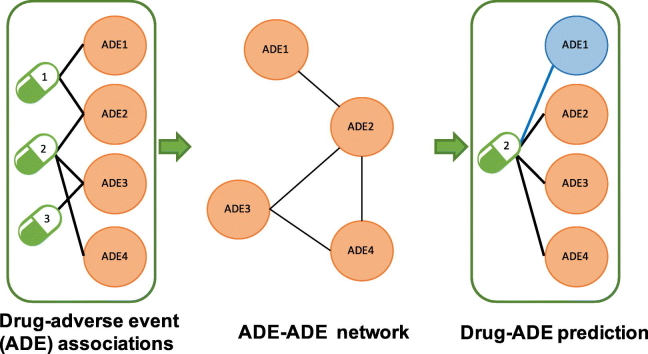
Abstract
Despite of their therapeutic effects, drug's exposure may have negative effects on human health such as adverse drug reaction (ADR) and side effects (SE).
Adverse drug events (ADEs), that correspond to an event occurring during the drug treatment (i.e. ADR and SE), is not necessarily caused by the drug itself, as this is the case with medical errors and social factors. Due to the complexity of the biological systems, not all ADEs are known for marketed drugs. Therefore, new and effective methods are needed to determine potential risks, including the development of computational strategies. We present an ADE association network based on 90,827 drug-ADE associations between 930 unique drug and 6221 unique ADE, on which we implemented a scoring system based on a pull-down approach for prediction of drug-ADE combination. Based on our network, ADEs proposed for three drugs, safinamide, sonidegib, rufinamide are further discussed. The model was able to identify, already known drug-ADE associations that are supported by the literature and FDA reports, and also to predict uncharacterized associations such as dopamine dysregulation syndrome, or nicotinic acid deficiency for the drugs safinamide and sonidegib respectively, illustrating the power of such integrative toxicological approach.
Abbreviations: AOP, adverse outcome pathway; ADR, adverse drug reaction; SE, side effect; ADE, adverse drug event; QSAR, Quantitative structure-activity relationships; wS, weighted score; pullS, pull-down score; FAERS, FDA Adverse Event Reporting System; FDA, Food and Drug Administration; LRT, Likelihood Ratio Test; MedDRA, Medical Dictionary for Regulatory Activities; PT, Preferred Term; SOC, System Organ Class; TAP–MS, tandem-affinity-purification method coupled to mass spectrometry; HMS-PCI, high-throughput mass spectrometric protein complex identification; PPAN, protein-protein association network
Keywords: Computational toxicology, Network science, System toxicology, Predictive toxicity, Adverse event network
Graphical abstract
Highlights
-
•
Effective computational strategies are needed to predict potential adverse drug events related to drug treatment.
-
•
A systems toxicological approach based on network science was developed to decipher adverse drug events associated to drugs.
-
•
Application to three drugs safinamide, sonidegib, rufinamide that allowed identification of new adverse drug events.
-
•
Integration of multi-data sources such as drug-target and drug-gene perturbations could improve drug safety prediction.
1. Introduction
The cost of developing a new drug is highly expensive and time consuming, and the success rate of investigational drugs being moved into effective therapies in clinic is declining. During the pre-marketing stages, many clinical trials fail due to lack of drug efficacy and safety concerns (Kola and Landis, 2004). Furthermore, even if a drug reaches the market, post-marketing studies can characterize some critical adverse drug reactions (ADR) not observed during the clinical studies. This is the case for some drugs described to cause teratogenicity and cardiovascular toxicity, that have been withdrawn from the market (C. for D.E. and Research, 2018; FitzGerald, 2004). Recently, some studies have reported liver and respiratory failure risk associated to the antioxidant drug Limbrel (O. of R. Affairs, 2019). Globally, lack of efficacy (therapeutic effect) and toxicity (adverse effect) have been identified as two major reasons for the drug attrition in late development stages and drug safety (Waring et al., 2015). According to the definition of WHO, adverse effects are divided into three groups: adverse drug event (ADE), adverse drug reaction (ADR) and side effect (SE). ADR and SE are unexpected effects that occurred during the use of drug at normal dose. The causality of both, ADR and SE, are related to the drug pharmacological properties. The main distinction between ADR and SE is that ADR (e.g. myocardial infarction) is always harmful for the patient, whereas SE (e.g. weight increase) may be beneficial for some patients (Thomas, 2018). For example, the drug mirtazapine used in anorexic patients, may be administered to some patients to solve weight problems, as one of the SEs cause weight gain (Hrdlicka et al., 2008). The third group of events belonging to adverse effects is the ADE (e.g. intentional product misuse). Adverse drug event corresponds to an event that occurs during the drug treatment, which is not necessarily caused by the drug itself. ADE include ADR, SE as well as other events (e.g. product issues, medical errors, social factors) (Schatz and Weber, n.d.; Definition—World Health Organization, n.d.).
Quantitative structure-activity relationships (QSAR) are computational methods widely used for drug safety prediction (Bloomingdale et al., 2017). Several studies have used such computational approach, for example to identify the correlation between cardiac adverse effects and drug properties (Frid and Matthews, 2010), to detect drug-hepatotoxicity associations (Zhu and Kruhlak, 2014), to predict SEs using combined canonical correlation analysis with network-based diffusion methods (Atias and Sharan, 2011), and to identify drugs that may cause ADRs (Hammann et al., 2010). Nevertheless, most of these studies do not explore potential mechanism of actions linking a drug to an ADE.
Today, with the acquisition of knowledge in drug-induced toxicities and ADRs through high throughput studies (LINCS, ToxCast) (Stathias et al., 2020; Dix et al., 2007) and databases (SIDER, Drug Central, ChemProt) (Ursu et al., 2019; Kuhn et al., 2016; Taboureau et al., 2011), development of innovative computational methods is feasible to decipher and predict biological targets associated to toxicity and ADE (Ciallella and Zhu, 2019; Audouze et al., 2010; Taboureau and Audouze, 2017; Hodos et al., 2016). For example, Bender et al. have developed a model to predict potential ADR(s) related to targets for drugs that are still on the market (Bender et al., 2007). Another study reported by Vogt et al. demonstrated the correlation between the genes perturbation and phenotype features at level of organ system. One of the outcomes was the strong linkage between gene GLB1 and Mucopolysaccharidosis type IV. In addition, they also found that the effect of drug was easier to cause organ damage compared to the influence of disease (Vogt et al., 2014).
With the development of sophisticated computational methods such as deep learning, a new paradigm is emerging to predict chemical toxicity (Uesawa, 2018). Although the initial reductionist drug design approach, i.e., one-target one-drug paradigm, has been the driven force for many years in drug discovery (Hopkins, 2008; Hopkins, 2007), the recent advances on molecular biology and genomics technologies are showing that drug action appears to be far more complex: a compound hits multiple targets, which are involved in complex cellular networks (Boezio et al., 2017; Wist et al., 2009).
Recently, network-based approaches have given new insights of drug action. Chen et al. performed an ADR-protein network, and identified 41 network modules related to specific ADRs (Chen et al., 2013). Campillos et al. constructed a drug-target-SE network, and demonstrated that drugs with similar side-effect profile have a similar protein target profile (Campillos et al., 2008). Oprea et al. included some tissue information on a similar network, and reported that a drug is more likely to cause SE in the organ/tissue where it is more likely to accumulate (Oprea et al., 2011). Scheiber et al. have developed a network-based model using the structural information of the drugs to predict ADEs (Scheiber et al., 2009). In this study, the authors show that compounds with similar chemical structures share ADE profile.
The objective of the present study was to develop an ADE-ADE network model based on drug-ADE information, in order to predict uncharacterized drug-ADE linkage, and to assess ADEs that are more often associated each other's. Such network may also suggest ADE complexes occurring when a new drug-ADE is observed. To perform this model, we took advantage of a protein-protein association network-based approach developed internally, previously applied, and experimentally validated (Audouze et al., 2010). Such models allowed to predict linkage between chemicals and proteins and diseases, therefore reflecting the complexity of a biological system (Audouze et al., 2010; Taboureau and Audouze, 2017; Audouze et al., 2014). A pull-down scoring system was implemented to automatically identify novel drug-ADE associations within the developed ADEs network. As an example, three case studies (i.e. safinamide, sonidegib, rufinamide) were used, and findings were validated through literature supports.
2. Materials and methods
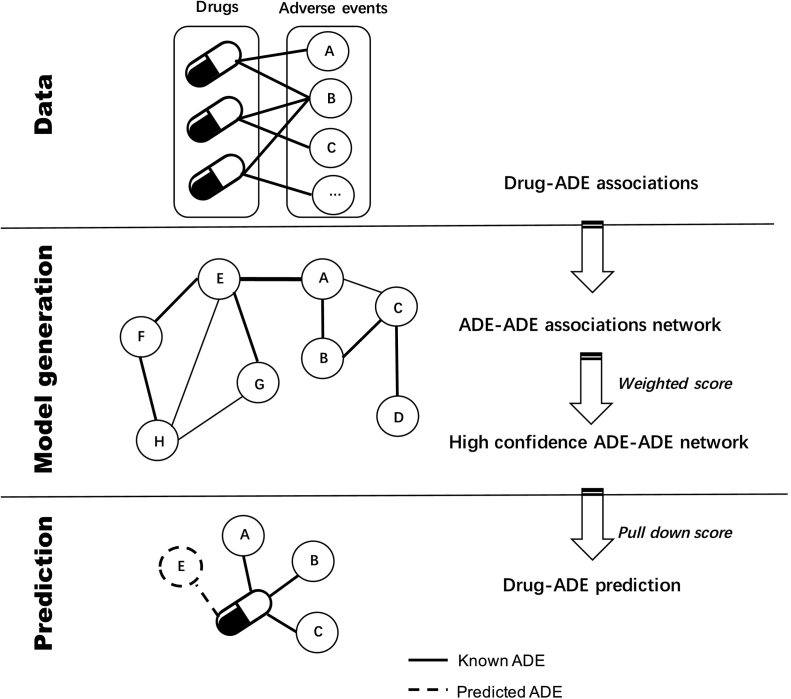
Due to the increasing concerns regarding drugs and their potential adverse events (ADE) during the pre-marketing period, computational strategy may help to decipher mode of action of drugs. Here we proposed a computational systems toxicology model to explore putative toxic effects of the compounds. A workflow of the strategy is shown in Fig. 1.
Fig. 1.
Workflow of the computational systems toxicology approach to predict adverse drug events (ADE) of drugs. Data: As a first step, drug-ADE associations were extracted from the DrugCentral database (http://drugcentral.org/ (accessed March 10, 2020). Model generation: An ADE-ADE network model was created based on the compiled data, in which two ADEs were connected if they shared at least one drug. For each ADE pair, a weighted score (wS) was calculated in order to highlight the most significant ADE-ADE associations. Prediction: for a given drug, the known ADEs were automatically screened against the ADE-ADE network. To quantify the prediction, and prioritize drug-ADE associations finding through the network, a pull-down score (pullS) was calculated between known ADE and its first order interacting ADEs present in the developed model.
2.1. Data set for model development
Drugs and their corresponding ADEs information were extracted from the DrugCentral V5.1.2 database (last access as of March 10, 2020) (Drug Central, n.d.). This database contains multiple types of data related to drugs (bioactivity profiles, chemical properties, clinical indication, pharmaceutical formulation, pharmaceutical exposure, …). Since 2018, the ADE data source Adverse Event Reporting System (FAERS) is included into the DrugCentral database (Ursu et al., 2019). FAERS was established in 1968 by Food and Drug Administration (FDA) to collect the ADE and medication error reports associated to the drugs on the market. The objective of FAERS is to support the FDA's post-marketing safety surveillance program for drugs and therapeutic biologic products (openFDA, n.d.).
In order to evaluate the associations between drugs and ADEs with disproportionally high reporting rates, the Likelihood Ratio Test (LRT) for safety signal detection method has been proposed by Huang et al. (Huang et al., 2011). LRT demonstrated the good power and sensitivity for searching the significant connection between ADE and drug (Huang et al., 2011; Huang et al., 2019). To create our model, the LRT values available in the DrugCentral database were considered. We only kept the drug-ADE associations with a LRT ratio ≥ 1 for the further analysis. The LRT ratio was calculated by following equation:
where the ‘likelihood ratio’ represents the score of an ADE for a specific drug, and the ‘likelihood ratio threshold’ is the threshold for a drug-event combination observed for this ADE. Based on the theory of LRT method, if the LRT ratio is ≥1, this ADE was considered to be associated with this specific drug (Huang et al., 2011).
2.2. Adverse drug reaction categorization with System Organ Class (SOC)
To classify the ADEs, the Medical Dictionary for Regulatory Activities (MedDRA) terminology was integrated into the network (MedDRA, n.d.). MedDRA (Version 22.1, last access as of March 10, 2020) is a standardized terminology used for all phases of development of medical products, from premarketing to post-marketing (Fig. S1) (Introductory Guide MedDRA Version 23.0, n.d.). All the ADEs in the developed network were in Preferred Term (PT). Preferred Term is a medical word or clinical expression to describe a concept in medical record (such as: therapeutic indication, pathology, disease…). We also integrated the System Organ Class (SOC) classification, that is the highest level in the MedDRA terminology. The aim of SOC was to represent the ADEs at the level of organs and body systems including other special categories (e.g. social factors, surgery, poison and injury). As a PT may be linked to more than one SOC, only the primary SOC of each PT was shown in our study (Introductory Guide MedDRA Version 23.0, n.d.).
2.3. Generating a high confidence ADE-ADE network
The relevant drug-ADE associations collected from the DrugCentral database were used to create the ADE-ADE network. The ADE model was generated based on the previously published and validated approach, the protein-protein association network (P-PAN) (Audouze et al., 2010; Audouze et al., 2014). The P-PAN was based under the assumption that nodes are linked to each other if they shared at least one component (i.e. proteins in the original model, ADEs in the present study). Here, the proposed network was constructed by representing each ADE with a node (for example the node ‘Vomiting’). Then, each ADE was connected to another ADE to form ADE's pair, if at least one overlapping drug between both ADE was identified. Therefore, ADEs are associated together in the drug space. Based on the distribution of the number of overlapping drugs between each pair of ADE, only ADE-ADE associations having at least three shared drugs were kept for further analysis (Audouze et al., 2010). In our network, the ADE-ADE links were converted into a non-redundant list of associations. That means, if the adverse event A and the adverse B are connected, the network may have two associations, A-B and B-A. Only one of them was retained in our model.
2.4. Weighted score
A weighted score (wS) was calculated for each ADE pair within the aim to select the most significant ADE-ADE associations, and to reduce noise in the model. The wS was calculated as the sum of weights for overlapping compounds between two ADEs, where weights were inversely proportional to the number of assigned ADE (Audouze et al., 2010; Audouze et al., 2014). The associations between ADE were kept if two ADE share at least three drugs. Only the top significant ADE pairs were loaded into Cytoscape V3.7.1, an open sourced software platform for visualization of the complex interacting ADE network (Saito et al., 2012).
2.5. Deciphering potential ADEs for a drug
To be able to detect new ADEs for a drug through the developed ADE network, we developed a ‘neighbor ADE procedure’ that is based on a multi-step network-neighbor's procedure developed previously (Audouze et al., 2010; Taboureau and Audouze, 2017). In the current study, we adapted it to ADE. We first listed ADEs that are known to be linked to one drug of interest, for which we aim to identify the novel potential ADE. Then, the developed ADE model was screened with the listed of known ADEs. This step allows to decipher the direct interacting ADEs as well as the surrounding ADEs. As a result, sub-network(s) containing the known ADE inputs and their first order interacting ADEs from ADE network was created. To quantify the associations between ADE input and ADEs from the developed model, we assigned a pull-down score (pullS) for each pair of ADE (Fig. S2). Originally, pullS was used for high quality scoring of protein-protein interaction data, and was developed from complex protein pull-down experiences (de Lichtenberg et al., 2005) (such as: tandem-affinity-purification method coupled to mass spectrometry (TAP–MS) assay (Gavin et al., 2002) and high-throughput mass spectrometric protein complex identification (HMS-PCI) assay (Ho et al., 2002)). This concept was therefore adapted to calculate pullS between input ADE and ADE from the model, following the equation:
where N1 was the number of drugs with ADE1, N2 was the number of drugs with ADE2, (N1 ∩ N2)was the number of common drugs with both ADEs (ADE1 and ADE2), (N1 ∪ N2) was the total number of drugs found with ADE1 and ADE2. Note that we only considered ADE pairs with N1-N2 ≥ 3, meaning that at least three drugs were shared between them. The lower the pullS value is, the more confident the ADE-ADE association is (see Fig. S2 for more details).
3. Results
3.1. Data compilation
ADE-drug associations and their corresponding LRT were downloaded from the DrugCentral database (v5.1.2, last access as of March 10, 2020). In total, we extracted 487,698 associations between 9998 unique ADE and 1524 unique drug. The maximum number of ADEs assigned to one drug ‘Methotrexate’ was 3598, and the maximum number of drugs associated with one ADE ‘Vomiting’ was 1044. To facilitate further analysis, we only kept the associations with LRT ratio≥1, reducing to 90,827 drug-ADE associations between 930 unique drug and 6221 unique ADE. The average number of ADEs for a drug was 7. The maximum number of ADE assigned to one drug ‘Methotrexate’ was 1575, and the maximum number of drugs associated with one ADE ‘Nausea’ was 409.
3.2. Generating the ADE-ADE network
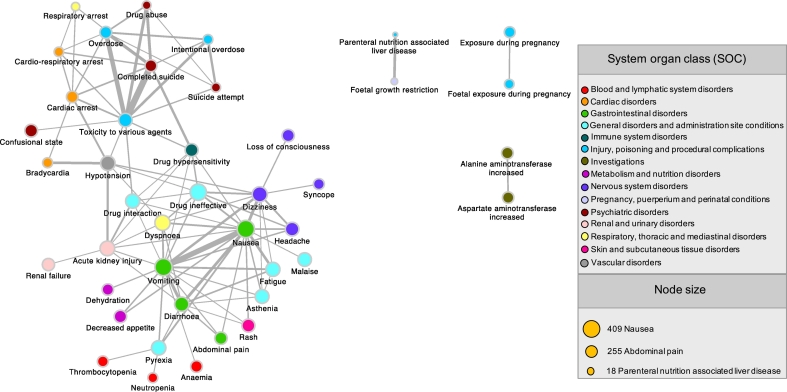
Using the selected drug-ADE associations, we developed a network model, using the previously established PPAN procedure (Audouze et al., 2010; Taboureau and Audouze, 2017; Audouze et al., 2014). The PPAN was based under the assumption that nodes (here each ADE is considered as a node) are connected to each other if there share at least one common interacting partner (e.g. a drug). Therefore, ADEs are associated together in the drug space. Based on the distribution of the number of overlapping drugs between each pair of ADE, only ADE-ADE associations having at least three shared drugs were kept for further analysis (Audouze et al., 2010). The final obtained ADEs network contains 1,515,860 ADE-ADE unique associations between 3721 ADEs (Fig. 2). The ADE-ADE association with the higher number of overlapping drugs was ‘nausea- vomiting’ with 363 drugs. To keep the most significant associations, we calculated a weighted score (wS) for each pair of nodes. Associations with the highest wS (value of 5.821) was ‘Completed suicide-Toxicity to various agents’ supported by the literature (Crome, 1993), and the smallest wS (value of 0.002) was ‘Drug interaction-Ovarian failure’ mentioned in a previous study (Anasti, 1998).
Fig. 2.
Representation of the top significant ADE-ADE associations based on the weighted score (wS). For clarity, only the top significant associations selected based on the weighted score (wS ≥ 2) are shown. Each node represents one unique ADE, colored by the organ system classification (SOC) to which it belongs. The size of the node is according to the number of drugs known to be linked to it. The width of each edge represents the wS, calculated based on the number of shared drugs between two ADEs. For example, ‘Nausea’ and ‘Vomiting’ share 363 drugs (wS of 5.07), and ‘Nausea’ and ‘Malaise’ have 266 common drugs (wS of 2.08).
In a last step, System Organ Classes (SOC) were integrated in the ADE-ADE network to facilitate further visualization, and exploration of targeted systems by drugs. In the developed ADE network, each ADE was classified into the 27 System Organ Classes using the Preferred Term (PT) (Fig. S1). Taking all the information into account, the maximum number of ADEs assigned to one SOC ‘Nervous system disorders’ is 442.
4. Case studies using the ADE-ADE network
The requirement of new or repurposed drugs to treat diseases such as neurological and oncological disorders is expected to increase rapidly in next five years. Due to the biological complexity in such diseases, it is necessary to improve patient safety during the preclinical phase (The Changing Landscape of Research and Development, n.d.). The developed ADE-ADE network model was used to assess potential uncharacterized ADEs for three marketing approved drugs by the FDA after 2000 e.g. safinamide, sonidegib, rufinamide. These three drugs were selected as they have very few known ADEs in DrugCentral (≤2) (Table 1). First, known ADEs for each drug were individually screened on the developed ADE model to identify potential ADE associations (using the pullS). As a next step, to evaluate the accuracy of the predictions, we manually evaluate the results using a two ways procedure: 1) Finding some literature-based evidence supporting the potential linkages between the most significant predicted ADEs; 2) Comparing the predicted ADEs with toxicity information reported in FDA documents.
Table 1.
Known adverse drug effects of three approved drugs. For each drug, ADE information was extracted from the DrugCentral database.
| Drug name | Adverse event in DrugCentral | SOC | Approval date in FDA | Indication |
|---|---|---|---|---|
| Safinamide | Parkinsonism hyperpyrexia syndrome | Nervous system disorders | March 21, 2017 | Parkinson's disease |
| Sonidegib | Febrile neutropenia | Blood and lymphatic system disorders | July 24, 2015 | Basal cell carcinoma of skin |
| Rufinamide | Status epilepticus | Nervous system disorders | Nov. 14, 2008 | Seizures associated with Lennox-Gastaut syndrome |
| Seizure | Nervous system disorders |
4.1. Case study 1: safinamide
Safinamide is a monoamine oxidase type B (MAO-B) inhibitor indicated as add-on treatment with levodopa for ‘off’ episode of Parkinson's disease (The Changing Landscape of Research and Development, n.d.). By inhibiting the metabolism of dopamine and glutamate release, safinamide makes the concentration of dopamine increased which lead to extend the time of ‘on’ episode. Meanwhile, safinamide would not be considered as effective treatment for Parkinson's disease as monotherapy (Cattaneo et al., 2016). By today, the unique known ADE in DrugCentral is ‘Parkinsonism hyperpyrexia syndrome’. Using the developed ADE-ADE network and the pull-down procedure, 52 potentials ADEs were identified, and among the most significant we were able to retrieve literature support, excepted for one ADE (Table S1) (Fig. 3a). The most significant predicted ADE was ‘Sudden_onset_of_sleep’. Exploring the literature with the PubMed database, this ADE has been reported in a study with patients treated with safinamide in 100 mg/day (Fabbri et al., 2015). Others ADEs were predicted for safinamide (‘Compulsive_shopping’, ‘Compulsive_sexual_behaviour’, ‘Gambling_disorder’ and ‘Impulse-control_disorder’, ‘Hypersexuality’, ‘Obsessive-compulsive_disorder’), that are part of various concerns about potential links between safinamide, impulse control problems, and compulsive and impulse behavior (FDA drug label, n.d.). Using safinamide with other antipsychotic drugs, such as dopamine antagonists may aggravate the symptom of ‘Parkinson's_disease’ and ‘Parkinsonism’, as indicated by the FDA (FDA drug label, n.d.). ‘Delusion’, ‘Jealous_delusion’ were identified in our model and also reported in clinical trials (CENTER FOR DRUG EVALUATION AND RESEARCH XADAGO (SAFINAMIDE) MEDICAL REVIEW(S), n.d.). Withdrawal effect was also predicted as a potential ADE for safinamide. Symptoms resembling to ‘Neuroleptic_malignant_syndrome’, which was reported with a rapid dose reduction or withdrawal of the drug support our suggestion (Marquet et al., 2012; Stocchi et al., 2012).
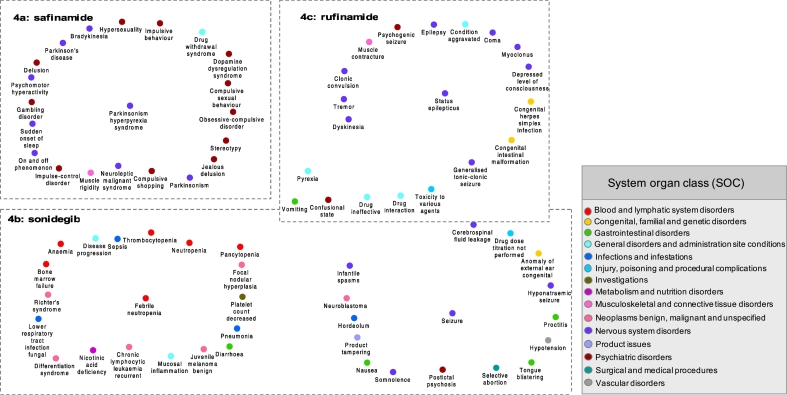
Fig. 3.
Visualization of the most significant ADEs predicted to be associated to safinamide (4a), sonidegib (4b) and rufinamide (4c). In each subnetwork, central nodes represent the known ADEs related to each drug (from the DrugCentral database). The predicted ADEs, using the developed ADE-ADE network model, are linked to each known ADE. The width of the edges is according to the pull-down score (pullS) that allow to prioritize the findings. Solid edges represent known-predicted ADEs associations, leading to identify an ADE for a drug that is supported by the literature and/or a FDA reports. Dash edges indicate novel ADE-ADE associations, allowing to reveal uncharacterized ADEs for a drug. For a better visualization of targeted systems, ADE from the model was classified with the System Organ Class (SOC), which is represented by colored nodes.
Our model was also capable to identify uncharacterized ADEs. The ADE ‘Dopamine_dysregulation_syndrome’, that is a dysfunction of the reward system with symptoms such as compulsive craving of a medication, was predicted with a significant score. Although no literature study allowed to confirm such linkage, a mechanism of action of safinamide is to inhibit catabolism of dopamine (FDA drug label, n.d.).
We went one step further and explored FDA reports. All ADEs identified by our model were compared to the observed ADEs during clinical trials and potential risk as described in FDA document (FDA drug label, n.d.). Interestingly, all ADEs mentioned in the FDA report were retrieved, and predicted by our computational network-based model (Table S2).
Nevertheless, the predicted ADE ‘Dopamine_dysregulation_syndrome’ was not previously reported in the FDA reports.
4.2. Case study 2: sonidegib
Sonidegib is a hedgehog signaling pathway inhibitor that plays an important role in the developmental process of embryo cells. This drug is used for treatment of basal cell carcinoma of skin, which is one type of malignant neoplasms (Katoh and Katoh, 2005). According to the DrugCentral database, only one known ADE exists for this drug that is ‘Febrile_neutropenia’. The developed ADE model was screened for ‘Febrile_neutropenia’, and among the 1750 potential ADEs, 20 ADEs had a significant score (Fig. 3b) (Table S3). Among them, no literature evidence between exposure to sonidegrib and ten predicted ADEs was found (Table S3). The most significant predicted ADE was ‘Nicotinic_acid_deficiency’. Nicotinic acid (also known as niacin) is a vitamin B3 used in prevention of basal cell carcinoma of skin (Totonchy and Leffell, 2017). Although there was no direct evidence to support this association sonidegib-niacin deficiency, this drug is known to be metabolized by the cytochrome CYP450 3A4, while niacin is an inhibitor of this enzyme (Gaudineau and Auclair, 2004). The ADE ‘Chronic lymphocytic leukaemia recurrent’ was also predicted. A published study revealed that perturbation for Hedgehog signaling pathway activates cancer stem cells in myeloid leukaemia (Zhao et al., 2009), leading potentially to ‘Richter's_syndrome’ (Collier et al., 2016) and may support our finding. Among the predicted ADEs with literature supports, ‘Lower_respiratory_tract_infection_fungal’ was significantly predicted. A publication reported that patients that received sonidegib in 800 mg, have shown the symptom of ‘pneumonia’ during the clinical trials (CENTER FOR DRUG EVALUATION AND RESEARCH, n.d.), which support our prediction. Another prediction, ‘Juvenile_melanoma_benign’ was supported by a FDA document that indicates that this ADE occurred in 3% of patient treated with 200 mg of sonidegib, and in 1% patients treated with 800 mg (CENTER FOR DRUG EVALUATION AND RESEARCH, n.d.). Other ADEs were deciphered, for which literature evidence were retrieved in different studies, and besides all these specific ADEs, more common ADEs (vomiting, diarrhea) were also identified by our approach (Table S3). Interestingly, some ADEs such as foetal death and teratogenicity (Table S4) were identified by our model. Such predictions are supported by the fact that this drug is known to affect early developmental signaling in the embryo, and therefore potentially invoke adverse pregnancy outcomes. When comparing our predictions with existing information in FDA pre-marketing document (FDA drug label ODOMZO(sonidegib), n.d.), all were retrieved, therefore supporting the good predictability of such computational approach (Table S4).
4.3. Case study 3: rufinamide
Rufinamide is an antiepileptic drug used for the treatment of the patient with Lennox-Gastaut syndrome, a severe form of epilepsy (called also seizure), usually occurring during infancy or early age (Heiskala, 1997). The mechanism of action of this drug is to modulate the inactivation of sodium channels. By screening independently, the two known ADEs ‘Seizure’ and ‘Status_epilepticus’ with the developed ADE network, 3095 potential unique ADEs were identified by our approach. Among them, 1721 were linked to ‘Status_epilepticus’ and 3088 to ‘Seizure’. The most significant predicted ADEs for each of the known ADEs are shown in Fig. 3c (Table S5). Among them, eight were not supported by the literature, and scientific publications could be retrieved for 27 rufinamide-ADE linkage.
Prediction of ADEs associated to ‘Seizure’: Among the high scored predicted ADEs, no literature evidence was found for seven of them. ‘Product_tampering’, could be indirectly supported by a study that demonstrated that antiepileptic treatment may induce seizure due to the dosage reduction of the drug (Bauer, 1996). Also, it has been reported that the predicted ADE ‘Postictal_psychosis’ is common after the occurrence of seizure (Morrow et al., 2006). Other ADEs were suggested as ‘Hyponatraemic_seizure’, that cause another predicted ADE ‘Cerebrospinal_fluid_leakage’, ‘Proctitis’; ‘Tongue blistering’ and ‘Hordeolum’. Several other significantly predicted ADEs were supported by the literature. ‘Infantile_spasms’, as rufinamide could be excreted in mother's milk (FDA drug label BANZEL(rufinamide), n.d.); ‘Confusional_state’ was supported by a clinical trial. It showed that rufinamide increased the risk of unusual thought or behavior (FDA drug label BANZEL(rufinamide), n.d.). Studies indicated a potential correlation between antiepileptic drug and risk of cancer (Singh et al., 2005). Other ADEs, less significant, were predicted such as ‘Drug_ineffective’. It could be explained by a non-adapted use of rufinamide in patient (Patsalos and Bourgeois, 2010; Kim et al., 2012). From a FDA perspective, one of the most significant prediction ‘Postictal_psychosis’, not supported by the literature, is mentioned in a report (FDA drug label BANZEL(rufinamide), n.d.). Also, rufinamide was indicated to cause hyperactivity reaction due to affection of multiple organs, leading to ‘Toxicity_to_various_agents’ (Guengerich, 2011) and was found in our network. Exposure to rufinamide may also affect the blood and lymphatic systems, leading to ‘Hypotension’. This result was noted in the FDA report, and observed during clinical trials (CENTER FOR DRUG EVALUATION AND RESEARCH, n.d.).
Prediction of ADEs associated to ‘Status_epilepticus’: Only one predicted ADE was not supported by the literature, ‘Psychogenic_seizure’, but a study shows that depression was considered as a risk factor inducing ‘Psychogenic_seizure’ in pediatric population (Devinsky et al., 2011). Among the most significant predicted ADEs supported by the literature and FDA reports, ‘Clonic_convulsion’ is known to occur in pediatric patients (4 to 17 years) at the dose of 45 mg/kg per day during clinical trials (FDA drug label BANZEL(rufinamide), n.d.). Changes of doses during treatment of rifunamide, may also lead to withdrawal effects (‘Muscle_contracture’ that is linked to ‘Generalised_tonic-clonic_seizure’, ‘Seizure’, ‘Condition_aggravated’, ‘Epilepsy’, ‘Myoclonus’) (FDA drug label BANZEL(rufinamide), n.d.; Conradsen et al., 2013). Therefore the FDA recommended to reduce progressively posology of rufinamide (FDA drug label BANZEL(rufinamide), n.d.). All predicted ADEs were compared with the ones described in the FDA report (FDA drug label BANZEL(rufinamide), n.d.), and common predicted ADEs (headache, dizziness, fatigue, somnolence, and nausea) were retrieved (Table S6).
5. Discussion
New and innovative methods are needed to predict putative uncharacterized ADEs from drug exposure. In the present study, we developed a network-based model using known information of drug-ADE associations, and demonstrated the predictive power of such approach to decipher uncharacterized ADEs for a specific drug. To our knowledge, the proposed ADE-ADE model is the first to provide a global mapping of ADE relationships in the drug space, without considering the structural information of the drugs. Integration of the SOC information into the ADE network allowed to shown that drugs does not affect only the targeted systems, but multiple systems leading to various pathways perturbations that may cause ADEs (Fig. 2, Fig. 3). Such findings may be explained by the fact that therapeutic targets might act in different cells and organs.
An exploration of the distribution of ADE-ADE associations in our model, shows that both the number of overlapping drugs between two connected ADEs, and the weighted scores were controlling most of the associations in the network, following the well know “richer get richer” effect (Hopkins, 2008). Such observation reflects the nature of scale free network that is generally related to biological network. The most significant ADE associations (based on the wS) were retrieved for common ADEs such as Nausea, Vomiting and Diarrhea, which is due to the high number of drugs linked to these ADEs. Associations with the lowest wS were between uncommon ADEs such as ‘Parkinsonism_hyperpyrexia_syndrome and Dopamine_dysregulation_syndrome’. This can be of interest for the assessment of drug-drug interactions that may cause more serious unknown ADEs.
The major limitation of our integrative systems toxicology approach is that the ADE predictions are limited to the ones present in the ADE model. Hence, drug effects that may be reported in the literature or in FDA reports (such as hematuria for rufinamide, or more complex effect as retinal degeneration and loss of photoreceptor cells reported for safinamide), but not in the DrugCentral database, cannot be predicted by the model. Such gaps could be paved when more data with LRT information will be available from diverse data sources.
In addition to the ADE predictions for drugs, the model allowed to point out another interesting issue, which is that some ADE occurrence is dose related. Some ADEs occurred depending to the posology such as “impulse behavior” for safinamide, ‘pneumonia’ for sonidegib and “severe seizure” for rufinamide. Therefore, clinical surveillance need to be done with the identification of suitable dose, especially for the drug with narrow therapeutic window (Role of systems pharmacology in understanding drug adverse events - Berger - 2011 - WIREs Systems Biology and Medicine - Wiley Online Library, n.d.). Overall, ADE appearance is dependent of the patient population type (age, gender, ethnicity…). For example, it is known that older patients have usually multiple illnesses, and therefore are exposed to polypharmacy treatment (Lavan and Gallagher, 2016). This may increase the risk of drug-drug interactions and multiple ADEs. Furthermore, it is important to consider the time of the start of medication, as the period of vulnerability is higher in newborn and infants due to their organ maturation. This population may have over reaction for the drug in terms of organ and result other underlying ADEs (FDA drug label BANZEL(rufinamide), n.d.; Wu and Peters, 2019). The different formulation (oral and IV) or the relation of an ADE to a prodrug, the active drug or metabolite are some other issues that may also cause different adverse effects (Uchegbu and Florence, 1996; Ma'ayan et al., 2007). So far, our ADE model does not differentiate between these points as our approach is limited to binary data (drugs associated or not with ADEs) compiled from the DrugCentral database. In the future, more specific ADE models could be developed when such diverse information will be accessible for a large set of drugs.
Such integrative toxicological modeling developed for drug attrition and pharmacovigilance, may be applied to other research areas such as the regulatory arena of toxicology that does not always embrace the complexity of biology. It could be considered more generally in chemical risk assessment. Gene-environment associations, life stage considerations, stressor-key event linkages related to adverse outcome pathways (AOPs) or evidences between the chemical exposome and proteins/pathways dysregulations could be also integrated in such predictive toxicology models to improve our understanding of the impact from chemical exposure to human health and toxicity (Wu et al., 2020; Carvaillo et al., 2019; Rugard et al., 2020; Jornod et al., 2020).
6. Conclusion
We propose a novel computational approach different from the existing ones, that integrate drugs information and calculate in a quantitative manner ADE-ADE linkage. The ability to make new findings, that is prediction of new potential ADEs for a drug, is illustrated with three case studies. Results indicate that the developed ADE-ADE network is powerful in the identification of ADEs associated to drugs and can guide further experimental studies during the drug development process. As a perspective, our network approach could help to extend the knowledge of the mechanisms of a drug related to ADEs with the integration of poly-pharmacological and poly-toxicological data. That would pave the way for effective medicine with patient safety.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
The authors would like to acknowledge the Université de Paris and Inserm.
Financial interest declaration
The authors declare they have no actual or potential competing financial interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crtox.2020.06.001.
Appendix A. Supplementary data
Fig. S1 The hierarchical structure of MedDRA and the list of 27 SOC.
Fig. S2 Examples of the pull-down score (pullS).
Table S1 Identified adverse effects for safinamide.
Table S2 ADEs reported in FDA pre-marketing reports for safinamide.
Table S3 Identified adverse effects for sonidegib.
Table S4 ADEs reported in FDA pre-marketing reports for sonidegib.
Table S5 Identified adverse effects for rufinamide.
Table S6 ADEs reported in FDA pre-marketing reports for rufinamide.
References
- Anasti J.N. Premature ovarian failure: an update. Fertil. Steril. 1998;70:1–15. doi: 10.1016/S0015-0282(98)00099-5. [DOI] [PubMed] [Google Scholar]
- Atias N., Sharan R. An algorithmic framework for predicting side effects of drugs. J. Comput. Biol. 2011;18:207–218. doi: 10.1089/cmb.2010.0255. [DOI] [PubMed] [Google Scholar]
- Audouze K., Juncker A.S., Roque F.J.S.S.A., Krysiak-Baltyn K., Weinhold N., Taboureau O., Jensen T.S., Brunak S. Deciphering diseases and biological targets for environmental chemicals using toxicogenomics networks. PLOS Computational Biology. 2010;6 doi: 10.1371/journal.pcbi.1000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audouze K., Tromelin A., Bon A.M.L., Belloir C., Petersen R.K., Kristiansen K., Brunak S., Taboureau O. Identification of odorant-receptor interactions by global mapping of the human odorome. PLoS One. 2014;9 doi: 10.1371/journal.pone.0093037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer J. Seizure-inducing effects of antiepileptic drugs: a review. Acta Neurol. Scand. 1996;94:367–377. doi: 10.1111/j.1600-0404.1996.tb00047.x. [DOI] [PubMed] [Google Scholar]
- Bender A., Scheiber J., Glick M., Davies J.W., Azzaoui K., Hamon J., Urban L., Whitebread S., Jenkins J.L. Analysis of pharmacology data and the prediction of adverse drug reactions and off-target effects from chemical structure. ChemMedChem. 2007;2:861–873. doi: 10.1002/cmdc.200700026. [DOI] [PubMed] [Google Scholar]
- Bloomingdale P., Housand C., Apgar J.F., Millard B.L., Mager D.E., Burke J.M., Shah D.K. Quantitative systems toxicology. Current Opinion in Toxicology. 2017;4:79–87. doi: 10.1016/j.cotox.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boezio B., Audouze K., Ducrot P., Taboureau O. Network-based approaches in pharmacology. Molecular Informatics. 2017;36:1700048. doi: 10.1002/minf.201700048. [DOI] [PubMed] [Google Scholar]
- C. for D.E. and Research, Thalidomide (marketed as Thalomid) Information, FDA. (2018). http://www.fda.gov/drugs/postmarket-drug-safety-information-patients-and-providers/thalidomide-marketed-thalomid-information (accessed March 10, 2020).
- Campillos M., Kuhn M., Gavin A.-C., Jensen L.J., Bork P. Drug target identification using side-effect similarity. Science. 2008;321:263–266. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- Carvaillo J.-C., Barouki R., Coumoul X., Audouze K. Linking bisphenol S to adverse outcome pathways using a combined text mining and systems biology approach. Environ. Health Perspect. 2019;127:47005. doi: 10.1289/EHP4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo C., Sardina M., Bonizzoni E. Safinamide as add-on therapy to levodopa in mid- to late-stage Parkinson’s disease fluctuating patients: post hoc analyses of studies 016 and SETTLE. J. Park. Dis. 2016;6:165–173. doi: 10.3233/JPD-150700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CENTER FOR DRUG EVALUATION AND RESEARCH MEDICAL REVIEW,SONIDEGIB.pdf. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2015/205266Orig1s000MedR.pdf (n.d.)
- CENTER FOR DRUG EVALUATION AND RESEARCH MEDICAL REVIEW(S),RUFINAMIDE. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2011/201367Orig1s000MedR.pdf (n.d.)
- CENTER FOR DRUG EVALUATION AND RESEARCH XADAGO (SAFINAMIDE) MEDICAL REVIEW(S), (n.d.). https://www.accessdata.fda.gov/drugsatfda_docs/nda/2017/207145Orig1s000MedR.pdf (accessed March 11, 2020).
- Chen X., Liu X., Jia X., Tan F., Yang R., Chen S., Liu L., Wang Y., Chen Y. Network characteristic analysis of ADR-related proteins and identification of ADR-ADR associations. Sci. Rep. 2013;3:1–7. doi: 10.1038/srep01744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciallella H.L., Zhu H. Advancing computational toxicology in the big data era by artificial intelligence: data-driven and mechanism-driven modeling for chemical toxicity. Chem. Res. Toxicol. 2019;32:536–547. doi: 10.1021/acs.chemrestox.8b00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier N.J., Ali F.R., Lear J.T. The safety and efficacy of sonidegib for the treatment of locally advanced basal cell carcinoma. Expert. Rev. Anticancer. Ther. 2016;16:1011–1018. doi: 10.1080/14737140.2016.1230020. [DOI] [PubMed] [Google Scholar]
- Conradsen I., Moldovan M., Jennum P., Wolf P., Farina D., Beniczky S. Dynamics of muscle activation during tonic–clonic seizures. Epilepsy Res. 2013;104:84–93. doi: 10.1016/j.eplepsyres.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Crome P. The toxicity of drugs used for suicide. Acta Psychiatr. Scand. 1993;87:33–37. doi: 10.1111/j.1600-0447.1993.tb05371.x. [DOI] [PubMed] [Google Scholar]
- de Lichtenberg U., Jensen L.J., Brunak S., Bork P. Dynamic complex formation during the yeast cell cycle. Science. 2005;307:724–727. doi: 10.1126/science.1105103. [DOI] [PubMed] [Google Scholar]
- Definition—World Health Organization, (n.d.). https://www.who.int/medicines/areas/quality_safety/safety_efficacy/trainingcourses/definitions.pdf (accessed March 10, 2020).
- Devinsky O., Gazzola D., LaFrance W.C. Differentiating between nonepileptic and epileptic seizures. Nat. Rev. Neurol. 2011;7:210–220. doi: 10.1038/nrneurol.2011.24. [DOI] [PubMed] [Google Scholar]
- Dix D.J., Houck K.A., Martin M.T., Richard A.M., Setzer R.W., Kavlock R.J. The ToxCast program for prioritizing toxicity testing of environmental chemicals. Toxicol. Sci. 2007;95:5–12. doi: 10.1093/toxsci/kfl103. [DOI] [PubMed] [Google Scholar]
- Drug Central, (n.d.). http://drugcentral.org/ (accessed March 10, 2020).
- Fabbri M., Rosa M.M., Abreu D., Ferreira J.J. Clinical pharmacology review of safinamide for the treatment of Parkinson’s disease. Neurodegenerative Disease Management. 2015;5:481–496. doi: 10.2217/nmt.15.46. [DOI] [PubMed] [Google Scholar]
- FDA drug label,XADAGO (safinamide) tablets, for oral use.pdf, (n.d.). https://www.accessdata.fda.gov/drugsatfda_docs/label/2017/207145lbl.pdf (accessed March 11, 2020).
- FDA drug label BANZEL(rufinamide), oral suspension, (n.d.). https://www.accessdata.fda.gov/drugsatfda_docs/label/2015/021911s012lbl.pdf (accessed March 11, 2020).
- FDA drug label ODOMZO(sonidegib), capsule, for oral use, (n.d.). https://www.accessdata.fda.gov/drugsatfda_docs/label/2017/205266s004lbl.pdf (accessed March 11, 2020).
- FitzGerald G.A. Coxibs and cardiovascular disease. N. Engl. J. Med. 2004;351:1709–1711. doi: 10.1056/NEJMp048288. [DOI] [PubMed] [Google Scholar]
- Frid A.A., Matthews E.J. Prediction of drug-related cardiac adverse effects in humans-B: use of QSAR programs for early detection of drug-induced cardiac toxicities. Regul. Toxicol. Pharmacol. 2010;56:276–289. doi: 10.1016/j.yrtph.2009.11.005. [DOI] [PubMed] [Google Scholar]
- Gaudineau C., Auclair K. Inhibition of human P450 enzymes by nicotinic acid and nicotinamide. Biochem. Biophys. Res. Commun. 2004;317:950–956. doi: 10.1016/j.bbrc.2004.03.137. [DOI] [PubMed] [Google Scholar]
- Gavin A.-C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.-M., Cruciat C.-M., Remor M., Höfert C., Schelder M., Brajenovic M., Ruffner H., Merino A., Klein K., Hudak M., Dickson D., Rudi T., Gnau V., Bauch A., Bastuck S., Huhse B., Leutwein C., Heurtier M.-A., Copley R.R., Edelmann A., Querfurth E., Rybin V., Drewes G., Raida M., Bouwmeester T., Bork P., Seraphin B., Kuster B., Neubauer G., Superti-Furga G. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Guengerich F.P. Mechanisms of drug toxicity and relevance to pharmaceutical development. Drug Metab Pharmacokinet. 2011;26:3–14. doi: 10.2133/dmpk.dmpk-10-rv-062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammann F., Gutmann H., Vogt N., Helma C., Drewe J. Prediction of adverse drug reactions using decision tree modeling. Clinical Pharmacology & Therapeutics. 2010;88:52–59. doi: 10.1038/clpt.2009.248. [DOI] [PubMed] [Google Scholar]
- Heiskala H. Community-based study of Lennox-Gastaut syndrome. Epilepsia. 1997;38:526–531. doi: 10.1111/j.1528-1157.1997.tb01136.x. [DOI] [PubMed] [Google Scholar]
- Ho Y., Gruhler A., Heilbut A., Bader G.D., Moore L., Adams S.-L., Millar A., Taylor P., Bennett K., Boutilier K., Yang L., Wolting C., Donaldson I., Schandorff S., Shewnarane J., Vo M., Taggart J., Goudreault M., Muskat B., Alfarano C., Dewar D., Lin Z., Michalickova K., Willems A.R., Sassi H., Nielsen P.A., Rasmussen K.J., Andersen J.R., Johansen L.E., Hansen L.H., Jespersen H., Podtelejnikov A., Nielsen E., Crawford J., Poulsen V., Sørensen B.D., Matthiesen J., Hendrickson R.C., Gleeson F., Pawson T., Moran M.F., Durocher D., Mann M., Hogue C.W.V., Figeys D., Tyers M. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Hodos R.A., Kidd B.A., Khader S., Readhead B.P., Dudley J.T. Computational approaches to drug repurposing and pharmacology. Wiley Interdiscip Rev Syst Biol Med. 2016;8:186–210. doi: 10.1002/wsbm.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins A.L. Network pharmacology. Nat. Biotechnol. 2007;25:1110–1111. doi: 10.1038/nbt1007-1110. [DOI] [PubMed] [Google Scholar]
- Hopkins A.L. Network pharmacology: the next paradigm in drug discovery. Nat. Chem. Biol. 2008;4:682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- Hrdlicka M., Beranova I., Zamecnikova R., Urbanek T. Mirtazapine in the treatment of adolescent anorexia nervosa. Eur Child Adolesc Psychiatry. 2008;17:187–189. doi: 10.1007/s00787-007-0670-8. [DOI] [PubMed] [Google Scholar]
- Huang L., Zalkikar J., Tiwari R.C. A likelihood ratio test based method for signal detection with application to FDA’s drug safety data. J. Am. Stat. Assoc. 2011;106:1230–1241. doi: 10.1198/jasa.2011.ap10243. [DOI] [Google Scholar]
- Huang L., Zalkikar J., Tiwari R. 2019. Likelihood-ratio-test methods for drug safety signal detection from multiple clinical datasets, Computational and Mathematical Methods in Medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Introductory Guide MedDRA Version 23.0, (n.d.). https://www.meddra.org/sites/default/files/guidance/file/intguide_23_0_english.pdf (accessed March 10, 2020).
- Jornod F., Rugard M., Tamisier L., Coumoul X., Andersen H.R., Barouki R., Audouze K. 2020. AOP4EUpest: Mapping of Pesticides in Adverse Outcome Pathways Using a Text Mining Tool, Bioinformatics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y., Katoh M. Hedgehog signaling pathway and gastric cancer. Cancer Biology & Therapy. 2005;4:1050–1054. doi: 10.4161/cbt.4.10.2184. [DOI] [PubMed] [Google Scholar]
- Kim J.Y., Lee C.G., Yu H.J., Nam S.H., Lee J., Lee M. The efficacy and tolerability of rufinamide in intractable pediatric epilepsy. J Epilepsy Res. 2012;2:33–37. doi: 10.14581/jer.12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kola I., Landis J. Can the pharmaceutical industry reduce attrition rates? Nat. Rev. Drug Discov. 2004;3:711–716. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- Kuhn M., Letunic I., Jensen L.J., Bork P. The SIDER database of drugs and side effects. Nucleic Acids Res. 2016;44:D1075–D1079. doi: 10.1093/nar/gkv1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavan A.H., Gallagher P. Predicting risk of adverse drug reactions in older adults. Ther Adv Drug Saf. 2016;7:11–22. doi: 10.1177/2042098615615472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma’ayan A., Jenkins S.L., Goldfarb J., Iyengar R. Network analysis of FDA approved drugs and their targets. Mount Sinai Journal of Medicine: A Journal of Translational and Personalized Medicine. 2007;74:27–32. doi: 10.1002/msj.20002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquet A., Kupas K., Johne A., Astruc B., Patat A., Krösser S., Kovar A. The effect of safinamide, a novel drug for Parkinson’s disease, on pressor response to oral tyramine: a randomized, double-blind, clinical trial. Clinical Pharmacology & Therapeutics. 2012;92:450–457. doi: 10.1038/clpt.2012.128. [DOI] [PubMed] [Google Scholar]
- MedDRA|, (n.d.). https://www.meddra.org/ (accessed March 10, 2020).
- Morrow E.M., Lafayette J.M., Bromfield E.B., Fricchione G. Postictal psychosis: presymptomatic risk factors and the need for further investigation of genetics and pharmacotherapy. Ann. General Psychiatry. 2006;5:9. doi: 10.1186/1744-859X-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O. of R. Affairs, Primus Announces a Voluntary Nationwide Recall of All Lots Within Expiry of Prescription Medical Food Limbrel® Due to Rare But Serious and Reversible Adverse Events While Seeking FDA's Cooperation to Restore Access for Patients with Medical Necessity, U.S. Food and Drug Administration. (2019). http://www.fda.gov/safety/recalls-market-withdrawals-safety-alerts/primus-announces-voluntary-nationwide-recall-all-lots-within-expiry-prescription-medical-food (accessed March 10, 2020).
- openFDA, (n.d.). https://open.fda.gov/data/faers/ (accessed March 10, 2020).
- Oprea T.I., Nielsen S.K., Ursu O., Yang J.J., Taboureau O., Mathias S.L., Kouskoumvekaki lrene, Sklar L.A., Bologa C.G. Associating drugs, targets and clinical outcomes into an integrated network affords a new platform for computer-aided drug repurposing. Mol Inform. 2011;30:100–111. doi: 10.1002/minf.201100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patsalos P.N., Bourgeois B.F.D. Cambridge University Press; 2010. The Epilepsy Prescriber’s Guide to Antiepileptic Drugs. [Google Scholar]
- Role of systems pharmacology in understanding drug adverse events - Berger - 2011 - WIREs Systems Biology and Medicine - Wiley Online Library, (n.d.). https://onlinelibrary.wiley.com/doi/full/10.1002/wsbm.114 (accessed March 11, 2020). [DOI] [PMC free article] [PubMed]
- Rugard M., Coumoul X., Carvaillo J.-C., Barouki R., Audouze K. Deciphering adverse outcome pathway network linked to bisphenol F using text mining and systems toxicology approaches. Toxicol. Sci. 2020;173:32–40. doi: 10.1093/toxsci/kfz214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito R., Smoot M.E., Ono K., Ruscheinski J., Wang P.-L., Lotia S., Pico A.R., Bader G.D., Ideker T. A travel guide to Cytoscape plugins. Nat. Methods. 2012;9:1069–1076. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz, S.N. and Weber, R.J. , Adverse drug reactions, Pharmacotherapy Self-Assessment Program (PSAP), 2015,Vol. 1 No. 1, pp. 5–21, (n.d.). https://www.accp.com/docs/bookstore/psap/2015B2.SampleChapter.pdf (accessed March 10, 2020).
- Scheiber J., Jenkins J.L., Sukuru S.C.K., Bender A., Mikhailov D., Milik M., Azzaoui K., Whitebread S., Hamon J., Urban L., Glick M., Davies J.W. Mapping adverse drug reactions in chemical space. J. Med. Chem. 2009;52:3103–3107. doi: 10.1021/jm801546k. [DOI] [PubMed] [Google Scholar]
- Singh G., Driever P.H., Sander J.W. Cancer risk in people with epilepsy: the role of antiepileptic drugs. Brain. 2005;128:7–17. doi: 10.1093/brain/awh363. [DOI] [PubMed] [Google Scholar]
- Stathias V., Turner J., Koleti A., Vidovic D., Cooper D., Fazel-Najafabadi M., Pilarczyk M., Terryn R., Chung C., Umeano A., Clarke D.J.B., Lachmann A., Evangelista J.E., Ma’ayan A., Medvedovic M., Schürer S.C. LINCS Data Portal 2.0: next generation access point for perturbation-response signatures. Nucleic Acids Res. 2020;48:D431–D439. doi: 10.1093/nar/gkz1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocchi F., Borgohain R., Onofrj M., Schapira A.H.V., Bhatt M., Lucini V., Giuliani R., Anand R. A randomized, double-blind, placebo-controlled trial of safinamide as add-on therapy in early Parkinson’s disease patients. Mov. Disord. 2012;27:106–112. doi: 10.1002/mds.23954. [DOI] [PubMed] [Google Scholar]
- Taboureau O., Audouze K. Human environmental disease network: a computational model to assess toxicology of contaminants. ALTEX - Alternatives to Animal Experimentation. 2017;34:289–300. doi: 10.14573/altex.1607201. [DOI] [PubMed] [Google Scholar]
- Taboureau O., Nielsen S.K., Audouze K., Weinhold N., Edsgärd D., Roque F.S., Kouskoumvekaki I., Bora A., Curpan R., Jensen T.S., Brunak S., Oprea T.I. ChemProt: a disease chemical biology database. Nucleic Acids Res. 2011;39:D367–D372. doi: 10.1093/nar/gkq906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Changing Landscape of Research and Development, (n.d.). https://www.iqvia.com/insights/the-iqvia-institute/reports/the-changing-landscape-of-research-and-development (accessed March 10, 2020).
- D. Thomas, Clinical Pharmacy Education, Practice and Research: Clinical Pharmacy, Drug Information, Pharmacovigilance, Pharmacoeconomics and Clinical Research, Elsevier, 2018.
- Totonchy M., Leffell D. Emerging concepts and recent advances in basal cell carcinoma. F1000Res. 2017;6:2085. doi: 10.12688/f1000research.11314.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchegbu I.F., Florence A.T. Adverse drug events related to dosage forms and delivery systems. Drug-Safety. 1996;14:39–67. doi: 10.2165/00002018-199614010-00005. [DOI] [PubMed] [Google Scholar]
- Uesawa Y. Quantitative structure–activity relationship analysis using deep learning based on a novel molecular image input technique. Bioorg. Med. Chem. Lett. 2018;28:3400–3403. doi: 10.1016/j.bmcl.2018.08.032. [DOI] [PubMed] [Google Scholar]
- Ursu O., Holmes J., Bologa C.G., Yang J.J., Mathias S.L., Stathias V., Nguyen D.-T., Schürer S., Oprea T. DrugCentral 2018: an update. Nucleic Acids Res. 2019;47:D963–D970. doi: 10.1093/nar/gky963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt I., Prinz J., Worf K., Campillos M. Systematic analysis of gene properties influencing organ system phenotypes in mammalian perturbations. Bioinformatics. 2014;30:3093–3100. doi: 10.1093/bioinformatics/btu487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring M.J., Arrowsmith J., Leach A.R., Leeson P.D., Mandrell S., Owen R.M., Pairaudeau G., Pennie W.D., Pickett S.D., Wang J., Wallace O., Weir A. An analysis of the attrition of drug candidates from four major pharmaceutical companies. Nat. Rev. Drug Discov. 2015;14:475–486. doi: 10.1038/nrd4609. [DOI] [PubMed] [Google Scholar]
- Wist A.D., Berger S.I., Iyengar R. Systems pharmacology and genome medicine: a future perspective. Genome Medicine. 2009;1:11. doi: 10.1186/gm11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Peters S.A. A retrospective evaluation of allometry, population pharmacokinetics, and physiologically-based pharmacokinetics for pediatric dosing using clearance as a surrogate. CPT Pharmacometrics Syst. Pharmacol. 2019;8:220–229. doi: 10.1002/psp4.12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Achebouche R., Audouze K. 2020. Computational Systems Biology As an Animal-free Approach to Characterize Toxicological Effects of Persistent Organic Pollutants, ALTEX. [DOI] [PubMed] [Google Scholar]
- Zhao C., Chen A., Jamieson C.H., Fereshteh M., Abrahamsson A., Blum J., Kwon H.Y., Kim J., Chute J.P., Rizzieri D., Munchhof M., VanArsdale T., Beachy P.A., Reya T. Hedgehog signalling is essential for maintenance of cancer stem cells in myeloid leukaemia. Nature. 2009;458:776–779. doi: 10.1038/nature07737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Kruhlak N.L. Construction and analysis of a human hepatotoxicity database suitable for QSAR modeling using post-market safety data. Toxicology. 2014;321:62–72. doi: 10.1016/j.tox.2014.03.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 The hierarchical structure of MedDRA and the list of 27 SOC.
Fig. S2 Examples of the pull-down score (pullS).
Table S1 Identified adverse effects for safinamide.
Table S2 ADEs reported in FDA pre-marketing reports for safinamide.
Table S3 Identified adverse effects for sonidegib.
Table S4 ADEs reported in FDA pre-marketing reports for sonidegib.
Table S5 Identified adverse effects for rufinamide.
Table S6 ADEs reported in FDA pre-marketing reports for rufinamide.