arena M. E. Comparison of Seven Structurally Related Coumarins on the Inhibition of Quorum Sensing of Pseudomonas Aeruginosa and Chromobacterium Violaceum. Bioorg. Chem. 2017, 73, 37–42. 10.1016/j.bioorg.2017.05.011. [DOI] [PubMed] [Google Scholar]
- Fothergill J. L.; Panagea S.; Hart C. A.; Walshaw M. J.; Pitt T. L.; Winstanley C. Widespread Pyocyanin Over-Production among Isolates of a Cystic Fibrosis Epidemic Strain. BMC Microbiol. 2007, 7, 45. 10.1186/1471-2180-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das T.; Kutty S. K.; Tavallaie R.; Ibugo A. I.; Panchompoo J.; Sehar S.; Aldous L.; Yeung A. W. S.; Thomas S. R.; Kumar N.; Gooding J. J.; Manefield M. Phenazine Virulence Factor Binding to Extracellular DNA Is Important for Pseudomonas Aeruginosa Biofilm Formation. Sci. Rep. 2015, 5, 8398. 10.1038/srep08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox C. D.; Adams P. Siderophore Activity of Pyoverdin for Pseudomonas Aeruginosa. Infect. Immun. 1985, 48, 130–138. 10.1128/iai.48.1.130-138.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peek M. E.; Bhatnagar A.; McCarty N. A.; Zughaier S. M. Pyoverdine, the Major Siderophore inPseudomonas aeruginosa, Evades NGAL Recognition. Interdiscip. Perspect. Infect. Dis. 2012, 2012, 1–10. 10.1155/2012/843509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejarano P. A.; Langeveld J. P.; Hudson B. G.; Noelken M. E. Degradation of Basement Membranes by Pseudomonas Aeruginosa Elastase. Infect. Immun. 1989, 57, 3783–3787. 10.1128/IAI.57.12.3783-3787.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.; He X.; Xie W.; Xiong J.; Sheng H.; Guo S.; Huang C.; Zhang D.; Zhang K. Elastase LasB of Pseudomonas Aeruginosa Promotes Biofilm Formation Partly through Rhamnolipid-Mediated Regulation. Can. J. Microbiol. 2014, 60, 227–235. 10.1139/cjm-2013-0667. [DOI] [PubMed] [Google Scholar]
- Kim H.-S.; Lee S.-H.; Byun Y.; Park H.-D. 6-Gingerol Reduces Pseudomonas Aeruginosa Biofilm Formation and Virulence via Quorum Sensing Inhibition. Sci. Rep. 2015, 5, 8656. 10.1038/srep08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey M. E.; Caiazza N. C.; O’Toole G. A. Rhamnolipid Surfactant Production Affects Biofilm Architecture in Pseudomonas Aeruginosa PAO1. J. Bacteriol. 2003, 185, 1027–1036. 10.1128/JB.185.3.1027-1036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packiavathy I. A. S. V.; Priya S.; Pandian S. K.; Ravi A. V. Inhibition of Biofilm Development of Uropathogens by Curcumin – An Anti-Quorum Sensing Agent from Curcuma Longa. Food Chem. 2014, 148, 453–460. 10.1016/j.foodchem.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Wang S.; Yu S.; Zhang Z.; Wei Q.; Yan L.; Ai G.; Liu H.; Ma L. Z. Coordination of Swarming Motility, Biosurfactant Synthesis, and Biofilm Matrix Exopolysaccharide Production in Pseudomonas Aeruginosa. Appl. Environ. Microbiol. 2014, 80, 6724–6732. 10.1128/AEM.01237-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H.; Deng Y.; Wang H.; Liu W.; Zhuang X.; Chu W. Tea Polyphenols as an Antivirulence Compound Disrupt Quorum-Sensing Regulated Pathogenicity of Pseudomonas Aeruginosa. Sci. Rep. 2015, 5, 16158. 10.1038/srep16158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goluszko P.; Nowicki B.; Goluszko E.; Nowicki S.; Kaul A.; Pham T. Association of Colony Variation in Serratia Marcescens with the Differential Expression of Protease and Type 1 Fimbriae. FEMS Microbiol. Lett. 1995, 133, 41–45. 10.1111/j.1574-6968.1995.tb07858.x. [DOI] [PubMed] [Google Scholar]
- Lau G. W.; Hassett D. J.; Ran H.; Kong F. The Role of Pyocyanin in Pseudomonas Aeruginosa Infection. Trends Mol. Med. 2004, 10, 599–606. 10.1016/j.molmed.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Liu G. Y.; Nizet V. Color Me Bad: Microbial Pigments as Virulence Factors. Trends Microbiol. 2009, 17, 406–413. 10.1016/j.tim.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan S.; Ravindran D.; Arunachalam K.; Arumugam V. R. Inhibition of Quorum Sensing-Dependent Biofilm and Virulence Genes Expression in Environmental Pathogen Serratia Marcescens by Petroselinic Acid. Antonie van Leeuwenhoek 2018, 111, 501–515. 10.1007/s10482-017-0971-y. [DOI] [PubMed] [Google Scholar]
- Kida Y.; Inoue H.; Shimizu T.; Kuwano K. Serratia Marcescens Serralysin Induces Inflammatory Responses through Protease-Activated Receptor 2. Infect. Immun. 2007, 75, 164–174. 10.1128/IAI.01239-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salini R.; Pandian S. K. Interference of Quorum Sensing in Urinary Pathogen Serratia Marcescens by Anethum Graveolens. Pathog. Dis. 2015, 73, ftv038. 10.1093/femspd/ftv038. [DOI] [PubMed] [Google Scholar]
- Alagely A.; Krediet C. J.; Ritchie K. B.; Teplitski M. Signaling-Mediated Cross-Talk Modulates Swarming and Biofilm Formation in a Coral Pathogen Serratia Marcescens. ISME J. 2011, 5, 1609–1620. 10.1038/ismej.2011.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompilio A.; Crocetta V.; De Nicola S.; Verginelli F.; Fiscarelli E.; Di Bonaventura G. Cooperative Pathogenicity in Cystic Fibrosis: Stenotrophomonas Maltophilia Modulates Pseudomonas Aeruginosa Virulence in Mixed Biofilm. Front. Microbiol. 2015, 6, 951. 10.3389/fmicb.2015.00951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickel J. C.; Ruseska I.; Wright J. B.; Costerton J. W. Tobramycin resistance of Pseudomonas aeruginosa cells growing as a biofilm on urinary catheter material. Antimicrob Agents Chemother 1985, 27, 619–624. 10.1128/AAC.27.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-H.; Kim Y.-G.; Cho H. S.; Ryu S. Y.; Cho M. H.; Lee J. Coumarins Reduce Biofilm Formation and the Virulence of Escherichia Coli O157:H7. Phytomedicine 2014, 21, 1037–1042. 10.1016/j.phymed.2014.04.008. [DOI] [PubMed] [Google Scholar]
- Ahmed M.; Bird S.; Wang F.; Palombo E. A. In Silico Investigation of Lactone and Thiolactone Inhibitors in Bacterial Quorum Sensing Using Molecular Modeling. Int. J. Chem. 2013, 5, 9–16. 10.5539/ijc.v5n4p9. [DOI] [Google Scholar]
- Gould T. A.; Schweizer H. P.; Churchill M. E. A. Structure of the Pseudomonas Aeruginosa Acyl-Homoserinelactone Synthase LasI. Mol. Microbiol. 2004, 53, 1135–1146. 10.1111/j.1365-2958.2004.04211.x. [DOI] [PubMed] [Google Scholar]
- Sybiya Vasantha Packiavathy I. A.; Agilandeswari P.; Musthafa K. S.; Karutha Pandian S.; Veera Ravi A. Antibiofilm and Quorum Sensing Inhibitory Potential of Cuminum Cyminum and Its Secondary Metabolite Methyl Eugenol against Gram Negative Bacterial Pathogens. Food Res. Int. 2012, 45, 85–92. 10.1016/j.foodres.2011.10.022. [DOI] [Google Scholar]
- Wade D. S.; Calfee M. W.; Rocha E. R.; Ling E. A.; Engstrom E.; Coleman J. P.; Pesci E. C. Regulation of Pseudomonas Quinolone Signal Synthesis in Pseudomonas Aeruginosa. J. Bacteriol. 2005, 187, 4372–4380. 10.1128/JB.187.13.4372-4380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath S.; Wade D. S.; Pesci E. C. Dueling Quorum Sensing Systems in Pseudomonas Aeruginosa Control the Production of the Pseudomonas Quinolone Signal (PQS). FEMS Microbiol. Lett. 2004, 230, 27–34. 10.1016/S0378-1097(03)00849-8. [DOI] [PubMed] [Google Scholar]
- Spencer J.; Murphy L. M.; Conners R.; Sessions R. B.; Gamblin S. J. Crystal Structure of the LasA Virulence Factor from Pseudomonas Aeruginosa: Substrate Specificity and Mechanism of M23 Metallopeptidases. J. Mol. Biol. 2010, 396, 908–923. 10.1016/j.jmb.2009.12.021. [DOI] [PubMed] [Google Scholar]
- Gupta P.; Sarkar A.; Sandhu P.; Daware A.; Das M. C.; Akhter Y.; Bhattacharjee S. Potentiation of antibiotic againstPseudomonas aeruginosabiofilm: a study with plumbagin and gentamicin. J. Appl. Microbiol. 2017, 123, 246–261. 10.1111/jam.13476. [DOI] [PubMed] [Google Scholar]
- Siddiqui S.; Ameen F.; Jahan I.; Nayeem S. M.; Tabish M. A Comprehensive Spectroscopic and Computational Investigation on the Binding of the Anti-Asthmatic Drug Triamcinolone with Serum Albumin. New J. Chem. 2019, 43, 4137–4151. 10.1039/C8NJ05486J. [DOI] [Google Scholar]
- Khan M. S.; Qais F. A.; Rehman M. T.; Ismail M. H.; Alokail M. S.; Altwaijry N.; Alafaleq N. O.; AlAjmi M. F.; Salem N.; Alqhatani R. Mechanistic Inhibition of Non-Enzymatic Glycation and Aldose Reductase Activity by Naringenin: Binding, Enzyme Kinetics and Molecular Docking Analysis. Int. J. Biol. Macromol. 2020, 159, 87–97. 10.1016/j.ijbiomac.2020.04.226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.