Abstract
Coronavirus disease (COVID-19) is an acute infectious disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Human SP-D (surfactant protein D) is known to interact with the spike protein of SARS-CoV, but its immune surveillance against SARS-CoV-2 is not known. The current study aimed to examine the potential of a recombinant fragment of human SP-D (rfhSP-D) as an inhibitor of replication and infection of SARS-CoV-2. The interaction of rfhSP-D with the spike protein of SARS-CoV-2 and human ACE-2 (angiotensin-converting enzyme 2) receptor was predicted via docking analysis. The inhibition of interaction between the spike protein and ACE-2 by rfhSP-D was confirmed using direct and indirect ELISA. The effect of rfhSP-D on replication and infectivity of SARS-CoV-2 from clinical samples was assessed by measuring the expression of RdRp gene of the virus using quantitative PCR. In silico interaction studies indicated that three amino acid residues in the receptor-binding domain of spike protein of SARS-CoV-2 were commonly involved in interacting with rfhSP-D and ACE-2. Studies using clinical samples of SARS-CoV-2–positive cases (asymptomatic, n = 7; symptomatic, n = 8) and negative control samples (n = 15) demonstrated that treatment with 1.67 μM rfhSP-D inhibited viral replication by ∼5.5-fold and was more efficient than remdesivir (100 μM) in Vero cells. An approximately two-fold reduction in viral infectivity was also observed after treatment with 1.67 μM rfhSP-D. These results conclusively demonstrate that the rfhSP-D mediated calcium independent interaction between the receptor-binding domain of the S1 subunit of the SARS-CoV-2 spike protein and human ACE-2, its host cell receptor, and significantly reduced SARS-CoV-2 infection and replication in vitro.
Keywords: SARS-CoV-2, COVID-19, surfactant protein D, spike protein, entry inhibitor
The coronavirus disease (COVID-19) pandemic, caused by the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) (1, 2), has affected ∼58 million people across the globe and has claimed more than 1 million lives within its first year (3). The SARS-CoV-2 spike protein (S protein) is cleaved into S1 subunit, which is involved in host receptor binding, and S2 subunit, which is involved in membrane fusion, by the host’s TMPRSS2 (transmembrane serine protease 2) (4). This priming of the S protein by host proteases enables it to bind with the ACE2 (angiotensin-converting enzyme 2) receptor on the nasopharyngeal epithelial cells, leading to its entry into the host cell (4). Although vaccines against the virus are being continually developed and trialed, therapeutic strategies to treat severe COVID-19 patients are limited and comprise remdesivir and dexamethasone (5).
The innate immune system plays a crucial role against SARS-CoV-2 infection; the majority of infected individuals purge the virus within a few days with minimal involvement of adaptive immune response (6). Collectins are a group of humoral pattern recognition receptors, of which human SP-D (pulmonary surfactant protein D) is known to act as a potent viral entry inhibitor, including against human immunodeficiency virus 1 and influenza A virus (7, 8). The primary structure of SP-D is characterized by an N-terminus that is involved in multimerization, a triple-helical collagenous region made up of Gly-X-Y repeats, an α-helical coiled-coil neck region, and a C-terminal C-type lectin or carbohydrate recognition domain (CRD) (9). The protective effects of SP-D against a range of bacterial, viral, and fungal pathogens, leading to their agglutination, growth inhibition, enhanced phagocytosis, neutralization, and modulation of immune responses, are well documented (9, 10).
During the SARS-CoV epidemic in 2002, elevated concentrations of SP-D were reported in the serum of the patients infected with highly pathogenic β-CoV SARS-CoV (11). Purified SP-D has been shown to bind to the receptor-binding domain (RBD) of the glycosylated S protein of SARS-CoV, which shares 74% homology with the RBD of SARS-CoV-2 (12). In addition, SP-D also binds α-CoV and HCoV-229E and inhibits infection in human bronchial epithelial cells (13). These mounting pieces of evidence encouraged exploration of the therapeutic potential of SP-D in patients with COVID-19.
In this study, we used a well-characterized recombinant fragment of human SP-D (rfhSP-D) comprising homotrimeric neck and CRD region to study its protective effect against SARS-CoV-2 infection. As the recombinant fragment has the advantages of a smaller size to reach the distal lung locations and higher resistance to proteases and collagenases over the full-length SP-D, we evaluated the interaction of rfhSP-D with RBD and the spike of SARS-CoV-2, and its inhibitory potential against infection and replication of SARS-CoV-2 in clinical samples.
Some of the results from this study have been previously reported in the form of a preprint (bioRxiv, [18 December 2020] https://doi.org/10.1101/2020.12.18.423415).
Methods
An additional detailed section for performing these experiments is provided as a supplement.
Clinical Samples
The clinical samples (throat and nasal swabs) (n = 15) (Table 1) were obtained from symptomatic contacts of lab-confirmed cases (category 2) (n = 2), hospitalized patients with severe acute respiratory infections (category 4) (n = 3), asymptomatic direct and high-risk contacts of lab-confirmed cases (category 5a) (n = 7), and hospitalized patients with symptomatic influenza-like illness (category 6) (n = 3), who had tested positive by RT-PCR for SARS-CoV-2. Samples (n = 15) that tested negative by RT-PCR for SARS-CoV-2 were used as control. The inclusion criteria for the cases were age (between 18 and 45 yr) and a cycle threshold value (Ct value) between 10 and 15 (18–45 yr and an undetectable viral load for control samples). The two groups showed no significant difference in their age and sex distribution. The 50% tissue culture infective dose (TCID50) of the clinical samples was estimated using a template that used the Spearman and Kärber method for calculation (14). The estimated TCID50 value was further confirmed via MTT assay. Briefly, 5 × 104 Vero cells in Vero growth media (minimal essential medium [MEM] Glutamax+10% FBS, 1% penicillin-streptomycin, and 1% sodium pyruvate; Gibco) were grown in a 96-well plate overnight. The clinical samples from the cases and controls at various dilutions were added to the cells and incubated for 1 hour, wells were washed with PBS twice, and fresh Vero growth medium was added to the cells and incubated (96 h; 37°C; 5% CO2). An MTT assay was performed to assess cell viability.
Table 1.
Characteristics of Clinical Samples Used in the Study
| Sample | Age (yr) | Sex | Ct Value for SARS-CoV-2 E Gene | Ct Value for SARS-CoV-2 RdRp Gene | Category |
|---|---|---|---|---|---|
| 1S | 32 | M | 18.5 | 15.46 | 2 |
| 2S | 41 | F | 17.48 | 15.36 | 6 |
| 3S | 44 | M | 13.46 | 12.01 | 6 |
| 4S | 39 | F | 13.22 | 12.67 | 2 |
| 5S | 36 | M | 12.08 | 14.55 | 6 |
| 6S | 42 | F | 12.28 | 9.4 | 4 |
| 7S | 37 | M | 16.26 | 15.42 | 4 |
| 8S | 28 | F | 17.15 | 15.62 | 4 |
| 9AS | 32 | M | 10.45 | 10.8 | 5a |
| 10AS | 31 | F | 12.32 | 12.4 | 5a |
| 11AS | 26 | M | 15.33 | 12.52 | 5a |
| 12AS | 35 | F | 10.59 | 12.4 | 5a |
| 13AS | 38 | M | 15.77 | 12.89 | 5a |
| 14AS | 29 | F | 18.07 | 10.72 | 5a |
| 15AS | 33 | M | 15.64 | 13.81 | 5a |
| 1C | 33 | M | Nd | Nd | Control |
| 2C | 39 | F | Nd | Nd | Control |
| 3C | 45 | M | Nd | Nd | Control |
| 4C | 39 | F | Nd | Nd | Control |
| 5C | 34 | M | Nd | Nd | Control |
| 6C | 44 | F | Nd | Nd | Control |
| 7C | 35 | M | Nd | Nd | Control |
| 8C | 30 | F | Nd | Nd | Control |
| 9C | 30 | M | Nd | Nd | Control |
| 10C | 32 | F | Nd | Nd | Control |
| 11C | 29 | M | Nd | Nd | Control |
| 12C | 33 | F | Nd | Nd | Control |
| 13C | 40 | M | Nd | Nd | Control |
| 14C | 32 | F | Nd | Nd | Control |
| 15C | 30 | M | Nd | Nd | Control |
Definition of abbreviations: Ct = cycle threshold; Nd = not detected; RdRp = RNA-dependent RNA polymerase; SARS-CoV-2 = severe acute respiratory syndrome coronavirus 2.
In Silico Analysis of rfhSP-D Interaction with SARS-CoV-2 S Protein and Human ACE-2
Structural information with respect to the molecular interactions between S protein and human ACE-2 (hACE-2) is available (15, 16). The rfhSP-D trimer (Protein Data Bank [pdb] identification 1PW9) was blind-docked with 1) RBD of S protein in open conformation (pdb identification 6VYB), and 2) dimeric hACE-2 (pdb identification 6VW1). Top 100 docked poses selected were further refined using the FireDock web server (17, 18) for calculating global free energy. The top five refined structures were filtered on the basis of interactions between the receptor-binding motif (RBM) of S protein, C-type lectin domain (aa 240–355) of rfhSP-D, and N-terminal of hACE-2.
The effect of binding of trimeric rfhSP-D to S protein and dimeric ACE-2 on ACE-2–S protein interaction was evaluated by further docking the complex of 1) S protein and rfhSP-D with hACE-2 and 2) hACE-2 and rfhSP-D with S protein. Patchdock web server (19, 20) was used for all docking experiments.
In Silico Interaction of Single-Residue rfhSP-D Mutants with S Protein and hACE-2
The importance of rfhSP-D residues predicted to interact with ACE-2 and S protein was further validated using single-residue mutations in the docked complex using the mCSM-PPI2 web server (21). Each of the rfhSP-D residues involved in interaction with virus-binding hotspot of ACE-2 and RBD of S protein were mutated to the standard amino acids, and its effect on the binding energy of the complex was assessed.
ELISA
The rfhSP-D used was expressed and purified from Escherichia coli, as described previously (22, 23). rfhSP-D binding to S protein or RBD was analyzed using the SARS-CoV-2 Inhibitor Screening Kit (EP-105; ACRO Biosystems).
Microtiter wells were coated with 0.3 μg/ml (0.54 nM) S protein and incubated with rfhSP-D (20, 10, and 5 μg/ml or 0.334, 0.167, and 0.083 μM), or full-length (FL) SP-D (20 μg/ml or 0.038 μM) for 1 hour at 37°C in PBS supplemented with 5 mM CaCl2. After blocking and washing, the wells were incubated with polyclonal or monoclonal antibodies against SP-D (1 mg/ml), respectively, and probed with their respective secondary antibodies conjugated with horseradish peroxidase (HRP). The binding was detected using 3,3′,5,5′-tetramethylbenzidine substrate, and absorbance was recorded at 450 nm. Similarly, to ascertain the calcium dependence of the interaction, a binding assay was performed with 10 mM EDTA.
rfhSP-D/FL SP-D and ACE-2 interaction was evaluated by coating rfhSP-D (0.1 μg/ml or 1.67 nM)/FL SP-D (0.1 μg/ml or 0.19 nM) and probing with biotinylated hACE-2 (0.12, 0.06, and 0.00 μg/ml or 0.52, 0.26, 0.0 nM), followed by the addition of streptavidin tagged with HRP.
To assess whether rfhSP-D inhibited the interaction between S protein/RBD and biotinylated hACE-2, rfhSP-D (5, 1, and 0 μg/ml or 83.5, 16.7, and 0.0 nM) was preincubated with the coated S protein/RBD, followed by the addition of biotinylated ACE-2. The S protein–hACE-2 binding was probed with the HRP-tagged streptavidin antibody. rfhSP-D (5 μg/ml or 83.5 nM) in 10 mM EDTA was also used.
Vero Cell Replication Assay
Vero cells (CCL–81; American Type Culture Collection) (5 × 104) were cultured in serum-free MEM. SARS-CoV-2–positive clinical samples (100 TCID50/well; multiplicity of infection [MOI], 0.01) were preincubated with rfhSP-D (0 μg/ml, 50 μg/ml, and 100 μg/ml or 0, 0.835, and 1.67 μM) in MEM containing 5 mM CaCl2 for 1 hour at room temperature and 1 hour at 4°C (7). SARS-CoV-2–negative clinical samples (volume equivalent to 100 TCID50 of the age- and sex-matched SARS-CoV-2–positive sample/well; MOI, 0.01) were used as a control. This pretreated or untreated virus was added to the cells and incubated for 1 hour at 37°C (5% CO2). After PBS washes, infection medium (MEM + 0.3% BSA) was added and incubated for 24 hours to assess viral replication. Total RNA was extracted from the cell pellet using the Perkin Elmer automated extractor. Real-time qPCR for SARS-CoV-2 was performed using Pathodetect kits (MyLabs). For the replication analysis, the Ct value for SARS-CoV-2 RdRp (RNA-dependent RNA polymerase) gene was used. Cells incubated with rfhSP-D without virus were used as the protein control.
Vero Cell Infection Assay
SARS-CoV-2–positive clinical samples (500 TCID50/well; MOI, 0.05) and SARS-CoV-2 negative clinical samples (equivalent volume) were treated with rfhSP-D (7). After adding the infection medium, Vero cells (5 × 105) were incubated for 2 hours and harvested, and real-time qPCR was performed, as described above.
Results
rfhSP-D Interacts with the S Protein of SARS-CoV-2 and Human ACE-2 In Silico
S protein is known to interact via the RBM (aa 455–508) in the RBD (aa 319–527) with virus-binding hotspot residues comprising Lys31, Glu35, and Lys353 of dimeric hACE-2 (15, 16). The structure of hACE2 receptor, co-crystallized with S protein of SARS-CoV-2, is available in RCSB (pdb identification 6VW1). The receptor (ACE-2) and ligand (S protein) were separated and docked to validate the docking protocol. The redocked complex of ACE-2 and S protein had root mean square deviation of 7.9 Å. The close agreement between the docked and crystal structures validated the docking protocol used in this study.
In the case of docked solutions for S protein and rfhSP-D (Figure E1 in the data supplement), the third-ranked docked pose with a binding energy of −20.63 kcal/mol exhibited rfhSP-D interactions with RBM residues Tyr449, Gln493, and Gln498, implying that rfhSP-D could bind to S protein in a manner that can inhibit ACE-2–S protein interaction (Table 2 and Figure 1). To ascertain this hypothesis, the complex of S protein with rfhSP-D was docked to ACE-2. S protein and rfhSP-D bound to ACE-2 via common interacting residues.
Table 2.
Results of Docking of S Protein, ACE-2, and rfhSP-D
| S. Number | Receptor | Ligand | Binding Energy (kcal/mol) | Interactions |
|
|---|---|---|---|---|---|
| Receptor* | Ligand† | ||||
| 1 | ACE-2 | S Protein | Crystal structure | Ser19 | Ala475 |
| Gln24 | Asn487 | ||||
| Lys31 | Phe456, Glu484, Tyr489, Gln493 | ||||
| His34 | Leu455, Tyr453 | ||||
| Glu35 | Gln493 | ||||
| Glu37 | Tyr505 | ||||
| Asp38 | Tyr449 | ||||
| Tyr41 | Thr500 | ||||
| Gln42 | Gln498 | ||||
| Met82 | Phe486‡ | ||||
| Tyr83 | Gly496, Asn487, Tyr489 | ||||
| Glu329 | Arg439 | ||||
| Lys353 | Tyr505, Gly502 | ||||
| Gly354 | Tyr505 | ||||
| 2 | rfhSP-D | S protein (open) | −20.63 | Gln219 | Gln493 |
| His220 | Tyr449 | ||||
| Ala223 | Gln493, Phe490 | ||||
| Ser226 | Ser494 | ||||
| Lys229 | Tyr449 | ||||
| Ser239 | Asn450 | ||||
| Gly241 | Asn448, Gln498 | ||||
| Glu242 | Gln498 | ||||
| Gln263 | Arg346 | ||||
| Thr308 | Arg466 | ||||
| 3 | ACE-2 | rfhSP-D | −24.30 | Ser19 | Ser328 |
| Asp30 | Thr255 | ||||
| Lys31 | Thr305 | ||||
| His34 | Gln258 | ||||
| Glu35 | Pro307, Gly309 | ||||
| Glu75 | Lys299 | ||||
| Gly319 | Ala275 | ||||
| Pro321 | Ala275 | ||||
| Gln552 | Ala274, Tyr314 | ||||
Definition of abbreviations: ACE-2 = angiotensin-converting enzyme 2; rfhSP-D = recombinant fragment of human SP-D; S protein = Spike protein.
The ACE2 residues in bold typeface interact with both S protein and rfhSP-D (docked structure).
The S protein residues in bold are predicted to be part of the common binding site for ACE-2 and rfhSP-D.
The structural coordinates of Phe486 are missing in the open conformation S protein (Protein Data Bank identification 6VYB).
Figure 1.
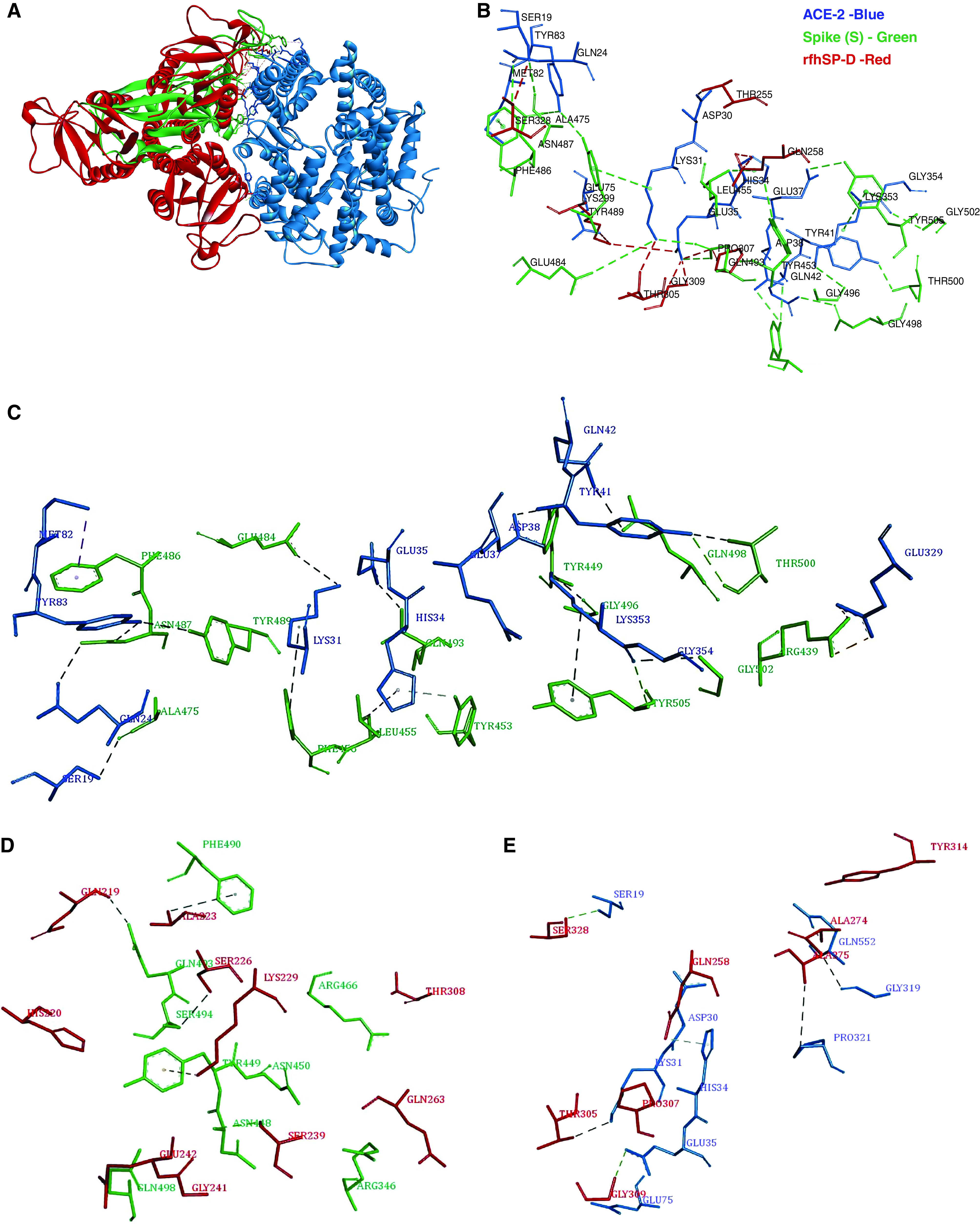
Tripartite interaction between spike protein (S protein) (green), recombinant fragment of human SP-D (surfactant protein D) (rfhSP-D) (red), and ACE-2 (angiotensin-converting enzyme 2) (blue) (A and B). ACE-2 residues Ser19, Lys31, Glu35 and His34 interact with both S protein and rfhSP-D. The interactions between S protein and ACE-2 are deduced from the crystal structure (Protein Data Bank identification 6VW1), and those between rfhSP-D and ACE-2 protein are based on docked complexes. B is a zoomed view of A. Individual intermolecular interactions between (C) S protein (green) and ACE-2 (blue), (D) S protein (green) and rfhSP-D (red), and (E) rfhSP-D (red) and ACE-2 (blue). The S protein residues Tyr449, Gln493, and Gln498 participate in intermolecular interactions with both ACE-2 and rfhSP-D.
The top-ranked docked structure of ACE-2 and rfhSP-D had a binding energy of −24.30 kcal/mol. In this pose, rfhSP-D interacted with the virus-binding hotspot residues Ser19, Lys31, His34, and Glu35 of ACE-2, implying that rfhSP-D could bind to ACE-2 in a manner that can inhibit ACE2–S protein interaction (Table 2 and Figure 1). To corroborate this postulate, the complex of ACE-2 with rfhSP-D was docked to S protein. The top-ranked pose of ACE-2–rfhSP-D complex docked with open S protein had a binding energy of −33.01 kcal/mol and several common interactions between rfhSP-D and ACE-2 with S protein (Figure E1). The docking experiments led us to infer that rfhSP-D could bind to both ACE-2 as well as S protein and prevent ACE-2–S protein interaction.
Single-Residue In Silico Mutants of rfhSP-D Displayed Lower Binding Affinities to S Protein and ACE-2
In silico mutational analysis was performed to evaluate the functional importance of residues of rfhSP-D that were identified to be involved in the intermolecular interactions with ACE-2 and S protein through docking studies (Table 2 and Figure 1). The rfhSP-D residues Ser328, Gly309, Pro307, Thr305, and Gln258 of the ACE-2–rfhSP-D complex and Lys229, Glu242, Gly241, His220, and Gln219 of S protein–rfhSP-D complex were individually substituted to standard amino acids. All the mutants demonstrated lower binding affinities, except for substitutions with similar physicochemical properties (Figure 2), corroborating the importance of the mutated residues in intermolecular interactions.
Figure 2.
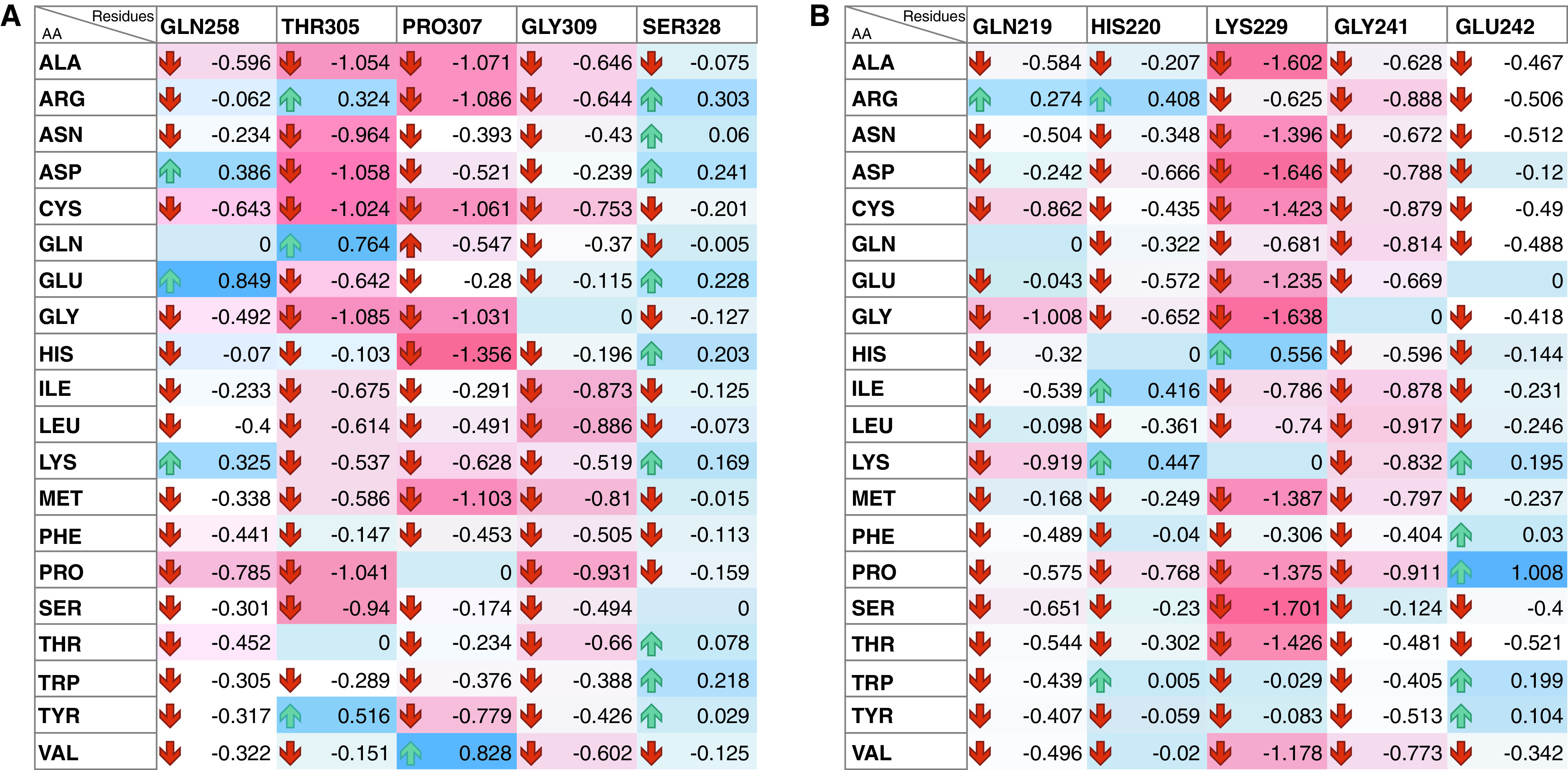
Heatmap representation of effect of single-residue mutations of rfhSP-D (y axis) on binding energy of docked rfhSP-D complexed with (A) ACE-2 and (B) S protein using mCSM-PPI2 web server. Each of the rfhSP-D residues involved in interaction with virus-binding hotspot of ACE-2 and the receptor-binding motif of the S protein were mutated to the standard amino acids, and its effect on binding energy of the complex was assessed. Most of the mutations led to decrease in the binding affinity (red cells). Few of the conserved substitutions led to increase in the binding affinity (blue cells), highlighting the likely functional importance of the mutated residues.
rfhSP-D Binds to the Immobilized S Protein of SARS-CoV-2 as Well as hACE-2
The possible binding between rfhSP-D and S protein hinted by the docking analysis was confirmed in vitro via an indirect ELISA. rfhSP-D was found to bind the immobilized S protein in a dose-dependent manner (Figure 3A). However, a significant difference in the absorbance was observed based on the specificity of the primary antibody used. S protein–rfhSP-D binding that was probed with the polyclonal antibody against SP-D reported a significantly higher absorbance when compared with the wells that were probed with a monoclonal antibody directed against the CRD of SP-D. This difference suggests the involvement of CRD of rfhSP-D with the S protein, and, therefore, the CRD was not available for interaction with the monoclonal antibody. S protein was also found to bind to the FL SP-D. The treatment of rfhSP-D with 10 mM EDTA did not significantly alter the binding of rfhSP-D and FL SP-D to the S protein (Figure 3B). Hence, rfhSP-D and FL SP-D bind to the S protein in a dose-dependent, but calcium-independent, manner. A similar parallel experiment revealed that rfhSP-D bound ACE-2 in a dose-dependent manner (Figure 3C).
Figure 3.
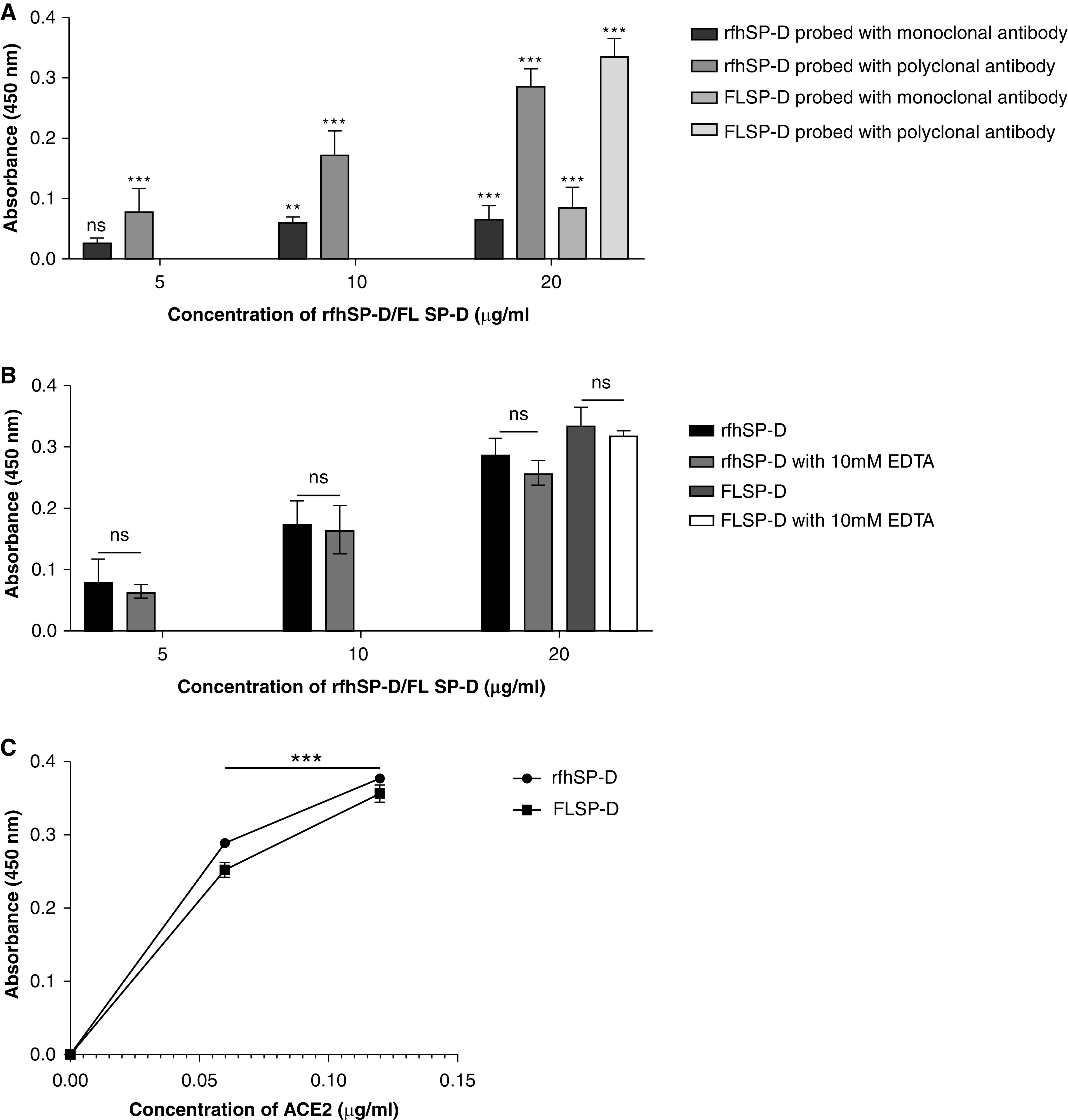
rfhSP-D binds to the immobilized S protein of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2); immobilized rfhSP-D binds to human ACE-2 (hACE-2) in a dose-dependent, but calcium-independent, manner. ELISA showing binding of rfhSP-D to the immobilized S protein. Microtiter wells were coated with 0.3 μg/ml (0.54 nM) of S protein. rfhSP-D (20, 10, and 5 μg/ml or 0.334, 0.167, and 0.083 μM in PBS with 5 mM CaCl2) were added to the wells. Full-length (FL) SP-D (20 μg/ml or 0.038 μM) was also used in a similar manner. BSA (20 μg/ml) was used as a non-specific protein control (mean of the normalized triplicates ± SEM = 0.07 ± 0.006 with polyclonal antibody and 0.025 ± 0.006 with monoclonal antibody). (A) S protein–SP-D binding was detected with either polyclonal or monoclonal antibodies against SP-D. (B) To assess the effect of calcium in the SP-D–S protein interaction, rfhSP-D (20, 10, and 5 μg/ml or 0.334, 0.167, and 0.083 μM) and FL SP-D (20 μg/ml or 0.038 μM), either with or without 10 mM EDTA, was used in a similar manner and probed with polyclonal antibodies against SP-D. (C) The binding of immobilized rfhSP-D to hACE-2 was assessed by coating microtiter wells with 0.1 μg/ml of FL SP-D (1.9 nM) or rfhSP-D (16.7 nM). BSA (0.1 μg/ml) was used as a non-specific protein control (mean of the normalized triplicates ± SEM = 0.006 ± 0.005). Decreasing concentration of hACE-2 (0.12, 0.06, and 0.00 μg/ml or 0.52, 0.26, and 0.0 nM) was added to the wells. The SP-D–hACE-2 binding was detected using streptavidin-HRP. The background was subtracted from all data points. The data were expressed as the mean of triplicates ± SD. Significance was determined using the two-way ANOVA (n = 3) test. **P < 0.05 and ***P < 0.0001. ns = no significance.
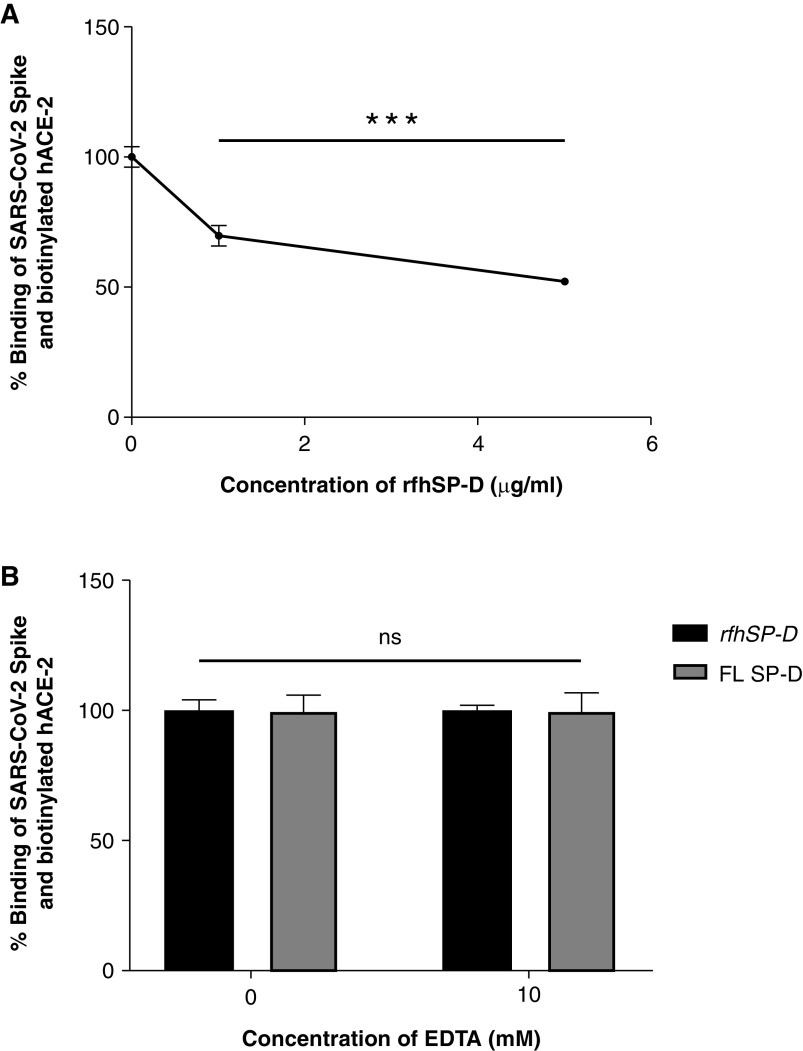
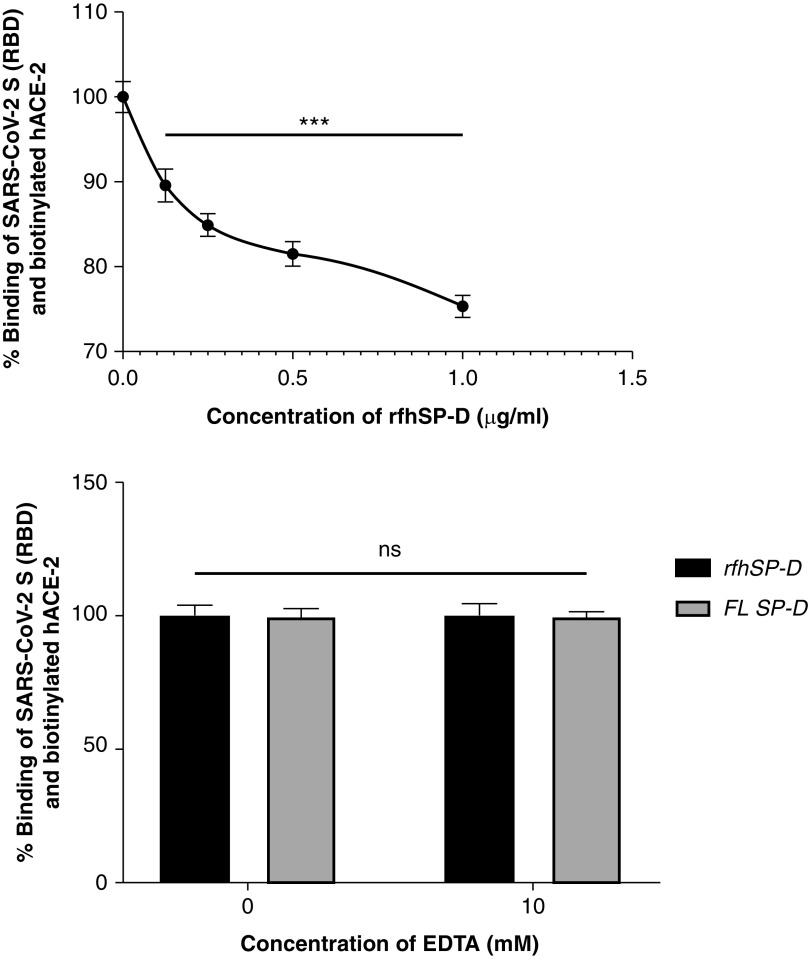
rfhSP-D Inhibits the Interaction of S Protein and Its RBD with Biotinylated hACE-2 in a Calcium-Independent Manner
Because rfhSP-D was found to bind to the S protein as well as ACE-2, and both rfhSP-D and ACE-2 were predicted to share the same binding site on the S protein, rfhSP-D–mediated inhibition of the interaction between the RBD of the S protein of SARS-CoV-2 and ACE-2 was assessed using a colorimetric ELISA.
The wells were coated with either the S protein or its RBD domain that was preincubated with rfhSP-D followed by addition of biotinylated hACE-2. The functionality and the range of the assay were initially assessed by verifying whether the assay could detect the binding of hACE-2 at concentrations of 0.12 μg/ml and 0.06 μg/ml. The binding occurred in a dose-dependent manner, confirming that the assay could detect binding between S protein or its RBD domain with hACE-2 at a concentration as low as 60 ng/ml (Figure E2). A decrease in binding between the S protein and hACE-2 was observed as the concentration of rfhSP-D increased (Figures 4 and 5). Approximately 50% decrease in S protein–hACE-2 binding was observed as rfhSP-D concentration increased fivefold (Figure 4A). A similar result was observed between the binding of the RBD of S protein and hACE-2. An eightfold increase in the concentration of rfhSP-D was found to decrease RBD–hACE-2 interaction by ∼25% (Figure 5A). No significant difference was observed when binding experiments were carried out with and without 10 mM EDTA in terms of rfhSP-D–mediated S protein/RBD–hACE-2 interaction (Figures 4B and 5B). Hence, rfhSP-D–mediated inhibition of the interaction between the RBD of the S protein or the S protein itself with biotinylated hACE-2 occurred in a calcium-independent manner.
Figure 4.
rfhSP-D inhibits the interaction between S protein of SARS-CoV-2 and biotinylated hACE-2 in a calcium-independent manner (A and B). Microtiter wells were coated with 0.3 μg/ml (0.54 nM) of S protein. (A) After blocking, rfhSP-D (5, 1, and 0 μg/ml or 83.5, 16.7, and 0.0 nM) was added and incubated for 1 hour, followed by probing with biotinylated hACE-2. (B) To assess the effect of calcium in the SP-D–mediated inhibition of S protein–hACE-2 interaction, 5 μg/ml or 83.5 nM of rfhSP-D and FL SP-D (5 μg/ml or 9.5 nM) with or without 10 mM EDTA was added. BSA (5 μg/ml) was used as nonspecific protein control (mean of the normalized triplicates ± SEM = 0.297 ± 0.005). S protein–hACE-2 binding was detected with streptavidin-HRP. Background was subtracted from all data points. The data were normalized with 100% S protein–hACE-2 binding being defined as the mean of the absorbance recorded from the control sample (0 μg/ml rfhSP-D). The data were presented as the mean of the normalized triplicates ± SEM. Significance was determined using the one-way ANOVA (n = 3). ***P < 0.0001.
Figure 5.
rfhSP-D inhibits the interaction between the receptor-binding domain (RBD) of the S protein of SARS-CoV-2 and biotinylated hACE-2 in a calcium-independent manner. Microtiter wells were coated with 0.1 μg/ml (2.5 nM) of S protein RBD. After blocking, decreasing concentrations of rfhSP-D (1, 0.5, 0.25, 0.125, and 0 μg/ml or 16.7, 8.35, 4.18, and 2.09 nM in PBS with 5 mM CaCl2) (A) were incubated for 1 hour, followed by probing with biotinylated hACE-2. To assess the effect of calcium in the rfhSP-D–mediated inhibition of S protein RBD–hACE-2 interaction (B), 5 μg/ml (83.5 nM) of rfhSP-D or FL SP-D (5 μg/ml or 9.5 nM) with or without 10 mM EDTA was used. BSA (5 μg/ml) was used as a nonspecific protein control (mean of the normalized triplicates ± SEM = 0.894 ± 0.006 [this data is not plotted in the figure]). S protein RBD–hACE-2 binding was detected using streptavidin-HRP. Background was subtracted from all data points. The data obtained were normalized with 100% S protein RBD–hACE-2 binding being defined as the mean of the absorbance recorded from the control sample (0 μg/ml rfhSP-D). The data were presented as the mean of the normalized triplicates ± SEM. Significance was determined using the one-way ANOVA (n = 3). ***P < 0.0001.
rfhSP-D Treatment Inhibits SARS-CoV-2 Infection and Replication
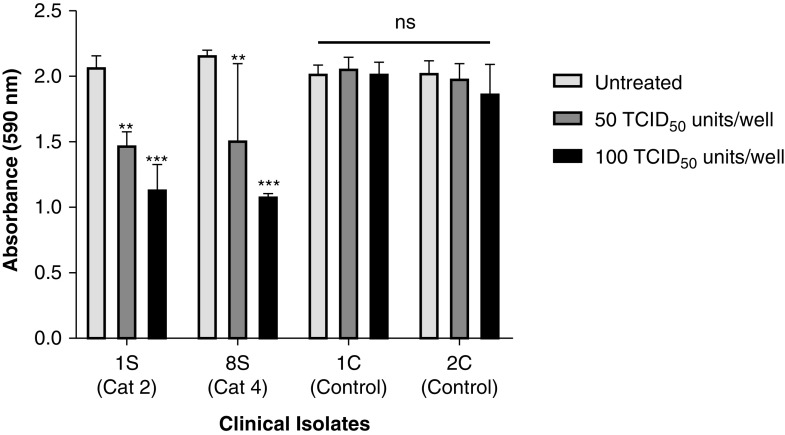
As rfhSP-D is known to induce apoptosis in cancer and immortalized cells (22, 24 –26), the effect of rfhSP-D on Vero cells was assessed using an MTT assay. rfhSP-D treatment had no significant effect on the viability of Vero cells (Figure E3). At the outset, the TCID50 values of the clinical samples were obtained by evaluating the cytopathic effects using an MTT assay. As expected, when Vero cells were challenged with 100 TCID50 or 50 TCID50 of viral samples from SARS-CoV-2 clinical samples, a 50% or 25% reduction in cell viability was observed, respectively, compared with the viability of uninfected Vero cells, confirming the assayed TCID50 values (Figure 6). The control samples showed no significant difference in the cell viability from the uninfected Vero cells when the control sample volumes equivalent to 100 TCID50 and 50 TCID50 of the matched clinical cases were used.
Figure 6.
Determination of 50% tissue culture infective dose (TCID50) value of the clinical samples in Vero cells using an MTT assay. Vero cells (5 × 104/well) were seeded in complete MEM in 96-well culture plates and grown overnight at 37°C with 5% CO2. Swab samples of 15 confirmed cases of coronavirus disease (COVID-19), including symptomatic contacts of laboratory-confirmed cases (category [Cat] 2) (n = 2), hospitalized patients with severe acute respiratory infections (Cat 4) (n = 3), asymptomatic direct and high-risk contacts of laboratory-confirmed cases (Cat 5a) (n = 7), and hospitalized patients with symptomatic influenza-like illness (Cat 6) (n = 3) who had tested positive by RT-PCR test for SARS-CoV-2, and 15 controls (at different dilutions/well) were added to the cells and incubated for 1 hour. The supernatants were removed, and the wells were washed twice with sterile PBS. Fresh complete MEM was added to the wells, and the cells were incubated for 96 hours. Viability of the cells was evaluated using an MTT assay. MTT (0.5 mg/ml)-containing medium was added to the wells for 4 hours. The supernatants were removed, and cells were lysed using DMSO. Absorbance was measured at 590 nm. The data obtained were normalized with 100% cell viability being defined as the mean of the absorbance recorded from the control sample (0 TCID50/well), and TCID50 units were evaluated in each sample. The same assay was used to validate the cytopathic effects of 100 TCID50 and 50 TCID50 units of the samples. Data for cytopathic effects of 100 TCID50 units for all 15 cases and 15 controls have been provided in the Table E1. The representative data for cases (n = 2) and controls (n = 2) are presented as the mean of the normalized triplicates ± SEM. Significance was determined using the two-way ANOVA (n = 3) test. **P < 0.01 and ***P < 0.0001.
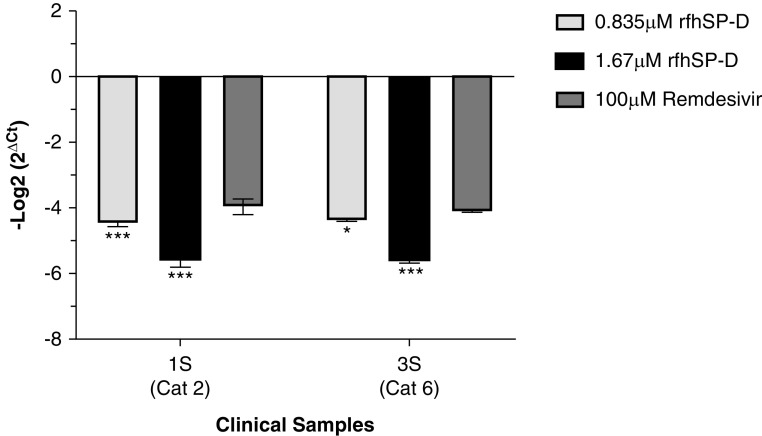
The effect of rfhSP-D on the replication of SARS-CoV-2 (100 TCID50/well; MOI, 0.01) in Vero cells was evaluated by measuring the concentrations of the RdRp gene of SARS-CoV-2 by real-time qPCR 24 hours after infection. Pretreatment of the positive samples (n = 15), comprising SARS-CoV-2 with rfhSP-D, led to a reduction in RdRp expression in a dose-dependent manner (Figure 7 and Table E1). The pretreatment of samples from all categories of SARS-CoV-2–positive cases (as representatives, the Figure 7 shows the data for 1S [category 2] and 3S [category 6]) with 0.835 μM rfhSP-D led to ∼4.5-fold reduction (−4.5 log2) of RdRp transcript compared with the untreated positive sample–challenged Vero cells. There was no significant difference in the Ct values of the RdRp gene from the untreated and control sample–treated Vero cells. Similarly, pretreatment with 1.67 μM rfhSP-D resulted in ∼5.5-fold reduction (−5.5 log2) of RdRp mRNA expression. Remdesivir, one of the antiviral drugs proposed for COVID-19, which functions by inhibiting viral RNA synthesis, was found to inhibit SARS-CoV-2 replication by ∼4-fold (−4 log2). Hence, rfhSP-D blocked SARS-CoV-2 infection in addition to inhibiting the replication of SARS-CoV-2 significantly better than remdesivir at both tested concentrations (0.835 μM and 1.67 μM rfhSP-D).
Figure 7.
rfhSP-D pretreatment of SARS-CoV-2 significantly inhibited its replication. Vero cells (5 × 104/well) were seeded in complete MEM in 96-well culture plates and grown overnight at 37°C under 5% CO2. Cells were washed with sterile PBS twice. SARS-CoV-2 clinical samples (100 TCID50/well; MOI 0.01), including symptomatic contacts of laboratory-confirmed cases (Cat 2) (n = 2), hospitalized patients with severe acute respiratory infections (Cat 4) (n = 3), asymptomatic direct and high-risk contacts of laboratory-confirmed cases (Cat 5a) (n = 7), and hospitalized patients with symptomatic influenza-like illness (Cat 6) (n = 3) who had tested positive by RT-PCR test for SARS-CoV-2, were preincubated with rfhSP-D (0 μg/ml, 50 μg/ml, and 100 μg/ml or 0, 0.835, and 1.67 μM) in MEM containing 5 mM CaCl2 for 1 hour at room temperature. The pretreated or untreated virus in the sample was added to the cells and incubated for 1 hour at 37°C under 5% CO2. The wells were washed with PBS twice, and infection medium (MEM + 0.3% BSA) was added to the cells and incubated for 24 hours at 37°C. The supernatants were collected, and RNA was extracted by Perkin Elmer automated extractor and subjected to qRT-PCR for SARS-CoV-2. For control samples, the volume of the sample taken was equivalent to the volume of the case sample (100 TCID50) in which no RdRp expression was detected. The relative expression of RdRp was calculated using rfhSP-D–untreated cells (0 μM rfhSP-D), infected with respective samples as the calibrator. Data are provided in Table E1, and data of representative cases (n = 2) are presented as the mean of triplicates (n = 3). Error bars represent ± SEM. Significance (compared with 100 μM remdesivir) was determined using the two-way ANOVA test. *P < 0.05 and ***P < 0.0001.
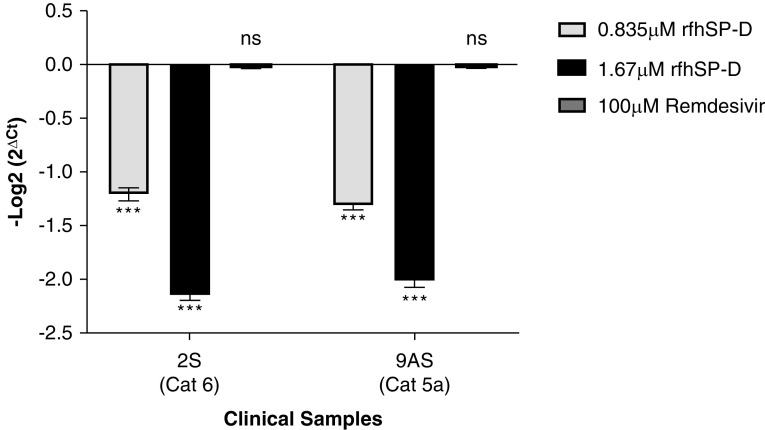
As rfhSP-D was found to interact with S protein and ACE-2, proteins that play an integral role in viral host cell recognition and entry, the role of rfhSP-D in viral infectivity was assessed in a similar manner to replication. Vero cells infected with SARS-CoV-2–positive samples (500 TCID50/well; MOI, 0.05) showed an rfhSP-D dose-dependent decrease in the expression levels of the RdRp gene 2 hours after infection (Figure 8 and Table E2). Clinical samples from all the categories of patients with SARS-CoV-2 (as representatives, Figure 8 shows the data from 2S [category 6] and 9AS [catagory 5a]) showed ∼1.25-fold reduction (−1.25 log2) or ∼2-fold reduction (−2 log2) in RdRp gene expression with the samples pretreated with either 0.835 μM or 1.67 μM, respectively, of rfhSP-D. Remdesivir was used as a control (remdesivir does not inhibit SARS-CoV-2 infection). Thus, pretreatment of SARS-CoV-2 in the clinical sample with rfhSP-D appears to make S protein unavailable to interact with the ACE-2 receptor on the host cell, thus reducing the infectivity of the virus and subsequent viral replication in a dose-dependent manner.
Figure 8.
rfhSP-D pretreatment of SARS-CoV-2 significantly inhibited its infectivity. Vero cells (5 × 105/well) were seeded in complete MEM in 12-well culture plates and grown overnight at 37°C under 5% CO2. Cells were washed with sterile PBS twice. SARS-CoV-2 clinical samples (500 TCID50/well, multiplicity of infection, 0.05), including symptomatic contacts of laboratory-confirmed cases (Cat 2) (n = 2), hospitalized patients with severe acute respiratory infections (Cat 4) (n = 3), asymptomatic direct and high-risk contacts of laboratory-confirmed cases (Cat 5a) (n = 7), and hospitalized patients with symptomatic influenza-like illness (Cat 6) (n = 3) who had tested positive by RT-PCR test for SARS-CoV-2, were preincubated with rfhSP-D (0 μg/ml, 50 μg/ml, and 100 μg/ml or 0, 0.835, and 1.67 μM) in MEM containing 5 mM CaCl2 for 1 hour at room temperature and 1 hour at 4°C. This pretreated or untreated virus-containing sample was added to the cells and incubated for 1 hour at 37°C under 5% CO2. The wells were washed with PBS twice, infection medium (MEM + 0.3% BSA) was added to the cells and incubated for 2 hours at 37°C under 5% CO2. The cells were scraped, and the media containing scraped cells were collected. RNA was extracted and subjected to RT-PCR for SARS-CoV-2. For control samples, the volume of the sample taken was equivalent to the volume of the case sample (500 TCID50); no RdRp expression was detected. The relative expression of RdRp was calculated by using rfhSP-D untreated cells (0 μM rfhSP-D) infected with respective samples as the calibrator. Data are provided in Table E1, and data for representative cases (n = 2) are presented as the mean of triplicates (n = 3). Error bars represent ± SEM. Significance (compared with control sample [cells + virus]) was determined using the two-way ANOVA test. ***P < 0.0001.
Discussion
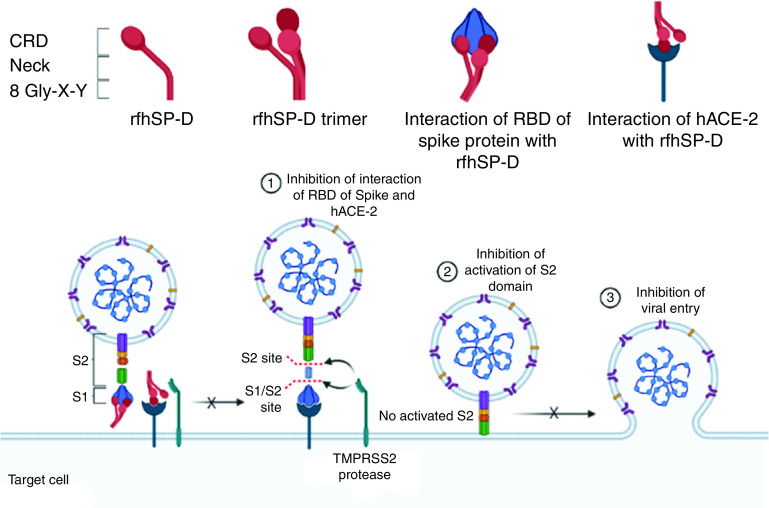
The present study explored the likely protective effect of rfhSP-D against SARS-CoV-2. As predicted by the docking study, rfhSP-D interacted with the S protein of SARS-CoV-2, its RBD, and ACE-2. Importantly, these interactions appear to have contributed to significant inhibition of infectivity and replication of SARS-CoV-2 present in the clinical samples derived from patients with asymptomatic, symptomatic, and severe COVID-19 (Figure 9).
Figure 9.
rfhSP-D binds spike/RBD and hACE-2 and inhibits SARS-CoV-2 infection and replication. Interaction of rfhSP-D with RBD/spike of SARS-CoV-2 and hACE-2 inhibit the interaction of RBD/spike and hACE-2, which is essential for viral entry into the host cells. This dual interaction of rfhSP-D plausibly causes the significant inhibition of infection and replication of SARS-CoV-2 in clinical samples. The figure was created using BioRender.com.
One of the first steps of the SARS-CoV-2 infection is the binding of the S protein to the host cell via the ACE-2 receptor (27). S1 protein is known to interact with the ACE-2 receptor via the RBM (455–508) in the RBD (aa 319–527), with virus-binding hotspot residues comprising Lys31, Glu35, and Lys353 of dimeric ACE-2 (15, 28, 29). Previously, SP-D interaction with the S protein of SARS-CoV, which shares 74% homology with the RBD of SARS-CoV-2 (12), has been reported. rfhSP-D is also known to bind to viral surface proteins such as hemagglutinin and neuraminidase of influenza A virus, gp120 of human immunodeficiency virus 1 (7, 30), and S protein of SARS-CoV (12). Thus, the possibility of rfhSP-D binding to the S protein of SARS-CoV-2 was examined.
In silico interaction of rfhSP-D with RBD of the S protein of SARS-CoV-2 revealed that Tyr449, Gln493, and Gln498 of RBD overlapped with the residues that are essential for the binding of the S protein to the target protein ACE-2. The binding of the S protein to rfhSP-D or FL SP-D was confirmed using an indirect ELISA. A comparatively lower absorbance with the monoclonal antibodies raised against the CRD region of human SP-D than the polyclonal antibodies could be attributed to the fact that the binding between rfhSP-D and the S protein occurred through the CRD region of rfhSP-D and FL SP-D. Furthermore, calcium independence suggested an involvement of protein–protein interaction. A significant inhibition of the S protein–ACE-2 interaction in presence of rfhSP-D suggested that rfhSP-D could interfere with the binding of the SARS-CoV-2 to the host cell, an essential step for infection to occur.
Clinical samples of SARS-CoV-2 were used to assess whether rfhSP-D modulated the infectivity and replication of the virus (isolation of the virus in the laboratory conditions may introduce alterations) in vitro. For assessing replication, qRT-PCR of the RdRp gene, which is essential for the replication of viral RNA, was measured. Remdesivir was used as a positive control for replication inhibition. Remdesivir, an adenosine analog, functions by incorporating itself into nascent viral RNA chains, which results in premature termination, thereby effectively inhibiting viral RNA synthesis (31). Downregulated RdRp expression in the remdesivir-treated samples clearly validated the platform for evaluating viral replication using clinical samples. A dose-dependent reduction of the RdRp mRNA expression in Vero cells challenged with rfhSP-D–pretreated SARS-CoV-2–positive clinical samples at a higher fold-change than remdesivir suggested a highly potent anti–SARS-CoV-2 activity of rfhSP-D.
Replication kinetic studies involving Vero cells infected with SARS-CoV-2 have demonstrated a significant synthesis of viral RNA at ⩾6 hours after infection (32). As such, any viral RNA detected 1 to 2 hours after infection could be considered to have come from the infecting viral particles and not from subsequent viral RNA synthesis or viral replication. Hence, to confirm whether rfhSP-D played a role in inhibiting SARS-CoV-2 infection, Vero cells were infected with SARS-CoV-2 clinical samples at a high concentration (500 TCID50; MOI, 0.05) for 2 hours. In accordance with the previous reports, lack of any significant effect of remdesivir on the Ct values of Vero cells, challenged with clinical samples, validated the assay format (33). Reduced RdRp transcripts in presence of rfhSP-D demonstrated the ability of rfhSP-D to act as an entry inhibitor against SARS-CoV-2. These results suggest that rfhSP-D is a potential candidate to be used as an S protein–based inhibitor against SARS-CoV-2 infection. With its established safety in vivo and therapeutic efficacy against several respiratory pathogens, rfhSP-D will effectively combat the nosocomial coinfections in patients with COVID-19.
There exists a dysregulated proinflammatory cytokine response without protective IFNs in response to SARS-CoV-2–mediated lung tissue damage, leading to acute respiratory distress syndrome. The concentrations of SP-D were significantly altered in the BAL of patients with acute respiratory distress syndrome and were strong predictors of poor prognosis (34, 35). Persistent complement activation leads to microangiopathy, leading to hypoxia in vital organs. The current therapeutic strategy comprises an antiviral such as remdesivir and immunosuppressants such as corticosteroids (which can potentially invite opportunistic infections). Importantly, there is a need to rapidly clear cell debris or damage-associated molecular patterns and polarize the immune response toward a protective one and regulate the complement activation. The rfhSP-D is capable of dampening the “cytokine storm” by rapid clearance of the virus-infected cells and strengthening the lung capacity by restoring homeostasis (36).
rfhSP-D has been previously shown to inhibit human immunodeficiency virus 1 entry as well as successfully thwart the cytokine storm in an ex vivo model of human vaginal tissue (37). SP-D has a compelling role in correcting lung pathophysiology and injury (38). It is likely that SP-D functions as an opsonin after binding to the S protein and helps in viral clearance. As a complement- and antibody-independent neutralizing agent against SARS-CoV-2, rfhSP-D may be a viable alternative as an inhalation formulation to control COVID-19 infection in immunocompromised/immunodeficient people and in individuals who can not be be vaccinated due to related health complications. Our promising results warrant further studies in COVID-19 animal models, such as mice humanized with human ACE-2 and Syrian hamsters (Mesocricetus auratus), to better understand the impact of rfhSP-D in the microenvironment of the respiratory system (39).
Acknowledgments
Ethics Statement
The project was approved by the institutional ethics committee of Institute of Liver and Biliary Sciences, Delhi, (IEC/2020/80/MA04) on July 20, 2020. The committee waived off the written informed consent in due consideration of the request, as these samples were stored in the facility; anonymized aliquots of the samples were provided for the study.
Acknowledgment
The authors thank Dr. Smita Mahale (Director, ICMR-NIRRH) and Dr. Shiv K. Sarin (Director, ILBS) for their support. The study was reviewed and recommended by ICMR Expert Review Committee. The authors also thank Dr. Nivedita Gupta and Dr. Raman Gangakhedkar of ICMR-ECD for their support. The study was reviewed by BIRAC-PACE expert panel.
Footnotes
Supported by an intramural grant from ICMR-NIRRH (accession number RA/1002/12–2020).
Author Contributions: T.M., B.B., P.M.V., and R.S. performed protein-related work. S.I.-T. and I.K. performed molecular modeling and mutational studies. T.M., S.R., R.A., D.M.T., S.K., E.G., and S.K.G. contributed to the clinical samples and infection assay. T.M., P.M.V., H.P., S.K.G., and U.K. analyzed and interpreted the data and prepared the manuscript. T.M., S.K.G., and U.K. performed conception and design. T.M., P.M.V., E.G., S.K.G., and U.K. performed data analysis. T.M. and U.K. performed interpretation. T.M., B.B., P.M.V., and U.K. drafted the first version of the manuscript. T.M., H.P., and U.K. reviewed and edited the manuscript.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2021-0005OC on March 30, 2021
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA, et al. Coronaviridae Study Group of the International Committee on Taxonomy of V. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. China Novel Coronavirus Investigating and Research Team. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. Geneva, Switzerland: World Health Organization; 2020. https://www.who.int/publications/m/item/weekly-epidemiological-update---24-november-2020 [Google Scholar]
- 4.Baughn LB, Sharma N, Elhaik E, Sekulic A, Bryce AH, Fonseca R. Targeting TMPRSS2 in SARS-CoV-2 infection. Mayo Clin Proc. 2020;95:1989–1999. doi: 10.1016/j.mayocp.2020.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.National Institutes of Health. Bethesda, MD: National Institutes of Health; https://www.covid19treatmentguidelines.nih.gov/therapeutic-management/ [Google Scholar]
- 6.Carmo A, Pereira-Vaz J, Mota V, Mendes A, Morais C, da Silva AC, et al. Clearance and persistence of SARS-CoV-2 RNA in patients with COVID-19. J Med Virol. 2020;92:2227–2231. doi: 10.1002/jmv.26103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Ahdal MN, Murugaiah V, Varghese PM, Abozaid SM, Saba I, Al-Qahtani AA, et al. Entry inhibition and modulation of pro-inflammatory immune response against influenza A virus by a recombinant truncated surfactant protein D. Front Immunol. 2018;9:1586. doi: 10.3389/fimmu.2018.01586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madsen J, Gaiha GD, Palaniyar N, Dong T, Mitchell DA, Clark HW. Surfactant Protein D modulates HIV infection of both T-cells and dendritic cells. PLoS One. 2013;8:e59047. doi: 10.1371/journal.pone.0059047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kishore U, Bernal AL, Kamran MF, Saxena S, Singh M, Sarma PU, et al. Surfactant proteins SP-A and SP-D in human health and disease. Arch Immunol Ther Exp (Warsz) 2005;53:399–417. [PubMed] [Google Scholar]
- 10.Murugaiah V, Tsolaki AG, Kishore U. Collectins: innate immune pattern recognition molecules. Adv Exp Med Biol. 2020;1204:75–127. doi: 10.1007/978-981-15-1580-4_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu YP, Liu ZH, Wei R, Pan SD, Mao NY, Chen B, et al. Elevated plasma surfactant protein D (SP-D) levels and a direct correlation with anti-severe acute respiratory syndrome coronavirus-specific IgG antibody in SARS patients. Scand J Immunol. 2009;69:508–515. doi: 10.1111/j.1365-3083.2009.02245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leth-Larsen R, Zhong F, Chow VTK, Holmskov U, Lu J. The SARS coronavirus spike glycoprotein is selectively recognized by lung surfactant protein D and activates macrophages. Immunobiology. 2007;212:201–211. doi: 10.1016/j.imbio.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Funk CJ, Wang J, Ito Y, Travanty EA, Voelker DR, Holmes KV, et al. Infection of human alveolar macrophages by human coronavirus strain 229E. J Gen Virol. 2012;93:494–503. doi: 10.1099/vir.0.038414-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Binder M. 2020 https://www.klinikum.uni-heidelberg.de/fileadmin/inst_hygiene/molekulare_virologie/Downloads/TCID50_calculator_v2_17-01-20_MB.xlsx
- 15.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 16.Xu C, Wang Y, Liu C, Zhang C, Han W, Hong X, et al. Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM. Sci Adv. 2021;7:eabe5575. doi: 10.1126/sciadv.abe5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrusier N, Nussinov R, Wolfson HJ. FireDock: fast interaction refinement in molecular docking. Proteins. 2007;69:139–159. doi: 10.1002/prot.21495. [DOI] [PubMed] [Google Scholar]
- 18.Mashiach E, Schneidman-Duhovny D, Andrusier N, Nussinov R, Wolfson HJ. FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 2008;36:W229–W332. doi: 10.1093/nar/gkn186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duhovny D, Nussinov R, Wolfson HJ. Efficient unbound docking of rigid molecules. Lect Notes Comput Sci. 2002;2452:185–200. [Google Scholar]
- 20.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodrigues CHM, Myung Y, Pires DEV, Ascher DB. mCSM-PPI2: predicting the effects of mutations on protein-protein interactions. Nucleic Acids Res. 2019;47:W338–W344. doi: 10.1093/nar/gkz383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murugaiah V, Agostinis C, Varghese PM, Belmonte B, Vieni S, Alaql FA, et al. Hyaluronic acid present in the tumor microenvironment can negate the pro-apototic effect of a recombinant fragment of human surfactant protein D on breast cancer cells. Front Immunol. 2020;11:1171. doi: 10.3389/fimmu.2020.01171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh M, Madan T, Waters P, Parida SK, Sarma PU, Kishore U. Protective effects of a recombinant fragment of human surfactant protein D in a murine model of pulmonary hypersensitivity induced by dust mite allergens. Immunol Lett. 2003;86:299–307. doi: 10.1016/s0165-2478(03)00033-6. [DOI] [PubMed] [Google Scholar]
- 24.Kaur A, Riaz MS, Murugaiah V, Varghese PM, Singh SK, Kishore U. A recombinant fragment of human surfactant protein D induces apoptosis in pancreatic cancer cell lines via fas-mediated pathway. Front Immunol. 2018;9:1126. doi: 10.3389/fimmu.2018.01126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar J, Murugaiah V, Sotiriadis G, Kaur A, Jeyaneethi J, Sturniolo I, et al. Surfactant protein D as a potential biomarker and therapeutic target in ovarian cancer. Front Oncol. 2019;9:542. doi: 10.3389/fonc.2019.00542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thakur G, Prakash G, Murthy V, Sable N, Menon S, Alrokayan SH, et al. Human SP-D acts as an innate immune surveillance molecule against androgen-responsive and androgen-resistant prostate cancer cells. Front Oncol. 2019;9:565. doi: 10.3389/fonc.2019.00565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benton DJ, Wrobel AG, Xu P, Roustan C, Martin SR, Rosenthal PB, et al. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature. 2020;588:327–330. doi: 10.1038/s41586-020-2772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lui I, Zhou XX, Lim SA, Elledge SK, Solomon P, Rettko NJ, et al. Trimeric SARS-CoV-2 Spike interacts with dimeric ACE2 with limited intra-Spike avidity [preprint] bioRxiv. 2020 https://www.biorxiv.org/content/10.1101/2020.05.21.109157v1 [Google Scholar]
- 30.Pandit H, Gopal S, Sonawani A, Yadav AK, Qaseem AS, Warke H, et al. Surfactant protein D inhibits HIV-1 infection of target cells via interference with gp120-CD4 interaction and modulates pro-inflammatory cytokine production. PLoS One. 2014;9:e102395. doi: 10.1371/journal.pone.0102395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eastman RT, Roth JS, Brimacombe KR, Simeonov A, Shen M, Patnaik S, et al. Remdesivir: a Review of its discovery and development leading to emergency use authorization for treatment of COVID-19. ACS Cent Sci. 2020;6:672–683. doi: 10.1021/acscentsci.0c00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogando NS, Dalebout TJ, Zevenhoven-Dobbe JC, Limpens RWAL, van der Meer Y, Caly L, et al. SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J Gen Virol. 2020;101:925–940. doi: 10.1099/jgv.0.001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greene KE, Wright JR, Steinberg KP, Ruzinski JT, Caldwell E, Wong WB, et al. Serial changes in surfactant-associated proteins in lung and serum before and after onset of ARDS. Am J Respir Crit Care Med. 1999;160:1843–1850. doi: 10.1164/ajrccm.160.6.9901117. [DOI] [PubMed] [Google Scholar]
- 35.Park J, Pabon M, Choi AMK, Siempos II, Fredenburgh LE, Baron RM, et al. Plasma surfactant protein-D as a diagnostic biomarker for acute respiratory distress syndrome: validation in US and Korean cohorts. BMC Pulm Med. 2017;17:204. doi: 10.1186/s12890-017-0532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vandivier RW, Ogden CA, Fadok VA, Hoffmann PR, Brown KK, Botto M, et al. Role of surfactant proteins A, D, and C1q in the clearance of apoptotic cells in vivo and in vitro: calreticulin and CD91 as a common collectin receptor complex. J Immunol. 2002;169:3978–3986. doi: 10.4049/jimmunol.169.7.3978. [DOI] [PubMed] [Google Scholar]
- 37.Pandit H, Kale K, Yamamoto H, Thakur G, Rokade S, Chakraborty P, et al. Surfactant protein D reverses the gene signature of transepithelial HIV-1 passage and restricts the viral transfer across the vaginal barrier. Front Immunol. 2019;10:264. doi: 10.3389/fimmu.2019.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knudsen L, Ochs M, Mackay R, Townsend P, Deb R, Mühlfeld C, et al. Truncated recombinant human SP-D attenuates emphysema and type II cell changes in SP-D deficient mice. Respir Res. 2007;8:70. doi: 10.1186/1465-9921-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muñoz-Fontela C, Dowling WE, Funnell SGP, Gsell PS, Riveros-Balta AX, Albrecht RA, et al. Animal models for COVID-19. Nature. 2020;586:509–515. doi: 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]