Abstract
People with myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) often report a high frequency of viral infections and flu-like symptoms during their disease course. Given that this reporting agrees with different immunological abnormalities and altered gene expression profiles observed in the disease, we aimed at answering whether the expression of the human angiotensin-converting enzyme 2 (ACE2), the major cell entry receptor for SARS-CoV-2, is also altered in these patients. In particular, a low expression of ACE2 could be indicative of a high risk of developing COVID-19. We then performed a meta-analysis of public data on CpG DNA methylation and gene expression of this enzyme and its homologous ACE protein in peripheral blood mononuclear cells and related subsets. We found that patients with ME/CFS have decreased methylation levels of four CpG probes in the ACE locus (cg09920557, cg19802564, cg21094739, and cg10468385) and of another probe in the promoter region of the ACE2 gene (cg08559914). We also found a decreased expression of ACE2 but not of ACE in patients when compared to healthy controls. Accordingly, in newly collected data, there was evidence for a significant higher proportion of samples with an ACE2 expression below the limit of detection in patients than healthy controls. Altogether, patients with ME/CFS can be at a higher COVID-19 risk and, if so, they should be considered a priority group for vaccination by public health authorities. To further support this conclusion, similar research is recommended for other human cell entry receptors and cell types, namely, those cells targeted by the virus.
Keywords: Myalgic encephalomyelitis/chronic fatigue syndrome, SARS-CoV-2, ACE2, Gene expression, DNA methylation
Myalgic encephalomyelitis/chronic fatigue syndrome, SARS-CoV-2, ACE2, Gene expression, DNA methylation
1. Introduction
Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS) is a multifactorial and complex disease characterized by two key symptoms: (1) persistent but unexplained fatigue that is not alleviated by rest; and (2) post-exertional malaise upon minimal physical or even mental effort [1, 2]. Although its cause remains unknown, a growing body of evidence strongly associates ME/CFS with several microbial and viral infections, as potential triggering factors [3, 4]. In addition, it is currently hypothesized that reactivations of dormant viral infections also play a role [5, 6] due to several immunological abnormalities [7, 8, 9]. On the molecular basis of the disease, peripheral blood mononuclear cells (PBMCs) have altered gene expression profiles [10], including a decreased abundance of the human angiotensin-converting enzyme 2 (ACE2) [11], the main receptor of the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) for cell invasion [12, 13, 14]. Altogether, this evidence raises the question about the COVID-19 risk in patients with ME/CFS.
As basic information, ACE2 is encoded by the X-linked ACE2 gene whose expression is predominant in the lungs, heart, skin, and kidneys [15, 16, 17, 18]. Its expression can also be detected in monocytes [19] and activated macrophages [20]. However, the percentage of ACE2-expressing cells is below 5% in the main immune-cell populations [20]. Accordingly, current RNA-Seq studies suggest a residual ACE2 expression in PBMCs from healthy controls [18]. ACE2 has an amino-acid sequence identity of 41% with its homologous angiotensin-converting enzyme (ACE) [21]. This sequence similarity increases to 61% at the nucleotide level [21]. The enzymes ACE and ACE2 are members of the renin-angiotensin-aldosterone system (RAAS), which regulates blood pressure and vascular resistance [22]. In particular, ACE and ACE2 have vasoconstriction and vasodilation effects, respectively. Given this counteracting effect, high ACE:ACE2 ratios are possible indicators of severe COVID-19 outcomes, linked to increased reactive oxygen species (ROS) production, vasoconstriction, and inflammation [23].
To answer our research question, we performed a meta-analysis of public DNA methylation and gene expression data of ACE2 and ACE in PBMCs. Similar study was conducted on the DNA methylation pattern of ACE2 in the same cell type from patients with systemic lupus erythematosus [24], an autoimmune disease whose symptoms overlap with the ones from ME/CFS [25]. To complement our findings, we also compared the mRNA levels of these two genes in PBMCs from a new cohort of female patients with ME/CFS and healthy women.
2. Materials and methods
2.1. Eligible diagnostic criteria of ME/CFS
In our meta-analysis, we selected public data from studies using either the 1994 Centre for Diseases Control criteria (1994 CDC/Fukuda) [1] or the 2003 Canadian Consensus Criteria (2003 CCC) [2] for the disease diagnosis. These criteria are defined by the presence of several key symptoms while excluding known medical conditions (e.g., multiple sclerosis or lupus) that can also explain fatigue. The choice of using these two criteria for study selection complies with the research standards set by the European Network on ME/CFS [26].
2.2. Analysis of published DNA methylation association studies
Our meta-analysis was based on six genome-wide DNA methylation association studies (Table 1), four of which [27, 28, 29, 30] were previously reviewed [31], and other two published after this review [32, 33]. Briefly, these studies aimed at identifying differentially methylated CpG dinucleotide sites between patients and healthy controls. Illumina methylation arrays were used to measure the respective DNA methylation levels with the exception of a single study (Table 1). In this study, the measurements were made by the reduced representation bisulfite sequencing [33].
Table 1.
Summary of the six DNA methylation studies under analysis.
| Reference | Sample type | ME/CFS patients |
Healthy controls, n | Technology (manufacturer) | NCBI GEO Accession number | ||
|---|---|---|---|---|---|---|---|
| n | Sample characteristics | Case definition | |||||
| [27] | CD4+ T cells | 25 | Female/male adults Mean age: 50 years old Mean BMI: not reported |
1994 CDC/Fukuda | 18 | Infinium HumanMethylation450K Array (Illumina) | NA |
| [28] | PBMC | 12 | Female adults Mean age: 41 years old Mean BMI: 23 kg/m2 |
1994 CDC/Fukuda & 2003 CCC | 12 | Infinium HumanMethylation450K Array (Illumina) | GSE59489 |
| [29] | PBMC | 49 | Female adults Mean age: 50 years old; Mean BMI: 23 kg/m2 |
1994 CDC/Fukuda &2003 CCC | 25 | Infinium HumanMethylation450 Array (Illumina) | GSE93266 |
| [30] | PBMC | 13 | Female adults Mean age: 50 years old Mean BMI: 26 kg/m2 |
1994 CDC/Fukuda & 2003 CCC | 12 | Methylation EPIC Array (Illumina) | GSE111183 |
| [32] | T lymphocytes | 61 | Female/male adults Mean age: 32 years old Mean BMI: 27 kg/m2 |
1994 CDC/Fukuda & 2003 CCC | 48 | Infinium HumanMethylation450K Array (Illumina) | GSE156792 |
| [33] | PBMC | 10 | Female/male adults Mean age: Not reported Mean BMI: not reported |
2003 CCC | 10 | Reduced representation Bisulfite sequencing | GSE153667 |
With respect to the exclusion criteria, one study excluded individuals who were taking beta-blockers or ACE inhibitors [30]. Three studies excluded participants who were treated with immunomodulatory effects or affecting the underlying DNA methylation levels at the time of data collection [28, 29, 32].
In four of the published DNA methylation studies, patients and healthy controls were matched for age, gender, and body mass index (Table 1) [28, 29, 30]. In two other studies, the matching was only based on age and gender [27, 33]. Ethnicity was also used for further matching [30, 32] or the same matching could be assumed in studies that only recruited white females [28, 29]. The DNA methylation levels were quantified in CD4+ T cells [27], PBMCs [28, 29, 30, 33], and T lymphocytes [32].
We conducted a joint analysis of the four array-based studies which made the data available [28, 29, 30, 32]. We first retrieved the data from all the CpG probes located in the coding regions and the transcription starting sites (TSS) of ACE and ACE2, respectively. We then restricted our data analysis to the 27 probes shared between the Infinium HumanMethylation450K and the Infinium HumanMethylationEPIC arrays (Supplementary Table 1).
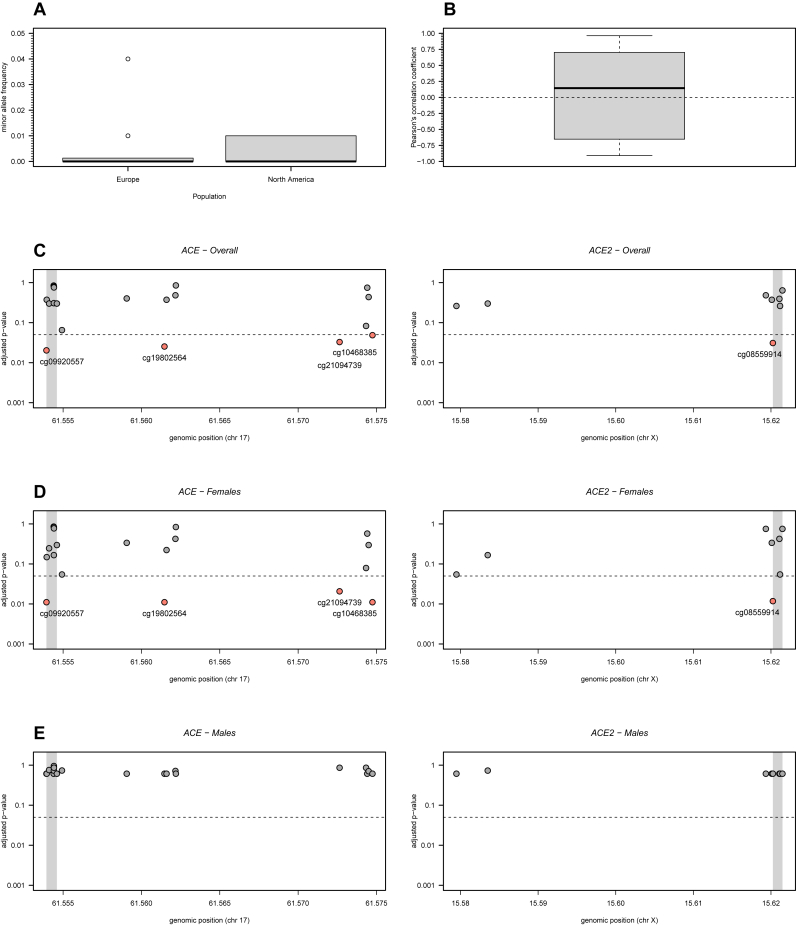
Before conducting the statistical analysis itself, we checked whether (1) the selected probes showed a high probability of detection, (2) they were not cross-reactive with other genomic regions, and (3) they were not affected by single nucleotide polymorphisms (SNPs) with high minor allele frequencies [34]. In the latter criterion, the SNPs included in the selected probes had a minor allele frequency less than 5% in Europeans and North Americans (Figure 1A; Supplementary Table 2) referring to the sampled populations of the studies. All probes passed the remaining basic quality control checks.
Figure 1.
DNA methylation analysis of 19 and 8 CpG probes located in the ACE and ACE2 genes, respectively. (A) Minor allele frequency in European and North American populations of SNPs located in the probes under analysis (see the respective data in Supplementary Table 2). (B) Boxplot of all possible Pearson's correlation coefficients (y axis) between the M-values of the probes under analysis. Horizontal dashed line represents the situation of lack of correlation. (C) Adjusted p-values for the overall association between each probe and ME/CFS. Adjusted p-values were calculated according to the Benjamini-Hochberg procedure with a false discovery rate of 5% (dashed line). Grey areas in the plots represent the TSS of the genes. (D) and (E) The same analyses as shown in C but for women and men separately.
We analyzed the M-values of a given probe instead of the respective β-values to ensure a good approximation of the Normal distribution to the data [35]. Briefly, the β-values were calculated as the proportion of the methylation signal relative to the total signal for a given probe. The M-values were finally obtained by applying a logit transformation to the β-values.
To analyze the M-values of each probe, we initially estimated a linear regression model where the respective covariates were the study indicator and the disease status of the participants. In this model, we included the main effects of the covariates and the interaction. The model parameters were then estimated by the maximum likelihood method. Note that the main effect of the disease status is usually seen as the pooled effect of this covariate across all studies, as done in meta-analysis.
We then simplified the model using a backward stepwise procedure based on Akaike's information criteria. Since the effect of the study indicator was significant for the data of each probe, we tested the association between ME/CFS and a given probe using a likelihood ratio test. In this test, we compared the model including the study indicator only with the best model including that covariate and the one associated with disease status (i.e., either the model only including the main effects or the model including both main effects and the interaction term).
To control for multiple testing, we adjusted the raw p-values using the Benjamini-Hochberg procedure [36]. This adjustment ensured a false discovery rate of 5% under the assumption of independent tests. Pearson's correlation coefficient was used to check the validity of this assumption (Figure 1B).
We also repeated the same association analysis for women and men separately. Note that three studies only recruited women [28, 29, 30] while the remaining study recruited both men and women [32]. In the latter study, there was no information available about the gender of each participant. In this case, we estimated this missing information using the function getSex of the R package minfi applied to the genome-wide DNA methylation data [37]. The resulting frequencies of men and women matched with those reported in the original study.
In the women-specific analysis, we performed the same association analysis as described above. In the men-specific analysis, we compared a linear regression model with the disease status as the single covariate against another model without that covariate, when analyzing data from each probe. The comparison was done by the likelihood ratio test whose p-values were then adjusted for multiple testing in the same way as described above.
Finally, for the study which did not share the respective data [27], we checked whether the reported differentially methylated CpG probes were located in either ACE or ACE2 (see Table 1 from this study). We did the same for the study based on the reduced representation bisulfite sequencing technology [33] (see Additional File 1 from this study).
2.3. Analysis of gene expression studies
Our meta-analysis of gene expression studies was focused on eight reports using microarray technology (Table 2) [11,[38], [39], [40], [41], [42], [43], [44]]. These studies complied with the Minimum Information about a Microarray Experiment (MIAME) standard [45] and, therefore, they were considered to have sufficient quality for their inclusion in the meta-analysis. In particular, these studies normalized the data which ensured comparability between different samples and between different measurements of the same genes.
Table 2.
Summary of the 8 microarray-based gene expression studies under analysis, ordered by the year of publication.
| Reference | Sample type | ME/CFS patients |
Healthy controls, n | Technology (Manufacturer) | ACE/ACE2 available | Data availability (NCBI GEO Assession number) | ||
|---|---|---|---|---|---|---|---|---|
| n | Sample characteristics | Case definition | ||||||
| [38] | PBMCs | 5 | Female adults Mean age: 42 years old Mean BMI: not reported |
1994 CDC/Fukuda | 5 | Atlas Glass Human 3.8 I Microarray (BD Biosciences Clontech) | No/No | No (NA) |
| [39] | PBMCs | 25 | Female/male adults Mean age: 41 years old Mean BMI: not reported |
1994 CDC/Fukuda | 25 | Custom microarray (Nimblegen) | Unclear | No (NA) |
| [40] | Whole blood | 25 | Female/male adults Mean age: 43 years old Mean BMI: not reported |
1994 CDC/Fukuda | 50 | GeneChip Human Genome U133 Plus 2.0 (Affymetrix) | Yes/Yes | No (NA) |
| [41] | Whole blood | 11 | Female/male adults Mean age: 34 years old Mean BMI: 20.3 kg/m2 |
1994 CDC/Fukuda | 11 | Custom microarray (NA) | Yes/No | Yes (NA)a |
| [42] | Muscle biopsies | 4 | Female/male adults Mean age: 45/37 years old Mean BMI: not reported |
1994 CDC/Fukuda | 5 | Operon V2.0 (CRIBI University of Padova) | Yes/Yes | No (NA) |
| [43] | PBMCs | 8 | Male adults Median age: 36 years old Mean BMI: not reported |
1994 CDC/Fukuda | 7 | GeneChip Human Genome U133 (Affymetrix) | Yes/Yes | Yes (GSE14577) |
| [11] | PBMCs | 37 | Female/male adults Mean age: 51 years old Mean BMI:29.4 kg/m2 |
1994 CDC/Fukuda | 25 | MWG 20K human Array (Biotech MWG) | Yes/Yes | No (NA) |
| [44] | PBMCs | 33 | Female/male adults Mean age: not reported Mean BMI: not reported |
1994 CDC/Fukuda | 21 | GeneChip Human Gene ST (Affymetrix) | Yes/No | No (NA) |
Data shared as a supplementary file in the online version of the study.
Gene expression of these studies was performed in PBMCs (5 studies), whole blood (2 studies) and muscle biopsies (one study). One study excluded participants who were taking any regular medication [43]. Another study reviewed the medications taken by the participants [11]. However, it was unclear which medications were considered as a part of the exclusion criteria. A third study reported that healthy controls were free from any medication at the time of sampling [41].
Three additional studies using microarray technology [46, 47, 48] were excluded from our meta-analysis due to unclear or ineligible case definitions of ME/CFS. We also excluded four RNA-seq studies [49, 50, 51, 52], because of insufficient reporting on the basic quality control checks. In particular, these studies did not report the percentage of reads that could be mapped onto the reference transcriptome, the percentage of the transcriptome covered, the average number of mapped reads per transcript, the relationship between the GC content and the mapped read distribution, as recommended elsewhere [53]. More importantly, given the high sequence homology between ACE and ACE2, these studies did not explain how their mapping algorithms dealt with reads that could be ambiguously mapped onto different locations in the transcriptome.
The selected studies were conducted in small cohorts of patients with ME/CFS (mean sample size = 18.5; range = 4–37) and healthy controls (mean sample size = 18.6; range = 5–50 individuals) (Table 2). In these studies, the patients and healthy controls were matched for age and gender. Different commercial and custom microarray technologies were used for the respective gene expression quantification. There was only one study in which the microarray did not include any probe in the genes of interest [38]. Another study used a custom array based on 9,522 genes from the RefSeq database, as available in August 2002 [39]. However, this study did not provide the list of genes included in the respective microarray. In terms of data sharing, one study made the data available in the GEO database [43] and another one within the respective publication [41]. The latter study used a custom microarray that measured the expression of stress-related genes including ACE but excluding ACE2.
Before conducting a meta-analysis of the available data, we first re-analyzed two studies where the normalized data were available [41, 43]. In the first study [41], we calculated the mean of the log2(fold-change) for ACE and the respective standard error. Note that the microarray used in this study did not include any probe in ACE2. In the second study [43], we initially calculated the mean and the respective standard error of the log2(fold-change) for each probe located in ACE and ACE2. We then pooled each pair of means for the same gene using the inverse-variance weighting method [54]. A third study reported the mean of the log2(fold-change) for ACE2 and the respective p-value using a two-tailed Student's test [11]. In this case, we determine the quantile of the t-distribution associated with half of the reported p-value, equated it to the test statistic, and solved the resulting equation as a function of the standard error. No information was available from this study concerning the expression levels of ACE.
Finally, we pooled the different estimates for the same gene from different studies using the inverse-variance weighting method [54].
2.4. Analysis of new RNA data on the ACE/ACE2 gene expression in ME/CFS
2.4.1. Study participants
Thirty-seven women with ME/CFS were recruited in 2020 from the outpatient clinic for immunodeficiencies at the Institute for Medical Immunology at the Charité-Universitätsmedizin Berlin, Germany. These patients were diagnosed according to the 2003 CCC while excluding other medical or neurological diseases which could explain fatigue [2]. Thirty-four women with self-reported healthy status were recruited from staff.
2.4.2. Experimental procedure for RNA isolation and expression
Consistently with previous studies of ME/CFS, the gene expression quantification was performed in PBMCs. These cells were isolated from heparinized whole blood by density gradient centrifugation using Biocoll Separating Solution (Merck Millipore). Total RNA was isolated and extracted from 2×106 PBMCs according to the manufacturer's instructions (NucleoSpin RNA Kit, Macherey-Nagel, cat. nr. 740955.50). Afterwards cDNA was prepared by reverse transcription (High-Capacity cDNA Reverse Transcription Kit, Applied Biosystems, cat. nr. 4368814) and real-time PCR was performed using TaqMan® Universal PCR Master Mix (cat. nr. 4305719) and TaqMan® Gene Expression Assays (cat. nr. 4331182) for ACE (Hs00174179_m1), ACE2 (Hs01085333_m1) and the housekeeping gene HPRT1 (Hs02800695_m1) (Applied Biosystems). The amplification of ACE and HPRT1 was based on 20 ng template cDNA. For the amplification of ACE2, this quantity was increased to 100 ng. All measurements were performed with the ABI7200 and software Step One Plus as absolute quantification according to manufacturer's instruction. Relative gene expression was analysed using the ΔCT method.
2.4.3. Statistical analysis
We first tested whether patients and healthy controls were matched for age using the Kolgomorov-Smirnov test for two independent samples. For statistical convenience, gene expression values were independently transformed for ACE and ACE2 using a Box-Cox transformation [55]. The parameter estimates of this transformation were 0.303 and 0.225 for ACE and ACE2, respectively. The transformed values for each gene were then analyzed as the outcome variable of a linear regression model specifying age and disease status of the participants as the respective covariates. The linear regression model was estimated using the maximum likelihood method. After estimating the models, we tested the normal distribution in the resulting residuals using the Shapiro-Wilk test. We also visually inspected the assumption of constant variance of the same residuals as a function of the covariates.
Note that we were unable to quantify the ACE2 expression in 11 patients due to cDNA material below the limit of detection. These problematic samples could be due to a lower expression of ACE2 in ME/CFS patients than in healthy controls. To test this hypothesis, we compared the respective proportion of samples below the limit of detection using the Pearson's χ2 test for two-way frequency tables.
The significance level of the statistical analysis was set at 5%.
2.4.4. Ethical approval
The protocol of this study was approved by the Ethics Committee of Charité-Universitätsmedizin Berlin in accordance with the 1964 Declaration of Helsinki and its later amendments (reference number EA2/067/20). All patients and healthy controls gave written informed consent to participate in the study.
2.5. Statistical software
We performed our statistical analysis in the R software version 4.0.3. In this analysis, we used the following Bioconductor packages: hgu133a.db, hgu133plus2.db, IlluminaHumanMethylation450kanno.ilmn12.hg19, and IlluminaHumanMethylationEPICanno.ilm10b2.hg19 to retrieve the annotation of the GeneChip HG-U133A, GeneChip U133 + 2, Infinium HumanMethylation450K Array and HumanMethylationEPIC arrays, respectively; minfi to estimate the sex of each individual from DNA methylation data [37]. The R scripts are freely available from the first and last authors upon request.
3. Results
3.1. Meta-analysis of ACE/ACE2 DNA methylation in ME/CFS patients
The oldest DNA methylation study [27] did not make the data available and hence, we screened the list of 120 differentially methylated probes (see table 1 from this study). Although located in 70 genes, these probes were neither located in ACE nor ACE2. We also screened the list of differentially methylated probes reported by the study based on the reduced representation bisulfite sequencing technology (see Additional File 1 from ref. [33]). Again, none of these probes was in the ACE or ACE2 loci.
For the four array-based studies [28, 29, 30, 32], we conducted a joint analysis of the respective data in accordance with a meta-analysis. We first observed that the M-values of the 27 probes under investigation tended to be uncorrelated with each other (Figure 1B). This observation supported the use of the Benjamini-Hochberg procedure to adjust the raw p-values under a multiple testing scenario.
The subsequent analysis suggested four CpG probes in ACE to be associated with ME/CFS (Figure 1C). The probe cg09920557 belongs to the TSS region of the gene while the remaining probes (cg19802564, cg21094739, and cg10468385) are located in the gene body. The best linear regression models for each probe included both the main effects of the study indicator and of the disease status and the respective interaction term (Supplementary Table 3). The statistical interaction between these two covariates could be seen when plotting the whole data set (Figure 2A). Although not significant, the estimated main effect of the disease status was negative for each of the significantly associated probes.
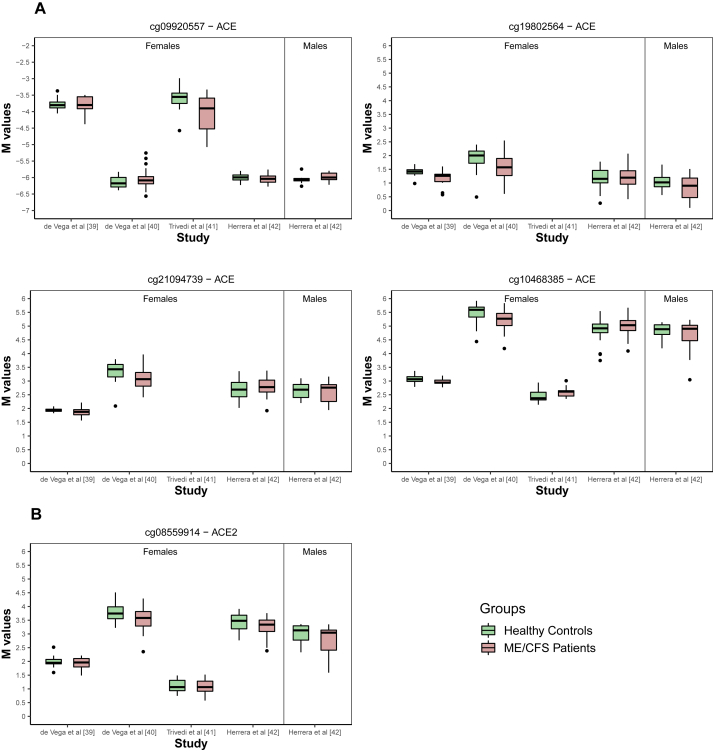
Figure 2.
Boxplots per study, group and gender of the M-values referring to probes identified in Figures 1C and 1D. (A) Significant probes located in ACE. (B) Significant probe located in ACE2.
Concerning the probes in ACE2, the only significant association with ME/CFS was obtained for cg08559914 located in the TSS region of the gene (Figure 1C). According to the best linear regression model for this probe, there was a negative association between the respective M-values and ME/CFS (coefficient estimate = -0.141 with a standard error of 0.048; Figure 2B and Supplementary Table 3). Given that a hypomethylated promoter region is typically indicative of an increased expression of the respective gene, this finding suggested an increased ACE2 expression in patients with ME/CFS.
We then repeated the same analysis for women and men separately. For women, we obtained the same disease associations, as described above (Figure 1D and Supplementary Table 3). For men, we did not find any significant associations, probably due to data from a single study [32] (Figure 1E).
3.2. Meta-analysis of ACE/ACE2 gene expression in ME/CFS patients
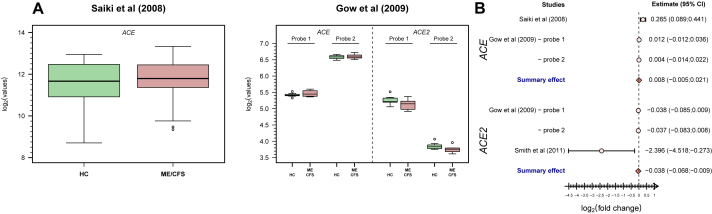
We first conducted a re-analysis of the two studies in which the expression levels of ACE or ACE2 were available for each participant (Figure 3A) [41,43]. In the first study [41], there was evidence for an increased expression of ACE in patients with ME/CFS (mean of the log2(fold-change) = 0.265; 95% CI=(0.089; 0.441)). In the second study [43], the means of the log2(fold-change) were estimated at 0.012 (95% CI=(-0.012; 0.036)) and 0.004 (95% CI=(-0.014; 0.022)) for the two probes in ACE. The corresponding estimates for the two probes in ACE2 were -0.038 (95% CI=(-0.085; 0.009)) and -0.037 (95% CI=(-0.083; 0.008)) (Figure 3A). The pooled estimates for this study were 0.007 (95% CI=(-0.006; 0.020)) and -0.038 (95% CI=(-0.067; -0.008)) for ACE and ACE2, respectively.
Figure 3.
Analysis of ACE/ACE2-related data from eligible microarray-based gene expression studies. (A) Boxplots of the data from these studies (Saiki et al (2008), ref. [41]; Gow et al (2009); ref. [43]). (B) Forest plot for the study-specific and pooled estimate of the mean of the log2(fold-change) between patients with ME/CFS and healthy controls using data shown in A.
Although not sharing the data, there was a study [11] that reported a significant negative association between ME/CFS and ACE2 expression (see online Supplementary Table 2 of this study). In this case, we obtained the following mean of the log2(fold-change) = -2.396 and 95% CI=(-4.518; -0.273).
We then pooled the estimates from different studies for the same gene: 0.008 (95% CI=(-0.005; 0.021)) and -0.038 (95% CI=(-0.068; -0.009)) for ACE and ACE2, respectively (Figure 3B). Therefore, our meta-analysis suggested a reduced expression of ACE2 but not of ACE in patients with ME/CFS when comparing to healthy controls.
Finally, the remaining gene expression studies neither shared the respective data nor reported any differential ACE/ACE2 expression between patients and healthy controls.
3.3. Analysis of ACE/ACE2 gene expression from a new female cohort
To complement our findings from the above meta-analysis, we measured the ACE and ACE2 mRNA levels in PBMCs from 37 women with ME/CFS (mean age = 41.1 years old) and 34 healthy women (mean age = 37.4 years old) (Table 3). Patients and healthy participants were matched for age (Kolmogorov-Smirnov test, p = 0.38). There was no information about the disease duration for 4 patients. The average disease duration for the remaining patients was 5.4 months in relation to the time of diagnosis (range = 0–24 months).
Table 3.
Summary statistics for the gene expression of ACE and ACE2 from the German female study participants where data of ACE2 were only available for 26 affected patients.
| Summary statistic | Healthy controls | ME/CFS patients |
|---|---|---|
| N | 34 | 37 |
| Mean age (range), years | 37.4 (23; 65) | 41.1 (19; 60) |
| Mean disease duration since diagnostic (range), months |
NA | 5.4 (0; 24) |
|
ACE | ||
| Geometric mean | 0.153 | 0.144 |
| Interquartile range |
0.087 |
0.073 |
|
ACE2 | ||
| Geometric mean | 0.002 | 0.001 |
| Interquartile range | 0.005 | 0.004 |
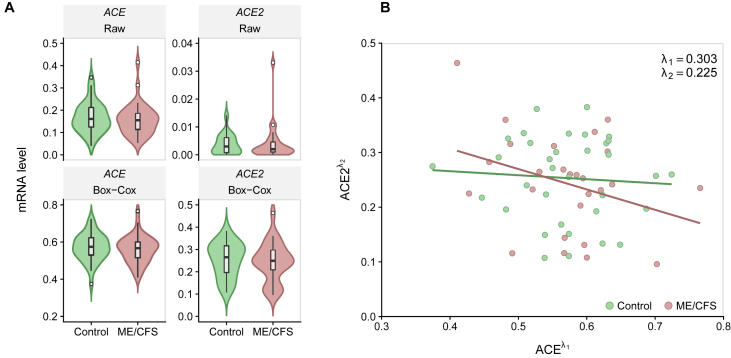
We observed higher mRNA levels of ACE than of ACE2 (Table 4, Figure 4A). There was no evidence for a significant correlation between ACE and ACE2 expression levels (Spearman's correlation coefficient = -0.120) (Figure 4B). In contrast to the above meta-analysis, we could not find a reduced expression of ACE2 in patients with ME/CFS using the complete case scenario (Table 4). However, there were 11 (29.7%) of the 37 samples from patients in which the expression level of ACE2 was below the limit of detection. This proportion of samples was significantly higher than that for healthy controls given that the expression of ACE2 could be quantified in all the samples (29.7% versus 0%; Pearson's χ2 test, p = 0.002). Consequently, we could not rule out that the patients with ME/CFS from this cohort have a decreased expression of ACE2 when compared to healthy controls. Finally, in accordance with our meta-analysis, there was no evidence of differential expression of ACE between patients and healthy controls from this cohort.
Table 4.
Analysis of the linear regression models for the Box-Cox-transformed ACE and ACE2 mRNA levels where data were only available for 26 ME/CFS patients.
| Analysis | Estimate (SE) | P-value |
|---|---|---|
| Box-Cox transformed ACE | ||
| (Intercept) | 0.541 (0.032) | <0.001 |
| Age | 0.001 (0.001) | 0.328 |
| Disease Status (ME/CFS) |
-0.013 (0.018) |
0.481 |
| Box-Cox transformed ACE2 | ||
| (Intercept) | 0.307 (0.038) | <0.001 |
| Age | -0.001 (0.001) | 0.137 |
| Disease Status (ME/CFS) | -0.006 (0.021) | 0.789 |
Figure 4.
Analysis of ACE and ACE2 expression levels from the German study. (A) Violin plots of ACE (left side) and ACE2 (right side) mRNA raw data (upper row) and transformed data using a Box-Cox transformation (lower row). (B) Scatterplot between the transformed ACE and ACE2 expression levels (Spearman's correlation coefficient = -0.120).
4. Discussion
In this work, we investigated potential differences in ACE/ACE2 DNA methylation and expression levels between patients with ME/CFS and healthy controls. With the identification of these differences, we expected to determine the health risk of patients with ME/CFS if infected by SARS-CoV-2. However, we stumbled upon hurdles related to (i) data unavailability for a possible re-analysis, (ii) availability of data derived from PBCMs and related subsets in which ACE2 is not particularly expressed, (iii) studies with unclear data quality, and (iv) studies using disease case definitions that are not recommended for research. As a consequence, we could not provide a more definite answer to our main research question.
Notwithstanding these difficulties, we could identify four CpG probes on ACE and another one on ACE2 with decreased DNA methylation levels in patients with ME/CFS. This finding suggested an increased expression of the respective genes. However, our meta-analysis of public data suggested the opposite. Such decrease in ACE2 expression was partially confirmed by new data in which there was a significant higher proportion of samples below the limit of detection in patients with ME/CFS than in healthy controls. Nonetheless, it was clear that ACE2 is not particularly expressed in PBMCs from both patients with ME/CFS and healthy controls, as mentioned in the introduction.
In general, ACE2 downregulation is known to occur after host-cell entry by SARS-CoV-2 [56]. This downregulation is particularly problematic in individuals affected by cardiovascular diseases, diabetes, and other medical conditions, due to their low ACE2 levels before the infection [57]. SARS-CoV-2 infection is then expected to further increase the ACE:ACE2 ratio, thus, promoting vasoconstriction, increased production of ROS and inflammation in patients with these co-morbidities [23]. In this scenario, a putative reduction of the ACE2 expression makes patients with ME/CFS similar to these patients with a high risk for COVID-19. As a consequence, patients with ME/CFS could be considered a priority group for vaccination by public health authorities. The fundamental question is then to know whether our findings based on PBMCs could recreate what occurs in pulmonary epithelial and endothelial cells, the main targets of SARS-CoV-2. Future research should be conducted to answer this question, as similarly done in past studies aiming at understanding how the gene expression profiles from PBMCs could mimick those present in other tissues affecting by a given disease [58, 59, 60].
Given the residual ACE2 expression in PBMCs under normal conditions, one is tempted to say that SARS-CoV-2 does not infect these cells. However, earlier studies on SARS-CoV-1 found this virus within T lymphocytes, macrophages, and dendritic cells [61]. More recently, an in vitro study was able to infect PBMCs with SARS-CoV-2 [62]. Monocytes are particularly susceptible to such infections. In this context, one cannot rule out that SARS-CoV-2 might use alternative receptors when infecting PBMCs.
Among the alternative receptors for SARS-CoV-2, the human transmembrane protease serine 2 (TMPRSS2) was suggested as a strong candidate [63] due to its role on SARS-CoV-1 infection [64, 65]. This protease seems to induce SARS-CoV-2 cell entry through endocytosis via a mechanism of ACE2 cleavage [14]. Another candidate receptor is the A disintegrin and metallopeptidase domain 17 protein (ADAM17) recognized by the immune system as a stress-response signal [66]. Like TMPRSS2, ADAM17 can also cleave ACE2 but with a reduced viral invasion efficiency [67].
With respect to the role of these proteases in ME/CFS, a targeted gene expression study analyzed ADAM17 and other stress-response proteins [41]. This study did not report any differential expression of this protease between patients with ME/CFS and healthy controls. However, this study is likely to be affected by a low statistical power due to small sample sizes for both groups. In addition, one of the selected DNA methylation studies suggested a decrease in the DNA methylation levels of one ADAM17-related CpG probe in patients with ME/CFS [30].
Dipeptidyl peptidase-4 (DPP4), also known as the lymphocyte cell surface protein CD26, was found to be the main receptor for the Middle East respiratory syndrome–related coronavirus [68, 69]. In contrast to ACE2, this surface protein is highly abundant in PBMCs including CD4+ and CD8+ T cells [18]. Bioinformatic analysis also suggested a strong interaction potential between this protein and SARS-CoV-2 [70,71]. Finally, DPP4 inhibitors were found to be protective against severe COVID-19 in patients with diabetes mellitus when compared to RAAS blockers [72]. After initial concerns, this finding combined with others suggested an interesting therapeutic avenue against COVID-19 using DPP4 blockers [73].
Interestingly, there is evidence for an increased proportion of natural killer cells and T cells expressing DPP4/CD26+ in patients with ME/CFS [7, 74]. However, the number of DPP4/CD26 molecules was significantly reduced in T lymphocytes and natural killer cells of these patients [74]. If DPP4 is indeed a relevant receptor for immune-cell invasion by SARS-CoV-2, research about this receptor should be prioritized when analyzing PBMCs from patients with ME/CFS.
Sialic acids were also hypothesized as binding receptors used by SARS-CoV-2, as reported for other human coronaviruses [75]. These acids are highly expressed in the epithelium cells of the lungs and oral cavity [76]. In vitro and in silico studies demonstrated the same binding potential for SARS-CoV-2 [77, 78, 79]. However, the ACE2 glycosylation inhibition studies suggested that sialic acids on ACE2 receptor prevent ACE2–virus interaction [80, 81]. Again, detailed research on these putative receptors could help to determine the health risk of patients with ME/CFS when infected by SARS-CoV-2.
It was suggested that the arousal state experienced by patients with ME/CFS protects them against microbial infections [82]. This suggestion came from a clinical trial where patients were treated with clonidine to decrease such a state. Treated patients got their symptoms worsened and had their inflammation markers increased during the trial. In contrast, basic epidemiological studies reported many patients with frequent viral infections and flu-like symptoms [3, 4, 83]. The question is how an infection by SARS-CoV-2 lies in this contrasting evidence. A possible answer can be given with the assistance of the so-called sustained arousal model of ME/CFS [84]. According to this model, a sustained arousal state promotes in the long-run deleterious alterations of different body systems, including the immune system. Similar prediction was made by a recent study discussing the natural history of ME/CFS [85]. If so, patients with longer disease durations are more likely to show these immunological alterations than patients at the early stages of the disease. However, we could not analyze the effect of disease duration on our results, because this variable was not available in the public data sets included in our meta-analyses.
Finally, our original idea was also to include a meta-analysis of ACE/ACE2 data from published genome-wide association studies on ME/CFS [11, 32, 86, 87, 88]. However, we could not materialize this idea, because such studies did not make their data publicly available. Nevertheless, evidence is scarce for a putative role of ACE/ACE2 polymorphisms on ME/CFS. Two studies reported many candidate SNPs for such association, but none was located in ACE or ACE2 [11,86]. Two other studies did not find any significant SNPs associated with ME/CFS [32, 88]. The most optimistic study reported thousands of SNPs related to the disease [87]. However, this study did not perform all the basic quality control checks [89].
5. Conclusions
Notwithstanding the low expression of ACE2 in PBMCs in general, there is evidence for a decreased expression of the gene in these cells from patients with ME/CFS. If PBMCs can qualitatively recreate what is occurring in the main cellular targets of SARS-CoV-2, then patients with this disease could be at a higher COVID-19 risk. In this regard, a recent preliminary report suggested that patients with ME/CFS got their symptoms worsened upon SARS-CoV-2 infection [91]. Altogether, these patients could be considered a priority group for vaccination against COVID-19, even though vaccines could trigger ME/CFS [92, 93] or even exacerbate ME/CFS symptoms as the case of the natural immunization by SARS-CoV-2. To further consolidate the existing evidence, future research should prioritize the collection of data from the main cellular targets in patients with ME/CFS. Further investigation should be also conducted on alternative SARS-CoV-2 receptors (i.e., DPP4 and sialic acids). At last, future research should also consider investigating putative sex differences in patients with ME/CFS given that, in general, men are more affected by COVID-19 than women [90].
Declarations
Author contribution statement
João Malato, André Fonseca, Anna D Grabowska, Luís Graça, Clara Cordeiro, Luís Nacul, Eliana M Lacerda, Jesus Castro-Marrero, Francisco Westermeier: Analyzed and interpreted the data; Wrote the paper.
Franziska Sotzny: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Sandra Bauer, Helma Freitag: Performed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Carmen Scheibenbogen: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Nuno Sepúlveda: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
João Malato and André Fonseca were fully funded by FCT – Fundação para a Ciência e Tecnologia, Portugal (ref.grant: SFRH/BD/149758/2019 and SFRH/BD/147629/2019, respectively). Nuno Sepúlveda and Clara Cordeiro were partially funded by FCT – Fundação para a Ciência e a Tecnologia, Portugal (ref. grant: UIDB/00006/2020). Luís Nacul and Eliana M Lacerda acknowledge the funding from the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH -Award Number: R01AI103629), and from the ME Association (Award number: PF8947) for their studies on ME/CFS.
Data availability statement
Data are available from the GEO database under the accession number GSE59489, GSE93266, GSE111183, GSE156792, GSE153667, GSE14577.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Contributor Information
Franziska Sotzny, Email: franziska.sotzny@charite.de.
Nuno Sepúlveda, Email: nunosep@gmail.com.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Fukuda K., Straus S.E., Hickie I., Sharpe M.C., Dobbins J.G., Komaroff A. The chronic fatigue syndrome: a comprehensive approach to its definition and study. Ann. Intern. Med. 1994;121:953–959. doi: 10.7326/0003-4819-121-12-199412150-00009. [DOI] [PubMed] [Google Scholar]
- 2.Carruthers B.M., Jain A.K., De Meirleir K.L., Peterson D.L., Klimas N.G., Lemer A.M., Bested A.C., Flor-Henry P., Joshi P., Powles A.C.P., Sherkey J.A., Van de Sande M.I. Myalgic encephalomyelitis/chronic fatigue syndrome: clinical working case definition, diagnostic and treatment protocols. J. Chronic Fatigue Syndrome. 2003;11:7–115. [Google Scholar]
- 3.Johnston S.C., Staines D.R., Marshall-Gradisnik S.M. Epidemiological characteristics of chronic fatigue syndrome/myalgic encephalomyelitis in Australian patients. Clin. Epidemiol. 2016;8:97–107. doi: 10.2147/CLEP.S96797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chu L., Valencia I.J., Garvert D.W., Montoya J.G. Onset patterns and course of myalgic encephalomyelitis/chronic fatigue syndrome. Front. Pediatr. 2019;7:12. doi: 10.3389/fped.2019.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rasa S., Nora-Krukle Z., Henning N., Eliassen E., Shikova E., Harrer T., Scheibenbogen C., Murovska M., Prusty B.K. Chronic viral infections in myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) J. Transl. Med. 2018;16 doi: 10.1186/s12967-018-1644-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ariza M.E. Myalgic encephalomyelitis/chronic fatigue syndrome: the human herpesviruses are back! Biomolecules. 2021;11:1–17. doi: 10.3390/biom11020185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klimas N.G., Salvato F.R., Morgan R., Fletcher M.A. Immunologic abnormalities in chronic fatigue syndrome. J. Clin. Microbiol. 1990;28:1403–1410. doi: 10.1128/jcm.28.6.1403-1410.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorusso L., Mikhaylova S.V., Capelli E., Ferrari D., Ngonga G.K., Ricevuti G. Immunological aspects of chronic fatigue syndrome. Autoimmun. Rev. 2009;8:287–291. doi: 10.1016/j.autrev.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Brenu E.W., van Driel M.L., Staines D.R., Ashton K.J., Ramos S.B., Keane J., Klimas N.G., Marshall-Gradisnik S.M. Immunological abnormalities as potential biomarkers in chronic fatigue syndrome/myalgic encephalomyelitis. J. Transl. Med. 2011;9 doi: 10.1186/1479-5876-9-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerr J.R. Gene profiling of patients with chronic fatigue syndrome/myalgic encephalomyelitis. Curr. Rheumatol. Rep. 2008;10:482–491. doi: 10.1007/s11926-008-0079-5. [DOI] [PubMed] [Google Scholar]
- 11.Smith A.K., Fang H., Whistler T., Unger E.R., Rajeevan M.S. Convergent genomic studies identify association of GRIK2 and NPAS2 with chronic fatigue syndrome. Neuropsychobiology. 2011;64:183–194. doi: 10.1159/000326692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W., Moore M.J., Vasllieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greeneugh T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ge X.Y., Li J.L., Lou Yang X., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C., Zhang Y.J., Luo C.M., Tan B., Wang N., Zhu Y., Crameri G., Zhang S.Y., Wang L.F., Daszak P., Shi Z.L. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamming I., Timens W., Bulthuis M.L.C., Lely A.T., Navis G.J., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.To K.F., Lo A.W.I. Exploring the pathogenesis of severe acute respiratory syndrome (SARS): the tissue distribution of the coronavirus (SARS-CoV) and its putative receptor, angiotensin-converting enzyme 2 (ACE2) J. Pathol. 2004;203:740–743. doi: 10.1002/path.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M.Y., Li L., Zhang Y., Wang X.S. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect. Dis. Poverty. 2020;9 doi: 10.1186/s40249-020-00662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Radzikowska U., Ding M., Tan G., Zhakparov D., Peng Y., Wawrzyniak P., Wang M., Li S., Morita H., Altunbulakli C., Reiger M., Neumann A.U., Lunjani N., Traidl-Hoffmann C., Nadeau K.C., O’Mahony L., Akdis C., Sokolowska M. Distribution of ACE2, CD147, CD26, and other SARS-CoV-2 associated molecules in tissues and immune cells in health and in asthma, COPD, obesity, hypertension, and COVID-19 risk factors. Allergy. 2020;75:2829–2845. doi: 10.1111/all.14429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rutkowska-Zapała M., Suski M., Szatanek R., Lenart M., Weglarczyk K., Olszanecki R., Grodzicki T., Strach M., Gasowski J., Siedlar M. Human monocyte subsets exhibit divergent angiotensin I-converting activity. Clin. Exp. Immunol. 2015;181:126–132. doi: 10.1111/cei.12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song X., Hu W., Yu H., Zhao L., Zhao Y., Zhao X., Xue H.H., Zhao Y. Little to no expression of angiotensin-converting enzyme-2 on most human peripheral blood immune cells but highly expressed on tissue macrophages. Cytometry. 2020:1–10. doi: 10.1002/cyto.a.24285. [DOI] [PubMed] [Google Scholar]
- 21.Tipnis S.R., Hooper N.M., Hyde R., Karran E., Christie G., Turner A.J. A human homolog of angiotensin-converting enzyme: cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 2000;275:33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- 22.Westermeier F., Bustamante M., Pavez M., García L., Chiong M., Ocaranza M.P., Lavandero S. Novel players in cardioprotection: insulin like growth factor-1, angiotensin-(1-7) and angiotensin-(1-9) Pharmacol. Res. 2015;101:41–55. doi: 10.1016/j.phrs.2015.06.018. [DOI] [PubMed] [Google Scholar]
- 23.Pagliaro P., Penna C. ACE/ACE2 ratio: a key also in 2019 coronavirus disease (Covid-19)? Front. Med. 2020;7:335. doi: 10.3389/fmed.2020.00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sawalha A.H., Zhao M., Coit P., Lu Q. Epigenetic dysregulation of ACE2 and interferon-regulated genes might suggest increased COVID-19 susceptibility and severity in lupus patients. Clin. Immunol. 2020;215:108410. doi: 10.1016/j.clim.2020.108410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blomberg J., Gottfries C.G., Elfaitouri A., Rizwan M., Rosén A. Infection elicited autoimmunity and Myalgic encephalomyelitis/chronic fatigue syndrome: an explanatory model. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pheby D.F.H., Araja D., Berkis U., Brenna E., Cullinan J., de Korwin J.-D., Gitto L., Hughes D.A., Hunter R.M., Trepel D., Wang-Steverding X. The development of a consistent europe-wide approach to investigating the economic impact of myalgic encephalomyelitis (ME/CFS): a report from the European Network on ME/CFS (EUROMENE) Healthcare. 2020;8:88. doi: 10.3390/healthcare8020088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brenu E.W., Staines D.R., Marshall-Gradisnik S.M. Methylation profile of CD4+ T cells in chronic fatigue syndrome/myalgic encephalomyelitis. J. Clin. Cell. Immunol. 2014;5:1–14. [Google Scholar]
- 28.De Vega W.C., Vernon S.D., McGowan P.O. DNA methylation modifications associated with Chronic Fatigue Syndrome. PloS One. 2014;9 doi: 10.1371/journal.pone.0104757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Vega W.C., Herrera S., Vernon S.D., McGowan P.O. Epigenetic modifications and glucocorticoid sensitivity in myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) BMC Med. Genomics. 2017;10 doi: 10.1186/s12920-017-0248-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trivedi M.S., Oltra E., Sarria L., Rose N., Beljanski V., Fletcher M.A., Klimas N.G., Nathanson L. Identification of myalgic encephalomyelitis/chronic fatigue syndrome-associated DNA methylation patterns. PloS One. 2018;13 doi: 10.1371/journal.pone.0201066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Almenar-Pérez E., Ovejero T., Sánchez-Fito T., Espejo J.A., Nathanson L., Oltra E. Epigenetic components of myalgic encephalomyelitis/chronic fatigue syndrome uncover potential transposable element activation. Clin. Ther. 2019;41:675–698. doi: 10.1016/j.clinthera.2019.02.012. [DOI] [PubMed] [Google Scholar]
- 32.Herrera S., de Vega W.C., Ashbrook D., Vernon S.D., McGowan P.O. Genome-epigenome interactions associated with myalgic encephalomyelitis/chronic fatigue syndrome. Epigenetics. 2018;13:1174–1190. doi: 10.1080/15592294.2018.1549769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Helliwell A.M., Sweetman E.C., Stockwell P.A., Edgar C.D., Chatterjee A., Tate W.P. Changes in DNA methylation profiles of myalgic encephalomyelitis/chronic fatigue syndrome patients reflect systemic dysfunctions. Clin. Epigenet. 2020;12:167. doi: 10.1186/s13148-020-00960-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dedeurwaerder S., Defrance M., Bizet M., Calonne E., Bontempi G., Fuks F.F. A comprehensive overview of Infinium HumanMethylation 450 data processing. Brief. Bioinform. 2013;15:929–941. doi: 10.1093/bib/bbt054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du P., Zhang X., Huang C.C., Jafari N., Kibbe W.A., Hou L., Lin S.M. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinf. 2010;11 doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- 37.Aryee M.J., Jaffe A.E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A.P., Hansen K.D., Irizarry R.A. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whistler T., Jones J.F., Unger E.R., Vernon S.D. Exercise responsive genes measured in peripheral blood of women with Chronic Fatigue Syndrome and matched control subjects. BMC Physiol. 2005;5 doi: 10.1186/1472-6793-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaushik N., Fear D., Richards S.C.M., McDermott C.R., Nuwaysir E.F., Kellam P., Harrison T.J., Wilkinson R.J., Tyrrell D.A.J., Holgate S.T., Kerr J.R. Gene expression in peripheral blood mononuclear cells from patients with chronic fatigue syndrome. J. Clin. Pathol. 2005;58:826–832. doi: 10.1136/jcp.2005.025718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kerr J.R., Petty R., Burke B., Gough J., Fear D., Sinclair L.I., Mattey D.L., Richards S.C.M., Montgomery J., Baldwin D.A., Kellam P., Harrison T.J., Griffin G.E., Main J., Enlander D., Nutt D.J., Holgate S.T. Gene expression subtypes in patients with chronic fatigue syndrome/myalgic encephalomyelitis. J. Infect. Dis. 2008;197:1171–1184. doi: 10.1086/533453. [DOI] [PubMed] [Google Scholar]
- 41.Saiki T., Kawai T., Morita K., Ohta M., Saito T., Rokutan K., Ban N. Identification of marker genes for differential diagnosis of chronic fatigue syndrome. Mol. Med. 2008;14:599–607. doi: 10.2119/2007-00059.Saiki. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pietrangelo T., Mancinelli R., Toniolo L., Montanari G., Vecchiet J., Fanò G., Fulle S. Transcription profile analysis of vastus lateralis muscle from patients with chronic fatigue syndrome. Int. J. Immunopathol. Pharmacol. 2009;22:795–807. doi: 10.1177/039463200902200326. [DOI] [PubMed] [Google Scholar]
- 43.Gow J.W., Hagan S., Herzyk P., Cannon C., Behan P.O., Chaudhuri A. A gene signature for post-infectious chronic fatigue syndrome. BMC Med. Genomics. 2009;2 doi: 10.1186/1755-8794-2-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeffrey M.G., Nathanson L., Aenlle K., Barnes Z.M., Baig M., Broderick G., Klimas N.G., Fletcher M.A., Craddock T.J.A. Treatment avenues in myalgic encephalomyelitis/chronic fatigue syndrome: a split-gender pharmacogenomic study of gene-expression modules. Clin. Ther. 2019;41:815–835.e6. doi: 10.1016/j.clinthera.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 45.Brazma A., Hingamp P., Quackenbush J., Sherlock G., Spellman P., Stoeckert C., Aach J., Ansorge W., Ball C.A., Causton H.C., Gaasterland T., Glenisson P., Holstege F.C.P., Kim I.F., Markowitz V., Matese J.C., Parkinson H., Robinson A., Sarkans U., Schulze-Kremer S., Stewart J., Taylor R., Vilo J., Vingron M. Minimum information about a microarray experiment (MIAME) - toward standards for microarray data. Nat. Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 46.Galbraith S., Cameron B., Li H., Lau D., Vollmer-Conna U., Lloyd A.R. Peripheral blood gene expression in postinfective fatigue syndrome following from three different triggering infections. J. Infect. Dis. 2011;204:1632–1640. doi: 10.1093/infdis/jir612. [DOI] [PubMed] [Google Scholar]
- 47.Vernon S.D., Unger E.R., Dimulescu I.M., Rajeevan M., Reeves W.C. Utility of the blood for gene expression profiling and biomarker discovery in chronic fatigue syndrome. Dis. Markers. 2002;18:193–199. doi: 10.1155/2002/892374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen C.B., Alsøe L., Lindvall J.M., Sulheim D., Fagermoen E., Winger A., Kaarbø M., Nilsen H., Wyller V.B. Whole blood gene expression in adolescent chronic fatigue syndrome: an exploratory cross-sectional study suggesting altered B cell differentiation and survival. J. Transl. Med. 2017;15 doi: 10.1186/s12967-017-1201-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bouquet J., Gardy J.L., Brown S., Pfeil J., Miller R.R., Morshed M., Avina-Zubieta A., Shojania K., McCabe M., Parker S., Uyaguari M., Federman S., Tang P., Steiner T., Otterstater M., Holt R., Moore R., Chiu C.Y., Patrick D.M. RNA-seq analysis of gene expression, viral pathogen, and B-cell/T-cell receptor signatures in complex chronic disease. Clin. Infect. Dis. 2017;64:476–481. doi: 10.1093/cid/ciw767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bouquet J., Li T., Gardy J.L., Kang X., Stevens S., Stevens J., VanNess M., Snell C., Potts J., Miller R.R., Morshed M., McCabe M., Parker S., Uyaguari M., Tang P., Steiner T., Chan W.S., De Souza A.M., Mattman A., Patrick D.M., Chiu C.Y. Whole blood human transcriptome and virome analysis of ME/CFS patients experiencing post-exertional malaise following cardiopulmonary exercise testing. PloS One. 2019;14 doi: 10.1371/journal.pone.0212193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sweetman E., Ryan M., Edgar C., Mackay A., Vallings R., Tate W. Changes in the transcriptome of circulating immune cells of a New Zealand cohort with myalgic encephalomyelitis/chronic fatigue syndrome. Int. J. Immunopathol. Pharmacol. 2019;33:1–8. doi: 10.1177/2058738418820402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raijmakers R.P.H., Jansen A.F.M., Keijmel S.P., Ter Horst R., Roerink M.E., Novakovic B., Joosten L.A.B., Van Der Meer J.W.M., Netea M.G., Bleeker-Rovers C.P. A possible role for mitochondrial-derived peptides humanin and MOTS-c in patients with Q fever fatigue syndrome and chronic fatigue syndrome. J. Transl. Med. 2019;17 doi: 10.1186/s12967-019-1906-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Conesa A., Madrigal P., Tarazona S., Gomez-Cabrero D., Cervera A., McPherson A., Szcześniak M.W., Gaffney D.J., Elo L.L., Zhang X., Mortazavi A. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17 doi: 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hartung J., Knapp G., Sinha B.K. first ed. John Wiley & Sons; New Jersey: 2008. Statistical Meta-Analysis with Applications. [Google Scholar]
- 55.Asar Ö., Ilk O., Dag O. Estimating Box-Cox power transformation parameter via goodness-of-fit tests. Commun. Stat. Simulat. Comput. 2017;46:91–105. [Google Scholar]
- 56.Datta P.K., Liu F., Fischer T., Rappaport J., Qin X. SARS-CoV-2 pandemic and research gaps: understanding SARS-CoV-2 interaction with the ACE2 receptor and implications for therapy. Theranostics. 2020;10:7448–7464. doi: 10.7150/thno.48076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Verdecchia P., Cavallini C., Spanevello A., Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection, Eur. J. Intern. Med. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gerling I.C., Ahokas R.A., Kamalov G., Zhao W., Bhattacharya S.K., Sun Y., Weber K.T. Gene expression profiles of peripheral blood mononuclear cells reveal transcriptional signatures as novel biomarkers of cardiac remodeling in rats with aldosteronism and hypertensive heart disease. JACC Hear. Fail. 2013;1:469–476. doi: 10.1016/S2213-1779(13)00374-0. [DOI] [PubMed] [Google Scholar]
- 59.Takamura T., Honda M., Sakai Y., Ando H., Shimizu A., Ota T., Sakurai M., Misu H., Kurita S., Matsuzawa-Nagata N., Uchikata M., Nakamura S., Matoba R., Tanino M., Matsubara Kichi, Kaneko S. Gene expression profiles in peripheral blood mononuclear cells reflect the pathophysiology of type 2 diabetes. Biochem. Biophys. Res. Commun. 2007;361:379–384. doi: 10.1016/j.bbrc.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 60.Manoel-Caetano F.S., Xavier D.J., Evangelista A.F., Takahashi P., Collares C.V., Puthier D., Foss-Freitas M.C., Foss M.C., Donadi E.A., Passos G.A., Sakamoto-Hojo E.T. Gene expression profiles displayed by peripheral blood mononuclear cells from patients with type 2 diabetes mellitus focusing on biological processes implicated on the pathogenesis of the disease. Gene. 2012;511:151–160. doi: 10.1016/j.gene.2012.09.090. [DOI] [PubMed] [Google Scholar]
- 61.Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Codo A.C., Davanzo G.G., de Monteiro L.B., de Souza G.F., Muraro S.P., Virgilio-da-Silva J.V., Prodonoff J.S., Carregari V.C., de Biagi Junior C.A.O., Crunfli F., Jimenez Restrepo J.L., Vendramini P.H., Reis-de-Oliveira G., Bispo dos Santos K., Toledo-Teixeira D.A., Parise P.L., Martini M.C., Marques R.E., Carmo H.R., Borin A., Coimbra L.D., Boldrini V.O., Brunetti N.S., Vieira A.S., Mansour E., Ulaf R.G., Bernardes A.F., Nunes T.A., Ribeiro L.C., Palma A.C., Agrela M.V., Moretti M.L., Sposito A.C., Pereira F.B., Velloso L.A., Vinolo M.A.R., Damasio A., Proença-Módena J.L., Carvalho R.F., Mori M.A., Martins-de-Souza D., Nakaya H.I., Farias A.S., Moraes-Vieira P.M. Elevated glucose levels favor SARS-CoV-2 infection and monocyte response through a HIF-1α/Glycolysis-Dependent Axis. Cell Metabol. 2020;32:437–446.e5. doi: 10.1016/j.cmet.2020.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., Worlock K.B., Yoshida M., Barnes J.L., Banovich N.E., Barbry P., Brazma A., Collin J., Desai T.J., Duong T.E., Eickelberg O., Falk C., Farzan M., Glass I., Gupta R.K., Haniffa M., Horvath P., Hubner N., Hung D., Kaminski N., Krasnow M., Kropski J.A., Kuhnemund M., Lako M., Lee H., Leroy S., Linnarson S., Lundeberg J., Meyer K.B., Miao Z., Misharin A.V., Nawijn M.C., Nikolic M.Z., Noseda M., Ordovas-Montanes J., Oudit G.Y., Pe’er D., Powell J., Quake S., Rajagopal J., Tata P.R., Rawlins E.L., Regev A., Reyfman P.A., Rozenblatt-Rosen O., Saeb-Parsy K., Samakovlis C., Schiller H.B., Schultze J.L., Seibold M.A., Seidman C.E., Seidman J.G., Shalek A.K., Shepherd D., Spence J., Spira A., Sun X., Teichmann S.A., Theis F.J., Tsankov A.M., Vallier L., van den Berge M., Whitsett J., Xavier R., Xu Y., Zaragosi L.E., Zerti D., Zhang H., Zhang K., Rojas M., Figueiredo F. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Glowacka I., Bertram S., Muller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K., Niemeyer D., Schneider H., Drosten C., Pohlmann S. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Düsterhöft S., Lokau J., Garbers C. The metalloprotease ADAM17 in inflammation and cancer. Pathol. Res. Pract. 2019;215 doi: 10.1016/j.prp.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 67.Heurich A., Hofmann-Winkler H., Gierer S., Liepold T., Jahn O., Pohlmann S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2014;88:1293–1307. doi: 10.1128/JVI.02202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Doremalen N., Miazgowicz K.L., Milne-Price S., Bushmaker T., Robertson S., Scott D., Kinne J., McLellan J.S., Zhu J., Munster V.J. Host species restriction of Middle East respiratory syndrome coronavirus through its receptor, dipeptidyl peptidase 4. J. Virol. 2014;88:9220–9232. doi: 10.1128/JVI.00676-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Widagdo W., Okba N.M.A., Li W., de Jong A., de Swart R.L., Begeman L., van den Brand J.M.A., Bosch B.-J., Haagmans B.L. Species-specific colocalization of Middle East respiratory syndrome coronavirus attachment and entry receptors. J. Virol. 2019;93:e00107–19. doi: 10.1128/JVI.00107-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li Y., Zhang Z., Yang L., Lian X., Xie Y., Li S., Xin S., Cao P., Lu J. The MERS-CoV receptor DPP4 as a candidate binding target of the SARS-CoV-2 spike. IScience. 2020;23 doi: 10.1016/j.isci.2020.101400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vankadari N., Wilce J.A. Emerging WuHan (COVID-19) coronavirus: glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerg. Microb. Infect. 2020;9:601–604. doi: 10.1080/22221751.2020.1739565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rhee S.Y., Lee J., Nam H., Kyoung D.S., Shin D.W., Kim D.J. Effects of a DPP-4 inhibitor and RAS blockade on clinical outcomes of patients with diabetes and COVID-19. Diabetes Metab. J. 2021;45:251–259. doi: 10.4093/dmj.2020.0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Scheen A.J. DPP-4 inhibition and COVID-19: from initial concerns to recent expectations. Diabetes Metab. 2021;47 doi: 10.1016/j.diabet.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fletcher M.A., Zeng X.R., Maher K., Levis S., Hurwitz B., Antoni M., Broderick G., Klimas N.G. Biomarkers in chronic fatigue syndrome: evaluation of natural killer cell function and dipeptidyl peptidase IV/CD26. PloS One. 2010;5 doi: 10.1371/journal.pone.0010817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun X.-L. The role of cell surface sialic acids for SARS-CoV-2 infection. Glycobiology. 2021 doi: 10.1093/glycob/cwab032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cross B.W., Ruhl S. Glycan recognition at the saliva – oral microbiome interface. Cell. Immunol. 2018;333:19–33. doi: 10.1016/j.cellimm.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Milanetti E., Miotto M., Di Rienzo L., Monti M., Gosti G., Ruocco G. In-Silico evidence for two receptors based strategy of SARS-CoV-2. BioRxiv. 2020:6197. doi: 10.3389/fmolb.2021.690655. 2020.03.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Awasthi M., Gulati S., Sarkar D.P., Tiwari S., Kateriya S., Ranjan P., Verma S.K. The sialoside-binding pocket of SARS-CoV-2 spike glycoprotein structurally resembles MERS-CoV. Viruses. 2020;12 doi: 10.3390/v12090909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Baker A.N., Richards S.J., Guy C.S., Congdon T.R., Hasan M., Zwetsloot A.J., Gallo A., Lewandowski J.R., Stansfeld P.J., Straube A., Walker M., Chessa S., Pergolizzi G., Dedola S., Field R.A., Gibson M.I. The SARS-COV-2 spike protein binds sialic acids and enables rapid detection in a lateral flow point of care diagnostic device. ACS Cent. Sci. 2020;6:2046–2052. doi: 10.1021/acscentsci.0c00855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chu H., Hu B., Huang X., Chai Y., Zhou D., Wang Y., Shuai H., Yang D., Hou Y., Zhang X., Yuen T.T.T., Cai J.P., Zhang A.J., Zhou J., Yuan S., To K.K.W., Chan I.H.Y., Sit K.Y., Foo D.C.C., Wong I.Y.H., Ng A.T.L., Cheung T.T., Law S.Y.K., Au W.K., Brindley M.A., Chen Z., Kok K.H., Chan J.F.W., Yuen K.Y. Host and viral determinants for efficient SARS-CoV-2 infection of the human lung. Nat. Commun. 2021;12 doi: 10.1038/s41467-020-20457-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang Q., Hughes T.A., Kelkar A., Yu X., Cheng K., Park S.J., Huang W.C., Lovell J.F., Neelamegham S. Inhibition of SARS-CoV-2 viral entry upon blocking N-and O-glycan elaboration. Elife. 2020;9:1–44. doi: 10.7554/eLife.61552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sulheim D., Fagermoen E., Winger A., Andersen A.M., Godang K., Muller F., Rowe P.C., Saul J.P., Skovlund E., Oie M.G., Bruun V.W. Disease mechanisms and clonidine treatment in adolescent chronic fatigue syndrome a combined cross-sectional and randomized clinical trial. JAMA Pediatr. 2014;168:351–360. doi: 10.1001/jamapediatrics.2013.4647. [DOI] [PubMed] [Google Scholar]
- 83.Slomko J., Newton J.L., Kujawski S., Tafil-Klawe M., Klawe J., Staines D., Marshall-Gradisnik S., Zalewski P. Prevalence and characteristics of chronic fatigue syndrome/myalgic encephalomyelitis (CFS/ME) in Poland: a cross-sectional study. BMJ Open. 2019;9 doi: 10.1136/bmjopen-2018-023955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wyller V.B., Eriksen H.R., Malterud K. Can sustained arousal explain the chronic fatigue syndrome? Behav. Brain Funct. 2009;5:1–10. doi: 10.1186/1744-9081-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nacul L., O’Boyle S., Palla L., Nacul F.E., Mudie K., Kingdon C.C., Cliff J.M., Clark T.G., Dockrell H.M., Lacerda E.M. How myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) progresses: the natural history of ME/CFS. Front. Neurol. 2020;11 doi: 10.3389/fneur.2020.00826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schlauch K.A., Khaiboullina S.F., De Meirleir K.L., Rawat S., Petereit J., Rizvanov A.A., Blatt N., Mijatovic T., Kulick D., Palotás A., Lombardi V.C. Genome-wide association analysis identifies genetic variations in subjects with myalgic encephalomyelitis/chronic fatigue syndrome. Transl. Psychiatry. 2016;6 doi: 10.1038/tp.2015.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Perez M., Jaundoo R., Hilton K., Del Alamo A., Gemayel K., Klimas N.G., Craddock T.J.A., Nathanson L. Genetic predisposition for immune system, hormone, and metabolic dysfunction in myalgic encephalomyelitis/chronic fatigue syndrome: a pilot study. Front. Pediatr. 2019;7 doi: 10.3389/fped.2019.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dibble J.J., McGrath S.J., Ponting C.P. Genetic risk factors of ME/CFS: a critical review. Hum. Mol. Genet. 2020;29:R118–R125. doi: 10.1093/hmg/ddaa169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Grabowska A.D., Lacerda E.M., Nacul L., Sepúlveda N. Review of the quality control checks performed by current genome-wide and targeted-genome association studies on myalgic encephalomyelitis/chronic fatigue syndrome. Front. Pediatr. 2020;8 doi: 10.3389/fped.2020.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gadi N., Wu S.C., Spihlman A.P., Moulton V.R. What’s sex got to do with COVID-19? Gender-based differences in the host immune response to coronaviruses. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.02147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.http://www.meaction.net/wp-content/uploads/2021/04/Report-on-the-impact-of-Covid-19-on-ME.pdf
- 92.Gherardi R.K., Crépeaux G., Authier F.J. Myalgia and chronic fatigue syndrome following immunization: macrophagic myofasciitis and animal studies support linkage to aluminum adjuvant persistency and diffusion in the immune system. Autoimmun. Rev. 2019;18:691–705. doi: 10.1016/j.autrev.2019.05.006. [DOI] [PubMed] [Google Scholar]
- 93.Phelan J., Grabowska A.D., Sepúlveda N. A potential antigenic mimicry between viral and human proteins linking Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS) with autoimmunity: the case of HPV immunization. Autoimmun. Rev. 2020;19 doi: 10.1016/j.autrev.2020.102487. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available from the GEO database under the accession number GSE59489, GSE93266, GSE111183, GSE156792, GSE153667, GSE14577.