Oculopharyngodistal myopathy (OPDM) is associated with trinucleotide repeat expansions in LRP12 or GIPC1, but the genetic basis of many cases remains unknown. Yu et al. identify GGC repeat expansions in NOTCH2NLC in Chinese patients with OPDM, and provide evidence for possible underlying pathogenic mechanisms.
Keywords: oculopharyngodistal myopathy, GGC repeat expansion, NOTCH2NLC, toxic protein gain-of-function mechanism, RNA gain-of-function mechanism
Abstract
Oculopharyngodistal myopathy (OPDM) is an adult-onset neuromuscular disease characterized by progressive ocular, facial, pharyngeal and distal limb muscle involvement. Trinucleotide repeat expansions in LRP12 or GIPC1 were recently reported to be associated with OPDM. However, a significant portion of OPDM patients have unknown genetic causes. In this study, long-read whole-genome sequencing and repeat-primed PCR were performed and we identified GGC repeat expansions in the NOTCH2NLC gene in 16.7% (4/24) of a cohort of Chinese OPDM patients, designated as OPDM type 3 (OPDM3). Methylation analysis indicated that methylation levels of the NOTCH2NLC gene were unaltered in OPDM3 patients, but increased significantly in asymptomatic carriers. Quantitative real-time PCR analysis indicated that NOTCH2NLC mRNA levels were increased in muscle but not in blood of OPDM3 patients. Immunofluorescence on OPDM muscle samples and expressing mutant NOTCH2NLC with (GGC)69 repeat expansions in HEK293 cells indicated that mutant NOTCH2NLC-polyglycine protein might be a major component of intranuclear inclusions, and contribute to toxicity in cultured cells. In addition, two RNA-binding proteins, hnRNP A/B and MBNL1, were both co-localized with p62 in intranuclear inclusions in OPDM muscle samples. These results indicated that a toxic protein gain-of-function mechanism and RNA gain-of-function mechanism may both play a vital role in the pathogenic processes of OPDM3. This study extended the spectrum of NOTCH2NLC repeat expansion-related diseases to a predominant myopathy phenotype presenting as OPDM, and provided evidence for possible pathogenesis of these diseases.
Introduction
Oculopharyngodistal myopathy (OPDM; MIM 164310) is a rare adult-onset neuromuscular disease with clinical features including progressive ocular, facial, pharyngeal and distal limb muscle weakness.1,2 Myopathological findings include rimmed vacuoles and chronic myopathic changes. Ultrastructural examination reveals tubulofilamentous inclusions in both sarcoplasm and nucleus.1,3–5 OPDM was first reported and named in 1977.5 Currently about 300 individuals with OPDM have been described worldwide.1–12
The diagnosis of OPDM used to depend on clinical manifestations, histopathological findings, and genetic exclusion of similar conditions. OPDM was reported to be either autosomal dominant or putative autosomal recessive inheritance. In 2019, Ishiura et al.7 identified that a CGG repeat expansion in the 5′-untranslated region (5′UTR) of the LDL receptor-related protein 12 (LRP12) gene was responsible for OPDM. In 2020, our group found that GGC repeat expansion in the 5′UTR of the GAIP/RGS19-interacting protein (GIPC1) was the second causative gene of OPDM.3 Herein, we suggest that the OPDM associated with LRP12 is designated as OPDM type 1 (OPDM1),13 and the OPDM associated with GIPC1 is designated as OPDM type 2 (OPDM2). However, OPDM1 was only identified in 25% and 4.17% of Japanese and Chinese OPDM cases, respectively, while OPDM2 accounted for 50% and 3.61% in a cohort of Chinese and Japanese cases, respectively.3,7 Therefore, there are still other genetic causes for OPDM waiting to be identified.
In this study, we further explored other underlying disease-causing genes in a cohort of OPDM patients who did not have the trinucleotide repeat expansion in either LRP12 or GIPC1. By using long-read whole-genome sequencing (LRS) on the Oxford Nanopore platform, we found that a GGC repeat expansion in the NOTCH2NLC gene (MIM: 618025), which had been reported as the causative gene of neuronal intranuclear inclusion disease (NIID) recently,7,14–16 was also associated with a proportion of OPDM patients, designated as OPDM type 3 (OPDM3). Additionally, we made a preliminary investigation of the pathogenesis of OPDM3.
Materials and methods
Study participants
Initially, a cohort of 24 unrelated OPDM cases from mainland China, including eight index patients with family history and 16 sporadic cases, were recruited from Peking University First Hospital. Among them, three index patients and nine sporadic cases were identified with GIPC1 mutation, and one index patient with a LRP12 mutation.3 Therefore, the remaining four affected families and seven sporadic cases were further investigated in this study. Family 1, Family 4, and sporadic Patient 1 have been reported previously.12 The diagnostic criteria of OPDM have been reported previously.3 Briefly, OPDM patients were diagnosed based on their typical clinical features, myopathological findings, and exclusion of GCN repeat expansion of the poly(A) binding protein nuclear 1 (PABPN1) gene (MIM: 602279),17 CTG repeat expansion of the DM1 protein kinase (DMPK) gene (MIM: 605377)18 and potential pathogenic variants in coding regions of known neuromuscular disorder-associated genes by repeat-primed PCR (RP-PCR) and next-generation sequencing via Nextera kits (Illumina). The clinical data and laboratory findings were collected. Four patients had brain MRI, and three had MRI for lower limb muscles. Genomic DNA of peripheral blood leucocytes was obtained from these OPDM patients with unknown genetic causes, their available family members, and 500 unaffected control subjects.
This study was approved by the Local Ethics Committee of Peking University First Hospital. All participants’ consent was obtained according to the Declaration of Helsinki for use of their remaining samples for research after the diagnosis.
Muscle biopsy, sural nerve biopsy, skin biopsy, immunofluorescence and electron microscopy study
Following written consent, all patients and one asymptomatic family member underwent muscle biopsy, meanwhile two patients (Patients S1 and S2) underwent sural nerve biopsy. Patient S1 and one asymptomatic family member (Subject F1-II7) underwent skin biopsy. Muscle biopsy, sural nerve biopsy and skin biopsy were conducted following a routine procedure as described previously.3,14,19 For histological examination, muscle samples were conducted on serially frozen sections (8 μm) using routine histological and histochemistry staining, including haematoxylin and eosin, and modified Gomori trichrome (mGT). Specimen of sural nerve biopsy was routinely processed for histological and ultrastructural examinations. Skin samples were fixed in formalin, embedded in paraffin and examined using haematoxylin and eosin staining. For immunofluorescence, the procedures were performed as previously described.3 Muscle sections were immunostained with primary antibodies against NOTCH2NL (Abcam, ab233287), hnRNP A/B (Santa Cruz, sc376411), MBNL1 (Santa Cruz, sc47740), p62 (Abcam, ab56416, anti-mouse), p62 (Abcam, ab194129, anti-guinea pig), and Glycine (Abcam ab9442), respectively. Skin sections were immunostained with primary antibody against p62 (Abcam, ab56416, anti-mouse). All the sections were subsequently incubated with Alexa Fluor488/555/647 secondary antibodies. Images were acquired with confocal microscopy (Nikon A1MP) at ×60 magnification. Electron microscopy analysis was also carried out as described previously.3
Whole-genome sequencing by short-read sequencing
For short-read sequencing, the DNA sequencing library was constructed using a MGIEasy DNA Library Prep Kit following the manufacturer’s instructions (BGI) to generate DNA nanoballs (DNB). Whole-genome sequencing (WGS) was carried out as in a previous study.3
Long-read whole-genome sequencing
LRS was first carried out on the four unrelated OPDM individuals (Patients F1-III8, F2-II4, F3-II1 and S1), using a PromethION sequencer (Oxford Nanopore Technologies) as in a previous study.3 The mean aligned coverage reached 14.07× to 19.74×. Library preparation was carried out using a 1D Genomic DNA ligation kit (SQKLSK109) according to the manufacturer’s protocol. For each individual, one PRO-002 (R9.4.1) flow cell was used. PromethION database calling was performed using guppy v3.3.0 (Oxford Nanopore Technologies), and only pass reads (qscore ≥ 7) were used for subsequent analysis. Subsequently, Patient S3, and an additional six members of Family 1 (Subjects F1-II2, F1-II3, F1-II5, F1-II7, F1-II8 and F1-III10), parents of Patient S2, and mother of Patient S3 were also sequenced using LRS on the same platform.
Short tandem repeats detection
By 2018, more than 40 nucleotide repeat expansion diseases (NREDs) had been identified, most of which affect the nervous system.20 In 2019, it reached more than 50.21 To identify the possible genetic mutations, 55 reported pathogenic NREDs were searched for subsequent analysis and named the NRED-LRS set.7,14,21–24
The pass reads from PromethION were aligned to the reference genome hg38 using ngmlr v0.2.7.25 For each repeat from 55 reported pathogenic NREDs, the repeat count of each read that aligned with the short tandem repeat (STR) locus was detected using RepeatHMM26 without the step of peak calling, which is not good at low coverage data.
Considering that nanopore reads have certain bias errors, the third quartile repeat count was defined as the individual’s estimated repeat count (ERC). According to the abovementioned description, close attention was paid to samples with ERC > 50. In particular, anomalies found at certain STR loci in several samples by RepeatHMM software were examined manually one-by-one using TRF software27 and IGV software28 to correct the repeat count.
Repeat-primed polymerase chain reaction
Genomic DNA from all patients and their available family members, and 500 unaffected control subjects were analysed by RP-PCR. RP-PCR was performed according to our previous study.14
Nanopore 5mC methylation modification calling
We called 5mC methylation using minimap229 and Nanopolish30 as previously reported. The methylation status upstream and downstream of the GGC repeats were compared between NOTCH2NLC-related OPDM patients and normal control subjects as well as NIID patients. Methylation status across expanded and non-expanded alleles was determined using the Wilcoxon Rank Sum Test.
Methylation analysis using methylation-specific polymerase chain reaction
Methylation-specific PCR (MSP) was used to investigate the CpG methylation status of promoter region of expanded GGC repeats in the 5′ UTR of NOTCH2NLC gene. RelaxGene Blood DNA System (Tiangen, DP349) and TIANamp Genomic DNA Kit (Tiangen, DP304) were used to extract genomic DNA from blood and skeletal muscle samples, respectively. DNA was converted using the DNA Bisulfite Conversion Kit (Tiangen, DP205). MSP was performed using the methylation-specific PCR Kit (Tiangen, Em101) with the following primers: methylation–specific-F: 5′-GAGGATCGTTTTTTTTTGTAAGTTTACGATTTC-3′; methylation-specific-R: 5′-AATCTACTACGAATCACTAACCCCG-3′; unmethylation-specific-F: 5′-GGATGAGGATTGTTTTTTTTTGTAAGTTTATGATTTT-3′; unmethylation-specific-R: 5′-CAAAAAAATCTACTACAAATCACTAACCCCA-3′. PCR conditions were as follows: 95°C for 5 min, followed by 35 cycles of 94°C for 20 s, 60°C for 30 s and 72°C for 20 s, and then 72°C for 5 min. PCR products were separated with 1.5% agarose gel and detected using Syngene GeneGenius (Syngene) and ImageJ software. The ratio of methylation level of NOTCH2NLC to that of unmethylation level was used to represent the relative methylation level.
Quantitative real-time polymerase chain reaction
Total RNA was extracted from peripheral blood leucocyte samples and skeletal muscle samples from OPDM3 patients, available family members and normal control subjects with TRIzol™ reagent (Invitrogen) following the manufacturer’s instructions. cDNA was synthesized using 500 ng RNA and the SuperScript III First-Strand Synthesis System (Thermo Fisher Scientific) according to the manufacturer’s instructions. To detect NOTCH2NLC mRNA expression levels in peripheral blood leucocytes from OPDM affected individuals and controls, the following primers were used with GAPDH as a reference gene: NOTCH2NLC-RT forward, 5′-GATCTTTCCAAAGAGAATTCTGTATCTC-3′; NOTCH2NLC-RT reverse, 5′-GAGAGCCACATGGCTGACTT-3′; GAPDH forward, 5′-CTGGGCTACACTGAGCACC-3′; GAPDH reverse, 5′-AAGTGGTCGTTGAGGGCAATG-3′. RT-qPCR was performed according to our previous study.3
Plasmid construction
The enhanced green fluorescent protein (EGFP) tagged pEGFP-N1-NOTCH2NLC wild-type [Wt: NOTCH2NLC-(GGC)9], and mutant [Mut: NOTCH2NLC-(GGC)69] were constructed as in a previous study.31 Wild-type and mutant NOTCH2NLC were created by oligonucleotide ligations and subcloned into the pEGFP-N1 vector.
Cell culture and transfection
HEK293 cells were cultured in Dulbecco’s modified Eagle medium (DMEM; Gibco) supplemented with 10% foetal bovine serum (FBS) (Gibco), 100 unit/ml penicillin-streptomycin in a humidified incubator at 37°C under 5% CO2/95% air. Transient transfection was performed using Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions.
Fluorescence microscopy
HEK293 cells were rinsed with 1× phosphate-buffered saline (PBS), fixed with 4% polyformaldehyde in 1× PBS at 48 h post-transfection with empty vector (Ctr) or wild-type, mutant NOTCH2NLC-EGFP. The samples were mounted using ProLongTM Gold Antifade mounting medium with DAPI (Invitrogen). Images were acquired at ×60 magnification using an inverted microscope ECLIPSE Ti2-E (Nikon).
Western blot analysis
Skeletal muscle tissues from one asymptomatic carrier and normal control subjects, as well as cultured cells were lysed with RIPA buffer [1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS (pH 7.4)] containing a cocktail of protease inhibitors (Roche). Lysates were analysed by western blotting with the corresponding specific antibodies.
Statistical analyses
GraphPad Prism version 8.0.2 for Windows (GraphPad Software, San Diego, California, USA) was used to perform statistical comparisons. Numerical variables and bar graphs with error bars were presented as mean ± standard deviation (SD). For differences between two groups of continuous variables, Student’s t-test and Wilcoxon rank sum test were used. Comparisons of categorical variables between groups were conducted with the Chi-square test or Fisher’s exact test, as appropriate. Differences were considered statistically significant if P < 0.05.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary material.
Results
GGC repeat expansions in the NOTCH2NLC gene in OPDM3
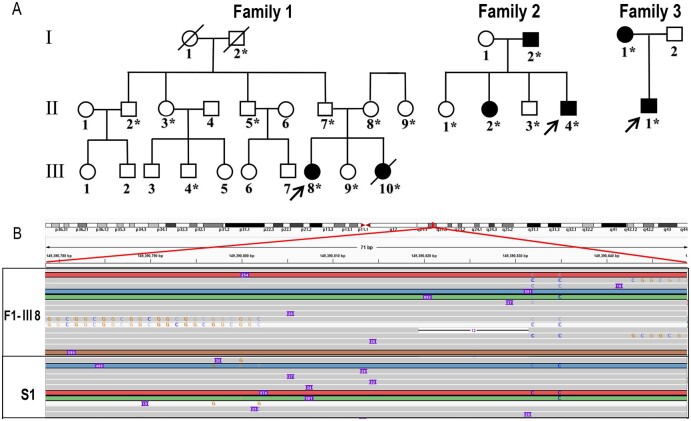
To determine the genetic causes for Patients F1-III8, F2-II4, F3-II1 and S1 with OPDM (Fig. 1A), WGS was first performed but it did not reveal potential pathogenic variants; then trinucleotide repeat expansions in LRP12 and GIPC1 were also excluded by RP-PCR analysis. Therefore, LRS was applied on these four patients using ONT PromethION sequencing machines. ONT PromethION sequencing data were analysed to search for known STR expansions in the NRED-LRS set as the first step. A heterozygous GGC repeat expansion in the NOTCH2NLC gene (MIM: 618025) was found in Patients F1-III8 and S1, and a heterozygous AAAAG repeat expansion in the RFC1 gene was found in Patients F2-II4 and F3-II1, respectively (Fig. 1B and Supplementary Table 1). The NOTCH2NLC gene was identified as the possible pathogenic gene, with more than 100 repeats in one allele of both patients. Because there are five homologous genes of NOTCH2NLC, we formed a list of single nucleotide polymorphisms (SNPs) of homologous genes and identified the STR region of the NOTCH2NLC gene using the SNPs list as previously reported.14RFC1 was excluded as a candidate gene since AAAAG repeat expansions were reported to be benign configurations, and the pathogenic mutation repeat unit was biallelic AAGGG32,33 (Supplementary Fig. 1A–C).
Figure 1.
Identification of GGC repeat expansion in the 5′UTR of the NOTCH2NLC gene in OPDM individuals. (A) Pedigree chart of three families with OPDM. The squares indicate males and the circles indicate females. A diagonal line through a symbol indicates a deceased individual. Affected individuals are indicated by filled symbols. All the asterisked individuals had available blood DNA. (B) Visualization on Integrative Genomics Viewer for Patients F1-III8 and S1 revealed DNA reads carrying the mutated repeat unit (chr1:149390780–149390837, hg38 version), corresponding to the GGC repeat expansions in the 5′UTR of the NOTCH2NLC gene.
To confirm the LRS results, RP-PCR analysis was then carried out. The electropherogram demonstrated a saw-tooth pattern in two familial patients (Patients F1-III8 and F1-III10) and a sporadic patient (Patient S1), but not in the other nine unaffected members in Family 1. This confirmed the GGC repeat expansion mutation of NOTCH2NLC in these patients, and also indicated co-segregation of this mutation in Family 1 (Fig. 2A and Supplementary Fig. 2A). Subsequently, RP-PCR analysis was performed on the remaining patients including the other three OPDM families, six sporadic OPDM patients, and 500 unaffected Chinese control subjects. GGC repeat expansions of NOTCH2NLC were identified in two more sporadic OPDM patients (Patients S2 and S3) (Supplementary Fig. 2B). None of the 500 normal Chinese control subjects showed repeat expansions in NOTCH2NLC.
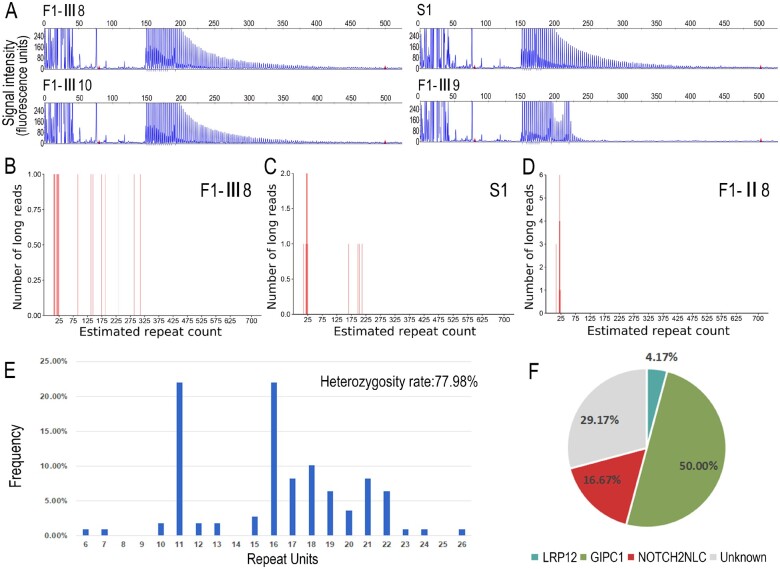
Figure 2.
Validation of GGC repeat expansions and repeat sizes in NOTCH2NLC among OPDM3 patients and normal control subjects. (A) Representative results of RP-PCR analysis showing GGC repeat expansions in Patients F1-III8, F1-III10 and S1. No repeat expansions were shown in one unaffected member Patient F1-III9. Experiments were conducted three times with reproducible results. LRS data showed estimated GGC repeat count of NOTCH2NLC reached more than 100 in Patients F1-III8 (B) and S1 (C) but no more than 25 in the unaffected member Patient F1-II8 (D). (E) Frequency distribution of GGC repeat units in NOTCH2NLC ranging from 6 to 26 among 109 normal control subjects. DNA samples from normal controls were amplified by NOTCH2NLC-specific primers and revealed by fragment analysis. Heterozygosity rate in normal controls was 77.98%. (F) Pie chart for the percentages of disease-causing gene mutations and unknown gene mutations in a cohort of Chinese OPDM patients. Disease-causing genes, trinucleotide repeat expansions in the 5′UTRs of LRP12, GIPC1 and NOTCH2NLC, accounted for 4.17%, 50% and 16.67% of cases in 24 unrelated Chinese OPDM patients, respectively, whereas unknown genetic cause accounted for 29.17% of cases in this cohort.
Furthermore, the GGC repeat sizes of NOTCH2NLC in two patients from Family 1 (Patients F1-III8 and F1-III10) and two sporadic patients (Patients S1 and S3) were determined by LRS. Unfortunately, one sporadic patient (Patient S2) sample did not contain enough DNA for LRS. The repeat expansions ranged from 128 to 198 times in the four patients (Fig. 2B, C and Supplementary Fig. 1D and E).
The GGC repeat sizes of the NOTCH2NLC gene ranged from 6 to 26 in 109 normal Chinese control subjects by fragment analysis, and the heterozygosity rate in normal control subjects was 77.98% (Fig. 2E). This result was consistent with the repeat size of 5–38 in 211 Chinese healthy control subjects identified previously.16 In conclusion, these results strongly suggested that GGC repeat expansions in NOTCH2NLC were associated with OPDM.
Intriguingly, a further LRS study in eight asymptomatic individuals (five asymptomatic individuals from Family 1, parents of Patient S2, and mother of Patient S3) showed that three asymptomatic members (Patients F1-II5 and F1-II7 and father of Patient S2) also carried heterozygous alleles of the GGC repeat expansions. Their repeat counts were more than 400 times (Supplementary Fig. 1F–H), while the other five asymptomatic family members carried normal alleles (Fig. 2D and Supplementary Fig. 1I–L). This suggests that the individuals with much higher GGC expansions might be paradoxically asymptomatic.
Clinical and pathological features
OPDM3 patients
In total, there were five OPDM patients including two patients from a family and three sporadic cases associated with GGC repeat expansions in the NOTCH2NLC gene. We named the OPDM associated with GGC repeat expansions in the NOTCH2NLC gene as OPDM3. In our cohort of 24 OPDM patients, trinucleotide repeat expansions in the LRP12, GIPC1, NOTCH2NLC and unknown gene(s) accounted for 4.17%, 50%, 16.67% and 29.17%, respectively (Fig. 2F). Detailed clinical characteristics and pathological changes of these patients are summarized in Table 1. A comparison of the clinical features between OPDM1, OPDM2, and OPDM3 are presented in Table 2.
Table 1.
The clinical features and GGC repeat size of five OPDM patients with NOTCH2NLC mutation
| Variables | Family 1 |
S1 | S2a | S3 | |
|---|---|---|---|---|---|
| F1-III8 | F1-III10a | ||||
| Sex | Female | Female | Male | Female | Male |
| Age of onset | 19 | 18 | 29 | 31 | 22 |
| Disease duration, years | 15 | 11 | 18 | 17 | 10 |
| Initial symptom | Weakness in bilateral legs | Weakness in bilateral legs | Weakness in bilateral legs | Binocular with diplopia | Weakness in bilateral legs |
| Clinical features | |||||
| Ptosis | + | + | + | + | + |
| External ophthalmoplegia | – | – | + | + | + |
| Dysphagia | + | + | + | + | + |
| Dysarthria | + | + | + | + | + |
| Facial weakness | + | + | + | + | + |
| Distal limb muscle weakness | + | + | + | + | + |
| Tremor | – | – | – | NA | – |
| Tendon reflexes | |||||
| Upper limb | Decreased | Decreased | Absent | Absent | Absent |
| Lower limb | Absent | Absent | Absent | Absent | Absent |
| Sensory disturbance | – | – | – | – | – |
| Leukodystrophy | + | NA | + | + | – |
| MMSE | 30 points | NA | 30 points | NA | 30 points |
| Serum creatine kinaseb (IU/l) | 1444 | 319 | 353 | 215 | 1140 |
| EMG pattern | MC | NC | NC | MC and NC | MC |
| Rimmed vacuoles | + | + | + | + | – |
| Repeat units | 187 | 128 | 198 | NA | 165 |
MC = myopathic change; MMSE = Mini-Mental State Examination; NA = not available; NC = neurogenic change.
Deceased individual.
Normal limits: 70–170 IU/l.
Table 2.
Comparison of clinical features between OPDM1, OPDM2 and OPDM3 in our cohort patients
| Variables |
OPDM1
(n = 1) |
OPDM2
(n = 12) |
OPDM3
(n = 4) |
P-value |
|---|---|---|---|---|
| Total patients, n | 2 | 19 | 5 | – |
| Sex, male/female | 2/0 | 10/9 | 2/3 | – |
| Age of onset, years | 30.5 ± 6.4 | 25.3 ± 6.7 | 23.8 ± 5.9 | 0.292 |
| Disease duration, years | 9 ± 4.2 | 12.8 ± 6.9a | 14.2 ± 3.6 | 0.932 |
| Clinical manifestations | ||||
| Ptosis | 2/2 (100%) | 19/19 (100%) | 5/5 (100%) | – |
| External ophthalmoplegia | 2/2 (100%) | 19/19 (100%) | 3/5 (60%) | – |
| Dysphagia | 2/2 (100%) | 19/19 (100%) | 5/5 (100%) | – |
| Dysarthria | 2/2 (100%) | 19/19 (100%) | 5/5 (100%) | – |
| Facial muscle weakness | 2/2 (100%) | 19/19 (100%) | 5/5 (100%) | – |
| Distal limb weakness | 2/2 (100%) | 19/19 (100%) | 5/5 (100%) | – |
| Tremor | 0/2 (0%) | 0/19 (0%) | 0/5 (0%) | – |
| White matter involvement | NA | NA | 3/4 (75%) | – |
| Serum CKb, IU/l (number of available patients) | 1211 (1) | 798.6 ± 671.7 (13) | 694.2 ± 558.5 (5) | 0.270 |
| EMG pattern (number of available patients) | MC (2) | MC (9); MC + NC (1) | MC (2); NC (2); MC + NC (1) | – |
| Rimmed vacuoles | 1/1 | 13/13 | 4/5 | – |
| Genetic cause | LRP12 | GIPC1 | NOTCH2NLC | – |
| Repeat expansion size (number of available patients) | 117.5 ± 12.02 (2) | 118.6 ± 17.6 (18) | 169.5 ± 30.9 (4) | – |
MC = myopathic change; NA = not available; NC = neurogenic change; OPDM1/2/3 = oculopharyngodistal myopathy type 1/type 2/type 3. P-value is shown as: OPDM2 versus OPDM3.
One patient lost to follow-up.
Normal limits: 70–170 IU/l.
Among these five OPDM3 patients, the mean age of onset was 23.8 ± 5.9 (ranging from 18 to 31 years). Disease duration ranged from 11 to 18 years. Four of five patients initially showed muscle weakness in bilateral legs, and the remaining patient had initial symptom of biocular exotropia with diplopia. All patients slowly developed the full OPDM phenotype with disease progression including ptosis, facial and bulbar muscle weakness, and predominant weakness of distal limbs. However, external ophthalmoplegia was observed in three of five patients in their final follow-up. No patients showed sensory impairment. The tendon reflexes were decreased or absent in all patients. All patients manifested steppage gait at their first visits. Patient F1-III10 died of aspiration pneumonia at 28 years old, and Patient S2 died of choking at 48 years old.
Based on laboratory examination, serum creatine kinase (CK) level was mildly or moderately elevated (215–1444 IU/l, normal: 25–195 IU/l) in all patients. EMG revealed myogenic changes in two patients, neurogenic changes in two patients, and both myogenic and neurogenic changes in one patient. The results of nerve conduction examination were available in four patients. Motor nerve conduction velocities were mildly reduced, but the amplitudes of compound muscle action potentials moderately decreased, indicating a mixed pattern of neuropathy in Patients F1-III8, S1 and S2. Furthermore, the motor nerves of lower limbs were more severely affected than those of upper limbs. The sensory nerve conduction velocities and action potentials were also affected in three patients. Detailed nerve electrophysiological data are listed in Supplementary Table 2.
Muscle MRI examinations were carried out in three OPDM3 patients. The muscles of the lower leg were more severely affected than those of the thigh (Fig. 3A–F). At the early or middle stage of the disease, the long head of the biceps femoris, adductor magnus, and semimembranosus were the most seriously affected by fatty replacement at the level of thigh, while the calf muscles exhibited severe involvement except for the anterior tibial muscle (Fig. 3A–D). With disease progression, severe fatty replacement was observed in both the thigh and calf muscles, although the gracilis, semitendinosus and short head of the biceps femoris were relatively slight (Fig. 3E–F). Muscle MRI of OPDM1, OPDM2 and OPDM3 patients showed a generally similar pattern of muscle involvement with regards to disease progression3 (data of muscle MRI of OPDM1 available on request).
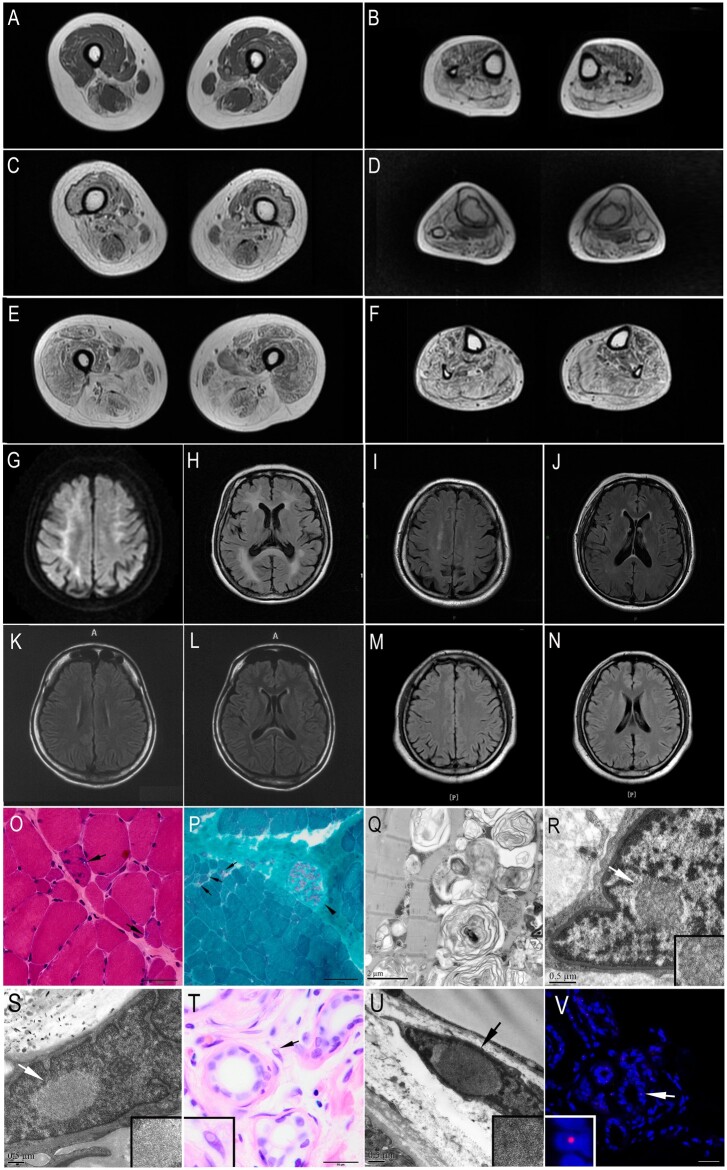
Figure 3.
MRI and pathological changes of the OPDM3 patients. (A–F) Muscle MRI of OPDM3 patients showed fatty infiltration of lower limb muscles, with the distal muscles (B, D and F calf level) more severely affected than the proximal muscles (A, C and E thigh level). (A and B) Muscle MRI of Patient F1-III10: moderate weakness with a disease duration of 6 years, ambulatory without support. (C and D) Muscle MRI of Patient F1-III8, moderate weakness with a disease duration of 9 years, ambulatory without support. (E and F) Muscle MRI of Patient S1: severe weakness with a disease duration of 11 years, ambulatory without support. (G–N) Brain MRIs of Patients S2 (G and H), S1 (I and J), F1-III8 (K and L), and S3 (M and N). (G) Hyperintense linear lesions in corticomedullary junctions in a diffuse weight image (DWI). (H) Severely diffuse leukoencephalopathy on T2-weighted fluid-attenuated inversion recovery (T2-FLAIR). (I–L) Mild white matter signal abnormalities on T2-FLAIR in periventricular white matter and splenium of corpus callosum after 18 years (Patient S1) and 15 years (Patient F1-III8) of the disease, respectively. (M and N) Normal brain MRIs on T2-FLAIR. (O) Haematoxylin and eosin, and (P) modified Gomori trichrome (mGT) stainings of muscle sections from Patient F1-III8, showing dystrophic change with variation in fibre size and endomysial fibrosis, and fibres with rimmed vacuoles (marked by arrow and shown at higher magnification). The intramuscular peripheral nerves were well myelinated (arrowhead) (P). (Q) Electron microscopy of muscle tissue from Subject F1-III10 revealed various myelin figures and autophagic vacuoles containing osmiophilic deposits and amorphous materials. (R and S) Electron microscopy of muscle sample from Patient S2 showed intranuclear inclusions contained filamentous aggregates (marked by arrowhead and shown at higher magnification). (T) Haematoxylin and eosin staining of skin sections from Patient S1 showed intranuclear inclusions in the fibroblasts (marked by arrow and shown at higher magnification). (U) Electron microscopy of skin sample from Patient S1 showed intranuclear inclusions. (V) Immunofluorescence on skin sections from Patient S1 showing p62 positive intranuclear inclusions in the sweat gland (marked by arrow and shown at higher magnification). Scale bars = 50 µm (O and T), 100 µm (P), 2 μm (Q), 0.5 µm (R, S and U) and 25 µm (V).
Brain MRIs were conducted in four patients (Patients F1-III8, S1, S2 and S3) (Fig. 3G–N). Severely diffuse leukoencephalopathy was observed in Patient S2 on T2-weighted fluid-attenuated inversion recovery (T2-FLAIR) with 12 years of disease duration, and hyperintense linear lesions were found in the corticomedullary junction on DWI (Fig. 3G and H). In Patients S1 and F1-III8, only mild white matter signal abnormalities were found on T2-FLAIR after 18 and 15 years of the disease, respectively, including periventricular white matter and splenium of corpus callosum (Fig. 3I–L). Brain MRI was normal in Patient S3 (Fig. 3M and N) with 10 years of disease duration. No cognitive impairments were identified in three patients (Patients F1-III8, S1 and S3) at the final visit, all of whom had 30 points in the Mini-Mental State Examination after 10 or more years of disease duration (Table 1).
Muscle biopsies of all cases showed fibre size variation and endomysial fibrosis to various degrees. Four of five patients had rimmed vacuoles on haematoxylin and eosin and modified Gomori trichrome staining (Fig. 3O and P). The intramuscular peripheral nerves were well myelinated in Patients F1-III8 and S1 (Fig. 3P). Electron microscopy of muscle samples revealed various myelin figures and autophagic vacuoles containing osmiophilic deposits and amorphous materials in Patient F1-III10 (Fig. 3Q), and intranuclear inclusions containing filamentous materials in Patient S2 (Fig. 3R and S). Additionally, skin biopsy of Patient S1 demonstrated the presence of intranuclear inclusions, which were p62 positive by immunofluorescence (Fig. 3T–V). Sural biopsies in Patients S1 and S2 showed the number of large myelinated fibres were mildly decreased, but the density of small fibres was decreased markedly. Some thin-myelinated fibres were observed (Supplementary Fig. 3).
Asymptomatic carriers
To further reinforce the finding that individuals with much higher GGC expansions could indeed be asymptomatic, the clinical and pathological examinations were further performed on two asymptomatic carriers. Clinically, Patient F1-II7 and the father of Patient S2 showed with normal muscle strength in trunk and four limbs [Medical Research Council (MRC) 5/5] without ocular, bulbar and facial muscle weakness on examination. They did not present with common NIID-related manifestations including cognition impairment, polyneuropathy, autonomic symptoms and tremor, either. For Patient F1-II7, the CK level (159 IU/l), and MRIs of brain and muscle were normal (Supplementary Fig. 4A–C). Results of peripheral nerve conduction studies and EMG revealed no abnormal findings. Pathologically, muscle tissues of Patient F1-II7 were completely normal without myopathy changes or rimmed vacuole (Supplementary Fig. 4D and E). Typical p62 deposits in nucleus was not observed in skin or muscle sample of Patient F1-II7 by immunofluorescence (Supplementary Fig. 4F and G).
Preliminary pathogenesis investigation of OPDM3
To investigate the methylation status of expanded GGC repeats and adjacent regions of NOTCH2NLC, we examined the 5-methylcytosine (5mC) modification using the ONT PromethION sequencing data obtained from three patients with OPDM, four patients with NIID and 10 normal control subjects. The methylation level around the GGC repeats was low and no significant differences were detected between OPDM3 individuals (Patients F1-III8, F1-III10 and S1) and normal control subjects (Fig. 4A), and NIID patients (Fig. 4B). Subsequently, 5mC modifications of both expanded and non-expanded alleles were examined in OPDM3 individuals (Patients F1-III8, F1-III10 and S1). No significant differences in methylation were detected (Supplementary Fig. 5A–D) but there tended to be hypermethylation in the asymptomatic carrier (Patient F1-II7) (Supplementary Fig. 5E).
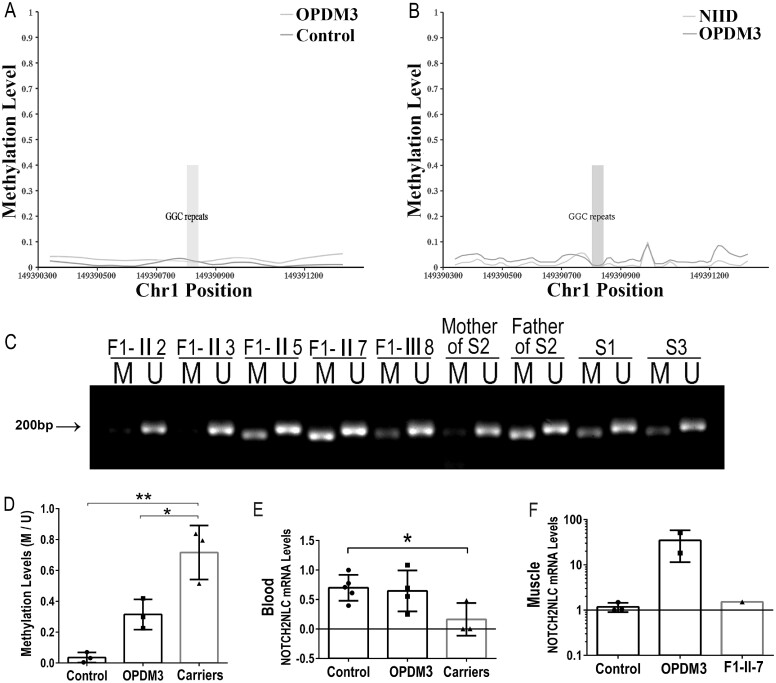
Figure 4.
Methylation and expression at the NOTCH2NLC locus. (A and B) Methylation status across the expanded GGC repeat region was determined using LRS data from three affected individuals (Patients F1-III8, F1-III10 and S1), 10 normal control subjects, and four NIID patients. (A) There were no significant differences between three affected individuals (Patients F1-III8, F1-III10 and S1) and 10 normal controls in methylation. (B) There were no significant differences in methylation between three affected individuals (Patients F1-III8, F1-III10 and S1) and four NIID patients. (C and D) Methylation analysis of the promoter region of expanded GGC repeats in the 5′UTR of the NOTCH2NLC gene in blood. Methylation levels were significantly increased in three asymptomatic carriers (Patients F-II5, F-II7 and father of Patient S2), compared with OPDM3 patients (Patients F-III8, S1 and S3) and controls (F-II2, F-II3 and mother of Patient S2). M = methylated; U = unmethylated. Ratio of methylation level of NOTCH2NLC to that of unmethylation level (M/U) was used to represent the relative methylation status. (E and F) Analysis of transcriptional levels of NOTCH2NLC mRNA in normal control subjects, OPDM3 patients and asymptomatic carriers by RT-qPCR in peripheral blood or muscle samples. No significant difference was detected between control subjects and OPDM3 patients (Patients F1-III8, F1-III10, S1 and S3), while asymptomatic carriers showed significantly lower NOTCH2NLC mRNA levels in peripheral blood (*P < 0.05) (E). The transcriptional levels of NOTCH2NLC mRNA were in an increasing trend in muscle samples (F) from two OPDM3 patients (Patients F1-III10 and S2).
To compare the methylation status of the promoter region of the NOTCH2NLC gene in different tissues and verify the 5mC modification results from LRS, we performed MSP with DNA from blood or muscle samples in OPDM3 patients (Patients F1-III10, S1, S2 and S3), asymptomatic carriers (Patients F1-II5 and F1-II7, father of Patient S2) and normal control subjects. The methylation status in the promoter region of the NOTCH2NLC gene was consistent in both blood and muscle DNA. The methylation levels of promoter region of the NOTCH2NLC gene were increased in the asymptomatic carriers (Patients F1-II5 and F1-II7 and the father of Patient S2) compared with OPDM3 patients or controls in both blood and muscle DNA (Fig. 4C, D and Supplementary Fig. 5F).
Transcriptional level of NOTCH2NLC
We conducted quantitative real-time PCR (RT-qPCR) to analyse the RNA transcription level that revealed no significant differences in mRNA levels between OPDM3 patients and age-matched controls in the peripheral blood samples (Fig. 4E). However, in muscle samples, mRNA levels were increased in two OPDM patients (Patients F1-III10 and S2) compared with controls (Fig. 4F). In the asymptomatic carriers (Patients F1-II5 and F1-II7, and father of Patient S2), the NOTCH2NLC mRNA levels were significantly lower in blood compared with control subjects (P < 0.05) (Fig. 4E). With only one muscle sample (Patient F1-II7) available from asymptomatic carriers, we did not observe significant changes in the NOTCH2NLC mRNA levels or the NOTCH2NL protein levels between the muscle tissue from controls and asymptomatic carriers (Fig. 4F and Supplementary Fig. 4H and I).
Toxic protein gain-of-function
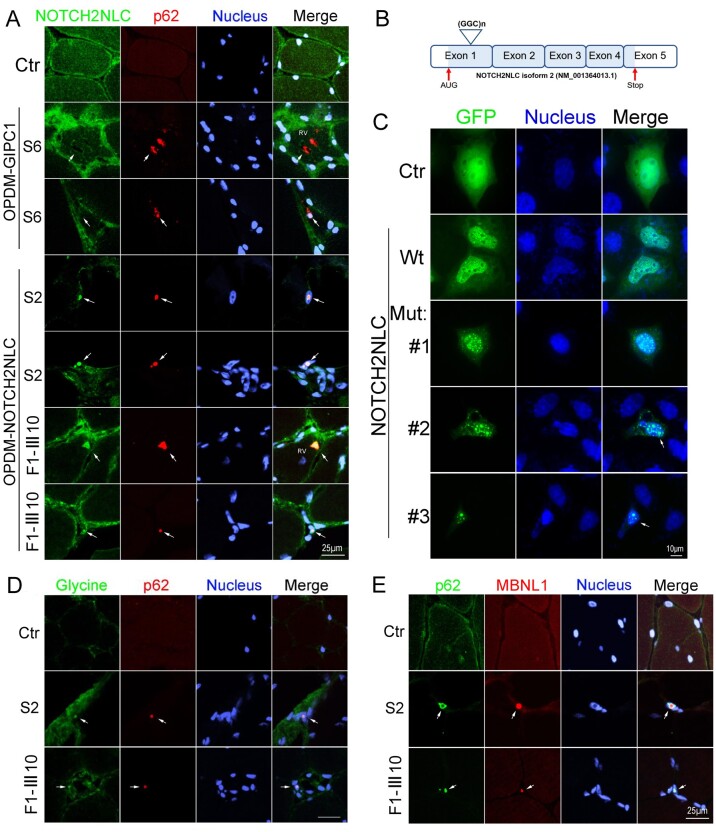
To examine the distribution of NOTCH2NLC protein in muscle fibres of OPDM3 patients, immunofluorescence was carried out on muscle sections from two OPDM3 patients (Patients S2 and F1-III10), an OPDM2 patient,3 and an age-matched control subject. The result indicated co-localization of NOTCH2NLC and p62 in the intranuclear inclusions, as well as in the rimmed vacuoles of OPDM3 patients but not in the control or the OPDM2 patient (Fig. 5A).
Figure 5.
Immunofluorescence on muscle biopsy samples and expression of NOTCH2NLC wild-type or mutant in HEK293 cells. (A) NOTCH2NLC distribution in skeletal muscle of control and OPDM patients. Immunofluorescence showing NOTCH2NLC and p62 co-localized in intranuclear inclusions and rimmed vacuoles of OPDM3 patients (Patients S2 and F1-III10), but not in control or GIPC1-affected OPDM patient (Patient S6), indicated by arrows. (B) Location of GGC repeat expansions in the coding region in the transcript isoform 2 (NM_001364013.1). (C) HEK293 cells were transfected with the control (GFP), NOTCH2NLC wild-type [NOTCH2NLC-(GGC)9] or mutant [NOTCH2NLC-(GGC)69] vectors and subjected to fluorescence observation at 48 h post-transfection. NOTCH2NLC wild-type (Wt) was primarily localized in the nucleus compared to the control, whereas the NOTCH2NLC mutant formed protein aggregates in the nucleus. Different stages of NOTCH2NLC-(GGC)69 aggregation were shown: diffuse small aggregates in Mutant 1, large aggregates in Mutants 2 and 3 (indicated by arrows). The apoptotic cell with condensed nuclei is indicated in Mutant 3. (D) Glycine and p62 co-localized in intranuclear inclusions of OPDM3 patients (Patients S2 and F1-III10), but not in the control (indicated by arrows). (E) RNA binding protein MBNL1 form aggregates in the intranuclear inclusions in OPDM3 patients. Scale bars = 25 μm (A, D and E), and 10 μm (C). Nuclei were counterstained with DAPI.
Since the NOTCH2NLC gene has two transcript isoforms and GGC repeat expansions located in the coding region in the transcript isoform 2 (NM_001364013.1), it is possible that the mutant NOTCH2NLC gene might be translated into a toxic NOTCH2NLC-polyglycine (polyG) protein. To explore this possibility, we constructed NOTCH2NLC wild-type [NOTCH2NLC-(GGC)9] and mutant [NOTCH2NLC-(GGC)69] expressing vectors under the transcript isoform 2 reading frame, resulting in the translation of NOTCH2NLC-polyG protein (Fig. 5B and Supplementary Fig. 6). The control (GFP), NOTCH2NLC wild-type or mutant vectors were transfected into HEK293 cells, and the results showed that NOTCH2NLC wild-type was majorly localized in the nucleus compared to the control. The NOTCH2NLC mutant formed protein aggregates in the nucleus, which recapitulated the major pathological features of NOTCH2NLC-related disorders. It should be noted that NOTCH2NLC-polyG protein showed progressive aggregation, from diffuse small aggregates to large aggregates in the nucleus, which contributed to toxicity and induced cell death (Fig. 5C). To check that the GGC repeat expansions might be translated into polyG, we performed immunofluorescence on muscle sections from two OPDM3 patients (Patients F1-III10 and S2) with anti-glycine and anti-p62 antibodies. The result indicated co-localization of glycine and p62 in the intranuclear inclusions of OPDM3 patients but not in the age-matched control subject (Fig. 5D).
Toxic RNA gain-of-function
RNA toxicity is reported to play an important role in the pathogenesis of NREDs,21,34 in which RNA binding proteins are sequestered into intranuclear inclusions. Here, immunofluorescence was carried out on muscle sections from two OPDM3 patients (Patients S2 and F1-III10) and an age-matched control subject. The results indicated that RNA binding protein hnRNP A/B or MBNL1 were co-localized with p62 in the intranuclear inclusions, in muscle sections from OPDM3 patients but not in the control (Fig. 5E and Supplementary Fig. 7A). Meanwhile, the co-localization of NOTCH2NLC, RNA binding protein MBNL1 and p62 in the intranuclear inclusions were also found in muscle sections from OPDM3 patients (Patients S2 and F1-III10) (Supplementary Fig. 7B). Subsequently, immunofluorescence was carried out on HEK293 cells expressing control (GFP), NOTCH2NLC wild-type or mutant. The results showed that p62 or MBNL1 was partially co-localized with intranuclear inclusions formed by the mutant NOTCH2NLC-polyG protein but not in cells expressing control (GFP) or NOTCH2NLC wild-type (Supplementary Fig. 7C and D). MBNL1 protein expression levels were then examined in HEK293 cells expressing control (GFP), NOTCH2NLC wild-type or mutant NOTCH2NLC. Western blot results indicated that MBNL1 protein levels were unaltered in cells expressing NOTCH2NLC wild-type or mutant compared with the control (Supplementary Fig. 7E and F).
Discussion
OPDM is an adult-onset hereditary muscle disease. Recent studies have shown that OPDM1 was associated with GGC repeat expansions in LRP12,7,13 and OPDM2 was associated with GGC repeat expansions in GIPC1.3 In the present study, we identified that GGC repeat expansions in the NOTCH2NLC gene accounted for 16.7% of patients in a cohort of 24 unrelated Chinese OPDM cases, but not in the 500 unaffected controls, suggesting that the NOTCH2NLC gene was the genetic cause of OPDM3.
Classic genetic linkage analysis was successfully used to identify causative repeat expansions in some large families.35 However, genetic linkage analysis was limited in our patients because there were not enough OPDM patients to explore the underlying genetic causes. Herein, we developed a set of reported pathogenic STRs based on LRS data to uncover possibly related genes. Since our NRED-LRS set can directly uncover possible pathogenic genomes in the known gene pool of expansions in the STR, it can be used for families with limited affected members and sporadic patients without family histories who are impracticable for classic linkage analysis. This method gives us a new strategy to make use of existing LRS resources.
The study broadened the clinical spectrum associated with GGC repeat expansions in NOTCH2NLC to myopathy, which was verified in a cohort of Japanese OPDM patients.36 The GGC repeat expansions in NOTCH2NLC were initially demonstrated to be the genetic cause of NIID.7,14–16 Then the clinical spectrum expanded to Alzheimer’s disease,16 parkinsonism-related disorders,16 essential tremor,37 adult leukoencephalopathy,38 frontotemporal dementia,39 and multiple system atrophy.40 In view of the possible causative relationship between NOTCH2NLC and these neurodegenerative diseases, the term NIID-related disorders (NIIDRD) was defined to describe NIID and other related neurodegenerative diseases caused by the GGC repeat expansions of NOTCH2NLC.16,40 Sone et al.15,41 and Tian et al.16 have reported a subgroup of muscle weakness-dominant NIID (NIID-M), but most patients were ascribed to neurogenic weakness. Symptoms of NIIDRD associated with central and peripheral nervous systems are very common, whereas a predominant myopathy phenotype has not been described or highlighted. Similarly, the GGC repeat expansions of LRP12 has been associated with OPDM1 and symptoms involved with various extraskeletal muscle organs including the central and peripheral nervous systems.42 In the present study, five OPDM3 patients exhibited definite muscle involvement supported by clinical, muscle imaging and muscle biopsy, while subclinical peripheral neuropathy was also observed in some of them. Therefore, we suggest that the clinical spectrum of NOTCH2NLC repeat expansion related diseases (NRERD) is extending to myopathy phenotype.13 Certain genes may cause distinct phenotypes through different mechanisms which may be related to the degree of involvement of different tissues and genetic backgrounds. In a similar scenario, the phenotype of mitochondrial diseases can extend from brain, spinal cord, and nerve to muscle.43 In fact, mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes (MELAS) has been considered as a differentiated disease of NIID.44
In our cohort of 24 OPDM patients, OPDM1, OPDM2 and OPDM3 accounted for 4.17%, 50%, and 16.67% of cases, respectively. Because of the scarcity of detailed clinical information of reported OPDM cases, we compared the clinical features of OPDM1, OPDM2 and OPDM3 in our cohort of patients (Table 2). The age of onset, weakness distribution, CK level, EMG changes and muscle involvement pattern on MRI showed a high similarity among the three subtypes. These OPDM3 patients did not present typical syndromes of adult-onset NIID, such as cognitive impairment, episodic encephalopathy, or parkinsonism. However, it is noteworthy that some OPDM3 patients with long disease duration showed white matter involvement, such as high signal lesions in the corticomedullary junction or the splenium of the corpus callosum that are typical changes of adult-onset NIID.45 Therefore, the subclinical white matter involvement may be a clinical feature of OPDM3 patients at the late stage of disease. Similarly, some patients with essential tremor associated with NOTCH2NLC presented with white matter lesions with disease development.46 Currently, our OPDM1 and OPDM2 patients have not been evaluated by MRI because they did not show CNS symptoms when they visited the myopathy clinics. However, one recent study reported that histological evidence of systemic NIID-like lesions with intranuclear inclusions were observed in OPDM1 patients who were associated with CGG repeat expansion in LRP12.42 Therefore, it is necessary to investigate the relationship between OPDM and leukoencephalopathy in more patients retrospectively and prospectively.
The size of the repeat expansion ranging from 128 to 198 in OPDM3 was not significantly different from the 118 to 517 repeats in NIID-M. Intriguingly, we found that the frequency of non-GGC/GGA/AGC/ACG repeat interruptions in OPDM3 was significantly higher than that in NIID (P = 0.018) (Table 3 and Supplementary Fig. 8). The trinucleotide repeats usually included AGC, ACG and GGA. The repeat numbers of AGC and ACG were not different between OPDM3 and NIID. GGA repeats were not present in OPDM3 patients but were significantly frequent in patients with weakness-dominant NIID or NOTCH2NLC-related essential tremor.15,37 These findings suggested a relationship between different repeat interruptions and phenotypes, i.e. NOTCH2NLC-related essential tremor interrupted by GGA repeats, but myopathy-dominant OPDM3 interrupted by non-GGA/AGC/ACG repeats. Similarly, the pure CAG repeat expansions in ATXN2 cause spinocerebellar ataxia type 247; however, similar length expansions that are interrupted with other codons can present atypically with parkinsonism or amyotrophic lateral sclerosis.48 Therefore, more precise screening of trinucleotide repeats should be conducted in NREDs to understand the phenotypic differences.
Table 3.
Comparison of frequencies of GGC, GGA, AGC and ACG repeats as well as other repeat interruptions between three OPDM3 patients and five NIID patients by LRS
| OPDM3 | NIID | P | |
|---|---|---|---|
| n | 3 | 5 | Null |
| GGC | 0.73 ± 0.11 | 0.77 ± 0.02 | 0.551 |
| GGA | 0.00 ± 0.00 | 0.02 ± 0.03 | 0.3 |
| AGC and ACG | 0.22 ± 0.09 | 0.10 ± 0.08 | 0.278 |
| Others | 0.06 ± 0.04 | 0.10 ± 0.05 | 0.018* |
OPDM3 = NOTCH2NLC affected OPDM.
P < 0.05.
It is proposed that there are three possible mechanisms underlying the pathogenesis of non-coding repeat expansion disorders, i.e. DNA hypermethylation, toxic GGC repeat translation, and RNA gain-of-function. First, the methylation status showed no significant differences between OPDM3 patients and age-matched controls, which is consistent with previous studies.15,16 Second, the commercial anti-NOTCH2NL antibody-labelled protein and anti-glycine antibody-labelled protein could co-localize with p62 in the intranuclear inclusions in muscle samples from OPDM3 patients, and co-localize with the NOTCH2NLC-polyG protein formed intranuclear aggregates in cellular model. This indicates that a transcript isoform 2 (NM_001364013.1) of NOTCH2NLC may use an upstream AUG start codon resulting in a polyG toxicity for OPDM3. Further study is needed to explore whether the transcript isoform 1 (NM_001364012.2) participates in the intranuclear aggregates. Third, RT-qPCR results suggested that elevated NOTCH2NLC mRNA levels in muscle samples from OPDM3 patients might result in RNA toxicity with the transcribed expanded GGC repeats as reported in FMR1.34,49 Additionally, our study showed that multiple RNA binding proteins were accumulated in the intranuclear inclusions in muscle samples and cultured cells, which further suggests that the toxic RNA gain-of-function mechanism may also play a role in the pathogenetic process of OPDM3. Consequently, toxic protein gain-of-function and RNA gain-of-function mechanism may both participate in the pathogenic processes of OPDM3 as observed in fragile X-associated tremor/ataxia syndrome (FXTAS).50–53 However, the mechanism of translating expanded trinucleotide repeats is not understood. Specifically, it is pivotal to question which pathological mechanism drives OPDM pathogenesis. Extensive experiments on cellular and animal models are needed for further investigation.
Interestingly, compared with their affected offspring, the carriers with much higher GGC repeats in NOTCH2NLC, were asymptomatic in the present study. Asymptomatic NOTCH2NLC GGC repeat expansions have not been reported before. Notably, these asymptomatic carriers showed hypermethylation in the promoter region of the NOTCH2NLC gene and significantly lower NOTCH2NLC mRNA levels in blood compared with control subjects. However, the NOTCH2NLC mRNA level and the NOTCH2NL protein level were unaltered in muscle sample from Patient F1-II7, which requires further study on more asymptomatic carriers when available. We speculate that it is possible that the haploinsufficiency of NOTCH2NLC may be non-pathogenic in asymptomatic carriers due to the existence of a wild allele. Additionally, its homologous, NOTCH2NLA and NOTCH2NLB, may also have some compensating function. This indicates that the disease-causing repeat number might have an upper limit, over which may result in hypermethylation (Fig. 4C and D) and thus restricting the transcription of the repeat region. However, considering small sample size in this study, more affected families and further experiments are needed.
In summary, we identify a GGC repeat expansion in the 5′UTR region of the NOTCH2NLC gene as the third pathogenic mutation of OPDM and discover a series of myopathy patients with expanded GGC repeats in NOTCH2NLC. We observed that asymptomatic GGC repeat expansion carriers present with hypermethylation in the NOTCH2NLC gene, which suggests that much higher GGC repeats may be non-pathogenic. This study extends the clinical spectrum associated with the NOTCH2NLC gene, and benefits the understanding of OPDM and NIID. Additionally, OPDM3 should be considered for differential diagnosis of muscle weakness dominant NIID.
Supplementary Material
Acknowledgements
We are indebted to the cooperation of the individuals and their families. We are grateful to Dr Nicolas Charlet-Berguerand (Institut de Génétique et de Biologie Moléculaire et Cellulaire) and Dr Dingfang Bu (Peking University First Hospital) for helpful suggestions. We thank Mr Lijun Chai (Peking University First Hospital) for his work in taking electron microscopy pictures, Ms Yuehuan Zuo and Ms Qiurong Zhang (Peking University First Hospital) for their work in preparations for pathological sections, Ms Xin Shi (Peking University First Hospital) for her work in electrophysiological examination.
Funding
The work was funded by the National Natural Science Foundation of China (No. 81571219, 82071409 and U20A20356 to Z.W. and No. 81801243 to Q.G.). Double thousand talents program of Jiangxi province (jxsq2019101021 to D.H.), Science and technology project of Jiangxi Health Committee (202110028 to D.H.), and Peking University Medicine Fund of Fostering Young Scholars’ Scientific & Technological Innovation (to J.D.).
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
Glossary
- LRS
long-read whole-genome sequencing; NIID = neuronal intranuclear inclusion disease
- NREDs
nucleotide repeat expansion diseases
- OPDM
oculopharyngodistal myopathy
- RP-PCR
repeat-primed polymerase chain reaction
- STR
short tandem repeat
References
- 1. Durmus H, Laval SH, Deymeer F, et al. Oculopharyngodistal myopathy is a distinct entity: Clinical and genetic features of 47 patients. Neurology. 2011;76:227–235. [DOI] [PubMed] [Google Scholar]
- 2. Minami N, Ikezoe K, Kuroda H, Nakabayashi H, Satoyoshi E, Nonaka IJND.. Oculopharyngodistal myopathy is genetically heterogeneous and most cases are distinct from oculopharyngeal muscular dystrophy. Neuromuscul Disord. 2001;11:699–702. [DOI] [PubMed] [Google Scholar]
- 3. Deng J, Yu J, Li P, et al. Expansion of GGC repeat in GIPC1 is associated with oculopharyngodistal myopathy. Am J Hum Genet. 2020;106(6):793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lu H, Luan X, Yuan Y, Dong M, Sun W, Yan C.. The clinical and myopathological features of oculopharyngodistal myopathy in a Chinese family. Neuropathology. 2008;28:599–603. [DOI] [PubMed] [Google Scholar]
- 5. Satoyoshi E, Kinoshita M.. Oculopharyngodistal myopathy. Arch Neurol. 1977;34:89–92. [DOI] [PubMed] [Google Scholar]
- 6. Amato AA, Jackson CE, Ridings LW, Barohn RJ.. Childhood-onset oculopharyngodistal myopathy with chronic intestinal pseudo-obstruction. Muscle Nerve. 1995;18:842–847. [DOI] [PubMed] [Google Scholar]
- 7. Ishiura H, Shibata S, Yoshimura J, et al. Noncoding CGG repeat expansions in neuronal intranuclear inclusion disease, oculopharyngodistal myopathy and an overlapping disease. Nat Genet. 2019;51:1222–1232. [DOI] [PubMed] [Google Scholar]
- 8. Jaspar HH, Bastiaensen LA, ter Laak HJ, Joosten EM, Horstink MW, Stadhouders AM.. Oculopharyngodistal myopathy with early onset and neurogenic features. Clin Neurol Neurosurg. 1977;80:272–282. [DOI] [PubMed] [Google Scholar]
- 9. Mignarri A, Carluccio MA, Malandrini A, et al. The first Italian patient with oculopharyngodistal myopathy: Case report and considerations on differential diagnosis. Neuromuscul Disord. 2012;22:759–762. [DOI] [PubMed] [Google Scholar]
- 10. Uyama E, Uchino M, Chateau D, Tomé FM.. Autosomal recessive oculopharyngodistal myopathy in light of distal myopathy with rimmed vacuoles and oculopharyngeal muscular dystrophy. Neuromuscul Disord. 1998;8:119–125. [DOI] [PubMed] [Google Scholar]
- 11. van der Sluijs BM, ter Laak HJ, Scheffer H, van der Maarel SM, van Engelen BG.. Autosomal recessive oculopharyngodistal myopathy: A distinct phenotypical, histological, and genetic entity. J Neurol Neurosurg Psychiatry. 2004;75:1499–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhao J, Liu J, Xiao J, et al. Clinical and muscle imaging findings in 14 mainland Chinese patients with oculopharyngodistal myopathy. PLoS One. 2015;10:e0128629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Westenberger A, Klein C.. Essential phenotypes of NOTCH2NLC-related repeat expansion disorder. Brain. 2020;143:5–8. [DOI] [PubMed] [Google Scholar]
- 14. Deng J, Gu M, Miao Y, et al. Long-read sequencing identified repeat expansions in the 5'UTR of the NOTCH2NLC gene from Chinese patients with neuronal intranuclear inclusion disease. J Med Genet. 2019;56:758–764. [DOI] [PubMed] [Google Scholar]
- 15. Sone J, Mitsuhashi S, Fujita A, et al. Long-read sequencing identifies GGC repeat expansions in NOTCH2NLC associated with neuronal intranuclear inclusion disease. Nat Genet. 2019;51:1215–1221. [DOI] [PubMed] [Google Scholar]
- 16. Tian Y, Wang JL, Huang W, et al. Expansion of human-specific GGC repeat in neuronal intranuclear inclusion disease-related disorders. Am J Hum Genet. 2019;105:166–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brais B, Bouchard JP, Xie YG, et al. Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy. Nat Genet. 1998;18:164–167. [DOI] [PubMed] [Google Scholar]
- 18. Brook JD, McCurrach ME, Harley HG, et al. Molecular basis of myotonic dystrophy: Expansion of a trinucleotide (CTG) repeat at the 3' end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. [DOI] [PubMed] [Google Scholar]
- 19. Wang Z, Hong D, Zhang W, et al. Severe sensory neuropathy in patients with adult-onset multiple acyl-CoA dehydrogenase deficiency. Neuromuscul Disord. 2016;26(2):170–175. [DOI] [PubMed] [Google Scholar]
- 20. Paulson H. Repeat expansion diseases. Handb Clin Neurol. 2018;147:105–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sznajder ŁJ, Swanson MS.. Short tandem repeat expansions and RNA-mediated pathogenesis in myotonic dystrophy. Int J Mol Sci. 2019;20:3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rafehi H, Szmulewicz DJ, Bennett MF, et al. Bioinformatics-based identification of expanded repeats: A non-reference intronic pentamer expansion in RFC1 causes CANVAS. Am J Hum Genet. 2019;105:151–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tang H, Kirkness EF, Lippert C, et al. Profiling of short-tandem-repeat disease alleles in 12,632 human whole genomes. Am J Hum Genet. 2017;101:700–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tazelaar GHP, Dekker AM, van Vugt JJFA, et al. Association of NIPA1 repeat expansions with amyotrophic lateral sclerosis in a large international cohort. Neurobiol Aging. 2019;74:234.e9–234.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sedlazeck FJ, Rescheneder P, Smolka M, et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat Methods. 2018;15:461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu Q, Zhang P, Wang D, Gu W, Wang K.. Interrogating the “unsequenceable” genomic trinucleotide repeat disorders by long-read sequencing. Genome Med. 2017;9:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Benson G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999;27(2):573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Robinson JT, Thorvaldsdóttir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Simpson JT, Workman RE, Zuzarte PC, David M, Dursi LJ, Timp W.. Detecting DNA cytosine methylation using nanopore sequencing. Nat Methods. 2017;14:407–410. [DOI] [PubMed] [Google Scholar]
- 31. Deng J, Wang P, Chen X, et al. FUS interacts with ATP synthase beta subunit and induces mitochondrial unfolded protein response in cellular and animal models. Proc Natl Acad Sci U S A. 2018;115(41):E9678–E9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Beecroft SJ, Cortese A, Sullivan R, et al. A Māori specific RFC1 pathogenic repeat configuration in CANVAS, likely due to a founder allele. Brain. 2020;143:2673–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cortese A, Simone R, Sullivan R, et al. Biallelic expansion of an intronic repeat in RFC1 is a common cause of late-onset ataxia. Nat Genet. 2019;51:649–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oostra BA, Willemsen R.. FMR1: A gene with three faces. Biochim Biophys Acta. 2009;1790:467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pulst SM. Genetic linkage analysis. Arch Neurol. 1999;56:667–672. [DOI] [PubMed] [Google Scholar]
- 36. Ogasawara M, Iida A, Kumutpongpanich T, et al. CGG expansion in NOTCH2NLC is associated with oculopharyngodistal myopathy with neurological manifestations. Acta Neuropathol Commun. 2020;8(1):204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sun QY, Xu Q, Tian Y, et al. Expansion of GGC repeat in the human-specific NOTCH2NLC gene is associated with essential tremor. Brain. 2020;143:222–233. [DOI] [PubMed] [Google Scholar]
- 38. Okubo M, Doi H, Fukai R, et al. GGC repeat expansion of NOTCH2NLC in adult patients with leukoencephalopathy. Ann Neurol. 2019;86:962‐968. [DOI] [PubMed] [Google Scholar]
- 39. Jiao B, Zhou L, Zhou Y, et al. Identification of expanded repeats in NOTCH2NLC in neurodegenerative dementias. Neurobiol Aging. 2020;89:142.e1–142.e7. [DOI] [PubMed] [Google Scholar]
- 40. Fang P, Yu Y, Yao S, et al. Repeat expansion scanning of the NOTCH2NLC gene in patients with multiple system atrophy. Ann Clin Transl Neurol. 2020;7:517–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sone J, Mori K, Inagaki T, et al. Clinicopathological features of adult-onset neuronal intranuclear inclusion disease. Brain. 2016;139:3170–3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Saito R, Shimizu H, Miura T, et al. Oculopharyngodistal myopathy with coexisting histology of systemic neuronal intranuclear inclusion disease: Clinicopathologic features of an autopsied patient harboring CGG repeat expansions in LRP12. Acta Neuropathol Commun. 2020;8(1):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. DiMauro S, Schon EA, Carelli V, Hirano M.. The clinical maze of mitochondrial neurology. Nat Rev Neurol. 2013;9:429–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liang H, Wang B, Li Q, et al. Clinical and pathological features in adult-onset NIID patients with cortical enhancement. J Neurol. 2020;267:3187–3198. [DOI] [PubMed] [Google Scholar]
- 45. Wang Y, Wang B, Wang L, et al. Diagnostic indicators for adult-onset neuronal intranuclear inclusion disease. Clin Neuropathol. 2020;39:7–18. [DOI] [PubMed] [Google Scholar]
- 46. Chen H, Lu L, Wang B, et al. Essential tremor as the early symptom of NOTCH2NLC gene-related repeat expansion disorder. Brain. 2020;143:e56. [DOI] [PubMed] [Google Scholar]
- 47. Ramos EM, Martins S, Alonso I, et al. Common origin of pure and interrupted repeat expansions in spinocerebellar ataxia type 2 (SCA2). Am J Med Genet B Neuropsychiatr Genet. 2010;153B:524–531. [DOI] [PubMed] [Google Scholar]
- 48. Yu Z, Zhu Y, Chen-Plotkin AS, et al. PolyQ repeat expansions in ATXN2 associated with ALS are CAA interrupted repeats. PLoS One. 2011;6:e17951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Glineburg MR, Todd PK, Charlet-Berguerand N, Sellier C.. Repeat-associated non-AUG (RAN) translation and other molecular mechanisms in fragile X tremor ataxia syndrome. Brain Res. 2018;1693(Pt A):43–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hagerman RJ, Berry-Kravis E, Hazlett HC, et al. Fragile X syndrome. Nat Rev Dis Primers. 2017;3:17065. [DOI] [PubMed] [Google Scholar]
- 51. Sellier C, Buijsen RAM, He F, et al. Translation of expanded CGG repeats into FMRpolyG is pathogenic and may contribute to fragile X tremor ataxia syndrome. Neuron. 2017;93:331–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sellier C, Freyermuth F, Tabet R, et al. Sequestration of DROSHA and DGCR8 by expanded CGG RNA repeats alters microRNA processing in fragile X-associated tremor/ataxia syndrome. Cell Rep. 2013;3:869–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zu T, Gibbens B, Doty NS, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A. 2011;108:260–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary material.