Abstract
Sound signals from the respiratory system are largely taken as tokens of human health. Early diagnosis of respiratory tract diseases is of great importance because, if delayed, it exerts irreversible effects on human health. The Coronavirus pandemic, which is deeply shaking the world, has revealed the importance of this diagnosis even more. During the pandemic, it has become the focus of researchers to differentiate symptoms from similar diseases such as influenza. Among these symptoms, the difference in cough sound played a distinctive role in research. Clinical data collected under the supervision of doctors in a reliable environment were used as the dataset consisting of 16 subjects suspected of COVID-19 with a specific patient demographic. Using the polymerase chain reaction test, the suspected subjects were divided into two groups as negative and positive. The negative and positive labels represent the patients with non-COVID and with a COVID-19 cough, respectively. Using the 3D plot or waterfall representation of the signal frequency spectrum, the salient features of the cough data are revealed. In this way, COVID-19 can be differentiated from other coughs by applying effective feature extraction and classification techniques. Power spectral density based on short-time Fourier transform and mel-frequency cepstral coefficients (MFCC) were chosen as the efficient feature extraction method. From among the classification techniques, the support vector machine (SVM) algorithm was applied to the processed signals in order to identify and classify COVID-19 cough. In terms of results evaluation, the cough of subjects with COVID-19 was detected with 95.86% classification accuracy thanks to the radial basis function (RBF) kernel function of SVM and the MFCC method. The diagnosis of COVID-19 coughs was performed with 98.6% and 91.7% sensitivity and specificity, respectively.
Keywords: COVID-19, Cough, Signal processing, STFT, MFCC, SVM
Introduction
Coronaviruses are a large family of viruses and a subset of Coronavirus ranging from the common cold virus to the cause of more serious diseases such as severe acute respiratory syndrome (SARS), Middle East respiratory syndrome (MERS), and COVID-19. They were discovered in the 1960s and continued to be studied until the mid-1980s (Tyrrell and Bynoe 1965; Visy et al. 1991). These viruses are common in mammals and birds in general, but seven Coronaviruses of human origin have also been identified (Zhou et al. 2020). The latest version of the virus, Coronavirus Acute Respiratory Syndrome 2, also known as SARS-CoV-2, became widespread among people in Wuhan, China, in December 2019 and was then transmitted to the whole world in a short time (Talaee et al. 2020).
As of December 25, 2020, approximately 1,8 million people have lost their lives due to this disaster. Despite great efforts, the lack of a definitive solution for this disease deeply concerns researchers and the medical world. Several ways to keep people away from this pandemic have been proposed. These measures are limited to social distancing, frequent hand washing, and not touching the face as much as possible. Based on recent research, the most common way this virus spreads is by the droplets released into the air when the person with COVID-19 coughs or sneezes (What is Coronavirus and|Johns Hopkins Medicine n.d). Since this virus behaves differently in subjects and has no fixed symptoms, it has been difficult for studies to reach the main goal of virus diagnosis. In fact, common symptoms of COVID-19 include headache, fever, cough, and fatigue (Schuller et al. 2020). Other symptoms include shortness of breath, muscle and sore throat, and sometimes diarrhea and vomiting (Clinical Manifestations and of Patients with Coronavirus Disease 2019 (COVID-19) in a Referral Center in Iran - PubMed n.d; Brown et al. 2020). Among these symptoms, shortness of breath and dry cough caused by respiratory failure are the major causes of death. Unfortunately, Coronavirus sometimes appears to affect other essential organs, namely the heart, kidneys, brain, and lungs (Sadegh Beigee et al. 2020). Despite all the information available about this virus, there are still many uncertainties about its behavior, virus-patient interaction, and pandemic process (Features n.d). Based on the studies, the elderly and chronic patients are the most affected by this virus (Yang et al. 2020; Abrishami et al. 2020; Coronavirus Disease n.d). According to a pioneering research, some COVID-19 patients are defined as virus carriers only. The data show that 81% of COVID-19 patients merely act as carriers without very serious symptoms (Features n.d). In the remaining percentage (19%), symptoms occur approximately 2–14 days after exposure to the virus. A study of 181 patients with COVID-19 found that the average incubation period was 5 days, and 97% of people had symptoms occurring on average 11.5 days after exposure to the virus (Lauer et al. 2019; When is someone infectious|FAQ n.d).
A detailed assessment of the situation of the COVID-19 infection, which was approximately 5 months old in May 2020, was carried out by Bchetnia et al. (2020). This study generally evaluated studies on COVID-19 and served as an information repository. Additionally, a comparative analysis of SARS-CoV, MERS-CoV, and SARS-CoV-2 was prominently shown. The life cycle of the COVID-19 and how it affects infected cells were also presented.
Clinical and radiological features of COVID-19 and its detailed effects on chronic kidney disease have been studied by medical researchers (Abrishami et al. 2020). It has been observed that individuals with kidney disease are vulnerable to severe COVID-19. In another medical study (Sadegh Beigee et al. 2020), damage to lung tissue biopsy samples of people who died due to COVID-19 was detected. Thus, it is hoped that when research is expanded, it will shed light on the development of effective treatment methods. The behavior changes of this virus and its effects on the human body constituted the main target of many other medical studies (Rajapakse and Dixit 2020; Guo et al. 2019; Oliveros et al. 2019; Paterson et al. 2020).
The general diagnostic method of this virus in the world is the PCR test. However, it seems that this method is not adequate to contain this global disease. The reasons for this are the limitation of the number of tests because of temporal factors, the low availability and high cost of clinical tests despite the great demands for them, and the fact that these tests are largely dependent on hospital and clinic visits (Subirana et al. 2020). This mandatory visit can actually be problematic, as the COVID-19 virus is thought to last for hours and even days on different surfaces (Doremalen et al. 2020; Opinion|Hospitals are overwhelmed because of the coronavirus n.d). Moreover, this face-to-face test endangers healthcare professionals and medical personnel and carries a serious risk. Since the appearance of this virus, test results were initially determined after as long as 10 days, so it was a huge waste of time. As time passed, faster types of tests evolved (Detect COVID-19 in as Little as 5 Minutes|Abbott Newsroom n.d), but the decrease in accuracy raised different concerns (FDA says Abbott’s 5-minute Covid-19 test may miss infected patients n.d). After understanding the virus behavior to some extent, two different methods, X-ray and CT scan, were used for diagnosis (Zhang et al. 2020; Narin et al. 2020; Li et al. 2019; Ye et al. 2019). These methods seem to be as successful as the PCR test (Subirana et al. 2020; Gietema et al. 2020; Adams et al. 2020) but also require a clinic or hospital visit.
In the age of technology, machine learning techniques can be used to control this pandemic. These techniques actually developed a model by adapting to the environment and based on experience without explicit programming (Shuja et al. 2020; Lan et al. 2018). The most important advantage of these techniques is staying away from hospital or clinic visit-based COVID-19 test centers (Imran et al. 2020). In short, they aim to make life easier within the scope of digital health thanks to computer science and engineering, as well as the information obtained in medicine (Schuller et al. 2020). As another example, the virus spread prediction can be facilitated by the application of artificial intelligence (AI) (Hu et al. 2020; Wang et al. 2020; Maghded et al. 2020). In (Schuller et al. 2020), the contribution of sound analysis to the diagnosis of COVID-19 was analysed. It was observed that the computer audition (AC) technique is an accessible and successful tool in diagnosis. These machine listening tools have just begun to be taken into account by researchers during the global epidemic. Based on technological information, it is predicted that the sensors of smartphones will also be useful in the early diagnosis of this virus (Maghded et al. 2020). In another cell phone-based research, a study that has the ability to make cheap, early and reliable diagnosis of COVID-19 has been conducted (Fozouni et al. 2020).
The work done in the field of COVID-19 machine learning is generally divided into two domains of image and sound analysis. Cough, a major symptom of COVID-19, is the basis of sound-based studies (Schuller et al. 2020; Belkacem et al. 2020). In fact, this symptom is an early harbinger of many human diseases (Chang et al. 2008; Higenbottam 2002; Windmon et al. 2019). Cough samples containing COVID-19 can be classified with acceptable success using machine learning and signal processing techniques (Brown et al. 2020). A statistically-intensive study has developed a model for the analysis of recorded COVID-19 cough sound signals (Bagad et al. 2020). In the proposed artificial intelligence model, findings that facilitate the diagnosis of COVID-19 were found in the statistical properties of cough sounds recorded on the phone. In another artificial intelligence study based on cough samples, a fast, reliable, and pre-screening-based diagnostic mechanism was proposed to prevent the spread of coronavirus and to reduce the workload of healthcare institutions (Chowdhury et al. 2021). This deep learning model can provide convenience by giving great confidence to patient candidates who suspect COVID-19 and different respiratory diseases. A crowdsourced data set consisting of 355 participants were analysed in detail using a deep neural network model. This pilot study has shown that COVID-19 detection is possible from cough and breath sounds. In the neural network model, an Area Under Curve (AUC) of 0.846 receiver operating characteristics was reached (Coppock et al. 2021). In a study focusing on two data sets, it has been shown that cough plays a very strong role in the detection of COVID-19 based on cough, breath, and speech sounds (Pahar and Niesler 2021). In the analysis of cough, breath and speech sounds, the highest AUC for COVID-19 classification was 0.93, 0.92, and 0.91, respectively. The importance of cough in the diagnosis of COVID-19 has become a common point of many studies. Based on this information and applying the deep learning workspace on validated samples, the cough analysis system and infection rapid detection platform were realized with promising AUC results (Andreu-Perez et al. 2021). Another comprehensive study has been focused on different types of cough and distinguished patients with COVID-19 from patients with asthma, bronchitis, and healthy individuals with a 95.04% success (Pal and Sankarasubbu 2021). In cough samples recorded using a smartphone app, COVID-19 was classified with a 97% AUC using the Resnet50 classifier due to the different pattern of cough (Laguarta et al. 2020).
It is an important and critical point that the dataset used to obtain information be safe (Shuja et al. 2020; Belkacem et al. 2020). Using these datasets, all researchers around the world have focused on a single target. It is aimed to expand and verify the data within the scope of cooperation by using open-source data in AI-based research by carrying out different studies on an engineering basis (Shuja et al. 2020). In fact, the possibility of easy access to open-source data is a key step in machine learning (Yates et al. 2018). These useful data lead to rich and scientific studies with high availability, accuracy, and clarity (Frazer et al. 2020).
In this study, the diagnosis of COVID-19 was made within the scope of classification using an appropriate combination of machine learning and “GitHub” open-source datasets (GitHub n.d). “Virufy” is a voluntary trustworthy organization to identify patterns caused by COVID-19 coughing noises. This organization offers free COVID-19 cough datasets as a pioneer in bringing innovation to human life, health, and the industry sector. The main goal of this study is to conduct an easy and early diagnosis based on the classification technique during this pandemic by focusing on cough sound samples. This model, based on computer algorithms and the analysis of cough samples, makes predictions and decisions by taking into account the training data. Thus, it is predicted that early diagnosis of the virus will be possible by using effective signal processing methods in this dataset consisting of positive and negative samples of COVID-19. As a result, thanks to the collaboration of technology and engineering, people with suspected infection will be less likely to seek clinical treatment. This research is a basic study in terms of the early diagnosis of COVID-19 using the machine learning technique. This pandemic has the capacity to offer encouraging and reliable information by making more and more detailed research in the academic and scientific field in a technological sense. The results showed that COVID-19 cough sound samples can be diagnosed with acceptable classification accuracy with software applications in homes and workplaces via smart tools (Dunne et al. 2020).
Methods and test protocol
COVID-19 dataset and subjects
The dataset was collected at the hospital or clinic under the supervision of physicians or medical personnel. The cough sound data discussed were recorded with the "Virufy" mobile app upon the request of Stanford University and made available on “GitHub”. After the preprocessing steps were carefully done on the data, the results obtained from the PCR test were labeled as positive or negative (GitHub n.d). The subject demographics are presented in detail in Table 1. A data pool of 121 segmentations obtained from cough samples of 16 subjects was created. As a result of this segmentation, there were a total of 73 non-COVID and 48 COVID-19 coughs. The segmentation preprocessing step contributes greatly to the analysis by clarifying the importance and predominance of infected regions (Shuja et al. 2020; Shan et al. 2020). In this segmentation, the cough time of each subject was taken into account as 1640 ms (ms), and the sampling frequency in the study was determined as 48,000.
Table 1.
Demographics of subjects suspected of COVID-19 (GitHub n.d)
| Age | Gender | Any chronic illness | Complaints and symptoms | PCR result |
|---|---|---|---|---|
| 53 | Male | None | None | Negative |
| 50 | Male | Congestive heart failure | Shortness of breath | Positive |
| 43 | Male | None | Sore throat | Negative |
| 65 | Male | Asthma or lung disease | Shortness of breath, worsening cough | Positive |
| 40 | Female | None | Sore throat, Loss of taste, Loss of smell | Positive |
| 66 | Female | Diabetes | None | Negative |
| 20 | Female | None | None | Negative |
| 17 | Female | None | Shortness of breath, Sore throat, Body aches | Negative |
| 47 | Male | None | Worsening cough | Negative |
| 53 | Male | None | Fever, chills, or sweating, Shortness of breath, worsening cough, Sore throat, Loss of taste, Loss of smell | Positive |
| 24 | Female | None | None | Positive |
| 51 | Male | Diabetes | Fever, chills, or sweating, worsening cough, Sore throat | Positive |
| 53 | Male | None | None | Negative |
| 31 | Male | None | Shortness of breath, worsening cough | Positive |
| 37 | Male | None | None | Negative |
| 24 | Female | None | Worsening cough | Negative |
Data processing
Figure 1 displays the flow chart that highlights the stages of the study in detail.
Fig. 1.
Flow diagram of data processing
Cough data STFT-based spectrogram analysis
One way to evaluate the frequency and phase behavior resulting from the change of signals over time is the short-time Fourier transform (STFT) (Sejdić et al. 2009). This technique divided a long signal into short and equal lengths and applies the Fourier transform to each segment separately (Kehtarnavaz 2008). For a random signal in the time domain, STFT is shown by Eq. (1) (He et al. 2016; Bajric et al. 2016).
| 1 |
where and are the continuous signal and windowing functions, respectively.
The visualization of this method (Krishnan et al. 2010), which provides a complete balance between time and frequency resolution (Mitrović et al. 2010), is defined as a spectrogram or waterfall plot which represents the square size of the coefficients of STFT (Sairamya et al. 2019).
Due to the unstable and unpredictable behavior of audio signals, the focus is on signal properties at a certain time by the windowing process. Thanks to these overlapping windows, the behavior of the unstable signal will resemble a knowing and stationary signal. The length of the sliding window is an important factor in this transformation (Manshouri et al. 2020). The purpose of the window is to actually select the time segment of the signal whose frequency properties are almost invariant (Boashash 2003). Considering the properties of soft behaving windows, the Hamming window (Harris 1987) was used in this study using the trial-and-error method (Manshouri and Kayikcioglu 2019). Furthermore, the size and overlap of the window were selected as 2400 and 600, respectively.
A spectrogram analysis was performed for each recording of COVID-19 and non-COVID cough sounds; then, the average was calculated for each stage. Finally, taking into account the spectrograms based on STFT for COVID-19 and non-COVID cough sounds, the difference in PSD was calculated for analysis. When spectrogram graphics were interpreted, time and frequency slices that best reflected PSD were considered in terms of COVID-19 positive and negative sample classification. The dominant time and frequency intervals reflecting PSD are represented in Table 2. Thus, taking into account these time and frequency intervals, the basic step was taken for the feature extraction method.
Table 2.
The dominant time and frequency intervals based on the spectrogram
| Time (s) | 2 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|
| Frequency intervals (Hz) |
140–340, 460–580 |
240–300 | 260– 620 | 340– 600 |
200–320, 480– 600 |
200–340 | 220–300, 460–600 | 220–320, 440–620 | 240– 340 |
STFT and MFCC feature extraction techniques
To give a general reason about choosing feature extraction methods, this choice was actually made as a result of the literature review in the field of audio signal analysis. Two techniques were used as feature extraction methods, namely STFT and mel-frequency cepstral coefficients (MFCC). STFT is widely used in the analysis of audio signals (Mitrović et al. 2010; Mateo and Talavera 2020). In the STFT method (Mehala and Dahiya n.d; Mehala and Dahiya n.d) the features were chosen from 13 effective frequency sub-bands and their corresponding times, aiming at a reduced number of feature extraction destinations. Using the trapezoidal digital integration (Trapz) of these effective time and frequency intervals, a (121* 13) dimensional feature vector was obtained.
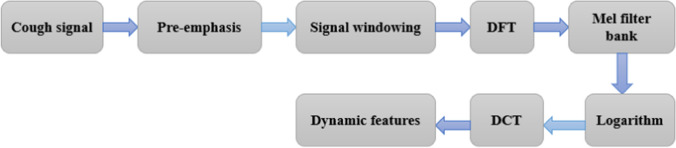
Another complex sound feature extraction technique to classify and distinguish COVID-19 and non-COVID samples has also been employed. The MFCC technique (Gonzalez 2013) is known to be compatible with the variation of the frequency band of the human ear (Hossan et al. 2010). In addition, MFCC is generally known as a technique with high success in recognizing sound systems (Winursito et al. 2018; Wang and Lawlor 2017). In a different study, MFCC has been shown to be a useful method for the differentiation of dry and wet coughs (Chatrzarrin et al. 2011). The working principle of MFCC can be briefly summarized as signal windowing, discrete Fourier transform (DFT) application, calculating the logarithm of the coefficients' magnitude, warping frequencies to a mel scale, and the application of discrete cosine transform (DCT) (Rao 2017). The detailed MFCC feature extraction flow chart is depicted in Fig. 2.
Fig. 2.
MFCC feature extraction flowchart
The mel spectrum is calculated based on the signal obtained from the Fourier transform passing through a series of band-pass filters. The mel approach from physical frequency () can be expressed as in Eq. (2).
| 2 |
In this technique, the Hamming window was chosen with a window size of 1024 and an overlap of 512. Finally, the size of the feature vector obtained from MFCC was obtained as (121 * 13).
SVM classification algorithm
In the Classification learner application (Train models to classify data using supervised machine learning - MATLAB n.d) of Matlab program, the obtained features were classified with the default parameters of the classifiers. Based on these results, it was determined that the SVM classifier is more successful than other methods. This supervised learning model is a powerful classification technique in the class determination of two-class problems. As a result of various scientific studies, it has been proven that this classifier is more successful than other classification methods (Hsu and Lin 2002; Joachims 1998). The advantages that distinguish SVM include its reliability, good generalization feature, strong theoretical basis, and clear geometric lines in terms of classification (Mavroforakis and Theodoridis 2006; El-Naqa et al. 2002). This classifier based on statistical functions minimizes structural risk possibilities (Romero et al. 2015). The working principle of SVM in linear or nonlinear problems follows almost the same basis. This essentially consists of discovering a hyperplane to distinguish positive and negative samples (Lin et al. 2008).
In the present study, linear and nonlinear SVM models were developed for cough-based COVID-19 diagnosis in the positively and negatively labeled problem. A brief description of linear and nonlinear SVM is given below.
Linear SVM
The linear SVM algorithm will be effective when the model created by the training dataset has a linear separable capability. The N-point training dataset is defined as , where is the class label and is the pattern to be classified. Now, the main problem is to build a decision function that can properly classify any x input. Ultimately, to solve the problem, the decision function is linearly defined in Eq. (3).
| 3 |
where represents the hyperplane normal vector and is a scalar. (El-Naqa et al. 2002; Romero et al. 2015). Ultimately, in a two-class problem, the training examples are separated by the hyperplane .
SVM chooses the plane with the maximum margin between the two classes from different hyperplanes for the training set (Burges 1998). The mathematical analysis of optimum hyperplane calculation is explained in detail in El-Naqa et al. (2002).
Nonlinear SVM and kernel functions
By using a nonlinear operator in the decision function, the linear SVM can be extended to a nonlinear classifier (El-Naqa et al. 2002; Burges 1998; Viitaniemi et al. 2015). In addition, this operator allows the analysis of data to be transferred to the multidimensional property area, also known as the “kernel trick” (Li et al. 2014).
Due to the different kernel functions, it is essential to choose the appropriate function (Lei 2017). The most common kernel function in the SVM classification method is defined as RBF. In this research, the two most commonly used kernel types in SVM studies were exploited, namely polynomial and RBF (Advances in Kernel Methods|The MIT Press n.d). These kernels were identified as in Eqs. (4) and (5).
| 4 |
| 5 |
are constants defining the kernel order and width, respectively. In this study, the parameter was chosen as 2. Hyper-parameters were automatically optimized using “fitcsvm” (Train support vector machine (SVM) classifier for one-class and binary classification - MATLAB fitcsvm n.d).
Classifier training
To make a skill assessment of the proposed model, leave-one-out (LOO) and hold-out (HO) cross-validation processes of the statistical techniques were used. In the cross-validation technique (Shultz et al. 2011), the dataset is divided into approximately equal-sized subsets or folds named k (Webb et al. 2011). If these subsets are shown as , the classifier learning algorithm is then applied times for, to , each time using the combination of all subsets except for as the training set and as the test set.
The LOO method, which is closely related to the jack-knife method (Efron 1982), is actually a special form of cross-validation. In this cross-validation method, where the number of folds is equal to the number of samples in the data set, a single sample is selected as the test set, and all other samples are selected as the training set, and this situation is used once for each time (Yuan et al. 2010; Cawley 2006).
The HO technique (Giresi et al. 2005) is a simple and comprehensive technique among cross-validation methods (Understanding 8 types of Cross-Validation|by Satyam Kumar | Towards Data Science n.d; Hold-out vs. Cross-validation in Machine Learning | by Eijaz Allibhai|Medium n.d). In this technique, the dataset is randomly divided into two sets, the training and the test set. After creating a model by training the training set, the proposed model will be evaluated in test validation data. The general separation for training and testing is 80% and 20%, respectively. In this study, training and test splitting was done as 50.41% and 49.58%. However, in the COVID-19 positive and negative two-class training data, two different separations were taken into account. This cross-validation process was repeated 1000 times.
Evaluating the performance of classification
A specific table called a confusion matrix is a useful tool for evaluating the classification performance. The parameters obtained from the elements of the complex matrix are accuracy, sensitivity, and specificity (Manshouri et al. 2020). These parameters were used for a detailed performance evaluation of the SVM classification technique. The mathematical equation of these parameters is presented in Eqs. (6), (7), (8). The non-COVID class was defined as the positive samples, and the COVID class as the negative samples. This matrix is defined in Table 3 in which P = Positive, N = Negative, TP = True Positive, FP = False Positive, TN = True Negative, and FN = False Negative.
| 6 |
| 7 |
| 8 |
Table 3.
Confusion matrix
| Predicted class | |||
|---|---|---|---|
| Non-COVID | COVID | ||
| True class | Non-COVID (P) | TP | FN |
| COVID (N) | FP | TN | |
Results
The spectrogram graphs depicting the difference in PSD of cough sounds with COVID-19 positive and negative labels from the time and frequency perspectives are illustrated in Figs. 3 and 4, respectively. The three-dimensional (3D) PSD graph reflecting this PSD is presented in Fig. 5. In short, this graph represents the PSD difference of the average cough sounds from COVID-19 and non-COVID. Focusing on the graph, the PSD difference in this cough sound analysis is evident up to 0.65 s. This distinction is useful in terms of the classification of COVID-19 positive and negative labeled cough sounds. Based on the information obtained from these graphs, effective time–frequency intervals were taken into account for feature extraction. SVM classification results using the STFT feature extraction technique for LOO and HO cross-validation are presented in Tables 4 and 5, respectively. The same tables are also calculated in the MFCC feature extraction method, as in Tables 6 and 7. Looking at the results in general, the classification of COVID-19 cough sounds seems successful with the suggested methods. The MFCC technique yields a better percentage of success compared to STFT feature extraction, and it seems that RBF is more successful than linear and polynomial among SVM types. In addition, the classification performance analysis parameters largely agreed with each other. In the MFCC feature extraction method, in terms of mel coefficients, the issue of how many overlapping windows will bring the highest success to the sum of 13 mel coefficients was considered. This description is given in detail in Fig. 6. The best accuracy achievement was obtained for 11 overlapping windows for almost three SVM classifier types in Fig. 6. The confusion matrix, including sensitivity, specificity, precision, and negative predictive value for the successful feature extraction method determined in Table 6 is displayed in Fig. 7. In Fig. 7, labels 1 and 0 indicate COVID-19 and non-COVID, respectively.
Fig. 3.

Spectrogram graph from a time perspective
Fig. 4.

Spectrogram graph from a frequency perspective
Fig. 5.

3D spectrogram
Table 4.
SVM classification results using STFT feature extraction technique for LOO cross-validation
| Accuracy | Sensitivity | Specificity | |
|---|---|---|---|
| Linear | 72.73 | 58.30 | 82.20 |
| Polynomial | 76.86 | 54.20 | 91.80 |
| RBF | 76.03 | 68.80 | 80.80 |
Table 5.
SVM classification results using STFT feature extraction technique for two training dataset’s splitting of HO cross-validation
| 36 Non-COVID & 24 COVID for training | 30 Non-COVID & 30 COVID for training | |||||
|---|---|---|---|---|---|---|
| Accuracy | Sensitivity | Specificity | Accuracy | Sensitivity | Specificity | |
| Linear | 72.79 | 70.41 | 75.19 | 71.65 | 53.13 | 85.11 |
| Polynomial | 72.97 | 76.57 | 73.00 | 71.13 | 54.08 | 84.13 |
| RBF | 70.10 | 63.84 | 74.36 | 68.05 | 48.33 | 87.60 |
Table 6.
SVM classification results using MFCC feature extraction technique LOO cross-validation
| Accuracy | Sensitivity | Specificity | |
|---|---|---|---|
| Linear | 86.78 | 81.25 | 90.41 |
| Polynomial | 90.08 | 83.33 | 94.52 |
| RBF | 94.21 | 89.58 | 97.26 |
Table 7.
SVM classification results using MFCC feature extraction technique for two training dataset’s splitting of HO cross-validation
| 36 Non-COVID & 24 COVID for training | 30 Non-COVID & 30 COVID for training | |||||
|---|---|---|---|---|---|---|
| Accuracy | Sensitivity | Specificity | Accuracy | Sensitivity | Specificity | |
| Linear | 81.16 | 80.03 | 82.60 | 79.97 | 63.91 | 90.33 |
| Polynomial | 87.31 | 87.64 | 87.83 | 87.07 | 75.62 | 93.65 |
| RBF | 87.70 | 90.68 | 87.02 | 87.61 | 75.96 | 94.61 |
Fig. 6.
Results of SVM based on the number of overlapping windows by considering 13 mel coefficients
Fig. 7.
Confusion matrix for the best classification result
Feature selection
Choosing the relevant features to select the most suitable model is a critical step in machine learning. A feature selection technique (Pudil et al. 1994) has many advantages, including simplifying the model for better understanding it, decreasing the size of the feature vector, and greatly reducing training time (Chandrashekar and Sahin 2014). Since the signals received for analysis often carry unnecessary information, it will be useful to remove these data without losing too much information (Bermingham et al. 2015). The presence of these irrelevant data deeply affects the accuracy of the model, while at the same time, it enables the model to be trained based on irrelevant features (Belkacem et al. 2020; Langley 1994; Zhao et al. 2010).
The sequential forward search (SFS) method, which is frequently used in the scientific field (Xue et al. 2016; Jain et al. 2000), was applied as a feature selection technique in this study. After performing SFS, a peak accuracy of 95.86% was observed on the dataset when using the best 9 features from among the 13 (Fig. 8). Also, as a result of the application of the SFS technique, the success rate of RBF kernel SVM classification increased from 94.21 to 95.86% (Pahar et al. 2020). For this successful feature selection, the confusion matrix is presented in Fig. 9.
Fig. 8.
RBF kernel SVM classification success percentage as a result of the SFS feature selection technique
Fig. 9.

Confusion matrix after SFS application for RBF kernel SVM
Discussion
This study was conducted for the diagnosis of COVID-19, taking into account cough sounds, based on machine learning technology. This research was defined as a machine learning study for the COVID-19 pandemic based on cough sounds without being present in any hospital or clinical setting. This machine learning model provides great advantages by recording the cough sound of subjects suspected of COVID-19 based on the application of signal processing and audio recording of smartphones (Imran et al. 2020; Hu et al. 2020; Laguarta et al. 2020; Pahar et al. 2020). One of the advantages of these studies is that they can be performed anywhere, at any time, and under easy conditions. These models are thought to be beneficial as a health aid tool when COVID-19 test kits are lacking. As stated from the outset of the pandemic, the easiest way to be protected from this dangerous virus is to stay at home and minimize interaction with people as much as possible. Taking this important issue into account, the proposed machine learning-based models protect healthcare professionals to a large extent (Schuller et al. 2020; Imran et al. 2020; Lalmuanawma et al. 2020; Bachtiger et al. 2020; Schaar et al. 2020). In fact, computer-based methods to prevent the rapid and latent spread of the Coronavirus are highly appreciated by researchers. Thanks to the application recommended in Imran et al. (2020), the virus spread tracking has been easily possible. This study has developed a pre-diagnose model for COVID-19 from cough samples using AI technology.
A small portion of the studies on COVID-19 has focused on machine learning-based cough sound classification, a topic which has recently come under the attention of researchers. One reason for this is that there is a limited number of open-access datasets based on cough sound. As these datasets are unveiled, they are quickly examined by researchers because the goal is to overcome the COVID-19 epidemic as soon as possible and return to normal life.
In a new and similar machine learning study in this area, two datasets were considered in the cough-based analysis of patients with COVID-19 and non-COVID-19. In this study, MFCC was chosen as the feature extraction method and it was observed that the k-NN classification algorithm was successful among seven different classification algorithms (Maleki 2021). Due to the shortage of a wide range of COVID-19 studies based on cough sound classification, we do not think it is very accurate to compare our proposed classification study with the few available classification studies in detail (Brown et al. 2020; Laguarta et al. 2020; Pahar et al. 2020; Sharma et al. 2020) because each study focuses on different datasets. The common goal is to be able to classify COVID-19 positive and negative labeled cough sounds with high success, using effective feature extraction and classification techniques through signal processing steps.
Using the STFT and MFCC feature extraction method, it has been observed that MFCC yields more successful results, as expected, in the COVID-19 positive and negative cough sound classification process (Huzaifah 2017). Although STFT is known as a simple linear time–frequency transformation and provides valuable information about the frequency position over time (Krishnan et al. 2010), it is sometimes possible to appear insufficient in terms of time–frequency resolution tradeoffs. Considering this issue, the appropriate method can be selected according to the area of use. The MFCC method takes an important place in the field of audio signal processing, thanks to producing effective features by reducing the margin of error of sound signals exposed to noise (Kamarulafizam et al. 2007). Due to the logarithmic structure of the frequency bands in the MFCC method, it is close to the human system response, which makes this method different from other methods (Janse et al. 2014). The disadvantage of the MFCC feature extraction method is that it does not provide the necessary accuracy in the analysis of noisy audio signals (Kohshelan 2014). In order to perform the classification process, the SVM classification method, which has gained momentum in this field was selected (Ismael and Şengür 2021; Singh et al. 2020; Loey et al. 2021). When the study is extended in terms of datasets, this classification technique should be reviewed for its use, because the SVM algorithm is sometimes not suited enough for large and noisy datasets. In addition, a balance must always be struck between the number of features and the number of training data samples to avoid underperforming the SVM algorithm. Moreover, understanding the model of the final SVM and also selecting an appropriate kernel function for the algorithm can be difficult (Ray 2019). Finally, it was confirmed that feature selection is an important step in all aspects of signal processing. The classification performance was improved by using the SFS feature selection technique in this study (Cen et al. 2016).
Conclusion
The main reason for the spread of Coronavirus can be the inadequacy of test kits, as well as the heavy cost and loss of time in determining clinical test results. In this study, we tried to find a solution to the COVID-19 epidemic using an open-access dataset and machine learning technology. Based on this technology, we have developed a model that can classify COVID-19 cough records obtained from smartphones. We knew that the dry cough symptom is an important factor in the diagnosis of COVID-19. Thus, we created the main framework of this research by taking cough-based studies into consideration. The most important goal was to provide a virtual test opportunity away from the clinical and hospital environment based on machine learning technology. Cough records of 16 subjects suspected of COVID-19 were analyzed to create the dataset. The obtained cough sounds became a ready-made dataset by the preprocessing and segmentation process. Considering the spectrogram graph based on STFT, we focused on effective features. Focusing on the PSD difference of negative and positively labeled cough sounds, strong features were prepared from dominant time and frequency ranges for the STFT feature extraction method. In addition, the MFCC feature extraction method was included in the analysis for the diagnosis of COVID-19 cough. Based on the work done for the diagnosis of COVID-19 cough sound, the MFCC feature extraction technique and RBF kernel SVM were selected as the method with the best percentage of success.
In the future, a stronger cough-based COVID-19 diagnostic study can be conducted by a larger dataset. Future studies can also increase the number of subjects and the classification accuracy using different feature extraction and classification methods. Moreover, the train and classification stages can be improved by using deep learning algorithms.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abrishami A, Khalili N, Dalili N, Tabari RK, Farjad R, Samavat S, Naghadehi AN, Haghighatkhah H, Nafar M, Sanei-Taheri M (2020) Clinical and radiologic characteristics of covid-19 in patients with ckd, Iran. J Kidney Dis 14 https://covid19.elsevierpure.com/en/publications/clinical-and-radiologic-characteristics-of-covid-19-in-patients-w Accessed from 27 Dec 2020 [PubMed]
- Adams HJA, Kwee TC, Kwee RM. Coronavirus disease 2019 and chest CT: do not put the sensitivity value in the isolation room and look beyond the numbers. Radiology. 2020;297:E236. doi: 10.1148/radiol.2020201709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Advances in Kernel Methods|The MIT Press, (n.d.). https://mitpress.mit.edu/books/advances-kernel-methods Accessed from 6 Jan 2021
- Andreu-Perez J, Perez-Espinosa H, Timonet E, Kiani M, Giron-Perez MI, Benitez-Trinidad AB, Jarchi D, Rosales A, Gkatzoulis N, Reyes-Galaviz OF, Torres A, Alberto Reyes-Garcia C, Ali Z, Rivas F. A generic deep learning based cough analysis system from clinically validated samples for point-of-need covid-19 test and severity levels. IEEE Trans Serv Comput. 2021 doi: 10.1109/TSC.2021.3061402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtiger P, Peters NS, Walsh SL. Machine learning for COVID-19—asking the right questions. Lancet Digit Heal. 2020;2:e391–e392. doi: 10.1016/S2589-7500(20)30162-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagad P, Dalmia A, Doshi J, Nagrani A, Bhamare P, Mahale A, Rane S, Agarwal N, Panicker R (2020) Cough against COVID: evidence of COVID-19 signature in cough sounds, ArXiv http://arxiv.org/abs/2009.08790 Accessed from 3 May 2021
- Bajric R, Zuber N, Skrimpas GA, Mijatovic N. Feature extraction using discrete wavelet transform for gear fault diagnosis of wind turbine gearbox. Shock Vib. 2016 doi: 10.1155/2016/6748469. [DOI] [Google Scholar]
- Bchetnia M, Girard C, Duchaine C, Laprise C. The outbreak of the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): a review of the current global status. J Infect Public Health. 2020;13:1601–1610. doi: 10.1016/j.jiph.2020.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkacem AN, Ouhbi S, Lakas A, Benkhelifa E, Chen C (2020) End-to-End AI-based point-of-care diagnosis system for classifying respiratory illnesses and early detection of COVID-19, ArXiv http://arxiv.org/abs/2006.15469 Accessed from 30 Dec 2020 [DOI] [PMC free article] [PubMed]
- Bermingham ML, Pong-Wong R, Spiliopoulou A, Hayward C, Rudan I, Campbell H, Wright AF, Wilson JF, Agakov F, Navarro P, Haley CS. Application of high-dimensional feature selection: evaluation for genomic prediction in man. Sci Rep. 2015;5:1–12. doi: 10.1038/srep10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boashash B. Heuristic formulation of time-frequency distributions. Amsterdam: Elsevier Ltd; 2003. pp. 29–57. [Google Scholar]
- Brown C, Chauhan J, Grammenos A, Han J, Hasthanasombat A, Spathis D, Xia T, Cicuta P, Mascolo C (2020) Exploring automatic diagnosis of COVID-19 from crowdsourced respiratory sound Data. In: Proc. ACM SIGKDD Int. Conf. Knowl. Discov. Data Min., Association for Computing Machinery, New York, NY, USA, pp. 3474–3484. 10.1145/3394486.3412865
- Burges CJC. A tutorial on support vector machines for pattern recognition. Data Min Knowl Discov. 1998;2:121–167. doi: 10.1023/A:1009715923555. [DOI] [Google Scholar]
- Cawley GC (2006) Leave-one-out cross-validation based model selection criteria for weighted LS-SVMs. In: IEEE Int. Conf. Neural Networks - Conf. Proc., Institute of Electrical and Electronics Engineers Inc., pp. 1661–1668. 10.1109/ijcnn.2006.246634.
- Cen H, Lu R, Zhu Q, Mendoza F. Nondestructive detection of chilling injury in cucumber fruit using hyperspectral imaging with feature selection and supervised classification. Postharvest Biol Technol. 2016;111:352–361. doi: 10.1016/j.postharvbio.2015.09.027. [DOI] [Google Scholar]
- Chandrashekar G, Sahin F. A survey on feature selection methods. Comput Electr Eng. 2014;40:16–28. doi: 10.1016/j.compeleceng.2013.11.024. [DOI] [Google Scholar]
- Chang AB, Redding GJ, Everard ML. Chronic wet cough: protracted bronchitis, chronic suppurative lung disease and bronchiectasis. Pediatr Pulmonol. 2008;43:519–531. doi: 10.1002/ppul.20821. [DOI] [PubMed] [Google Scholar]
- Chatrzarrin H, Arcelus A, Goubran R, Knoefel F (2011) Feature extraction for the differentiation of dry and wet cough sounds. In: MeMeA 2011–2011 IEEE Int. Symp. Med. Meas. Appl. Proc., IEEE Computer Society, pp. 162–166. 10.1109/MeMeA.2011.5966670
- Chowdhury MEH, Ibtehaz N, Rahman T, Mekki YMS, Qibalwey Y, Mahmud S, Ezeddin M, Zughaier S, Al-Maadeed SASA (2021) QUCoughScope: an artificially intelligent mobile application to detect asymptomatic COVID-19 patients using cough and breathing sounds http://arxiv.org/abs/2103.12063 Accessed from 3 May 2021
- Clinical Manifestations of Patients with Coronavirus Disease 2019 (COVID-19) in a Referral Center in Iran - PubMed, (n.d.). https://pubmed.ncbi.nlm.nih.gov/33262799/ Accessed 27 Dec 2020 [PMC free article] [PubMed]
- Coppock H, Gaskell A, Tzirakis P, Baird A, Jones L, Schuller B. End-to-end convolutional neural network enables COVID-19 detection from breath and cough audio: a pilot study. BMJ Innov. 2021;7:356–362. doi: 10.1136/bmjinnov-2021-000668. [DOI] [PubMed] [Google Scholar]
- Coronavirus disease 2019 (COVID-19) - Symptoms and causes - Mayo Clinic, (n.d.). https://www.mayoclinic.org/diseases-conditions/coronavirus/symptoms-causes/syc-20479963 Accessed from 27 Dec 2020
- Detect COVID-19 in as Little as 5 Minutes|Abbott Newsroom, (n.d.). https://www.abbott.com/corpnewsroom/diagnostics-testing/detect-covid-19-in-as-little-as-5-minutes.html Accessed from 29 Dec 2020
- Dunne R, Morris T, Harper S, Report S (2020) High accuracy classication of COVID-19 coughs using Mel-frequency cepstral coecients and a Convolutional Neural Network with a use case for smart home devices 10.21203/rs.3.rs-63796/v1
- Efron B. The jackknife, the bootstrap and other resampling plans. Soc Ind Appl Math. 1982 doi: 10.1137/1.9781611970319. [DOI] [Google Scholar]
- El-Naqa I, Yang Y, Wernick MN, Galatsanos NP, Nishikawa RM. A support vector machine approach for detection of microcalcifications. IEEE Trans Med Imaging. 2002;21:1552–1563. doi: 10.1109/TMI.2002.806569. [DOI] [PubMed] [Google Scholar]
- FDA says Abbott’s 5-minute Covid-19 test may miss infected patients, (n.d.). https://www.statnews.com/2020/05/15/fda-says-abbotts-5-minute-covid-19-test-may-miss-infected-patients/ Accessed from 29 Dec 2020
- Features, Evaluation, and Treatment of Coronavirus - PubMed, (n.d.). https://pubmed.ncbi.nlm.nih.gov/32150360/ Accessed from 27 Dec 2020
- Fozouni P, Son S, Díaz M, de León Derby GJ, Knott CNG, D’Ambrosio MV, Zhao C, Switz NA, Kumar GR, Stephens SI, Boehm D, Tsou CL, Shu J, Bhuiya A, Armstrong M, Harris AR, Chen PY, Osterloh JM, Meyer-Franke A, Joehnk B, Walcott K, Sil A, Langelier C, Pollard KS, Crawford ED, Puschnik AS, Phelps M, Kistler A, DeRisi JL, Doudna JA, Fletcher DA, Ott M. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell. 2020 doi: 10.1016/j.cell.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer JS, Shard A, Herdman J. Involvement of the open-source community in combating the worldwide COVID-19 pandemic: a review. J Med Eng Technol. 2020 doi: 10.1080/03091902.2020.1757772. [DOI] [PubMed] [Google Scholar]
- Gietema HA, Zelis N, Nobel JM, Lambriksi LJG, Alphen LBV, Lashof AMLO, Wildberger JE, Nelissen IC, Stassen PM. CT in relation to rt-PCR in diagnosing covid-19 in the netherlands: a prospective study. PLoS ONE. 2020 doi: 10.1371/journal.pone.0235844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giresi PG, Stevenson EJ, Theilhaber J, Koncarevic A, Parkington J, Fielding RA, Kandarian SC. Identification of a molecular signature of sarcopenia. Physiol Genom. 2005;21:253–263. doi: 10.1152/physiolgenomics.00249.2004. [DOI] [PubMed] [Google Scholar]
- GitHub - virufy/virufy_data, (n.d.). https://github.com/virufy/virufy_data Accessed from 30 Dec 2020
- Gonzalez R. The Era of interactive media. New York: Springer; 2013. Better than MFCC audio classification features; pp. 291–301. [Google Scholar]
- Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, Jin HJ, Sen Tan K, Wang DY, Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak- A n update on the status. Mil Med Res. 2020 doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris FJ. Handbook of digital signal processing. Amsterdam: Elsevier; 1987. Multirate FIR filters for interpolating and desampling; pp. 173–287. [Google Scholar]
- He A, Yu J, Wei G, Chen Y, Wu H, Tang Z. Short-time Fourier transform and decision tree-based pattern recognition for gas identification using temperature modulated microhotplate gas sensors. J Sens. 2016 doi: 10.1155/2016/7603931. [DOI] [Google Scholar]
- Higenbottam T. Chronic cough and the cough reflex in common lung diseases. Pulm Pharmacol Ther. 2002 doi: 10.1006/pupt.2002.0341. [DOI] [PubMed] [Google Scholar]
- Hold-out vs. Cross-validation in Machine Learning|by Eijaz Allibhai | Medium, (n.d.). https://medium.com/@eijaz/holdout-vs-cross-validation-in-machine-learning-7637112d3f8f Accessed from 7 Jan 2021
- Hossan MA, Memon S, Gregory MA (2010) A novel approach for MFCC feature extraction. In: 4th Int. Conf. Signal Process. Commun. Syst. ICSPCS’2010 – Proc, 10.1109/ICSPCS.2010.5709752.
- Hsu CW, Lin CJ. A comparison of methods for multiclass support vector machines. IEEE Trans Neural Netw. 2002;13:415–425. doi: 10.1109/72.991427. [DOI] [PubMed] [Google Scholar]
- Hu Z, Ge Q, Li S, Jin L, Xiong M (2020) Artificial intelligence forecasting of Covid-19 in China, ArXiv http://arxiv.org/abs/2002.07112 Accessed from 29 Dec 2020
- Huzaifah M (2017) Comparison of time-frequency representations for environmental sound classification using convolutional neural networks http://arxiv.org/abs/1706.07156 Accessed from 10 Jan 2021
- Imran A, Posokhova I, Qureshi HN, Masood U, Riaz MS, Ali K, John CN, Hussain MI, Nabeel M. AI4COVID-19: AI enabled preliminary diagnosis for COVID-19 from cough samples via an app. Inform Med Unlocked. 2020;20:100378. doi: 10.1016/j.imu.2020.100378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismael AM, Şengür A. Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Syst Appl. 2021;164:114054. doi: 10.1016/j.eswa.2020.114054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain AK, Duin RPW, Mao J. Statistical pattern recognition: a review. IEEE Trans Pattern Anal Mach Intell. 2000;22:4–37. doi: 10.1109/34.824819. [DOI] [Google Scholar]
- Janse PV, Magre SB, Kurzekar P, Deshmukh R (2014) A comparative study between MFCC and DWT feature extraction technique, @article{Janse2014ACS, Title={A Comp. Study between MFCC DWT Featur. Extr. Tech. Author={P. V. Janse AnInternational J Eng Res Technol 3
- Joachims T. Text categorization with support vector machines: learning with many relevant features. Berlin, Heidelberg: Springer; 1998. pp. 137–142. [Google Scholar]
- Kamarulafizam I, Salleh SH, Najeb JM, Ariff AK, Chowdhury A. Heart sound analysis using MFCC and time frequency distribution. New York: Springer Verlag; 2007. pp. 402–405. [Google Scholar]
- Kehtarnavaz N. Digital signal processing syststem design. Amsterdam: Elsevier; 2008. Frequency domain processing; pp. 175–196. [Google Scholar]
- Kohshelan N. Wahid, improvement of audio feature extraction techniques in traditional indian musical instrument. Adv Intell Syst Comput. 2014;287:507–516. doi: 10.1007/978-3-319-07692-8_48. [DOI] [Google Scholar]
- Krishnan S, Umapathy K, Ghoraani B. Audio signal processing using time-frequency approaches: coding, classification, fingerprinting, and watermarking. EURASIP J Adv Signal Process. 2010;2010:28. doi: 10.1155/2010/451695. [DOI] [Google Scholar]
- Laguarta J, Hueto F, Subirana B. COVID-19 artificial intelligence diagnosis using only cough recordings. IEEE Open J Eng Med Biol. 2020 doi: 10.1109/ojemb.2020.3026928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalmuanawma S, Hussain J, Chhakchhuak L. Applications of machine learning and artificial intelligence for Covid-19 (SARS-CoV-2) pandemic: a review. Chaos Solitons Fractals. 2020;139:110059. doi: 10.1016/j.chaos.2020.110059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan K, Tong Wang D, Fong S, Sheng Liu L, Wong KKL, Dey N. A survey of data mining and deep learning in bioinformatics. J Med Syst. 2018;42:1–20. doi: 10.1007/s10916-018-1003-9. [DOI] [PubMed] [Google Scholar]
- Langley P (1994) Selection of relevant features in machine learning, www.aaai.org Accessed from 22 Mar 2019
- Lauer SA, Grantz KH, Bi Q, Jones FK, Zheng Q, Meredith HR, Azman AS, Reich NG, Lessler J, The incubation period of coronavirus disease (CoVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2019;172(2020):577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y. intelligent fault diagnosis and remaining useful life prediction of rotating machinery. Amsterdam: Elsevier; 2017. Individual intelligent method-based fault diagnosis; pp. 67–174. [Google Scholar]
- Li Y, Xia L. Coronavirus disease (COVID-19): role of chest CT in diagnosis and management. Am J Roentgenol. 2019;214(2020):1280–1286. doi: 10.2214/AJR.20.22954. [DOI] [PubMed] [Google Scholar]
- Li X, Chen X, Yan Y, Wei W, Wang Z, Li X, Chen X, Yan Y, Wei W, Wang ZJ. Classification of eeg signals using a multiple kernel learning support vector machine. Sensors. 2014;14:12784–12802. doi: 10.3390/s140712784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-PP, Wang C-HH, Wu T-LL, Jeng S-KK, Chen J-HH. Support vector machine for EEG signal classification during listening to emotional music. IEEE. 2008 doi: 10.1109/MMSP.2008.4665061. [DOI] [Google Scholar]
- Loey M, Manogaran G, Taha MHN, Khalifa NEM. A hybrid deep transfer learning model with machine learning methods for face mask detection in the era of the COVID-19 pandemic. Meas J Int Meas Confed. 2021;167:108288. doi: 10.1016/j.measurement.2020.108288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maghded HS, Ghafoor KZ, Sadiq AS, Curran K, Rawat DB, Rabie K (2020) A novel AI-enabled framework to diagnose coronavirus COVID-19 using smartphone embedded sensors: design study. In: Proc. - 2020 IEEE 21st Int. Conf. Inf. Reuse Integr. Data Sci. IRI 2020, Institute of Electrical and Electronics Engineers Inc., pp. 180–187. 10.1109/IRI49571.2020.00033.
- Maleki M (2021) Diagnosis of COVID-19 and Non-COVID-19 patients by classifying only a single cough sound http://arxiv.org/abs/2102.04880 Accessed from 3 May 2021 [DOI] [PMC free article] [PubMed]
- Manshouri N, Kayikcioglu T. A comprehensive analysis of 2D&3D video watching of EEG signals by increasing PLSR and SVM classification results. Comput J. 2019 doi: 10.1093/comjnl/bxz043. [DOI] [Google Scholar]
- Manshouri N, Maleki M, Kayikcioglu T. An EEG-based stereoscopic research of the PSD differences in pre and post 2D&3D movies watching. Biomed Signal Process Control. 2020 doi: 10.1016/j.bspc.2019.101642. [DOI] [Google Scholar]
- Mateo C, Talavera JA. Bridging the gap between the short-time Fourier transform (STFT), wavelets, the constant-Q transform and multi-resolution STFT. Signal Image Video Process. 2020;14:1535–1543. doi: 10.1007/s11760-020-01701-8. [DOI] [Google Scholar]
- Mavroforakis ME, Theodoridis S. A geometric approach to support vector machine (SVM) classification. IEEE Trans Neural Netw. 2006;17:671–682. doi: 10.1109/TNN.2006.873281. [DOI] [PubMed] [Google Scholar]
- Mehala N, Dahiya R (n.d.) A Comparative study of FFT, STFT and wavelet techniques for induction machine fault diagnostic analysis. https://pdfs.semanticscholar.org/3b7a/7c7a2bb4b8bccca373c1fd5c555d13a339cf.pdf Accessed from 23 May 2019
- Mitrović D, Zeppelzauer M, Breiteneder C. Features for content-based audio retrieval. Amsterdam: Elsevier; 2010. pp. 71–150. [Google Scholar]
- Narin A, Kaya C, Pamuk Z (2020) Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. ArXiv http://arxiv.org/abs/2003.10849 Accessed from 29 Dec 2020 [DOI] [PMC free article] [PubMed]
- Oliveros E, Brailovsky Y, Scully P, Nikolou E, Rajani R, Grapsa J. Coronavirus disease 2019 and heart failure: a multiparametric approach. Card Fail Rev. 2020 doi: 10.15420/cfr.2020.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opinion|Hospitals are overwhelmed because of the coronavirus (n.d.) Here’s how to help. - The Washington Post https://www.washingtonpost.com/opinions/2020/03/15/hospitals-are-overwhelmed-because-coronavirus-heres-how-help/ Accessed from 29 Dec 2020
- Pahar M, Niesler T (2021) Machine learning based COVID-19 detection from smartphone recordings: cough, breath and speech, http://arxiv.org/abs/2104.02477 Accessed from 4 May 2021
- Pahar M, Klopper M, Warren R, Niesler T (2020) COVID-19 cough classification using machine learning and global smartphone recordings, http://arxiv.org/abs/2012.01926 Accessed from 30 Dec 2020 [DOI] [PMC free article] [PubMed]
- Pal A, Sankarasubbu M (2021) Pay attention to the cough. In: Proc. 36th Annu. ACM Symp. Appl. Comput., ACM, New York, NY, USA, pp. 620–628. 10.1145/3412841.3441943
- Paterson RW, Brown RL, Benjamin L, Nortley R, Wiethoff S, Bharucha T, Jayaseelan DL, Kumar G, Raftopoulos RE, Zambreanu L, Vivekanandam V, Khoo A, Geraldes R, Chinthapalli K, Boyd E, Tuzlali H, Price G, Christofi G, Morrow J, McNamara P, McLoughlin B, Lim ST, Mehta PR, Levee V, Keddie S, Yong W, Trip SA, Foulkes AJM, Hotton G, Miller TD, Everitt AD, Carswell C, Davies NWS, Yoong M, Attwell D, Sreedharan J, Silber E, Schott JM, Chandratheva A, Perry RJ, Simister R, Checkley A, Longley N, Farmer SF, Carletti F, Houlihan C, Thom M, Lunn MP, Spillane J, Howard R, Vincent A, Werring DJ, Hoskote C, Jäger HR, Manji H, Zandi MS. The emerging spectrum of COVID-19 neurology: clinical, radiological and laboratory findings. Brain. 2020;143:3104–3120. doi: 10.1093/brain/awaa240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pudil P, Novovičová J, Kittler J. Floating search methods in feature selection. Pattern Recognit Lett. 1994;15:1119–1125. doi: 10.1016/0167-8655(94)90127-9. [DOI] [Google Scholar]
- Rajapakse N, Dixit D. Human and novel coronavirus infections in children: a review. Paediatr Int Child Health. 2020 doi: 10.1080/20469047.2020.1781356. [DOI] [PubMed] [Google Scholar]
- Rao KS, Manjunath KE. Speech recognition using articulatory and excitation source features. Cham: Springer International Publishing; 2017. [Google Scholar]
- Ray S (2019) A quick review of machine learning algorithms. In: Proc. Int. Conf. Mach. Learn. Big Data, Cloud Parallel Comput. Trends, Prespectives Prospect. Com. 2019, Institute of Electrical and Electronics Engineers Inc., pp. 35–39. 10.1109/COMITCon.2019.8862451
- Romero R, Iglesias EL, Borrajo L. A linear-RBF multikernel SVM to classify big text corpora. Biomed Res Int. 2015 doi: 10.1155/2015/878291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadegh Beigee F, Pourabdollah Toutkaboni M, Khalili N, Nadji SA, Dorudinia A, Rezaei M, Askari E, Farzanegan B, Marjani M, Rafiezadeh A. Diffuse alveolar damage and thrombotic microangiopathy are the main histopathological findings in lung tissue biopsy samples of COVID-19 patients. Pathol Res Pract. 2020;216:153228. doi: 10.1016/j.prp.2020.153228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sairamya NJ, Susmitha L, Thomas George S, Subathra MSP. Intelligent data analysis for biomedical applications challenges solution. Amsterdam: Elsevier; 2019. Hybrid approach for classification of electroencephalographic signals using time-frequency images with wavelets and texture features; pp. 253–273. [Google Scholar]
- Schuller BW, Schuller DM, Qian K, Liu J, Zheng H, Li X (2020) COVID-19 and computer audition: an overview on what speech & sound analysis could contribute in the SARS-CoV-2 corona crisis, ArXiv http://arxiv.org/abs/2003.11117 Accessed 16 Dec 2020 [DOI] [PMC free article] [PubMed]
- Sejdić E, Djurović I, Jiang J. Time-frequency feature representation using energy concentration: an overview of recent advances. Digit Signal Process A Rev J. 2009;19:153–183. doi: 10.1016/j.dsp.2007.12.004. [DOI] [Google Scholar]
- Shan F, Gao Y, Wang J, Shi W, Shi N, Han M, Xue Z, Shen D, Shi Y (2020) Lung infection quantification of COVID-19 in CT images with deep learning, ArXiv
- Sharma N, Krishnan P, Kumar R, Ramoji S, Chetupalli SR, Ghosh PK, Ganapathy S (2020) Coswara -- a database of breathing, cough, and voice sounds for COVID-19 diagnosis, ArXiv http://arxiv.org/abs/2005.10548 Accessed from 10 Jan 2021
- Shuja J, Alanazi E, Alasmary W, Alashaikh A. COVID-19 open source data sets: a comprehensive survey. Appl Intell. 2020 doi: 10.1007/s10489-020-01862-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz TR, Fahlman SE, Craw S, Andritsos P, Tsaparas P, Silva R, Drummond C, Ling CX, Sheng VS, Drummond C, Lanzi PL, Gama J, Wiegand RP, Sen P, Namata G, Bilgic M, Getoor L, He J, Jain S, Stephan F, Jain S, Stephan F, Sammut C, Harries M, Sammut C, Ting KM, Pfahringer B, Case J, Jain S, Wagstaff KL, Nijssen S, Wirth A, Ling CX, Sheng VS, Zhang X, Sammut C, Cancedda N, Renders J-M, Michelucci P, Oblinger D, Keogh E, Mueen A. Encyclopedia of machine learning. US: Springer; 2011. Cross-validation; pp. 249–249. [Google Scholar]
- Singh V, Poonia RC, Kumar S, Dass P, Agarwal P, Bhatnagar V, Raja L. Prediction of COVID-19 corona virus pandemic based on time series data using support vector machine. J Discret Math Sci Cryptogr. 2020;23:1583–1597. doi: 10.1080/09720529.2020.1784535. [DOI] [Google Scholar]
- Subirana B, Hueto F, Rajasekaran P, Laguarta J, Puig S, Malvehy J, Mitja O, Trilla A, Moreno CI, Valle JFM, González AEM, Vizmanos B, Sarma S (2020) Hi sigma, do I have the coronavirus?: call for a new artificial intelligence approach to support health care professionals dealing with The COVID-19 pandemic, ArXiv http://arxiv.org/abs/2004.06510 Accessed from 16 Dec 2020
- Talaee N, Varahram M, Jamaati H, Salimi A, Attarchi M, Kazempour Dizaji M, Sadr M, Hassani S, Farzanegan B, Monjazebi F, Seyedmehdi SM. Stress and burnout in health care workers during COVID-19 pandemic: validation of a questionnaire. J Public Heal. 2020 doi: 10.1007/s10389-020-01313-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Train models to classify data using supervised machine learning - MATLAB, (n.d.). https://www.mathworks.com/help/stats/classificationlearner-app.html Accessed from 23 Apr 2021
- Train support vector machine (SVM) classifier for one-class and binary classification - MATLAB fitcsvm, (n.d.). https://www.mathworks.com/help/stats/fitcsvm.html Accessed from 8 Jan 2021
- Tyrrell DAJ, Bynoe ML. Cultivation of a novel type of common-cold virus in organ cultures. Br Med J. 1965;1:1467–1470. doi: 10.1136/bmj.1.5448.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Understanding 8 types of Cross-Validation | by Satyam Kumar | Towards Data Science, (n.d.). https://towardsdatascience.com/understanding-8-types-of-cross-validation-80c935a4976d Accessed from 7 Jan 2021
- van der Schaar M, Alaa AM, Floto A, Gimson A, Scholtes S, Wood A, McKinney E, Jarrett D, Lio P, Ercole A. How artificial intelligence and machine learning can help healthcare systems respond to COVID-19. Mach Learn. 2020 doi: 10.1007/s10994-020-05928-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N, Bushmaker T, Morris DH, Holbrook MG, Gamble A, Williamson BN, Tamin A, Harcourt JL, Thornburg NJ, Gerber SI, Lloyd-Smith JO, de Wit E, Munster VJ. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med. 2020;382:1564–1567. doi: 10.1056/nejmc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viitaniemi V, Sjöberg M, Koskela M, Ishikawa S, Laaksonen J. Advances in independent component analysis and learning Machines. Amsterdam: Elsevier; 2015. Advances in visual concept detection: TEN years of TRECVID; pp. 249–278. [Google Scholar]
- Visy JM, Le Coz P, Chadefaux B, Fressinaud C, Woimant F, Marquet J, Zittoun J, Visy JM, Vallat JM, Haguenau M. Homocystinuria due to 5, 10–methylenetetra–hydrofolate reductase deficiency revealed by stroke in adult siblings. Neurology. 1991;41:1313–1315. doi: 10.1212/wnl.41.8.1313. [DOI] [PubMed] [Google Scholar]
- Wang Y, Lawlor B (2017) Speaker recognition based on MFCC and BP neural networks, In: 2017 28th Irish Signals Syst. Conf. ISSC 2017, Institute of Electrical and Electronics Engineers Inc., 10.1109/ISSC.2017.7983644
- Wang S, Kang B, Ma J, Zeng X, Xiao M, Guo J, Cai M, Yang J, Li Y, Meng X, Xu B. A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID-19) MedRxiv. 2020 doi: 10.1101/2020.02.14.20023028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb GI, Sammut C, Perlich C, Horváth T, Wrobel S, Korb KB, Noble WS, Leslie C, Lagoudakis MG, Quadrianto N, Buntine WL, Quadrianto N, Buntine WL, Getoor L, Namata G, Getoor L, Han XJJ, Ting J-A, Vijayakumar S, Schaal S, De Raedt L. Encyclopedia of machine learning. US: Springer; 2011. Leave-one-out cross-validation; pp. 600–601. [Google Scholar]
- What is coronavirus? | Johns Hopkins Medicine, (n.d.). https://www.hopkinsmedicine.org/health/conditions-and-diseases/coronavirus Accessed from 25 Dec 2020
- When is someone infectious? | FAQ, (n.d.). https://covid19.nj.gov/faqs/coronavirus-information/about-the-virus/when-is-someone-infectious Accessed from 27 Dec 2020
- Windmon A, Minakshi M, Bharti P, Chellappan S, Johansson M, Jenkins BA, Athilingam PR. TussisWatch: a smart-phone system to identify cough episodes as early symptoms of chronic obstructive pulmonary disease and congestive heart failure. IEEE J Biomed Heal Inform. 2019;23:1566–1573. doi: 10.1109/JBHI.2018.2872038. [DOI] [PubMed] [Google Scholar]
- Winursito A, Hidayat R, Bejo A (2018) Improvement of MFCC feature extraction accuracy using PCA in Indonesian speech recognition, In: 2018 Int. Conf. Inf. Commun. Technol. ICOIACT 2018, Institute of Electrical and Electronics Engineers Inc., pp. 379–383. 10.1109/ICOIACT.2018.8350748
- Xue B, Zhang M, Browne WN, Yao X. A Survey on evolutionary computation approaches to feature selection. IEEE Trans Evol Comput. 2016;20:606–626. doi: 10.1109/TEVC.2015.2504420. [DOI] [Google Scholar]
- Yang X, Yu Y, Xu J, Shu H, Xia J, Liu H, Wu Y, Zhang L, Yu Z, Fang M, Yu T, Wang Y, Pan S, Zou X, Yuan S, Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates EJ, Yates LC, Harvey H. Machine learning “red dot”: open-source, cloud, deep convolutional neural networks in chest radiograph binary normality classification. Clin Radiol. 2018;73:827–831. doi: 10.1016/j.crad.2018.05.015. [DOI] [PubMed] [Google Scholar]
- Ye Z, Zhang Y, Wang Y, Huang Z, Song B. Chest CT manifestations of new coronavirus disease (COVID-19): a pictorial review. Eur Radiol. 2019;30(2020):4381–4389. doi: 10.1007/s00330-020-06801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Li YM, Liu CL, Zha XF. Lecture Notes in Computer Science (Including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics) Berlin, Heidelberg: Springer; 2010. Leave-one-out cross-validation based model selection for manifold regularization; pp. 457–464. [Google Scholar]
- Zhang J, Xie Y, Pang G, Liao Z, Verjans J, Li W, Sun Z, He J, Li Y, Shen C, Xia Y (2011) Viral pneumonia screening on chest X-ray images using confidence-aware anomaly detection, IEEE Trans. Med. Imaging 1–1. http://arxiv.org/abs/2003.12338 Accessed from 29 Dec 2020 [DOI] [PMC free article] [PubMed]
- Zhao Z, Morstatter F, Sharma S, Alelyani S, Anand A, Liu H (2010) Advancing feature selection research, https://eprints.kku.edu.sa/65/ Accessed from 22 Mar 2019
- Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. Addendum: a pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 doi: 10.1038/s41586-020-2951-z. [DOI] [PMC free article] [PubMed] [Google Scholar]