Abstract
Background
Coccidioidomycosis is often diagnosed with a collection of tests that measure a patient’s ability to mount an immune response to the fungus (antibody-based diagnostics) utilizing fungal protein preparations. Here we present an antigen-based assay that detects and quantifies coccidioidal chitinase-1 (CTS1) in diagnostic antigen preparations with potential for use in human serum.
Methods
An inhibition-based enzyme-linked immunoassay (ELISA) was developed that utilizes a monoclonal antibody specific for coccidioidal CTS1. CTS1 was quantified in commercial antigen preparations using recombinant CTS1 as a standard. Sera from 192 individuals from an endemic area were tested which included 78 patients (40.6%) with proven or probable coccidioidomycosis.
Results
The quantity of CTS1 in diagnostic commercial antigen preparations from different suppliers varied. Temporal constraints of availability of different lots of commercial antigens does not allow for immediate comparison of lot-to-lot variability. Assay results from patient serum samples correlated with low- and high-titer serology from patients with a coccidioidomycosis diagnosis. Further analysis suggested that patient derived anti-CTS1 antibodies may overlap with the mouse monoclonal antibody used in the assay. This unexpected overlap in CTS1 binding suggests the assay can detect antigen, antibody, or both, which contributes to its high level of clinical sensitivity of 89.74% and specificity of 94.90%.
Conclusions
The CTS1 inhibition ELISA described in this report is a promising tool to aid in quality control of antigens used in the diagnosis of coccidioidomycosis. Further optimization is needed to harness its utility as a diagnostic tool to aid in diagnosis and disease monitoring of coccidioidomycosis.
Keywords: Valley Fever, coccidioidomycosis, diagnostic, ELISA
Diagnosis of coccidioidomycosis often relies on the host’s immune response. Here we present an antigen-based assay that detects and quantifies coccidioidal chitinase-1 in diagnostic antigen preparations. In human serum, the assay also detects host antibodies, increasing its clinical sensitivity
Coccidioidomycosis, or Valley Fever (VF), is an infection caused by the pathogenic fungi Coccidioides immitis and Coccidioides posadasii. Coccidioides spp. inhabit arid areas throughout the Americas [1–3] including Arizona and California, both of which have experienced a significant increase in incidence since 2014 [4–6]. Infection may be asymptomatic or manifest as a pneumonia difficult to distinguish from community-acquired viral or bacterial pneumonia, which may lead to inappropriate treatment [7, 8]. In a small but significant number of cases, extra-pulmonary dissemination of the fungus occurs resulting in significant morbidity, need for long-term antifungal therapy, and sometimes in death [9, 10].
Coccidioides spp. are dimorphic fungi that exist in two forms dependent on their environment. In the soil, Coccidioides grow as septate mycelia composed of arthroconidia that become aerosolized upon disruption and are easily inhaled into the lungs of a human or animal host [11]. In the host, the fungus transforms into its spherule growth form that undergoes maturation, internally dividing and producing endospores [11]. Production of chitin, a major component in the cell wall of spherules, increases significantly during the beginning of the endosporulation process [12, 13]. This surge is then diminished with the progression of endospore formation and release, at which time chitinases are detectable [9, 13]. Among these is chitinase-1 (CTS1), a 48-kDa protein that plays a role in weakening the spherule cell wall prior to endospore release [14–16]. The presence of CTS1 is shown to decrease in vitro shortly after endosporulation due to an upregulation of proteases [9, 17], so the process of spherule growth and endosporulation can restart.
CTS1 is also known as the “CF” antigen used in serodiagnostics, namely complement fixation (CF) assays and its immunodiffusion (ID) correspondent (IDCF) [15, 16]. CTS1 is also utilized in enzyme immunoassay (EIA) formats [18]. EIA and ID can distinguish between IgM and IgG antibodies, while CF can quantify the antibody response and measure disease progression and/or regression [8, 19]. The sensitivity and specificity of these assays in diagnosis have been evaluated by multiple groups [20–26], and although helpful, the sensitivity of serology is still lacking, especially in early infection and in immunosuppressed individuals [9, 24, 27]. Specificity can also be a problem. Several groups have recombinantly produced and characterized CTS1 in an effort to increase sensitivity and specificity for detecting antibodies in patients [18, 28–31]. Still, it can take weeks to months after onset of symptoms to detect an immune response [9, 24, 32]. Unlike many serologic assays for infectious diseases, IgG is not a marker of distant infection but instead used to diagnose recent or latent infection [25]. As disease resolves, the antibodies detected by clinical assays wane. Alternatively, resolution of fungal infection may be incomplete (latent) or unconfirmed due to a long period of detectable antibody [9, 33]. Thus, while serology continues to be a mainstay diagnostic tool, it is an incomplete and imperfect approach.
Morphological and growth-based diagnostics such as microscopy and culture, respectively, exist, but both are lacking in sensitivity and the latter poses a risk to laboratory personnel despite being considered the gold-standard of diagnosis [8, 34]. Polymerase chain reaction (PCR) for Coccidioides has since been developed but demonstrates a sensitivity similar to that of culture [35, 36]. Further, detection of coccidioidal antigens has been approached using spherule lysate [37], and more recently using galactomannan [38]. No follow up evaluations of the former have been explored, but the latter has shown value in diagnosing more severe-forms of disease such as coccidioidal meningitis [38, 39]. A recent publication affirmed the modest incremental value of performing the Coccidioides galactomannan antigen assay, though the assay was positive for only a minority of non-disseminated cases [26]. This experience has therefore highlighted the need for a sensitive antigen-based diagnostic test that detects and/or measures direct biomarkers from Coccidioides spp. as opposed to a patient response. In this report, we present the development of a new enzyme-linked immunoassay (ELISA) that measures coccidioidal CTS1 antigen in commercial antigen preparations. While its ability to exclusively quantify antigen in human serum is obfuscated due to interference from endogenous antibodies, the clinical sensitivity to detect active infection appears superior to existing serologic methods.
METHODS
Production and Purification of Recombinant CTS1
The CTS1 gene was provided to us as a kind gift from Dr. Mitch Magee (Arizona State University). We cloned CTS1 into pcDNA3.1 V5/HisA, verified its identity by sequence analysis (Supplementary Figure 1) and transfected it into 293F cells (Thermo Freestyle Expression System). Seven-day supernatants were harvested and recombinant CTS1 (rCTS1) was purified on a nickel column via a C-terminal histidine tag. Purified protein was quantified using a BCA protein assay kit (Thermo Scientific) and frozen at -80°C in 250ul aliquots at 1 mg/ml.
Monoclonal Antibody Generation and Purification
Mice were immunized and boosted with rCTS1 mixed with Magic Mouse Adjuvant (Creative Diagnostics) under an IACUC approved protocol at Mayo Clinic. Anti-CTS1 antibody titers were monitored by rCTS1-coated 96-well plates. When a sufficient antibody titer was reached (>1:32 000), mice were sacrificed and spleens were processed into single cell suspensions. Splenocytes were fused with myeloma cells (P3X63Ag8.653) by a standard hybridoma generation technique [40]. Briefly, splenocytes were fused with P3X63Ag8.653 myeloma cells at a ratio of 1 splenocyte: 2 myeloma cells using 50% polyethylene glycol solution (Sigma-Aldrich). The fused cells were resuspended in hypoxanthine-aminopterin-thymidine (HAT) selective medium (Sigma-Aldrich) and plated at 50 000 splenocytes per well in a 96-well plate. Plates were incubated at 37°C with 5% CO2 for ten days. Supernatant was collected and tested by indirect ELISA for antibodies against rCTS1. Positive wells were subcloned by limiting dilution of one cell per well and re-screened using the same procedure after ten days. Positive subclones were cultured in 10% FBS cDMEM for antibody purification by protein A/G (Thermo Scientific) chromatography. Multiple mAbs against rCTS1 were identified, but one in particular, 2F11, performed well in our inhibition assay.
Western Blot
rCTS1 was either treated with PNGaseF or untreated and subjected to SDS-PAGE under reducing conditions using 12% polyacrylamide gels at 140 V for 60 minutes (Bio-Rad) followed by staining with Coomassie Blue dye. For Western blot analysis, electrophoresed proteins were transferred to a PVDF membrane using a western blot apparatus (Bio-Rad). After transfer, membranes were blocked in 1% BSA for at least one hour followed by incubation with anti-CTS1 mouse monoclonal antibody 2F11 at 2 ug/ml in 1% BSA in PBS for 1 hour. Membranes were then washed three times with tris-buffered saline, 0.1% Tween-20 (TBST) followed by addition of peroxidase-conjugated goat anti-mouse IgG antibody (Jackson ImmunoResearch) at a dilution of 1:5000 for 45 minutes. Membranes were subsequently washed four times with TBST followed by addition of nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl-phosphate (Thermo Scientific) substrate for development.
Inhibition ELISA
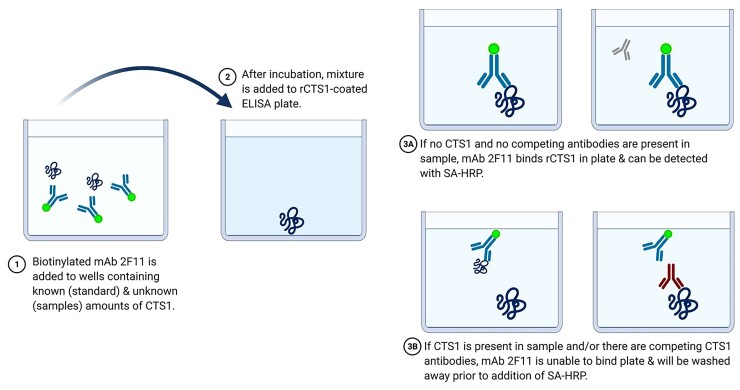
The first step of the inhibition assay requires pre-incubating a biofluid potentially containing Coccidioides-produced CTS1 with a calibrated concentration of biotinylated 2F11 anti-CTS1 mAb. Then, the solution is transferred to rCTS1-coated ELISA plates (Figure 1). A sample that does not contain any CTS1 antigen or anti-CTS1 antibodies would result in 2F11 mAb binding to rCTS1 coated on the plate, whereas a sample containing CTS1 antigen or anti-CTS1 antibodies that overlap binding of 2F11 would inhibit 2F11 mAb from binding to plate-bound rCTS1. A standard of rCTS1 was run with each test so that CTS1 in the biofluid could be compared at an appropriate dilution along the linear portion of the standard curve. Results for biofluids that may contain anti-CTS1 antibodies were reported in EIA units in place of utilizing the standard curve, which quantifies detection of antigen alone.
Figure 1.
Diagram of inhibition assay used for quantification of CTS1. Biotinylated monoclonal antibody 2F11 is incubated with patient serum at different dilutions for one hour. After incubation, mixture is added to an ELISA plate that has been coated with rCTS1 and allowed to incubate. If CTS1 antigen and/or anti-CTS1 antibodies are present in the sample being tested, the antibody will be blocked from being able to bind rCTS1 in the ELISA plate, generating low signal. If CTS1 antigen and anti-CTS1 antibodies are not present, the antibody will bind rCTS1 in the ELISA plate, generating high signal. Schematic was created with BioRender.com.
Commercial antigen preparations and human sera were tested undiluted and at 2-fold dilutions in 1% BSA. The assay was performed using a 96-well flat bottom plate (Corning) coated with rCTS1 at 2 ug/ml for 75 minutes at 37°C followed by blocking overnight in 1% BSA in 1X PBS at 4°C. The following day, rCTS1 standard was spiked in a 96-well U-bottom plate (Corning) at a known concentration (16 ug/ml) into 1% BSA or normal donor serum followed by ten 2-fold dilutions into 1% BSA. Biotinylated 2F11 mAb was added to either commercial antigen preparations or sera at an equal volume such that the final concentration of 2F11 was 0.1 ug/ml, final dilution of standard 8ug/ml, and final dilution of samples 2-fold greater than starting dilution (eg, starting dilution of 1:2 became 1:4). Samples were mixed by gently tapping the 96-well U-bottom plates followed by incubation at room temperature for one hour. Samples were then transferred to rCTS1-coated ELISA plates and incubated an additional hour. ELISA plates were then washed three times with 1X PBS, 0.05% Tween-20 (PBST), followed by addition of Streptavidin-HRP (BD Biosciences) at a dilution of 1:5000 and incubated for 45 minutes. The plates were washed three times in PBST, then developed with 3,3′,5,5′ -Tetramethylbenzidine substrate (BD Biosciences) for ten minutes. Sulfuric acid 0.16 M was added to stop development and the plate was read at 450 nm.
Assay Limits and Precision
A standard curve for quantification of CTS1 was generated by spiking a known concentration of rCTS1 into either 1% BSA or normal donor serum followed by two-fold dilutions into 1% BSA. The limit of blank (LOB) and limit of detection (LOD) were determined by measuring the standard curve in triplicate. The LOB was calculated by taking the mean OD value of triplicate blank samples and subtracting 1.645 times their standard deviation [41]. Subtraction instead of addition of standard deviation was used due to the reverse nature of our standard curve (low optical density corresponds to high concentration of CTS1 while high optical density corresponds to low concentration of CTS1). The LOD was determined by using the LOB and replicates of the CTS1 standard with concentrations that approached the LOB with the following equation: LOD = LOB – 1.645(SDlow concentration sample) [41]. Once again, subtraction of SD was utilized in place of addition due to the reverse nature of our standard curve. The LOD optical density value was then back calculated to a concentration of CTS1 to account for minor day-to-day variation. This process was repeated across three days for both rCTS1 spiked into 1% BSA and normal donor serum, with dilutions into 1% BSA. The average LOD concentration is reported. The back-calculated concentrations from these replicates were used to calculate intra- and inter-assay precision of the CTS1 standard (Supplementary Table 1). For human serum tested, in place of back-calculation, OD450 values were normalized by dividing the patient OD450 value by the average of the negative control per plate. Normalized values were multiplied by a factor of 100 to convert data into more comprehensible integers, termed EIA units. Dilution was accounted for by dividing by n, where n is the exponent of dilution in 2n. Division instead of multiplication of dilution factor is utilized due to the reverse nature of our assay. The numbers reported are EIA units, which are arbitrary to our assay and do not discriminate if antigen or antibody is being detected.
Clinical Specimens
Human sera were obtained under ASU IRB 0601000548 and Mayo Clinic IRB 12-000965. Samples were randomly selected from those sent for at least one routine Coccidioides-related diagnostic test and were collected between May-September 2018 and stored <−20°C until use. Samples were tested in dilution replicates of 1:2–1:32. If dilution replicate did not allow for quantification within the assay limits, samples were re-tested at higher dilutions. The status of each patient at time of sample collection was reviewed and categorized as “proven”, “probable”, or “not coccidioidomycosis” using European Organization for Research and Treatment of Cancer (EORTC) and Mycoses Study Group (MSG) criteria for endemic mycoses, with clarifying criteria for “probable” based on our published clinical composite reference standard [25, 42]. Any patient that was not classified as “proven”, “probable” or “not coccidioidomycosis” was categorized as “possible”. Briefly, patients who had Coccidioides identified by culture, histopathology, or PCR were classified as “proven”. Patients with concurrent clinical findings (including either radiology findings [25] or symptoms [25]) along with positive serology (antibodies) for Coccidioides were classified as “probable”. Our group classified anyone with either relevant clinical findings or positive serology, but not both, as “possible” coccidioidomycosis, however there is not a clear consensus about what criteria must be met for this classification [42]. Since the true nature of the “possible” category cannot be known, these patients were excluded from the sensitivity and specificity analysis. Any patient who did not have “proven”, “probable”, or “possible” coccidioidomycosis, or was diagnosed with a different illness, was classified as “not coccidioidomycosis.”
Patient Consent Statement
This study does not include factors necessitating patient consent.
Statistical Analysis
Receiver operating characteristic analysis was used to determine the cutoff for positivity as well as estimate sensitivity and specificity.
RESULTS
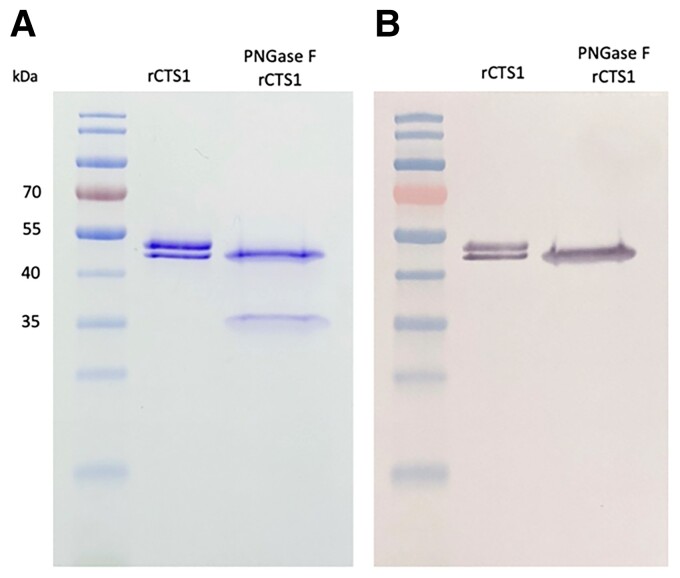
Recombinant CTS1 was electrophoresed with and without pre-treatment with PNGase F (Figure 2A). rCTS1 appears as a doublet band, which is presumed to be a glycosylation since rCTS1 appears as a single band after deglycosylation with PNGase F (∼35 kDa). Although multiple mAbs were identified from anti-CTS1-secreting mouse hybridomas, one mAb in particular, 2F11, demonstrated binding in both ELISA and western blotting. A western blot of 2F11 reacting with rCTS1 is shown in Figure 2B. This antibody was used to develop the inhibition assay. Dilutions of rCTS1 were used in an empirical approach to establish the limit of blank, which was then used to calculate the limit of detection of Coccidioides rCTS1, 155 ng/ml (SD 0.022 µg/ml).
Figure 2.
(A) Coomassie blue-stained SDS-PAGE of rCTS1 and PNGase F-treated rCTS1. (B) Western blot probed with 2F11 mAb.
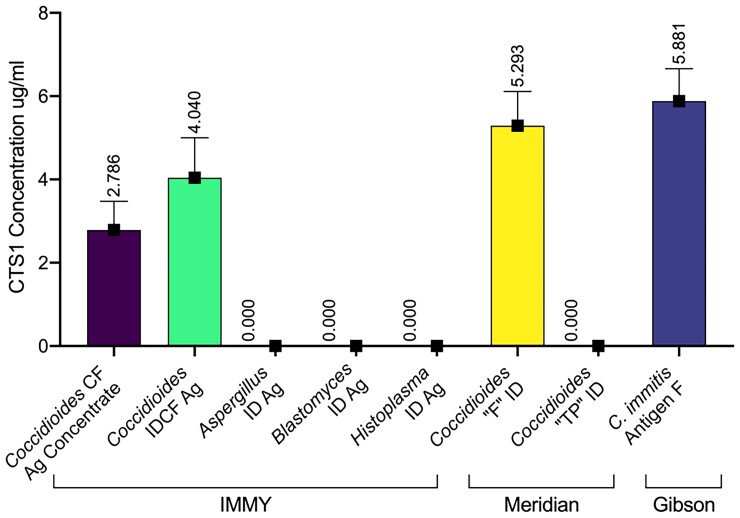
The first step of characterizing the assay was to test it against commercially available antigen preparations. These included Coccidioides CF Antigen Concentrate (IMMY, Norman, OK), Coccidioides IDCF Antigen (IMMY, Norman, OK), Coccidioides “F” Antigen for Immunodiffusion (Meridian Biosciences, Cincinnati, OH), and Coccidioides immitis Antigen F (Gibson Bioscience, Lexington, KY). The quantity of CTS1 in each was 2.79 ug/ml, 4.04 ug/ml, 5.29 ug/ml, and 5.88 ug/ml, respectively (Figure 3). Commercially available antigen preparations for Aspergillus, Blastomyces, and Histoplasma ID assays (IMMY, Norman, OK) were also tested, but no CTS1 was detected in non-coccidioidal antigen preparations, demonstrating the specificity of 2F11 mAb (Figure 3).
Figure 3.
Quantification of CTS1 in different commercially available antigen preparations used in fungal diagnostics. Brackets at the bottom designate manufacturers of each antigen preparation. For values below 0.155 ug/ml (the analytical limit of detection), the assigned value was zero.
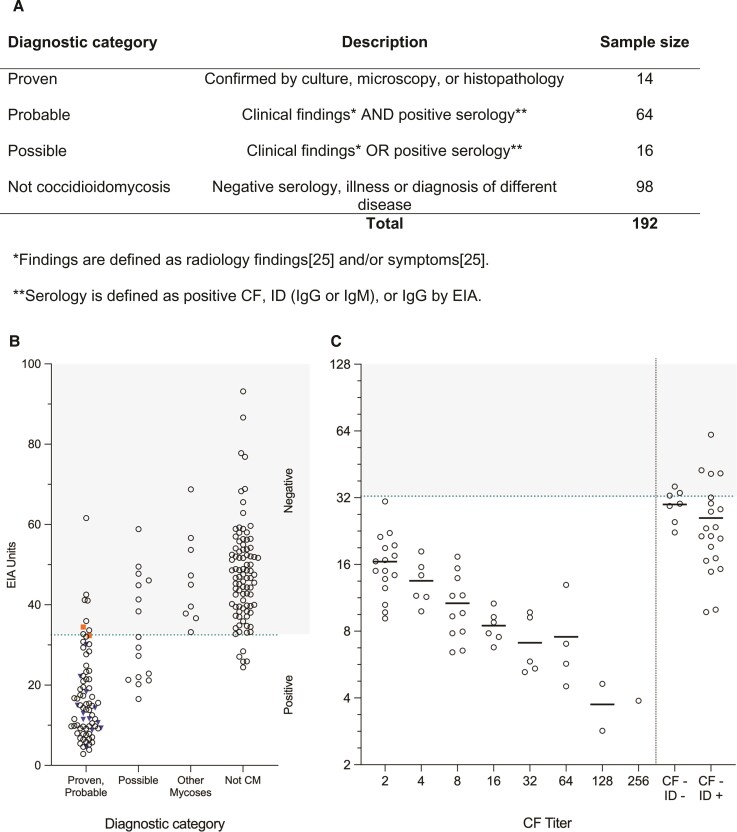
The same inhibition ELISA format was used to test 192 pre-existing serum samples, of which 78 (40.6%) patients had proven or probable coccidioidomycosis, 16 (8.3%) had possible coccidioidomycosis, and the remaining 98 (51.0%) did not have coccidioidomycosis (Figure 4A). Of the 98 samples classified as not coccidioidomycosis, 9 (4.7%) were diagnosed with a different fungal infection (Aspergillus n = 6, by Aspergillus galactomannan antigen, and Candida n = 3, by fungal culture). Mean EIA units of dilution replicates for all samples tested are shown in Figure 4B. Of the 78 patients with proven or probable coccidioidomycosis, 51 had a positive complement fixation titer ranging from 1:2 to 1:256. The mean EIA units of these patients in relation to CF titer, as well as in patients without a CF titer, is shown in Figure 4C. Importantly, four patient serum samples that did not have CF titers and were negative for ID show reactivity in our assay (Figure 4C).
Figure 4.
(A) Composition of serum samples tested and sorted by diagnosis category. (B) Calculated EIA units in all patient serum tested, separated by diagnostic category. Blue inverted triangles represent patients with proven diagnosis, while orange squares indicate patients with proven coccidioidomycosis whose disease appeared to be resolved at time of specimen collection (no symptoms and negative serology, see Supplementary Table 2). A dotted line represents the cutoff value for positivity at 32.5 units, with any value above this categorized as a negative result (shaded area). (C) Correlation of EIA units with serologic antibody titers determined by complement fixation. Individual dots on the left side of the line show EIA units determined by ELISA in 51 proven and probable patients with a positive CF titer (grouped by CF titer on x-axis). Individual dots on the right side of the line show EIA units in 27 proven and probable patients with a negative CF titer and either positive or negative ID results. Average EIA units by titer are represented with a horizontal black line.
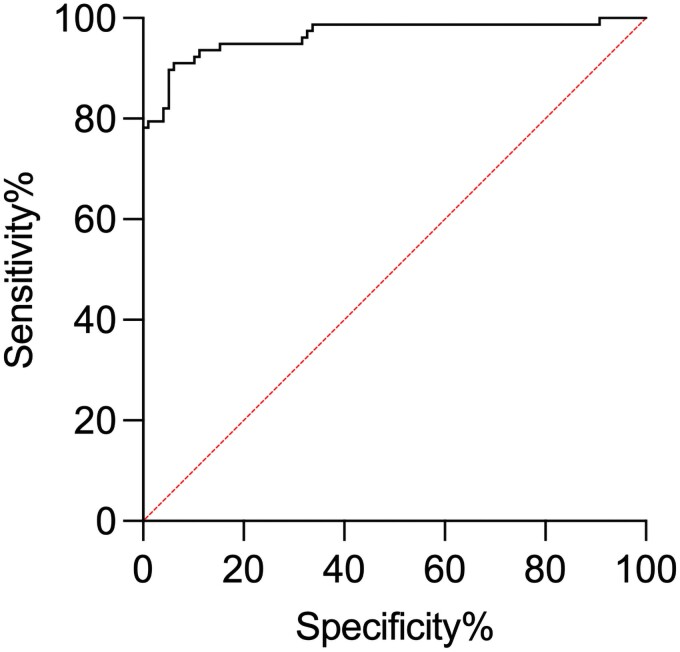
A receiver operating characteristic curve (ROC) was plotted using patients with proven or probable coccidioidomycosis as true positives and not coccidioidomycosis patients as true negatives (Figure 5) while excluding the possible group, as their status is unknown. The area under the receiver operating characteristic curve was 0.9652 (SE, 0.01441; 95%CI,.9370–.9934; P < .0001) using a positive cutoff value of 32.5 EIA units, resulting in a sensitivity and specificity of 89.74% and 94.90%, respectively.
Figure 5.
Receiver operating characteristic (ROC) plot for the diagnosis of coccidioidomycosis with use of the inhibition ELISA. Patients with proven and probable coccidioidomycosis represent the true positive group while patients categorized as not coccidioidomycosis make up the true negative group. Patients classified as possible coccidioidomycosis were excluded from this analysis. The area under the curve (AUC) was 0.9652 (SE, 0.01441; 95% CI,.9370–.9939; P < .0001). With a cutoff of32.5 EIA units, the sensitivity is 89.74% and the specificity is 94.90%.
DISCUSSION
Current diagnostic methods for coccidioidomycosis rely on a constellation of clinical, radiologic and laboratory findings that may be conflicting, and in the case of serodiagnostics may be inconsistent [11, 20, 24, 25, 43]. However, their collective use is the current accepted standard used to inform clinical decisions, and often repeated testing over time is needed to clarify patient status. Further complicating diagnosis is the possibility that early treatment of coccidioidomycosis may abrogate serocoversion [44], which can make definitive diagnosis nearly impossible in some patients with other co-morbidities that have overlapping symptoms. Another group did not report this phenomenon [45]. Although a comparison cannot be made between the two studies, the different experience of each group highlights the challenges of antibody-based methods as a component of diagnosis. Yet, various CTS1 antigen preparations have been used in antibody-based diagnostic assays for over six decades. Although CTS1 may be an accepted serologic target, antibody responses in individuals infected with Coccidioides spp. are often delayed or even absent, especially in immunocompromised patients [24]. However, if a fungal antigen could be detected in a biofluid, a more definitive diagnosis could be made, independent of the host immune response.
Complexity and inconsistency in the current diagnostic testing regimen make evaluation of a new diagnostic tool challenging. Several groups have proposed criteria to classify patients with varying certainties of coccidioidomycosis infection to aid with this challenge. We opted to categorize the patients included in this study using the general definitions offered by the EORTC and the MSG [42] who provide criteria for proven, probable, and not coccidioidomycosis. The remaining patients who did not fit these criteria were categorized as possible coccidioidomycosis and were excluded from our sensitivity and specificity analysis since their true status is unknown. The results show that the use of our EIA provides sensitivity and specificity comparable only to the use of multiple existing serodiagnostic tests [24–26]. While our assay measures CTS1 antigen, potential interference from anti-CTS1 antibodies in patient sera may interfere with absolute CTS1 antigen quantification. Nonetheless, our assay was positive for 4 sera from patients who had negative CF and ID results, demonstrating the value of the assay in detecting active infection. Our updated results suggest that, for human specimens, the assay yields a positive result from either CTS1 antigen or antibodies against CTS1, providing a distinct advantage over other assays. This feature was borne out in the clinical performance of the assay, whereby this assay alone was as good as the composite clinical standard, and better than any other single serologic approach. Some patients take many weeks to become antibody-positive using current tests [24] and are given one or more courses of unnecessary antibiotics, posing risk for patients and contributing to antimicrobial resistance.
Another use of this assay is to measure the CTS1 present in antigen preparations, and potential lot to lot variability. While the relative amounts of CTS1 between commercial products is somewhat irrelevant, it is essential to have consistent reagents for diagnostic methods. Importantly, Histoplasma and Blastomyces antigen ID preparations from IMMY did not cross-react on our assay, demonstrating a critical feature of analytical specificity of the assay. Nine patients had other fungal infections and were negative by the assay, suggesting that the assay has good clinical specificity. More samples from patients with other fungal infections will need to be tested to further characterize clinical specificity of the assay.
We must also acknowledge that the samples tested in this study are a single point in time from individuals at different stages of disease with a wide range in time since diagnosis, time since treatment initiation, and/or time since symptom resolution. In some cases, the course of disease led to evidence that would categorize the patient as a proven case. We categorized patient samples with information available at the time of blood draw. Additional context from future events is available in Supplementary Table 2 for thirty patients. In one example, Patient 2 was characterized as probable due to innumerable pulmonary nodules and positive serology by CF (1:2), ID, and EIA. The patient’s serum was quantified at 9.13 EIA units at that time. The patient was initiated on antifungal treatment 4–5 weeks later, however 11 months later returned a positive PCR, confirming proven infection. It would be difficult to know whether this patient was sufficiently treated, given this presentation. Using the CTS1 EIA in this report, trending of antigenemia and/or seroreactivity may correlate with clinical response and/or response to treatment. This assay is more likely to be sensitive in early infection, detecting antigen, as well as sustaining sensitivity for active disease by also detecting antibody. To investigate this possibility, we plan to test samples longitudinally and correlate course of disease with treatment and other diagnostic tests. A longitudinal study will also allow us to investigate the potential utility of this assay for earlier diagnosis, which would be helpful in the management of patients. Assessment of the performance of this assay using specimen types other than serum, such as cerebrospinal fluid, will also be considered in future studies.
The one other antigen assay described for Coccidioides was recently shown to be positive in only 38.6% of sera from immunocompetent patients, and only 37.1% of pulmonary cases [26]. That assay is also positive more often in disseminated cases, which typically have other indicators of infection. The CTS1 detection assay reported here has the potential for similar or better performance vs. serologic assays. Although additional prospective studies are needed to further evaluate and define the role of this test in the clinical laboratory, it may hold promise as a useful diagnostic tool for a disease that can be challenging to diagnose.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online (http://academic.oup.com/ofid). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Supplementary Material
Contributor Information
Francisca J Grill, School of Life Sciences, Arizona State University, Tempe, AZ, USA.
Thomas E Grys, Department of Laboratory Medicine and Pathology, Mayo Clinic, Phoenix, AZ, USA.
Marie F Grill, Department of Neurology, Mayo Clinic, Phoenix, AZ, USA.
Alexa Roeder, School of Life Sciences, Arizona State University, Tempe, AZ, USA.
Janis E Blair, Division of Infectious Diseases, Mayo Clinic, Phoenix, AZ, USA.
Douglas F Lake, School of Life Sciences, Arizona State University, Tempe, AZ, USA.
Notes
Acknowledgements. We would like to thank the Mayo Clinic Department of Laboratory Medicine and Pathology for their support in acquiring samples to use in this study.
Financial support. This work was supported by the Arizona Biomedical Research Commission [ADHS16-162513] and the Mayo Clinic Center for Individualized Medicine
Potential conflicts of interest. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Author Contributions. Francisca J. Grill: Investigation, Methodology, Data Collection, Retrospective Chart Review, Writing – Original Draft and Editing; Thomas E. Grys: Conceptualization, Methodology – Sample Classification, Resources, Writing – Reviewing and Editing; Marie F. Grill: Methodology – Sample Classification, Writing – Reviewing and Editing; Alexa J. Roeder: Sample Collection, Retrospective Chart Review; Janis E. Blair: Methodology – Sample Classification, Writing – Reviewing and Editing; Douglas F. Lake: Conceptualization, Methodology, Resources, Supervision, Writing – Reviewing and Editing.
References
- 1. Hector RF, Laniado-Laborin R. Coccidioidomycosis- A fungal disease of the Americas. PLoS Med 2005; 2:0015–18. doi: 10.1371/journal.pmed.0020002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Saubolle MA, McKellar PP, Sussland D. Epidemiologic, clinical, and diagnostic aspects of coccidioidomycosis. J Clin Microbiol 2007; 45:26–30. doi: 10.1128/JCM.02230-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stewart ER, Thompson GR. Update on the epidemiology of coccidioidomycosis. Curr Fungal Infect Rep 2016; 10:141–6. doi: 10.1007/s12281-016-0266-1 [DOI] [Google Scholar]
- 4.https://www.cdc.gov/fungal/diseases/coccidioidomycosis/statistics.html Centers for Disease Control and Prevention. Valley Fever (Coccidioidomycosis) Statistics [Internet]. [cited 2020 Mar 2]. Available from:
- 5. Wilson L, Ting J, Lin H, et al. . The rise of valley fever: Prevalence and cost burden of coccidioidomycosis infection in California. Int J Environ Res Public Health 2019; 16. doi: 10.3390/ijerph16071113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grizzle AJ, Wilson L, Nix DE, Galgiani JN. Clinical and economic burden of valley fever in arizona: an incidence-based cost-of-illness analysis. Open Forum Infect Dis 2021; 8:1–9. doi: 10.1093/ofid/ofaa623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blair JE, Chang YHH, Cheng MR, et al. . Characteristics of patients with mild to moderate primary pulmonary coccidioidomycosis. Emerg Infect Dis 2014; 20:983–90. doi: 10.3201/eid2006.131842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Malo J, Luraschi-Monjagatta C, Wolk DM, Thompson R, Hage CA, Knox KS. Update on the diagnosis of pulmonary coccidioidomycosis. Ann Am Thorac Soc 2014; 11:243–53. doi: 10.1513/AnnalsATS.201308-286FR [DOI] [PubMed] [Google Scholar]
- 9. Pappagianis D, Zimmer BL. Serology of coccidioidomycosis. Clin Microbiol Rev 1990; 3:247–68. doi: 10.1128/CMR.3.3.247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Adam RD, Elliott SP, Taljanovic MS. The spectrum and presentation of disseminated coccidioidomycosis. Am J Med. Elsevier Inc.; 2009; 122:770–7. doi: 10.1016/j.amjmed.2008.12.024 [DOI] [PubMed] [Google Scholar]
- 11. Nguyen C, Barker BM, Hoover S, et al. . Recent advances in our understanding of the environmental, epidemiological, immunological, and clinical dimensions of Coccidioidomycosis. Clin Microbiol Rev 2013; 26:505–25. doi: 10.1128/CMR.00005-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hector RF, Pappagianis D. Enzymatic degradation of the walls of spherules of Coccidioides immitis. Exp Mycol 1982; 6:136–52. doi: 10.1016/0147-5975(82)90088-3 [DOI] [Google Scholar]
- 13. Cole GT, Hung CY. The parasitic cell wall of Coccidioides immitis. Med Mycol Suppl 2001; 39:31–40. doi: 10.1080/mmy.39.1.31.40 [DOI] [PubMed] [Google Scholar]
- 14. Johnson SM, Zimmermann CR, Pappagianis D. Amino-terminal sequence analysis of the Coccidioides immitis chitinase/immunodiffusion-complement fixation protein. Infect Immun 1993; 61:3090–92. doi: 10.1128/iai.61.7.3090-3092.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pishko EJ, Kirkland TN, Cole GT. Isolation and characterization of two chitinase-encoding genes (ctsl, cts2) from the fungus Coccidioides immitis. Pathology 1995; 167:173–7. [DOI] [PubMed] [Google Scholar]
- 16. Johnson SM, Pappagianis D. The coccidioidal complement fixation and immunodiffusion-complement fixation antigen is a chitinase. Infect Immun 1992; 60:2588–92. doi: 10.1128/iai.60.7.2588-2592.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Resnick S, Pappagianis D, McKerrow JH. Proteinase production by the parasitic cycle of the pathogenic fungus Coccidioides immitis. Infect Immun 1987; 55:2807–15. doi: 10.1128/iai.55.11.2807-2815.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Johnson SM, Zimmermann CR, Pappagianis D. Use of a recombinant Coccidioides immitis complement fixation antigen- chitinase in conventional serological assays. J Clin Microbiol 1996; 34:3160–4. doi: 10.1128/jcm.34.12.3160-3164.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McHardy IH, Dinh BTN, Waldman S, et al. . Coccidioidomycosis complement fixation titer trends in the age of antifungals. J Clin Microbiol 2018; 56:1–11. doi: 10.1128/JCM.01318-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Johnson JE, Jeffery B, Huppert M. Evaluation of five commercially available immunodiffusion kits for detection of Coccidioides immitis and Histoplasma capsulatum antibodies. J Clin Microbiol 1984; 20:530–2. doi: 10.1128/jcm.20.3.530-532.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaufman L, Sekhon AS, Moledina N, Jalbert M, Pappagianis D. Comparative evaluation of commercial premier EIA and microimmunodiffusion and complement fixation tests for Coccidioides immitis antibodies. J Clin Microbiol 1995; 33:618–9. doi: 10.1128/jcm.33.3.618-619.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lindsley MD, Ahn Y, McCotter O, et al. . Evaluation of the specificity of two enzyme immunoassays for coccidioidomycosis by using sera from a region of endemicity and a region of nonendemicity. Clin Vaccine Immunol 2015; 22:1090–5. doi: 10.1128/CVI.00375-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kirsch EJ, Greene RT, Prahl A, et al. . Evaluation of Coccidioides antigen detection in dogs with coccidioidomycosis. Clin Vaccine Immunol 2012; 19:343–5. doi: 10.1128/CVI.05631-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Blair JE, Coakley B, Santelli AC, Hentz JG, Wengenack NL. Serologic testing for symptomatic coccidioidomycosis in immunocompetent and immunosuppressed hosts. Mycopathologia 2006; 162:317–24. doi: 10.1007/s11046-006-0062-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grys TE, Brighton A, Chang YH, Liesman R, LaSalle CB, Blair JE. Comparison of two FDA-cleared EIA assays for the detection of Coccidioides antibodies against a composite clinical standard. Med Mycol 2019; 57:595–600. doi: 10.1093/mmy/myy094 [DOI] [PubMed] [Google Scholar]
- 26. Kassis C, Durkin M, Holbrook E, Myers R, Wheat L. Advances in Diagnosis of Progressive Pulmonary and Disseminated Coccidioidomycosis. Clin Infect Dis 2021; 72:968–75. doi: 10.1093/cid/ciaa188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rutala PJ, Smith JW. Coccidioidomycosis in potentially compromised hosts: the effect of immunosuppressive therapy in dissemination. Am J Med Sci 1978:283–95. doi: 10.1097/00000441-197805000-00006 [DOI] [PubMed] [Google Scholar]
- 28. Zhu Y, Yang C, Magee DM, Cox RA. Molecular cloning and characterization of Coccidioides immitis antigen 2 cDNA. Infect Immun 1996; 64:2695–9. doi: 10.1128/iai.64.7.2695-2699.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zimmermann RC, Johnson SM, Martens GW, White AG, Pappagianis D. Cloning and expression of the complement fixation antigen-chitinase of Coccidioides immitis. Infect Immun 1996; 64:4967–75. doi: 10.1128/iai.64.12.4967-4975.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang MC, Magee DM, Cox RA. Mapping of a Coccidioides immitis-specific epitope that reacts with complement-fixing antibody. Infect Immun 1997; 65:4068–74. doi: 10.1128/iai.65.10.4068-4074.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Peng T, Zong Y, Johnson MD, Menghani S V, Lewis ML, Galgiani JN. A quantitative enzyme-linked immunoassay (ELISA) to approximate complement-fixing antibody titers in serum from patients with coccidioidomycosis. Diagn Microbiol Infect Dis. Elsevier Inc 2021; 99:115198. doi: 10.1016/j.diagmicrobio.2020.115198 [DOI] [PubMed] [Google Scholar]
- 32. Galgiani JN, Grace GM, Lundergan LL. New serologic tests for early detection of coccidioidomycosis. J Infect Dis 1991; 163:671–4. doi: 10.1093/infdis/163.3.671 [DOI] [PubMed] [Google Scholar]
- 33. Smith CE, Saito MT, Beard RR, Kepp RM, Clark RW, Eddie BU. Serological tests in the diagnosis and prognosis of coccidioidomycosis. Am J Epidemiol 1950; 52:1–21. doi: 10.1093/oxfordjournals.aje.a119404 [DOI] [PubMed] [Google Scholar]
- 34. DiTomasso JP, Ampel NM, Sobonya RE, Bloom JW. Bronchoscopic diagnosis of pulmonary coccidioidomycosis comparison of cytology, culture, and transbronchial biopsy. Diagn Microbiol Infect Dis 1994; 18:83–7. doi: 10.1016/0732-8893(94)90070-1 [DOI] [PubMed] [Google Scholar]
- 35. Vucicevic D, Blair JE, Binnicker MJ, et al. . The utility of Coccidioides polymerase chain reaction testing in the clinical setting. Mycopathologia 2010; 170:345–51. doi: 10.1007/s11046-010-9327-0 [DOI] [PubMed] [Google Scholar]
- 36. Binnicker MJ, Buckwalter SP, Eisberner JJ, et al. . Detection of Coccidioides species in clinical specimens by real-time PCR. J Clin Microbiol 2007; 45:173–8. doi: 10.1128/JCM.01776-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Galgiani JN, Grace GM, Lundergan LL. New serologic tests for early detection of coccidioidomycosis. J Infect Dis 1991; 163:671–4. doi: 10.1093/infdis/163.3.671 [DOI] [PubMed] [Google Scholar]
- 38. Durkin M, Connolly P, Kuberski T, et al. . Diagnosis of coccidioidomycosis with use of the Coccidioides antigen enzyme immunoassay. Clin Infect Dis 2008; 47:e69–73. doi: 10.1086/592073 [DOI] [PubMed] [Google Scholar]
- 39. Kassis C, Zaidi S, Kuberski T, et al. . Role of coccidioides antigen testing in the cerebrospinal fluid for the diagnosis of coccidioidal meningitis. Clin Infect Dis 2015; 61:1521–6. doi: 10.1093/cid/civ585 [DOI] [PubMed] [Google Scholar]
- 40. Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 1975; 256:495–7. doi: 10.1038/256495a0 [DOI] [PubMed] [Google Scholar]
- 41. Armbruster DA, Pry T. Limit of blank, limit of detection and limit of quantitation. Clin Biochem Rev 2008; 29:S49–52. [PMC free article] [PubMed] [Google Scholar]
- 42. Peter Donnelly J, Chen SC, Kauffman CA, et al. . Revision and update of the consensus definitions of invasive fungal disease from the european organization for research and treatment of cancer and the mycoses study group education and research consortium. Clin Infect Dis 2020; 71:1367–76. doi: 10.1093/cid/ciz1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Blair JE, Mendoza N, Force S, Chang YHH, Grys TE. Clinical specificity of the enzyme immunoassay test for coccidioidomycosis varies according to the reason for its performance. Clin Vaccine Immunol 2013; 20:95–8. doi: 10.1128/CVI.00531-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Thompson GR, Lunetta JM, Johnson SM, et al. . Early treatment with fluconazole may abrogate the development of IgG antibodies in coccidioidomycosis. Clin Infect Dis 2011; 53. doi: 10.1093/cid/cir466 [DOI] [PubMed] [Google Scholar]
- 45. Deresinki SC, Stevens DA. Coccidioidomycosis in compromised hosts. Medicine (Baltimore) 1975; 54:377–95. doi: 10.1097/00005792-197509000-00002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.